Abstract
The study of microbial interactions in mixed cultures remains an important conceptual and methodological challenge for which transcriptome analysis could prove to be the essential method for improving our understanding. However, the use of whole-genome DNA chips is often restricted to the pure culture of the species for which the chips were designed. In this study, massive cross-hybridization was observed between the foreign cDNA and the specific Lactococcus lactis DNA chip. A very simple method is proposed to considerably decrease this nonspecific hybridization, consisting of adding the microbial partner's DNA. A correlation was established between the resulting cross-hybridization and the phylogenetic distance between the microbial partners. The response of L. lactis to the presence of Saccharomyces cerevisiae was analyzed during the exponential growth phase in fermentors under defined growth conditions. Although no differences between growth kinetics were observed for the pure and the mixed cultures of L. lactis, the mRNA levels of 158 genes were significantly modified. More particularly, a strong reorientation of pyrimidine metabolism was observed when L. lactis was grown in mixed cultures. These changes in transcript abundance were demonstrated to be regulated by the ethanol produced by the yeast and were confirmed by an independent method (quantitative reverse transcription-PCR).
In technological food processes as well as in natural ecosystems, the biological transformations that are observed are generally the result of the activities of complex microbial flora. Moreover, the global activity of a mixed microbial population is determined by the presence and functions of each species, which are strongly determined by the interactions among the different partners. However, the current knowledge of microbial physiology is based generally on pure culture studies, conditions for which are different than those encountered in complex ecosystems. In consequence, performing mixed culture studies is an essential way to get closer to the reality of complex populations.
Microarray technology provides a powerful tool for giving an overview of cell responses to environmental changes at the transcriptional level. However, the application of this technology to the study of heterogeneous microbial populations is still a conceptual and methodological challenge. Generally, the biochips used with mixed cultures are not pangenomic and serve only rarely for transcript detection. Most of them are devoted to the detection of microbial species in complex ecosystems through ribosomal DNA sequences or to the detection of a reduced number of DNA sequences without quantifying their expression levels (5, 27, 34, 41, 42). Some articles mention the use of DNA biochips for mRNA quantification, but these chips are mostly restricted to a limited number of mRNAs (8, 12, 32). Only very recently has the use of pangenomic biochips to study mixed cultures been mentioned (15).
The major obstacle to the utilization of microarray tests for transcriptome analysis of a multiple species population is the cross-hybridization of the partner species' DNA on the spots of the microarray test, which is defined for a specific organism. RNA extracted from mixed cultures comes from each species in the population and is reverse transcribed and labeled, and the resulting mixture of cDNA is hybridized with the microarray. In consequence, the specific signal of cDNA coming from the organism of interest is corrupted by the hybridization with foreign cDNA. Differential extraction protocols could be an interesting way, in the future, to reduce this cross-hybridization, but in this case, the methodological improvements should be more or less species specific. In this paper, we describe a new and simple method which significantly reduced the noise resulting from the cross-hybridization of other species' cDNA with Lactococcus lactis microarrays.
L. lactis is considered a model lactic acid bacterium, encountered extensively in numerous food fermentation processes, particularly in cheese production, industrial processes, and natural ecosystems. DNA arrays for L. lactis strain IL-1403, the first lactococcal strain to be sequenced (6), have recently been used for transcriptome analysis of L. lactis pure cultures (4, 19, 30, 31). However, in most cases, L. lactis lives together with multiple microorganisms such as lactobacilli, corynebacteria, and yeasts (7, 11, 24, 26, 28, 33, 35). It is critical, then, to be able to study its behavior in mixed cultures. On the other hand, Saccharomyces cerevisiae is the model yeast, well characterized and studied, that coexists with lactic acid bacteria in ecosystems such as kefir. Thus, yeasts and lactic acid bacteria often share the same environment, but little is known about their possible interactions. In our study, L. lactis pure cultures and L. lactis/S. cerevisiae mixed cultures grown in fermentors under well-defined culture conditions were compared. The macroscopic behavior of each culture was linked to the measured nutritional and environmental parameters. Finally, the proposed microarray method improvement enabled the complete transcriptome analysis of a bacterium in a mixed culture to be undertaken.
MATERIALS AND METHODS
Organisms and growth conditions.
The microorganisms used throughout this work were L. lactis strain IL-1403, Lactobacillus plantarum WCFS1, Corynebacterium glutamicum ATCC 13032, and Saccharomyces cerevisiae CEN-PK905. They were all grown, except for C. glutamicum, in a semisynthetic medium containing glucose (20 g·liter−1), yeast extract (10 g·liter−1), KH2PO4 (9 g·liter−1), K2HPO4 (7.5 g·liter−1), MgCl2 (0.2 g·liter−1), and ergosterol (6 mg·liter−1) dissolved in pure ethanol and sterilized by filtration (0.2 μm; Sartorius Co.) under anaerobic conditions in a nitrogen atmosphere in a 2-liter fermentor (Setric Génie Industriel, Toulouse, France) at an agitation speed of 350 rpm. The temperature in the fermentor was maintained at 34°C, and the pH was maintained at 6.6, with the automatic addition of KOH (10 N). Cells from precultures grown on the same medium were used for inoculation. Precultured cells were harvested during their exponential growth phase and concentrated in order to obtain an initial optical density at 580 nm (OD580) of 0.01 to 0.02 in the fermentor. C. glutamicum was grown in LB medium supplemented with glucose (20 g·liter−1) under the same conditions but with aeration.
In order to analyze the effect of the yeast fermentation products on some of the L. lactis transcript levels, pulse experiments consisting of the addition of ethanol (30 mM) or sodium carbonate (30 mM) at 2.5-h intervals to L. lactis batch cultures were carried out. Concentrated solutions of sodium carbonate (160 g·liter−1) and pure ethanol were sterilized by filtration (0.2 μm [Sartorius]) and added to the fermentor with a syringe.
Fermentation analysis.
Bacterial growth was estimated by spectrophotometric measurement at 580 nm and by a plate count on the medium used in the bioreactors supplemented with agarose (20 g·liter−1). (One optical density unit was equivalent to 0.30 and 0.24 g of dry matter per liter and to 7 × 108 and 5 × 107 CFU per liter for L. lactis and S. cerevisiae, respectively.) Differences in colony morphology enabled the two species to be counted separately in mixed cultures. Protein precipitation of the samples was performed with a solution of barium hydroxide and zinc sulfate, 0.3 M, prior to the measurement of substrate (glucose) and fermentation product (lactate, formate, acetate, glycerol, and ethanol) concentrations by high-pressure liquid chromatography (9). Concentrations of ethanol and acetic acid in the medium were determined by gas chromatography, using a Poraplot Q column (25 m by 0.53 mm), with nitrogen as the carrier gas and flame ionization detection (5890A; Hewlett Packard) under the following conditions: an injection temperature of 220°C, an initial oven temperature of 210°C to a final temperature of 245°C at a rate of 30°C·min−1, and an isotherm of 5 min, a flow rate of carrier gas of 20 ml·min−1, and an injected volume of 0.1 μl. The CO2 concentration was not measured in the medium but was estimated according to the theoretical stoichiometry of 1 mol of CO2 produced by a mole of ethanol. Concentrations of nucleotide precursor in the supernatant cultures were measured as described recently with a limit of detection of 1 μM (20).
Phylogenetic tree.
