Abstract
Between 2001 and 2006, the incidence of human Campylobacter infections decreased by 10 and 27% in Scotland and the Grampian region of Scotland, respectively. Contemporaneous collection and analyses of human and retail-chicken isolates from Grampian were carried out over a 10-week period in 2001 and again in 2006 in order to determine whether the fall in the incidence of human infections was related to the retail-chicken exposure route. Rates of carriage of Campylobacter on chicken carcasses from retail outlets in Grampian in 2001 and 2006 were estimated. Chicken-derived Campylobacter isolates from 2001 (n = 84) and 2006 (n = 105) and human-derived isolates from patients with clinical cases of infection in 2001 (n = 172) and 2006 (n = 119) were typed by multilocus sequence typing. We found no evidence for statistically significant changes in prevalence and counts per carcass. We found by rarefaction that although the degree of diversity in humans tended to be higher than that in chickens, these differences were not significant. The genetic distance between chicken and human isolates from 2001 according to sequence type, clonal complex (CC), or allele composition was not significant, whereas the distances between 2006 isolates at the CC and allele levels were significant. This difference was attributable to a lower proportion of CC-21's being found in retail-chicken isolates from 2006 than in chicken isolates from 2001. We conclude that human exposure to Campylobacter via retail chicken is important and that changes in the population structure of campylobacters in this reservoir need to be taken into account in investigating human infection.
The microbial pathogens of the genus Campylobacter are the single largest cause of sporadic bacterial gastrointestinal infection worldwide (6). The majority of Campylobacter infections are due to Campylobacter jejuni (80 to 85%), with the remainder due predominantly to C. coli. In Scotland, the incidence of Campylobacter infection in 2005 was 90.2 cases/100,000 people (29), while in the United States it was 12.7 cases/100,000 people (9). However, the real disease burden is substantially higher since it has been estimated that only 1 in 7 cases in the United Kingdom (64) and 1 in 38 in the United States (36) are reported.
The incidence of human campylobacteriosis in the United Kingdom increased steadily from the first reports in the 1970s (7) until it peaked in 2000 (26). There was a reduction in the occurrence of infection post-2000 (39), and between 2001 and 2006, the incidence rate dropped by 10.1% across Scotland and 27.1% in the Grampian region of Scotland. Reasons for this decline in infection are unknown. A 30% decrease in the United States between 1998 and 2005 has also been reported (8), and it has been suggested that this reduction may be due to the introduction of the Hazard Analysis and Critical Control Points program in 1997 (18). An important reservoir of Campylobacter is chicken (17). The 50% infective dose for chickens is 103 CFU (10), and at slaughter, approximately 60% of United Kingdom broiler flocks are positive for Campylobacter (3). Due to the coprophagic behavior of chickens, fecal shedding is a significant factor in the spread of Campylobacter throughout a large flock (45). In experimentally challenged birds, the bacterial load has been shown to reach 109 CFU/g of cecal content (63); however, in broilers, numbers in the range of 105 to 109 CFU/g of cecal content have been commonly observed (4). It has been reported that 20% of birds at retail sale harbor campylobacters at >105 CFU/carcass (27). A recent study showed a prevalence of 87% on retail chicken breast, with a mean count of 1.9 ×103 CFU/fillet (30). The 50% infective dose for humans is approximately 900 cells (57); thus, the potential for human exposure to the bacteria through cross contamination, poor hygiene, or undercooking in the kitchen is high (12). From April to June 2001, the United Kingdom Food Standards Agency (FSA) conducted a survey of retail chicken (20) and found that 52 and 89% of chickens in England and Scotland, respectively, were contaminated with this pathogen. Evidence to support the link between chicken consumption and human infection was demonstrated in the Belgian dioxin crisis of 1999 (61), when chicken feed containing high levels of dioxins resulted in the withdrawal of chicken and eggs from retail sale. The outcome was a 40% decrease in human Campylobacter infection until the chicken ban was lifted, when incidence rates returned to former levels. During the ban, only Belgian poultry was removed from sale but imported poultry was still available. Hence, the 40% of infections attributable to poultry is likely to be an underestimate. Further, Vellinga and Van Loock reported that approximately 20% of cases were associated with non-food-borne sources (61). In the United States, food-borne disease information shows that 80% of Campylobacter infections are considered to be food borne (36).
The consumption of chicken has been demonstrated to be a major risk factor for acquiring campylobacteriosis (47, 56), as has consumption of raw or rare chicken (23, 25), although the deliberate consumption of raw or rare chicken does not occur in the United Kingdom. Further, chicken products such as liver (53) and chicken consumed in a restaurant (51) have also been implicated as significant risk factors. Interestingly, another study has shown that the handling and consumption of chicken in the home may be protective (2), as these factors are significantly associated with a decrease in the risk of becoming ill, possibly due to an acquired immunity. Since most cases of Campylobacter infection are sporadic (49), it is difficult to trace infection pathways. There is well-established diversity within C. jejuni and C. coli strains that has been displayed at the genotypic and phenotypic levels (62). Scientific agreement on suitable typing methods for Campylobacter exists, to an extent, but such methods are not as universally adopted as, for example, methods of serotyping and phage typing for Salmonella species. Multilocus sequence typing (MLST) (31) has proven to be a suitable tool for the characterization of a number of bacterial species, such as Streptococcus, Yersinia, and Staphylococcus spp. (1, 16, 58), and is increasingly used for C. jejuni (13, 14, 34). MLST enables the estimation of the genetic diversity and the structure of a bacterial population by using the DNA sequences of seven housekeeping genes. The evolutionary conservation of these genes makes them convenient markers for identifying variation that is accumulating slowly in a population.
