Abstract
The anoxic metabolism of cholesterol was studied in the denitrifying bacterium Sterolibacterium denitrificans, which was grown with cholesterol and nitrate. Cholest-4-en-3-one was identified before as the product of cholesterol dehydrogenase/isomerase, the first enzyme of the pathway. The postulated second enzyme, cholest-4-en-3-one-Δ1-dehydrogenase, was partially purified, and its N-terminal amino acid sequence and tryptic peptide sequences were determined. Based on this information, the corresponding gene was amplified and cloned and the His-tagged recombinant protein was overproduced, purified, and characterized. The recombinant enzyme catalyzes the expected Δ1-desaturation (cholest-4-en-3-one to cholesta-1,4-dien-3-one) under anoxic conditions. It contains approximately one molecule of FAD per 62-kDa subunit and forms high molecular aggregates in the absence of detergents. The enzyme accepts various artificial electron acceptors, including dichlorophenol indophenol and methylene blue. It oxidizes not only cholest-4-en-3-one, but also progesterone (with highest catalytic efficiency, androst-4-en-3,17-dione, testosterone, 19-nortestosterone, and cholest-5-en-3-one. Two steroids, corticosterone and estrone, act as competitive inhibitors. The dehydrogenase resembles 3-ketosteroid-Δ1-dehydrogenases from other organisms (highest amino acid sequence identity with that from Pseudoalteromonas haloplanktis), with some interesting differences. Due to its catalytic properties, the enzyme may be useful in steroid transformations.
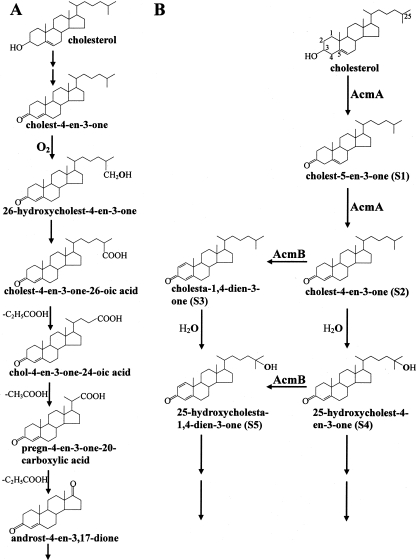
The microbial metabolism of cholesterol and related steroids has attracted great attention because of its potential impact on pharmaceutical and clinical applications (14, 26). Detailed studies have been conducted on the oxic metabolism of cholesterol, as shown in Fig. 1A (12). The pathway for oxic cholesterol metabolism starts with the transformation of cholesterol to cholest-4-en-3-one by a bifunctional cholesterol dehydrogenase (oxidase) (8, 14). The subsequent reaction involves the introduction of a double bond between C-1 and C-2 to produce cholesta-1,4-dien-3-one. So far the enzyme that catalyzes such a reaction in oxic cholesterol metabolism has not been reported. However, 3-ketosteroid-Δ1-dehydrogenase (KSTD), which catalyzes the same type of reaction, has been reported for oxic metabolism of other steroids lacking the aliphatic side chain on the C-17 position, e.g., testosterone (4, 10, 16, 24, 25). Further metabolism of cholesterol requires a monoxygenase catalyzed hydroxylation of the side chain to an alcohol group, followed by its oxidation to a carboxyl group, and subsequent repeated cycles of β-oxidation to degrade the side chain. Recently, genes involved in the oxic cholesterol metabolism by Rhodococcus sp. strain RHA1 were discovered (26).
FIG. 1.
The initial steps in the microbial cholesterol metabolism. (A) Established oxic pathway. (B) Proposed initial steps in the anoxic pathway, as studied in S. denitrificans.
The denitrifying bacterium Sterolibacterium denitrificans, our model organism in this study, is one of a few bacteria that can completely mineralize cholesterol to CO2 in the absence of oxygen (7, 23). We have recently identified the initial products of anoxic cholesterol metabolism (3). The pathway starts with the oxidation of cholesterol to cholest-4-en-3-one (S2; Fig. 1B). This intermediate is further oxidized to the corresponding 1,2-dehydro derivative, cholesta-1,4-dien-3-one (S3). In a third reaction, the tertiary C-25 atom of the side chain becomes hydroxylated with water as the oxygen donor, providing a first example for such unprecedented, but expected oxygen-independent hydroxylation reactions (Fig. 1B).
Based on these findings, we searched for the genes coding for the initial enzymes of the pathway by using a reverse genetics approach (Chiang et al., submitted for publication). A 43-kbp chromosomal DNA fragment was identified and sequenced, which carried one open reading frame (acmB [anoxic cholesterol metabolism enzyme B]) that showed high sequence similarity to KSTD from several bacteria. AcmB therefore may be a good candidate for the second enzyme in the anoxic pathway (transformation of S2 to S3; Fig. 1B). To substantiate the role of acmB in the anoxic cholesterol metabolism, we cloned acmB and expressed it in Escherichia coli. Here, we show that the recombinant His-tagged AcmB (AcmBhis) is a cholest-4-en-3-one-Δ1-dehydrogenase that catalyzes the Δ1 desaturation under anoxic condition.
MATERIALS AND METHODS
Materials and bacterial strains.
