Abstract
The western corn rootworm remains one of the most important pests of corn in the United States despite the use of many pest management tools. Cry3A, the first coleopteran-active Bacillus thuringiensis toxin isolated, has not been useful for control of the corn rootworm pest complex. Modification of Cry3A so that it contained a chymotrypsin/cathepsin G protease recognition site in the loop between α-helix 3 and α-helix 4 of domain I, however, resulted in consistent activity of the toxin (“mCry3A”) against neonate western corn rootworm. In vitro chymotrypsin digests showed that there was a substantial difference between the enzyme sensitivity of mCry3A and the enzyme sensitivity of Cry3A, with mCry3A rapidly converted from a 67-kDa form to a ∼55-kDa form. The introduced protease site was also recognized in vivo, where the ∼55-kDa form of mCry3A toxin was rapidly generated and associated with the membrane fraction. After a point mutation in mcry3A that resulted in the elimination of the native domain I chymotrypsin site (C terminal to the introduced chymotrypsin/cathepsin G protease site of mCry3A), the in vitro and in vivo digestion patterns remained the same, demonstrating that the introduced site was required for the enhanced activity. Also, 55-kDa mCry3A generated by cleavage with chymotrypsin exhibited specific binding to western corn rootworm brush border membrane, whereas untreated 67-kDa mCry3A did not. These data indicate that the mCry3A toxicity for corn rootworm larvae was due to the introduction of a chymotrypsin/cathepsin G site, which enhanced cleavage and subsequent binding of the activated toxin to midgut cells.
The dominant pest control challenge in United States maize (Zea mays L.) cultivation continues to be the corn rootworm complex (35, 41). Control of the western corn rootworm (WCR), Diabrotica virgifera virgifera LeConte, has been particularly troublesome (18, 35). In modern agriculture, the use of transgenic crop systems with toxins derived from Bacillus thuringiensis has provided efficacious insect control and has often resulted in decreased application of traditional insecticides (5, 50). Until recently, however, this technology has not been useful for control of corn rootworms due to the lack of potency of any rootworm-active principal from B. thuringiensis toxins. A variant of the Cry3Bb1 B. thuringiensis toxin is active against corn rootworms in transgenic maize, and it had a significantly lower root damage rating than the negative control in 3 years of field trials (54). Also, activity against WCR has been reported (4, 36) for paired B. thuringiensis toxins with molecular masses of approximately 14 and 44 kDa, which had a significantly lower root damage rating than the negative controls in field trials as well. Cry3A, the first coleopteran-active B. thuringiensis toxin isolated, has not been useful for control of WCR or related species in the corn rootworm pest complex. Cry3A was found to be at least 2,000-fold less toxic (51) to larvae of Diabrotica undecempunctata howardi Barber (southern corn rootworm) than to Leptinotarsa decemlineata (Say) (Colorado potato beetle), and other workers have reported very low toxicity of this protein for Diabrotica spp. (20, 30).
The mode of action of Cry3A toxin involves steps similar to those described for Cry1 toxins (47), but there are some differences. The midgut of coleopteran insects is slightly acidic or neutral (22, 45), conditions in which Cry3A shows only limited solubility (22, 56). It has been hypothesized (32) that the presence of gut surfactants may ameliorate solubility-limiting conditions in vivo and hence may be important for the bioavailability of toxins such as Cry3A (7). Also, while Cry1 toxin processing by target insects (27) (involving removal of 27 to 29 amino acids at the N terminus and 500 to 600 amino acids at the C terminus) is clearly an “activation” step, this designation is less well established for Cry3 toxins. Cry3A lacks the large C-terminal domain present in Cry1 toxins and is processed by the action of microbial proteases, which involves removal of 57 N-terminal amino acids (8). Removal of an additional 101 amino acids occurs following in vitro digestion with exogenous enzymes and certain insect gut extracts (for a review, see reference 42), but it has not been demonstrated to occur in vivo in a target pest. Finally, while specific binding interactions between Cry1 toxins and insect brush border receptor proteins have been well characterized (for a review, see reference 47), Cry3 toxins display a high level of nonspecific binding (7, 51). Although measurable specific binding of Cry3A to intact coleopteran brush border membrane vesicles (BBMV) has been reported in some cases (28, 51), individual receptor interactions in vitro are not well documented.
Importantly, serine protease activity has been implicated in the ability of Tenebrio gut extract to process B. thuringiensis Cry3A (8). It has been proposed that chymotrypsin processing of Cry3A may increase its solubility and functional binding, thereby facilitating its coleopteran toxicity (7). Other groups have also found that the processed form of Cry3A exhibits specific binding to coleopteran brush border (33, 46). Exogenous chymotrypsin cleaves Cry3A into stable 55- and 49-kDa fragments, as well as smaller polypeptides having molecular masses of 6 to 11 kDa (7).
It is commonly believed that coleopteran insects depend on cysteine proteases for the majority of digestive protease activity (6, 38); however, digestive serine protease activity has been identified in at least seven different families of Coleoptera (10, 11, 19, 40, 43, 53, 58). Such activity has also been identified in extracts from second-instar WCR larvae (17), but the contribution and role of this serine protease activity in larvae remain uncertain.
Here, we provide evidence that neonate WCR larvae possess enzymatic activity which recognizes a chymotrypsin/cathepsin G site. This target sequence, AAPF, is useful for characterizing these members of the broader chymotrypsin family of serine proteases (2, 52). We designed a modified Cry3A toxin (mCry3A) (GenBank accession number AX712174) with a chymotrypsin/cathepsin G site in domain I (Fig. 1) which displays consistently greater biological activity than Cry3A. mCry3A can be processed more rapidly and bind in a specific fashion to isolated membranes from neonate WCR, supporting the hypothesis that the introduced chymotrypsin/cathepsin G protease site is responsible for the increased biological activity.
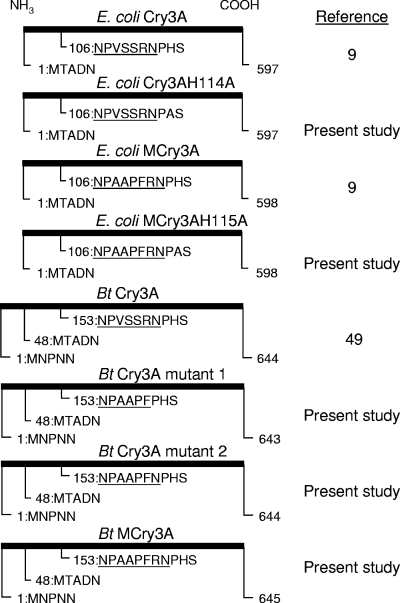
FIG. 1.
Location of introduced chymotrypsin/cathepsin G recognition sequence in domain I of E. coli-produced mCry3A. The secondary structure determined for the Cry3A crystal structure has been described previously (26). The arrows below the sequences indicate native trypsin and chymotrypsin sites described by Carroll and Ellar (8) and Carroll et al. (7), respectively. Shading indicates the introduced recognition sequence, as well as the histidine residue targeted for removal of the native chymotrypsin site by alanine substitution.
