Abstract
A sequential batch reactor (SBR) and a membrane bioreactor (MBR) were inoculated with the same sludge from a municipal wastewater treatment plant, supplemented with ammonium, and operated in parallel for 84 days. It was investigated whether the functional stability of the nitrification process corresponded with a static ammonia-oxidizing bacterial (AOB) community. The SBR provided complete nitrification during nearly the whole experimental run, whereas the MBR showed a buildup of 0 to 2 mg nitrite-N liter−1 from day 45 until day 84. Based on the denaturing gradient gel electrophoresis profiles, two novel approaches were introduced to characterize and quantify the community dynamics and interspecies abundance ratios: (i) the rate of change [Δt(week)] parameter and (ii) the Pareto-Lorenz curve distribution pattern. During the whole sampling period, it was observed that neither of the reactor types maintained a static microbial community and that the SBR evolved more gradually than the MBR, particularly with respect to AOB (i.e., average weekly community changes of 12.6% ± 5.2% for the SBR and 24.6% ± 14.3% for the MBR). Based on the Pareto-Lorenz curves, it was observed that only a small group of AOB species played a numerically dominant role in the nitritation of both reactors, and this was true especially for the MBR. The remaining less dominant species were speculated to constitute a reserve of AOB which can proliferate to replace the dominant species. The value of these parameters in terms of tools to assist the operation of activated-sludge systems is discussed.
Fernandez et al. (12) demonstrated that ecosystem stability encompasses a broad spectrum of definitions. These definitions resolve into two components: the measurable functional properties of the ecosystem and the change over time of the community composition. Frequently, only the functional parameters are evaluated (37), but recently attempts were made to relate (dys)functioning with the structure of a microbial community (13, 25, 46, 49). It was found that highly dynamic communities can still maintain a stable ecosystem function (12, 25).
Curtis and Sloan (8) postulated that the relationship between structure and function in a microbial community can be evaluated only through an understanding of the source of diversity from which the community is drawn, named the global reservoir or metacommunity. Up to now, diversity has been mainly measured statically, based on indices such as Shannon and Simpson indices. By adding Lorenz curves, a nonambiguous graphical presentation of species evenness can be provided (31, 35, 42), investigating interspecies abundance ratios and thus the internal community structure. Besides the importance of the community diversity and composition at one time point, the dynamics of microbial communities can play an important role in the functionality of a system (49). Nevertheless, due to infrequent sampling, the dynamics of functionally stable microbial communities has not yet been quantified in terms of a rate of change on a short-term basis (weeks).
Nitrification is the succession of the conversion of ammonium to nitrite (nitritation) by the ammonia-oxidizing bacteria (AOB) and the subsequent oxidation to nitrate (nitratation) by nitrite-oxidizing bacteria (NOB) (14). Ammonia oxidation is often a rate-limiting step in nitrogen removal in wastewater treatment plants (WWTP), as the AOB are influenced by various environmental factors such as light, dissolved oxygen, pH, and substrate concentrations (2, 19, 47, 48) and easily inhibited by various organic compounds or heavy metals (24, 26, 34). Curtis and Sloan (8) postulated that autotrophic ammonia oxidation in WWTP is governed by a small metacommunity, leading to easy replicate sampling of the microbially diverse community, which is thought to be stable. Therefore, the AOB form an ideal model group to study functional and microbial stability.
This paper describes a monitoring study of the physicochemical parameters, the floc structures, and the nitrifying bacterial communities of a sequential batch reactor (SBR) and a membrane bioreactor (MBR) operated in parallel. Both reactors were inoculated with the same sludge from a municipal WWTP and received the same ammonium-enriched water with equal volumetric loading rates. Other environmental parameters were kept the same as well. It was investigated whether the functional stability of the nitrification process corresponded with a static bacterial community. Hereto, the bacterial reactor dynamics were characterized by a rate of change as a function of time, Δt(week), and the internal community structure was evaluated by Pareto-Lorenz evenness distribution curves.
MATERIALS AND METHODS
Experimental setup and sampling.
Two simultaneously run reactors were followed for 84 consecutive days. At day 0, the reactors were inoculated with sludge obtained from a full-scale municipal WWTP (Ossemeersen, Gent, Belgium). The influent consisted of NH4Cl (volumetric loading rate between day 1 and day 17, gradually from 15 to 100 mg N liter−1 day−1; volumetric loading rate from day 18 to 84, fixed at 100 mg N liter−1 day−1) supplemented with 0.5 g day−1 Nutriflok as a source of macro- and micronutrients (Avecom, Beernem, Belgium) and 0.17 g day−1 KH2PO4-P. The first reactor was a 37.5-liter SBR. Daily, two complete cycles (12 h in total) were performed, consisting of (i) a 10-h aeration phase (feeding during the first 0.5 h at 25 liter h−1), (ii) an 85-min settling phase, (iii) a 5-min decanting phase, (iv) a 15-min safety phase, and (v) a 15-min restarting phase for the aeration and pH control (prior to feeding). The second reactor was a 25-liter MBR (Solis Engineering bv, Lelystad, The Netherlands) equipped with three double-deck flat-sheet membranes (Kubota, Osaka, Japan). The liquid feed of the MBR was gradually administered throughout the whole day (0.7 liter h−1). Each of the reactors had a separate acid-base pH controlling system to maintain the pH between 7.2 and 7.6. For both reactors, the hydraulic residence time was 1.5 days. The solids retention time was 1,050 days, since no sludge was removed (except for volatile suspended solids [VSS] determination, fluorescence in situ hybridization [FISH] analysis, and DNA extraction).