The phylogenetic distances between the various species used in the study were estimated by using their rRNA homologies. Multiple alignments of rRNA sequences (16S for prokaryotes and 18S for the yeast) of Lactococcus lactis, Lactobacillus plantarum, C. glutamicum, and S. cerevisiae were performed with ClustalW version 1.8 (36). The phylogenetic tree was then constructed with PHYLIP version 3.65 (Joseph Felsenstein, University of Washington), with distance and neighbor-joining methods. Branch lengths are reported in Fig. 1.
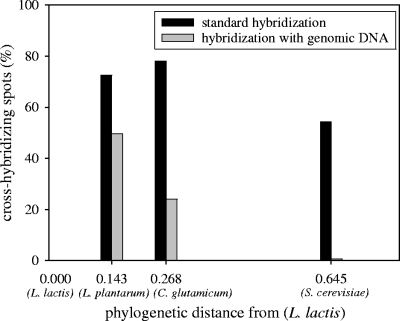
FIG. 1.
Effect of the addition of unlabeled genomic DNA from partner species on the percentages of labeled cDNA cross-hybridizing spots, by L. lactis IL-1403 DNA microarrays, relative to the phylogenetic distances between L. lactis and the partner species.
Biochips of Lactococcus lactis.
L. lactis IL-1403-specific PCR products (mean length, 535 bp) were provided by Eurogentec and spotted in duplicate on glass slides by the Biochips Platform (Toulouse Génopôle, France). Two thousand three of the 2,310 open reading frames identified on the genome were effectively available.
RNA extraction.
Frozen cell pellets corresponding to 6 mg of dry weight were dropped into a precooled 5-ml Teflon vessel. For the coculture, 6 mg of cell dry weight containing L. lactis and S. cerevisiae was estimated from the plate count. A 7-mm bead made of tungsten carbide was added. The flask was then shaken at 2,600 rpm for 2 min in a Mikro-Dismembrator unit (Braun, Melsungen). Powder was resuspended in 4 ml of RLT buffer (Qiagen) supplied with the beta-mercaptoethanol. The steps of RNA extraction that followed were done according to Qiagen's RNeasy protocol, including DNase I treatment. RNA quality and quantification were analyzed using a Bioanalyser 2100 model (Agilent Technologies).
Genomic DNA extraction.
DNA from S. cerevisiae, Lactobacillus plantarum, and C. glutamicum was extracted using zymolyase or lysozyme digestion and EDTA/sodium dodecyl sulfate treatment in Tris-EDTA buffer. DNA was then purified by using isopropanol and ethanol 70% concentrations. The final DNA pellet was treated with RNase and partially digested with Sau3AI.
Reverse transcription and labeling.
RNA from pure or mixed cultures was reverse transcribed and labeled with a cyanine dye (Cy3 for the reference and Cy5 for the test condition) in a direct process using a LabelStar array kit (Qiagen). For the biochip specificity, the L. lactis RNA was labeled with Cy5, and the other species' RNA was labeled with Cy3, and both were hybridized on the same biochip.
For the transcriptome analysis, the mixed culture (test condition, 2.5 h of culture) was compared to the pure culture (reference, 2.5 h of culture) on the same slide at the same time. The mRNA level changes that occurred between 2 and 3 h flanking the test condition (2.5 h) in the pure culture were also determined on another slide. RNA (30 μg) was denatured and then combined with reverse transcription (RT) buffer, a mixture of deoxynucleoside triphosphates (0.5 mM final concentration for dATP, dTTP, dCTP; 0.08 mM final concentration for dCTP), random hexamer primers (5 μl; Invitrogen), Cy3- or Cy5-labeled dCTP (0.02 mM final concentration), RNase inhibitor (20 units; Qiagen), and LabelStar reverse transcriptase (Qiagen). The mixture was incubated for 2 h at 37°C. The reaction was stopped by adding LS solution (Qiagen), and cDNA was purified by using MinElute columns (Qiagen). Samples (10 μl) were collected for each reaction.
An indirect labeling technique adapted for bacterial organisms with high GC percentages was used for C. glutamicum as previously described (14). Aminoallyl-modified dUTP nucleotides (aa-dUTP) were incorporated during a first-strand RT (RT reaction). Each RT reaction was performed in 30-μl volumes containing 30 μg total RNA, 2 μl of random hexamer primers (Amersham), 0.8 mM dATP, 0.8 mM dCTP, 0.8 mM dGTP, 0.2 mM dTTP, 0.6 mM aa-dUTP (Sigma-Aldrich), 300 units of SuperscriptII reverse transcriptase (Invitrogen), 6 μl of Superscript IIA buffer (5×), 3 μl 100 mM dithiothreitol, and 0.4 μl of sterile water. Total RNA and hexamer primers were heated to 70°C for 10 min to denature the RNA and chilled on ice for 5 min to anneal the primers. Subsequently, the other components were added to the reaction mixture. The RT reaction was carried out for 3 h at 42°C. The solution was treated with 1.25 μl of RNaseH (1.5 U·μl−1) for 15 min at 37°C to digest RNA and for 10 min at 70°C to denature the enzymes. The cDNA was purified with columns provided by a MinElute PCR purification kit (Qiagen). The columns were washed with 70% ethanol, and the cDNA was eluted with 10 μl of EB buffer (LabelStar kit; Qiagen). Subsequently, 1 μl of 0.5 M sodium bicarbonate (pH 9.0) was added to the preparation. The cDNA was transferred to aliquots of Cy3 and Cy5 monofunctional NHS-esters (Amersham Biosciences Europe) and incubated for 60 min in the dark. To remove unincorporated fluorophores, the probes were purified again with a MinElute PCR purification kit, following the manufacturer's protocol.
Hybridization of labeled cDNA and scans of the glass slides.
Experiments were carried out at the Biochips platform of Toulouse with an automatic hybridization chamber (Discovery; Ventana Medical System, Inc.). Prehybridization occurred with a freshly prepared solution of 1% bovine serum albumin, 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% sodium dodecyl sulfate over 1.5 h at 42, 47 or 50°C. After slides were automatically washed, according to the manufacturer's instructions, the slides were hybridized for 8 h at 42 or 47°C or for 5 h at 50°C in 200 μl of ChipHybe buffer (Ventana Medical System, Inc.) containing 3 μl of Cy3-labeled cDNA and 3 μl of Cy5-labeled cDNA. Unlabeled genomic DNA (20 μl at 20 μg·μl−1) was added to the solution when necessary, as stated previously. After hybridization, the slides were washed twice in RiboWash (Discovery; Ventana Medical System, Inc.), then for 2 min in 0.2× SSC and quickly rinsed in isopropanol and then dried by centrifugation for 5 min at 3,000 × g. Fluorescence signals were captured with a laser scanner (GenePix 4000A; Axon Instrument, CA) and analyzed with GenePix version 3.01 software.
Statistical analysis.
Data were further processed by statistical methods using BioPlot software developed by S. Sokol (Biochips Platform of Toulouse; http://biopuce.insa-toulouse.fr/). Three methodologically independent repetitions were made for each experiment.
For the biochip specificity, spot intensity significance was estimated from the fluorescence measured on empty spots. A cutoff threshold (T) was calculated with the 562 empty spots, using the formula T = Kempty + 3·σ, where Kempty is the mean of the log intensity of all empty spots, and σ is the corresponding standard deviation. Intensities beyond this threshold were considered significant and evidence of cross-hybridizing. Spots with insignificant signals as well as empty spots were subtracted from each biochip before normalization.
