Abstract
Our objectives were to quantify the Salmonella enterica burdens in harvest-ready cattle and to identify specific at-risk populations of cattle most likely to harbor multiply resistant S. enterica. Hide swabs were collected in abattoirs from three cohorts of cattle (feedlot origin cattle that had achieved desirable harvest characteristics and dairy- and beef-type cows harvested because of poor productivity). Feces were collected from two cohorts housed in feedlots (cattle that had achieved desirable harvest characteristics and animals identified for salvage recovery because of poor productivity). Facilities were visited on four occasions over a 12-month period. Salmonella enterica isolates were recovered, and organisms were quantified using standard microbiological methodologies. Susceptibility to antimicrobial drugs and serotype were determined for one S. enterica isolate per sample. Salmonella enterica was recovered from 55.6% of 1,681 samples. The prevalences on hides and in feces were 69.6% and 30.3%, respectively. The concentrations of S. enterica organisms averaged (as determined by the most probable number technique) 1.82 log10/100 cm2 of hides and 0.75 log10/g of feces. None of the isolates recovered from cattle that had achieved desirable harvest characteristics were resistant to four or more drugs. For isolates recovered from animals with poor productivity characteristics, 6.5% were resistant to four or more drugs. Twenty-two serovars were identified, with the most common being Salmonella enterica serovar Anatum (25.5%), Salmonella enterica serovar Montevideo (22.2%), and Salmonella enterica serovar Cerro (12.5%). High-level resistance, i.e., resistance to four or more drugs, was clustered within a few relatively uncommon serovars. These results demonstrate that even though S. enterica isolates are readily recoverable from harvest-ready cattle, multiply resistant variants are rare and are associated with specific serovars in cattle harvested because of poor productivity characteristics.
Salmonella enterica is an important cause of food-borne disease, with an estimated 1.4 million illnesses and 500 deaths attributed to salmonellosis in the United States annually (20). Of all salmonellosis events, the vast majority, in excess of 90%, are believed to result from contaminated foods or beverages (20, 21, 25). Poultry, pork, and fresh produce are traditionally viewed as the principal vehicles of human salmonellosis, presumably attributable to fecal contamination (16, 21). Salmonella enterica organisms are, however, common inhabitants of the gastrointestinal tracts of all animals, including cattle, and as a consequence, beef and dairy products can also serve as vehicles for human exposure to this organism (4, 5, 10, 22).
Not all S. enterica serovars appear equally likely to cause disease. Of the more than 2,000 serovars identified, only 5 serovars (Salmonella enterica serovars Typhimurium, Enteritidis, Newport, Javiana, and Heidelberg) account for approximately 56% of human salmonellosis cases reported to the U.S. Centers for Disease Control and Prevention (7), and an additional 25 serovars account for the majority of the remaining human illnesses. Furthermore, some serovars, such as Salmonella serovar Newport and Salmonella serovar Typhimurium, appear more likely to harbor a diverse ensemble of genetic determinants that convey resistance to a variety of antimicrobial drugs (6, 14). There is a growing body of evidence that disease caused by multiply resistant variants is more severe than disease caused by broadly susceptible S. enterica (15, 24). Presumably, this is not a factor of the genes encoding resistance per se; rather, S. enterica strains that harbor these genes are more likely to possess additional virulence determinants.
The factors that contribute to the coselection of resistance and virulence determinants are not certain. Since antimicrobial drug use provides a selection pressure that favors those isolates resistant to that drug and since more-virulent strains are by definition more likely associated with disease, it seems logical to speculate that antimicrobial use at least in part selects for resistant variants and multiply resistant variants when resistance determinants are linked. If so, such selection could occur either in the animal reservoir or in human salmonellosis patients, although some argue that multiply resistant S. enterica strains arise primarily as a direct consequence of antimicrobial use in food animals (12, 15). Regardless of the origin of multiply resistant S. enterica, it is certain that at least a portion of human salmonellosis cases are attributable to the consumption of beef products. Well-publicized outbreaks of human salmonellosis caused by multiply resistant S. enterica strains associated with ground beef (Salmonella serovar Typhimurium DT104 and Salmonella serovar Newport) have been reported (4, 25). In one instance, beef produced from dairy-type cattle was implicated in an outbreak of multiply resistant Salmonella serovar Newport (5). Because specific animal types are often associated with documented outbreaks of salmonellosis, it is conceivable that multiply resistant S. enterica strains might be clustered within definable populations of harvested cattle. If so, further research of these populations that is designed to better understand the factors that contribute to their elevated risk as well as the development of targeted strategies to mitigate their risk may be warranted. The objectives of this study, therefore, were to quantify S. enterica burdens in harvest-ready cattle and to identify specific at-risk populations of cattle most likely to harbor multiply resistant S. enterica.
MATERIALS AND METHODS
Study design.
A cross-sectional observational study was conducted in which samples were collected from two levels of production: abattoir and feedlot. Samples included hide swabs collected from cattle in four abattoirs and pen floor feces collected in six feedlots. Animals presented for harvest at abattoirs were classified into three cohorts that included feedlot origin cattle that had achieved desirable harvest characteristics, such as body weight and composition, and dairy-type and beef-type cows harvested because of poor productivity characteristics, such as failure to reproduce, injury, or mastitis. Cattle in feedlots were classified into two cohorts that included cattle that had achieved desirable harvest characteristics, as described above, and animals identified for salvage recovery because of poor productivity characteristics, such as injury or disease considered refractory to therapeutic intervention. Samples were collected from all four abattoirs during each of the four seasons. All six feedlots were visited during winter, spring, and summer. However, only two of the six feedlots were visited in the fall.