Physicochemical parameters.
The nitrite and nitrate contents of the effluent were analyzed by ion chromatography as previously described (49). Ammonium (colorimetric), suspended solids (SS), and VSS were analyzed by standard methods (18).
Floc size distribution.
Floc size distribution was measured with a CIS-100 apparatus (Ankersmid, Edegem, Belgium), which combines a laser channel performing size measurements based on the time-of-transition principle with an image-analyzing channel allowing characterization of the dynamic size and shape. Equipment operation was performed according to the procedure described by Govoreanu et al. (17).
FISH analysis.
For FISH analysis, activated-sludge samples were fixed according to the protocol of Amann et al. (1). FISH assays were performed with fluorescence-labeled rRNA-targeted oligonucleotide probes by use of the method of Biesterfeld et al. (3). For the AOB species, the Cy3-labeled probe NSO1225 (36) was applied, and for the Nitrospira spp., the Alexa488-labeled probe S-G-Ntspa-0662-a-A-18 (9) was applied. Hybridizations were performed using the probe mixture and a hybridization buffer with a formamide concentration of 35%. Hybridized samples were visualized with a Nikon Eclipse TE300 epifluorescence microscope equipped with a Bio-Rad Radiance 2000 confocal system. Further image processing was performed using ImageJ 1.36b software freely available at http://rsb.info.nih.gov/ij.
Nucleic acid extraction.
Mixed liquor samples of 50 ml were centrifuged at 3,000 × g for 2 min, and supernatants were discarded. Nucleic acids were extracted from 0.5 g of the centrifuged sludge samples by using a low-pH hot phenol extraction procedure as previously described (44). The DNA concentration was measured spectrophotometrically at 260 nm (NanoDrop ND-1000 spectrophotometer; Isogen Life Science, Sint-Pieters-Leeuw, Belgium) and set to 5 ng μl−1 using DNase- and RNase-free filter-sterilized water (Sigma-Aldrich Chemie, Steinheim, Germany).
PCR amplification.
PCR was performed with primers targeting all bacteria (P338f with GC-clamp and P518r) to obtain DNA amplicons for further analysis of the total bacterial community by denaturing gradient gel electrophoresis (DGGE) (39). To study the AOB community, a nested PCR approach was used. The first round made use of the primers CTO189AB, CTO189C, and CTO653r to amplify the AOB specifically (27). The second round was performed with total bacterial primers (P338f with GC-clamp and P518r). PCRs were performed according to the protocol of Boon et al. (4). In all PCR runs, a positive control (1 μl of purified DNA from strain Nitrosomonas europaea ATCC 19718) and a negative control (1 μl of DNase- and RNase-free filter-sterilized water [Sigma-Aldrich Chemie, Steinheim, Germany]) were incorporated. After each PCR run, the size of the amplicon was verified by running it next to a low-range DNA Massruler (Fermentas, Burlington, ON, Canada) on a 1% agarose gel.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) was performed on an ABI Prism SDS 7000 (PE Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands). To evaluate the variation of the qRT-PCR technique, all amplification reactions were carried out in triplicate with SYBR green PCR master mix (Applied Biosystems). The following primer sets were used: (i) for the AOB, amoA-1F and amoA-2R (41); (ii) for the Nitrobacter spp., FGPS872 and FGPS1269 (10); and (iii) for the Nitrospira spp., NSR1113F and NSR 1264R (11). An oligonucleotide concentration of 150 nM and a DNA template volume of 5 μl were added to 20 μl of PCR master mix in MicroAmp optical 96-well reaction plates (Applied Biosystems). The real-time PCR protocol for all primer sets was as follows: 2 min at 50°C (for enzyme activation, as recommended by the manufacturer), 10 min at 95°C, and 40 cycles consisting of 1 min at 95°C, 1 min at the respective annealing temperatures, and 1 min at 60°C. The respective annealing temperatures were 54°C for the amoA-1F and amoA-2R primer set, 50°C for the FGPS872 and FGPS1269 primer set, and 65°C for the NSR1113F and NSR 1264R primer set. For the amoA, FGPS, and NSR standard curves, the respective standard sequences originated from Nitrosomonas europaea ATCC 19718, Nitrobacter winogradskyi ATCC 25391T, and Nitrospira moscoviensis NCMIB 13793T. In all experiments, appropriate negative controls containing no template DNA were subjected to the same procedure to exclude or detect any possible contamination or carryover. For the primer sets applied in this research, the respective amplification specificities for sludge samples were previously demonstrated by cloning (15). For each primer set, a standard curve was constructed by diluting a PCR-amplified and cloned standard sequence. Furthermore, melting curves were constructed by constant fluorescent measurement during a final 60°C-to-95°C heating at 0.1°C s−1 and routinely checked to confirm the purity of the amplified products. For the recalculation from gene copy numbers to numbers of cells, it was assumed that there were two copies of the amoA gene per AOB cell and one copy of the 16S rRNA gene per NOB cell (11).
DGGE analysis.
A Bio-Rad DGene system (Hercules, CA) was used for running 8% (wt/vol) polyacrylamide DGGE gels with a denaturing gradient ranging from 45 to 60% (100% denaturing contains 7 mol liter−1 urea and 40% formamide) as described previously (4). The desired DGGE bands were cut out, cloned with the pCR 2.1-TOPO cloning kit (Invitrogen, Carlsbad, CA) according to the manual instructions, and sequenced by IIT Biotech-Bioservice (Bielefeld, Germany). DNA sequence analysis was performed using the BLAST server of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) with the BLAST algorithm and specifically with the BLASTN program.