MATERIALS AND METHODS
Gene cloning and plasmid construction.
A synthetic truncated cry3A gene based on the native sequence (49) was constructed using maize-preferred codons (39). This gene served as a template for overlap extension PCR (21, 37) to construct the mcry3A gene (9), comprising a nucleotide sequence beginning at the Met 48 codon (49), which also encodes the chymotrypsin/cathepsin G recognition sequence AAPF (52) inserted in domain I between amino acids 107 and 111 of the truncated Cry3A toxin (Fig. 2). The mcry3A gene was designed with maize codon optimization for the benefit of future transformation requirements. The mcry3A gene encoding the inserted AAPF actually encodes substitution of three amino acids (V108A, S109A, and S110P), followed by insertion of F111, making mCry3A one amino acid longer than Cry3A (598 and 597 residues, respectively) (Fig. 2). Three primer pairs were used to insert the nucleotide sequence encoding the chymotrypsin/cathepsin G recognition site into the maize-optimized cry3A gene by the overlap PCR method (21, 37). These primers were designated primer pairs 1 to 3, and their sequences are as follows (sense strand/antisense strand): primer pair 1, 5′-GGATCCACCATGACGGCCGAC-3′/5′-GAACGGTGCAGCGGGGTTCTTCTGCCAGC-3′; primer pair 2, 5′-GCTGCACCGTTCCGCAACCCCCACAGCCA-3′/5′-TCTAGACCCACGTTGTACCAC-3′; and primer pair 3, 5′-GGATCCACCATGACGGCCGAC-3′/5′-TCTAGACCCACGTTGTACCAC-3′. Primer pairs 1 and 2 generated two unique PCR products. Equal parts of these products were combined, and primer pair 3 was used to join the products to generate one PCR fragment that was cloned back into the original template. The mcry3A gene was then transferred into a pUC-derived vector following restriction digestion with BamHI and SacI. This vector contained the COLE I origin of replication, a constitutive Cry1Ac promoter (48), and a beta-lactamase ampicillin resistance gene for expression in Escherichia coli.
FIG. 2.
Cry3A protein variants used. The first five amino acids indicate the relative positions of N-terminal start site variants, while 9 to 11 amino acids are shown for the targeted mutation region in the domain I loop between α-helix 3 and α-helix 4, extending into α-helix 4. Underlining indicates the loop region. Bt, B. thuringiensis.
Point mutations in the synthetic truncated mcry3A and cry3A genes were made in order to generate E. coli-expressed toxin proteins in which the native (preexisting) domain I chymotrypsin site was removed (Fig. 1 and 2). mCry3A H115A and Cry3A H114A mutations were introduced using the QuikChange method (Stratagene). The correct nucleotide sequence was confirmed for each entire gene following transformation of the mutated plasmid constructs into E. coli.
In addition, to obtain mcry3A expression in B. thuringiensis and to examine the importance of the introduced AAPF recognition sequence in various altered loop regions between α-helices 3 and 4 of domain I, a full-length native cry3Aa gene (49) was mutated by overlap extension PCR as described above with the following primer pairs: primer pair 4, 5′-GCGGCACCATTTCCACATAGCCAGGGG-3′/5′-GAGCTCTTAATTATTCACTGGAATAAAT-3′; primer pair 5, 5′-AAATGGTGCCGCAGGATTTTTTTGCCATG-3′/5′-GGATCCACCATGATAAGAAAGGGAGG-3′; and primer pair 6, 5′-GGATCCACCATGATAAGAAAGGGAGG-3′/5′-GAGCTCTTAATTATTCACTGGAATAAAT-3′. The end product encoded a protein designated B. thuringiensis Cry3A mutant 1 (Fig. 2 and Table 1). This product was then subcloned into a B. thuringiensis/E. coli pUC18-derived shuttle vector following restriction digestion with BamHI and SacI. This vector contained the replication origin and an erythromycin resistance gene as described previously for pHT 370 (1), along with an upstream Cry1Ac promoter sequence (48) for expression in B. thuringiensis. Next, QuikChange (Stratagene) site-directed mutagenesis was used to generate a gene for B. thuringiensis Cry3A mutant 2 (Fig. 2 and Table 1). Finally, a second round of QuikChange mutagenesis was performed to obtain a gene encoding B. thuringiensis mCry3A (Fig. 2 and Table 1), which was equivalent to the mCry3A protein generated in E. coli plus the first 47 residues present in the native full-length B. thuringiensis Cry3A sequence (49). B. thuringiensis cry3A mutant 1, B. thuringiensis cry3A mutant 2, and B. thuringiensis mcry3A in the shuttle vector were electroporated into an acrystalliferous strain of B. thuringiensis using a Bio-Rad Gene Pulser (Bio-Rad Laboratories, Hercules, CA). The conditions used were those described previously (29), except that the resistance was 200 Ω.
TABLE 1.
Insecticidal activity of B. thuringiensis Cry3A crystal proteins containing the AAPF mutation with first-instar WCR larvae
| Samplea | α-Helix 3/α-helix 4 loop regionb | % Mortality (mean ± SD)c |
|---|---|---|
| Cry3A | NPVSS_RNP | 34 ± 23 A |
| Cry3A mutant 1 | NPAAPF__P | 93 ± 4 B |
| Cry3A mutant 2 | NPAAPF_NP | 72 ± 13 C |
| mCry3A | NPAAPFRNP | 92 ± 7 B |
| Chymotrypsin-treated mCry3A | —d | 68 ± 7 C |
| Buffer alone | — | 7 ± 7 D |
All toxins were purified from B. thuringiensis as crystal preparations. The concentration of each toxin was 100 μg/ml in 50 mM NaHCO3 (pH 10.0) buffer.
The first N corresponds to N153 of B. thuringiensis Cry3A (49) for the unprocessed toxins.
Mortality values were transformed using the arcsine square root of the proportion transformation for analysis; actual means are shown. Means followed by the same letter are not significantly different (P > 0.05, Student-Newman-Keul test; n = 6).
—, not applicable.
Toxin preparation.
E. coli-generated toxins used for in vitro and in vivo mode-of-action studies were isolated from inclusion bodies using the B-PER bacterial protein extraction reagent protocol (Pierce, Rockford, IL) according to the manufacturer's instructions. Inclusion body pellets were then washed with distilled water an additional three times and solubilized in 50 mM NaHCO3 (pH 10.0) with mild shaking for 30 min at 37°C. These inclusion body preparations typically produced a single dominant band at about 67 kDa.
Chymotrypsinized stocks of toxins (1,000 μg/ml) were typically obtained by digestion with 100 μg/ml α-chymotrypsin (Sigma, St. Louis, MO) for 16 to 24 h at 37°C. To reduce the amount of residual chymotrypsin in subsequent studies, toxin stocks were dialyzed with Spectrum Micro DispoDialyzer tubing (molecular weight cutoff, 60,000) into 50 mM NaHCO3 (pH 10.0) buffer with 2 mM dithiothreitol for 24 h at 4°C and then into 50 mM NaHCO3 (pH 10.0) buffer alone. The protein in solution was quantified using the bicinchoninic acid (BCA) assay (Pierce).