In this study, we analyzed four collections of isolates: those collected from retail chicken in 2001 and 2006 and those collected from patients diagnosed with campylobacteriosis in 2001 and 2006. The aim was to determine if a change in rates of exposure to contaminated chicken meat was associated with the decline of human campylobacteriosis between 2001 and 2006. Firstly, we investigated whether the prevalence and numbers of Campylobacter CFU in retail chicken changed over this 5-year period. Secondly, we investigated the diversity of strains within and among these collections to establish changes over time. Finally, we calculated genetic distances between the collections at the levels of the MLST sequence type (ST), the clonal complex (CC), and the allele profile and established whether significant changes occurred during the 5-year period.
MATERIALS AND METHODS
Data sets.
The four study groups were retail-chicken isolates from 2001 (n = 84) and 2006 (n = 105) and clinical isolates from Grampian from the same years (n, 172 and 119, respectively).
Sampling of retail chicken.
Retail chicken was sampled according to the protocol of the 2001 FSA study of chicken for retail sale (20) at the same sites around Grampian as those in the FSA study. In that study from 2001, 747 samples were taken from around Scotland, 84 of which were from the Grampian area. Briefly, in 2006, chicken meat (114 samples) was purchased from the same 10 supermarket outlets and 16 butcher stores in the Grampian area over a 10-week period. Chicken samples included breasts, drumsticks, wings, thighs, legs, and whole birds (sizes small, medium, and large). A small proportion of organic and free-range chicken portions were selected in order to represent the comparatively small market share these products occupy. The ratio of 80% fresh to 20% frozen meat was maintained, in line with the ratio in the 2001 study. Multiple samples were taken from the same retailer at the same time, depending on the size of the outlet. More chicken was purchased from larger supermarkets, while fewer samples were obtained from butchers. Again, this process was an imitation of the procedures of the 2001 study. For two chicken samples from 2006, more than one isolate was used due to the detection of phenotypically different colonies, suggesting multiple-strain carriage in the meat.
Isolation of strains.
To each 500-g portion of chicken, 300 ml of enrichment broth was added. Enrichment broth contained, per liter, 950 ml of nutrient broth (DM180D; MAST Group Ltd., United Kingdom), 50 ml of defibrinated horse blood (E & O Laboratories Ltd., United Kingdom), 10 ml of Campylobacter growth supplement (containing 250 mg of iron II sulfite per liter, 250 mg of sodium metabisulfite per liter, and 250 mg of sodium pyruvate per liter), and the following Campylobacter selective supplements: polymyxin B (2.5 IU), rifampin (5 μg/ml), amphotericin (2 μg/ml), cefoperazone (15 μg/ml), and trimethoprim (10 μg/ml). Chicken samples were incubated at room temperature for 1 h, after which the broth was decanted. From this wash, 100 μl was plated onto modified charcoal cefoperazone deoxycolate agar (mCCDA [CM0739; Oxoid Ltd, United Kingdom]), and then two further decimal dilutions (two- and threefold) were created and also plated onto mCCDA for colony enumeration. Plates were incubated at 37°C microaerophilically (in an atmosphere of 2% H2, 5% CO2, 5% O2, and 88% N2) for 48 h before colonies were enumerated. Following the direct plating, the remaining wash was further incubated under the same conditions, after which 100 μl of this enrichment broth from each sample was plated onto mCCDA and further incubated microaerophilically at 37°C for 48 h for sensitive detection.
Campylobacter isolates from Grampian from the 2001 retail chicken survey (20) were obtained from FSA Scotland. Clinical isolates from 2001 and 2006 were obtained from NHS Grampian from storage at −80°C. The isolates had been obtained from samples submitted by patients with campylobacteriosis in Grampian. These patients were all infected in the Grampian area of Scotland, and the isolates were stored by the local hospital. Following thawing at room temperature, isolates were plated onto mCCDA and incubated microaerophilically for 48 h.
Genus and species confirmation.
Genus confirmation of Campylobacter spp. from all four data sets was achieved by a latex agglutination test. Bacterial DNA from latex test-positive strains was prepared via a DNA release method using Chelex resin (catalogue no. 142-1253; Bio-Rad, United States) and identified to the species level by a multiplex PCR speciation assay as previously described (28).
MLST.
MLST PCR and sequencing reactions were carried out at the University of Aberdeen by the methods of Dingle et al. (15). Sequencing was carried out at the University of Oxford. Fragments of the seven housekeeping genes (aspA, encoding aspartase A; glnA, encoding glutamine synthetase; gltA, encoding citrate synthase; glyA, encoding serine hydroxymethyl transferase; pgm, encoding phosphoglucomutase; tkt, encoding transketolase; and uncA, encoding the ATP synthase subunit) were amplified by PCR. Primers for five of the loci (aspA, glnA, gltA, glyA, and pgm) were used as previously described (40). Primers for the loci tkt and uncA were tktJDF (GCWGATATTTTAASKGTTTTAAGTTATC), tktJDR (TGACTKCCTTCAAGCTCTC), uncAJDF (TGTTGCMATWGGWCAAAAGC), and uncAJDR (CTTTGTCCRCGTTCAAGTTG), whose sequences were derived from sequences obtained from amplification with primer pairs previously described (40). The amplicon sizes were 653 bp (tkt) and 660 bp (uncA). Sequences were assembled from the chromatograms by using the Staden software package (55), and allele profiles and STs were assigned using the public Campylobacter MLST profile database (http://www.pubmlst.org). The seven sequences are each assigned an allele number, the combination of which yields an ST, and related STs can be clustered into appropriate CCs.