The obtained DGGE patterns were subsequently processed using BioNumerics software version 2.0 (Applied Maths, Sint-Martens-Latem, Belgium). A matrix of similarities for the densitometric curves of the band patterns was calculated based on the Pearson product-moment correlation coefficients and used to perform moving-window analysis (49) by plotting the correlation between day x and day x − 7. This way, each data point in the graph is in itself a weekly comparison. The Δt(week) values were calculated as the averages and standard deviations for the respective moving-window curve data points subtracted from the 100% similarity value. The higher the changes between the DGGE profiles of day x and day x − 7, the lower the moving-window curve data point and the higher the Δt(week) values will be.
In order to graphically represent the evenness of the AOB communities, Lorenz distribution curves (31) were set up based on the DGGE profiles as previously described (35). Briefly, for each DGGE lane, the respective bands are ranked from high to low based on their intensities. Consecutively, the cumulative proportions of species are used as the x axis, and the y axis is represented by their respective cumulative proportions of intensities. Mathematically, this yields a convex curve. The more the Lorenz curve deviates from the theoretical perfect evenness line (i.e., the 45° diagonal), the less evenness can be observed in the structure of the studied community. A lower evenness means that a smaller fraction of the different species is present in dominant numbers. In this study, the Lorenz curves were also evaluated based on the Pareto principle (40). According to this principle, the cumulative y axis value (in casu the proportion of intensities) corresponding with the 20% level on the x axis (in casu the cumulative proportion of species) is evaluated. In economics, from which the Pareto principle originates, it was observed that a mere 20% of a country's population possesses 80% of its wealth (40). We refer to Lorenz curves which were evaluated on the 20% level of the x axis as “Pareto-Lorenz curves.”
Reproducibility of the DGGE method.
The low-pH hot phenol nucleic acid extraction procedure was successfully applied previously to detect different kinds of ammonia oxidizers and both Nitrobacter and Nitrospira (nitrite-oxidizing) species (38). To test the reproducibility of the sampling, extraction, PCR, and DGGE, triplicate sludge samples were analyzed. Similarity values above 97% (data not shown) were noted between DGGE samples originating from the same reactor on the same day. This demonstrated the high reproducibility of the molecular methods.
Nucleotide sequence accession numbers.
Sequences from the DGGE analysis described above were deposited in the GenBank database under accession numbers EF446325 to EF446336.
RESULTS
Physicochemical analyses.
The concentrations of the nitrogen compounds (influent N, NH4+, NO2−, and NO3−) and mass balances were followed closely during the experimental run of an SBR and an MBR to evaluate the extents of nitrification. For both reactors, the conversion of ammonium to nitrite and further to nitrate was complete and stable over the course of the sampling period from day 1 to day 40 (data not shown). Afterwards, the SBR maintained complete ammonium-to-nitrate conversion, but the MBR showed incomplete nitrification during the second half of the sampling period, as demonstrated by a residual nitrite-N concentration of around 1 mg liter−1 effluent. Daily measurements of the dissolved oxygen of both reactors showed that this parameter did not limit nitrification (in casu between 6.0 ± 0.5 mg O2 liter−1). An important physicochemical parameter reflecting a difference between the reactors was the VSS (see Fig. S1 in the supplemental material). Starting from ca. 3.75 g liter−1, the VSS of both reactors decreased over time. However, this trend was more pronounced for the SBR, which halved its initial VSS by day 40 to attain 1.4 g liter−1 by day 84. For the MBR, it decreased to 2.4 g liter−1 on day 84. A total of 0.3 g of VSS was removed from each bioreactor over the course of the experiment for analysis. Starting from the same inoculum with a sludge volume index (SVI) of 112 ml (g SS)−1, on day 84 the SVI of the SBR had decreased to 73 ml (g SS)−1 and the SVI of the MBR had risen to 205 ml (g SS)−1, reflecting a distinct difference in floc-settling properties between the two reactors.
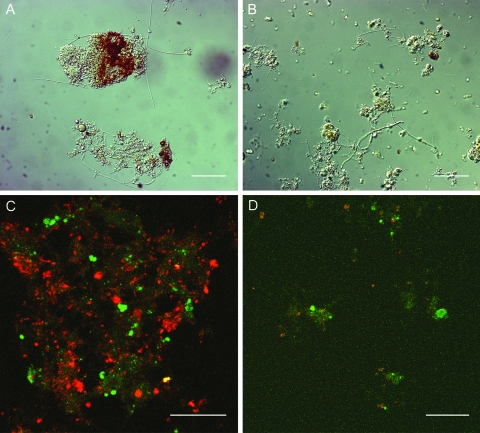
For both reactors, the distribution patterns of the floc diameters were established at the end of the sampling period (see Fig. S2 in the supplemental material). For the SBR, the floc diameter was restricted to 300 μm, and 50% of the flocs had diameters above 165 μm. The MBR floc diameter maximum was 40 μm, and 50% of the flocs were smaller than 13 μm. Based on the steepness of the floc distribution curves, it was concluded that the SBR flocs were more of the same diameter (between 100 μm and 300 μm) and that only 10% of the flocs had a diameter below 100 μm. On the other hand, all flocs of the MBR ranged between 1 and 40 μm. To substantiate this observation, light microscope images were taken for both reactors on day 84 (Fig. 1A and B). These images supported the activated-sludge floc distribution measurements by indicating large and compact flocs in a clear suspension for the SBR, while the MBR's activated sludge consisted of dispersed microflocs.