Sample collection.
Hide swab samples were collected poststunning and prior to any antimicrobial intervention in abattoirs that specialize in harvesting cows culled for poor productivity and abattoirs that process solely feedlot origin cattle. A single hide swab of 1,000 cm2 was collected from the perineum region of the hides of 45 animals per cohort per visit using sterile sponges (SpongeSicle; Biotrace International Inc., Bothell, WA) hydrated with Butterfield's solution. On one occasion, 46 samples were similarly collected from dairy breed cows. Pen floor fecal samples were collected from six feedlots. Within each feedlot, samples were collected from a pen of cattle deemed to have reached a desirable harvest weight and composition and a pen of cattle identified for salvage recovery because of poor productivity characteristics. Fifteen freshly voided, well-defined fecal pats per pen were identified, and approximately 150 g of feces was collected into specimen containers using disposable plastic spoons. All samples were kept cool and transported on the day of collection to Texas Tech University, Lubbock, TX.
Microbial analysis.
Upon arrival at the laboratory, samples were removed from the transport cooler and then refrigerated at approximately 4°C. Initially, 1 ml of hide diluent or 1 g of feces was suspended in separate 9-ml aliquots of tetrathionate (TT) broth and 9 ml of buffered peptone water. One milliliter of the inoculated buffered peptone water was placed in 9 ml of Rappaport-Vassiliadis (RV) broth. Each TT and RV tube was incubated at 42°C for 24 h, and the original sample was refrigerated at 4°C. After inoculation, a separate loop from each TT broth tube and RV broth tube was streaked onto a xylose-lysine-tergitol 4 (XLT4) agar plate that was divided into halves. These plates were incubated for 24 h at 37°C, and a presumptively positive or negative result was reported.
Samples from which presumptively positive colonies were identified (yellow or red with black centers; approximately 48 h after the initial inoculation of the RV and TT broth) were subjected to a most-probable-number (MPN) technique using a 3-by-5 dilution scheme, i.e., five dilutions with three tubes within each dilution. One milliliter of each sample was placed in three 9-ml tubes of RV broth (100 MPN tubes). Serial dilutions were performed to obtain RV broth tubes with MPN of 10−1 to 10−4, which were incubated at 37°C for 18 h. Following incubation, the contents of the tubes were streaked onto XLT4 plates and incubated at 37°C for 24 h. After incubation, the XLT4 plates were inspected for positive growth (colonies with black centers), and the corresponding tubes were recorded as positive. A freely available MPN calculator was used to calculate the MPN of organisms per unit of substrate (100 cm2 of hide or 1 g of feces) (http://www.i2workout.com/mcuriale/mpn/index.html).
S. enterica isolates were forwarded to the National Veterinary Services Laboratory in Ames, IA, for determination of the serovars.
Susceptibility testing.
Isolates were transferred to 10 ml of brain heart infusion broth, vortexed, and incubated at 37°C for 24 h. The broth was transferred and streaked for isolation onto Trypticase soy agar plates and then incubated at 37°C for 24 h. Three to five well-isolated colonies were collected using a sterile swab, transferred into 4 ml of sterile deionized water, and adjusted to a 0.5 McFarland standard. Ten microliters of the standardized inoculum was then transferred into 10 ml Mueller-Hinton broth and used to inoculate the 96-well microbroth dilution plates (Sensititre, catalog number CMV1AGNF; TREK Diagnostics, Cleveland, OH) (see Table A1). At least two plates from each lot of 96-well plates were evaluated with Escherichia coli ATCC 23742 for quality control purposes. The MIC was reported as the lowest concentration that inhibited growth; if, however, the isolate grew at the greatest concentration, the MIC was arbitrarily set at double the greatest concentration included on the plate. Isolates were classified as susceptible or resistant based on available breakpoints (8, 3).
TABLE A1.
Panel of antimicrobial drugs to which susceptibility was determined and the dilution ranges and breakpoints used to determine the resistance of Salmonella enterica
| Antimicrobial(s) | Dilution range(s) (μg/ml)
|
Breakpoint(s) (μg/ml) | |
|---|---|---|---|
| Minimum | Maximum | ||
| Amikacin | 0.5 | 64.0 | ≥64.0 |
| Gentamicin | 0.25 | 16.0 | ≥16.0 |
| Kanamycin | 8.0 | 64.0 | ≥64.0 |
| Streptomycin | 32.0 | 64.0 | ≥64.0 |
| Ampicillin | 1.0 | 32.0 | ≥32.0 |
| Amoxicillin and clavulanic acid | 1.0 and 0.5 | 32.0 and 16.0 | ≥32.0 and ≥16.0 |
| Cefoxitin | 0.5 | 35.0 | ≥32.0 |
| Ceftiofur | 0.12 | 8.0 | ≥8.0 |
| Ceftriaxone | 0.25 | 64.0 | ≥64.0 |
| Ciprofloxacin | 0.015 | 4.0 | ≥4.0 |
| Nalidixic acid | 0.5 | 32.0 | ≥32.0 |
| Sulfisoxazole | 16.0 | 256.0 | ≥256.0 |
| Trimethoprim and sulfamethoxazole | 0.12 and 2.38 | 4.0 and 76.0 | ≥4.0 and ≥76.0 |
| Chloramphenicol | 2.0 | 32.0 | ≥32.0 |
| Tetracycline | 4.0 | 32.0 | ≥16.0 |
Statistical analysis.