The chemicals used were of analytical grade and were purchased as reported before (3). Sterolibacterium denitrificans Chol-1ST (DSMZ13999) (23) was obtained from the Deutsche Sammlung für Mikroorganismen und Zellkulturen (Braunschweig, Germany). Primers were purchased from Biomers (Ulm, Germany).
Bacterial cultures and preparation of cell extract.
Large-scale cultivation was performed in a 200-liter fermentor. S. denitrificans was grown anaerobically as described before (3). E. coli cells that produced the recombinant AcmBhis were grown as described below. All steps used for preparation of cell extracts were performed at 4°C. Frozen cells were suspended in twice the volume of 20 mM 3-morpholinopropanesulfonic acid (MOPS)-K+ (pH 7.9) containing 0.1 mg of DNase I ml−1. Cells were broken by passing the cell suspension through a French pressure cell (American Instruments, Silver Spring, MD) twice at 137 MPa. The soluble protein fraction was obtained by centrifugation at 100,000 × g for 1.5 h. Protein was determined by the Bradford method (2).
Cloning and expression of the acmB gene coding for cholest-4-en-3-one-Δ1-dehydrogenase.
The acmB gene in S. denitrificans was cloned in E. coli with a pEXP5-CT/TOPO TA expression kit according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany). Primers were designed to remove the native stop codon and place the gene in frame with the DNA coding for a C-terminal peptide containing six histidines. The standard protocol was used to isolate chromosomal DNA from S. denitrificans cells (1). The gene was amplified from genomic DNA of S. denitrificans by using Taq DNA polymerase and the following primer pair: ATGAGCATCGAAACCAACACATATGACGT (AcmB_for, forward primer based on the N-terminal amino acid sequence of AcmB)/CTTGCTCGCGGCGATATGGTTGGCCGCG (AcmB_rev, reverse primer based on the C-terminal amino acid sequence of AcmB). The following PCR program was used: 95°C for 5 min followed by 40 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 3 min, and then 72°C for 5 min. The recombinant plasmid (pYRE2) was transformed into One Shot TOP 10 chemically competent E. coli cells (Invitrogen, Karlsruhe, Germany) and E. coli K38/pGP1-2 cells (a kind gift from J. Heider, Darmstadt, Germany) (22) for maintenance and overexpression, respectively. The cloned PCR amplicon was verified by DNA sequencing and restriction analysis. For gene overexpression experiments, E. coli K38/pGP1-2 containing pYRE2 was incubated in LB medium containing ampicillin (100 mg liter−1) and kanamycin (50 mg liter−1) at 30°C until the optical density at 578 nm (OD578) reached 0.7. The culture was heat shocked at 42°C for 30 min and then incubated at 37°C for further 2 h. E. coli cells were harvested at OD578 of 1.4 and stored at −70°C. The soluble protein fraction from frozen cells was prepared as described above.
Purification of AcmBhis from E. coli K38/pGP1-2.
A Ni2+-chelating Sepharose affinity column (50 ml; Amersham Biosciences Europa, Freiburg, Germany) was equilibrated with 150 ml of 20 mM MOPS-K+ buffer (pH 7.9) containing 0.25 M KCl (buffer A). The protein sample (100 ml of soluble protein fraction) was applied to the column followed by washing with buffer A containing 0.15 M imidazole. The fusion protein was subsequently eluted with buffer A containing 0.35 M imidazole. The flow rate was 1 ml min−1, and the eluate was collected in 2-ml fractions. The salts were removed from the combined active fractions by using a prepacked PD-10 desalting column (8.3 ml) according to the manufacturer's instructions (Amersham Biosciences Europa).
Gel filtration chromatography.
The native mass was estimated by using gel filtration chromatography and ferritin (450 kDa), catalase (240 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (67 kDa), and ovalbumin (45 kDa) as the molecular mass standards. The recombinant AcmBhis purified by Ni2+-chelating Sepharose affinity column was concentrated to 2 ml by ultrafiltration (30-kDa-cutoff membrane; Amicon, Witten, Germany) and applied to a 289-ml Superdex 200 HiLoad 26/60 column (Amersham Biosciences Europa). The column was equilibrated with 2 bed volumes of equilibration buffer containing 20 mM MOPS-K+ (pH 7.9) and 0.1 M KCl. The purified recombinant protein was loaded onto the column and eluted at a flow rate of 1.0 ml min−1, and protein elution was monitored at 280 nm.
Partial purification of cholest-4-en-3-one-Δ1-dehydrogenase (AcmB) from S. denitrificans.
AcmB was purified from 50 g of S. denitrificans cells grown anaerobically with cholesterol and nitrate. The basic buffer (buffer B) used in the following purification steps was 20 mM MOPS-K+ (pH 7.9). The enzyme activity was tested by the thin-layer chromatography (TLC) method (see below).
(i) DEAE-Sepharose.
A column with a 70-ml bed volume (Amersham Biosciences Europa) was equilibrated with 2 volumes of buffer B. The soluble protein fraction (100 ml) was loaded onto the column, followed by washing with 2 volumes of buffer B. The flowthrough and the wash were collected in 3-ml fractions. The flow rate was 2 ml min−1.
(ii) Ammonium sulfate precipitation.