FIG. 1.
Light microscopic images (magnification, ×400) of the SBR (A) and MBR (B) flocs on day 84, and confocal microscopic images (magnification, ×400) of FISH analysis of the SBR (C) and MBR (D) flocs focused on the AOB (red) and Nitrospira (NOB, green) communities on day 84. Scale bars, 50 μm.
Amounts and localizations of AOB and NOB in both reactors.
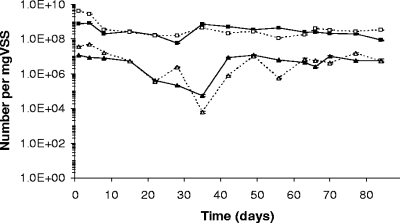
The numbers of cells of AOB and NOB per mg VSS were determined with qRT-PCR (Fig. 2). By day 35, the amount of AOB per mg VSS decreased by about 3 log units from what was seen at day 1. After day 35, a rapid rise in the amount of AOB was noted. Afterwards, a constant number of 6 log units of AOB species per mg VSS was maintained. Concerning the NOB subgroup, members of the Nitrobacter group were not detected (<2 log units of cells per mg VSS), whereas the Nitrospira group was present in high amounts (8 to 9 log units per mg VSS) and remained constant during the complete sampling period. Thus, an abundance of Nitrospira approximately 100-fold higher than that for the AOB was observed for both the SBR and the MBR.
FIG. 2.
Number of AOB (triangles) and Nitrospira (squares) for the SBR (full line) and the MBR (dashed line) as measured by qRT-PCR. Standard deviations (n = 3) are indicated but are very small.
FISH analysis was applied to visualize the interindividual positioning of the AOB and the Nitrospira organisms in the floc structure (Fig. 1C and D). Apart from the differences in floc size, it was noted that both clusters and individual AOB and Nitrospira organisms were present in the SBR and MBR.
Total bacterial DGGE analysis.
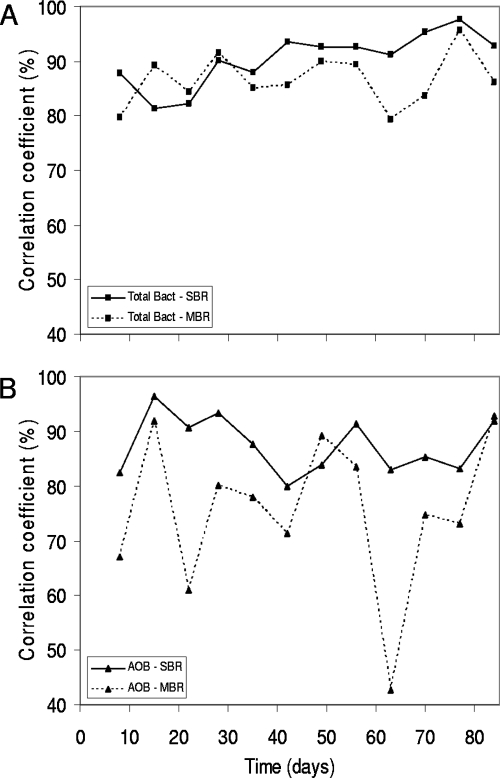
The DGGE analysis of all bacteria for both reactors showed more than 45 bands, indicating a very large number of species (see Fig. S3a and b in the supplemental material). Weekly deviations were established with the moving-window analysis (Fig. 3A). The SBR demonstrated weekly shifts of between 10 and 20% during the first month and changes of up to 10% during the last 2 months. However, the MBR shifted up to 20% during the whole sampling period. Average values for these weekly deviations were calculated and expressed as rates of change [Δt(week)]. It was concluded that the total bacterial community changed more rapidly in the MBR [Δt(week), 13.4% ± 4.8%] than in the SBR [Δt(week), 9.6% ± 4.9%].
FIG. 3.
Moving-window analysis based on DGGE profiles for the SBR (full line) and MBR (dashed line) of the total bacteria (A) and the AOB subgroup (B). Each data point in the graph is in itself a weekly comparison, as it represents the correlation between the samples of day x and day x − 7.
AOB DGGE analysis.
By using AOB-specific primers, a DNA-based DGGE comparison was made to monitor the development of the community structure of the ammonia oxidizers from day 0 to day 84 (see Fig. S3c and d in the supplemental material). In total, about 15 different bands were visible, of which the 12 most dominant bands (designated A to L in Fig. S3c and d in the supplemental material) were sequenced. According to the Ribosomal Database Project II (http://rdp.cme.msu.edu), DGGE bands A to K were all classified as belonging to the genus Nitrosomonas. Table 1 lists for each DGGE band the corresponding closest match via the NCBI database (http://www.ncbi.nlm.nih.gov) with indications of similarity percentages and sources of origin. Sequence analysis revealed that band L had 100% similarity to the uncultured Ferribacterium sp. partial 16S rRNA gene (AJ890204) found in a lab-scale wastewater reactor by DGGE. Therefore, band L can be considered as a result of CTO primer nonspecificity and was excluded from further data processing.
TABLE 1.