Data were entered into an electronic spreadsheet and analyzed using commercially available statistical analysis software (SAS System for Windows, release 9.1.3., SAS Institute, Cary, NC). Descriptive statistics were generated and presented in graphic and tabular formats. Within each sample type, an analysis of the Salmonella prevalence and load was performed using logistic and linear mixed-model methodology, respectively. Model variations attributable to various independent variables (season and cohort) were evaluated for significance. The location of the sample collection was forced into the models as a random variable to account for within-location clustering of the outcome. A significance level (α) of 0.10 was used for the determination of statistical significance.
RESULTS
A total of 1,681 samples were collected and were comprised of 1,081 hide swab samples from the four abattoirs and 600 fecal samples from the feedlots; S. enterica was recovered from 55.6% (n = 934) of 1,681 samples. The prevalence of S. enterica organisms on hide swab samples was 69.6% (n = 752), with a least-square mean (and median) concentration (MPN) in positive samples of log10 1.73 (1.32)/100 cm2, respectively. Salmonella enterica was recovered from 30.3% (n = 182) of fecal samples, with a least-square mean (and median) concentration (MPN) in positive samples of log10 0.71 (−0.21)/g, respectively. The burdens of S. enterica by cohort are presented in Table 1.
TABLE 1.
Least-square mean prevalences and concentrations of Salmonella enterica organisms and 95% confidence intervals by cohort, averaged across seasons
| Cohort | Source | Sample type | Burden of Salmonella entericaa
|
|
|---|---|---|---|---|
| Prevalence (%) (95% CI) | Concnb (95% CI) | |||
| Feedlot origin, desirable harvest wt | Abattoir | Hide swab | 81.4 (64.4-91.4) | 2.32 (1.87-2.77) |
| Dairy-type cows, poor productivity | Abattoir | Hide swab | 71.2 (63.7-77.7) | 1.24 (0.79-1.70) |
| Beef-type cows, poor productivity | Abattoir | Hide swab | 59.0 (35.2-79.3) | 1.48 (1.01-1.95) |
| Feedlot cattle, desirable harvest wt | Feedlot | Feces | 31.6 (15.5-53.7) | 1.03 (−0.22-2.27) |
| Feedlot cattle, poor productivity | Feedlot | Feces | 27.2 (20.3-35.4) | 0.48 (−0.77-1.72) |
CI, confidence interval.
Concentrations are either log10 MPN/100 cm2 of hide or log10 MPN/g of feces.
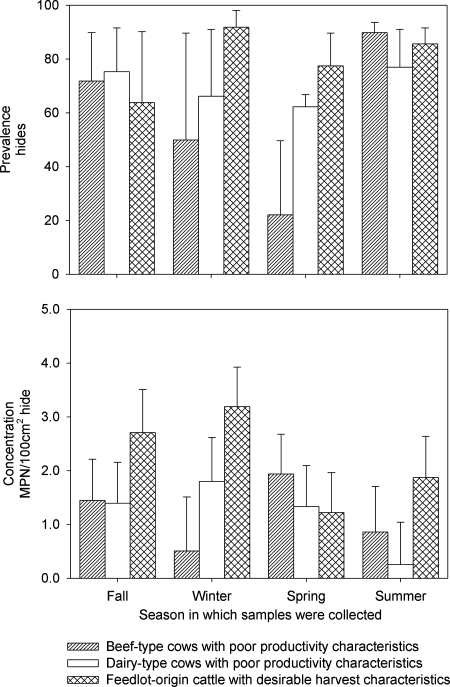
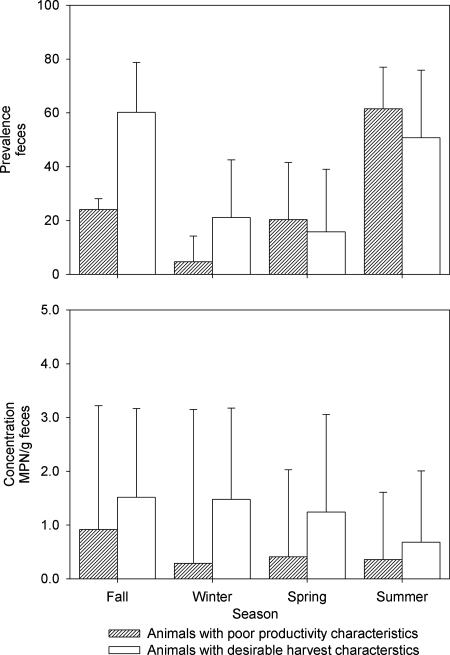
The prevalence of S. enterica organisms on the hides of animals presented for harvest did not vary by animal type (P = 0.47) or season (P = 0.39). The Salmonella enterica concentrations on the hides of cattle in abattoirs varied between cohorts and over time (P < 0.01) (Fig. 1, bottom). When season was held as a constant, the concentration of S. enterica organisms on the hides from which Salmonella was recovered was greater for feedlot origin cattle than for both beef-type (P = 0.05) and dairy-type (P = 0.01) cows. The S. enterica concentrations did not vary significantly between beef- and dairy-type cows presented for harvest (P = 0.23). When animal type was held as a constant, the S. enterica concentration on the hides during the fall was greater than that during the winter (P < 0.01), the spring concentration was greater than both the summer (P < 0.01) and winter (P < 0.01) concentrations, and the summer concentration was greater than the winter (P = 0.06) concentration; other pairwise comparisons were not significantly different (P > 0.10). Neither the prevalence nor the concentration of S. enterica organisms in the feces of animals in feedlots varied significantly by animal type (P = 0.61 and 0.23, respectively) or season (P = 0.11 and 0.71, respectively) (Fig. 2).
