Abstract
In recent years increasing attention has been given to the potential health effects of fungal exposure in indoor environments. We used large-scale sequencing of the fungal internal transcribed spacer region (ITS) of nuclear ribosomal DNA to describe the mycoflora of two office buildings over the four seasons. DNA sequencing was complemented by cultivation, ergosterol determination, and quantitative PCR analyses. Sequences of 1,339 clones were clustered into 394 nonredundant fungal operational taxonomical units containing sequences from 18 fungal subclasses. The observed flora differed markedly from that recovered by cultivation, the major differences being the near absence of several typical indoor mold genera such as Penicillium and Aspergillus spp. and a high prevalence of basidiomycetes in clone libraries. A total of 55% of the total diversity constituted of unidentifiable ITS sequences, some of which may represent novel fungal species. Dominant species were Cladosporium cladosporioides and C. herbarum, Cryptococcus victoriae, Leptosphaerulina americana and L. chartarum, Aureobasidium pullulans, Thekopsora areolata, Phaeococcomyces nigricans, Macrophoma sp., and several Malassezia species. Seasonal differences were observed for community composition, with ascomycetous molds and basidiomycetous yeasts predominating in the winter and spring and Agaricomycetidae basidiomycetes predominating in the fall. The comparison of methods suggested that the cloning, cultivation, and quantitative PCR methods complemented each other, generating a more comprehensive picture of fungal flora than any of the methods would give alone. The current restrictions of the methods are discussed.
Moisture enables reproduction of atypical fungi and bacteria in building materials (59), whereas dampness and mold observations have been associated with building-related health problems (2, 3, 30). Several microbial indicators have been suggested to associate with moisture damage and/or health complaints (13, 27, 33, 39, 46, 58), yet a consensus regarding their causative role in relation to the health effects is lacking. The potential harmfulness of microbial contamination is suggested to be dependent on the species content and concurrently occurring species combinations, as well as on the material and growth conditions in which the microbial proliferation occurred (19, 31, 53, 59, 71).
Traditionally, indoor fungi have been studied by plate cultivation, and practically all information on their occurrence and prevalence rely on viable studies. However, the restrictions of cultivation are known to bias the picture of fungal community structure both qualitatively and quantitatively (24). The percentage of cultivable fungi varies from as low as <1% to as high as 100%, depending on the organism and studied material (11, 48, 70). Different species have distinct growth requirements and in mixed cultures fast growth and chemical competition lead to overgrowth and displacement or to growth inhibition of the more fastidious species. Moreover, the number, dispersion potential and survival of fungal spores vary between species (8). Fungal identification may require days to weeks, and experienced personnel are needed to reliably accomplish morphological identification. Some organisms can only be identified at genus or group level, and some may remain morphologically indistinguishable under laboratory conditions.
The main benefits of using DNA as an identification target instead of cultivation-based methods are the speed, accuracy, and analytical sensitivity of detection and the possibility to detect and identify dead or dormant organisms (1, 43). The latter benefit is especially valuable in indoor studies since the main exposure hazards relating to indoor microbial contamination are not dependent on viability (5, 26). Molecular methods most often used in fungal studies include conventional or quantitative PCR (qPCR) specific for fungal species or groups (15, 18, 23, 75), universal fungal PCR combined with denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis (4, 62, 65), terminal or conventional restriction fragment length polymorphism analysis (7, 15), and simple labeling-based fingerprinting methods (36). Furthermore, universal fungal PCR combined with cloning and restriction fragment length polymorphism analysis and/or DNA sequencing of cloned fragments has been used (4, 7, 21, 34, 37), as well as probing methods (25, 44, 80). Some of these methods have also been used to detect indoor fungi (14, 15, 23, 75, 80). qPCR has provided valuable information on the occurrence of the most common indoor fungi, and this method shows great promise in giving fast quantitative data on the occurrence of the studied organisms (45). Apart from cloning and sequencing of amplified fungal DNA, or in some cases DGGE followed by sequencing, these methods do not produce detailed information on previously unknown or unexpected taxa.
Since the early 1990s, numerous studies on bacterial communities based on small subunit ribosomal DNA (rDNA) sequencing have revealed a vast diversity of uncultivable and novel prokaryotic taxa (17, 20, 40). More recently, the same phenomenon has been observed in fungal community studies. Vandenkoornhuyse et al. (72) obtained 49 mostly novel fungal phylotypes from plant roots and detected five putatively novel fungal groups. Similar results have been obtained by others (34, 47, 49, 63). In these studies the theoretical diversity in the studied environment was estimated to comprise at least 100 and in some studies even more than 500 fungal phylotypes. To the best of our knowledge, the fungi in indoor environments have not previously been studied by using rDNA sequencing.
In the present study, large-scale rDNA sequencing was used to characterize the fungal flora in the indoor dust of two nonindustrial buildings, one of which was moisture damaged. The internal transcribed spacer (ITS) region of fungal nuclear rDNA was chosen as the target. This region is the best-annotated gene locus for fungi with more than 39,000 full-length or nearly full-length sequences currently available, and it provides useful, phylogenetically informative sequence characters for fungi, especially at lower taxonomic levels. Sequence analysis was carried out side by side with cultivation. In addition, the quantities of seven indoor fungi known to be common from previous studies were determined by qPCR analysis, and ergosterol measurement was conducted as a proxy for total fungal biomass. Our aim was to investigate (i) fungal diversity in indoor dust using sequencing of rDNA clone libraries; (ii) differences in the fungal diversity obtained by clone libraries and by standard cultivation; (iii) congruence of the qPCR, clone library, and culture results; and (iv) differences in the fungal flora obtained by sequencing of clone libraries between a moisture-damaged and an undamaged building in each season.
MATERIALS AND METHODS
Buildings.
The study was performed in two nursing home buildings located in two small towns about 100 km apart in central Finland. Both buildings were brick-framed, were built on a ground slab, had two floors in addition to a partly below-grade basement, and had a mechanical exhaust ventilation system with passive flow of intake air. Both buildings were inspected by a trained civil engineer for signs of moisture damage.
One of the buildings, defined here as the moisture-damaged or index building, was built around 1920; no substantial repairs or structural modifications had been done since that time. There was moisture and microbial damage on the floor and external walls in the basement. Local moisture and microbial damage was also observed on the first and second floors. The staff who worked in the building reported building-related symptoms and indoor air problems. The control building was built around 1940 and underwent a thorough renovation in 1982. No visible signs of moisture or microbial damage were seen, except for minor signs of water damage in the washroom. The staff who used this building did not report any problems with the building or indoor air quality.
Sample description and sampling.
Indoor dust was collected from both buildings over 1 year in four separate time periods corresponding to the four North European seasons, resulting in a total of eight samples, four from each building. Sampling was done in office rooms located on the second floors of both buildings. There was no visible mold in the office rooms. Settled dust was collected from smooth surfaces, including mainly large areas of uncarpeted floor and tables, using a vacuum cleaner (Miele S371; Miele & Cie, KG, Gütersloh, Germany) twice a week during 1 to 3 months. After collection, the dust bag was cut open, and the accumulated house dust was vacuumed into a chamber. The dust was subsequently sieved through a sterile strainer with pore size of about 1 by 1 mm and divided into aliquots for analyses. Microbes were cultured on the same or next day, and aliquots of the dust were stored at −20°C for the other analyses.
Culturing.
Dust samples of 1 g were suspended into 9 ml of buffer (distilled water with 42.5 mg of KH2PO4/liter, 250 mg of MgSO4·7H2O/liter, 8 mg of NaOH/liter, and 0.02% Tween 80 detergent), and the sample was shaken for 60 min at room temperature. The suspension was filtered with a metal strainer, with a mesh size of 1 mm, and serial tenfold dilutions were made in the same buffer. Aliquots (100 μl) were plated on 2% malt extract agar (59) and dichloran-glycerol agar (DG18; Oxoid, Ltd., Basingstoke, Hampshire, England), supplemented with 100 ng of chloramphenicol/ml for determination of the mesophilic and xerophilic fungi, respectively (60). Samples were incubated in the dark at 25°C for 7 days. After incubation, the colonies were counted and identified to the genus level by using conventional micromorphological methods. Aspergillus species, including A. ochraceus, A. niger, A. versicolor, A. penicilloides, and A. sydowii were identified and quantified at the species level. The identifications of representative isolates of each colony type were verified by ITS sequence analysis.
For sequencing of the ribosomal ITS region, representatives of identified pure cultures from the index building were grown in standard 96-well culture plates in 1 ml of nutrient broth (Oxoid) under agitation (200 rpm) at 25°C for 48 h. Representatives of fungal strains are deposited in culture collection of National Public Health Institute of Finland and are available on request (Helena Rintala, Environmental Microbiology Laboratory).
DNA extraction.
Total DNA from dust samples was isolated separately for cloning experiments and qPCR because the addition of an internal reference standard was required at the extraction step for qPCR analysis. For cloning, DNA was isolated from 25 mg of dust in duplicate by using a GenElute plant genomic DNA miniprep kit (Sigma-Aldrich Chemie Gmbh, Steinheim, Germany). The sample was weighed and transferred to a 2-ml screw cap tube with 0.5 g of 0.1-mm glass beads (BioSpec Products, Inc., Bartlesville, OK), 400 μl of lysis solution was added, and the cells were disrupted with Mini Beadbeater-8 (BioSpec) at maximum speed for 1 min. DNA was recovered according to the manufacturer's instructions, purified by using a Wizard DNA Clean-Up column (Promega, Madison, WI), and eluted in 50 μl of nuclease-free water. Replicates were pooled to give 100 μl of DNA originating from 50 mg of dust.