The active DEAE pool was further fractionated by ammonium sulfate precipitation. The protein sample was precipitated with ammonium sulfate at 50% saturation, followed by centrifugation at 20,000 × g for 20 min. The supernatant was discarded, and the protein pellet was redissolved in 50 ml of buffer B containing 0.5 M ammonium sulfate.
(iii) Hydrophobic interaction chromatography.
A 25-ml butyl-Sepharose 4 fast flow column (Amersham Biosciences Europa) was equilibrated with 2 volumes of buffer B containing 500 mM ammonium sulfate. The protein sample from the ammonium sulfate treatment was applied to the column, followed by washing with buffer B containing 0.5 M ammonium sulfate. The column was further washed sequentially with buffer B containing 0.1 M ammonium sulfate and buffer B. The active protein was eluted with buffer B containing 0.3% Tween 20. The flow rate was 2 ml min−1, and the eluate was collected in 2-ml fractions. The active fractions were pooled (50 ml) and concentrated to 2.5 ml with ultrafiltration (30-kDa-cutoff membrane).
(iv) Gel filtration chromatography.
A 289-ml Superdex 200 HiLoad 26/60 column was equilibrated with 2 bed volumes of buffer B containing 0.1 M KCl. The protein sample (2.5 ml) was loaded onto the column and was then eluted at a flow rate of 0.4 ml min−1. Fractions of 3 ml were collected, and the dominant protein peak showing activity was concentrated to 1 ml by ultrafiltration (30-kDa-cutoff membrane).
Enzyme assays.
AcmB activity was routinely measured at 37°C under anoxic conditions.
(i) TLC assay.
AcmB dehydrogenase activity was measured by monitoring the formation of cholesta-1,4-dien-3-one. The assay mixture (0.6 ml) containing soluble protein fractions from S. denitrificans (0.2 to 5.0 mg), 100 mM MOPS-K+ (pH 7.9), 5 mM 2,6-dichlorophenolindophenol (DCPIP) and 0.5 mM cholest-4-en-3-one was incubated for 2 h with shaking. Assay mixtures were first extracted twice by an equal volume of ethyl acetate, and the ethyl acetate fraction was concentrated under vacuum. The extracted products were separated on silica gel aluminum TLC plates (silica gel 60 F254; Merck, Darmstadt, Germany). The solvent system used was n-hexane-ethyl acetate (65:35 [vol/vol]). The product was visualized under UV light at 254 nm.
Electron acceptor specificity of purified recombinant AcmBhis was tested by TLC assay under anoxic conditions by incubating the following electron acceptors (5 mM) with 0.5 mM cholest-4-en-3-one, 90 μg enzyme, and 100 mM MOPS-K+ (pH 7.9) in a final volume of 0.8 ml: NaNO3, K3[Fe(CN)6], NAD+, NADP+, phenazine methosulfate, DCPIP, methylene blue, and methyl viologen. After 2 h of incubation, the steroids were extracted and separated on TLC plates as described above. The position corresponding to the substrate was scraped off carefully from the TLC plate and extracted with 0.6 ml ethyl acetate three times. The ethyl acetate fractions were combined and evaporated to dryness. Acetonitrile (0.5 ml) was used to redissolve the residual substrate. The amount of residual steroid substrate was determined spectrophotometrically at 238 nm with cholest-4-en-3-one (5 to 20 μg) as the standard.
(ii) Spectrophotometric assays.
The substrate-dependent reduction of DCPIP was followed at 600 nm (ɛ600 11,000 M−1 cm−1 [pH 6.0]). The assay mixtures (0.5 ml) contained enzyme (1.2 to 5.6 μg), 100 mM Na+-phosphate (pH 6.0), 100 μM DCPIP, and 100 μM steroid substrates dissolved in 2-propanol (10 μl). The apparent Km and Vmax values of different steroid substrates were determined by varying the substrate concentration (1 to 500 μM) and keeping a fixed concentration of DCPIP (100 μM). Apparent Km and Vmax values were obtained by fitting Michaelis-Mention curve to the data using the Prism GraphPad software package (Graphpad Software, San Diego, CA). Buffers used to determine the pH optimum were 100 mM Na+-phosphate (pH 5.0 to 6.0), 100 mM MOPS-KOH (pH 6.5 to 7.9), and 100 mM glycine-NaOH (pH 8.5 to 10.0). To determine any inhibitory effect, the assay was started by addition of 100 μM cholest-4-en-3-one after incubation for 3 min with the tested compound. The type of inhibition was determined by incubating purified AcmBhis with the inhibitor (10 to 100 μM) for 3 min before starting the reaction with the substrate (cholest-4-en-3-one; 25 to 200 μM).
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel eletrophoresis (SDS-PAGE; 10% polyacrylamide) was performed with a discontinuous buffer system (13). Protein bands were visualized by Coomassie blue staining. The following molecular mass protein standards were used: phosphorylase B, 97 kDa; bovine serum albumin, 67 kDa; ovalbumin, 45 kDa; lactate dehydrogenase, 34 kDa; carbonic anhydrase, 29 kDa; and lysozyme, 14 kDa.
N-terminal labeling of native cholest-4-en-3-one-Δ1-dehydrogenase.