The 12 AOB DGGE bands with corresponding closest matches via the NCBI database and indication of similarity percentages and sources of origina
| Band | Closest match | % Similarity (no. of similar bp/total bp) | Origin |
|---|---|---|---|
| A | Uncultured Nitrosomonas sp. (AY543074) | 99 (196/197) | Lab-scale biological aerated filter, Newcastle, United Kingdom |
| B | Uncultured bacterium (AB176884) | 100 (197/197) | Activated-sludge sewage system, Tokyo, Japan |
| C | Uncultured Nitrosomonas sp. (AY583659) | 100 (197/197) | Treated wastewater, lower Seine River, France |
| D | Uncultured Nitrosomonas sp. (AY583659) | 98 (195/197) | Treated wastewater, lower Seine River, France |
| E | Uncultured betaproteobacterium (AJ299048) | 98 (195/197) | Ammonia oxidizer enrichment from freshwater sediment, Lake Drontemeer, The Netherlands |
| F | Nitrosomonas sp. isolate (AJ621026) | 100 (197/197) | Freshwater sediment, Lake Drontemeer, The Netherlands |
| G | Nitrosomonas ureae Nm10T (AF272414) | 100 (197/197) | Soil, Sardinia, Italy |
| H | Uncultured bacterium (AB239537) | 100 (197/197) | River sediment, Niida River, Japan |
| I | Uncultured bacterium (AM295532) | 100 (197/197) | Marine aquaculture biofilm, Rehovot, Israel |
| J | Uncultured bacterium (AB239537) | 99 (196/197) | River sediment, Niida River, Japan |
| K | Uncultured betaproteobacterium (AJ299052) | 99 (196/197) | Ammonia oxidizer enrichment from freshwater sediment, Lake Drontemeer, The Netherlands |
Closest matches by BLASTN; NCBI database, http://www.ncbi.nlm.nih.gov.
Moving-window analysis (Fig. 3B) revealed that the AOB community of the SBR gradually changed over time (weekly changes of up to 20%) but that the MBR AOB community shifts were more pronounced, as demonstrated by the larger fluctuations in the moving-window analysis plot of the MBR (up to 40% correlation and thus 60% change). As for the total bacterial community, Δt(week) values were calculated. The SBR had a Δt(week) value of 12.6% ± 5.2% and the MBR a Δt(week) value of 24.6% ± 14.3% for their respective AOB subgroups.
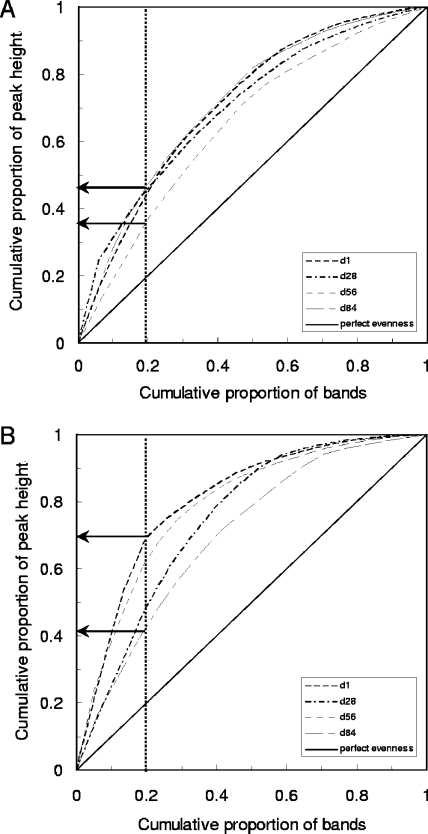
To asses the interspecies abundance ratios of the AOB, Pareto-Lorenz curve distribution patterns of the AOB DGGE profiles were plotted based on the numbers of bands and their intensities (Fig. 4). Over time, it was observed for the SBR reactor that 20% of the bands (number based) corresponded with 35 to 45% (on average 40%) of the cumulative band intensities. For the MBR reactor, 20% of the bands corresponded with 40 to 70% (on average 55%) of the cumulative band intensities. Thus, for both reactor types it was observed that overall only a small group of AOB species was numerically dominant and that this dominance was even more pronounced for the MBR.
FIG. 4.
Pareto-Lorenz distribution curves based on AOB PCR-DGGE analysis on days 1, 28, 56, and 84 of the SBR (A) and the MBR (B). The dashed vertical line at the 0.2 x axis level is plotted to evaluate the range of the Pareto values.
DISCUSSION
This research evaluated differences in physicochemical parameters and microbial community compositions and dynamics of an SBR and an MBR inoculated with the same sludge and operated in parallel for 84 days. The SBR provided complete nitrification during the whole experiment, whereas the MBR had a buildup of 0 to 2 mg nitrite-N liter−1 from day 45 to day 84. Between the SBR and MBR, the floc sizes, floc size distributions, and settling rates of the sludges showed large differences, but the amounts of AOB and NOB were highly comparable. Taking into account a startup-related adaptation period, neither reactor had an AOB community which was stable over a period of more than 1 month (Fig. 3; also see Fig. S3 in the supplemental material). Therefore, it could be concluded that a static nitrifying community is not essential to maintain full nitrification. Furthermore, this research attempted to characterize and quantify these bacterial community changes on a short-term basis (weeks) by (i) applying moving-window analysis to determine the rate of community change and (ii) introducing Pareto-Lorenz curves to evaluate possible changes in terms of the community evenness.