For qPCR, dust samples were extracted by a rapid bead-milling method (22). Briefly, 5 mg of dust supplemented with Geotrichum candidum conidia as an internal standard and 90 μl of AE buffer (Qiagen, Valencia, CA) were suspended with 0.3 g of glass beads and 100 and 300 μl of lysis and binding buffer, respectively, from an Elu-Quik DNA purification kit (Whatman Plc, Brentford, United Kingdom). The tubes were shaken in a Mini Beadbeater-8 at maximum speed for 1 min and centrifuged at 8,000 × g for 1 min. The supernatants were further purified by using a DNeasy kit (Qiagen).
For extraction of the genomic DNA from fungal pure cultures, liquid cultures were centrifuged and harvested by using MasterPure yeast DNA purification kit (Epicenter, Madison, WI) according to the manufacturer's instructions.
PCR amplification.
Universal PCR amplification of the fungal nuclear ribosomal ITS region was carried out by using the following primer pair: novel forward primer Fun18Sf (TTGCTCTTCAACGAGGAAT, positioned at nucleotides 1558 to 1575 in Saccharomyces cerevisiae 18S rRNA gene [GenBank accession no. J01353]) (J. Hultman, unpublished data) and the ITS4 (78). The primers amplify a fragment containing ca. 240 bp at the 3′ end of the 18S gene, the entire ITS region and also ca. 50 bp at the 5′ end of the 28S gene. Ten replicates in a final volume of 50 μl were prepared using 1× DyNAzyme PCR buffer (Finnzymes, Espoo, Finland), 200 μM concentrations of each deoxynucleoside triphosphate (dNTP), 0.2 μM concentrations of each primer, 5% dimethyl sulfoxide, 0.5 mM betaine, 1 U of DyNAzyme DNA polymerase (Finnzymes), 0.05 U of Pfu polymerase (Promega), and 2 μl of DNA template. Thermocycling conditions were as follows: 95°C for 3 min; 6 cycles of 96°C for 30 s, 50°C for 45 s, and 72°C for 3 min; 19 cycles of 96°C for 30 s, 50°C for 45 s, and 72°C for 1 min; and a final extension at 72°C for 10 min. For some dust samples, supplementary cloning was needed to gain libraries of the desired size. Consequently, a seminested PCR protocol was required. Replicate products of the primary PCR were combined, purified, and concentrated 10 times, and 10 μl of concentrate was used as a template. The primer pair ITS1 (78) and ITS4 and the above-mentioned PCR method were used. DNA from pure cultures was amplified with the Fun18Sf-ITS4 primer pair in the above-mentioned PCR conditions, excluding the extension of elongation time in the first six cycles. The replicate PCR products were combined, visualized in 1% agarose gels with ethidium bromide staining, and purified with MultiScreen 384PCR plates (Millipore, Billerica, MA) or Wizard PCR Preps (Promega).
Cloning and sequencing of ITS fragments.
Immediately before ligation, the purified PCR products were incubated in 72°C for 30 min in the following reaction mix: DyNAzyme PCR buffer, dNTPs, and DyNAzyme DNA polymerase in the concentrations mentioned above. Using a PCR cloning kit (Qiagen) according to the manufacturer's instructions, 4 μl of PCR products was ligated and cloned into an Escherichia coli plasmid library. Aliquots of cloned cultures were stored in 15% glycerol at −80°C, and plasmids were extracted from the remaining culture by using MultiScreen 96PLASMID plates (Millipore). Inserts were reamplified with universal forward and reverse vector primers. PCR products were visualized in 1% agarose gel with ethidium bromide staining and purified with MultiScreen 384PCR plates. Purified fragments were sequenced by using a BigDye terminator cycle sequencing kit with vector primers or the primers Fun18Sf and ITS4 for pure-culture ITS products. Sequencing reactions were run on an ABI 3700 automated DNA sequencer (Applied Biosystems, Foster City, CA).
Sequence analysis and OTU definition.
Sequences were edited, and identical sequences were clustered by using the Pregap and Gap4 programs of the Staden Package (66). Full-length sequences, excluding the Fun18Sf and ITS4 primer sites, were aligned against the EMBL DNA databases by using Fasta3.4 (52) and subsequently clustered into operational taxonomical units (OTU) using CLUSTAL W (69) with a gap open penalty of 5 and a gap extension penalty of 0.5. More than 99% sequence similarity within the cluster and uniform match with EMBL database sequences were used to define OTU. Sequences of nonfungal origin were excluded from further analyses. Because eukaryotic ITS sequences of public databases seldom extend in the 5′ direction over the 18S gene, most of this gene was excluded from OTU sequences in subsequent analyses. All representative OTU sequences were manually checked for chimerism by visual inspection of the CLUSTAL W alignments of ITS1 and ITS2 fragments. This was followed by separated database searches of the fragments for unevenly clustering OTU types. High sequence similarity with taxonomically diverging reference sequences was considered to support chimerism. The occurrence of certain OTU types in more than one library was considered strong support for nonchimerism. When ≥99% sequence similarity with a database sequence over the entire ITS region was observed, the determination of chimerism was considered impracticable. A species name was assigned to OTU with more than 99% similarity with a reference sequence. Sequences with less than 99% similarity to a database match were considered unknown but were affiliated with fungal subclass level groups when possible by the taxonomy of closest Fasta hits. The Dictionary of the Fungi (38) served as the source of taxonomic references for fungal species.
Statistical analyses.
Two nonparametric richness estimators, Chao1 and ACE (9, 10), were used to calculate theoretical diversity and coverage of libraries. The species diversities of the index building and reference building for each season were then compared by dividing the estimated diversity value of the index building by that of the reference building. To compare the content of two libraries on the OTU level, the similarity coefficient (S) was determined for sample pairs as follows: S = 2 C/(A + B), where A and B are the numbers of observed OTU in libraries A and B, respectively, and C is the number of OTU present in both libraries (50).
qPCR.
Earlier developed and described methods (6, 22) for preparing conidia or spore suspensions from fungal cultures, extracting DNA, performing qPCR analyses, and preparing standard calibration curves were used. Coextracted DNA from Geotrichum candidum was used as an exogenous internal reference, and therefore 10 μl of a 2 × 108 conidia per ml reference suspension of G. candidum was added to the dust samples before bead beating. All primer and probe sequences used in the assays, as well as known species comprising the assay groups, are available online (http://www.epa.gov/nerlcwww/moldtech.htm). Primers and probes were synthesized commercially (Applied Biosystems, Cheshire, United Kingdom).
qPCR assays for the target organism and G. candidum reference DNA in the extracts were prepared by using Universal Master Mix, a 2× concentrated mixture of optimized buffer components, AmpliTaq Gold DNA polymerase, AmpErase UNG, dNTPs, passive reference dye (Applied Biosystems), and 2 mg of bovine serum albumin (New England Biolabs, Inc., Beverly, MA) ml−1. Appropriate primers and probes in a final concentration of 1.0 μM (primers) and 80 nM (probes) and 2.5 μl of template were used, and the final volume was 12 μl. PCR was performed in an Applied Biosystems Prism model 7000 instrument. Methods for estimating the amplification factors and extrapolating spore or conidia equivalents for the assays using standard curves have also been described (6). For calibration curves, serially diluted log target conidia or spore equivalents versus delta threshold cycle values (ΔCT = CT,target − CT,reference) were plotted. The numbers of spores or conidia detected in dust samples were calculated by using the comparative threshold cycle method as follows. First, the comparative target level was calculated using the equation: Ae−ΔΔCT, where Ae is the amplification efficiency, and ΔΔCT is ΔCT,sample − ΔCT,calibrator [(CT,target − CT,reference)sample − (CT,target − CT,reference)calibrator]. The ΔCT,calibrator value was obtained from the calibration curve as the mean ΔCT value of all dilutions. The log number of cells in the sample was then obtained by multiplying the comparative target level by the amount of cells in the calibrator, which was the mean log cells in all dilutions of the calibration curve. The concentrations of seven fungal species or groups, selected based on information obtained from cultivation, were analyzed by using qPCR assays. These were Cclad1 for Cladosporium cladosporioides serovar 1; Eamst for teleomorphic states of members of the Aspergillus glaucus group, including Eurotium amstelodami, E. chevalieri, E. herbariorum, E. rubrum, and E. repens; Tviri for Trichoderma viride, T. atroviride, and T. koningii; Stac for Stachybotrys chartarum; Asydo3 for Aspergillus sydowii; PenGrp2 for Penicillium species, including P. crustosum, P. camembertii, P. commune, P. echinulatum, and P. solitum; and Pdigi for Penicillium digitatum. Dust sample extracts were analyzed for potential inhibition of qPCR reactions by performing serial 10-fold dilutions of DNA extracts of Geotrichum-spiked reference samples. Analyses of negative (containing AE buffer only) and positive (mixed positive control conidia suspensions) control samples were performed in parallel with the assays.
Analysis of ergosterol content of dust samples.