As the WCR larval bioassay is often subject to much variation, our experience led us to use B. thuringiensis-generated crystals as a source of toxin for bioassays. Toxins were isolated from crystal preparations of sporulated, lysed cultures grown in T3 medium for 72 to 96 h. T3 medium contains 3 g/liter Difco tryptone (Becton Dickinson Co., Franklin Lakes, NJ), 2 g/liter Difco tryptose, 1.5 g/liter Difco yeast extract, 250 μM MnCl2, and 50 mM Na2HPO4 (pH 6.8). As determined by microscopic examination of cultures during sporulation and after lysis, B. thuringiensis cry3A mutant 1, cry3A mutant 2, and mcry3A each produced crystalline inclusions, as did the Cry3A-producing strain (data not shown). To isolate crystals, 100-ml cultures were shaken vigorously, decanted from the foam (enriched in spores) 10 times in 500-ml Erlenmeyer flasks, and then washed thoroughly as described previously (25). Final crystalline suspensions were washed three times with 10 volumes of 25 mM NaH2PO4-10 mM NaCl (pH 7.0) buffer following centrifugation at 25,000 × g at 4°C. Toxin was solubilized in 50 mM NaHCO3 (pH 10.0) buffer for 1 h at 37°C with shaking and then centrifuged as described above. The resulting toxin preparations produced a doublet with molecular masses of about 73 and 67 kDa, in accord with the known alternative translation initiation codons in the same reading frame present in the native cry3A sequence (34). The protein in solution was quantified using the BCA assay (Pierce).
Biotinylation of toxins.
E. coli-generated toxins (≥1 mg/ml) were biotinylated using a biotin labeling kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions, except that reactions were carried out in NaHCO3 (pH 10) buffer because of the limited solubility of the 67-kDa form of Cry3A toxins below pH 10. Nonreacted d-biotinoyl-ɛ-aminocaproic acid-N-hydroxy-succinimide ester reagent was removed by gel filtration on a prepared Sephadex G-25 column performed according to the instructions in the manual (Roche Applied Science).
Artificial diet bioassays.
The insecticidal activity of mCry3A toxins against neonate WCR larvae was tested using artificial diet bioassays. Eggs of WCR in soil were obtained from Crop Characteristics, Inc. (Farmington, MN) or from French Agricultural Research Services (Lamberton, MN) and were typically incubated for 14 days at 28 to 30°C for hatching. Bioassays with toxin isolated and solubilized from B. thuringiensis crystals were conducted using a diet incorporation method (31), with 200 μl of a toxin solution mixed with an equal volume of 2× molten diet to obtain a final concentration of 100 μg/ml for a single high-dose study with B. thuringiensis Cry3A, B. thuringiensis Cry3A mutants 1 and 2, B. thuringiensis mCry3A, and chymotrypsin-treated B. thuringiensis mCry3A (obtained by digestion of B. thuringiensis mCry3A with 100 μg/ml α-chymotrypsin as described previously for E. coli mCry3A). The artificial diet used was the diet described by Marrone et al. (31), with the following modifications: agarose (Cambrex Bioscience Rockland, Inc., Rockland, ME) was substituted for phytagar, formalin was omitted, and antibiotics were added as described by Chen and Stacy (9). The solidified diet mixture was transferred into 47-mm sterilized petri dishes (Millipore, Billerica, MA), and then 20 neonate corn rootworm larvae were added. The petri dishes were kept at room temperature in the dark, and mortality was recorded after 6 days. Mortality values from six replicates were transformed using the arcsine square root of the proportion, and a one-way analysis of variance was performed. Significant differences between means were then determined using the Student-Newman-Keul test. In addition, 5, 10, 20, 60, 100, and 200 μg/ml (final concentrations) of B. thuringiensis Cry3A or B. thuringiensis mCry3A (35 larvae per dose) were used for a dose-response assay, and mortality was recorded after 6 days. The 50% lethal concentration (LC50) was determined using probit analysis (15) with the EPA probit analysis program (version 1.5; Washington, DC).
In vitro processing of toxins.
In vitro processing of E. coli-generated Cry3A or mCry3A protein was examined by incubation of toxin substrates (1,000 μg/ml) with 100 μg/ml α-chymotrypsin (Sigma) in 50 mM NaHCO3 buffer (pH 10) at 37°C. Aliquots were removed for up to 20 h and immediately quenched with 2× Complete protease inhibitor cocktail (Roche Applied Science) on ice, followed by addition of Laemmli sample buffer (24) and incubation at 100°C. Samples were separated by 12.5% Phastgel sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (Amersham Biosciences, Piscataway, NJ) and stained with 0.02% Phastgel Blue R (Amersham Biosciences).
N-terminal sequence analysis.
To generate 55-kDa chymotrypsinized mCry3A and mCry3A H115A for N-terminal sequence analysis, digestion was carried out as described above with 20 to 40 μg toxin, and 5 to 10 μg of product was loaded in each lane. Gels were transferred to a Problott polyvinylidene difluoride membrane (Applied Biosystems, Foster City, CA) and stained with 0.025% Coomassie brilliant blue with 1% acetic acid and 40% methanol for ≤1 min. The blots were dried, and toxin bands were cut out and sent to ProSeq, Inc. (Boxford, MA) for protein microsequencing (carried out using an optimized Edman degradation approach).
In vivo processing and stability assays.
To determine the processing and stability of mCry3A or Cry3A toxin in vivo, WCR first-instar larvae (20 larvae/treatment) were allowed to feed on E. coli-generated toxin incorporated into WCR diet at a concentration of 600 μg/ml. After periods ranging from 25 min to 72 h, larvae were then collected for preparation of whole-body homogenate (WBH) in 20 μl of 50 mM Tris-10 mM NaCl (pH 9.0) with 2× Complete protease inhibitor cocktail (Roche Applied Science) on ice. A small glass rod was used to mechanically disrupt the sample in a 600-μl microcentrifuge tube, and this was followed by a freeze-thaw cycle in a dry ice bath; this procedure was then repeated to generate the WBH. The homogenates were centrifuged (25,000 × g) for 25 min at 4°C, and the supernatants were removed. The WBH pellet fractions were resuspended in 10 volumes of 50 mM Tris-10 mM NaCl (pH 9.0) with Complete protease inhibitor (Roche Applied Science) and recentrifuged. The resuspension and centrifugation step was repeated once to obtain the final washed pellet fractions, which were resuspended in Laemmli sample buffer (24) containing 2× Complete protease inhibitor cocktail. In addition, insect frass generated during the experiment was collected by dissolution in Laemmli sample buffer (24) with 2× Complete protease inhibitor cocktail (Roche Applied Science) using manual repipetting action over the assay chamber petri plate surface. To examine if in vivo processing was due to WCR chymotrypsin-like activity, the chymotrypsin inhibitor α-chymostatin (a specific inhibitor of alpha-, beta-, gamma-, or delta-chymotrypsin; Sigma) was incorporated into the diet (at concentrations up to 100 μg/ml) in some experiments; the amount of inhibitor used was not toxic to the larvae over the assay period and did not adversely affect feeding of the larvae. All samples were separated by SDS-PAGE (12.5%) with the Phastgel system (Amersham Biosciences), transferred to nitrocellulose membranes, and blocked with 2% bovine serum albumin in TPBS buffer, which consisted of 0.05% Tween 20 (Sigma) in phosphate-buffered saline (PBS) (8 mM NaH2PO4, 2 mM KH2PO4, 150 mM NaCl; pH 7.4). Blots derived from WBH containing unlabeled toxin were incubated with primary antibody (rabbit polyclonal anti-Cry3A) for 80 min at room temperature, washed, and then incubated with secondary antibody conjugate (goat anti-rabbit horseradish peroxidase; Kirkegaard & Perry Laboratories, Gaithersburg, MD) for 1.75 h at room temperature. Blots derived from WBH containing biotinylated toxin were incubated with streptavidin-conjugated peroxidase (Roche Applied Science). Labeled or unlabeled toxin bands were then visualized using the SuperSignal West Pico chemiluminescence kit (Pierce).