The SBR maintained complete nitrification during the entire experimental run. The MBR showed prolonged small nitrite effluent concentrations (0 to 2 mg NO2−-N liter−1) starting from around day 40, indicating incomplete nitratation. This lack of tight coupling between nitritation and nitratation could be linked with a decrease in the MBR sludge floc size (Fig. 1; also see Fig. S2 in the supplemental material). Both floc size distribution and light microscopic analyses indicated floc diameters in the SBR more than 10 times larger than those in the studied MBR, which had a microfloc structure. The lack of good floc structuring in the MBR resulted in sludge bulking, as supported by an SVI of 205 ml (g SS)−1 on day 84. Previously, floc size was reported to play an important role in maintaining complete nitrification, and MBR flocs are known to be smaller than flocs of a conventional activated-sludge WWTP (33), probably due to the lack of a settling and selection phase. In this research, confocal FISH analysis demonstrated the structural similarity of the AOB and Nitrospira clusters, i.e., distinct clusters of AOB and NOB in the flocs, for both SBR and MBR reactor sludges (Fig. 1C and D). Conclusively, it was demonstrated that flocs smaller than 40 μm in diameter can still contain distinct clusters of AOB and NOB but that larger floc sizes favor nitritation-nitratation tight coupling.
In this research, fairly comparable amounts of AOB and NOB were detected in both reactor types. Per mg VSS, a constant level of ca. 6 to 7 log units of AOB (the temporary 2- to 3-log unit drop was not taken into account) and 8 to 9 log units of Nitrospira cells were observed (Fig. 2). With qRT-PCR, AOB are reported to range in number between 5 and 8 log units and Nitrospira cells between 4 and 8 log units per mg VSS in WWTP samples (20, 21, 30). Comparing AOB (also amoA gene based) and Nitrospira cell numbers in the same WWTP samples, Harms et al. (21) reported ca. 10 times higher Nitrospira levels, whereas in the study of Layton et al. (30), the Nitrospira levels ranged from 100 times higher to 2 times lower than the AOB levels. In this research, it was observed that (i) even at startup, AOB values were evaluated as rather high and Nitrospira levels as very high, (ii) no further rise of either subgroup was noted though selective growth substrates were administered, and (iii) internal community dynamics were present. Therefore, this could indicate the existence of a maximal support capacity of AOB and NOB for this municipal WWTP sludge when a volumetric loading rate of 100 mg N liter−1 day−1 was administered; moreover, this finding was independent of the applied reactor configuration.
A static community composition was not observed in the SBR or the MBR. Nevertheless, the stability of the nitrification reaction was maintained. The rate of change of the bacterial communities was visualized by moving-window analysis based on the DGGE profiles (Fig. 3). The moving-window analysis technique has already shown to be useful for monitoring WWTP community shifts (49). In this research, the rate of change was quantified by introducing the “change over time” or “Δt value,” reported as an average value with its standard deviation. To enable comparison of Δt values, the time window is indicated between brackets. Here, Δt(week) values were applied to characterize short-term changes. The SBR was observed to change more gradually than the MBR both for the total bacterial (SBR, 9.6% ± 4.9%; MBR, 13.4% ± 4.8%) and for the AOB (SBR, 12.6% ± 5.2%; MBR, 24.6% ± 14.3%) levels, probably due to their different floc characteristics. The operational value of this parameter needs to be corroborated by studies on other and larger reactor types. However, it is of interest that a pragmatic analysis of the DGGE data as performed by moving-window analysis can visualize large differences in microbial community dynamics between two reactors inoculated with the same sludge.
To interpret the interspecies abundance, Pareto-Lorenz evenness distribution curves were plotted. These curves were based on the numbers of bands and the band intensities of the AOB DGGE profiles as described by Mertens et al. (35). It should be noted that the introduction of bias due to preferential amplification is considered as a possible disadvantage when applying conventional PCR (45). However, according to more-recent studies, this bias of preferential amplification may be overestimated, and the band intensity of 16S rRNA genes in a DGGE gel may correspond at least semiquantitatively with the abundance of the corresponding species (22, 23). Nevertheless, extra care should be taken when using nested PCR with primers having specificity issues, as nonspecific amplification might influence the intensity of specific bands. Therefore, it was decided in this research to work with the relative instead of the absolute band intensities. Over time, it was observed for the SBR reactor that 20% of the bands (number based) corresponded with on average 40% of the cumulative intensity of the bands. For the MBR reactor, 20% of the bands corresponded with on average 55% of the cumulative band intensity. This internal structuring of the highly dynamic AOB community indicated that at each moment only a small group of species played a numerically dominant role and that this dominance was even more pronounced for the MBR. The remaining less dominant DGGE bands are speculated to constitute a reserve of Nitrosomonas species which can proliferate to replace the dominant species. Just as for the rate of change, for this parameter as well more data need to be collected for other and larger reactors in order to substantiate the value for practicing this approach in interpreting the microbial ecology of activated-sludge communities.
Up to now, shifts in bacterial compositions have been reported on one hand as a result of changing environmental factors influencing the investigated processes (6, 46); on the other hand (i.e., under constant environmental factors), the sampling occurred infrequently (12), without visualizing short-term shifts. The DGGE monitoring of this study also reported dynamics of the relative interspecies abundance, without measurable environmental changes and based on weekly sampling. The latter observation was constantly noted, as visualized by the DGGE fingerprints and moving-window analysis. This correlates with findings of a study on anaerobic reactors performed earlier (12) and of research on activated-sludge SBRs (25), but our research examined the shifts on a short-term basis (2 to 3 weeks). Furthermore, it should be noted that this study was performed on 25-liter lab-scale reactors in order to control for most of the environmental factors influencing the nitrification process. Natural ecosystems as well as full-scale sewage treatment plants have larger dimensions and are often confronted with newly arriving allochthonous organisms. As it still remains unknown whether these effects lead to a more stable or more dynamic community structure, extrapolation of the observed results to natural ecosystems was not intended in this research phase.