Statistical analysis.
Campylobacter prevalences in retail chicken in 2001 and 2006 were compared by the chi-square test using formulas in Microsoft Excel. A bootstrap method using Poptools enabled the determination of 95th percentiles from the counts obtained from both chicken data sets. This method involves sampling with replacement the original count data and determining the average count. This process was repeated 10,000 times using Poptools, and from the resulting data, a global average as well as 95th percentiles was obtained.
Data for STs, CCs, and alleles were compared using Fisher's exact test (http://home.clara.net/sisa/twoby2.htm) to determine whether particular genotypes were more common among clinical isolates than among isolates from chicken hosts. Briefly, common types were identified and odds ratios (OR) were calculated (5). OR were defined as the odds of a particular CC, ST, or allele's occurring in a clinical isolate compared to the odds of that same type's being found in a chicken isolate. A larger value indicated a higher proportion in humans. For multiple comparisons, P values obtained from Fisher's exact test were modified by sequential Bonferroni correction (52) in order to determine statistical significance. As there were so many subtypes, only those occurring more than 10 times were included. Further, significant differences could not be found with fewer than 10 isolates.
MLST clonal groups were examined using the eBURST package (http://eburst.mlst.net/). This algorithm assigns all related STs to CCs and predicts the theoretical founder ST.
Rarefaction analysis was used to estimate maximum levels of diversity in each of the four collections of strains and to compare all chicken strains with all clinical strains and all strains from 2001 to all strains from 2006. Rarefaction was performed using the method described previously (24) and incorporated the MLST data at the levels of CC, ST, and allele. To compare the degrees of diversity of strains in any two collections (e.g., chicken isolates from 2001 and chicken isolates from 2006), the following procedure was carried out. Rarefaction was performed for both collections, and the difference in diversity was determined by subtraction. Testing for significance was carried out using a randomization test (32). Briefly, the data from the two collections were randomized in Excel using Poptools (http://www.cse.csiro.au/poptools/), the rarefaction analysis was then rerun, and the difference in diversity was calculated. This process was then repeated 10,000 times using the Monte Carlo Excel add-in @RISK (Palisade Ivybridge, United Kingdom). The posterior distribution of diversity differences obtained was then compared with the diversity difference calculated for the two original collections to obtain the level of significance (P value).
Standardized genetic distances between the 2001 chicken, 2006 chicken, 2001 clinical, and 2006 clinical Campylobacter populations were determined using the method of Nei (42) and applied to genetic locus data by the method of Manly (33). The level of significance was obtained using randomization tests (10,000 iterations) by the same technique described for the rarefaction analysis above. An analysis of molecular variance was carried out in GenIEx, and fixation index statistics were obtained (37) using Arlequin version 2.000 (http://anthropologie.unige.ch/arlequin/).
RESULTS
Prevalence and concentration of Campylobacter in chicken in Grampian.
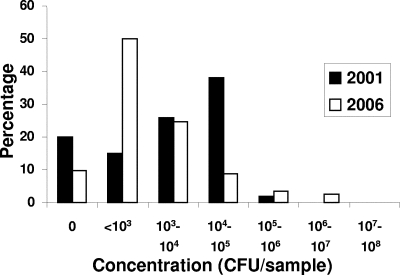
In 2006, 103 (90.4%) of 114 chicken samples contained thermotolerant Campylobacter. This prevalence was higher than that observed in 2001, 68 (80.9%) of 84, by the FSA survey (20). The difference, however, was found by the chi-square test not to be significant (P = 0.06). The average concentration of Campylobacter in chicken meat as obtained by the bootstrap method was 5.3 × 104 CFU/sample in 2006 (2.5 percentile, 1.5 × 104 CFU, and 97.5 percentile, 1.1 × 105 CFU), compared to 1.6 × 104 CFU/sample in 2001 (2.5 percentile, 1.0 × 104 CFU, and 97.5 percentile, 2.3 × 104 CFU) (Fig. 1). In two particular meat samples, phenotypically different colony morphologies were observed. The corresponding colonies were treated as distinct isolates and screened by MLST individually.
FIG. 1.
Concentrations of Campylobacter in retail chicken during 2001 and 2006.
Species identification of Campylobacter.
It was found that 92.7% of isolates were C. jejuni and 7.3% were C. coli. In each of the four data sets, the proportions of C. jejuni and C. coli were 94.1 and 5.9% (2001 chicken isolates); 95.9 and 4.1.% (2001 clinical isolates); 85.7 and 14.3% (2006 chicken isolates); and 93.3 and 6.7% (2006 clinical isolates), respectively.
MLST analysis. (i) STs.