For the transcriptome analysis, the cross-hybridizing spots were removed from further analysis. Normalization by all spot means was applied, excluding insignificant and empty spots of the slide. The normalized log intensity of each remaining spot (Ns) was then calculated by subtracting the mean of the log intensity of all those spots (Knorm) from the log intensity of each spot (Ms), according to the formula Ns = Ms − Knorm. Student's test was then applied to each normalized spot log intensity in order to detect significant differences between the two wavelengths (P < 0.05). Cutoff thresholds have been arbitrarily set so that ratios within the interval of 0.91 to 1.1 were excluded from analysis. Genes with false discovery rate values of >0.1 were excluded from analysis.
Real-time PCR.
Four independent RNA extractions from four biological replicates were tested with four independent real-time PCR experiments.
RNA extraction and purification including DNase treatment was performed as described above. Then, cDNAs were synthesized using a random primer (10 ng·μl−1 final concentration; Invitrogen), each deoxynucleoside triphosphate (0.3 mM), and superscript II transcriptase (6 U·μl−1; Invitrogen) incubated for 1 h at 42°C. The reaction was stopped by incubation for 15 min at 70°C. RNase H (0.05 U; Invitrogen) was added before purification of cDNA by Microspin G25 columns (Amersham Biosciences).
Primers for real-time PCR were designed to have a length of about 20 to 24 bases, a GC content of more than 50%, and a melting temperature of about 60°C, and the length of the PCR products ranged between 90 and 150 bp. Beacon designer Bio-Rad software was used to select primer sequences. The specificity of the primers for the genes of interest relative to those of the entire L. lactis genome sequence was controlled by using NCBI BLAST software. Primers were purchased from Sigma or Eurogentec.
Real-time PCR were carried out on a MyIQ unit with SYBR Green Supermix (BioRad) in 96-well plates. After cDNA was diluted, 5 μl was added to 20 μl of PCR mixture (12.5 μl SYBR Green Supermix, 4 μl of primer at 5 μM, and 3.5 μl of RNase-free water). Three or four dilutions of cDNA were performed to determine the efficiencies of real-time PCR. A negative control in which cDNA was replaced by water was systematically included. Thermal cycling conditions were denaturation at 95°C for 3 min and 40 repeats of 95°C for 15 s and 60°C for 45 s and an annealing step where fluorescence measurements were recorded. The melting curve was performed from 60 to 95°C (0.05°C·s−1). The threshold cycle value was determined with a baseline set manually at 70 relative fluorescence units above that of the background. The PCR efficiency ranged between 85 and 110%. The butB gene was chosen as a suitable internal control gene to normalize the results from the biochip data and after checking by RT-PCR that its mRNA level was unaffected during the pulse experiments. The Pfaffl method (29) was used to calculate the change in transcript abundance normalized to that of the butB gene and relative to that of the sample collected before the pulse.
RESULTS
Biochip specificity.
In order to measure the specificity of the L. lactis IL-1403 biochip, the slides were hybridized with mixed labeled cDNA coming from L. lactis IL-1403 (Cy5-labeled reference cells) and from pure cultures of one of the other species (Cy3-labeled L. plantarum, C. glutamicum, and S. cerevisiae test condition isolates). As expected, cDNA coming from species other than L. lactis IL-1403 hybridized on the L. lactis-specific DNA microarrays (Fig. 1), so that 72.5% of the spots gave out significant fluorescence intensity signals with labeled cDNA from L. plantarum, 78.1% with labeled cDNA from C. glutamicum, and 54.2% with labeled cDNA from S. cerevisiae, indicating a strong interspecies hybridization.
Hybridizations were made under the same conditions but with different hybridization temperatures ranging from 42°C to 65°C, in order to reduce the nonspecific signal. The temperature increase was not efficient enough to improve the biochip specificity (data not shown) and led to some technical disadvantages, like the necessity to perform manual hybridizations. Moreover, fluorescence intensity decreased with high temperature, leading to a loss of data; thus, it was not pertinent to further increase the hybridization temperature.
Effect of adding genomic DNA to cross-hybridization.
A method was developed to reduce the interspecies hybridization and be able to use the L. lactis microarrays in binary cocultures. The addition of unlabeled genomic DNA of the partner species to the cDNA coming from the partner allowed the signal stemming from cross-hybridizations to be significantly decreased. The efficiency of the method seemed to be correlated to the phylogenetic distance between L. lactis and the partner species (Fig. 1). While 49.6% of the spots still emitted significant fluorescence intensity with labeled cDNA from L. plantarum, only 24.1% and 0.3% gave out fluorescence with labeled cDNA from C. glutamicum and S. cerevisiae, respectively. The dye swap was tested (L. lactis cDNA was labeled with Cy3, and the other species' cDNA was labeled with Cy5), and no significant differences between the two methods were observed.
The addition of foreign, unlabeled genomic DNA may also diminish the specific fluorescence from the labeled L. lactis IL-1403 cDNA. Hybridization of L. lactis cDNA on microarrays without yeast genomic DNA gave out 1,743 spots with significant signal (87% of the entire biochip) and 1,336 spots with significant signal when yeast genomic DNA was added (67% of the entire biochip); thus, 1.30-fold fewer spots were produced with yeast genomic DNA. However, the addition of yeast genomic DNA significantly diminished the labeled S. cerevisiae cDNA signal from 54% to 0.3% (Fig. 1), which is a 163-fold reduction. Finally, the effect of unlabeled yeast genomic DNA on cDNA hybridization was around 125-fold lower with L. lactis cDNA than with S. cerevisiae cDNA.
Metabolic analysis.
In order to study the interactions between L. lactis and S. cerevisiae at macroscopic and transcriptome levels, three types of cultures were compared, pure cultures of L. lactis IL-1403 and S. cerevisiae CEN-PK905 and mixed culture in which both species shared the same environment.
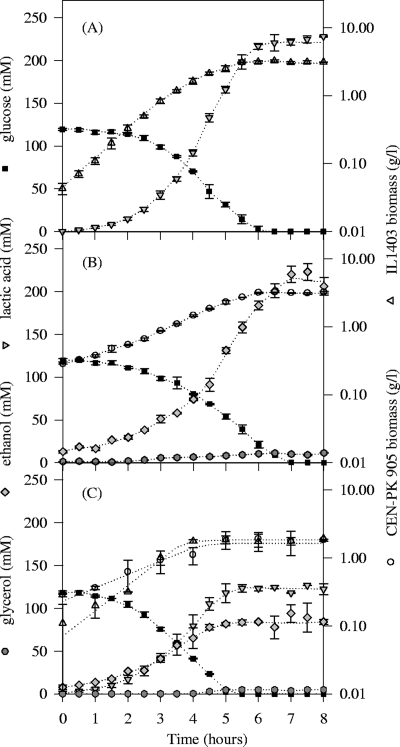
L. lactis IL-1403 exhibited a homolactic metabolism at the expense of glucose, which was exhausted at 6 h of culture (Fig. 2A). Under the same conditions, S. cerevisiae CEN-PK905 consumed the glucose in 7 h and produced ethanol and low amounts of glycerol (Fig. 2B). The product yields observed for these two pure cultures fitted the theoretical yields of the homolactic metabolism and the anaerobic yeast metabolism (Table 1). Levels of CO2, not quantified here, were also produced by S. cerevisiae, with a theoretical yield given by the metabolic pathways of 1 mole of CO2 per mole of ethanol. The initial biomass concentrations in the mixed culture were chosen to reach similar concentrations of lactococci and yeast at the end of growth. The stationary phase was reached at 5 h of culture, while the products formed were intermediate concentrations of lactic acid and ethanol and trace amounts of glycerol (Fig. 2C).