Alternatively, second-instar WCR larvae (five larvae/treatment) were allowed to feed on biotinylated toxins incorporated into WCR diet for to 20 h, and midguts were dissected into 30 μl of 0.5× PBS with 2× Complete protease inhibitor mixture (Roche Applied Science). Dissected midguts were homogenized and centrifuged, and the pellets were washed in buffer (as described above for first-instar larva WBH samples) prior to SDS-PAGE and electrophoretic transfer.
BBMV binding assays.
BBMV were prepared from first-instar larva WBH by the differential magnesium precipitation method (57). The final pellet was resuspended in PBS, and the protein concentration was determined using the BCA protein assay reagent (Pierce). For qualitative estimation of competitive binding, biotinylated mCry3A (15 nM) or biotinylated chymotrypsinized mCry3A toxin (27 nM) was incubated with 10 μg BBMV in the presence or absence of a 100-fold excess of the corresponding unlabeled toxin. The binding buffer used consisted of 0.1% bovine serum albumin in PBS with 2× Complete protease inhibitor mixture (Roche Applied Science). After 1 h of incubation at room temperature, reaction mixtures were centrifuged at 25,000 × g for 25 min at 4°C. The pellets were washed once with binding buffer and recentrifuged. The final pellets were resuspended in 50 mM NaHCO3 buffer (pH 10) with 2× Complete protease inhibitor mixture and then mixed with 2× Laemmli sample buffer (24) with 2× Complete protease inhibitor mixture prior to SDS-PAGE. For visualization, samples were transferred to a nitrocellulose membrane as described above. Bound biotinylated toxin was detected after probing with streptavidin-conjugated peroxidase (Roche Applied Science) and the SuperSignal West Pico chemiluminescence kit (Pierce).
RESULTS
B. thuringiensis mCry3A and Cry3A mutants 1 and 2 are more biologically active than Cry3A.
The introduction of a chymotrypsin/cathepsin G site into the domain I loop region between α-helix 3 and α-helix 4 of Cry3A (Fig. 1, 2) resulted in the mCry3A protein, which was reproducibly active against WCR larvae. The results of experiments conducted with protein isolated from B. thuringiensis crystals were consistent, demonstrating that B. thuringiensis mCry3A had significantly greater activity than B. thuringiensis Cry3A (Table 1). Also, two other B. thuringiensis mCry3A-related proteins (B. thuringiensis Cry3A mutants 1 and 2), made by positioning the introduced AAPF sequence in slightly different environments in the loop region between α-helix 3 and α-helix 4, exhibited significantly greater bioactivity than B. thuringiensis Cry3A (Table 1). The data for these B. thuringiensis mCry3A-related proteins, therefore, also demonstrated that the presence of the native trypsin site (due to deletion of an Arg in the loop region) was not necessary for biological activity that was significantly greater than that of Cry3A, but mCry3A and Cry3A mutant 1 performed the best (Table 1). We also found that B. thuringiensis mCry3A exhibited significantly higher toxicity for first-instar WCR larvae whether it was unprocessed or pretreated with chymotrypsin (Table 1). In a dose-response assay, the LC50 of B. thuringiensis mCry3A was estimated to be 65 μg/ml (95% confidence interval, 28 to 171 μg/ml), whereas no LC50 could be determined for B. thuringiensis Cry3A. The mortality response due to B. thuringiensis Cry3A was quite flat and was never more than 41% (the mortality at a concentration of 10 μg/ml) in this assay. The mortality due to B. thuringiensis Cry3A was 35% at a concentration of 100 μg/ml, for example, which was similar to the results shown for B. thuringiensis Cry3A in Table 1. In other bioassay experiments we found that E. coli-produced mCry3A and biotinylated mCry3A, used for mode-of-action studies, were also biologically active but were more variable with respect to the range of mortality produced (data not shown).
mCry3A is processed more readily by α-chymotrypsin in vitro.
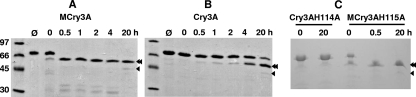
E. coli-produced mCry3A and Cry3A showed substantially different patterns of susceptibility to in vitro chymotrypsin digestion (the enzyme concentration was 100 μg/ml). mCry3A was much more susceptible to the action of chymotrypsin and was fully converted to a ∼55-kDa form in 30 min or less, whereas less than 50% of Cry3A was processed even after 4 h of incubation (Fig. 3A and B). In separate digestion experiments, we observed similar degrees of processing for mCry3A even when only 10 μg/ml enzyme was used (data not shown). As previously shown for B. thuringiensis Cry3A (7, 33), a 49-kDa form of either the mCry3A or Cry3A protein was also formed in this work (Fig. 3). The 49-kDa form, however, was only a minor proteolytic product, likely due to the different protein preparation and milder digestion conditions used in the present study. N-terminal sequencing of E. coli-produced in vitro chymotrypsinized mCry3A revealed the sequence SQGRIRELFSQ, indicating that the stable 55-kDa processed sequence begins at Ser-116 (Fig. 1 and 2). This sequence result matches what was observed previously for chymotrypsin processing of Cry3A, when the native chymotrypsin site was established (7). Chymotrypsinized mCry3A was soluble at 4°C to a concentration of at least 1.5 mg/ml in 25 mM NaCl-25 mM HEPES (pH 8.0) or in PBS, in contrast to Cry3A and mCry3A, each of which was soluble only to a concentration of about 0.25 mg/ml in 20 mM Tris (pH 7.0) at 4°C (data not shown).
FIG. 3.
In vitro processing of mCry3A, Cry3A, and H→A mutants by chymotrypsin. All toxins were produced in E. coli. (A and B) mCry3A digest (A) and Cry3A digest (B). The lane on the left contained molecular weight markers (10−3), as indicated on the left; lane φ contained the no-enzyme control at zero time; and lanes 0, 0.5, 1, 2, 4, and 20 contained 100 μg/ml chymotrypsin digest obtained at 0, 0.5, 1, 2, 4, and 20 h (∼500 ng/lane), respectively. (C) Cry3A H114A and mCry3A H115A digests. Lanes 0 and 20 contained 100 μg/ml chymotrypsin digest of Cry3A H114A (∼250 ng/lane) obtained at 0 and 20 h, respectively; and lanes 0, 0.5, and 20 contained 100 μg/ml chymotrypsin digest of mCry3A H115A (∼250 ng/lane) obtained at 0, 0.5, and 20 h, respectively. The double and single arrowheads indicate the positions of the ∼55- and ∼49-kDa chymotrypsinized forms of the toxins, respectively. Bands were detected by Coomassie blue staining after SDS-PAGE.