Two replicate samples of 5 mg of dust were prepared according to the method of Sebastian and Larsson (64) with minor modifications. In our study hexane was used instead of heptane, and the samples were diluted to 500 μl of solvent prior to the analysis by high-resolution gas chromatography-mass spectrometry. The analysis of ergosterol was performed with a Micromass AutoSpec Ultima magnetic sector mass spectrometer connected to an HP 6890 gas chromatograph with a DB-5MS fused-silica capillary column (60 m by 0.25 mm by 0.25 μm) from J&W Scientific (Agilent Technologies, Santa Clara, CA). The injection volume of the samples was 2 μl, the temperature of the injector was 270°C, helium was used as the carrier gas at a constant pressure of 200 kPa, and the gas chromatograph operated in splitless mode. The temperature program of the gas chromatograph was as follows: 170°C for 1 min, 30°C/min to 260°C, 10°C/min to 300°C, and holding at 300°C for 20 min. Positive electron ionization was performed at an ionization voltage of 35 eV, and the temperature of the ion source was 270°C. The selected ion monitoring method was used to monitor the two fragment ions of silylated ergosterol, and 7-dehydrocholesterol was used as the internal standard. The ions monitored were m/z 337.2895 and 338.2929 for ergosterol and m/z 325.2895 and 326.2929 for 7-dehydrocholesterol. The mass resolution of the instrument was 10,000.
Nucleotide sequence accession numbers.
Representatives of produced fungal DNA sequences were submitted to the EMBL Nucleotide Sequence Database (http:www.ebi.ac.uk/embl) and assigned accession numbers AM901670 to AM901704 and AM901705 to AM902098 for the sequences of cultivated strains and clones, respectively.
RESULTS
Structure of ITS clone libraries.
Full-length ITS sequences were obtained from 1,578 clones. Of these, 1,339 originated from fungi, 223 originated from plants, and 11 originated from algae, and 30 were chimeric. On average, 168 fungal sequences were sampled from each library (Table 1). Sequences were clustered into 394 OTU based on ≥99% sequence similarity and a uniform database match. Of these, 77% occurred in only one sample, and 61% were singletons. The lowest numbers of OTU were obtained from winter samples from both buildings. The OTU number increased steadily over the year from winter to fall for both buildings. The estimated species diversity in samples as determined by ACE and Chao1 analyses varied from around 100 OTU in winter to ca. 300 to 400 in the fall. The corresponding coverage values varied between 21 and 77%, being highest in the winter samples estimated by the Chao1 method and lowest in the summer and fall samples estimated by ACE (Table 1). Plant sequences were found mainly in spring and to lesser extent in summer samples, probably originating from pollen particles.
TABLE 1.
rDNA clone library sizes and estimates of theoretical diversity and sampling coverage over a 1-year period in index and reference buildingsa
| Sample | Library size (clones) | No. of OTU | Singletons (%) | ACE
|
Chao1
|
||
|---|---|---|---|---|---|---|---|
| %C | TD | %C | TD | ||||
| Index | |||||||
| Winter | 141 | 50 | 46 | 46 | 109 | 71 | 70 |
| Spring | 180 | 70 | 71 | 24 | 295 | 29 | 242 |
| Summer | 225 | 81 | 69 | 25 | 327 | 49 | 167 |
| Fall | 141 | 91 | 74 | 22 | 418 | 35 | 264 |
| Reference | |||||||
| Winter | 174 | 47 | 39 | 59 | 79 | 77 | 61 |
| Spring | 152 | 61 | 61 | 31 | 194 | 50 | 123 |
| Summer | 170 | 69 | 74 | 21 | 333 | 25 | 277 |
| Fall | 157 | 92 | 72 | 23 | 393 | 28 | 334 |
| Both buildings (all seasons) | 1,339 | 394 | 61 | 30 | 1,319 | 46 | 858 |
%C, % coverage; TD, corresponding theoretical diversity.
Fungal diversity analysis of clone libraries.
A total of 161 OTU (41%) were identified at species level by ≥99% similarity with named database sequences. An additional 17 sequences had equally high matches, but the closest database relative was incompletely named or annotated as an environmental clone. A total of 44 OTU were 100% identical to a reference sequence. In 18 cases more than one reference sequence gave equal similarities, and in several other cases the sequence similarity was ≥99% with more than one reference sequence. OTU with ≥99% database match were observed to be more abundant (on average, 5.2 clones per OTU) and to occur more frequently among the samples (on average, in 1.8 libraries) than the OTU with <99% similarity with the reference sequence. The latter contained, on average, 2.3 clones and occurred in 1.2 samples.
OTU with <99% similarity to the database sequence represent uncultured species or species whose ITS region has not been previously sequenced. To clarify their phylogenetic position, sequences were aligned together with database representatives from different fungal subclasses, and tree construction was attempted. Due to the hypervariable nature of the ITS region the tree branches near the root were short and poorly supported by bootstrap analysis (data not shown), and the treeing procedure was not considered to give reliable information about the phylogenetic relationships of the OTU. Instead, OTU were affiliated to the subclass level according to the phylogeny of the closest database relatives. This classification of sequence types is the suggested method, especially in the case of sequences whose similarity to closest relatives is low. All recovered OTU types with EMBL nucleotide database identity codes, their closest database references, taxonomic affiliation, and incidence in samples, along with corresponding information of cultivated strains are available (see Table S1 in the supplemental material).
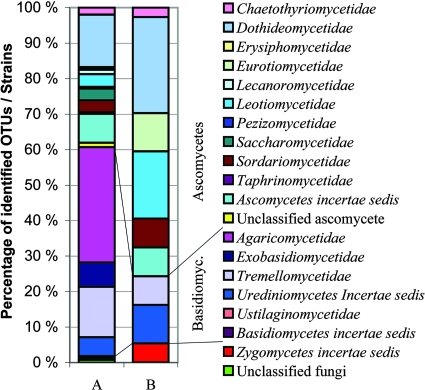
Of the total 394 sequence types, 234 were of basidiomycetous affinity and 155 of ascomycetous affinity. Two representatives of zygomycota and three sequences of unknown fungal origin were also found. Representatives of 18 fungal subclasses were found (Fig. 1). The proportions of abundant OTU types were similar in both buildings; about 10 sequence types occurred in clone libraries of either building in 10 or more clones (Table 2). Most of these OTU were abundant in both buildings, and about one-third were also detected by culture. A majority of the abundant OTU were named at species level.
FIG. 1.
Comparison of the total diversity obtained by cultivation and clone library analysis from two buildings over 1 year. (A) Cumulative diversity at subclass level in clone libraries. The number of OTU is 394. (B) Diversity of cultivated strains at subclass level. The number of strains is 37.
TABLE 2.
Frequencies of the most abundant OTU types in the index and reference buildings
| OTU type
|
Best FASTA match
|
%Da | %Rb | ||||
|---|---|---|---|---|---|---|---|
| ID no. | EMBL accession no. | Taxon | Subclass | EMBL accession no. | % Similarity | ||
| 366 | AM901959 | Cladosporium magnusianumc | Dothideomycetidae | AF455404 | 100 | 9.9 | 5.9 |
| 368 | AM901891 | Cladosporium tenuissimumd | Dothideomycetidae | AF393691 | 100 | 6.2 | 2.9 |
| 249 | AM901845 | Cryptococcus victoriae | Tremellomycetidae | AF444645 | 100 | 3.9 | 4.7 |
| 205 to 207 | AM901735, AM902074, AM901925 | Malassezia restricta | Exobasidiomycetidae | AY387145 | 98.6-99.6 | 3.1 | 4.4 |
| 232 | AM901924 | Thekopsora areolata | Uredinales incertae sedis | DQ087229 | 99.4 | 4.6 | 2.7 |
| 212 | AM901708 | Malassezia restricta | Exobasidiomycetidae | AJ437695 | 96.9 | 2.8 | 2.3 |
| 276 | AM901015 | Leptosphaerulina chartarum | Dothideomycetidae | DQ384571 | 99.8 | 3.8 | 1.2 |
| 256 | AM902094 | Saccharomyces cerevisiae | Saccharomycetidae | U53879 | 99.9 | 0.9 | 3.9 |
| 230 | AM901970 | Thekopsora areolata | Uredinales incertae sedis | DQ087230 | 99.7 | 0 | 4.5 |
| 290 | AM901867 | Leptosphaerulina trifoliie | Dothideomycetidae | AY131203 | 99.8 | 1.4 | 2.3 |
| 342 | AM901731 | Aureobasidium pullulans | Dothideomycetidae | AM160630 | 100 | 2.5 | 1.1 |
| 210 | AM902048 | Malassezia restricta | Exobasidiomycetidae | AY743636 | 100 | 1.4 | 2 |
| 311 | AM901933 | Macrophoma sp. | Dothideomycetidae | DQ279798 | 95.5 | 2 | 0.8 |
| 361 | AM901734 | Hypogymnia physodes | Lecanoromycetidae | AF058036 | 99.5 | 0.6 | 1.4 |
| 296 | AM901755 | Phaeosphaeriaceae sp. | Dothideomycetidae | AY465459 | 96.5 | 0 | 1.7 |
| 229 | AM902053 | Chrysomyxa arctostaphyli | Uredinales incertae sedis | L76488 | 92.6 | 0 | 1.5 |
%D, percentage of clones in the water-damaged index building.
%R, percentage of clones in the reference building.
Equal match with Cladosporium magnusianum, C. herbarum, C. magnusianum, Cladophialophora minourae, and Mycosphaerella macrospora.
Equal match with Cladosporium tenuissimum, C. cladosporioides, C. cucumerinum, and C. oxysporum.
Equal match with L. trifolii and L. americana.
