Abstract
The conversion of amino acids into volatile and nonvolatile compounds by lactic acid bacteria in cheese is thought to represent the rate-limiting step in the development of mature flavor and aroma. Because amino acid breakdown by microbes often entails the reversible action of enzymes involved in biosynthetic pathways, our group investigated the genetics of amino acid biosynthesis in Lactobacillus helveticus CNRZ 32, a commercial cheese flavor adjunct that reduces bitterness and intensifies flavor notes. Most lactic acid bacteria are auxotrophic for several amino acids, and L. helveticus CNRZ 32 requires 14 amino acids. The reconstruction of amino acid biosynthetic pathways from a draft-quality genome sequence for L. helveticus CNRZ 32 revealed that amino acid auxotrophy in this species was due primarily to gene absence rather than point mutations, insertions, or small deletions, with good agreement between gene content and phenotypic amino acid requirements. One exception involved the phenotypic requirement for Asp (or Asn), which genome predictions suggested could be alleviated by citrate catabolism. This prediction was confirmed by the growth of L. helveticus CNRZ 32 after the addition of citrate to a chemically defined medium that lacked Asp and Asn. Genome analysis also predicted that L. helveticus CNRZ 32 possessed ornithine decarboxylase activity and would therefore catalyze the conversion of ornithine to putrescine, a volatile biogenic amine. However, experiments to confirm ornithine decarboxylase activity in L. helveticus CNRZ 32 by the use of several methods were unsuccessful, which indicated that this bacterium likely does not contribute to putrescine production in cheese.
Flavor development in Cheddar and other bacterium-ripened cheeses is a dynamic and complex biochemical process that requires lactic acid bacteria (LAB) and enzymes. The LAB that contribute to this process include deliberately added starter cultures and adjunct cultures as well as nonstarter LAB that enter the cheese through the milk or the processing environment. Collectively, these microbes influence flavor development through several basic mechanisms that include lactose fermentation, conversion of milk proteins (primarily caseins) into peptides and free amino acids, catabolism of amino acids into volatile aroma compounds, lipase/esterase activity, and citrate catabolism (6). In particular, the conversion of amino acids into volatile and nonvolatile compounds by LAB in cheese is thought to represent the rate-limiting step in the development of mature flavor and aroma (28). The conversion of amino acids into volatile cheese flavor compounds in cheese may be catalyzed by starter, adjunct, and nonstarter cultures of LAB and may also occur via biochemical interactions between different bacteria (13). The basis for culture interactions is not fully understood, but LAB are typically auxotrophic for several amino acids, and amino acid breakdown by LAB often involves the reversible action of enzymes involved in anabolic pathways (28). Thus, the interplay that occurs between LAB in amino acid catabolism may be a reflection of the “incomplete” enzymology associated with amino acid metabolism among the various bacteria in cheese. Because the primary sequences of many enzymes involved in amino acid anabolic and catabolic reactions are relatively well conserved, access to genome sequence information should enhance our ability to predict, and test, pathways for amino acid metabolism in cheese.
Our group is interested in amino acid catabolism by Lactobacillus helveticus CNRZ 32, a commercial cheese flavor adjunct used to intensify flavor and reduce bitterness in several cheese varieties. As a species, L. helveticus has more extensive amino acid requirements than most LAB (10), and previous analysis of the amino acid requirements for L. helveticus CNRZ 32 by single-amino-acid omission in a chemically defined medium (CDM) demonstrated that this strain was auxotrophic for Arg, Glu, His, Ile, Leu, Lys, Met, Phe, Pro, Thr, Trp, Tyr, Val, and either Asp or Asn (4). In this study, we used a ninefold draft-quality genome sequence for L. helveticus CNRZ 32 to reconstruct amino acid biosynthetic pathways in this organism. Our analysis showed that amino acid auxotrophy in this species was due primarily to gene absence and revealed good agreement between gene content and phenotypic amino acid requirements. In addition, experiments confirmed a genome-based prediction that Asp (or Asn) auxotrophy could be alleviated by the addition of citrate, but results did not support another prediction that L. helveticus CNRZ 32 could catalyze the conversion of ornithine to putrescine (1,4-diaminobutane or butanediamine), a volatile biogenic amine.
MATERIALS AND METHODS
Microorganisms.
L. helveticus CNRZ 32 was obtained from the laboratory collection of James L. Steele (University of Wisconsin—Madison), and the putrescine-producing strain Lactobacillus sp. strain ATCC 33222 (previously designated Lactobacillus sp. strain 30a [7]) was purchased from The American Type Culture Collection (Manassas, VA). Stocks of each culture were maintained at −80°C in sterile nonfat milk with 11% (wt/vol) glycerol, and working cultures were prepared from frozen stocks by two sequential transfers (1% inoculation) in de Man Rogosa and Sharpe (MRS) broth medium (Difco Laboratories, Sparks, MD) incubated overnight at 37°C without shaking.
Gene identification.
A draft-quality genome sequence (2.28 Mbp of sequence data on seven total contigs with greater-than-ninefold average sequence redundancy and predicted to encompass more than 99.9% of the genome) for L. helveticus CNRZ 32 (18) was assembled using SeqMan II sequence analysis software (version 5.08; DNAStar, Madison, WI) and screened for genes encoding enzymes involved in amino acid biosynthesis. To accomplish this, enzymes associated with each pathway were first determined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (12) and then protein sequences for each enzyme were collected from the nearest phylogenetic relative, using ERGO bioinformatics software (Integrated Genomics, Chicago, IL). A FASTA file for the genome sequence was exported into BioEdit software (Ibis Therapeutics, Carlsbad, CA), and then the presence or absence of a gene or a gene fragment encoding each protein in the L. helveticus CNRZ 32 genome was analyzed using BLAST tools in BioEdit. When necessary, independent PCR experiments were performed to collect additional DNA sequence information to ensure that all of the genes described in this study were represented by at least a fourfold level of unambiguous, bidirectional nucleotide sequence data.