FIG. 2.
Evolution of biomass (g·liter−1) substrates and fermentation product concentrations (mM) during the first 8 h of the pure culture of L. lactis IL-1403 (A), the pure culture of S. cerevisiae CEN-PK905 (B), and the mixed culture (C).
TABLE 1.
Fermentation values calculated from pure cultures and mixed culture of both species
| Fermentation parametera | Fermentation value ± SDb
|
||
|---|---|---|---|
| L. lactis IL-1403 pure culture | S. cerevisiae CEN-PK905 pure culture | Mixed culture | |
| μIL-1403 (h−1) | 0.92 ± 0.04 | 1.01 ± 0.08 | |
| μCEN-PK905 (h−1) | 0.45 ± 0.01 | 0.45 ± 0.01 | |
| qglucose (mmol·g−1·h−1) | 25.6 ± 0.9 | 15.8 ± 0.1 | |
| vlactate (mmol·g−1·h−1) | 49.3 ± 1.4 | 42.1 ± 0.2c | |
| vethanol (mmol·g−1·h−1) | 26.6 ± 0.5 | 22.8 ± 1.0d | |
| Yglucose,IL-1403 (g·mmol−1) | 0.03 ± 0.00 | ||
| Yglucose, CEN-PK905 (g·mmol−1) | 0.02 ± 0.00 | ||
| Yglucose, lactate (cmol·cmol−1) | 0.93 ± 0.03 | ||
| Yglucose, ethanol (cmol·cmol−1) | 0.56 ± 0.05 | ||
| Yglucose, glycerol (cmol·cmol−1) | 0.09 ± 0.05 | ||
Abbreviations: μIL-1403, specific growth rate of L. lactis IL-1403; μCEN-PK905, specific growth rate of S. cerevisiae CEN-PK905; qglucose, specific glucose consumption rate; vlactate, specific lactate production rate; vethanol, specific ethanol production rate; Yglucose, IL-1403, biomass yield of L. lactis IL-1403 relative to that of glucose; Yglucose, CEN-PK905, biomass yield of S. cerevisiae CEN-PK905 relative to that of glucose; Yglucose, lactate, lactate yield relative to that of glucose; Yglucose, ethanol, ethanol yield relative to that of glucose; Yglucose, glycerol, glycerol yield relative to that of glucose.
Values ± standard deviations (SD) were derived from cultures performed three times.
Calculated taking into account the L. lactis IL-1403 biomass only.
Calculated taking into account the S. cerevisiae CEN-PK905 biomass only.
The maximal specific growth rates of L. lactis (μIL-1403) in pure and mixed cultures were similar, around 0.9 to 1.0 h−1 (Table 1). Furthermore, the maximum lactic acid-specific production rate (νlactate) was similar under both culture conditions. The exponential growth phase was slightly shorter in the mixed culture than in the pure culture, since the growth rate started to decrease at about 3 to 4 h. The maximal specific growth rate of S. cerevisiae (μCEN-PK905) and the maximum ethanol-specific production rate (νethanol) were similar in pure and mixed cultures at around 0.45 h−1 and 23 to 27 mmol·g−1·h−1, respectively. As for L. lactis, the exponential growth phase of S. cerevisiae was shorter in mixed culture (4 h) than in pure culture (5.5 h).
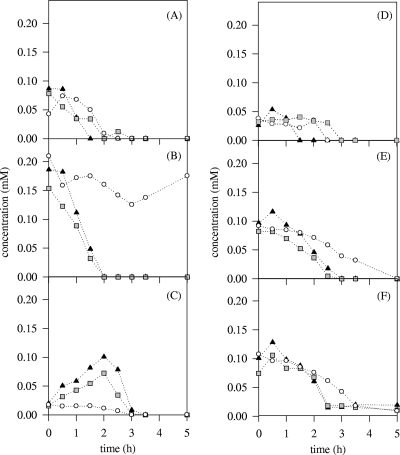
Besides the classical fermentation substrates and product concentrations, purine and pyrimidine precursor concentrations in the culture supernatants were analyzed. The variations observed were widely above the limit of detection of the method (1 μM). The yeast extract-containing medium harbored various nucleobase precursor concentrations, from 0.2 mM for adenosine to undetectable concentrations, with most around 0.1 mM (Fig. 3). Pure cultures of S. cerevisiae showed decreased concentrations of adenine, hypoxanthine, xanthine, uracil, and uridine, which were totally exhausted from the medium at around 2.5, 3, 2.5, 5, and 3.5 h of culture, respectively. Adenosine concentration did not show strong variations during the culture. The adenine, adenosine, xanthine, uracil, and uridine concentrations in pure cultures of L. lactis IL-1403 decreased until exhaustion during the first 1.5, 2, 1.5, 3, and 2.5 h, respectively (Fig. 3). Hypoxanthine concentration increased during 2 h of culture and then decreased, becoming undetectable within the next hour. For the above-mentioned nucleobase precursors, the concentration profiles obtained with mixed cultures were similar to those obtained with pure cultures of L. lactis IL-1403, except for xanthine.
FIG. 3.
Evolution of extracellular adenine (A), adenosine (B), hypoxanthine (C), xanthine (D), uracil (E), and uridine (F) concentrations during the first 5 h of pure L. lactis IL-1403 (▴) or S. cerevisiae CEN-PK905 (○) cultures and of the mixed culture (□). The limit of detection is 1 μM.
The initial guanine concentration in the culture medium was about 0.01 mM, and hence its profile could not be accurately determined (data not shown). Finally, guanosine, cytosine, and cytidine were not detected in the samples with our chromatographic method, indicating that they were at concentrations lower than the method threshold in the culture medium.
Transcriptome analysis.
Samples for transcriptome analysis were taken during exponential growth phase of each culture, e.g., at 2.5 h, and labeled. The goal of the experiment was to compare L. lactis IL-1403 behavior during pure cultures and cultures mixed with S. cerevisiae CEN-PK905. Hybridizations were performed with IL-1403 biochips in the presence of unlabeled genomic DNA of S. cerevisiae CEN-PK905 in order to limit cross-hybridization. Spots with cross-hybridization representing 0.3% of the genome (see above) were excluded from the analysis. One hundred seventy-four spots showed a significant variation in fluorescence intensity, according to our statistical test, between these two conditions; i.e., at the time of sampling, the mRNA abundance amounts of these L. lactis genes were different in mixed cultures compared to those in pure cultures. However, care should be taken when analyzing transcriptome data from two parallel cultures, since the variations of transcript level observed could be either specific to the comparison of the two culture conditions or linked to a difference between the dynamic culture progress of the two cultures. In this example of mixed culture, the growth of two species led to conjointly accelerated substrate consumption and product formation.