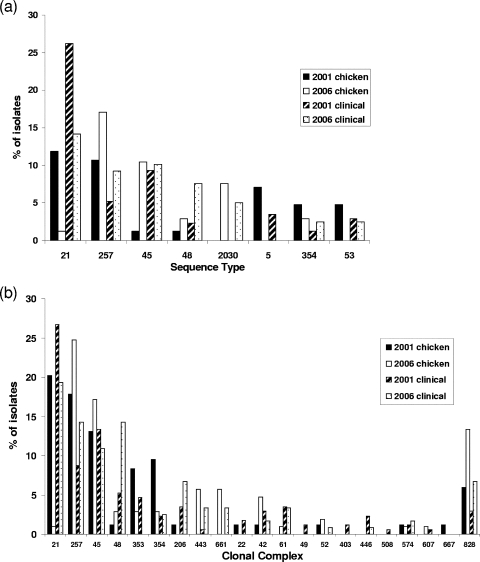
A total of 480 isolates were analyzed by MLST, yielding 131 STs, 35 of which were novel. In each data set, certain common STs and CCs appeared to encompass the majority of isolates (Fig. 2). In the 2001 chicken collection, ST-21 (11.9%) and ST-257 (10.7%) were the most common (Fig. 2a). In 2006 chicken isolates, the distribution of STs was different, with ST-21 appearing less frequently, ST-45, ST-354, ST-48, and ST-2030 appearing more commonly, and ST-257 being most prevalent (17.1%). Clinical isolates from 2001 were dominated by ST-21 (26.2% of the total). The common STs found among human isolates from 2006 displayed a distribution pattern consistent with that among 2001 clinical isolates, with ST-21 again being the most prevalent (14.2% of the total).
FIG. 2.
Common Campylobacter STs (those represented by >10 isolates in each data set) (a) and all defined CCs by host and year (b).
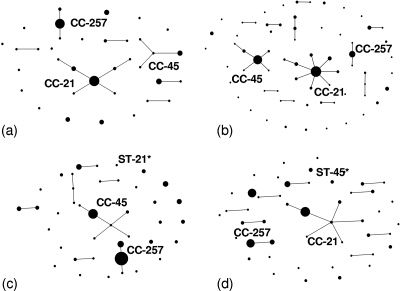
The eBURST population snapshots (Fig. 3) of the four data sets illustrate the diverse nature of Campylobacter (i.e., very few clusters appear). Aside from the major CCs, the degree of clustering was minimal, with 14.4% of STs being singletons (occurring only once). Throughout the four data sets, 6.9% of STs were unique to chicken and 45.8% were found only in humans. In the 2001 sets, 5.1% of STs were unique to chicken hosts and 13.7% were unique to humans. In the 2006 sets, 8% of STs were found only in chickens and 8.5% were found only in humans. Due to the diversity of types found, only OR for types occurring in numbers of 10 or higher are presented (Table 1).
FIG. 3.
eBURST diagrams showing population snapshots of each of the four collections of isolates: 2001 chicken isolates (a), 2001 clinical isolates (b), 2006 chicken isolates (c), and 2006 clinical isolates (d). The size of each circle is representative of the number of isolates with that particular ST. Asterisks indicate that the STs displayed are the only STs from their respective complexes.
TABLE 1.
Comparisons of common STs and CCs in the different data sets and ORa
| Comparison | ST | OR | 95% CI | P value | CC | OR | 95% CI | P value |
|---|---|---|---|---|---|---|---|---|
| All clinical and all chicken isolates | 21 | 2.14 | 1.06-4.34 | 0.27 | 21 | 2.73* | 1.61-4.60 | 0.0008 |
| 45 | 1.57 | 0.78-3.17 | 1.76 | 45 | 0.83 | 0.49-1.40 | 10.40 | |
| 48 | 2.16 | 0.70-3.17 | 1.73 | 48 | 3.76* | 1.42-10.0 | 0.03 | |
| 61 | 6.69 | 0.84-52.70 | 0.40 | 257 | 0.42* | 0.26-0.70 | 0.005 | |
| 257 | 0.44 | 0.24-0.82 | 0.08 | |||||
| 2001 clinical and 2001 chicken isolates | 5 | 0.47 | 0.15-1.50 | 2.11 | 21 | 1.35 | 0.74-2.50 | 2.60 |
| 21 | 1.09 | 0.49-2.41 | 6.59 | 45 | 1.13 | 0.53-2.40 | 5.98 | |
| 45 | 8.51 | 1.11-65.32 | 0.12 | 257 | 0.40 | 0.19-0.80 | 0.17 | |
| 257 | 0.46 | 0.18-1.21 | 1.17 | 353 | 0.52 | 0.19-1.40 | 12.22 | |
| 2006 clinical and 2006 chicken isolates | 21 | 11.66* | 3.96-34.35 | 0.03 | 21 | 24.92* | 3.30-188 | 0.00003 |
| 45 | 0.96 | 0.40-6.19 | 8.19 | 45 | 0.59 | 0.28-1.30 | 1.56 | |
| 48 | 2.78 | 0.73-10.56 | 1.30 | 257 | 0.48 | 0.25-0.90 | 0.26 | |
| 257 | 0.49 | 0.22-1.10 | 0.78 | 828 | 0.43 | 0.18-1.10 | 0.65 |
OR are defined as the ratio of the odds of having a proportion of one type in the clinical host to the odds of that type's occurring in the chicken host. 95% CI, 95% confidence interval; *, statistically significant using Fisher's exact test with sequential Bonferroni correction for multiple comparisons.
Comparing common chicken and clinical isolates from 2001 and 2006 combined (Table 1) showed that none of the STs were found to be overrepresented in either host by Fisher's exact test. Comparing 2001 clinical and chicken isolates also revealed no statistical evidence of a particular type's being either over- or underrepresented in a particular host. In the 2006 sets, however, ST-21 was found more commonly in the human host (OR = 11.66), being almost absent from retail chicken.