As observed for mCry3A, in vitro chymotrypsin digestion of the E. coli-produced mCry3A H115A protein resulted in rapid conversion to the 55-kDa product, whereas the Cry3A H114A protein appeared to be not susceptible to this enzyme (Fig. 3C). Processing of mCry3A H115A was so rapid (before proteolytic quenching took place) that the time zero aliquot contained some 55-kDa product; this band was not present in the stock sample without added enzyme (data not shown). N-terminal sequencing of E. coli-produced in vitro chymotrypsinized mCry3A H115A resulted in the sequence RNPASQGRIRE, which supported the hypothesis that processing occurred directly after the introduced AAPF mutation of mCry3A (Fig. 1 and 2).
mCry3A shows facilitated processing by WCR in vivo.
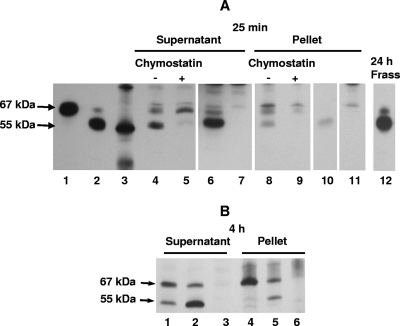
Following ingestion of biotinylated mCry3A for 25 min or less, first-instar WCR larvae quickly converted much of the full-length protein to a ∼55-kDa form similar to that observed after in vitro chymotrypsin digestion. Both the 67- and 55-kDa forms could bind to WCR membranes and were present in supernatant and washed pellet fractions (Fig. 4A). When the biotinylated 55-kDa form of mCry3A was fed directly to WCR larvae, it was stable and also bound to the membrane fraction (Fig. 4A, lanes 6 and 10). The same overall patterns of relative processing and binding of mCry3A were also apparent when other larvae were allowed to feed for 24 h or for up to 72 h (data not shown). In addition, the 55-kDa processed form was found in the recovered frass (Fig. 4A, lane 12), indicating that it likely can be formed without preliminary membrane binding of the 67-kDa form and further supporting the hypothesis that it remains stable during passage through the gut. The in vivo processing could be inhibited by inclusion of chymostatin in the diet, both for mCry3A (Fig. 4A, lanes 5 and 9) and for mCry3A H115A (data not shown), which supports the hypothesis that chymotrypsin/cathepsin G-like enzymatic activity is present in the neonate WCR gut.
FIG. 4.
In vivo processing and binding of mCry3A or Cry3A toxin in first-instar WCR larvae. Toxins were produced in E. coli. (A) Western blot for feeding study with biotinylated toxins. Lanes 1 and 2, biotinylated mCry3A and biotinylated chymotrypsinized mCry3A toxin stocks, respectively; lane 3, biotinylated markers (∼97, 58, and 40 kDa); lanes 4 to 7, supernatants of whole-body homogenates after 25 min of feeding on the WCR diet with biotinylated mCry3A, biotinylated mCry3A plus 100 μg/ml chymostatin, biotinylated-chymotrypsinized-mCry3A, and PBS alone, respectively; lanes 8 to 11, pellets of whole-body homogenates after 25 min of feeding on the WCR diet with biotinylated mCry3A, biotinylated mCry3A plus 100 μg/ml chymostatin, biotinylated-chymotrypsinized-mCry3A, and PBS alone, respectively; lane 12, frass extract recovered after 24 h of feeding on the WCR diet with biotinylated mCry3A. (B) Western blot for feeding study with unlabeled mCry3A and Cry3A. Lanes 1 to 3, supernatants of whole-body homogenates after 4 h of feeding on the WCR diet with Cry3A, mCry3A, and NaHCO3 buffer (pH 10) alone, respectively; lanes 4 to 6, pellets of whole-body homogenates after 4 h of feeding on the WCR diet with Cry3A, mCry3A, and NaHCO3 buffer (pH 10) alone, respectively.
After 4 h of feeding, more rapid processing of 67-kDa mCry3A than of 67-kDa Cry3A was observed. While the 67-kDa form of either toxin bound to WCR membranes, increased binding of the 55-kDa toxin formed was seen for mCry3A compared to Cry3A (Fig. 4B). This increased binding of the 55-kDa mCry3A generated in vivo was consistent (regardless of whether labeled or unlabeled mCry3A had been fed to the insects). In 10 separate experiments involving WCR larvae fed toxin for ≤24 h, the amount of binding of the 55-kDa form of toxin was larger for mCry3A samples than for Cry3A samples (data not shown).
Examination of gut homogenates from second-instar WCR larvae fed biotinylated mCry3A for 20 h indicated that processing to a stable 55-kDa toxin and strong binding of this toxin to membranes (by association with the pellet fraction) also occurred at this stage (Fig. 5). These data confirm that the observed processing and binding were not restricted to conditions generated by the whole-body homogenization technique used for the smaller first-instar larvae.
FIG. 5.
In vivo processing and binding of biotinylated mCry3A toxin in second-instar WCR larvae after 20 h of feeding. Toxins were produced in E. coli. Western blot results are shown. Unmarked lanes, biotinylated mCry3A (left lane) and biotinylated chymotrypsinized mCry3A (right lane) toxin stocks; supernatant lanes, gut homogenate supernatants after feeding on the WCR diet with biotinylated mCry3A (left lane) or PBS alone (right lane); pellet lanes, gut homogenate pellets after feeding on the WCR diet with biotinylated mCry3A (left lane) or PBS alone (right lane).
Chymotrypsinized mCry3A shows specific binding to WCR brush border.
After incubation of biotinylated 67-kDa mCry3A with BBMV, the toxin did not show specific binding, as a 100-fold excess of unlabeled toxin was not able to reduce the amount of label that bound (Fig. 6A). The amount of biotinylated mCry3A bound actually increased greatly when excess unlabeled toxin was present, which can best be explained by a large amount of unlabeled mCry3A bound and toxin aggregates which may have formed at the membrane surface. In contrast, incubation of biotinylated chymotrypsinized mCry3A with a 100-fold excess of unlabeled toxin resulted in a large reduction in bound label for the 55- and 49-kDa forms of toxin present (Fig. 6B), in agreement with the hypothesis that there is specific binding of the processed forms of the toxin.
FIG. 6.