Partially purified native AcmB protein in equilibration buffer containing 50 mM Tris-HCl (pH 8.2) and 100 mM KCl (1 ml) was treated with 5N-(succinimidyloxycarbonylmethyl)tris(2,4,6-trimethoxyphenyl) phosphonium bromide (TMPP) under conditions allowing the labeling of only the N-terminal amino group of proteins. After N-terminal TMPP labeling, the sample volume was reduced to 20 μl using Vivaspin microconcentrator (30-kDa-cutoff membrane) (Vivascience, Hannover, Germany). The sample was separated by SDS-PAGE (12% polyacrylamide). The proteins in the gel were precipitated by using 200 ml of the mixture containing 50% ethanol and 3% phosphoric acid for 2 h. After three washes with 500 ml distilled water, the gel was stained by the colloidal Coomassie method (17) and then cut into 2-mm-thick slices. The gel slices between 50 and 70 kDa were digested with trypsin, and the resulting tryptic peptides were extracted and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
LC-MS/MS.
Nanoscale LC-MS/MS analysis of the tryptic peptides was performed using an Agilent 1100 series high-performance liquid chromatography (HPLC)-Chip/MS system (Agilent Technologies, Palo Alto, CA) coupled to an HCT Ultra ion trap (Bruker Daltonics, Bremen, Germany). The voltage applied to the capillary cap was optimized to −1,850 V. For MS/MS experiments, the system was operated with automatic switching between MS and MS/MS modes. The three most abundant peptides, preferring doubly charged ions, were selected on each MS spectrum for further isolation and fragmentation. The MS/MS scanning was performed in the ultrascan resolution mode at a scan rate of 26,000 m/z s−1. A total of six scans were averaged to obtain an MS/MS spectrum. The complete system was fully controlled by ChemStation (Agilent Technologies) and EsquireControl (Bruker Daltonics) softwares.
Analysis of the flavin cofactor.
The flavin cofactor from the purified AcmBhis (1.9 mg ml−1) was extracted and identified as described previously (15). The eluate was monitored at 450 nm in this study. Under these conditions, the retention times were 6.3 min for flavin adenine dinucleotide (FAD), 10.2 min for flavin mononucleotide (FMN), and 28.0 min for riboflavin. The amount of extracted flavin cofactor was determined by comparison of the peak area with that of flavin standard (0.5 to 2.0 μg of FAD).
GC-EI-MS.
Gas chromatography-electron impact mass spectrometry (GC-EI-MS) analysis was performed as follows. MS analyses were performed on a Agilent Technologies 6890N gas chromatograph equipped with a split/splitless programmed temperature injector and an HP-5MS fused silica column (30 m by 0.25 mm; film thickness, 0.25 m) connected to a Agilent 5975 inert MSD quadrupole spectrometer. The mass spectrometer was operated in electron impact (EI) mode at 70 eV, and spectra were recovered over a mass range of m/z 50 to 500 with a cycle time of 1.6 scan s−1. The oven temperature was programmed from 180°C to 300°C at 6°C min−1 and then held isothermal for 10 min. The other conditions were as follows: helium split, 1:10; constant flow, 1.5 ml min−1; transfer line, 280°C; and MS source temperature, 230°C. Samples were injected using the split mode with a ratio of 1:10 via an autoinjector, and the temperature of the injector was 280°C.
UV-visible spectroscopy.
The UV-visible absorption spectrum of purified recombinant AcmBhis protein was obtained by using a CARY 100 Bio UV-visible spectrometer (Varian Deutschland, Darmstadt, Germany). The concentration of the recombinant enzyme was 2.4 μM in 20 mM MOPS-K+ (pH 7.9).
Nucleotide sequence accession number.
The sequence data reported in this study have been deposited in the GenBank database under accession no. EU004090.
RESULTS
Cholest-4-en-3-one-Δ1-dehydrogenase (AcmB) activity in cell extract from S. denitrificans and partial sequence determination of the partially purified protein.
The soluble protein fraction from S. denitrificans grown anaerobically on cholesterol transformed cholest-4-en-3-one (S2) to cholesta-1,4-dien-3-one (S3) (Fig. 1B). The reaction took place in the presence of DCPIP as an artificial electron acceptor. Based on this enzyme assay the enzyme was purified in four steps. SDS-PAGE analysis of the active enzyme pool after the final fractionation revealed four protein bands (data not shown). The amino acid sequences of tryptic peptides from the dominant protein in the active pool exhibited high sequence similarity (24% coverage) to 3-ketosterol-Δ1-dehydrogenase from Comamonas testosteroni (Q7WSH6). Thus, AcmB likely represents the target enzyme. The N-terminal amino acid sequence (SIETNTYDVIVVGSGAGAMLAAAR) of the putative cholest-4-en-3-one-Δ1-dehydrogenase was determined by LC-MS/MS using a strategy of N-terminal protein derivatization as described in Materials and Methods.
Heterologous overexpression of the acmB gene.
Recent studies on the genes involved in anoxic cholesterol metabolism by S. denitrificans revealed several ORFs that might play a role in that metabolic route (Chiang et al., submitted). A putative gene, acmB, encodes a protein of 61.4 kDa that is highly similar to KSTD from several bacteria. In addition, the determined N-terminal amino acid sequence and the amino acid sequences of the tryptic peptides of native AcmB purified from S. denitrificans were identical to those deduced from the acmB gene.