In order to discard possible nonspecific responses from the 174 spots showing a significant variation between the mixed and pure L. lactis IL-1403 cultures, a second set of experiments was done to analyze the dynamic variations due to the culture progression. The culture progression effect was estimated by the quantification of the mRNA abundance ratio surrounding the time for which the comparison (the mixed culture versus the pure culture) was performed. Labeled L. lactis cDNA samples collected after 2 and 3 h from the pure culture were hybridized simultaneously on the same biochip. Statistical analysis showed that 109 spots exhibited significant signal variations between these two cultivation times (Table 2), among which, 16 spots overlapped with the 174 spots identified in the previous comparison. Consequently, it was considered that the 158 (174 minus 16) remaining L. lactis open reading frames corresponded to genes for which the transcript level specifically varied due to the presence of S. cerevisiae in the culture. Data are available online at the GEO database (accession number GPL6052).
TABLE 2.
Change in mRNA abundances between 2 and 3 h of pure culturea
| Functional category | Underexpressed genes
|
Overexpressed genes
|
||||
|---|---|---|---|---|---|---|
| Gene | Intensity ratio | P value | Gene | Intensity ratio | P value | |
| Amino acid biosynthesis | gltD | 2.11 | 0.025 | |||
| hom | 1.79 | 0.03 | ||||
| Cell envelope | plpD | 0.65 | 0.017 | |||
| Cellular processes | ahpC | 1.67 | 0.032 | |||
| ftsY | 1.66 | 0.041 | ||||
| Central intermediary metabolism | dexB | 2.77 | 0.006 | |||
| lnbA | 3.02 | 0.050 | ||||
| ypcD | 1.89 | 0.006 | ||||
| Energy metabolism | frdC | 2.12 | 0.021 | |||
| gnd | 1.77 | 0.005 | ||||
| ldh | 1.52 | 0.017 | ||||
| pdhD | 2.37 | 0.031 | ||||
| scrK | 2.26 | 0.034 | ||||
| ypbG | 2.11 | 0.014 | ||||
| ypdB | 3.02 | 0.014 | ||||
| ypgB | 2.49 | 0.032 | ||||
| Fatty acid and phospholipid metabolism | fabF | 1.94 | 0.039 | |||
| Purines, pyrimidines, nucleosides, and nucleotides | carB | 1.54 | 0.044 | |||
| Regulatory functions | lacR | 0.08 | 0.029 | yecE | 1.76 | 0.040 |
| Translation | infA | 0.44 | 0.050 | pepC | 3.77 | 0.023 |
| rplC | 0.55 | 0.030 | pepM | 1.81 | 0.032 | |
| rplQ | 0.53 | 0.038 | proS | 1.90 | 0.035 | |
| yphL | 0.59 | 0.020 | yqEL | 2.15 | 0.037 | |
| rpmE | 0.47 | 0.016 | ||||
| rpsL | 0.58 | 0.003 | ||||
| Transport and binding proteins | zitS | 0.65 | 0.001 | msmK | 2.00 | 0.018 |
| optF | 1.95 | 0.0004 | ||||
| ptnD | 2.99 | 0.045 | ||||
| ypcH | 4.49 | 0.012 | ||||
| ypdA | 2.76 | 0.025 | ||||
| Unknown | ybcH | 0.65 | 0.013 | ychH | 2.14 | 0.021 |
| ywaB | 0.58 | 0.033 | yihF | 2.06 | 0.016 | |
| ykiF | 2.06 | 0.046 | ||||
| yqaC | 2.04 | 0.048 | ||||
| yrjD | 2.14 | 0.019 | ||||
| ymgG | 1.59 | 0.006 | ||||
Genes for which mRNA abundances have changed between 2 and 3 h of pure culture of L. lactis IL-1403. Intensity ratios and P values are shown.
These 158 genes were classified by functional categories following the classification of Bolotin et al. (6). All the functional categories were represented. The change ratios of normalized signals between mixed and pure cultures varied from 0.18 to 2.83. The selected genes were fairly distributed among genes for which the mRNA level diminished (54%) and those genes for which the mRNA level increased (46%) in mixed culture compared to that in pure culture. These genes were involved in various metabolic pathways such as amino acid biosynthesis or degradation, transporters, and nucleoside and nucleotide metabolism (Table 3).
TABLE 3.
Change in gene mRNA abundances in mixed cultures versus pure culturesa
| Functional category | Underexpressed genes
|
Overexpressed genes
|
||||
|---|---|---|---|---|---|---|
| Gene | Intensity ratio | P value | Gene | Intensity ratio | P value | |
| Amino acid biosynthesis | metB2 | 0.39 | 0.030 | aroE | 1.82 | 0.020 |
| thrC | 0.63 | 0.006 | ilvC | 1.66 | 0.025 | |
| leuB | 1.90 | 0.005 | ||||
| trpB | 1.98 | 0.007 | ||||
| Biosynthesis of cofactors, prosthetic groups, and carriers | dfpB | 1.80 | 0.040 | |||
| thiM | 1.74 | 0.019 | ||||
| Cell envelope | acmB | 0.54 | 0.022 | pbp2A | 1.56 | 0.008 |
| pbp1B | 0.055 | 0.048 | ycbF | 2.69 | 0.033 | |
| plpA | 0.64 | 0.014 | ||||
| Cellular processes | sipL | 2.10 | 0.028 | |||
| Energy metabolism | adhE | 0.32 | 0.026 | citC | 2.51 | 0.032 |
| atpE | 0.64 | 0.001 | fbp | 1.95 | 0.036 | |
| pdhA | 0.41 | 0.041 | noxB | 1.52 | 0.032 | |
| pta | 0.59 | 0.011 | ypdD | 1.60 | 0.016 | |
| yjiB | 0.64 | 0.021 | ||||
| Fatty acid and phospholipid metabolism | fabl | 0.36 | 0.015 | |||
| thiL | 0.44 | 0.009 | ||||
| Other categories | clpP | 0.18 | 0.006 | dinF | 1.42 | 0.026 |
| clpE | 0.38 | 0.035 | pi103 | 2.26 | 0.008 | |
| pi359 | 0.64 | 0.016 | pi242 | 1.85 | 0.041 | |
| ps122 | 1.71 | 0.037 | ||||
| yebB | 2.04 | 0.002 | ||||
| Purines, pyrimidines, nucleosides, and nucleotides | carA | 0.65 | 0.020 | deoD | 1.774 | 0.007 |
| carB | 0.044 | 0.012 | purL | 2.15 | 0.023 | |
| pydB | 0.28 | 0.010 | pyrG | 2.02 | 0.039 | |
| pyrB | 0.37 | 0.021 | ||||
| pyrE | 0.33 | 0.025 | ||||
| pyrZ | 0.63 | 0.02 | ||||
| rmlB | 0.63 | 0.017 | ||||
| pyrC | 0.40 | 0.009 | ||||
| Regulatory functions | codY | 0.48 | 0.002 | llrG | 1.88 | 0.026 |
| glnR | 0.51 | 0.036 | yjfE | 1.55 | 0.045 | |
| rliB | 0.43 | 0.006 | ||||
| typA | 0.49 | 0.049 | ||||
| Replication | polA | 1.83 | 0.014 | |||
| Transcription | rpoA | 1.96 | 0.019 | |||
| trmD | 1.79 | 0.002 | ||||
| Translation | metS | 0.56 | 0.015 | argS | 1.89 | 0.005 |
| pepO | 0.66 | 0.017 | cysS | 2.39 | 0.013 | |
| rplF | 0.45 | 0.045 | infC | 1.50 | 0.042 | |
| rpsH | 0.55 | 0.040 | leuS | 1.58 | 0.023 | |
| serS | 0.60 | 0.041 | ppiA | 1.70 | 0.003 | |
| tuf | 0.44 | 0.030 | prfA | 2.14 | 0.010 | |
| rplV | 2.06 | 0.036 | ||||
| thrS | 1.67 | 0.046 | ||||
| yugD | 2.12 | 0.023 | ||||
| Transport and binding proteins | mtsB | 0.50 | 0.037 | arcD1 | 1.88 | 0.023 |
| pyrP | 0.23 | 0.015 | pstC | 2.32 | 0.044 | |
| ychE | 0.66 | 0.037 | rbsA | 1.57 | 0.041 | |
| yogJ | 0.37 | 0.047 | ynaD | 1.57 | 0.007 | |
| ypcG | 0.36 | 0.020 | ||||
| Unknown | ybeC | 0.60 | 0.018 | ydiB | 1.50 | 0.027 |
| yhgE | 0.53 | 0.026 | ygiK | 1.60 | 0.032 | |
| yicE | 0.65 | 0.008 | yqgC | 1.82 | 0.045 | |
| yniC | 0.49 | 0.033 | yfdC | 2.30 | 0.025 | |
| ypdC | 0.60 | 0.036 | ythA | 1.54 | 0.041 | |
| yqgA | 0.48 | 0.016 | ybgD | 1.73 | 0.037 | |
| ytcC | 0.41 | 0.012 | ydgI | 1.93 | 0.028 | |
| ytdB | 0.52 | 0.025 | yjhE | 2.83 | 0.046 | |
| ytdC | 0.45 | 0.035 | ybfB | 1.50 | 0.028 | |
| yvdE | 0.57 | 0.034 | yqaD | 1.55 | 0.018 | |
| ywaI | 0.57 | 0.016 | ynbD | 1.79 | 0.013 | |
| yudK | 2.07 | 0.006 | ||||
Genes for which mRNA abundances have changed in mixed cultures compared to those in pure cultures of L. lactis IL-1403. Intensity ratios and P values are shown.