Asp/Asn auxotrophy in L. helveticus CNRZ 32.
A CDM was prepared according to the method described by Christensen and Steele (4), with the following modifications: one sample lacked citrate, Asp, and Asn (CDM); a second sample contained 2 g per liter sodium citrate (Cit) but lacked Asp or Asn (CDM plus Cit); and a third sample, which served as the positive control, lacked Cit but contained 200 mg per liter of l-aspartic acid and 400 mg per liter of l-asparagine (CDM plus Asp/Asn). Medium ingredients were obtained from Sigma-Aldrich, Inc. (St. Louis, MO), except for sodium acetate (trihydrate), manganese sulfate (monohydrate), sodium chloride, and glucose, which were obtained from Mallinckrodt, Inc. (Hazelwood, MO); potassium phosphate (dibasic) was obtained from Fisher Scientific, Inc. (Pittsburgh, PA); and potassium phosphate (monobasic) was obtained from MP Biomedicals, Inc. (Solon, OH). After all ingredients except the vitamin solution were added and solubilized, the medium was adjusted to pH 6.5 and autoclaved at 121°C for 10 min. The vitamin solution was sterilized by passage through a 0.2-μm cellulose acetate membrane (VWR International, West Chester, PA) and aseptically added to the cooled medium immediately prior to inoculation.
To eliminate carryover of any essential nutrients (10), 10-ml working cultures of L. helveticus CNRZ 32 were harvested by centrifugation (5,500 rpm for 15 min) and washed twice in 10 ml of sterile sodium phosphate (50 mM, pH 7.0). Cell samples were split into three equal parts, and each sample was washed in one of the variations of CDM. Then, a 1% dilution of the cells was made in a fresh 10-ml tube of the corresponding CDM. Growth in each medium was determined by measuring absorbance spectrophotometrically at 660 nm (A660) after 0, 19, 24, 48, and 55 h of incubation.
Screening for ornithine decarboxylase activity.
Several methods were employed to detect putrescine production by L. helveticus CNRZ 32. Lactobacillus sp. strain ATCC 33222, a bacterium previously shown to rapidly convert ornithine to putrescine (7), was used as a positive control for this activity in some assays. The first technique involved using an improved decarboxylase medium, described by Bover-Cid and Holzapfel (3), which is reported to have a limit of detection for biogenic amine production of 350 mg per liter. Next, thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC) were used to obtain greater sensitivity (10 and 3 mg per liter, respectively) in assays to detect putrescine production from ornithine. TLC was performed as described by Garcia-Moruno et al. (7), and HPLC was done as outlined by Hernandez-Jover et al. (11). Both procedures required a dansyl chloride derivation step with the L. helveticus CNRZ 32 supernatant, which was collected after 3 and 7 days of incubation at 30°C in MRS medium supplemented with 3 mM l-ornithine.
Finally, 13C-labeled nuclear magnetic resonance (NMR) analysis of the L. helveticus CNRZ 32 supernatant was performed to gain greater assay sensitivity (7.6 mg per liter) without the need for sample derivation. Prior to NMR analysis, a 300 mM solution of uniformly labeled l-[U-13C5]ornithine (Cambridge Isotope Laboratories Inc., Andover, MA) and a 2 mM solution of pyridoxal 5-phosphate (Sigma-Aldrich) were prepared in sterile, double-distilled H2O and filter sterilized through a 0.2-μm cellulose acetate membrane (VWR International). The 13C-labeled ornithine solution was stored in the dark at 4°C until needed.
In an effort to promote the induction of ornithine decarboxylase activity before NMR analysis (3), L. helveticus CNRZ 32 and Lactobacillus sp. strain ATCC 33222 were subcultured twice in MRS containing 0.1% l-ornithine monohydrochloride and 0.005% pyridoxal-5-phosphate. Cell samples were prepared in three 1.5-ml sterile centrifuge tubes. One tube served as the positive control and contained 1 ml of MRS, a 1% inoculation of Lactobacillus sp. strain ATCC 33222, and 100 μl of l-[U-13C5]ornithine solution. The cell-free negative control contained 1 ml of MRS and 100 μl of l-[U-13C5]ornithine solution. The test sample consisted of 1 ml of MRS, 1% L. helveticus CNRZ 32, and 100 μl of l-[U-13C5]ornithine solution. The tubes were incubated for 3 days at 37°C or for 7 days at 30°C and then centrifuged at 12,000 rpm for 5 min, and 0.6 ml of sample supernatant was transferred to 5-mm high-pressure NMR tubes (Wilmad-Labglass, Buena, NJ). To address the possibility that ornithine decarboxylase activity might be induced at low pH, similar experiments were performed using cells suspended in MRS buffered to a pH of 5.1 or 6.0 (5).
All NMR spectra were collected on a Bruker DMX400 NMR spectrometer (Bruker Analytik GmbH, Ettlingen, Germany) operated at a carbon frequency of 100.6 MHz. The probe temperature ranged between 13 and 37°C. NMR spectra were referenced for carbon by a capillary insert tube that contained chloroform-d (Sigma Chemical Company, St. Louis, MO). Five thousand scans were recorded for each sample. In addition, 50,000 scans were run with L. helveticus CNRZ 32 samples to increase the sensitivity for putrescine detection. 13C-labeled chemical shifts for ornithine and putrescine were identified by NMR analysis of unlabeled standards.
RESULTS
Amino acid biosynthetic pathways.