The difficulty of having several repeats during such long-term experiments is a common problem when studying wastewater treatment systems. Often, single reactors are set up and compared over longer time periods (7, 16, 28, 29, 38, 50). In another study, we ran the reactor setup in triplicate for more than 100 days, and both functional and microbial community structure dynamics were highly reproducible (unpublished data). These observations are in agreement with other studies showing minor differences between appropriately repeated lab-scale setups (5, 25, 32, 43).
As observed in this research, floc structure is an important parameter to obtain complete nitrification. However, stability of the microbial community is not essential to maintain the functional stability of the nitrification process. Moving-window analysis and the derived rate of change values were demonstrated to be valuable tools for monitoring microbial community dynamics. Finally, Pareto-Lorenz curve distribution patterns facilitated the visualization of the evenness of the bacterial communities. This way, it was demonstrated that only a small group of AOB species played a numerically dominant role in the studied nitrifying reactors. The remaining less dominant species were speculated to constitute a reserve of AOB which can proliferate to replace the dominant species.
Supplementary Material
Acknowledgments
This work was mainly funded by a Ph.D. grant (no. 41428) from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) and also by G.O.A. contract grant no. 1205073 of the “Ministerie van de Vlaamse Gemeenschap, Bestuur Wetenschappelijk Onderzoek” (Belgium).
We thank Greet Van de Velde, Aleksandra Ziembiñska, Tijs Decuypere, Veerle De Schepper, and Winnok De Vos for their practical assistance in the completion of this research. Furthermore, Peter Aelterman, Tom Defoirdt, and Joke Geets are gratefully appreciated for their suggestions to improve the manuscript.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R., W. Ludwig, and K. -H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthonisen, A. C., R. C. Loehr, T. B. S. Prakasam, and E. G. Srinath. 1976. Inhibition of nitrification by ammonia and nitrous acid. J. Water Pollut. Control Fed. 48:835-852. [PubMed] [Google Scholar]
- 3.Biesterfeld, S., L. Figueroa, M. Hernandez, and P. Russell. 2001. Quantification of nitrifying bacterial populations in a full-scale nitrifying trickling filter using fluorescent in situ hybridization. Water Environ. Res. 73:329-338. [DOI] [PubMed] [Google Scholar]
- 4.Boon, N., W. De Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 5.Boon, N., E. M. Top, W. Verstraete, and S. D. Siciliano. 2003. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl. Environ. Microbiol. 69:1511-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucher, D., L. Jardillier, and D. Debroas. 2006. Succession of bacterial community composition over two consecutive years in two aquatic systems: a natural lake and a lake-reservoir. FEMS Microbiol. Ecol. 55:79-97. [DOI] [PubMed] [Google Scholar]
- 7.Briones, A. M., B. J. Daugherty, L. T. Angenent, K. D. Rausch, M. E. Tumbleson, and L. Raskin. 2007. Microbial diversity and dynamics in multi- and single-compartment anaerobic bioreactors processing sulfate-rich waste streams. Environ. Microbiol. 9:93-106. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 9.Daims, H., P. H. Nielsen, J. L. Nielsen, S. Juretschko, and M. Wagner. 2000. Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology. Water Sci. Technol. 41:85-90. [Google Scholar]
- 10.Degrange, V., and R. Bardin. 1995. Detection and counting of Nitrobacter populations in soil by PCR. Appl. Environ. Microbiol. 61:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dionisi, H. M., A. C. Layton, G. Harms, I. R. Gregory, K. G. Robinson, and G. S. Sayler. 2002. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez, A., S. Y. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, A. S., S. A. Hashsham, S. L. Dollhopf, L. Raskin, O. Glagoleva, F. B. Dazzo, R. F. Hickey, C. S. Criddle, and J. M. Tiedje. 2000. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl. Environ. Microbiol. 66:4058-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Focht, D. D., and W. Verstraete. 1977. Biochemical ecology of nitrification and denitrification. Adv. Microb. Ecol. 1:135-214. [Google Scholar]
- 15.Geets, J., M. De Cooman, L. Wittebolle, K. Heylen, B. Vanparys, P. De Vos, W. Verstraete, and N. Boon. 2007. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 75:211-221. [DOI] [PubMed] [Google Scholar]
- 16.Gentile, M. E., C. M. Jessup, J. L. Nyman, and C. S. Criddle. 2007. Correlation of functional instability and community dynamics in denitrifying dispersed-growth reactors. Appl. Environ. Microbiol. 73:680-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govoreanu, R., H. Saveyn, P. Van der Meeren, and P. A. Vanrolleghem. 2004. Simultaneous determination of activated sludge floc size distribution by different techniques. Water Sci. Technol. 50(12):39-46. [PubMed] [Google Scholar]
- 18.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton. 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, DC.
- 19.Guerrero, M. A., and R. D. Jones. 1996. Photoinhibition of marine nitrifying bacteria. 1. Wavelength-dependent response. Mar. Ecol. Prog. Ser. 141:183-192. [Google Scholar]
- 20.Hall, S. J., P. Hugenholtz, N. Siyambalapitiya, J. Keller, and L. L. Blackall. 2002. The development and use of real-time PCR for the quantification of nitrifiers in activated sludge. Water Sci. Technol. 46:267-272. [PubMed] [Google Scholar]
- 21.Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 22.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities. In J. D. van Elsas, E. M. H. Wellington, and J. T. Trevors (ed.), Modern soil microbiology. Marcel Dekker, New York, NY.
