Abstract
Lactococcus lactis is a primary constituent of many starter cultures used for the manufacturing of fermented dairy products, but the species also occurs in various nondairy niches such as (fermented) plant material. Three genome sequences of L. lactis dairy strains (IL-1403, SK11, and MG1363) are publicly available. An extensive molecular and phenotypic diversity analysis was now performed on two L. lactis plant isolates. Diagnostic sequencing of their genomes resulted in over 2.5 Mb of sequence for each strain. A high synteny was found with the genome of L. lactis IL-1403, which was used as a template for contig mapping and locating deletions and insertions in the plant L. lactis genomes. Numerous genes were identified that do not have homologs in the published genome sequences of dairy L. lactis strains. Adaptation to growth on substrates derived from plant cell walls is evident from the presence of gene sets for the degradation of complex plant polymers such as xylan, arabinan, glucans, and fructans but also for the uptake and conversion of typical plant cell wall degradation products such as α-galactosides, β-glucosides, arabinose, xylose, galacturonate, glucuronate, and gluconate. Further niche-specific differences are found in genes for defense (nisin biosynthesis), stress response (nonribosomal peptide synthesis and various transporters), and exopolysaccharide biosynthesis, as well as the expected differences in various mobile elements such as prophages, plasmids, restriction-modification systems, and insertion sequence elements. Many of these genes were identified for the first time in Lactococcus lactis. In most cases good correspondence was found with the phenotypic characteristics of these two strains.
Lactococcus lactis is a primary constituent of many starter cultures used for the manufacturing of fermented dairy products. Because of its tremendous industrial importance, numerous studies have been dedicated to the elucidation of the physiology and molecular biology of many traits relevant for industrial applications, including the production of flavor compounds, vitamins and other nutraceuticals, and exopolysaccharides relevant for texture development (33, 60, 68). Moreover, due to the availability of a vast molecular toolbox for genetic engineering and recent genome sequencing efforts, L. lactis has gained a strong position as a model organism for low-GC gram-positive bacteria and lactic acid bacteria in particular (7, 39, 43). Much of the biochemical and genetic research has been conducted with a limited number of strains, mainly the plasmid-cured strains IL-1403 and MG1363, which originate from dairy fermentations.
(Fermenting) plant material is a second important ecosystem occupied by L. lactis, where it typically occurs as an early colonizer that is later replaced by species that are more tolerant of low pH values (30, 31). Most plant-associated strains belong to Lactococcus lactis subsp. lactis, whereas Lactococcus lactis subsp. cremoris is typically found in dairy fermentations (30, 31). Fermenting plant material comprises a broad array of highly variable niches with respect to chemical composition, as for instance the availability of carbohydrates other than lactose as growth substrates. Moreover, protein concentrations are typically much lower than those observed in the dairy environment. As a result, strains isolated from fermenting plant material do not harvest amino acids through proteolysis but depend on amino acid biosynthesis and consequently exhibit fewer amino acid auxotrophies than do dairy isolates (2). Therefore, it can be anticipated that strains adapted to the plant ecological niche will exhibit large metabolic differences and their metabolic diversity will most certainly exceed that of dairy strains. Recently it has been shown that strains isolated from a nondairy environment exhibit flavor-forming activities that may be beneficial to dairy fermentation, as exemplified by the production of the key flavor fusel aldehydes as a result of a unique α-keto acid decarboxylase activity (58, 59). Moreover, it was shown that some nondairy L. lactis strains produce the enzyme glutamate dehydrogenase, which converts glutamate to α-ketoglutarate (62). This compound, α-ketoglutarate, is the acceptor of the amino group in aminotransferase reactions, the first step in the production of flavor compounds from amino acid, and present at rate-limiting concentrations in cheese (62).
Several studies have addressed the biodiversity of L. lactis using a variety of molecular approaches as well as extended phenotyping, and this has revealed an unusual population structure (30, 46, 50, 68). A recent extensive genotypic and phenotypic diversity analysis of a large strain collection of dairy and nondairy origins confirmed the existence of two major genomic lineages (50). Most nondairy isolates belong to the lineage containing strains of L. lactis subsp. lactis, and it was shown that nondairy isolates represent molecular diversity not found within the dairy strains. Therefore, our present view on genomic and metabolic diversity of L. lactis may be limited and is likely to be highly biased toward dairy isolates, as only the genomes of strains L. lactis subsp. lactis IL-1403 (7), L. lactis subsp. cremoris SK11 (39), and L. lactis subsp. cremoris MG1363 (71) of dairy origin have been sequenced. In the current study we report the low-coverage (threefold) diagnostic sequencing of two L. lactis subsp. lactis strains isolated from (fermenting) plant material. Unique genes and gene clusters found within these isolates are discussed, and predicted phenotypes are validated experimentally. The results provide a first view of the gene pool present within the species and yield insight into the molecular basis of adaptation to their propagation in fermenting plant material.