(ii) CCs.
STs were grouped into 21 defined CCs (Fig. 2b and Fig. 3); however, 28 STs could not be grouped into any previously defined clonal groups. The most frequently occurring complexes were CC-21, CC-45, and CC-257, which were present in all four data sets. The 2001 chicken data set showed CC-21 to be the most common clone, accounting for 20.2%. Similarly, this clone was common among the 2001 clinical isolates (26.7%) and the 2006 clinical isolates (19.3%). CC-45 and CC-257 also had similar distributions, appearing frequently in all four data sets. The groups CC-353 and CC-354 were noticeably common among 2001 chicken isolates (8.3 and 9.5%, respectively). Finally, the C. coli complex CC-828 was represented in both hosts, with its highest frequency among 2006 chicken isolates (13.3%).
Comparing common CCs among all clinical and chicken isolates (Table 1) showed that CC-21 (OR = 2.73) and CC-48 (OR = 3.76) were significantly more prevalent in clinical strains than chicken strains by Fisher's exact test but that CC-257 (OR = 0.42) was significantly more prevalent in chickens. Comparing 2001 clinical and chicken isolates showed that none of the CCs were overrepresented in either host. Analyzing 2006 data showed that CC-21 was more prevalent in humans (OR = 24.92), with CC-21 being virtually absent from the chicken samples tested.
Rarefaction analysis.
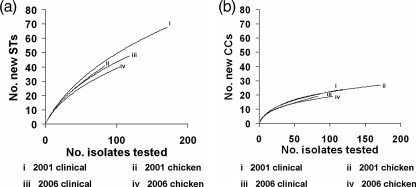
Figure 4 shows rarefaction curves for STs and CCs. The curves with the greatest gradients (2001 chicken and 2001 clinical isolates) correspond to the greatest degrees of diversity. Hence, Fig. 4 shows that the clinical isolates were on average more diverse than the chicken isolates from both 2001 and 2006 based on STs and CCs. However, these differences were found not to be statistically significant (P > 0.05). This analysis was also performed using allele profiles (data not shown), and again no significant differences were found.
FIG. 4.
Rarefaction curves showing diversity by ST (a) and CC (b) for each of the four data sets.
Genetic distances.
The genetic distance between each of the four populations was calculated at three levels, STs, CCs, and allele profiles (Table 2), by the method of Nei (42). Results from the analysis of molecular variance were also obtained and are available upon request. No significant differences between clinical isolates from 2001 and 2006 were found when these isolates were compared at all three levels. However, significant differences in the retail-chicken isolates at all three levels were found, suggesting that there had been a significant change in the genetic population structure in this host. Further, in comparing the difference between clinical and chicken isolates from 2001 and that between clinical and chicken isolates from 2006, significant differences were found at the CC and allele levels, suggesting that the genetic distance between clinical and chicken samples had increased between 2001 and 2006.
TABLE 2.
Genetic distances calculated at the ST, CC, and allelic profile levels
| Comparison | Distance at level ofa:
|
||
|---|---|---|---|
| ST | CC | Allele profile | |
| 2001 chicken isolates and 2006 chicken isolates | 0.904* | 0.379* | 0.303* |
| 2001 chicken isolates and 2001 clinical isolates | 0.372 | 0.111 | 0.094 |
| 2001 chicken isolates and 2006 clinical isolates | 0.581 | 0.181 | 0.109 |
| 2006 chicken isolates and 2001 clinical isolates | 0.792* | 0.720* | 0.560* |
| 2001 clinical isolates and 2006 clinical isolates | 0.238 | 0.129 | 0.050 |
| 2006 chicken isolates and 2006 clinical isolates | 0.383* | 0.429* | 0.407* |
| (2006 chicken and clinical isolates) and (2001 chicken and clinical isolates) | 0.010 | 0.318* | 0.313* |
DISCUSSION
Despite the reduction in human infection between 2001 and 2006, Campylobacter prevalence and concentrations in retail chicken have not decreased. We found that the prevalence of thermotolerant Campylobacter in retail chicken in the Grampian area of Scotland had increased, though not significantly so, from 2001 (20) to 2006. The bootstrap confidence intervals for the chicken counts obtained in 2001 and 2006 overlap, indicating that the overall load of Campylobacter on poultry had not changed. The elevated counts in 2006 were due predominantly to elevated levels in three samples (Fig. 1), and consequentially, this apparent increase in concentration should be taken cautiously, as shown for Escherichia coli O157 concentrations shed by cattle (46). These data suggest that it is unlikely that the fall in human campylobacteriosis can be explained solely by a change in the concentration and prevalence of the pathogen in retail chicken.
The reduction in ST-21 in retail chicken may be a contributing factor in the decline of human infection, as this ST has frequently been associated with human campylobacteriosis. Further, it is possible that the subtypes that were more prevalent in 2006 retail chicken may be less pathogenic to humans. The relationship between the MLST genotype and the strain pathogenicity is, however, controversial (59). MLST provides an insight into population structure by using the core genes. However, the robustness of the technique for the attribution of disease to particular sources can be backed up using antigenic genes such as the flagellin gene (flaA) or genes for major outer membrane proteins in addition to the seven housekeeping genes. The striking reduction in not only ST-21 but also its respective complex (CC-21) has been observed in other studies. A previous chicken broiler study in 2000 (University of Aberdeen, unpublished data) found high levels of CC-21, whereas an ongoing study shows a subsequent decline in the numbers of members of this CC (University of Aberdeen, unpublished data). These results provide evidence to suggest that the decline observed in the present study was real and not a sampling artifact.