Specific binding of biotinylated chymotrypsinized mCry3A to WCR first-instar larva BBMV. Toxins were produced in E. coli. (A) Western blot for incubation with 67-kDa mCry3A. Lanes 1 to 3, biotinylated mCry3A incubated with BBMV alone; lanes 4 to 6, biotinylated mCry3A with 100-fold molar excess of unlabeled mCry3A. (B) Western blot for incubation with chymotrypsinized mCry3A. Lanes 1 to 3, biotinylated chymotrypsinized mCry3A incubated with BBMV alone; lanes 4 to 6, biotinylated chymotrypsinized mCry3A with 100-fold molar excess of unlabeled chymotrypsinized mCry3A. Each lane shows the results of a separate experiment.
DISCUSSION
While Cry3A has exhibited activity against a number of coleopteran pests (including the Colorado potato beetle) both in diet bioassays (3, 8, 13, 20, 23, 30) and in planta (12, 16, 44), it has not been useful for control of WCR or related species of the corn rootworm pest complex (20, 30, 51). In general, in vitro Cry3A has shown a small amount of specific binding to coleopteran brush border compared to the Cry1 binding to lepidopteran brush border (7, 33, 47, 51). Our data suggest that like Cry3A, the mCry3A protein also binds predominantly in a nonspecific fashion to the target pest brush border membrane. The prevalence of nonspecific binding of 67-kDa Cry3A has led to the conclusion that chymotrypsin-like processing of the toxin is actually necessary before specific binding can occur (7, 33).
There is good evidence that there is chymotrypsin/cathepsin G-like enzymatic activity in WCR larvae. Our data suggest that addition of the chymotrypsin/cathepsin G AAPF recognition site to the domain I loop region between α-helix 3 and α-helix 4 permits mCry3A to be processed to the same stable form as chymotrypsinized Cry3A, but in a facilitated and more rapid fashion (Fig. 3 to 5). This in vivo processing can be inhibited by inclusion of chymostatin (a specific inhibitor of alpha-, beta-, gamma-, or delta-chymotrypsin) in the diet, for both mCry3A (Fig. 4A, lanes 5 and 9) and mCry3A H115A (data not shown), supporting the hypothesis that there is chymotrypsin/cathepsin G-like enzymatic activity in WCR larvae. In addition, preliminary in vitro digests of homogenized midgut from second-instar WCR larvae recognized the colorimetric substrate N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (CalBiochem) compared to buffer or no-substrate controls, which also supported the hypothesis that there is larval WCR chymotrypsin/cathepsin G-like enzymatic activity (data not shown).
The facilitated mCry3A processing was likely due to the fact that the introduced chymotrypsin/cathepsin G site was in a more exposed position for interaction with the enzyme than the native chymotrypsin site (Fig. 1). This would allow the initial processing of mCry3A to occur at the introduced F111 (as observed for the in vitro chymotrypsin-processed mCry3A H115A protein), followed by rapid further processing at the subsequently exposed native H115 chymotrypsin cleavage site of mCry3A. Facilitated generation of the 55-kDa form of mCry3A is the most reasonable explanation for its consistent insecticidal effect with WCR larvae. We found that B. thuringiensis mCry3A exhibits significantly higher toxicity with first-instar WCR larvae whether it is unprocessed or pretreated with chymotrypsin (Table 1). Our data also show that the 55-kDa form is stable in the insect and can rapidly bind to WCR membranes when they are presented in vivo. In addition, although we found no evidence of specific binding of the 67-kDa mCry3A to intact BBMV, the 55-kDa mCry3A did bind specifically, with a 100-fold molar excess of unlabeled toxin resulting in a large reduction in bound label (Fig. 6B). No specific binding proteins could be found, however, when ligand blotting was used with either the 67- or 55-kDa mCry3A (data not shown). Therefore, overall, these data support the hypothesis that proteolytic processing is an activation step for mCry3A (and Cry3A); however, removal of all potential domain I processing sites for both toxins with concomitant loss of biological activity is needed to confirm this.
A variant of Cry3Bb1 protein which shows activity against WCR in planta has been described (54). Wild-type Cry3Bb1 had a lower level of activity with rootworms, and the previously described variant consisted of a collection of several mutations which were reported to increase the activity eightfold. Wild-type Cry3A shares only about 68% amino acid sequence identity (55) with Cry3Bb1 (formerly designated CryIIIB2) and has shown no or only sporadic activity with corn rootworms (20, 30, 51). The mutations in the Cry3Bb1 variant (54) did not include mutations similar to those that were introduced in this study to generate mCry3A. Therefore, our approach to generating a B. thuringiensis toxin active against WCR differs both in the type of mutation introduced and in the activity of the starting molecule.
Importantly, the B. thuringiensis mCry3A protein and two variants each exhibited significantly greater bioactivity than Cry3A (Table 1). This supports the hypothesis that the chymotrypsin recognition sequence is essential for the increased biological activity compared to the activity of Cry3A. Furthermore, the data suggest that neither trypsinlike processing nor the action of a rootworm cysteine protease that prefers substrates with an arginine residue in the P1 position (14) is essential for the bioactivity of mCry3A (since the two mCry3A-like variants lacked the Arg residue in this domain I loop region).
Finally, we found that the mutation in mCry3A did not adversely affect the activity against Colorado potato beetle larvae in an artificial diet bioassay, where in three separate experiments E. coli-lysed culture supernatants containing either mCry3A or Cry3A caused high mortality (corrected mortality, 100% ± 0% or 94% ± 6% [mean ± standard deviation], respectively). Therefore, the introduced mutation essentially broadened the spectrum of activity of Cry3A.
Acknowledgments
This work was supported by Syngenta Biotechnology, Inc.
We thank Jared Conville and Chris Campbell for assistance with WCR insect culture maintenance, Milan Jucovic for help with the mutagenesis strategy, and Hope Hart for helpful discussions and preliminary BBMV binding results. The support of Greg Warren is also acknowledged.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, A. J., N. D. Rawlings, and J. F. Woessner. 1998. Handbook of proteolytic enzymes. Academic Press, New York, NY.
- 3.Bauer, L. S. 1990. Response of the cottonwood leaf beetle (Coleoptera: Chrysomelidae) to Bacillus thuringiensis var. san diego. Environ. Entomol. 19:428-431. [Google Scholar]
- 4.Baum, J. A., C.-R. Chu, M. Rupar, G. R. Brown, W. P. Donovan, J. E. Huesing, O. Ilagan, T. M. Malvar, M. Pleau, M. Walters, and T. Vaughn. 2004. Binary toxins from Bacillus thuringiensis active against the western corn rootworm, Diabrotica virgifera virgifera LeConte. Appl. Environ. Microbiol. 70:4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betz, F. S., B. G. Hammond, and R. L. Fuchs. 2000. Safety and advantages of Bacillus thuringiensis-protected plants to control insect pests. Regul. Toxicol. Pharmacol. 32:156-173. [DOI] [PubMed] [Google Scholar]
- 6.Bown, D. P., H. S. Wilkinson, M. A. Jongsma, and J. A. Gatehouse. 2004. Characterisation of cysteine proteinases responsible for digestive proteolysis in guts of larval western corn rootworm (Diabrotica virgifera) by expression in the yeast Pichia pastoris. Insect Biochem. Mol. Biol. 34:305-320. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, J., D. Convents, J. Van Damme, A. Boets, J. Van Rie, and D. J. Ellar. 1997. Intramolecular proteolytic cleavage of Bacillus thuringiensis Cry3A δ-endotoxin may facilitate its coleopteran toxicity. J. Invertebr. Pathol. 70:41-49. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, J., J. Li, and D. J. Ellar. 1989. Proteolytic processing of a coleopteran-specific δ-endotoxin produced by Bacillus thuringiensis var. tenebrionis. Biochem. J. 261:99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, E., and C. Stacy. April 2006. Modified Cry3A toxins and nucleic acid sequences coding therefor. U.S. patent 7,030,295.