(A preliminary report of this work appeared in reference 68.)
MATERIALS AND METHODS
Bacterial isolates and media.
Lactococcus lactis subsp. lactis strains KF147 and KF282 were obtained from W. Kelly (31); KF147 was isolated from mung bean sprouts, and KF282 was isolated from mustard and cress. Strains were maintained in M17 broth (Oxoid Ltd., Basingstoke, Hampshire, England) with 0.5% (wt/vol) glucose as a carbon source (GM17). Serial transfer was minimized to prevent the occurrence of mutations as a result of adaptation to laboratory media and conditions.
Fermentation tests.
The ability to ferment citrate was measured with whey permeate calcium citrate Castione agar as described previously (22) but with milk replaced by whey permeate. The fermentation substrate range with respect to mono/oligosaccharide range was determined with the API 50 CHL assay (bioMérieux, Marcy l'Etoile, France) as described in reference 50. In the current paper, these data were used for experimental validation of the phenotypes predicted from comparative genome analysis. The ability to ferment other carbohydrates and complex polysaccharides was tested by growing the strains at 30°C in 96-well microplates filled with 250 μl GM17 medium per well. Subsequently, these cultures were used to inoculate 250 μl M17 containing 0.5% or 1% of a carbohydrate carbon source. All incubations were carried out at 30°C and in quadruplicate. Growth was analyzed by determining the turbidity at 600 nm and scored as negative (−), weak (+/−), or positive (+), compared with GM17.
Nisin assay.
Nisin concentrations in supernatants of overnight culture were determined by a plate diffusion method (65) using Micrococcus flavus DSM1790 as the assay organism. Standards were prepared from nisin (Sigma; 2.5% pure nisin).
DNA isolation and sequencing.
Total DNA of the isolates KF147 and KF282 was isolated as described previously (55). The total DNA was sequenced using a minimal shotgun sequencing approach. Four different genomic libraries were constructed: three plasmid libraries with average insert sizes of 0.8, 1.8, and 5 kb, respectively, and a fosmid library with inserts of 30 to 40 kb. Over 8 Mb was sequenced from these libraries, corresponding to approximately threefold coverage, assuming a genome size of 2.75 Mb (GATC Biotech, Konstanz, Germany). Contigs were assembled using the Seqman Genome Software of DNAstar (Madison, WI). Contigs were mapped to the template genome of L. lactis IL-1403 using Projector 2 (http://bioinformatics.biol.rug.nl/websoftware/projector2/) (67). Projector 2 settings were as follows: BLAST analysis; size of chopped fragments, 250 nucleotides (nt); minimum size contig, 1,000 nt; minimum size remaining fragment, 250 nt; cutoff for repeats, 1E-20; maximum length deviation mapped versus original template, 20%. Repeats (e.g., rRNA and insertion sequence [IS] elements) are filtered out before template matching. Statistics of sequencing and template mapping are summarized in Table 1. Interesting novel genes/gene clusters from strain KF147 contigs were resequenced by primer walking and PCR on fosmid clones, to close gaps, improve sequence quality, merge contigs, and correct frameshifts (GATC Biotech AG, Konstanz, Germany).
TABLE 1.
Initial shotgun sequencing and template mapping statistics
| Property | Strain KF147 | Strain KF282 |
|---|---|---|
| No. of total sequence reads | 13,464 | 11,878 |
| % G+C | 34.9 | 34.9 |
| No. of contigs | ||
| >1,000 nt | 544 | 610 |
| Mapped to IL-1403 | 402 | 418 |
| Not mapped to IL-1403 | 142 | 192 |
| Total no. of kb in contigs | ||
| Total assembled (nonredundant) in contigs of >1,000 nta | 2,344 | 2,321 |
| Mapped to IL-1403 | 1,658 | 1,666 |
| Not mapped to IL-1403 (no. of kb actually matching with >90% identity)b | 686 (163) | 655 (118) |
Approximately 200 to 250 kb in each genome was present in contigs of <1,000 nt and was not analyzed in detail.