From their growth studies with a CDM, Christensen and Steele (4) determined that L. helveticus CNRZ 32 was prototrophic for only 5 amino acids (Ala, Gln, Cys, Ser, and Gly) and auxotrophic for 13 amino acids (Arg, Glu, His, Ile, Leu, Lys, Met, Phe, Pro, Thr, Trp, Tyr, and Val) and could interconvert Asn and Asp (hence, only one of these amino acids was required). Primary sequences for many enzymes involved in amino acid biosynthesis show a relatively high degree of sequence similarity, so BLAST tools were used to search the L. helveticus CNRZ 32 genome and to manually annotate genes involved in these pathways. No arbitrary cutoff value was used to determine the statistical significance of protein alignments during manual annotation (represented in the BLAST output by an expected [E] value), but all of the genes identified in this work gave an E value of less than 1.5e−51 and most (>75%) had an E value smaller than 1e−70 (data not shown). As shown in Table 1, the reconstruction of amino acid biosynthetic pathways from the L. helveticus CNRZ 32 genome showed good agreement between gene content and phenotypic amino acid requirements and revealed that amino acid auxotrophy in this species was due primarily to gene absence (e.g., Arg, Glu, His, Ile, Leu, Lys, Phe, Pro, Thr, Trp, Tyr, and Val) rather than point mutations, insertions, or small deletions (e.g., of Asp/Asn and Met).
TABLE 1.
Genetics and predicted enzymology of amino acid biosynthesis in L. helveticus CNRZ 32
| Amino acid(s) | Gene(s) presenta | Predicted product | Pathway | Essentialb |
|---|---|---|---|---|
| Ala | araT | Aromatic aminotransferase (EC 2.6.1.57) | Completec | No |
| bcaT | Branched-chain aminotransferase (EC 2.6.1.42) | |||
| ataA-C | Aminotransferase (EC 2.6.1.-) | |||
| Arg | argFd* | Ornithine carbamoyltransferase (EC 2.1.3.3) | Incomplete | Yes |
| arcCd* | Carbamate kinase (EC 2.7.2.2) | |||
| arcAd | Arginine deiminase (EC 3.5.3.6) | |||
| carA† | Carbamoyl-phosphate synthase A (EC 6.3.4.16) | |||
| carB† | Carbamoyl-phosphate synthase B (EC 6.3.4.16) | |||
| odcI | Ornithine decarboxylase (EC 4.1.1.17) | |||
| odiC | Ornithine decarboxylase (EC 4.1.1.17) | |||
| Asn, Asp | asnA | Aspartate-ammonia ligase (EC 6.3.1.1) | Incomplete | Asn or Asp |
| asnB | Asparagine synthase (EC 6.3.5.4) | |||
| ans | Asparaginase (EC 3.5.1.1) | |||
| aspC* | Aspartic aminotransferase (EC 2.6.1.1) | |||
| asd* | Aspartate-semialdehyde dehydrogenase (EC 1.2.1.11) | |||
| ppcd | Phosphoenolpyruvate carboxylase (EC 4.1.1.31) | |||
| Glu | glnA | Glutamate-ammonia ligase (EC 6.3.1.2) | Incomplete | Yes |
| aspC | Aspartic aminotransferase (EC 2.6.1.1) | |||
| Gln | glnA | Glutamate-ammonia ligase (EC 6.3.1.2) | From Glu | No |
| Cys | cysK* | O-Acetylserine (thiol) lyase (EC 2.5.1.47) | Complete | No |
| cbl* | Cystathionine-beta-lyase (EC 4.4.1.8) | |||
| cysE* | Serine acetyl-transferase (EC 2.3.1.30) | |||
| Met | metA* | Homoserine-O-succinyl transferase (EC 2.3.1.46) | Incomplete | Yes |
| cysK2d* | O-Acetylserine (thiol) lyase (EC 2.5.1.47) | |||
| metE | 5-Methyltetrahydropteroyl(Glu3)-homocysteine | |||
| methyltransferase (EC 2.1.1.14) | ||||
| patCd | Cystathionine-gamma-lyase (EC 4.4.1.1) | |||
| Lys | lysA* | Diaminopimelate decarboxylase (EC 4.1.1.20) | Completec | Yes |
| dapD* | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (EC 2.3.1.117) | |||
| dapE* | Succinyl-diaminopimelate desuccinylase (EC 3.5.1.18) | |||
| dapA* | Dihydrodipicolinate synthase (EC 4.2.1.52) | |||
| dapB* | Dihydrodipicolinate reductase (EC 1.3.1.26) | |||
| aspC* | Aspartic aminotransferase (EC 2.6.1.1) | |||
| asd* | Aspartate-semialdehyde dehydrogenase (EC 1.2.1.11) | |||
| lysC† | Aspartate kinase (EC 2.7.2.4) | |||
| dapF† | Diaminopimelate epimerase (EC 5.1.1.7) | |||
| ataA-C | Aminotransferase (EC 2.6.1.-) | |||
| Ile, Leu, Val | bcaT | Branched-chain aminotransferase (EC 2.6.1.42) | Incomplete | Yes |
| ilvB | Acetolactate synthase (EC 2.2.1.6) | |||
| Phe, Tyr, Pro | araT | Aromatic aminotransferase (EC 2.6.1.57) | Incomplete | Yes |
| aspC | Aspartic aminotransferase (EC 2.6.1.1) | |||
| His | none | Incomplete | Yes | |
| Pro | none | Incomplete | Yes | |
| Ser | glyA | Glycine hydroxymethyltransferase (EC 2.1.2.1) | Complete | No |
| glyK | Glycerate kinase (EC 2.7.1.31) | |||
| serC* | Phosphoserine transaminase (EC 2.6.1.52) | |||
| serA* | Phosphoglycerate dehydrogenase (EC 1.1.1.95) | |||
| ycsE | Phosphoserine phosphatase (EC 3.1.3.3) | |||
| sdaBd† | l-Serine ammonia-lyase B (EC 4.3.1.17) | |||
| sdaA† | l-Serine ammonia-lyase A (EC 4.3.1.17) | |||
| Gly | glyA | Glycine hydroxymethyltransferase (EC 2.1.2.1) | From Ser | No |
| Thr | none | Incomplete | Yes |
Genes for the same amino acid with a shared symbol (* or †) are adjacent to one another in an operon-like structure in the genome and are listed in predicted order of sequential transcription.