The acmB gene was cloned and expressed in E. coli as a C-terminal His-tagged fusion protein. The cholest-4-en-3-one-Δ1-dehydrogenase activity was measured spectrophotometrically assay monitoring the reduction of DCPIP. The recombinant protein was purified from the soluble protein fraction of E. coli by one Ni2+-chelating affinity chromatography step. SDS-PAGE analysis of the active protein pool revealed a single protein band with an apparent molecular mass of ca. 60 kDa (data not shown). This molecular mass agrees well with that deduced from the acmB gene (61.4 kDa).
Functional analysis of the recombinant AcmBhis.
The recombinant protein catalyzed the transformation of cholest-4-en-3-one to cholesta-1,4-dien-3-one in the presence of DCPIP under anoxic conditions. The identity of the reaction product was confirmed by TLC, HPLC, and GC-EI-MS. Its EI mass spectrum (data not shown) matched exactly with that of cholesta-1,4-dien-3-one in a spectral database (www.aist.go.jp/RIODB/SDBS).
Catalytic properties and stability.
To facilitate the kinetic measurements with purified recombinant AcmBhis, cholest-4-en-3-one oxidation with artificial electron acceptors was followed by monitoring product formation. DCPIP and methylene blue were the best (Table 1). DCPIP was thus chosen for all further experiments and for spectrophotometric assays.
TABLE 1.
Utilization of electron acceptors of the recombinant enzymea
| Artificial electron acceptor | Residual cholest-4-en-3-one concn (μM) |
|---|---|
| Negative control | 442 ± 13 |
| NaNO3 | 383 ± 10 |
| K3[Fe(CN)6] | 99 ± 8 |
| NAD+ | 307 ± 15 |
| NADP+ | 326 ± 19 |
| Phenazine methosulfate | 263 ± 13 |
| DCPIP | 36 ± 5 |
| Methylene blue | 38 ± 11 |
| Methyl viologen | 340 ± 18 |
The assay mixtures containing 90 μg purified enzyme, 0.5 mM cholest-4-en-3-one, and 5 mM each of different electron acceptors were incubated at 37°C for 2 h under anoxic conditions. Samples were treated and measured as described in Materials and Methods. The negative control was without addition of any electron acceptor. Data are averages ± standard deviations of three experimental measurements.
The recombinant enzyme had a pH optimum of 6.0 and a temperature optimum of 40°C (data not shown). Purified AcmBhis retained activity without significant loss when kept at 4°C for 2 weeks or frozen for several months in 20 mM MOPS-K+ (pH 7.9). In addition, no decrease in activity was observed after 20 h of exposure to air at 4°C, thus indicating that purified AcmBhis is not oxygen labile. The recombinant enzyme had a specific activity of 67 μmol min−1 mg protein−1 and an apparent Km for the natural substrate cholest-4-en-3-one of 42 ± 6 μM (Table 2). The turnover rate per monomer was 69 ± 4 s−1, and the catalytic efficiency, kcat/Km, was calculated to be 1.6 × 106 s−1 M−1 (Table 2).
TABLE 2.
Substrate spectrum of the recombinant AcmBhisa
| Substrate | Km (μM) | kcat (s−1) | kcat/km (s−1 M−1) |
|---|---|---|---|
| Cholest-4-en-3-one | 42 ± 6 | 69 ± 4 | 1.6 × 106 |
| Androst-4-en-3,17-dione | 100 ± 12 | 82 ± 5 | 8.0 × 105 |
| Progesterone | 4 ± 1 | 43 ± 2 | 1.2 × 107 |
| Testosterone | 15 ± 3 | 33 ± 2 | 2.2 × 106 |
| 19-Nortestosterone | 50 ± 9 | 26 ± 1 | 5.2 × 105 |
| Cholest-5-en-3-one | 9 ± 2 | 14 ± 0 | 1.6 × 106 |
| Androsta-1,4-dien-3,17-dione | <1 | ||
| 5α-Cholestane | <1 | ||
| Cholesterol | <1 | ||
| Cholesta-3,5-diene | <1 | ||
| Cortisone | <1 | ||
| Corticosterone | <1 | ||
| Estrone | <1 |
The apparent Km values and kcat values (which refer to monomers of 62 kDa) were measured in triplicate at 37°C. The assay mixture contained 100 μM DCPIP, 2.8 μg enzyme, 100 mM Na+-phosphate (pH 6.0), and different concentrations of the respective substrate (1 to 500 μM).
Substrate specificity.
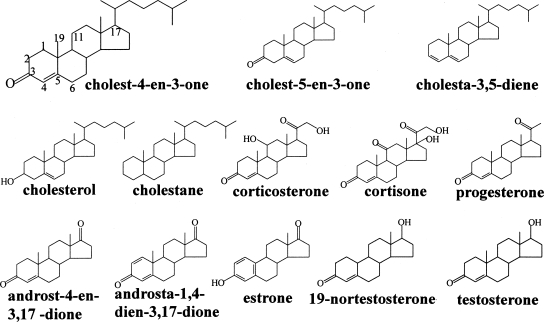
The substrate specificity of the recombinant AcmBhis was tested by screening various steroids (Table 2), whose structures are shown in Fig. 2. The enzyme showed activity with steroids having a carbonyl group at position C-3. 3-Hydroxysteroids such as cholesterol and estrone could not serve as substrates. Moreover, the enzyme showed no activity with cortisone and corticosterone, thus suggesting that a functional group (carbonyl or hydroxyl group) at the C-11 position of the steroid substrates may hinder the dehydrogenase activity. In addition, the enzyme was also inactive with steroids lacking a functional group at the C-3 position such as 5α-cholestane and cholesta-3,5-diene. However, a C = C structure at C4 position or a methyl group at C-19 position of steroid substrates was not necessary for the activity since the enzyme was active toward cholest-5-en-3-one and 19-nortestosterone (Table 2). In summary, progesterone is an excellent substrate for the enzyme, even much better than the natural substrate.