FIG. 1.
Least-square mean estimates of prevalences (%) (top) and concentrations (log10 MPN/100 cm2) (bottom) of Salmonella enterica organisms on hides of beef-type and dairy-type cows removed from the herd because of poor productivity and feedlot origin cattle that had achieved desirable harvest characteristics, by season. Error bars represent the upper 95% confidence interval.
FIG. 2.
Least-square mean estimates of prevalences (%) (top) and concentrations (log10 MPN/g) (bottom) of Salmonella enterica organisms in feces of feedlot cattle removed from the herd because of poor productivity and feedlot cattle that had achieved desirable harvest characteristics, by season. Error bars represent the upper 95% confidence interval.
Twenty-two serovars were successfully reported for 762 isolates of the 934 S. enterica isolates tested (Table 2). Of the serovars recovered from hides, Salmonella enterica serovar Anatum (22.9% of hide origin serovars), S. enterica serovar Montevideo (22.9%), S. enterica serovar Cerro (14.9%), S. enterica serovar Mbandaka (10.0%), S. enterica serovar Muenster (6.5%), and S. enterica serovar Kentucky (5.7%) were the most common. Similarly, the most common serovars recovered from feces were Salmonella serovar Anatum (32.5% of fecal-origin serovars), Salmonella serovar Montevideo (19.6%), Salmonella serovar Mbandaka (14.7%), Salmonella serovar Kentucky (16.0%), S. enterica serovar Reading (4.9%), and Salmonella serovar Cerro (4.3%). One Salmonella serovar Newport isolate was recovered (from a fecal sample of an animal identified with poor productivity characteristics), but no Salmonella serovar Typhimurium strains were recovered.
TABLE 2.
Frequencies of serovar recovery by sample type, mean and median numbers of drugs to which the serovars were resistant, and occurrence of ACSSuT R-type and MDR-AmpC phenotypes
| Serotype | Proportion of isolates (n = 762) found:
|
No. of drugs to which the serovar was resistant
|
Phenotype present (% of isolates with phenotype)
|
|||
|---|---|---|---|---|---|---|
| On hides | In fecesa | Mean | Median | ACSSuT | MDR-AmpC | |
| Anatum | 22.9 | 32.5 | 0.6 | 1.0 | No | No |
| Montevideo | 22.9 | 19.6 | 0.6 | 0.0 | Yes (0.6) | No |
| Cerro | 14.9 | 4.3 | 0.4 | 0.0 | No | No |
| Mbandaka | 10.0 | 14.7 | 0.4 | 0.0 | No | No |
| Muenster | 6.5 | 0.61 | 1.2 | 1.0 | Yes (5.0) | Yes (5.0) |
| Kentucky | 5.7 | 16.0 | 0.6 | 1.0 | No | No |
| Agona | 3.8 | 0.61 | 2.7 | 2.0 | Yes (8.3) | Yes (8.3) |
| Braenderup | 3.0 | NR | 0.3 | 0.0 | No | No |
| Muenchen | 1.8 | 0.61 | 0.5 | 0.0 | No | No |
| Reading | 1.8 | 4.9 | 6.5 | 7.0 | Yes (73.7) | Yes (66.7) |
| Meleagridis | 1.7 | 3.7 | 0.4 | 0.0 | No | No |
| Fresno | 1.0 | NR | 0.8 | 1.0 | No | No |
| Give | 0.83 | NR | 0.4 | 0.0 | No | No |
| Havana | 0.83 | NR | 0.1 | 0.0 | No | No |
| Panama | 0.83 | NR | 1.2 | 1.0 | No | No |
| Infantis | 0.50 | 0.61 | 0.8 | 0.5 | No | No |
| Paratyphi B variant L.(+) tartrate(+) | 0.33 | NR | 4.5 | 4.5 | Yes (50.0) | Yes (50.0) |
| Senftenberg | 0.33 | NR | 0.3 | 0.5 | No | No |
| Heidelberg | 0.17 | NR | 3.0 | 3.0 | No | No |
| Minnesota | 0.17 | 0.61 | 0.5 | 0.5 | No | No |
| Newport | NR | 0.61 | 0.0 | 0.0 | No | No |
| Kiambu | NR | 0.61 | 0.0 | 0.0 | No | No |
NR, not recovered.