Several abundant ascomycetous OTU sequences were members of the subclass Dothideomycetidae. These were Leptosphaerulina trifolii/americana and L. chartarum, Aureobasidium pullulans, two Cladosporium OTU, one identical to the sequence of the closely related species Cladosporium herbarum, C. magnusianum, Cladophialophora minourae, and Mycosphaerella macrospora and the other identical to Cladosporium cladosporioides, C. tenuissimum, C. cucumerinum, and C. oxysporum. Sequence relatives of Phaeococcomyces nigricans and Macrophoma sp. were also frequently encountered. Other abundant ascomycete sequences originated from S. cerevisiae (subclass Saccharomycetidae) and Hypogymnia physodes (Lecanoromycetidae) (Table 2).
Only 9 of the 234 basidiomycetous sequence types were abundant in our study material. Two of these were closely related to diverging strains of Thekopsora areolata, and one was related to Chrysomyxa arctostaphyli (Urediniomycetidae). Cryptococcus victoriae (Tremellomycetidae) was most abundant of the several cryptococcal species observed. Five sequences originated from strains of Malassezia restricta (subclass Exobasidiomycetidae), which was prevalent in both buildings in most seasons. The OTU related to various Malassezia species and strains accounted for 14% of all clones (Table 2). Basidiomycetous sequences belonging to Agaricomycetidae (corresponding roughly to the Homobasidiomycetes, fruiting body-producing basidiomycete fungi), although representing the majority of OTU types, were mainly present as singletons and occurred mostly in samples taken in summer and fall.
Seasonal variation of indoor fungal flora.
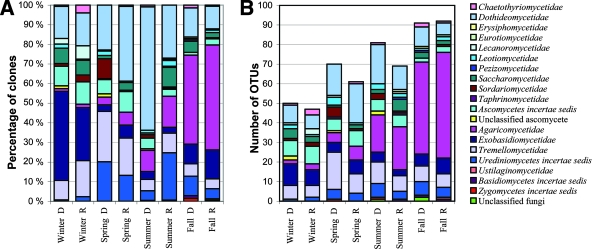
The most prominent structural change in the molecular diversity of fungi was the constant increase in diversity of the OTU belonging to Agaricomycetidae basidiomycetes at the expense of ascomycetes and yeast-like basidiomycetes from winter to fall (Fig. 2B; see Table S1 in the supplemental material for detailed information on the species content of the subclasses). This increase was also evident in the clone numbers (Fig. 2A). In the winter samples, yeasts represented ca. 60% of all clones, whereas in other seasons their proportion was 20 to 30%. The level of ascomycetous diversity was stable from winter to summer, with about 30 OTU types being found in each sample. Most of these had closest relatives in the subclasses Dothideomycetidae and Saccharomycetidae or Ascomycetes incertae sedis. A peak in Dothideomycetidae was seen during the summer in the index building, mainly due to the abundance of Cladosporium spp. In the fall, both the diversity and the relative proportion of clones belonging to these groups decreased significantly (Fig. 2).
FIG. 2.
Seasonal diversity profiles in index and reference building. (A) Relative proportions of clones affiliated with fungal subclasses. (B) Cumulative diversity in form of number of different OTU affiliated with fungal subclasses. D, sample from the water damaged index building; R, sample from the reference building.
In winter samples, sequences similar to species of the yeast-like genera Malassezia and Cryptococcus, and to a lesser extent Filobasidium, Ustilago, Rhodosporidium, Rhodotorula, Cystofilobasidium, and Mrakia accounted for most of the basidiomycetous diversity. In the spring and summer, the subclass Exobasidiomycetidae was largely replaced by Tremellomycetidae yeasts and Thekopsora areolata rust and other Urediniomycetes in both buildings. The diversity of Exobasidiomycetidae increased significantly again in fall samples, nearly reaching the diversity of winter samples (Fig. 2B).
Filamentous basidiomycete species in winter samples included a few clones of the saprophytic Psathyrella candolleana, the polypore Trametes versicolor, and two unknown species related to the corticoid fungus Athelia arachnoidea (anamorph Fibularhizoctonia carotae). The diversity of filamentous basidiomycete species was also relatively low in the spring samples, consisting mainly of Agaricomycetidae polypores and Uredinales rusts. A wide variety of OTU with highest similarity to mycorrhizal, decomposer, and phytopathogenic basidiomycetes such as Amanita, Collybia, Chrysomyxa, Gymnopus, Hypholoma, Melampsoridium, Tricholoma, Dermocybe, Polyporus, and Trametes appeared in the summer and especially the fall samples (Fig. 2). Unknown sequences with 70 to 90% similarity to known fungi were most abundant in fall samples, where they accounted for almost 30% of all OTU compared to the 20% for other seasons.
Similarity coefficients were calculated for pairwise comparisons of pooled clone libraries from the two buildings for all four seasons. The coefficients ranged from 0.154 to 0.23, the most similar species diversity being found in spring-summer and summer-fall sample pairs (data not shown).
The total viable concentrations of fungi in dust showed strong variation between samples (8 × 103 to 4 × 105 CFU g−1). Most of this variation was explained by elevated numbers of nonsporulating fungi and yeasts in some samples. Without these two groups the concentrations varied between 4 × 103 and 5 × 104 CFU g−1. Elevated numbers (5 × 104 to 3 × 105 CFU g−1) of yeasts occurred in winter and spring samples from the reference building and in the winter sample of the index building. Cultivation was performed in parallel on DG18 and malt extract agar, and the higher count was used (Table 3). In contrast to clone analysis, the fungal communities identified by culture were most diverse in winter for both buildings. The concentrations of culturable fungi were highest during the spring in both buildings. The prevalent groups in addition to yeasts and nonsporulating isolates were species of Cladosporium, Penicillium, and Sphaeropsidales. Penicillium spp. occurred mainly in the samples taken in winter and spring. The total concentrations of culturable fungi were higher in the reference building, mostly due to the higher concentrations of yeasts in the samples.
TABLE 3.
Concentrations of culturable fungi and ergosterol in dust
| Genus and/or group | Representative strain(s) | Concn (CFU g−1) of culturable fungia
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Winter
|
Spring
|
Summer
|
Fall
|
||||||
| D | R | D | R | D | R | D | R | ||
| Sphaeropsidales | BF45, BF12 | 3,600 | 1,400 | 9,009 | 9,009 | 10,811 | 18,957 | ||
| Cladosporium | BF87, BF72 | 45 | 1,400 | 13,514 | 4,505 | 11,712 | 4,500 | 995 | 14,218 |
| Penicillium | BF101, BF79, BF14, BF18, BF11 | 4,500 | 9,100 | 4,505 | 4,505 | 135 | 2,703 | 995 | 2,370 |
| Acremonium | BF100 | 1,400 | 4,505 | 900 | 498 | 4,739 | |||
| Aspergillus spp. | 3,245 | 2,700 | 4,550 | 45 | 450 | 452 | |||
| A. ochraeus | BF83 | 45 | |||||||
| A. niger | 45 | ||||||||
| A. versicolor | BF23 | 45 | 452 | ||||||
| A. penicilloides | BF8c | 1,800 | |||||||
| A. sydowii | BF7 | 4,505 | |||||||
| Aspergillus sp. | —b | 1,800 | 2,700 | 450 | |||||
| Aureobasidium | BF69, BF13 | 900 | 45 | 1,802 | 901 | 3,620 | |||
| Mucor | BF68, BF81 | 450 | 0 | 4,739 | |||||
| Rhinocladiella | — | 45 | 90 | 4,505 | |||||
| Fusarium | BF22 | 45 | 4,500 | ||||||
| Eurotium | BF78, BF106 | 45 | 2,252 | 1,422 | |||||
| Exophiala | BF16 | 450 | 901 | 450 | |||||
| Scopulariopsis | BF44 | 450 | 900 | ||||||
| Chaetomium | — | 910 | |||||||
| Stachybotrys | BF15 | 900 | |||||||
| Paecilomyces | — | 450 | |||||||
| Absidia | 45 | ||||||||
| Trichoderma | 45 | ||||||||
| Very rare or unidentifiable isolates | BF105, BF102 | 4,505 | 1,800 | 498 | 905 | ||||
| Nonsporulating isolates | BF62, BF19, BF75, BF104 | 1,400 | 6,800 | 211,712 | 4,505 | 1,351 | 1,350 | 995 | 4,739 |
| Yeasts | BF73, BF108, BF9, BF109, BF98, BF92, BF10 | 8,200 | 100,100 | 50,000 | 382,883 | 1,351 | 4,950 | 2,985 | 48,341 |
| Total | 25,630 | 127,990 | 303,290 | 415,363 | 27,160 | 15,303 | 8,388 | 103,080 | |
D, sample from water-damaged index building; R, sample from reference building. Numbers in boldface indicate a value from DG18 agar plate, other than from MEA. The ITS region of the colonies from index building were sequenced. The ergosterol concentrations were as follows: winter D, 2.2 ng/mg; winter R, 0.5 ng/mg; spring D, 3.7 ng/mg; spring R, 3.5 ng/mg; summer D, 3.3 ng/mg; summer R, 5.7 ng/mg; fall D, 1.6 ng/mg; fall R, 1.6 ng/mg.
—, all representative isolates of the colony type were either contaminated or a sufficiently good quality DNA sequence could not be obtained.
The ITS region of the representative isolate had greatest sequence similarity with the ITS region of A. restrictus (97.5%).
The ergosterol concentrations of house dust varied between 1.6 and 3.3 ng and 0.5 and 5.7 ng mg−1 of dust in the index and reference buildings, respectively. The highest concentrations of ergosterol in both index and reference buildings were observed during the spring and the summer. Ergosterol concentrations were higher in the index than in the reference building during the winter (Table 3).