ST-45 was found frequently in both human and chicken isolates but was underrepresented in 2001 chicken isolates. CC-45, however, was more common, comprising 6 of 11 individual STs, again emphasizing the degree of diversity of subtypes, but these observations were found not to be significant. It has been shown before that strains in this complex are predominant in farm chickens (13). Other than CC-21, CC-257 was identified frequently and was more common in chickens than in humans. Conversely, CC-48 was more common in humans.
It was found that the genetic distance between 2001 chicken and 2001 human isolates was not significant at the levels of STs, CCs, and allele profiles but that in 2006 the distance was significant at the levels of CCs and allele profiles. This difference in 2006 is attributable to the reduction of CC-21 in retail chicken. When the genetic analysis was recalculated without CC-21 representation, the genetic distance based on CCs and allele profiles was reduced considerably and was shown to be reduced significantly (data not shown). This finding suggests that there has been an increase in genetic distance between the chicken and clinical isolates at the levels of CCs and alleles. These results support the hypothesis that chickens may now be less important as a source of human infection than in 2001, potentially explaining the fall in numbers of human cases.
Among all of the 480 isolates examined here, 131 STs were observed. The rarefaction analysis (Fig. 4) demonstrates that although the degree of diversity in humans appears to be higher than that in chickens, this difference is not significant as shown at the ST, CC, and allele levels. A higher degree of diversity in clinical isolates may be expected since humans are probably infected from a variety of sources. Further, overseas travel, in which contact with strains not commonly found in the United Kingdom is likely, has been linked with human infection. It is interesting that the ST rarefaction curves are much steeper than the corresponding CC examples. This finding suggests that, if sampling were extended, more new STs than new CCs would be found. However, it is likely that the isolates we have sampled represent most of the major STs, and the campylobacters in the Grampian region are not an isolated population.
It is possible that the decrease in human incidence can be explained by more effective cooking of chicken and improved overall hygiene in the home (reducing the potential for cross contamination) or possibly by a change in the method of reporting human infection, but there is no evidence available to support these theories. It has also been hypothesized that human populations are acquiring immunity to certain Campylobacter subtypes, and this is a possible reason for the decline in infection. It has been further suggested that older people (>40 years of age) are less likely to become infected with common Campylobacter serotypes (38), suggesting acquired immunity.
Attributing human infection to its origin can be achieved by case control studies, risk and exposure assessment, subtyping, or outbreak investigations (43). As most infections are described as being sporadic (41), outbreak investigations are rare. Source attribution ideally requires representative isolates obtained throughout the production process. Microbial source tracking has been carried out with Salmonella (22), as specific serotypes and phage types are commonly found in specific hosts. It is much more difficult to accurately attribute sources to cases of Campylobacter infection, as the host specificities of types are poorly understood. However, developments in this area are occurring (35), as a degree of host association with particular MLST types has been shown to exist (21, 35, 54). Here, we concentrated on poultry, which is regarded as the single most important source of human campylobacteriosis, to determine if it was associated with the fall in human incidence.
We observed a marked decrease in the occurrence of ST-21 in the chicken host, from a high level in 2001 to the virtual absence of this ST in 2006. The decrease in this subtype was concurrent with an increase in ST-45, ST-257, and ST-2030 in chickens, as well as an increase in these STs in humans. These results can be compared to changes in the prevalence of Salmonella enterica in the United States, where phage type 4 appears to have displaced phage types 8 and 13a (11). Further, the dominant serovar causing human Salmonella infections in the United Kingdom changes over time (11). In our Campylobacter clinical isolate collections, we also found a decrease in ST-21, raising the interesting possibility that, similar to the case of Salmonella, as the frequency of infection with a particular type increases, the immunity of the recipient population also increases, with a resulting decline in infection caused by that type. Genetic distances (Table 2) demonstrated that the distribution of chicken strains had changed to a greater extent than that of human strains between the two years tested. It is not clear why this should be the case. There is considerable evidence that humans are infected via a number of different pathways and that secondary infections are relatively rare. However, for chickens, which are generally reared in large broiler houses, there is much opportunity for bird-to-bird transmission, resulting in higher prevalence within positive flocks at the point of slaughter. The source of infection in chickens is likely to be the environment surrounding the broiler farm (44); an external vector, e.g., water (48); neighboring farm or wild animals (60); or a previously positive flock (50). As a result, the contamination in the broiler is more likely to be representative of the local environment (or the strains that are circulating in the local environment). Hence, the change in genotypes in retail chicken observed in this study may reflect changes in the genotypes in the environmental reservoirs in the vicinities of the broiler houses.
The FSA is currently working with the food industry to halve the number of United Kingdom chickens that are positive for Campylobacter by 2010 (19). This study demonstrates that although determining the prevalence and concentration of Campylobacter in retail chicken is important, subtyping is essential for attributing human infection to specific strains. The identification of Campylobacter to not only the species but also the ST level allows comparisons among time periods and hosts to be made. This study suggests that, similar to Salmonella, specific Campylobacter strains do not always remain dominant in a reservoir and may be subject to displacement by other clones.