As determined by Christensen and Steele (4).
Pathway completion assumes activity for one or more transamination reactions is provided by the aromatic-chain (araT), aspartic (aspC), or branched-chain (bcaT) aminotransferase or by one of three other predicted amino acid aminotransferases (ataA-C) whose genes were identified in the L. helveticus CNRZ 32 genome.
Probable pseudogene; the product is predicted to lack biological activity.
Alleviation of Asp auxotrophy in the CDM by citrate.
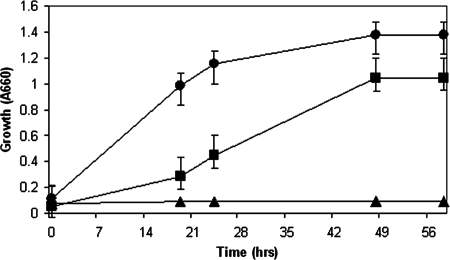
As expected, L. helveticus CNRZ 32 cells incubated in CDM did not show any significant growth (Fig. 1). This observation was expected because the absence of citrate and Asn eliminated each of the predicted pathways for Asp formation by this bacterium (Fig. 2). In contrast, cells incubated in CDM plus Cit grew to an A660 of 1.0 ± 0.1 within 48 h, while cells inoculated in CDM plus Asp/Asn grew to a slightly higher final cell density (A660, 1.4 ± 0.2) (Fig. 1). Interestingly, growth curves for L. helveticus CNRZ 32 grown in CDM plus Cit showed an extended lag phase compared to growth curves of cells incubated in CDM plus Asp/Asn (Fig. 1). The basis for this observation is unknown, but the difference could reflect the later induction of genes for citrate utilization in CDM plus Cit or a low level of in vivo activity for one or more of those enzymes. We recently showed that citrate gene transcription in L. helveticus CNRZ 32 was differentially regulated in milk compared to that in a complex laboratory medium (17), and similar studies could be performed to evaluate gene regulation during growth in CDM plus Cit.
FIG. 1.
Growth of L. helveticus CNRZ 32 in CDM supplemented with Asp and Asn (•), in CDM with citrate (▪), or in CDM without Asp, Asn, and citrate (▴). Values represent the means ± standard errors of the means from triplicate experiments.
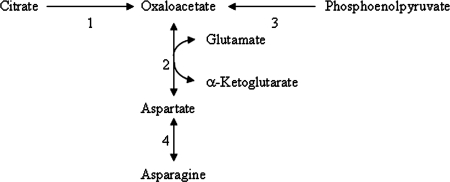
FIG. 2.
Predicted pathways for Asp and Asn biosynthesis in L. helveticus CNRZ 32. Enzymes involved in these conversions include citrate lyase (1), aspartate aminotransferase (2), phosphoenolpyruvate carboxylase (3), and asparaginase (4). Reversible reactions are indicated by a double-headed arrow.
Ornithine decarboxylase activity.
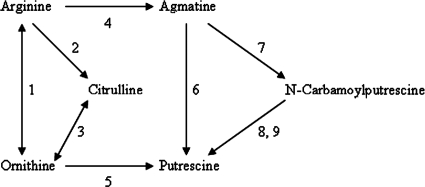
The formation of putrescine by lactobacilli has been shown by Arena and Manca de Nadra (1) to proceed via the conversion of ornithine, agmatine, or N-carbamoylputrescine (Fig. 3). Protein homology searches against the translated protein database for the L. helveticus CNRZ 32 genome sequence did not detect any orthologs to arginase (EC 3.5.3.1), arginine decarboxylase (EC 4.1.1.19), agmatine deiminase (EC 3.5.3.12), agmatinase (EC 3.5.3.11), N-carbamoylputrescine hydrolase (EC 2.1.3.6), or N-carbamoylputrescine amidase (EC 3.5.1.53). Moreover, arginine deiminase and ornithine transcarbamylase are pseudogenes in L. helveticus CNRZ 32 (Table 1). Therefore, we predicted that this bacterium cannot produce putrescine from Arg, but the presence of two genes encoding paralogs to ornithine decarboxylase in the genome (Table 1) suggested that the bacterium may be able to produce putrescine from exogenously supplied ornithine (Fig. 3). To test this hypothesis, we assayed L. helveticus CNRZ 32 for the ability to convert ornithine into putrescine, using several different techniques.
FIG. 3.
Potential pathways for putrescine synthesis by lactobacilli. Enzymes that may be involved in these conversions include 1, arginase (EC 3.5.3.1); 2, arginine deiminase (EC 3.5.3.6); 3, ornithine carbamoyltransferase (EC 2.1.3.3); 4, arginine decarboxylase (EC 4.1.1.19); 5, ornithine decarboxylase (EC 4.1.1.17); 6, agmatine deiminase (EC 3.5.3.12); 7, agmatinase (EC 3.5.3.11); 8, N-carbamoylputrescine hydrolase (EC 2.1.3.6); and 9, N-carbamoylputrescine amidase (EC 3.5.1.53). Reversible reactions are indicated by a double-headed arrow. (Adapted from reference 1 with permission from Blackwell Publishing.)