FIG. 2.
Structures of different steroids that were tested by using the spectrophotometric assay with DCPIP. The carbon numbering system of steroids is as shown for cholest-4-en-3-one.
Inhibition and inactivation.
The recombinant AcmBhis was extremely sensitive to Ag+; 1.5 μM of AgNO3 completely inactivated the dehydrogenase activity. Also, 100 μM of Cu2+ completely inactivated the enzyme. The enzyme was sensitive to neither thiol reagents such as iodoacetamide (500 μM) nor metal chelators such as EDTA (500 μM). Corticosterone and estrone strongly inhibited the enzyme activity. According to Dixon plot analysis, the two compounds act as competitive inhibitors. The Ki value for corticosterone was 28 ± 10 μM, and that for estrone was 68 ± 18 μM by fitting the following modified Michaelis-Mention equation (5) to the data: ν = [E]0[S]kcat/[S] + Km(1 + [I]/Ki).
Other steroids such as androsta-1,4-dien-3,17-dione, 5α-cholestane, cholesterol, cholesta-3,5-diene, and cortisone (50 μM) exhibited no inhibitory effects, when 100 μM substrate (cholest-4-en-3-one) was used (for the structures, see Fig. 2).
Molecular properties.
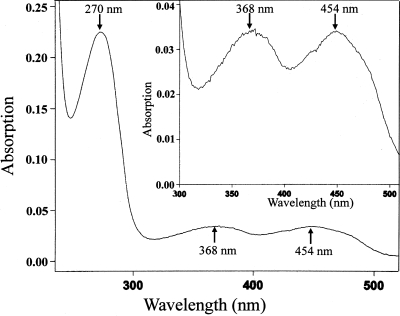
The UV-visible spectrum of the oxidized AcmBhis showed an absorption spectrum typical for flavoproteins (10, 16). Three dominant peaks were observed at 270, 368, and 454 nm as well as a shoulder at around 470 nm (Fig. 3). The flavin cofactor was readily dissociated from the holoprotein by treatment with perchloric acid. The free flavin cofactor was identified by HPLC as FAD (data not shown). The amount of FAD was also determined by HPLC to be 0.65 mol FAD per mol of the recombinant enzyme based on the molecular mass of 62.2 kDa. In addition, taking into account the UV-visible spectrum of the oxidized AcmBhis (Fig. 3), 1 mol of enzyme contained 1.21 mol of FAD on the basis of the molar absorption coefficient for free FAD of 11.3 mM−1 cm−1 at 450 nm and the protein concentration (2.4 μM) determined by the Bradford method.
FIG. 3.
UV-visible absorption spectrum of the purified recombinant enzyme. The concentration of the recombinant enzyme was 2.4 μM in 20 mM MOPS-K+ (pH 7.9). The molar absorption coefficient for free FAD at 450 nm is 11.3 mM−1 cm−1 (pH 7).
Gel filtration chromatography was applied to determine the molecular mass of the recombinant AcmBhis. The active enzyme resulting from the affinity chromatography appeared in the void volume, indicating that AcmBhis forms a massive aggregate (>600 kDa). To find out whether the native cholest-4-en-3-one-Δ1-dehydrogenase also exists as aggregates in the parent strain S. denitrificans, the soluble cell protein was fractionated via the same gel filtration column. Here also, the enzyme activity was found exclusively in the void volume.
DISCUSSION
Catalytic properties.
The purified cholest-4-en-3-one-Δ1-dehydrogenase from S. denitrificans catalyzes the expected oxidation of cholest-4-en-3-one to cholesta-1,4-dien-3-one. The enzyme was competitively inhibited by corticosterone and estrone. Interestingly, cortisone, which is highly similar to corticosterone, is neither a substrate nor an inhibitor for AcmB. It is unknown why a hydroxyl group at C-11 position (corticosterone in this study) allows binding of the steroid as inhibitor, whereas the corresponding carbonyl group on the same carbon (cortisone in this study) does not. The enzyme accepts various artificial electron acceptors. However, the natural electron acceptor remains to be identified.
In general, all steroids, which could serve as substrates for the enzyme, have in common a carbonyl group at the C-3 position. The importance of the 3-keto group is supported by the observation that cholesterol, 5α-cholestane, and cholesta-3,5-diene are not substrates for AcmB. This was also reported for other KSTDs (4, 10). Therefore, it is tempting to postulate an important role of the carbonyl group and the corresponding binding sites of the enzyme (discussed by Kadode et al. [11]).