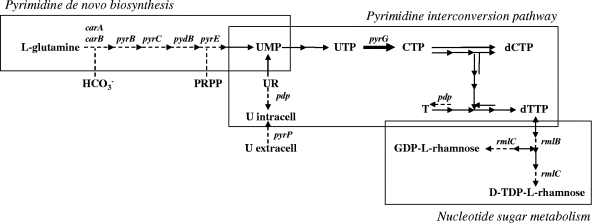
Among the identified transcriptome responses, the lowering of the mRNA level linked with the pyrimidine metabolism appeared to be the major event (Table 2). The pyrimidine de novo biosynthesis pathway (Fig. 4) consists of six enzymatic reactions (eight genes), leading to the synthesis of uridine-monophosphate (UMP) from l-glutamine and 5-phosphorybosyl-pyrophosphate (PRPP) (1, 2, 13, 21, 22, 38; for a review, see reference 18). Six of the eight genes that are known to be in the pyrimidine de novo biosynthesis pathway (carA, carB, pydB [also named pyrDb in L. lactis MG1363], pyrB, pyrC, and pyrE) and the four genes related to the pyrimidine metabolism (pyrZ, encoding the electron transfer domain of the dihydroorotate dehydrogenase; pdp, encoding pyrimidine-nucleoside phosphorylase; rmlB and rmlC, involved in thymidine sugar metabolism) exhibited a lower mRNA level in mixed culture than that in the pure culture. The pyrP gene encoding a uracil transporter (the “transport and binding proteins” category) also showed a lower transcript level in the mixed culture.
FIG. 4.
Simplified representation of the pyrimidine nucleotide metabolism in Lactococcus lactis. Only reactions relevant to this study are included in the figure. Genes whose mRNA abundances differed between pure and mixed culture are indicated. Dotted arrows indicate a lower mRNA abundance, while bold arrows indicate a higher mRNA abundance of genes in mixed culture than in pure culture. PRPP, phosphoribosyl-pyrophosphate.
The only four genes of the purine, pyrimidine, nucleoside, and nucleotide metabolism in which the amounts of mRNA were higher in mixed culture than in pure culture (pyrG, purH, purL, and deoB) were related to the purine metabolism, except for pyrG, which is responsible for the synthesis of the enzyme catalyzing the conversion from UTP to CTP (39).
RT-PCR analysis after a pulse in a pure culture of L. lactis.
The main difference between the physicochemical environment of pure L. lactis culture and that of mixed cultures was the presence of ethanol and CO2 produced by S. cerevisiae. These metabolic products may have affected gene expression. In order to define the role of those chemicals in the modification of pyrimidine metabolism, pulse experiments, consisting of the addition of ethanol or carbonate to pure batch cultures of L. lactis IL-1403, were conducted. The concentration of these products was chosen to be identical to the ethanol concentration observed with the mixed culture at the time of sampling. Total RNA was extracted 15 min before (2 h 15 min) and 15 min after the pulse (2 h 45 min) and reverse-transcribed, and real-time quantitative PCR was performed with primers specific to the carB, pydB, pyrE, pyrG, pyrP, and pyrR genes.
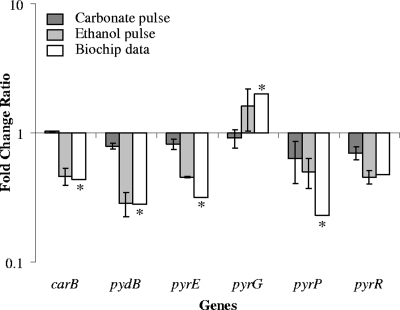
The measured ratios of mRNA levels for most of those genes were close to 1 after carbonate addition, and a very weak mRNA level decrease was observed only for pyrE, pyrP and pyrR (Fig. 5). After the addition of ethanol, the change ratio of carB, pydB, pyrE, pyrP, and pyrR was lower than onefold (from about 0.2 to 0.5), while the pyrG mRNA level seemed to be about 1.5-fold increased.
FIG. 5.
Comparison of transcript level ratios of pyrimidine de novo synthesis genes measured with real-time PCR after ethanol or carbonate pulses or with biochips for the comparison between mixed L. lactis/S. cerevisiae cultures and pure L. lactis cultures. Asterisks indicate P values of <0.05 in biochip experiments.
DISCUSSION
The measurement of biochip hybridization specificity using labeled S. cerevisiae cDNA on an L. lactis biochip showed that a big part of the L. lactis designed spots hybridized with the unspecific labeled S. cerevisiae cDNA. With this standard hybridization condition, the biochip was clearly not usable for mixed cultures. Since the possibility of using specific biochips for a mixed culture remained, until now, both a methodological constraint and a challenge to further knowledge acquisition, attempts were made to decrease the cross-hybridizations of foreign DNA on the specific L. lactis microarrays. The temperature, known to have a positive effect on the hybridization specificity by increasing the stringency (40), was unable to significantly reduce the cross-hybridization signal.