Acknowledgments
We offer thanks to the University of Aberdeen and FSA Scotland for funding.
Thanks to the FSA for the 2001 chicken isolates and to T. M. S. Reid (NHS Grampian) for clinical isolates. Additionally, we are grateful to Martin Maiden's research group at the University of Oxford for carrying out DNA sequencing.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adak, G. K., J. M. Cowden, S. Nicholas, and H. S. Evans. 1995. The Public Health Laboratory Service national case-control study of primary indigenous sporadic cases of campylobacter infection. Epidemiol. Infect. 115:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advisory Committee on the Microbiological Safety of Food. 2005. Second report on Campylobacter. Advisory Committee on the Microbiological Safety of Food, London, United Kingdom.
- 4.Berrang, M. E., R. J. Buhr, and J. A. Cason. 2000. Campylobacter recovery from external and internal organs of commercial broiler carcass prior to scalding. Poult. Sci. 79:286-290. [DOI] [PubMed] [Google Scholar]
- 5.Bland, J. M., and D. G. Altman. 2000. Statistics notes. The odds ratio. BMJ 320:1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl. 2):S103-S105. [DOI] [PubMed] [Google Scholar]
- 7.Butzler, J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868-876. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, United States, 2005. Morb. Mortal. Wkly. Rep. 55:392-395. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2005. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food. Morb. Mortal. Wkly. Rep. 54:352-356. [PubMed] [Google Scholar]
- 10.Chen, L., H. Geys, S. Cawthraw, A. Havelaar, and P. Teunis. 2006. Dose response for infectivity of several strains of Campylobacter jejuni in chickens. Risk Anal. 26:1613-1621. [DOI] [PubMed] [Google Scholar]
- 11.Cogan, T. A., and T. J. Humphrey. 2003. The rise and fall of Salmonella enteritidis in the UK. J. Appl. Microbiol. 94(Suppl.):114S-119S. [DOI] [PubMed] [Google Scholar]
- 12.Cogan, T. A., J. Slader, S. F. Bloomfield, and T. J. Humphrey. 2002. Achieving hygiene in the domestic kitchen: the effectiveness of commonly used cleaning procedures. J. Appl. Microbiol. 92:885-892. [DOI] [PubMed] [Google Scholar]
- 13.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144(Pt. 11):3049-3060. [DOI] [PubMed] [Google Scholar]
- 17.Evans, S. J., and A. R. Sayers. 2000. A longitudinal study of campylobacter infection of broiler flocks in Great Britain. Prev. Vet. Med. 46:209-223. [DOI] [PubMed] [Google Scholar]
- 18.FoodNet. 2006. FoodNet surveillance report for 2004 (final report). Centers for Disease Control and Prevention, Atlanta, GA.
- 19.Food Standards Agency. 11 August 2005, posting date. Agency consults on campylobacter baseline target. Food Standards Agency, London, United Kingdom. http://www.food.gov.uk/news/newsarchive/2005/aug/baseline.
- 20.Food Standards Agency. 27 February 2003, posting date. UK-wide survey of Salmonella and Campylobacter contamination of fresh and frozen chicken on retail sale. Food Standards Agency, London, United Kingdom. http://www.food.gov.uk/multimedia/webpage/111802.
- 21.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 22.Hald, T., D. Vose, H. C. Wegener, and T. Koupeev. 2004. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 24:255-269. [DOI] [PubMed] [Google Scholar]
- 23.Harris, N. V., N. S. Weiss, and C. M. Nolan. 1986. The role of poultry and meats in the etiology of Campylobacter jejuni/coli enteritis. Am. J. Public Health 76:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heck, K. L., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 25.Hopkins, R. S., and A. S. Scott. 1983. Handling raw chicken as a source for sporadic Campylobacter jejuni infections. J. Infect. Dis. 148:770. [DOI] [PubMed] [Google Scholar]
- 26.Jones, K. 2001. The Campylobacter conundrum. Trends Microbiol. 9:365-366. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen, F., R. Bailey, S. Williams, P. Henderson, D. R. Wareing, F. J. Bolton, J. A. Frost, L. Ward, and T. J. Humphrey. 2002. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int. J. Food Microbiol. 76:151-164. [DOI] [PubMed] [Google Scholar]
- 28.Klena, J. D., C. T. Parker, K. Knibb, J. C. Ibbitt, P. M. Devane, S. T. Horn, W. G. Miller, and M. E. Konkel. 2004. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42:5549-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locking, M., L. Browning, A. Smith-Palmer, and S. Brownlie (ed.). 10 January 2006, posting date. Gastro-intestinal and foodborne infections. HPS Wkly. Rep. 40(2006/01):2-4. http://www.documents.hps.scot.nhs.uk/ewr/pdf2006/0601.pdf. [Google Scholar]
- 30.Luber, P., and E. Bartelt. 2007. Enumeration of Campylobacter spp. on the surface and within chicken breast fillets. J. Appl. Microbiol. 102:313-318. [DOI] [PubMed] [Google Scholar]
- 31.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manly, B. F. J. 2007. Randomization, bootstrap and Monte Carlo methods in biology. Chapman and Hall/CRC, Boca Raton, FL.
- 33.Manly, B. F. J. 1985. Statistics of natural selection and animal populations. Chapman and Hall, London, United Kindom.