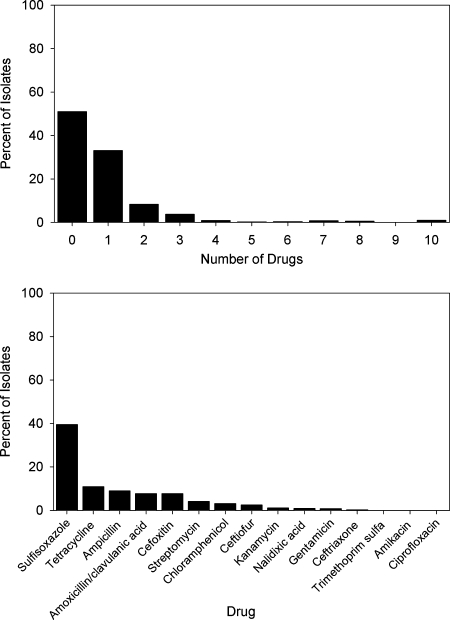
Of the 934 S. enterica isolates, 51.1% (n = 477) (Fig. 3) were pansusceptible, 33.1% (n = 309) were resistant to one drug, 8.35% (n = 78) were resistant to two drugs, 3.75% (n = 35) were resistant to three drugs, and 3.75% (n = 35) were resistant to four or more antimicrobial drugs. The most common resistance observed was to sulfisoxazole (39.5% of isolates), followed by tetracycline (10.9%) and ampicillin (8.89%). Ciprofloxacin resistance was not detected, and resistance to ceftriaxone was observed in 0.21% (n = 2) of isolates.
FIG. 3.
Percentages of Salmonella enterica isolates by number of drugs (top) and drugs (bottom) to which they were resistant.
Of the isolates recovered from fecal and hide swab samples, 6.59 and 3.06%, respectively, were resistant to four or more drugs (Table 3). Across sample types, however, 0.0% of isolates recovered from animals that had reached a desirable harvest weight and 6.51% of isolates from animals removed from the herd for poor productivity (either dairy-type cows, beef-type cows, or feedlot animals) were resistant to four or more drugs (P was <0.01 in the comparison of the two groups). Specific patterns of coresistance were evident in the data (Table 4). Of the isolates resistant to four or more drugs, 62.9% (n = 22) were coresistant to ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline (ACSSuT R type). Further, of these ACSSuT R-type isolates, 77.3% (n = 17) were of the multidrug resistance (MDR)-AmpC phenotype in that they were also coresistant to a potentiated β-lactam (amoxicillin potentiated with clavulanic acid) and an expanded-spectrum cephalosporin (the MDR-AmpC phenotype is defined as coresistance to ACSSuT, a potentiated β-lactam, and an expanded-spectrum cephalosporin [14]; all isolates were resistant to ceftiofur, and two isolates were also resistant to ceftriaxone). There were no isolates singly resistant to ceftiofur. The mean and median numbers of drugs to which ceftiofur-resistant isolates were resistant were 7.7 and 8, respectively, whereas ceftiofur-susceptible isolates were typically resistant to 0.7 and 0 drug (mean and median, respectively). There was substantial agreement beyond chance (κ = 0.75) between ceftiofur resistance and the ACSSuT R type. Of the 23 ceftiofur-resistant isolates, 17 also were of the ACSSuT R type, and these 17 were also of the MDR-AmpC phenotype. The ACSSuT R type was overrepresented in a few serovars; Salmonella serovar Reading isolates were 99 times (95% confidence interval = 42 to 227; P < 0.01) more likely to be of the ACSSuT R type than the isolates of other serovars. Salmonella enterica serovar Agona isolates were also overrepresented in that they were 16 times (95% confidence interval = 3.1 to 85; P = 0.01) more likely to be of the ACSSuT R type than the isolates of other serovars (excluding Salmonella serovar Reading).
TABLE 3.
Percentages of isolates by number of drugs to which they were resistant and cohort
| Cohort | Source | Sample type | No. of isolates | % of isolates that were resistant to the following no. of drugs:
|
|||
|---|---|---|---|---|---|---|---|
| 0 | 2 or more | 3 or more | 4 or more | ||||
| Beef-type cows, poor productivity | Abattoir | Hide swab | 210 | 43.3 | 30.0 | 12.4 | 4.3 |
| Dairy-type cows, poor productivity | Abattoir | Hide swab | 243 | 42.0 | 14.0 | 11.1 | 5.8 |
| Feedlot origin, desirable harvest wt | Abattoir | Hide swab | 299 | 54.5 | 8.4 | 1.0 | 0.0 |
| Feedlot cattle, poor productivity | Feedlot | Feces | 85 | 63.5 | 21.2 | 16.5 | 14.1 |
| Feedlot cattle, desirable harvest wt | Feedlot | Feces | 97 | 69.1 | 8.3 | 0.0 | 0.0 |
TABLE 4.