- 24.Juliastuti, S. R., J. Baeyens, C. Creemers, D. Bixio, and E. Lodewyckx. 2003. The inhibitory effects of heavy metals and organic compounds on the net maximum specific growth rate of the autotrophic biomass in activated sludge. J. Hazard. Mater. 100:271-283. [DOI] [PubMed] [Google Scholar]
- 25.Kaewpipat, K., and C. P. L. Grady. 2002. Microbial population dynamics in laboratory-scale activated sludge reactors. Water Sci. Technol. 46(1-2):19-27. [PubMed] [Google Scholar]
- 26.Keener, W. K., and D. J. Arp. 1993. Kinetic studies of ammonia monooxygenase inhibition in Nitrosomonas europaea by hydrocarbons and halogenated hydrocarbons in an optimized whole-cell assay. Appl. Environ. Microbiol. 59:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurisu, F., H. Satoh, T. Mino, and T. Matsuo. 2002. Microbial community analysis of thermophilic contact oxidation process by using ribosomal RNA approaches and the quinone profile method. Water Res. 36:429-438. [DOI] [PubMed] [Google Scholar]
- 29.LaPara, T. M., C. G. Klatt, and R. Chen. 2006. Adaptations in bacterial catabolic enzyme activity and community structure in membrane-coupled bioreactors fed simple synthetic wastewater. J. Biotechnol. 121:368- 380. [DOI] [PubMed] [Google Scholar]
- 30.Layton, A. C., H. Dionisi, H. W. Kuo, K. G. Robinson, V. M. Garrett, A. Meyers, and G. S. Sayler. 2005. Emergence of competitive dominant ammonia-oxidizing bacterial populations in a full-scale industrial wastewater treatment plant. Appl. Environ. Microbiol. 71:1105-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz, M. O. 1905. Methods of measuring concentration of wealth. J. Am. Stat. Assoc. 9:209-219. [Google Scholar]
- 32.Lozada, M., E. L. M. Figuerola, R. F. Itria, and L. Erijman. 2006. Replicability of dominant bacterial populations after long-term surfactant-enrichment in lab-scale activated sludge. Environ. Microbiol. 8:625-638. [DOI] [PubMed] [Google Scholar]
- 33.Manser, R., W. Gujer, and H. Siegrist. 2005. Membrane bioreactor versus conventional activated sludge system: population dynamics of nitrifiers. Water Sci. Technol. 52:417-425. [PubMed] [Google Scholar]
- 34.McCarty, G. W. 1999. Modes of action of nitrification inhibitors. Biol. Fertil. Soils 29:1-9. [Google Scholar]
- 35.Mertens, B., N. Boon, and W. Verstraete. 2005. Stereospecific effect of hexachlorocyclohexane on activity and structure of soil methanotrophic communities. Environ. Microbiol. 7:660-669. [DOI] [PubMed] [Google Scholar]
- 36.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan-Sagastume, F., and D. G. Allen. 2003. Effects of temperature transient conditions on aerobic biological treatment of wastewater. Water Res. 37:3590-3601. [DOI] [PubMed] [Google Scholar]
- 38.Mota, C., M. A. Head, J. A. Ridenoure, J. J. Cheng, and F. L. de los Reyes III. 2005. Effects of aeration cycles on nitrifying bacterial populations and nitrogen removal in intermittently aerated reactors. Appl. Environ. Microbiol. 71:8565-8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Øvreås, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pareto, V. 1897. Le cours d'économie politique. Macmillan, London, United Kingdom.
- 41.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rousseau, R., P. Van Hecke, D. Nijssen, and J. Bogaert. 1999. The relationship between diversity profiles, evenness and species richness based on partial ordering. Environ. Ecol. Stat. 6:211-223. [Google Scholar]
- 43.Saikaly, P. E., P. G. Stroot, and D. B. Oerther. 2005. Use of 16S rRNA gene terminal restriction fragment analysis to assess the impact of solids retention time on the bacterial diversity of activated sludge. Appl. Environ. Microbiol. 71:5814-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terahara, T., T. Hoshino, S. Tsuneda, A. Hirata, and Y. Inamori. 2004. Monitoring the microbial population dynamics at the start-up stage of waste-water treatment reactor by terminal restriction fragment length polymorphism analysis based on 16S rDNA and rRNA gene sequences. J. Biosci. Bioeng. 98:425-428. [DOI] [PubMed] [Google Scholar]
- 47.Tokutomi, T. 2004. Operation of a nitrite-type airlift reactor at low DO concentration. Water Sci. Technol. 49(5-6):81-88. [PubMed] [Google Scholar]
- 48.Villaverde, S., M. T. Fernandez, M. A. Uruena, and F. FdzPolanco. 1997. Influence of substrate concentration on the growth and activity of a nitrifying biofilm in a submerged biofilter. Environ. Technol. 18:921-928. [Google Scholar]
- 49.Wittebolle, L., N. Boon, B. Vanparys, K. Heylen, P. De Vos, and W. Verstraete. 2005. Failure of the ammonia oxidation process in two pharmaceutical wastewater treatment plants is linked to shifts in the bacterial communities. J. Appl. Microbiol. 99:997-1006. [DOI] [PubMed] [Google Scholar]
- 50.Yan, X., Z. M. Xu, X. X. Feng, Y. D. Liu, B. B. Liu, X. J. Zhang, C. G. Zhu, and L. P. Zhao. 2007. Cloning of environmental genomic fragments as physical markers for monitoring microbial populations in coking wastewater treatment system. Microb. Ecol. 53:163-172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.