- 34.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy, N. D., F. M. Colles, K. E. Dingle, M. C. Bagnall, G. Manning, M. C. J. Maiden, and D. Falush. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg. Infect. Dis. 13:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michalakis, Y., and L. Excoffier. 1996. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142:1061-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, G., G. M. Dunn, T. M. Reid, I. D. Ogden, and N. J. Strachan. 2005. Does age acquired immunity confer selective protection to common serotypes of Campylobacter jejuni? BMC Infect. Dis. 5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, G., G. M. Dunn, A. Smith-Palmer, I. D. Ogden, and N. J. Strachan. 2004. Human campylobacteriosis in Scotland: seasonality, regional trends and bursts of infection. Epidemiol. Infect. 132:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, J. E., D. Corcoran, J. S. Dooley, S. Fanning, B. Lucey, M. Matsuda, D. A. McDowell, F. Megraud, B. C. Millar, R. O'Mahony, L. O'Riordan, M. O'Rourke, J. R. Rao, P. J. Rooney, A. Sails, and P. Whyte. 2005. Campylobacter. Vet. Res. 36:351-382. [DOI] [PubMed] [Google Scholar]
- 42.Nei, M. 1975. Molecular population genetics and evolution. North-Holland Publishing Company, Amsterdam, The Netherlands.
- 43.Newell, D. G. 2006. Network for the Prevention and Control of Zoonoses. Annual report. Veterinary Laboratories Agency, London, United Kingdom. http://www.medvetnet.org/cms/templates/doc.php?id=60. Accessed 1 June 2007.
- 44.Newell, D. G. 2001. The molecular epidemiology of campylobacters in poultry and poultry meat and use to develop intervention strategies. Final report FS3033. Food Standards Agency, London, United Kingdom.
- 45.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogden, I. D., M. MacRae, and N. J. Strachan. 2004. Is the prevalence and shedding concentrations of E. coli O157 in beef cattle in Scotland seasonal? FEMS Microbiol. Lett. 233:297-300. [DOI] [PubMed] [Google Scholar]
- 47.Oosterom, J., C. H. den Uyl, J. R. Banffer, and J. Huisman. 1984. Epidemiological investigations on Campylobacter jejuni in households with a primary infection. J. Hyg. (London) 93:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson, A. D., M. Greenwood, T. D. Healing, D. Rollins, M. Shahamat, J. Donaldson, and R. R. Colwell. 1993. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl. Environ. Microbiol. 59:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pebody, R. G., M. J. Ryan, and P. G. Wall. 1997. Outbreaks of campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33-R37. [PubMed] [Google Scholar]
- 50.Petersen, L., and A. Wedderkopp. 2001. Evidence that certain clones of Campylobacter jejuni persist during successive broiler flock rotations. Appl. Environ. Microbiol. 67:2739-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues, L. C., J. M. Cowden, J. G. Wheeler, D. Sethi, P. G. Wall, P. Cumberland, D. S. Tompkins, M. J. Hudson, J. A. Roberts, and P. J. Roderick. 2001. The study of infectious intestinal disease in England: risk factors for cases of infectious intestinal disease with Campylobacter jejuni infection. Epidemiol. Infect. 127:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salkind, N. J. 2007. Encyclopedia of measurement and statistics. Sage Publications, Thousand Oaks, CA.
- 53.Schorr, D., H. Schmid, H. L. Rieder, A. Baumgartner, H. Vorkauf, and A. Burnens. 1994. Risk factors for Campylobacter enteritis in Switzerland. Zentralbl. Hyg. Umweltmed. 196:327-337. [PubMed] [Google Scholar]
- 54.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2006. Campylobacter jejuni multilocus sequence types in humans, northwest England, 2003-2004. Emerg. Infect. Dis. 12:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 56.Studahl, A., and Y. Andersson. 2000. Risk factors for indigenous campylobacter infection: a Swedish case-control study. Epidemiol. Infect. 125:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teunis, P. F. M., O. G. van der Heijden, J. W. B. van der Giessen, and A. H. Havelaar. 30 April 1996, posting date. RIVM report no. 284550002. The dose-response relation in human volunteers for gastro-intestinal pathogens. RIVM, Bilthoven, The Netherlands. http://www.rivm.nl/bibliotheek/rapporten/284550002.pdf.
- 58.Thomas, J. C., M. R. Vargas, M. Miragaia, S. J. Peacock, G. L. Archer, and M. C. Enright. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45:616-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner, K. M., and E. J. Feil. 2007. The secret life of the multilocus sequence type. Int. J. Antimicrob. Agents 29:129-135. [DOI] [PubMed] [Google Scholar]
- 60.van de Giessen, A. W., J. J. Tilburg, W. S. Ritmeester, and J. van der Plas. 1998. Reduction of campylobacter infections in broiler flocks by application of hygiene measures. Epidemiol. Infect. 121:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vellinga, A., and F. Van Loock. 2002. The dioxin crisis as experiment to determine poultry-related campylobacter enteritis. Emerg. Infect. Dis. 8:19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wassenaar, T. M., B. A. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139(Pt. 6):1171-1175. [DOI] [PubMed] [Google Scholar]
- 64.Wheeler, J. G., D. Sethi, J. M. Cowden, P. G. Wall, D. S. Tompkins, M. J. Hudson, and P. J. Roderick. 1999. On behalf of the Infectious Intestinal Disease Executive. Br. Med. J. 318:1046. [DOI] [PMC free article] [PubMed] [Google Scholar]