The first method for this purpose used an improved decarboxylase medium described by Bover-Cid and Holzapfel (3). As expected, colonies from the positive-control strain, Lactobacillus sp. strain ATCC 33222, gave a faint purple color indicative of ornithine decarboxylase activity (with a detection limit of 350 mg per liter), but colonies of L. helveticus CNRZ 32 did not produce any purple color on the agar (data not shown). Subsequent efforts to detect ornithine decarboxylase activity in L. helveticus CNRZ 32 by TLC or HPLC also proved unsuccessful (data not shown). The limits of detection of biogenic amines by TLC and HPLC are reported to be 10 and 3 mg per liter, respectively (11, 16), but both of these techniques require a derivation (dansyl chloride) step that theoretically can reduce assay sensitivity.
To overcome the need for sample derivation, experiments to detect putrescine production from ornithine were performed using 13C-labeled NMR analysis. The limit of detection with unlabeled putrescine for our instrument and conditions is approximately 0.08 mM (based on the detection of 8.7 mM unlabeled putrescine and considering that the natural abundance of 13C is 1%) or 7.6 mg per liter. Moreover, putrescine can be readily distinguished from ornithine with 13C-labeled NMR spectra by the presence of a peak at 24 ppm (Fig. 4A). As shown in Fig. 4B, 13C-labeled NMR scans for the positive control, Lactobacillus sp. strain ATCC 33222, showed two major peaks; one was a chemical shift peak at 24 ppm (putrescine), and the other peak was at 39 ppm. The cell-free l-[U-13C5]ornithine solution (negative control) showed five major peaks at 23, 27, 39, and 54 ppm and a carbonyl signal at 174 ppm (Fig. 4C). The 13C-labeled NMR scans from L. helveticus CNRZ 32 supernatant (Fig. 4D) were highly similar to that obtained for the negative control. Similar results were also obtained from samples incubated at 30°C for 7 days, after 50,000 scans, or from cells suspended in MRS buffered to a pH of 5.1 or 6.0 (data not shown).
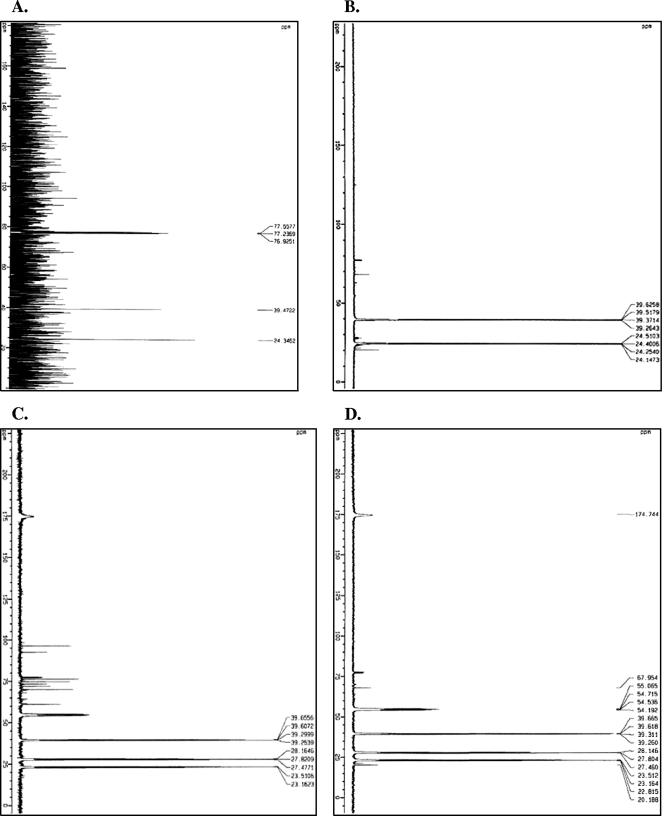
FIG. 4.
13C-labeled NMR scans of putrescine and ornithine samples. (A) 13C-labeled NMR scans of 8.7 mM putrescine in MRS broth (putrescine was unlabeled, so natural 13C is shown, which results in higher background noise); (B) putrescine production by Lactobacillus sp. strain ATCC 33222 incubated at 37°C for 3 days in MRS with l-[U-13C5]ornithine; (C) 13C-labeled NMR scans of a cell-free control incubated at 37°C for 3 days in MRS with l-[U-13C5]ornithine; (D) assay for putrescine production by L. helveticus CNRZ 32 after incubation at 37°C for 3 days in MRS with l-[U-13C5]ornithine. Peaks show resonating 13C atoms within a compound in parts per million after 1,000 (A) and 5,000 (B, C, and D) NMR scans at room temperature. Carbon reference chloroform-d was added by capillary insertion and is indicated by the peak at 77 ppm. Peaks from about 60 to 100 ppm in panel C are probably due to residual alcohol from cleaning.
DISCUSSION
As expected, the reconstruction of amino acid biosynthetic pathways from a draft-quality genome sequence for L. helveticus CNRZ 32 showed good agreement between gene content and phenotypic amino acid requirements (Table 1). For example, genes encoding enzymes for the anabolism of all five nonessential amino acids (Ala, Gln, Cys, Ser, and Gly) were identified in L. helveticus CNRZ 32 (Table 1). In the case of Ala biosynthesis, the formation of this amino acid from pyruvate may be catalyzed by more than one of the six predicted aminotransferases, including the branched-chain and aromatic amino acid aminotransferases (22, 23) that were detected in the genome sequence. In contrast, the pathway for Lys biosynthesis was theoretically functional, yet this amino acid is required for growth in a CDM. This discrepancy is likely due to the absence of a clear ortholog for succinyldiaminopimelate transaminase (EC 2.6.1.17), whose activity is apparently not provided by any of the predicted aminotransferase genes found in L. helveticus CNRZ 32 (Table 1, footnote c).