Among the seven fungi analyzed by qPCR, the Cladosporium cladosporioides, Trichoderma viride, and Eurotium amstelodami group were found in all samples except in the winter sample from the index building. Despite repeated DNA isolations and PCR analyses, this sample did not yield an amplification result for any of the qPCR assays or for the internal control (Table 4). The concentrations of the three above-mentioned fungi were higher in the index building than in the reference building in all seasons, with the highest concentration in the summer. Stachybotrys chartarum was only detected in the winter sample from the reference site. The other fungi analyzed, i.e., Aspergillus sydowii, Penicillium digitatum, and Penicillium group 2, were not found in the samples by qPCR.
TABLE 4.
Concentrations of fungi in dust analyzed by qPCR
| Assay | Concn (spores g−1) of fungia
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Winter De | Winter R | Spring D | Spring R | Summer D | Summer R | Fall D | Fall R | |
| C. cladosporioidesb | i | 31,000 | 441,500 | 173,700 | 3,228,500 | 1,044,400 | 76,900 | 71,100 |
| E. amstelodamic | i | 59,600 | 528,600 | 13,800 | 568,200 | 30,100 | 26,000 | 19,600 |
| S. chartarum | i | 7,900 | ||||||
| T. virided | i | 47,800 | 46,500 | 11,300 | 128,900 | 52,700 | 31,600 | 4,900 |
D, sample from water-damaged index building; R, sample from reference building.
The assay detects C. cladosporioides serovar 1.
The assay detects Eurotium amstelodami, E. chevalieri, E. herbariorum, E. rubrum, and E. repens.
The assay detects Trichoderma viride, T. atroviride, and T. koningii; A. sydowii; P. digitatum, and Penicillium group 2 (Penicillium crustosum, P. camembertii, P. commune, P. echinulatum, and P. solitum) were also tested being negative for all samples.
i, The sample showed PCR inhibition.
Differences in fungal flora of the index and reference buildings.
The ratio of species diversity in the index building versus the control building ranged from 1.0 to 1.5. It was highest in the spring and winter (1.5 and 1.4, respectively) and close to 1 in the summer and fall. On the subclass level, the overall fungal diversity was fairly similar in both buildings both in terms of the number of clones and in terms of the number of OTU belonging to each group (Fig. 2).
In contrast to the relatively uniform fungal diversities at the subclass level, remarkable variation was observed in the two target buildings at the species level, since ca. 40% of all OTU were found only from the index building, 40% were found only from the reference building, and 20% were found from both buildings. No OTU types unique to either building were found that were present throughout the year. The similarity coefficients calculated by pairwise comparisons of clone libraries ranged from 0.2 to 0.33, being highest in the fall and lowest in the winter (data not shown).
The samples taken during the winter season from the water-damaged building and the reference building harbored different sequence varieties except for the most abundant OTU types listed in Table 2. The OTU sequences from the species Cryptococcus albidosimilis and C. niccombsii and OTU related to Arthrobotrys superba, Cephalotheca sulfurea, Phaeoseptoria sp., Phialophora sp., and Phaeotheca fissurella were present in multiple clones in the winter sample and were unique to the index building. Corresponding findings from the reference building included sequences from Capnobotryella sp., Cordyceps sinensis, Cryptococcus friedmannii, cf. Pseudocladosporium sp., Exophiala salmonis, Rhodotorula slooffiae, and Trimmatostroma abietina and OTU related to Coniosporium sp., Cryptococcus elinovii, Exophiala sp., Neofusicoccum corticosae, Phaeosphaeriaceae sp., and Phialocephala fluminis. During the winter, the proportion of unknown sequences with less than 99% similarity to any known DNA sequence was considerably higher in the damaged compared to the reference building (52% versus 32%). In other seasons, the proportions were similar (data not shown).
Diversity of isolated colonies.
In all, 72 pure cultures were obtained, comprising 23 fungal genera, groups, or species based on microscopic identification. ITS sequence analysis divided these strains into 35 fungal species or strains representing nine subclasses (Fig. 1 and Table S1 in the supplemental material). More than 90% of the cultivated strains had similarity value of ≥99% to an EMBL database reference. The identification by sequence analysis corresponded well with morphological identification. The sequences of 42% of the strains were also found in clone libraries. Cladosporium, yeasts, and nonsporulating isolates were found in all samples. Other common fungi were Penicillium, Sphaeropsidales group, Eurotium, Acremonium, and Aureobasidium spp. (Table 3). Many of the nonsporulating isolates did not grow in liquid culture or were not amplified in PCR analyses. The isolates that could be sequenced had ITS regions most similar to Arthrinium sp., Coniothyrium cereale, Epicoccum nigrum, Geomyces pannorum, and Leptosphaerulina chartarum. One morphologically unidentifiable strain was identified to be Botrytis elliptica by ITS sequencing. One strain identified as Aureobasidium by morphology represented the related species Hormonema carpetanum on the basis of sequence analysis. Detailed information about the cultured isolates is presented in elsewhere (see Table S1 in the supplemental material).
DISCUSSION
The present results demonstrate that considerably more diverse microbial flora is observed from the same material by PCR and clone library sequencing compared to traditional cultivation. The fungal diversity in indoor dust by the former method was high, consisting of nearly 400 OTU types, 45% of which were identified at the species level. The theoretical diversity calculated by using nonparametric estimators was about 100 to 400 OTU per building per season, a richness comparable to that found in soil and plant organs by the same methodology (47, 49). In addition to the overall high diversity of fungi in both buildings, a constant increase in fungal diversity from winter through spring and summer to fall was observed. This is explained by the amplification and diversification of natural outdoor fungal source communities in accordance with their annual succession. With certain exceptions (39), this kind of seasonal variation has not been observed in viable counts or diversity from house dust samples (28, 56). The present findings reflected the various sources of microbes in indoor environments. The largest diversity in cold seasons originated from the ustilaginous basidiomycete subclass Exobasidiomycetidae, consisting mainly of Malassezia yeasts that belong to human skin flora (51). In general, winter samples were dominated by yeasts, and all other seasons were dominated by filamentous fungal species. Sequences belonging to or closely related to various phylloplane and saprotrophic fungi, including Aureobasidium pullulans, Leptosphaerulina sp., Cladosporium herbarum and other Cladosporium spp., Macrophoma sp., Candida sp., Cryptococcus sp., and Rhodotorula sp. were observed in both buildings throughout the year, and these were among the most abundant fungi in the studied material. Also, several DNA sequences belonging to or related to phytopathogenic, mycorrhizal, and lichenous fungi in various subclasses were detected, emphasizing the strong influence of outdoor fungal flora on indoor microbiota. In the summer and fall samples the outdoor load of OTU belonging to decomposing filamentous basidiomycete species increased and finally crowded out some of the ascomycetous diversity in our clone libraries. This predictable phenomenon also explained most of the observed seasonal increase in the total diversity.
Except for Cladosporium spp., which were repeatedly encountered in all clone libraries, few typical indoor air molds were observed in our clone libraries, although these were shown to be present in the material by cultivation techniques. According to the literature, the genera most commonly found in house dust by cultivation are Cladosporium, Alternaria, Aspergillus, Penicillium, and yeasts (11, 12, 16, 39, 54, 74). Moreover, Aureobasidium, Mucor, Botrytis, Wallemia, Fusarium, Eurotium, Rhizopus, Ulocladium, Epicoccum, basidiomycetes, and nonsporulating or unidentifiable isolates have been frequently isolated from dust (11, 12, 56, 73, 74). The Sphaeropsidales group, Cladosporium, Penicillium, Acremonium, and Aspergillus were the major components of culturable mycoflora in our material. Clone sequences originating from Alternaria, Penicillium, and Fusarium were found, but none of these was abundant or prevalent in the material. Sequences from Aureobasidium pullulans were present in all clone libraries, and the genus was also present in substantial amounts in about half of the samples as determined by cultivation. Isolates placed as being Sphaeropsidales group by culture were prevalent and were annotated to represent Phoma herbarum or Phoma sp. based on their ITS sequences. Corresponding sequences were also found in clone libraries in most samples.
In cultivation studies dustborne yeasts have not usually been characterized further, and yet their prevalence in most studies is high (11, 12, 56, 74, 79). In our study, the cultivation results showed that yeasts were abundant especially in the reference building, but this difference between buildings was not discernible in clone libraries. Yeasts were predominant in winter samples in clone libraries but were present in considerable amounts during other seasons as well. In a cultivation study conducted in The Netherlands the predominant yeast species were Rhodotorula glutinis, R. minuta, R. mugilaginosa, Cryptococcus albidus, and C. laurentii (74). Here, the yeast floras consisted of several species of the genera Candida, Cryptococcus, Filobasidium, Mrakia, Pichia, Rhodotorula, Malassezia, and Trichosporon, as well as the species Udeniomyces pannonicus, Saccharomyces cerevisiae, Debaryomyces hansenii, and Phaeococcomyces nigricans. In our study most of the yeast species were detected by clone library analysis only. Basidiomycetes were not quantified by cultivation in our study but accounted for half of the diversity and also about half of the total clone number in our libraries.
In addition to the above-mentioned and several other well-characterized species and groups, we observed a large variety of previously unknown sequence types originating from fungi that have not been cultivated or whose ITS sequences have not been deposited in public databases. These unknown sequence types occurred, on average, in lower counts in clone libraries and also in a smaller number of samples than the known species. The OTU similarities with known sequences were, on average, highest in the winter samples and gradually decreased toward fall. Similarity coefficients were, on average, higher between buildings than between seasons. This suggests that the main factor behind the observed high fungal diversity was the seasonally fluctuating outdoor fungal load.