- 10.Christeller, J. T., B. D. Shaw, S. E. Gardiner, and J. Dymock. 1989. Partial purification and characterization of the major midgut proteases of grass grub larvae (Costelytra zealandica, Coleoptera: Scarabaeidae). Insect Biochem. 19:221-231. [Google Scholar]
- 11.Colepicolo-Neto, P., E. J. H. Bechara, C. Ferreira, and W. R. Terra. 1987. Digestive enzymes in close and distant genera of a same family: properties of midgut hydrolases from luminescent Pyrophorus divergens (Coleoptera: Elateridae) larvae. Comp. Biochem. Physiol. 87:755-759. [Google Scholar]
- 12.Coombs, J. J., D. S. Douches, W. B. Li, E. J. Grafius, and W. L. Pett. 2003. Field evaluation of natural, engineered, and combined resistance mechanisms in potato for control of colorado potato beetle. J. Am. Soc. Hortic. Sci. 128:219-224. [Google Scholar]
- 13.Donovan, W. P., J. M. Gonzalez, Jr., M. P. Gilbert, and C. Dankocsik. 1988. Isolation and characterization of EG2158, a new strain of Bacillus thuringiensis toxic to coleopteran larvae, and nucleotide sequence of the toxin gene. Mol. Gen. Genet. 214:365-372. [DOI] [PubMed] [Google Scholar]
- 14.Fabrick, J., C. Behnke, T. Czapla, K. Bala, A. G. Rao, K. J. Kramer, and G. R. Reeck. 2002. Effects of a potato cysteine proteinase inhibitor on midgut proteolytic enzyme activity and growth of the southern corn rootworm, Diabrotica undecempunctata howardi (Coleoptera: Chrysomelidae). Insect Biochem. Mol. Biol. 32:405-415. [DOI] [PubMed] [Google Scholar]
- 15.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, England.
- 16.Génissel, A., J.-C. Leplé, N. Millet, S. Augustin, L. Jouanin, and G. Pilate. 2003. High tolerance against Chrsomela tremulae of transgenic poplar plants expressing a synthetic cry3Aa gene from Bacillus thuringiensis ssp. tenebrionis. Mol. Breed. 11:103-110. [Google Scholar]
- 17.Gillikin, J. W., S. Bevilacqua, and J. S. Graham. 1992. Partial characterization of digestive tract proteinases from western corn rootworm larvae, Diabrotica virgifera. Arch. Insect Biochem. Physiol. 19:285-298. [Google Scholar]
- 18.Godfrey, L. D., L. J. Meinke, and R. J. Wright. 1993. Field corn vegetative and reproductive biomass accumulation: response to western corn rootworm (Coleoptera: Chrysomelidae) root injury. J. Econ. Entomol. 86:1557-1573. [Google Scholar]
- 19.Hernández, C. A., M. Pujol, J. Alfonso-Rubí, R. Armas, Y. Coll, M. Pérez, A. González, M. Ruiz, P. Castañera, and F. Ortego. 2003. Proteolytic gut activities in the rice water weevil, Lissorhoptrus brevirostris Suffrian (Coleoptera: Curculionidae). Arch. Insect Biochem. Physiol. 53:19-29. [DOI] [PubMed] [Google Scholar]
- 20.Herrnstadt, C., G. G. Soares, E. R. Wilcox, and D. L. Edwards. 1986. A new strain of Bacillus thuringiensis with activity against coleopteran insects. Bio/Technology 4:305-308. [Google Scholar]
- 21.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Koller, C. N., L. S. Bauer, and R. M. Hollingworth. 1992. Characterization of the pH-mediated solubility of Bacillus thuringiensis var. san diego native δ-endotoxin crystals. Biochem. Biophys. Res. Commun. 184:692-699. [DOI] [PubMed] [Google Scholar]
- 23.Krieg, V. A., A. M. Huger, G. A. Langenbruch, and W. Schnetter. 1984. New results on Bacillus thuringiensis var. tenebrionis with special regard to its effect on the Colorado beetle. Anz. Schaedlkd. Pflanzenschutz Umweltschutz 57:145-150. [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. K., R. E. Milne, A. Z. Ge, and D. H. Dean. 1992. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis δ-endotoxin. J. Biol. Chem. 267:3115-3121. [PubMed] [Google Scholar]
- 26.Li, J., J. Caroll, and D. J. Ellar. 1991. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature 353:815-821. [DOI] [PubMed] [Google Scholar]
- 27.Lightwood, D. J., D. J. Ellar, and P. Jarrett. 2000. Role of proteolysis in determining potency of Bacillus thuringiensis Cry1Ac δ-endotoxin. Appl. Environ. Microbiol. 66:5174-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loseva, O., M. Ibrahim, M. Candas, C. N. Koller, L. S. Bauer, and L. A. Bulla, Jr. 2002. Changes in protease activity and Cry3Aa toxin binding in the Colorado potato beetle: implications for insect resistance to Bacillus thuringiensis toxins. Insect Biochem. Mol. Biol. 32:567-577. [DOI] [PubMed] [Google Scholar]
- 29.Macaluso, A., and A. M. Mettus. 1991. Efficient transformation of Bacillus thuringiensis requires nonmethylated plasmid DNA. J. Bacteriol. 173:1353-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacIntosh, S. C., T. B. Stone, S. R. Sims, P. L. Hunst, J. T. Greenplate, P. G. Marrone, F. J. Perlak, D. A. Fischoff, and R. L. Fuchs. 1990. Specificity and efficacy of purified Bacillus thuringiensis proteins against agronomically important insects. J. Invertebr. Pathol. 56:258-266. [DOI] [PubMed] [Google Scholar]
- 31.Marrone, P. G., F. D. Ferri, T. R. Mosley, and L. J. Meinke. 1985. Improvements in laboratory rearing of the southern corn rootworm, Diabrotica undecimpuncta howardi Barber (Coleoptera: Chrysomelidae), on an artificial diet and corn. J. Econ. Entomol. 78:290-293. [Google Scholar]
- 32.Martin, M. M., and J. S. Martin. 1984. Surfactants: their role in preventing the precipitation of proteins by tannins in insect guts. Oecologia 61:342-345. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Ramirez, A. C., and M. D. Real. 1996. Proteolytic processing of Bacillus thuringiensis CryIIIA toxin and specific binding to brush-border membrane vesicles of Leptinotarsa decemlineata (Colorado potato beetle). Pestic. Biochem. Physiol. 54:115-122. [Google Scholar]
- 34.McPherson, S. A., F. J. Perlak, R. L. Fuchs, P. G. Marrone, P. B. Lavrik, and D. Fischoff. 1988. Characterization of the coleopteran-specific protein gene of Bacillus thuringiensis var. tenebrionis. Bio/Technology 6:61-66. [Google Scholar]
- 35.Metcalf, R. L. 1986. Foreword, p. vii-xv. In J. L. Krysan and T. A. Miller (ed.), Methods for the study of the pest Diabrotica. Springer-Verlag, New York, NY.