With respect to amino acid auxotrophy in L. helveticus CNRZ 32, most requirements were traced to the absence of several and sometimes all of the genes needed for biosynthesis. For example, no genes for the biosynthesis of His, Pro, or Thr were detected in this strain, while aromatic and branched-chain amino acid biosynthetic pathways were represented by only one or two genes (Table 1). These findings conflict with the results of Morishita et al. (14), who noted that the auxotrophy in L. helveticus ATCC 15009 for Pro, aromatic, and branched-chain amino acids could be reversed by single-step mutations and thus were presumably due to minor genetic lesions. This difference is interesting because L. helveticus ATCC 15009 and CNRZ 32 are reported to possess almost identical amino acid requirements (4, 14). The bases for these observations are unclear, but one explanation could be that the predominant mechanisms for gene inactivation differ between L. helveticus ATCC 15009 and those of CNRZ 32 (e.g., point mutations versus deletions) and that the similarities between amino acid requirements in both strains are a reflection of their adaptation to a milk environment. Another possibility is that because the L. helveticus CNRZ 32 genome is not complete, there are additional genes for these (and perhaps other) pathways in this organism that have not yet been captured in the sequence assembly. However, the advanced state of the genome assembly and methods used to identify genes associated with amino acid biosynthesis suggest that the latter possibility is unlikely.
While genetic analysis indicated that amino acid auxotrophy in L. helveticus CNRZ 32 was due primarily to large-scale gene absence, the requirements for Met and Asp did appear to be due to a single pseudogene in each pathway (Table 1). For example, Met anabolism from Cys appeared to be blocked by nonsense mutations in patC, a gene predicted to encode cystathionine γ-lyase, while auxotrophy for Asp appeared to be due to insertional inactivation of the gene encoding phosphoenolpyruvate carboxylase (ppc) by the insertion element ISL2. Phosphoenolpyruvate carboxylase catalyzes the production of oxaloacetate from phosphoenolpyruvate and CO2, and Asp biosynthesis occurs via AspC-mediated transamination of oxaloacetate. However, citrate catabolism by L. helveticus was recently reported by Torino et al. (24), and analysis of the L. helveticus CNRZ 32 genome identified genes encoding the citrate lyase (EC 4.1.3.6) enzyme complex and a putative citrate transporter (17). In theory, these enzymes should allow the bacterium to produce oxaloacetate and, hence, Asp and Asn from citrate rather than phosphoenolpyruvate and circumvent the need for a functional Ppc enzyme (15).
As shown in Fig. 1, experiments to test this prediction found that the growth of L. helveticus CNRZ 32 in CDM was restored by the addition of citrate. These results and the observation that growth is supported in CDM plus Asp/Asn confirmed that L. helveticus CNRZ 32 is dependent on Asp or Asn for growth and that citrate can alleviate Asp/Asn auxotrophy in this strain. These findings underscore the usefulness of genotype analysis as a tool for physiological discovery and support our hypothesis that citrate permease and citrate lyase activities in L. helveticus CNRZ 32 can generate the oxaloacetate needed for Asp biosynthesis (Fig. 2).
The production of Asp from oxaloacetate also results in the formation of α-ketoglutarate (α-KG), a compound known to favorably enhance aroma development in cheese (21). This results from the use of α-KG as the amino group acceptor by aminotransferases, which carry out the first step of many amino acid catabolic reactions in LAB (15). The contribution of citrate catabolism and Asp biosynthesis to α-KG production and aroma in the development of cheese by L. helveticus CNRZ 32 is unknown and would require future analysis of gene expression and enzyme activities for proteins involved in these conversions. Nonetheless, results from CDM growth studies indicated that L. helveticus CNRZ 32 does possess at least one mechanism for α-KG production.
The presence of remnant enzymes from some truncated pathways, such as the aminotransferases used in the synthesis of aromatic or branched-chain amino acids, suggests there is strong selective pressure to retain the activities provided by these enzymes. Transamination is a common theme in amino acid biochemistry, and while many of these enzymes exhibit some degree of overlapping specificity, evolutionary pressure to retain several paralogs is more likely a reflection of individual enzyme specificity than gross functional redundancy. In any event, previous work has suggested that the “remnant” aminotransferases do contribute to amino acid breakdown by L. helveticus CNRZ 32 (8, 9) and that the α-keto acids generated by these enzymes may, depending on enzyme activity and substrate availability, be further catabolized by other bacteria present in cheese.
The potential for metabolic cross-feeding is also apparent in remnants from the pathway for Arg biosynthesis, where auxotrophy in L. helveticus CNRZ 32 results from a combination of gene absence and the presence of pseudogenes (Table 1). One of the theoretical, and undesirable, end products of Arg catabolism is putrescine, a vasoactive amine that can impart a putrid odor to food (27). To our knowledge, the production of bioactive amines by L. helveticus has not been reported, but this attribute has been noted in a few strains of the closely related species L. acidophilus (19). Moreover, the L. helveticus CNRZ 32 genome contained two genes, odcI and odiC (Table 1), as well as one copy of the potABCD operon. The odcI and odiC genes were predicted to encode ornithine decarboxylase, which catalyzes the conversion of ornithine to putrescine, while potABCD shows homology to a spermidine/putrescine ABC transport system that may accommodate ornithine uptake in lactobacilli (25). The presence of these genes in L. helveticus CNRZ 32 suggested that this strain might be able to convert ornithine, which is produced by many LAB from Arg (2), into putrescine, a biogenic amine characterized by a putrid aroma (1, 19, 27).
In contrast to data for citrate catabolism, experiments to confirm ornithine decarboxylase activity in L. helveticus CNRZ 32 proved unsuccessful. After initial experiments using chromogenic agar gave negative results, the cells were assayed using TLC and HPLC, but again no evidence for biogenic amine formation was noted. Both chromatographic procedures provide substantially greater sensitivity than that available with the improved decarboxylase medium. The limit of detection for the agar with HPLC was reported to be 350 mg per liter, while that of TLC is about 0.1 mM (∼10 mg per liter), and the HPLC method is about threefold more sensitive than TLC (3, 7, 11, 16). However, a derivation step is required for both the TLC and the HPLC methods, so the absence of detectable putrescine may theoretically be due to inefficient derivation.