In order not to miss part of the diversity, which was expected to be high on the basis of previous studies (29, 47, 49, 55), we used large-scale sequencing instead of screening clones or PCR products by methods such as DGGE or amplified ribosomal DNA restriction analysis. For the same reason, a strict 99% OTU threshold was used. The observed high diversity of fungi at various genetic levels supported these decisions. It was observed, however, that even the 99% similarity threshold may not be high enough to differentiate between some closely related species.
The amounts of ergosterol measured in the house dust samples in the present study are comparable to those reported by Saraf et al. (61), who measured ergosterol concentrations of between 2 and 16.5 ng mg−1 in 17 floor dust samples. The highest ergosterol concentrations in both buildings were observed during the spring and summer seasons, which presumably reflects the influence of fungi originating from the outdoor air. The low concentrations observed during the fall were unexpected, since the effect of outdoor air should also have been seen then (39), and abundant fungal diversity was readily visible in clone library results. The observed seasonal trend was supported by the summed qPCR spore counts of the assayed fungi. The amounts of culturable fungi also roughly followed the concentrations of ergosterol except for the summer sample of the reference building, which had low viable counts compared to the measured ergosterol level. The highest concentrations of culturable fungi in our study were observed during the spring. Evidently, the importance of the season on the culturable fungal counts of house dust is not straightforward, since the results of Koch et al. (39) showed the highest concentrations of viable fungi during the summer months, especially August.
Differences between studied buildings.
In subarctic climates indoor sampling is generally recommended to be done during the cold season since the outdoor airborne spore load is then very low due to frozen soil, and the effect of indoor sources becomes most visible (57). During winter, we observed an elevated ergosterol concentration and somewhat higher culturable diversity in dust sampled from the index building than in dust from the reference building. Using clone-library analyses an increased diversity and larger proportion of unknown fungi was observed for the index building compared to the reference building in winter, but—based on the knowledge of moisture-indicating species (59)—the details of the clone data for fungal flora of winter samples did not clearly refer to a difference in the moisture conditions of the buildings. However, it is difficult to interpret the differences between buildings for such a small sample number.
In combined seasonal libraries the qualitative difference between the buildings was noticeable at the OTU level but became indeterminate when higher taxonomic groups or morphological groups were examined. Most OTU sporadically occurred in one or two seasons, and none of the observed prevalent species was unique to just one building. The observed difference in the fungal diversity between the two buildings nevertheless most likely relates to real differences in microbial source populations rather than to low sample coverages, since the highest number of common OTU in the buildings was observed in the fall when the diversity was high, library coverages were low, and the highest proportion of unknown species was present. The opposite was found in winter when the fungal flora was least similar between the buildings, the coverage values were highest, and the highest number of identifiable species was present.
Considering fungal species that might contribute to building related illnesses, some potentially toxic or allergenic fungi were found by ITS clone library sequencing, yet their occurrence was sporadic in most cases. Stachybotrys echinata, Trichosporon sp., and Mucor hiemalis were found only in the index building. Stachybotrys echinata, which is often found growing in wet cellulose materials, may produce cytotoxic trichothecene mycotoxins, as well as several griseofulvins (32). Trichosporon sp. has been associated with hypersensitivity pneumonitis (67). Furthermore, Mucor hiemalis is a common food spoilage mold that produces toxic ergoline alkaloids (41). The potentially allergenic Alternaria tenuissima (58) was detected from the reference building only.
Comparison of methods.
The observed fungal diversity was substantially higher in all seasons when analyzed by the direct cloning approach compared to the traditional culturing method. Using cloning, the number of OTU was on average seven times higher than the number of identified culturable fungal species or groups in the sample (see Table S1 in the supplemental material). The sequences of almost half of the cultivated fungal strains were found in clone libraries. In the majority of the cases, the morphological identification at genus level corresponded to the sequence analysis. The community composition observed by the two methods diverged profoundly both qualitatively and quantitatively. Species affiliated with subclasses Ustilaginomycetidae, Exobasidiomycetidae, Agaricomycetidae, Taphrinomycetidae, Saccharomycetidae, Pezizomycetidae, Lecanoromycetidae, and Erysiphomycetidae were only detected and identified by cloning, whereas subclasses Dothideomycetidae, Eurotiomycetidae, and Leotiomycetidae were accentuated in the cultured diversity, and sequences affiliated with Eurotiomycetidae were practically missing from clone libraries.
Based on cultivation results that were obtained in the beginning of the study and on previous experience with indoor dust, seven mold species or groups were picked for quantitative analysis by qPCR. The occurrence of these did not correlate well with either cultivation or clone library analysis except for Cladosporium cladosporioides, which was found to be the most prevalent species with all of the methods used. The spore counts from the qPCR assay Cclad1 for C. cladosporioides were 2 to 3 orders of magnitude higher than the culturable Cladosporium counts. Comparable ratios have been encountered before in studies comparing viable counts with qPCR results in dust (45) and viable counts in air samples using direct microscopic counts (70). The Cclad1 assay probe matched with the major Cladosporium isolate and two OTU sequences in clone libraries. According to the database comparisons, this assay lacks the ability to differentiate between Cladosporium species C. cladosporioides, C. tenuissimum, C. cucumerinum, and C. oxysporum due to the high sequence similarity of these species' ITS regions.
High CFU counts of Penicillium were observed using cultivation, but this group was not detected by qPCR. Here, the absence of assayed Penicillia, as well as Aspergillus sydowii, in qPCR was explained by single mismatches in the probe sequences compared to the actual sequences of cultivated strains of the named species. Single clones of Penicillium commune and P. crustosum were found in the clone libraries. The unexpected absence or rareness of Penicillium, as well as some other fast-growing abundantly sporulating fungi has been encountered before in several studies utilizing different PCR primer sets with clone library sequencing (29, 37, 42, 49). It seems that, for some unknown reason, cloning methods detect these fungi in much lower numbers than do traditional methods. On the other hand, it is probable that cultivation methods overestimate the proportions of Penicillium and other ubiquitous fast-growing fungi since on plate culture conditions these may compete by overgrowth with the more fastidious organisms (8).
Concerning other species studied using qPCR, two of the quantified fungi, T. viride and E. amstelodami, showed high spore counts but were absent from all clone libraries, as well as plate cultures of most of the samples. When these species were encountered in culture, the viable counts were about 500 times lower than the qPCR result, which suggests low spore viability under the studied conditions. In our study setup, a long collection period, exposure to daylight, and desiccation during the collection probably affected the cultivation results of the more sensitive species. Comparison of the methods is also inherently affected by the fact that CFU often consist of chains of spores or clumps of conidia, whereas qPCR measures single conidia. In addition, dust can be inhomogeneous, and since the sample was divided into aliquots for the different analyses, the separate aliquots may have harbored slightly different flora, although the dust was homogenized before it was divided into aliquots. qPCR inhibition was observed in one sample (the index winter sample). This effect was not removed by further purification or dilution of the samples.
Although offering major advantages to microbial community analysis, DNA-based methods inevitably also bias the results. The profound differences in the relative contributions of diverse taxa such as Penicillium and basidiomycetes to the dust mycoflora revealed by DNA- versus culture-based methods underscore the practical importance of the biases inherent in each of these methods. Technological factors such as the extractability of genomic DNA, the rDNA copy number, primer matching and performance, PCR amplification efficiency of the target region, and the cloning efficiency of the amplified PCR fragments may vary between microbial species and affect the OTU frequencies in clone libraries (75). Besides differential amplification of target species, another considerable source of bias is artificial diversity mainly originating from PCR errors and chimeric PCR products (35, 68, 76). In the present study PCR conditions were optimized to produce as small a bias as possible in this regard. The proportion of chimeric PCR products was significantly lower among sequences originating from simple PCR than from seminested PCR (0.02% versus 8.7%, respectively), a finding that is consistent with previous studies reporting a positive correlation between frequency of chimeric products and PCR cycle number (68, 77).
The Fun18Sf-ITS4 primer pair has been designed to be fungal specific in silico and in microbial pure cultures. It provides robust PCR amplification result from fungal pure cultures and environmental samples and has produced ITS libraries free of plant sequences when used in fungal community analysis of waste composting process (Hultman et al., unpublished data). In the present study, however, this primer pair amplified plant and, to a lesser extent, algal DNA. This behavior was unexpected and calls for reevaluation of the primer performance and required stringency conditions before the primer pair is further used in samples that harbor plant material.
The comparison of clone sequencing with qPCR and cultivation in the present study supported the idea that clone libraries do not reliably reflect the original frequencies of species in a sample. Nevertheless, the inherent differences of the methods we used complicated the comparison of the results. For instance, the comparison of the cultivation and library cloning methods with qPCR appeared partly irrelevant since the widely used U.S. Environmental Protection Agency qPCR probe set was not suitable for some of the local fungal strains. It is also possible that the ITS sequences in clone libraries are not always correctly identified by similarity searches against databases due to incomplete coverage of available reference sequences or due to falsely annotated reference sequences. For this, corresponding sequence analysis of cultivated strains and sequence alignment with clone sequences is crucial for comparing the results.
Despite the discussed methodological limitations, the present study supported the idea that the cloning and sequencing method offers a very sensitive and accurate tool, especially for the detection of rare or uncultivable fungi. This method may provide a useful account of total fungal diversity in the screening of poorly known environments. However, it is not suitable as a sole method for quantitative analyses of fungal communities.