The enzyme was sensitive to thiol reagents such as AgNO3 and CuCl2, suggesting that thiol groups may play an important role in the catalysis by AcmB. On the contrary, iodoacetamide did not inhibit the enzyme activity, thus apparently excluding the involvement of thiol groups. In addition, no conserved cysteine residues were found in amino acid sequences of AcmB and related KSTDs.
Potential application.
The double-bond formation between C-1 and C-2 of 3-ketosteroids is of pharmaceutical interest. Examples are the production of 1-dehydro analogues of some adrenal cortical steroids such as prednisone and prednisolone (4). The enzyme KSTD carrying out such a reaction has been characterized from a variety of microorganisms during the past two decades (4, 10, 16, 20, 24, 25). However, these studies focused only on steroids without an aliphatic side chain at the C-17 position. Although the same Δ1-desaturation reaction takes place during the (an)oxic metabolism of cholesterol (oxidation of cholest-4-en-3-one to cholesta-1,4-dien-3-one; Fig. 1A and B), the corresponding enzyme has not been characterized (3, 12).
Molecular properties.
The enzyme is highly similar to KSTDs from several microorganisms (highest amino acid sequence identity [56%] with that from Pseudoalteromonas haloplanktis [YP_340631]). These enzymes are flavoproteins, and they possess near their N termini a highly conserved FAD-binding domain, which comprises the G-X-G-X-X-G(A) sequence as an adenine binding motif (6, 20, 24). Furthermore, a conserved glutamate residue is found near the FAD-binding motif and was reported also to be involved in the binding of FAD (18). In addition, this consensus FAD-binding domain of AcmB is preceded (residues 9 to 12) and followed (residues 28 to 35) by many hydrophobic amino acids, resulting in a typical βαβ structure (16, 21). A similar motif is found in other flavoproteins such as succinate dehydrogenase from Shewanella pealeana (Fig. 4). In many succinate dehydrogenases, FAD is covalently bound to a highly conserved histidine residue located about 20 residues downstream of the adenine-binding motif (19). Despite the high similarity (54% amino acid sequence identity) between the AcmB and succinate dehydrogenase from S. pealeana, the conserved histidine residue is lacking in AcmB and the KSTDs (Fig. 4).
FIG. 4.
Partial sequence alignment of the deduced amino acid sequence of the acmB gene from S. denitrificans with FAD-dependent 3-KSTD from Comamonas testosteroni (BAC81692), Pseudoalteromonas haloplanktis (YP_340631), Ralstonia eutropha (YP_728801), Nocardia farcinica (YP_116669), and succinate dehydrogenase (flavoprotein subunit) from Shewanella pealeana (ZP_01603424). Conserved amino acids are highlighted by letters in black, dark gray, and light gray, depending on their similarity from high to low. A conserved consensus sequence for the proposed FAD-binding region, G-X-G-X-X-G(A)-(X)17-E, is underlined.
Comparison with other KSTDs.
Despite the overall sequence similarity and similar catalyzed reaction, AcmB differs from KSTDs in several aspects. First, cortisone serves as a substrate for many characterized KSTDs (4, 10), while it does not for AcmB. Second, these flavoproteins may utilize quinones as physiological electron acceptors. However, they react differently toward various artificial electron acceptors. For instance, phenazine methosulfate is an excellent electron acceptor for KSTD from Nocardia corallina, while K3[Fe(CN)6], NAD+, and NADP+ cannot be used at all (10). For AcmB, DCPIP, methylene blue, and K3[Fe(CN)6] are highly efficient electron acceptors, whereas NAD+, NADP+, and phenazine methosulfate are modest ones. Third, the pH optimum is around pH 6 for AcmB and about pH 10 for KSTDs from N. corallina and Arthrobacter simplex (4, 10). Fourth, KSTDs from N. corallina and A. simplex are monomeric proteins (4, 10), whereas AcmB forms soluble oligomeric aggregates, as is well known for some enzyme families: e.g., the members of the aldehyde dehydrogenase enzyme family (9). The aggregation of AcmB may result from hydrophobic interactions between the monomers. However, it cannot be excluded that formation of monomers may be triggered by a small amount of detergent or other compounds in vivo.
Acknowledgments
This work was supported by the Deutscher Akademischer Austauschdienst (DAAD) by awarding a fellowship to Y.-R.C.
We thank Nasser Gad'on, Freiburg, for expert technical assistance and Christine Schaeffer, Strasbourg, for expert analysis in gas chromatography-electron impact mass spectrometry.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 2.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chiang, Y. R., W. Ismail, M. Müller, and G. Fuchs. 2007. Initial steps in the anoxic metabolism of cholesterol by the denitrifying Sterolibacterium denitrificans. J. Biol. Chem. 282:13240-13249. [DOI] [PubMed] [Google Scholar]
- 4.Choi, K. P., I. Molnár, M. Yamashita, and Y. Murooka. 1995. Purification and characterization of the 3-ketosteroid-Δ1-dehydrogenase of Arthrobacter simplex produced in Streptomyces lividans. J. Biochem. 117:1043-1049. [DOI] [PubMed] [Google Scholar]
- 5.Fersht, A. 1999. The basic equation of enzyme kinetics, p. 103-131. In M. R. Julet (ed.), Structure and mechanism in protein science. W. H. Freeman and Company, New York, NY.