To resolve this question, we turned to the 13C-labeled NMR assay, a method that does not require sample derivation. Visual examination of the chemical shifts of the putrescine standard (Fig. 4A) showed that the peak which most clearly distinguishes putrescine from l-ornithine occurs at 24 ppm. This peak is caused by the second and third carbons of putrescine and was clearly present in samples from the Lactobacillus sp. strain ATCC 33222 positive control (Fig. 4B), even though the carbon-carbon splitting is dividing the signal. Except for the peak at 39 ppm, the five chemical shifts that describe l-ornithine (Fig. 4C and D) are entirely missing from Fig. 4B, suggesting that Lactobacillus sp. strain ATCC 33222 converted most of this substrate into putrescine. As expected, the negative controls (Fig. 4C) simply showed the presence of labeled ornithine, thereby ruling out false positives.
13C-labeled NMR spectra from L. helveticus CNRZ 32 samples (Fig. 4D) looked very similar to those from the negative control (Fig. 4C). The limit of detection with unlabeled putrescine for our instrument and conditions is approximately 0.08 mM; putrescine has an odor threshold of 0.1 mM in water and 0.5 mM in a food matrix (2% soybean flour solution [27]). Based on these values and the absence of detectable putrescine in L. helveticus CNRZ 32 samples incubated with 13C-labeled ornithine under neutral or acidic conditions, we conclude that ornithine decarboxylase activity in this strain is unlikely to affect the odor or flavor of cheese products containing the bacterium. It remains possible, however, that L. helveticus CNRZ 32 is able to generate putrescine at a concentration below the limit of detection for the 13C-labeled NMR assay or that it might produce detectable levels under different growth conditions, including conditions found in ripening cheese. Previous work has shown that lactobacilli produce putrescine under low-pH (26) conditions, and production of this compound was detected with Lactobacillus sp. strain ATCC 33222, the positive control (7, 26). Since putrescine production by L. helveticus CNRZ 32 was not detected under similar conditions and this bacterium has never (to our knowledge) been associated with the development of putrid flavor in cheese, a more plausible conclusion is that either L. helveticus CNRZ 32 does not possess ornithine decarboxylase activity (and odiC and odcI therefore encode an alternate and as-yet-unknown enzyme function) or else the potABCD operon (or some other transport systems in this cell) does not support ornithine uptake. Recent discovery of the potABCD operon, two ornithine decarboxylases, and the atypical potBCAD operon in the genome sequence of L. delbrueckii subsp. bulgaricus ATCC 11842 was interpreted to reflect a physiological need for polyamines in this species (25). Our finding that a closely related bacterium (L. helveticus CNRZ 32) with similar gene content does not convert ornithine to putrescine indicates that this hypothesis requires further exploration to confirm biological activity. This suggestion was also supported by amino acid alignments between predicted proteins from L. helveticus and the most closely related homolog for which enzyme activity has been experimentally established, i.e., ornithine decarboxylase from Lactobacillus sp. strain ATCC 33222 (26), which shows 46 to 48% amino acid identity (65 to 66% similarity) to the predicted proteins in L. helveticus CNRZ 32, and PotD from Escherichia coli (20), which has 40% amino acid identity (57% similarity) to the L. helveticus protein.
Protein alignment for ornithine decarboxylase confirmed that all residues for GTP and pyridoxal phosphate binding were conserved in both L. helveticus proteins and that most amino acids that contribute to the active site were also present. However, the active enzyme in Lactobacillus sp. strain ATCC 33222 exists as a dimer or dodecadimer, six residues (Arg120 to Phe125) that come from the adjacent monomer to fill in the active site (26) are not conserved in the L. helveticus (or L. delbrueckii) enzyme, and the amino-terminal region that would house them shows very little similarity to that of the Lactobacillus sp. strain ATCC 33222 enzyme. In similar fashion, residues associated with spermidine or putrescine binding by the E. coli PotD protein (20) are also not completely conserved in the L. helveticus (or L. delbrueckii) PotD enzymes. Whether these differences have any implication for enzyme function is unknown, but they do reinforce the need for biological validation of genome-derived metabolic predictions.
In summary, this study provided examples of both the value and the pitfalls associated with phenotypic predictions from genome sequence information for bacteria. First, genome sequence analysis showed good agreement between gene content and phenotypic amino acid requirements and provided fundamental insight into the genetic basis for auxotrophy. Additionally, experiments confirmed the genome sequence prediction that citrate can alleviate Asp/Asn auxotrophy in L. helveticus CNRZ 32 grown in CDM and revealed a possible mechanism for α-KG production by L. helveticus CNRZ 32. However, our inability to substantiate putrescine synthesis from ornithine by this bacterium underscored the need to experimentally confirm genome sequence predictions.
Acknowledgments
We thank Victor Haroldsen for technical contributions to the study.
This work was supported by Dairy Management, Inc., through the Wisconsin Center for Dairy Research and the Western Dairy Center, and by Chr. Hansen, Inc. The research was also supported by the Utah Agricultural Experiment Station and is approved as UAES Journal Paper number 7893.
Footnotes
Published ahead of print on 9 November 2007.
REFERENCES
- 1.Arena, M. E., and M. C. Manca de Nadra. 2001. Biogenic amine production by Lactobacillus. J. Appl. Microbiol. 90:158-162. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson, L. 1998. Lactic acid bacteria: classification and physiology, p. 1-72. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria: microbiology and functional aspects, 2nd ed. Marcel Dekker, Inc., New York, NY.