Conclusions.
To our knowledge, this is the first time that the fungal flora of indoor dust has been characterized using sequencing of rDNA clone libraries. Based on our data, the answers to the issues posed in the first part of the present study can be stated as follows. (i) The diversity of the fungal flora in indoor dust is wide, comprising many basidiomycetous species in addition to the ascomycetes found by cultivation in earlier studies. (ii) The fungal flora observed by the cultivation and clone sequencing methods differs markedly. Diversity was found to be wider and more evenly distributed between fungal groups using the clone sequencing. Uncultivable fungi and species whose detection would be challenging by cultivation were detected by clone analysis. On the other hand, both methods failed to detect some taxa readily detected by the other. (iii) qPCR did not support the findings in clone libraries or culture. This calls into question the reliability of characterizations of microbial populations based on a single method. (iv) Seasonal variation of fungal communities can be observed by clone analysis. Trends in the occurrence of fungal subclass level groups and morphological types were detected. The differences between the water-damaged index building and undamaged reference building occurred at the species level, but no fungi characteristic to either building were found. Drawing further conclusions about the relevance of the results in this respect would require more prominent differences between the buildings or a much larger data set.
Supplementary Material
Acknowledgments
This study was financially supported by the Finnish Technology Agency (Tekes) and was part of the FINE technology program.
We thank Mari Koski and Kirsi Lipponen for skillful technical assistance and Martin Romantschuk for critical review of the manuscript.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bornehag, C. G., J. Sundell, S. Bonini, A. Custovic, P. Malmberg, S. Skerfving, T. Sigsgaard, A. Verhoeff, et al. 2004. Dampness in buildings as a risk factor for health effects, EUROEXPO: a multidisciplinary review of the literature (1998-2000) on dampness and mite exposure in buildings and health effects. Indoor Air 14:243-257. [DOI] [PubMed] [Google Scholar]
- 3.Bornehag, C. G., G. Blomquist, F. Gyntelberg, B. Jarvholm, P. Malmberg, L. Nordvall, A. Nielsen, G. Pershagen, and J. Sundell. 2001. Dampness in buildings and health: Nordic interdisciplinary review of the scientific evidence on associations between exposure to “dampness” in buildings and health effects (NORDDAMP). Indoor Air 11:72-86. [DOI] [PubMed] [Google Scholar]
- 4.Bougoure, D. S., and J. W. Cairney. 2005. Assemblages of ericoid mycorrhizal and other root-associated fungi from Epacris pulchella (Ericaceae) as determined by culturing and direct DNA extraction from roots. Environ. Microbiol. 7:819-827. [DOI] [PubMed] [Google Scholar]
- 5.Brasel, T. L., D. R. Douglas, S. C. Wilson, and D. C. Straus. 2005. Detection of airborne Stachybotrys chartarum macrocyclic trichothecene mycotoxins on particulates smaller than conidia. Appl. Environ. Microbiol. 71:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkman, N. E., R. A. Haugland, L. J. Wymer, M. Byappanahalli, R. L. Whitman, and S. J. Vesper. 2003. Evaluation of a rapid, quantitative real-time PCR method for enumeration of pathogenic Candida cells in water. Appl. Environ. Microbiol. 69:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchan, A., S. Y. Newell, J. I. Moreta, and M. A. Moran. 2002. Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern U.S. salt marsh. Microb. Ecol. 43:329-340. [DOI] [PubMed] [Google Scholar]
- 8.Carlile, M. J., S. C. Watkinson, and G. W. Gooday. 2001. The fungi. Academic Press, London, England.
- 9.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 10.Chao, A., M. Ma, and M. C. K. Yang. 1993. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrics 43:783-791. [Google Scholar]
- 11.Chao, H. J., D. K. Milton, J. Schwartz, and H. A. Burge. 2002. Dustborne fungi in large office buildings. Mycopathologia 154:93-106. [DOI] [PubMed] [Google Scholar]
- 12.Chew, G. L., C. Rogers, H. A. Burge, M. L. Muilenberg, and D. R. Gold. 2003. Dustborne and airborne fungal propagules represent a different spectrum of fungi with differing relations to home characteristics. Allergy 58:13-20. [DOI] [PubMed] [Google Scholar]
- 13.Cooley, J. D., W. C. Wong, C. A. Jumper, and D. C. Straus. 1998. Correlation between the prevalence of certain fungi and sick building syndrome. Occup. Environ. Med. 55:579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, T. R., B. Roop, D. Betancourt, and M. Y. Menetrez. 2005. A simple multiplex polymerase chain reaction assay for the identification of four environmentally relevant fungal contaminants. J. Microbiol. Methods 61:9-16. [DOI] [PubMed] [Google Scholar]
- 15.Dean, T. R., M. Kohan, D. Betancourt, and M. Y. Menetrez. 2005. A simple polymerase chain reaction/restriction fragment length polymorphism assay capable of identifying medically relevant filamentous fungi. Mol. Biotechnol. 31:21-28. [DOI] [PubMed] [Google Scholar]
- 16.Dillon, H. K., J. D. Miller, W. G. Sorenson, J. Douwes, and R. R. Jacobs. 1999. Review of methods applicable to the assessment of mold exposure to children. Environ. Health Perspect. 107(Suppl. 3):473-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gachon, C., and P. Saindrenan. 2004. Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by Alternaria brassicicola and Botrytis cinerea. Plant Physiol. Biochem. 42:367-371. [DOI] [PubMed] [Google Scholar]
- 19.Garrett, M. H., P. R. Rayment, M. A. Hooper, M. J. Abramson, and B. M. Hooper. 1998. Indoor airborne fungal spores, house dampness and associations with environmental factors and respiratory health in children. Clin. Exp. Allergy 28:459-467. [DOI] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 21.Guo, L. D., K. D. Hyde, and E. C. Liew. 2001. Detection and taxonomic placement of endophytic fungi within frond tissues of Livistona chinensis based on rDNA sequences. Mol. Phylogenet. Evol. 20:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Haugland, R. A., N. Brinkman, and S. J. Vesper. 2002. Evaluation of rapid DNA extraction methods for the quantitative detection of fungi using real-time PCR analysis. J. Microbiol. Methods 50:319-323. [DOI] [PubMed] [Google Scholar]
- 23.Haugland, R. A., M. Varma, L. J. Wymer, and S. J. Vesper. 2004. Quantitative PCR analysis of selected Aspergillus, Penicillium, and Paecilomyces species. Syst. Appl. Microbiol. 27:198-210. [DOI] [PubMed] [Google Scholar]
- 24.Hawksworth, D. L. 1991. The fungal dimension of biodiversity: magnitude, significance and conservation. Mycol. Res. 95:641-655. [Google Scholar]
- 25.Hendolin, P. H., L. Paulin, P. Koukila-Kahkola, V. J. Anttila, H. Malmberg, M. Richardson, and J. Ylikoski. 2000. Panfungal PCR and multiplex liquid hybridization for detection of fungi in tissue specimens. J. Clin. Microbiol. 38:4186-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirvonen, M. R., M. Ruotsalainen, K. Savolainen, and A. Nevalainen. 1997. Effect of viability of actinomycete spores on their ability to stimulate production of nitric oxide and reactive oxygen species in RAW264.7 macrophages. Toxicology 124:105-114. [DOI] [PubMed] [Google Scholar]
- 27.Hope, A. P., and R. A. Simon. 2007. Excess dampness and mold growth in homes: an evidence-based review of the aeroirritant effect and its potential causes. Allergy Asthma Proc. 28:262-270. [DOI] [PubMed] [Google Scholar]
- 28.Horner, W. E., A. G. Worthan, and P. R. Morey. 2004. Air- and dustborne mycoflora in houses free of water damage and fungal growth. Appl. Environ. Microbiol. 70:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt, J., L. Boddy, P. F. Randerson, and H. J. Rogers. 2004. An evaluation of 18S rDNA approaches for the study of fungal diversity in grassland soils. Microb. Ecol. 47:385-395. [DOI] [PubMed] [Google Scholar]
- 30.Husman, T. 1996. Health effects of indoor-air microorganisms. Scand. J. Work Environ. Health 22:5-13. [DOI] [PubMed] [Google Scholar]
- 31.Huttunen, K., J. Pelkonen, K. F. Nielsen, U. Nuutinen, J. Jussila, and M. R. Hirvonen. 2004. Synergistic interaction in simultaneous exposure to Streptomyces californicus and Stachybotrys chartarum. Environ. Health Perspect. 112:659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarvis, B. B., Y. Zhou, J. Jiang, S. Wang, W. G. Sorenson, E. L. Hintikka, M. Nikulin, P. Parikka, R. A. Etzel, and D. G. Dearborn. 1996. Toxigenic molds in water-damaged buildings: dechlorogriseofulvins from Memnoniella echinata. J. Nat. Prod. 59:553-554. [DOI] [PubMed] [Google Scholar]
- 33.Johanning, E., R. Biagini, D. Hull, P. Morey, B. Jarvis, and P. Landsbergis. 1996. Health and immunology study following exposure to toxigenic fungi (Stachybotrys chartarum) in a water-damaged office environment. Int. Arch. Occup. Environ. Health 68:207-218. [DOI] [PubMed] [Google Scholar]
- 34.Jumpponen, A., and L. C. Johnson. 2005. Can rDNA analyses of diverse fungal communities in soil and roots detect effects of environmental manipulations? A case study from tallgrass prairie. Mycologia 97:1177-1194. [DOI] [PubMed] [Google Scholar]
- 35.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy, N., E. Brodie, J. Connolly, and N. Clipson. 2006. Seasonal influences on fungal community structure in unimproved and improved upland grassland soils. Can. J. Microbiol. 52:689-694. [DOI] [PubMed] [Google Scholar]
- 37.Kernaghan, G., L. Sigler, and D. Khasa. 2003. Mycorrhizal and root endophytic fungi of containerized Picea glauca seedlings assessed by rDNA sequence analysis. Microb. Ecol. 45:128-136. [DOI] [PubMed] [Google Scholar]
- 38.Kirk, P. M., A. Aptroot, G. Ainsworth, and CABI Bioscience. 2001. Ainsworth & Bisby's dictionary of the fungi. CAB International, Wallingford, United Kingdom.