The addition of genomic DNA of the partner species enabled us to obtain a good specificity of the biochip. The efficiency of the method seemed to be correlated with the phylogenetic distance between the microbial partners. Since the sequence divergence can be considered to be similar between ribosomal and other genomic sequences, the estimation of the phylogenetic distance between the partners was based on 16S and 18S ribosomal sequence alignment. The further the partner species was from L. lactis, the more efficiently the genomic DNA addition reduced unspecific signals (Fig. 1). Some additives were added frequently prior to hybridization on biochips, in order to decrease the nonspecific fixation of the cDNA on the support; among them, for example, were proteins such as bovine serum albumin or genomic DNA (mostly salmon sperm DNA). In our method-improvement strategy, the added DNA was not a random nonspecific DNA but the DNA of the species being cultivated with our model bacterium. This method enabled the microarray cross-hybridization between S. cerevisiae cDNA and that of L. lactis to be reduced from 54% to less than 1%, or in other words, more than 99% of the L. lactis spots could now be analyzed. The method proposed to decrease this nonspecific hybridization in binary cultures can be extended to more complex ecosystems. Thus, this approach can be considered to be universal in principle, but in practice, it will not be applicable for closely related genera or for species of the same genera.
The interactions between L. lactis IL-1403 and S. cerevisiae CEN-PK905 were investigated using comparisons of transcriptome and some metabolic features of pure and mixed cultures. Biochip analysis was made with methodological replicates in order to evaluate the method's precision, avoiding artifacts due to biological interactions. The strategy, with regard to the culture medium and the environmental parameters, was chosen to avoid any evident interactions between the two partners, such as medium acidification. The macroscopic data observed were in agreement with those in the literature and particularly with the specific growth rates of the two species and their productions rates (10, 23, 37). The growth period in each culture started with an exponential growth phase, in which the environmental conditions allow the cells to grow at a constant and maximum rate. Nevertheless, there were differences in substrate and product concentrations between pure and mixed cultures during this exponential growth phase. For example, after 3 h of culture, at the end of the exponential growth phase, there was about 40 mM of lactic acid in both cultures but higher ethanol concentration in the mixed culture and an increased amount of glucose consumed under these conditions. Moreover, one cannot exclude the accumulation of other undetected metabolites during the cultures. Thus, even if no major kinetic variations were observed between the mixed and pure cultures, there were some differences in substrate and metabolite concentrations.
It is well known that the diminished growth rate, whatever the cause of the slowing down, has large implications for global gene expression, affecting functions such as amino acid transporters, sugar metabolism, large and small ribosomal subunits, etc. (30, 31). On the other hand, it is expected that very few genes will show variation of expression during exponential growth phases in which specific rates are comparable. Indeed, the comparative analysis of transcript abundance amounts between 2 and 3 h in L. lactis pure culture showed only 109 spots with a significant variation of fluorescence intensity (5% of the entire biochip; Table 2) compared to about 900 spots at the onset of the growth decrease (31). Nevertheless, this result indicates that even between two samples taken during an exponential growth phase, the mRNA level of certain genes can vary significantly. It was then considered that these differences in transcript abundance amounts between 2 and 3 h were due to the natural progression of the culture and then would be expected equally between the same samples taken in the mixed culture. In consequence, these variations were nonspecific to the L. lactis-yeast interactions and the corresponding genes considered to be false positive or negative genes were discarded from the comparative analysis of the pure and mixed cultures. This is a crucial problem when comparing gene expression in two different cultures: differentiating between those variations in mRNA levels which can be specifically correlated to the planned modification of the culture and those which are probably linked to time course differences between the two cultures.
Most of the genes linked to the pyrimidine metabolism and exhibiting a lower mRNA abundance in mixed culture were PyrR regulated (2, 18, 21, 22). The others were independently regulated (pdp, rmlB, rmlC, and pyrG). These results naturally led us to formulate two nonexclusive hypotheses: the yeast may produce pyrimidines in coculture, or there may be an earlier depletion of purines in coculture. Both phenomena could provoke a modification of the relative amounts of intracellular purine/pyrimidine and trigger the inhibition of pyrimidine synthesis to restore balance.
The estimation of purine and pyrimidine precursors in the extracellular medium did not give a satisfactory explanation for this strong variation of transcript levels. Globally, L. lactis used nucleobase precursors faster than the yeast, and thus, the evolution of their concentrations in mixed culture was very close to that observed for pure L. lactis cultures (Fig. 3). The only significant difference between the pure L. lactis culture and the mixed cultures was observed for xanthine. In opposition to the hypothesis suggested above, this purine was apparently less consumed in the mixed culture than in the L. lactis pure culture.
Since the nucleobase metabolism did not seem to be responsible for the observed transcriptome change of the corresponding genes, the factors affecting the pyr mRNA abundance remained to be found. Regarding fermentation dynamics, the main difference between the pure and the mixed cultures was that the yeast produced ethanol and carbonate in mixed culture. Those chemical compounds were absent from the L. lactis pure culture, since it exhibited a homolactic metabolism under these growth conditions. Carbonate is a substrate for the carbamoyl-phosphate synthase, encoded by carA and carB, catalyzing the first reaction of the de novo pyrimidine biosynthesis pathway. This reaction leads to the transformation of l-glutamine and carbonate to l-glutamate and carbamoyl-phosphate (25). So, a variation in carbonate concentration might have an effect on pathway flux and/or the regulation of genes involved in de novo pyrimidine biosynthesis, as demonstrated recently with L. plantarum (3). Thus, we investigated separately the effect of carbonate and ethanol on the mRNA levels of L. lactis genes involved in pyrimidine metabolism by RT-quantitative PCR. The pulses did not change the observed kinetic parameters (data not shown), indicating that the observations for pulse experiments are similar to what took place in mixed cultures.
While biochip measurement showed an increased amount of the pyrG mRNA in mixed cultures, neither ethanol nor carbonate significantly altered the transcript abundance ratio when they were pulsed into the medium. It is more probable that control of the pyrG expression responds to the intracellular CTP concentration (16, 17). Decreased expression of the de novo pyrimidine biosynthesis genes could have led indirectly to a diminished intracellular pool of CTP, thereby inducing higher expression of pyrG. A consequent time shift response to the decrease of the intracellular CTP pool could explain, therefore, that the change of mRNA level may not have time to occur within the 15 min after the pulse. Surprisingly, the addition of carbonate to the medium had very little influence on the transcription level of these genes (Fig. 5), since no significant differences were observed before and after the addition of carbonate. Only for pyrP and pyrR could carbonate accumulation contribute to the diminished transcription profiles. On the other hand, the addition of ethanol led to a decreased transcript abundance for the carB, pydB, pyrE, and pyrR genes, to the same extent as revealed previously by DNA microarrays, indicating that ethanol accumulation could account for the totality of this transcriptome change in the mixed culture.
Thus, ethanol production appeared to be the main vector of the observed interspecies interaction. Ethanol stress on Lactococci spp. is rarely mentioned in the literature, especially regarding transcription analysis. Further research may provide useful data with which to understand the mechanisms triggering such transcriptional changes and the extent of the effect that ethanol has on L. lactis.
Acknowledgments
We thank Lidwine Trouilh and Sergei Sokol (Biochips Platform, Genopole Toulouse) for technical assistance and help with the biochip analysis and Marie-Odile Loret for nucleoside/nucleotide analysis.