- 6.Florin, C., T. Köhler, M. Grandguillot, and P. Plesiat. 1996. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: gene and protein characterization. J. Bacteriol. 178:3322-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harder, J., and C. Probian. 1997. Anaerobic mineralization of cholesterol by a novel type of denitrifying bacterium. Arch. Microbiol. 167:269-274. [DOI] [PubMed] [Google Scholar]
- 8.Horinouchi, S., H. Ishizuka, and T. Beppu. 1991. Cloning, nucleotide sequence, and transcriptional analysis of the NAD(P)-dependent cholesterol dehydrogenase gene from a Nocardia sp. and its hyperexpression in Streptomyces spp. Appl. Environ. Microbiol. 57:1386-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichihara, K., Y. Noda, C. Tanaka, and M. Kusunose. 1986. Purification of aldehyde dehydrogenase reconstitutively active in fatty alcohol oxidation from rabbit intestinal microsomes. Biochim. Biophys. Acta 878:419-425. [PubMed] [Google Scholar]
- 10.Itagaki, E., T. Wakabayashi, and T. Hatta. 1990. Purification and characterization of 3-ketosteroid-Δ1-dehydrogenase from Nocardia corallina. Biochim. Biophys. Acta 1038:60-67. [DOI] [PubMed] [Google Scholar]
- 11.Kadode, M., H. Matsushita, K. Suzuki, and E. Itagaki. 1994. Essential arginine residue(s) of 3-ketosteroid-Δ1-dehydrogenase, p. 339-342. In K. Yagi (ed.), Flavins and flavoproteins. Walter de Gruyter & Co., Berlin, Germany.
- 12.Kieslich, K. 1985. Microbial side-chain degradation of sterols. J. Basic Microbiol. 25:461-474. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.MacLachlan, J., A. T. L. Wotherspoon, R. O. Ansell, and C. J. W. Brooks. 2000. Cholesterol oxidase: sources, physical properties and analytical applications. J. Steroid Biochem. Mol. Biol. 72:169-195. [DOI] [PubMed] [Google Scholar]
- 15.Mohamed, M. E., A. Zaar, C. Ebenau-Jehle, and G. Fuchs. 2001. Reinvestigation of a new type of aerobic benzoate metabolism in the proteobacterium Azoarcus evansii. J. Bacteriol. 183:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morii, S., C. Fujii, T. Miyoshi, M. Iwami, and E. Itagaki. 1998. 3-Ketosteroid-Δ1-dehydrogenase of Rhodococcus rhodochrous: sequencing of the genomic DNA and hyperexpression, purification, and characterization of the recombinant enzyme. J. Biochem. 124:1026-1032. [DOI] [PubMed] [Google Scholar]
- 17.Neuhoff, V., R. Stamm, I. Pardowitz, N. Arold, W. Ehrhardt, and D. Taube. 1990. Essential problems in quantification of proteins following colloidal staining with Coomassie brilliant blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11:101-117. [DOI] [PubMed] [Google Scholar]
- 18.Nishiya, Y., and T. Imanaka. 1996. Analysis of interaction between the Arthrobacter sarcosine oxidase and the coenzyme flavin adenine dinucleotide by site-directed mutagenesis. Appl. Environ. Microbiol. 62:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearling, S. L., A. C. Black, F. D. C. Manson, B. Ward, S. K. Chapman, and G. A. Reid. 1992. Sequence of the gene encoding flavocytochrome c from Shewanella putrefaciens: a tetraheme flavoenzyme that is a soluble fumarate reductase related to the membrane-bound enzymes from other bacteria. Biochemistry 31:12132-12140. [DOI] [PubMed] [Google Scholar]
- 20.Plesiat, P., M. Grandguillot, S. Harayama, S. Vragar, and Y. Michel-Briand. 1991. Cloning, sequencing, and expression of the Pseudomonas testosteroni gene encoding 3-ketosteroid-Δ1-dehydrogenase. J. Bacteriol. 173:7219-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreuder, H. A., J. M. Van der Laan, W.-G. J. Hol, and J. Drenth. 1991. The structure of p-hydroxybenzoate hydroxylase, p. 31-64. In F. Müller (ed.), Chemistry and biochemistry of flavoproteins, vol. II. CRC Press, Boca Raton, FL. [Google Scholar]
- 22.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarlera, S., and E. B. M. Denner. 2003. Sterolibacterium denitrificans gen. nov., sp. nov., a novel cholesterol-oxidizing, denitrifying member of the β-Proteobacteria. Int. J. Syst. Evol. Microbiol. 53:1085-1091. [DOI] [PubMed] [Google Scholar]
- 24.Van der Geize, R., G. I. Hessels, and L. Dijkhuizen. 2002. Molecular and functional characterization of the kstD gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid-Δ1-dehydrogenase isoenzyme. Microbiology 148:3285-3292. [DOI] [PubMed] [Google Scholar]
- 25.Van der Geize, R., G. I. Hessels, R. Van Gerwen, J. W. Vrijbloed, P. Van der Meijden, and L. Dijkhuizen. 2000. Targeted disruption of the kstD gene encoding a 3-ketosteroid-Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl. Environ. Microbiol. 66:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Geize, R., K. Yam, T. Heuser, M. H. Wilbrink, H. Hara, M. C. Anderton, E. Sim, L. Dijkhuizen, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 104:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]