- 39.Koch, A., K. J. Heilemann, W. Bischof, J. Heinrich, and H. E. Wichmann. 2000. Indoor viable mold spores: a comparison between two cities, Erfurt (eastern Germany) and Hamburg (western Germany). Allergy 55:176-180. [DOI] [PubMed] [Google Scholar]
- 40.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lugauskas, A., J. Repeckiene, and H. Novosinskas. 2005. Micromycetes, producers of toxins, detected on stored vegetables. Ann. Agric. Environ. Med. 12:253-260. [PubMed] [Google Scholar]
- 42.Lydolph, M. C., J. Jacobsen, P. Arctander, M. T. Gilbert, D. A. Gilichinsky, A. J. Hansen, E. Willerslev, and L. Lange. 2005. Beringian paleoecology inferred from permafrost-preserved fungal DNA. Appl. Environ. Microbiol. 71:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacNeil, L., T. Kauri, and W. Robertson. 1995. Molecular techniques and their potential application in monitoring the microbiological quality of indoor air. Can. J. Microbiol. 41:657-665. [DOI] [PubMed] [Google Scholar]
- 44.Martin, C., D. Roberts, M. van Der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 38:3735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meklin, T., R. A. Haugland, T. Reponen, M. Varma, Z. Lummus, D. Bernstein, L. J. Wymer, and S. J. Vesper. 2004. Quantitative PCR analysis of house dust can reveal abnormal mold conditions. J. Environ. Monit. 6:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, J. D., P. D. Haisley, and J. H. Reinhardt. 2000. Air sampling results in relation to extent of fungal colonization of building materials in some water-damaged buildings. Indoor Air 10:146-151. [DOI] [PubMed] [Google Scholar]
- 47.Neubert, K., K. Mendgen, H. Brinkmann, and S. G. Wirsel. 2006. Only a few fungal species dominate highly diverse mycofloras associated with the common reed. Appl. Environ. Microbiol. 72:1118-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niemeier, R. T., S. K. Sivasubramani, T. Reponen, and S. A. Grinshpun. 2006. Assessment of fungal contamination in moldy homes: comparison of different methods. J. Occup. Environ. Hyg. 3:262-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Brien, H. E., J. L. Parrent, J. A. Jackson, J. M. Moncalvo, and R. Vilgalys. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71:5544-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odum, E. P. 1971. Principles and concepts pertaining to organization at the community level, p. 140-161. In E. P. Odum (ed.), Fundamentals of ecology. W. B. Saunders, Philadelphia, PA.
- 51.Paulino, L. C., C. H. Tseng, B. E. Strober, and M. J. Blaser. 2006. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 44:2933-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penttinen, P., J. Pelkonen, K. Huttunen, and M. R. Hirvonen. 2006. Cocultivation of Streptomyces californicus and Stachybotrys chartarum stimulates the production of cytostatic compound(s) with immunotoxic properties. Toxicol. Appl. Pharmacol. 217:342-351. [DOI] [PubMed] [Google Scholar]
- 54.Piecková, E., and K. Wilkins. 2004. Airway toxicity of house dust and its fungal composition. Ann. Agric. Environ. Med. 11:67-73. [PubMed] [Google Scholar]
- 55.Pitkäranta, M., H. Rintala, M. Hänninen, and L. Paulin. 2005. Characterization of fungal flora from moisture damaged building material by rDNA sequencing and culture, p. 375-383. In E. Johanning (ed.), Bioaerosols, fungi, bacteria, mycotoxins, and human health: proceedings of the 5th international conference. Boyd Publishing, Albany, NY.
- 56.Ren, P., T. M. Jankun, and B. P. Leaderer. 1999. Comparisons of seasonal fungal prevalence in indoor and outdoor air and in house dusts of dwellings in one Northeast American county. J. Expo. Anal. Environ. Epidemiol. 9:560-568. [DOI] [PubMed] [Google Scholar]
- 57.Reponen, T., A. Nevalainen, M. Jantunen, M. Pellikka, and P. Kalliokoski. 1992. Normal range criteria for indoor air bacteria and fungal spores in subarctic climate. Indoor Air 2:26-31. [Google Scholar]
- 58.Saenz-de-Santamaria, M., I. Postigo, A. Gutierrez-Rodriguez, G. Cardona, J. A. Guisantes, J. Asturias, and J. Martinez. 2006. The major allergen of Alternaria alternata (Alta1) is expressed in other members of the Pleosporaceae family. Mycoses 49:91-95. [DOI] [PubMed] [Google Scholar]
- 59.Samson, R. A., B. Flannigan, M. E. Flannigan, A. P. Verhoeff, O. C. G. Adan, and E. S. Hoekstra. 1994. Recommendations, p. 529-538. In R. A. Samson and B. B. Flannigan (ed.), Health implications of fungi in indoor environments (air quality monographs), vol. 2. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 60.Samson, R. A., E. S. Hoekstra, J. C. Frisvad, and O. Filtenborg. 1996. Introduction to food-borne fungi. Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
- 61.Saraf, A., L. Larsson, H. Burge, and D. Milton. 1997. Quantification of ergosterol and 3-hydroxy fatty acids in settled house dust by gas chromatography-mass spectrometry: comparison with fungal culture and determination of endotoxin by a Limulus amebocyte lysate assay. Appl. Environ. Microbiol. 63:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schabereiter-Gurtner, C., G. Pinar, W. Lubitz, and S. Rolleke. 2001. Analysis of fungal communities on historical church window glass by denaturing gradient gel electrophoresis and phylogenetic 18S rDNA sequence analysis. J. Microbiol. Methods 47:345-354. [DOI] [PubMed] [Google Scholar]
- 63.Schadt, C. W., A. P. Martin, D. A. Lipson, and S. K. Schmidt. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359-1361. [DOI] [PubMed] [Google Scholar]
- 64.Sebastian, A., and L. Larsson. 2003. Characterization of the microbial community in indoor environments: a chemical-analytical approach. Appl. Environ. Microbiol. 69:3103-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 67.Sugita, T., R. Ikeda, and A. Nishikawa. 2004. Analysis of Trichosporon isolates obtained from the houses of patients with summer-type hypersensitivity pneumonitis. J. Clin. Microbiol. 42:5467-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toivola, M., S. Alm, T. Reponen, S. Kolari, and A. Nevalainen. 2002. Personal exposures and microenvironmental concentrations of particles and bioaerosols. J. Environ. Monit. 4:166-174. [DOI] [PubMed] [Google Scholar]
- 71.Tuomi, T., K. Reijula, T. Johnsson, K. Hemminki, E. L. Hintikka, O. Lindroos, S. Kalso, P. Koukila-Kahkola, H. Mussalo-Rauhamaa, and T. Haahtela. 2000. Mycotoxins in crude building materials from water-damaged buildings. Appl. Environ. Microbiol. 66:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vandenkoornhuyse, P., S. L. Baldauf, C. Leyval, J. Straczek, and J. P. Young. 2002. Extensive fungal diversity in plant roots. Science 295:2051. [DOI] [PubMed] [Google Scholar]
- 73.Verhoeff, A. P., E. S. van Reenen-Hoekstra, R. A. Samson, B. Brunekreef, and J. H. van Wijnen. 1994. Fungal propagules in house dust. I. Comparison of analytic methods and their value as estimators of potential exposure. Allergy 49:533-539. [DOI] [PubMed] [Google Scholar]
- 74.Verhoeff, A. P., J. H. van Wijnen, E. S. van Reenen-Hoekstra, R. A. Samson, R. T. van Strien, and B. Brunekreef. 1994. Fungal propagules in house dust. II. Relation with residential characteristics and respiratory symptoms. Allergy 49:540-547. [DOI] [PubMed] [Google Scholar]
- 75.Vesper, S. J., L. J. Wymer, T. Meklin, M. Varma, R. Stott, M. Richardson, and R. A. Haugland. 2005. Comparison of populations of mould species in homes in the UK and USA using mould-specific quantitative PCR. Lett. Appl. Microbiol. 41:367-373. [DOI] [PubMed] [Google Scholar]
- 76.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 77.Wang, G. C., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 78.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315-322. In D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY.
- 79.Wickman, M., S. Gravesen, S. L. Nordvall, G. Pershagen, and J. Sundell. 1992. Indoor viable dust-bound microfungi in relation to residential characteristics, living habits, and symptoms in atopic and control children. J. Allergy Clin. Immunol. 89:752-759. [DOI] [PubMed] [Google Scholar]
- 80.Zeng, Q. Y., A. Rasmuson-Lestander, and X. R. Wang. 2004. Extensive set of mitochondrial LSU rDNA-based oligonucleotide probes for the detection of common airborne fungi. FEMS Microbiol. Lett. 237:79-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.