- 3.Bover-Cid, S., and W. H. Holzapfel. 1999. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53:33-41. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, J. E., and J. L. Steele. 2003. Impaired growth rates in milk of Lactobacillus helveticus peptidase mutants can be overcome by use of amino acid supplements. J. Bacteriol. 185:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley, E. G., and J. L. Steele. 2005. Succinate production and citrate catabolism by Cheddar cheese nonstarter lactobacilli. J. Appl. Microbiol. 98:14-23. [DOI] [PubMed] [Google Scholar]
- 6.Fox, P. F., and J. M. Wallace. 1997. Formation of flavor compounds in cheese. Adv. Appl. Microbiol. 45:17-85. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Moruno, E., A. V. Carrascosa, and R. Munoz. 2005. A rapid and inexpensive method for the determination of biogenic amines from bacterial cultures by thin-layer chromatography. J. Food Prot. 68:625-629. [DOI] [PubMed] [Google Scholar]
- 8.Gummalla, S., and J. R. Broadbent. 1999. Tryptophan catabolism by Lactobacillus casei and Lactobacillus helveticus cheese flavor adjuncts. J. Dairy Sci. 82:2070-2077. [DOI] [PubMed] [Google Scholar]
- 9.Gummalla, S., and J. R. Broadbent. 2001. Tyrosine and phenylalanine catabolism by Lactobacillus cheese flavor adjuncts. J. Dairy Sci. 84:1011-1019. [DOI] [PubMed] [Google Scholar]
- 10.Hebert, E. M., R. R. Raya, and G. S. De Giori. 2000. Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062. Appl. Environ. Microbiol. 66:5316-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Jover, T., M. Izquierdo-Pulido, M. T. Veciana-Nogues, and M. C. Vidal-Carou. 1996. Ion-pair high-performance liquid chromatographic determination of biogenic amines in meat and meat products. J. Agric. Food Chem. 44:2710-2715. [Google Scholar]
- 12.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieronczyk, A., S. Skeie, T. Langsrud, and M. Yvon. 2003. Cooperation between Lactococcus lactis and nonstarter lactobacilli in the formation of cheese aroma from amino acids. Appl. Environ. Microbiol. 69:734-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morishita, T., Y. Deguchi, M. Yajima, T. Sakurai, and T. Yura. 1981. Multiple nutritional requirements of lactobacilli: genetic lesions affecting amino acid biosynthetic pathways. J. Bacteriol. 148:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson, D. L., and M. M. Cox. 2005. Amino acid oxidation and the production of urea, p. 656-685 and 860-861. In D. L. Nelson and M. M. Cox (ed.), Lehninger principles of biochemistry, 4th ed. W. H. Freeman and Company, New York, NY.
- 16.Shakila, R. J., T. S. Vasundhara, and K. V. Kumudavally. 2001. A comparison of the TLC-densitometry and HPLC method for the determination of biogenic amines in fish and fishery products. Food Chem. 75:255-259. [Google Scholar]
- 17.Smeianov, V. V., P. Wechter, J. R. Broadbent, J. E. Hughes, B. Rodriguez, T. K. Christensen, Y. Ardo, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sridhar, V. R., J. E. Hughes, D. L. Welker, J. R. Broadbent, and J. L. Steele. 2005. Identification of endopeptidase genes from the genomic sequence of Lactobacillus helveticus CNRZ32 and the role of these genes in hydrolysis of model bitter peptides. Appl. Environ. Microbiol. 71:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straub, B. W., M. Kicherer, S. M. Schilcher, and W. P. Hammes. 1995. The formation of biogenic amines by fermentation organisms. Z. Lebensm. Unters. Forsch. 201:79-82. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama, S., Y. Matsuo, K. Maenaka, D. G. Vassylyev, M. Matsushima, K. Kashiwagi, K. Igarashi, and K. Morikawa. 1996. The 1.8-Å x-ray structure of the Escherichia coli PotD protein complexed with spermidine and the mechanism of polyamine binding. Protein Sci. 5:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanous, C., A. Gori, L. Rijnen, E. Chambellon, and M. Yvon. 2005. Pathways for α-ketoglutarate formation by Lactococcus lactis and their role in amino acid catabolism. Int. Dairy J. 15:759-770. [Google Scholar]
- 22.Tan-Wilson, A. L. 1983. Alanine biosynthesis and the general transaminases, p. 19-34. In K. M. Herrmann and R. L. Somerville (ed.), Amino acids: biosynthesis and genetic regulation. Addison-Wesley Publishing Co., Reading, MA.
- 23.Thage, B. V., F. P. Rattray, M. W. Laustsen, Y. Ardo, V. Barkholt, and U. Houlberg. 2004. Purification and characterization of a branched-chain amino acid aminotransferase from Lactobacillus paracasei subsp. paracasei CHCC 2115. J. Appl. Microbiol. 96:593-602. [DOI] [PubMed] [Google Scholar]
- 24.Torino, M. I., M. P. Taranto, and G. Font de Valdez. 2005. Citrate catabolism and production of acetate and succinate by Lactobacillus helveticus ATCC 15807. Appl. Microbiol. Biotechnol. 69:79-85. [DOI] [PubMed] [Google Scholar]
- 25.Van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J.-M. Batto, T. Walunas, J.-F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2005. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitali, J., D. Carroll, R. G. Chaudhry, and M. L. Hackert. 1999. Three-dimensional structure of the Gly121Tyr dimeric form of ornithine decarboxylase from Lactobacillus 30a. Acta Crystallogr. D 55:1978-1985. [DOI] [PubMed] [Google Scholar]
- 27.Wang, L. C., B. W. Thomas, K. Warner, W. J. Wolf, and W. F. Kwolek. 1975. Apparent odor thresholds of polyamines in water and 2% soybean flour dispersions. J. Food Sci. 40:274-276. [Google Scholar]
- 28.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-202. [Google Scholar]