This work was supported by an ANR (French Research National Agency) grant under the Genoferment 2E.11 PNRA program.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Andersen, P. S., P. J. Gildsig Jansen, and K. Hammer. 1994. Two different dihydroorotate dehydrogenases in Lactococcus lactis. J. Bacteriol. 176:3975-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, P. S., J. Martinussen, and K. Hammer. 1996. Sequence analysis and identification of the pyrKDbF operon from Lactococcus lactis including a novel gene, pyrK, involved in pyrimidine biosynthesis. J. Bacteriol. 178:5005-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arsene-Ploetze, F., V. Kugler, J. Martinussen, and F. Bringel. 2006. Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine-dependent regulator PyrR1. J. Bacteriol. 188:8607-8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrière, C., M. Veiga-da-Cunha, N. Pons, E. Guédon, S. A. van Hijum, J. Kok, O. P. Kuipers, D. S. Ehrlich, and P. Renault. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodrossy, L., N. Stralis-Pavese, M. Konrad-Koszler, A. Weilharter, T. G. Reichenauer, D. Schofer, and A. Sessitsch. 2006. mRNA-based parallel detection of active methanotroph populations by use of a diagnostic microarray. Appl. Environ. Microbiol. 72:1672-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis spp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callon, C., L. Millet, and M. C. Montel. 2004. Diversity of lactic acid bacteria isolated from AOC Salers cheese. J. Dairy Res. 71:231-244. [DOI] [PubMed] [Google Scholar]
- 8.Cho, J.-C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocaign-Bousquet, M., and N. D. Lindley. 1995. Pyruvate overflow and carbon flux within the central metabolic pathways of Corynebacterium glutamicum during growth on lactate. Enzyme Microb. Technol. 17:260-267. [Google Scholar]
- 10.Cocaign-Bousquet, M., C. Garrigues, P. Loubière, and N. D. Lindley. 1996. Physiology of pyruvate metabolism in Lactococcus lactis. Antonie Leeuwenhoek 70:253-267. [DOI] [PubMed] [Google Scholar]
- 11.Corsetti, A., J. Rossi, and M. Gobbetti. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Dennis, P., E. A. Edwards, S. N. Liss, and R. Fulthorpe. 2003. Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl. Environ. Microbiol. 69:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elagoz, A., A. Abdi, J. C. Hubert, and B. Kammerer. 1996. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene 183:37-43. [DOI] [PubMed] [Google Scholar]
- 14.Hüser, A. T., A. Becker, I. Brune, M. Dondrup, J. Kalinowski, J. Plassmeier, A. Puhler, I. Wiegrabe, and A. Tauch. 2003. Development of a Corynebacterium glutamicum DNA microarray and validation by genome-wide expression profiling during growth with propionate as carbon source. J. Biotechnol. 106:269-286. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, M. R., S. B. Conners, C. I. Montero, C. J. Chou, K. R. Shockley, and R. M. Kelly. 2006. The Thermotoga maritima phenotype is impacted by syntrophic interaction with Methanococcus jannaschii in hyperthermophilic coculture. Appl. Environ. Microbiol. 72:811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jørgensen, C. M., K. Hammer, and J. Martinussen. 2003. CTP limitation increases expression of CTP synthase in Lactococcus lactis. J. Bacteriol. 185:6562-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jørgensen, C. M., K. Hammer, P. R. Jensen, and J. Martinussen. 2004. Expression of the pyrG gene determines the pool sizes of CTP and dCTP in Lactococcus lactis. Eur. J. Biochem. 271:2438-2445. [DOI] [PubMed] [Google Scholar]
- 18.Kilstrup, M., K. Hammer, P. Ruhdal Jensen, and J. Martinussen. 2005. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 29:555-590. [DOI] [PubMed] [Google Scholar]
- 19.Larsen, R., T. G. Kloosterman, J. Kok, and O. P. Kuipers. 2006. GlnR-mediated regulation of nitrogen metabolism in Lactococcus lactis. J. Bacteriol. 188:4978-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loret, M. O., L. Pedersen, and J. François. 2007. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast 24:47-60. [DOI] [PubMed] [Google Scholar]
- 21.Martinussen, J., and K. Hammer. 1998. The carB gene encoding the large subunit of carbamoylphosphate synthetase from Lactococcus lactis is transcribed monocistronically. J. Bacteriol. 180:4380-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 183:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Møller, K., L. Olsson, and J. Piskur. 2001. Ability for anaerobic growth is not sufficient for development of the petite phenotype in Saccharomyces kluyveri. J. Bacteriol. 183:2485-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narvhusa, J. A., and T. H. Gadaga. 2003. The role of interaction between yeasts and lactic acid bacteria in African fermented milks: a review. Int. J. Food Microbiol. 86:51-60. [DOI] [PubMed] [Google Scholar]
- 25.Nicoloff, H., A. Elagöz, F. Arsene-Ploetze, B. Kammerer, J. Martinussen, and F. Bringel. 2005. Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 187:2093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouadghiri, M., M. Amar, M. Vancanneyt, and J. Swings. 2005. Biodiversity of lactic acid bacteria in Moroccan soft white cheese (Jben). FEMS Microbiol. Lett. 251:267-271. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, C., E. M. Bik, M. B. Eisen, P. B. Eckburg, T. R. Sana, P. K. Wolber, D. A. Relman, and P. O. Brown. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patrignani, F., R. Lanciotti, J. M. Mathara, M. E. Guerzoni, and W. H. Holzapfel. 2006. Potential of functional strains, isolated from traditional Maasai milk, as starters for the production of fermented milks. Int. J. Food Microbiol. 107:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raynaud, S., R. Perrin, M. Cocaign-Bousquet, and P. Loubière. 2005. Metabolic and transcriptomic adaptation of Lactococcus lactis subsp. lactis biovar diacetylactis in response to autoacidification and temperature downshift in skim milk. Appl. Environ. Microbiol. 71:8016-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redon, E., P. Loubière, and M. Cocaign-Bousquet. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J. Bacteriol. 187:3589-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee, S. K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakala, R. M., H. Hayashidani, Y. Kato, T. Hirata, Y. Makino, A. Fukushima, T. Yamada, C. Kaneuchi, and M. Ogawa. 2002. Change in the composition of the microflora on vacuum-packaged beef during chiller storage. Int. J. Food Microbiol. 74:87-99. [DOI] [PubMed] [Google Scholar]
- 34.Sessitsch, A., E. Hackl, P. Wenzl, A. Kilian, T. Kostic, N. Stralis-Pavese, B. T. Sandjong, and L. Bodrossy. 2006. Diagnostic microbial microarrays in soil ecology. New Phytol. 171:719-735. [DOI] [PubMed] [Google Scholar]
- 35.Simova, E., D. Beshkova, A. Angelov, T. Hristozova, G. Frengova, and Z. Spasov. 2002. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J. Ind. Microbiol. Biotechnol. 28:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visser, W., W. A. Scheffers, W. H. Batenburg-Van der Vegte, and J. P. Van Dijken. 1990. Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56:3785-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadskov-Hansen, S. L. L., J. Martinussen, and K. Hammer. 2000. The pyrH gene of Lactococcus lactis subsp. cremoris encoding UMP kinase is transcribed as part of an operon including the frr1 gene encoding ribosomal recycling factor 1. Gene 241:157-166. [DOI] [PubMed] [Google Scholar]
- 39.Wadskov-Hansen, S. L. L., M. Willemoes, J. Martinussen, K. Hammer, J. Neuhard, and S. Larsen. 2001. Cloning and verification of the Lactococcus lactis pyrG gene and characterization of the gene product, CTP synthase. J. Biol. Chem. 276:38002-38009. [DOI] [PubMed] [Google Scholar]
- 40.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, L., X. Liu, C. W. Schadt, and J. Zhou. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]