Frequencies of antimicrobial drug resistance phenotypes observed by number of drugs to which the isolates were resistant and their sources
| No. of drugs to which isolates were resistant (no. of isolates) | Resistance phenotypea | % of all isolates | Source(s)b |
|---|---|---|---|
| 0 (477) | None (pansusceptible) | 51.1 | 1, 2, 3, 4, 5 |
| 1 (309) | Su | 28.3 | 1, 2, 3, 4, 5 |
| T | 2.4 | 1, 3, 4, 5 | |
| A | 1.5 | 1, 2, 3, 4 | |
| S | 0.43 | 2, 4 | |
| Fox | 0.32 | 1, 2 | |
| Nal | 0.11 | 1 | |
| 2 (78) | Su T | 5.1 | 1, 2, 3, 4, 5 |
| S Su | 0.86 | 2, 3, 4 | |
| Fox Aug | 0.86 | 1, 2 | |
| A Su | 0.64 | 1, 2, 3 | |
| Su Cef | 0.32 | 3 | |
| Nal Su | 0.32 | 3 | |
| C S | 0.11 | 2 | |
| A Fox | 0.11 | 4 | |
| 3 (35) | A Fox Aug | 3.4 | 1, 2, 3, 4 |
| S T Kan | 0.11 | 1 | |
| Su T TrSu | 0.11 | 4 | |
| S T Aug | 0.11 | 3 | |
| 4 (8) | A Su Fox Aug | 0.43 | 1, 2 |
| C S Su T | 0.21 | 4 | |
| C Su T Nal | 0.11 | 4 | |
| A Su T Fox | 0.11 | 4 | |
| 5 (2) | A Su Fox Aug Cef | 0.11 | 2 |
| A C Su T Aug | 0.11 | 1 | |
| 6 (3) | C Su T TrSu Fox Aug | 0.11 | 4 |
| A Su TrSu Fox Aug Cef | 0.11 | 2 | |
| A C S Su T Aug | 0.11 | 4 | |
| 7 (7) | A C S Su T Fox Aug | 0.43 | 1, 2, 4 |
| A C S Su T Aug Cef | 0.21 | 4 | |
| A C Su TrSu Fox Aug Cef | 0.11 | 2 | |
| 8 (5) | A C S Su T Fox Aug Cef | 0.54 | 1, 4 |
| 9 (1) | A C S Su T TrSu Fox Aug Cef | 0.11 | 2 |
| 10 (9) | A C S Su T Fox Aug Cef Kan Gen | 0.75 | 1, 2 |
| A C S Su T Fox Aug Cef Axo Kan | 0.21 | 2 |
A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; T, tetracycline; Fox, cefoxitin; Aug, amoxicillin-clavulanic acid; Nal, nalidixic acid; TrSu, trimethoprim-sulfisoxazole; Cef, ceftiofur; Axo, ceftriaxone; Kan, kanamycin; Gen, gentamicin.
1, hide swab collected in an abattoir from beef-type cows removed from the herd because of poor productivity; 2, hide swab collected in an abattoir from dairy-type cows removed from the herd because of poor productivity; 3, hide swab collected in an abattoir from feedlot origin cattle that had achieved desirable harvest characteristics; 4, fecal sample collected from feedlot cattle removed from the herd because of poor productivity; 5, fecal sample collected from feedlot cattle that had achieved desirable harvest characteristics.
DISCUSSION
Salmonella enterica was readily recovered from the populations of cattle enrolled in the study described herein; all of the populations were eligible for harvest. These estimates are generally greater than those reported elsewhere (1, 9) but somewhat consistent with studies of cattle from the southern High Plains of the United States (2, 13, 18). In our study, the estimate of prevalence was greater on hides than in feces, and this phenomenon has been reported previously (18). Ostensibly, this observation may imply that hides are more likely to be S. enterica positive than feces; we believe, however, that it is instead a consequence of the size of the sampled area (1,000 cm2 versus ∼150 g of feces), the subsampling of the sample substrate within the laboratory (approximately 1.1 ml from 25 ml of diluent versus approximately 1.1 g from 150 g of feces), and the sensitivity of the microbiological methods for recovering S. enterica from the substrate (i.e., diluent versus feces). Furthermore, evidence from previous studies indicates that E. coli O157:H7 is not uniformly distributed in feces (11) and that S. enterica is not uniformly distributed on hides (23), and if multiple samples per animal are cultured, the estimate of prevalence increases dramatically. It is, therefore, inevitable that some animals are incorrectly classified as S. enterica negative when only a relatively small proportion of feces or hide has been sampled (i.e., 150 g of feces and 1,000 cm2 represents a small proportion of the total daily fecal production or surface area). In other words, the sampling design employed in the study described herein almost certainly underestimated prevalence (both on hides and in feces). Consequently, had we increased the amount sampled, more uniformly distributed the organism in the substrate, or inoculated a greater volume, we expect that the estimates of prevalence would have been greater (11, 17, 23). After accounting for the lack of sensitivity inherent in the design, it seems most likely that S. enterica is ubiquitous among cattle populations from the southern High Plains.