- 36.Moellenbeck, D. J., M. L. Peters, J. W. Bing, J. R. Rouse, L. S. Higgins, L. Sims, T. Nevshemal, L. Marshall, R. T. Ellis, P. G. Bystrak, B. A. Lang, J. L. Stewart, K. Kouba, V. Sondag, V. Gustafson, K. Nour, D. Xu, J. Swenson, J. Zhang, T. Czapla, G. Schwab, S. Jayne, B. A. Stockhoff, K. Narva, H. E. Schnepf, S. J. Stelman, C. Poutre, M. Koziel, and N. Duck. 2001. Insecticidal proteins from Bacillus thuringiensis protect corn from corn rootworms. Nat. Biotechnol. 19:668-672. [DOI] [PubMed] [Google Scholar]
- 37.Mullis, K. B., and F. A. Faloona. 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155:335-350. [DOI] [PubMed] [Google Scholar]
- 38.Murdock, L. L., G. Brookhart, P. E. Dunn, D. E. Foard, S. Kelley, L. Kitch, R. E. Shade, R. H. Shukle, and J. L. Wolfson. 1987. Cysteine digestive proteinases in Coleoptera. Comp. Biochem. Physiol. 87B:783-787. [Google Scholar]
- 39.Murray, E. E., J. Lotzer, and M. Eberle. 1989. Codon usage in plant genes. Nucleic Acids Res. 17:477-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novillo, C., P. Castañera, and F. Ortego. 1997. Characterization and distribution of chymotrypsin-like and other digestive proteases in Colorado potato beetle larvae. Arch. Insect Biochem. Physiol. 36:181-201. [Google Scholar]
- 41.Oehme, F. W., and J. A. Pickerell. 2003. Genetically engineered corn rootworm resistance: potential for reduction of human health effects from pesticides. Biomed. Environ. Sci. 16:17-28. [PubMed] [Google Scholar]
- 42.Oppert, B. 1999. Protease interactions with Bacillus thuringiensis insecticidal toxins. Arch. Insect Biochem. Physiol. 42:1-12. [DOI] [PubMed] [Google Scholar]
- 43.Oppert, B., K. Hartzer, and M. Zuercher. 2002. Digestive proteinases in Lasioderma serricorne (Coleoptera:Anobiidae). Bull. Entomol. Res. 92:331-336. [DOI] [PubMed] [Google Scholar]
- 44.Perlak, F. J., T. B. Stone, Y. M. Muskopf, L. J. Peterson, G. B. Parker, S. L. McPherson, J. Wyman, S. Love, G. Reed, D. Biever, and D. A. Fischoff. 1993. Genetically improved potatoes: protection from Colorado potato beetles. Plant Mol. Biol. 22:313-321. [DOI] [PubMed] [Google Scholar]
- 45.Purcell, J. P., J. T. Greenplate, and R. D. Sammons. 1992. Examination of midgut luminal proteinase activities in six economically important insects. Insect Biochem. Mol. Biol. 22:41-47. [Google Scholar]
- 46.Rausell, C., I. García-Robles, J. Sánchez, C. Muñoz-Garay, A. C. Martínez-Ramírez, M. D. Real, and A. Bravo. 2004. Role of toxin activation on binding and pore formation activity of the Bacillus thuringiensis Cry3 toxins in membranes of Leptinotarsa decemlineata (Say). Biochim. Biophys. Acta 1660:99-105. [DOI] [PubMed] [Google Scholar]
- 47.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Ziegler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnepf, H. E., H. C. Wong, and H. R. Whiteley. 1985. The amino acid sequence of a crystal protein from Bacillus thuringiensis deduced from the DNA base sequence. J. Biol. Chem. 260:6264-6272. [PubMed] [Google Scholar]
- 49.Sekar, V., D. V. Thompson, M. J. Maroney, R. G. Bookland, and M. J. Adang. 1987. Molecular cloning and characterization of the insecticidal crystal protein gene of Bacillus thuringiensis var. tenebrionis. Proc. Natl. Acad. Sci. USA 84:7036-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelton, A. M., J.-Z. Zhao, and R. T. Rousch. 2002. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annu. Rev. Entomol. 47:845-881. [DOI] [PubMed] [Google Scholar]
- 51.Slaney, A. C., H. L. Robbins, and L. English. 1992. Mode of action of Bacillus thuringiensis toxin CryIIIA: an analysis of toxicity in Leptinotarsa decemlineata (Say) and Diabrotica undecempunctata Howardi Barber. Insect Biochem. Mol. Biol. 22:9-18. [Google Scholar]
- 52.Tanaka, T., Y. Minematsu, C. F. Reilly, J. Travis, and J. C. Powers. 1985. Human leukocyte cathepsin G. Subsite mapping with 4-nitroanilides, chemical modification, and effect of possible cofactors. Biochemistry 24:2040-2047. [DOI] [PubMed] [Google Scholar]
- 53.Vaje, S., D. Mossakowski, and D. Gabel. 1984. Temporal, intra- and interspecific variation of proteolytic enzymes in carabid-beetles. Insect Biochem. 14:313-320. [Google Scholar]
- 54.Vaughn, T., T. Cavato, G. Brar, T. Coombe, T. DeGooyer, S. Ford, M. Groth, A. Howe, S. Johnson, K. Kolacz, C. Pilcher, J. Purcell, C. Romano, L. English, and J. Pershing. 2005. A method of controlling corn rootworm feeding using a Bacillus thuringiensis protein expressed in transgenic maize. Crop Sci. 45:931-938. [Google Scholar]
- 55.Von Tersch, M. A., S. L. Slatin, C. A. Kulesza, and L. H. English. 1994. Membrane-permeabilizing activities of Bacillus thuringiensis coleopteran-active toxin CryIIIB2 and CryIIIB2 domain I peptide. Appl. Environ. Microbiol. 60:3711-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walters, F. S., C. A. Kulesza, A. T. Phillips, and L. H. English. 1994. A stable oligomer of Bacillus thuringiensis delta-endotoxin, Cry IIIA. Insect Biochem. Mol. Biol. 24:963-968. [Google Scholar]
- 57.Wolfersberger, M., P. Luethy, A. Maurer, P. Parenti, F. V. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation of brush border membrane vesicles (BBMV) from larval lepidopteran midgut. Comp. Biochem. Physiol. 86A:301-308. [Google Scholar]
- 58.Zhu, Y.-C., and J. E. Baker. 2000. Molecular cloning and characterization of a midgut chymotrypsin-like enzyme from the lesser grain borer, Rhyzopertha dominica. Arch. Insect Biochem. Physiol. 43:173-184. [DOI] [PubMed] [Google Scholar]