Despite other reports of seasonal variation in S. enterica prevalence, we did not detect statistically significant variation in S. enterica prevalence from season to season. However, there may have been biologically significant variation in the concentrations of S. enterica organisms on hides across cattle type and season. Seasonal variation, however, is of limited inferential value because it is uncertain how representative the seasons included in the study described herein were of typical seasons. For example, the spring sampling time frame was unusually dry, yet the summer period of this study was unusually wet. Regardless of what may or may not be concluded from the observed seasonal variation in loads, when season was held as a constant, feedlot origin cattle had greater S. enterica loads on hides than cows (either of the beef type or dairy type). Furthermore, there was some evidence that within fecal samples, animals that had achieved desirable harvest characteristics may have had greater concentrations than those removed from the herd. Despite the greatest concentrations being observed in cattle that had achieved desirable harvest characteristics to satisfy market demands (i.e., feedlot origin cattle in abattoirs as well as those in feedlots), the S. enterica isolates recovered from these cohorts were of serovars rarely observed in either human or animal disease. Further, they were, at least phenotypically, broadly susceptible to the panel of antimicrobial drugs and presumably unlikely to be a source for resistance determinants for other bacteria. It seems likely, therefore, that the vast majority of S. enterica serovars carried by the productive feedlot cattle that had successfully achieved desirable harvest characteristics are of limited public or animal health importance, and they may even represent nonpathogenic variants or commensal organisms.
Only samples in which presumptively positive colonies were observed were subjected to enumeration methodologies. Presumptively positive identification required approximately 48 h from the time of initial laboratory processing to the inoculation of the MPN tubes. While samples were held at 4°C, it is possible that the concentration of bacteria declined over time. If this is so, then we underestimated concentration, but this is at present uncertain without the appropriate evaluation of temperature-, time-, and substrate-dependent changes in Salmonella concentration. Regardless, storage conditions were consistent across all samples, so within-substrate changes in concentration, if any, should have been consistent from cohort to cohort and season to season.
While high-level resistance was rare in this study, it was clustered within a few serovars and in those animals that had been removed from the herd for poor productivity characteristics. Productivity characteristics that warrant removal from the herd vary by production type but in general include injury, lameness, disease deemed refractory to continued treatment, and failure to reproduce (in cows). The proportion of cattle that are removed from the herd varies by production type and may range from 25% per year in dairies to approximately 1% per cohort in feedlots. Antimicrobial therapy is indicated for some illnesses that may ultimately lead to a decision to cull the animal from the herd, such as respiratory disease in feedlot cattle or metritis in dairy cows. Historical data were not available for the animals included in this study, but it is probable that at least some of the animals removed from the herds because of poor productivity were administered one or more courses of therapy with injectable antimicrobial drugs prior to sample collection. In contrast, however, feedlot origin cattle that have reached a desirable harvest weight and body composition have done so because of acceptable performance characteristics and were unlikely to have required injectable drug therapy, at least in their recent history. It is possible, therefore, that prior antimicrobial therapy may have contributed to the clustering of highly resistant S. enterica strains in cattle culled from the herd. Antimicrobial drug use per se cannot, however, explain all of the clustering, as the vast majority of feedlot cattle in the study area, including those sampled in our study, were administered subtherapeutic doses of tylosin and monensin to improve the efficiency of production. It seems likely, therefore, that any effect that antimicrobial drug use exerts on the selection of resistant variants may be bacterium and drug specific and also influenced by both animal- and group-level factors.
The so-called MDR-AmpC phenotype was first described for Salmonella serovar Newport (14), and the broad ensemble of resistance determinants is typically thought to be plasmid borne. In our study, we recovered only one Salmonella serovar Newport isolate; it was recovered during the summer from the feces of a feedlot animal removed from the herd for poor productivity and was pansusceptible. In the study described herein, the ACSSuT R-type and MDR-AmpC phenotypes were observed in five and four serovars, respectively, and were most commonly observed in Salmonella serovar Reading isolates. Two phenotypes were observed for Salmonella serovar Reading isolates: they were either broadly susceptible or broadly resistant. The mean (median) numbers of drugs to which the isolates were resistant were 2 (1) for non-ACSSuT R types and 8.1 (8) for phenotypically ACSSuT R-type isolates. Further, strong agreement between resistance to ceftiofur and the ACSSuT R type was observed across the five serovars displaying the ACSSuT R type, and this observation is consistent with earlier observations from our laboratory (19).
The ACSSuT and MDR-AmpC phenotypes were recovered only from samples collected from animals identified with poor productivity characteristics (either in feedlots or cow herds) but were never identified in the feedlot animals (feces collected in feedlots or hide swab samples collected in abattoirs) that had achieved desirable harvest characteristics. In one feedlot, Salmonella serovar Reading was recovered from the feces of both cohorts; however, degrees of resistance (broadly susceptible or broadly resistant) varied by cohort in that broadly resistant variants were recovered only from animals that were removed from the herd because of poor productivity characteristics. More research is needed to determine if movement from a productive cohort to a poor-productivity cohort was associated with the acquisition of the MDR-AmpC phenotype-encoding plasmid by strains of a few serovars or if the animals themselves acquired a distinct subpopulation of the serovar already harboring the plasmid. In other words, it is uncertain if the observed serovar-specific clustering of high-level resistance was associated with the dissemination of either a mobile/mobilizable plasmid or an S. enterica clone.
Acknowledgments
This study was funded in part by the Beef Checkoff program through the Cattlemen's Beef Board and the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (grant 2004-35212-14864).
APPENDIX
The antimicrobial drugs to which susceptibility was determined and the dilution ranges and breakpoints used to determine the resistance of Salmonella enterica are shown in Table A1.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Bacon, R. T., J. N. Sofos, K. E. Belk, D. R. Hyatt, and G. C. Smith. 2002. Prevalence and antibiotic susceptibility of Salmonella isolated from beef animal hides and carcasses. J. Food Prot. 65:284-290. [DOI] [PubMed] [Google Scholar]
- 2.Barham, A. R., B. L. Barham, A. K. Johnson, D. M. Allen, J. R. Blanton, Jr., and M. F. Miller. 2002. Effects of the transportation of beef cattle from the feedyard to the packing plant on prevalence levels of Escherichia coli O157 and Salmonella spp. J. Food Prot. 65:280-283. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2006. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): 2003 human isolates final report. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA.
- 4.Centers for Disease Control and Prevention. 2006. Multistate outbreak of Salmonella typhimurium infections associated with eating ground beef—United States, 2004. Morb. Mortal. Wkly. Rep. 55:180-182. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Outbreak of multidrug-resistant Salmonella Newport—United States, January-April 2002. Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2007. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2004. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA.
- 7.Centers for Disease Control and Prevention. 2007. Salmonella surveillance: annual summary, 2005. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA.
- 8.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Dargatz, D. A., P. J. Fedorka-Cray, S. R. Ladely, C. A. Kopral, K. E. Ferris, and M. L. Headrick. 2003. Prevalence and antimicrobial susceptibility of Salmonella spp. isolates from US cattle in feedlots in 1999 and 2000. J. Appl. Microbiol. 95:753-761. [DOI] [PubMed] [Google Scholar]
- 10.Dechet, A. M., E. Scallan, K. Gensheimer, R. Hoekstra, J. Gunderman-King, J. Lockett, D. Wrigley, W. Chege, and J. Sobel. 2006. Outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium definitive type 104 infection linked to commercial ground beef, northeastern United States, 2003-2004. Clin. Infect. Dis. 42:747-752. [DOI] [PubMed] [Google Scholar]
- 11.Echeverry, A., G. H. Loneragan, B. A. Wagner, and M. M. Brashears. 2005. Effect of intensity of fecal pat sampling on estimates of Escherichia coli O157 prevalence. Am. J. Vet. Res. 66:2023-2027. [DOI] [PubMed] [Google Scholar]
- 12.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 13.Fluckey, W. M., W. G. Loneragan, R. Warner, and M. M. Brashears. 2007. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J. Food Prot. 70:551-556. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, A., J. Fontana, C. Crowe, B. Bolstorff, A. Stout, S. Van Duyne, M. P. Hoekstra, J. M. Whichard, T. J. Barrett, and F. J. Angulo. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 15.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Molbak. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg. Infect. Dis. 8:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen, F., R. Bailey, S. Williams, P. Henderson, D. R. Wareing, F. J. Bolton, J. A. Frost, L. Ward, and T. J. Humphrey. 2002. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int. J. Food Microbiol. 76:151-164. [DOI] [PubMed] [Google Scholar]
- 17.Keen, J. E., and R. O. Elder. 2002. Isolation of shiga-toxigenic Escherichia coli O157 from hide surfaces and the oral cavity of finished beef feedlot cattle. J. Am. Vet. Med. Assoc. 220:756-763. [DOI] [PubMed] [Google Scholar]
- 18.Loneragan, G. H., and M. M. Brashears. 2005. Effects of using retention-pond water for dust abatement on performance of feedlot steers and carriage of Escherichia coli O157 and Salmonella spp. J. Am. Vet. Med. Assoc. 226:1378-1383. [DOI] [PubMed] [Google Scholar]
- 19.Lowrance, T. C., G. H. Loneragan, D. J. Kunze, T. M. Platt, S. E. Ives, H. M. Scott, B. Norby, A. Echeverry, and M. M. Brashears. 2007. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 68:501-507. [DOI] [PubMed] [Google Scholar]
- 20.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proctor, M. E., M. Hamacher, M. L. Tortorello, J. R. Archer, and J. P. Davis. 2001. Multistate outbreak of Salmonella serovar Muenchen infections associated with alfalfa sprouts grown from seeds pretreated with calcium hypochlorite. J. Clin. Microbiol. 39:3461-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez, A., P. Pangloli, H. A. Richards, J. R. Mount, and F. A. Draughon. 2006. Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 69:2576-2580. [DOI] [PubMed] [Google Scholar]
- 23.Stephens, T. P., G. H. Loneragan, T. W. Thompson, A. Sridhara, L. A. Branham, S. Pitchiah, and M. M. Brashears. 2007. Distribution of Escherichia coli O157 and Salmonella on hide surfaces, the oral cavity, and in feces of feedlot cattle. J. Food Prot. 70:1346-1349. [DOI] [PubMed] [Google Scholar]
- 24.Varma, J. K., K. Molbak, T. J. Barrett, J. L. Beebe, T. F. Jones, T. Rabatsky-Ehr, K. E. Smith, D. J. Vugia, H. G. Chang, and F. J. Angulo. 2005. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J. Infect. Dis. 191:554-561. [DOI] [PubMed] [Google Scholar]
- 25.Zansky, S., B. Wallace, D. Schoonmaker-Bopp, P. Smith, F. Ramsey, J. Painter, A. Gupta, P. Kalluri, and S. Noviello. 2002. From the Centers for Disease Control and Prevention. Outbreak of multi-drug resistant Salmonella Newport—United States, January-April 2002. JAMA 288:951-953. [PubMed] [Google Scholar]