Many nonmapped contigs actually contained additional segments that were highly matched (>90% identity) with the L. lactis IL-1403 genome (total matched kb in parentheses). These matches were not found by Projector 2 because the contigs either (i) contained repeats such as rRNA and IS elements (these are filtered out before mapping), (ii) contained a large deletion but were otherwise >90% identical to IL-1403 (exceeding maximum allowed length deviation), or (iii) contained a large insertion relative to IL-1403.
Sequence analysis.
Automatic open reading frame calling was performed with Glimmer (15) and Pedant-Pro (Biomax Informatics AG, Martinsreid, Germany). Annotation was first performed automatically using the Pedant-Pro integrated annotation package, which includes BLASTP/BLASTN analysis (1) and homology analysis against the protein family database PFAM (3) and the orthologous gene database COG (63). Protein and nucleotide sequences of the L. lactis strains KF147 and KF282 were compared to each other, to the published genomes of L. lactis IL-1403 (7) and L. lactis SK11 (39), to the ERGO genome database (http://ergo.integratedgenomics.com/ERGO), and to the GenBank nonredundant database of NCBI (ftp://ftp.ncbi.nih.gov/BLAST/db/FASTA/), using different types of homology tools such as BLAST and CD-hit from the Entrez package of NCBI (http://www.ncbi.nlm.nih.gov/entrez/). Sequence similarity was detected with BLAST, while multiple sequence alignments were made with Clustal W (64). Improved manual annotation was performed using PFAM (3, 21), InterproScan (49), and the ERGO Bioinformatics Suite (48). Carbohydrate-active enzymes were identified using the CAZy database (11) (http://www.cazy.org). Transmembrane helices were predicted with TMHMM 2 (35) and signal peptides with SignalP 3.0 (5, 45). Sortase-dependent LPxTG-type peptidoglycan anchors were searched for using a hidden Markov model (6).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of selected novel regions of L. lactis KF147 (about 250 kb; summarized in Table S1 in the supplemental material) have been submitted to the EMBL/GenBank/DDBJ databases and are available under accession numbers EU255902 to EU255918. Detailed annotation of the contigs is available in Table S2 in the supplemental material.
RESULTS AND DISCUSSION
Phenotypic analysis.
A variety of phenotypic properties of L. lactis strains IL-1403 (dairy), SK11 (cheese), KF147 (mung bean), and KF282 (mustard and cress) were analyzed and compared. Growth tests on various mono- and oligosaccharides showed that the plant isolates grew on a wider range of sugar substrates than either IL-1403 or SK11 did (Table 2). For instance, only the two plant isolates grew on l-arabinose, d-xylose, mannitol, sucrose, gluconate, or glucuronate, while strain KF147 was the only one to grow on melibiose, raffinose, or galacturonate. Furthermore, only the plant isolates grew on galacto-oligosaccharides and some fructans. These phenotypic traits were specifically targeted for genotypic analysis in the present study and in particular for strain KF147 since it apparently has the broadest substrate growth range.
TABLE 2.
Growth characteristics of L. lactis strains on carbohydratesa
| Carbohydrate |
L. lactis strain code (source)
|
Carbohydrate typeb | |||
|---|---|---|---|---|---|
| KF147 (mung bean) | KF282 (mustard/cress) | IL-1403 (dairy) | SK11 (dairy [cheese]) | ||
| Organic acid: citrate | − | − | − | − | Tricarboxylic acid |
| Mono/oligosaccharides | |||||
| d-Arabinose | − | − | − | − | Pentose |
| l-Arabinose | + | + | − | − | Pentose |
| Ribose | + | + | + | − | Pentose |
| d-Xylose | + | + | − | − | Pentose |
| l-Xylose | − | − | − | − | Pentose |
| Galactose | + | + | + | + | Hexose |
| Glucose | + | + | + | + | Hexose |
| Fructose | + | + | + | + | Hexose |
| Mannose | + | + | + | + | Hexose |
| Rhamnose | − | − | − | − | Hexose |
| Mannitol | + | + | − | − | Hexose |
| N-Acetylglucosamine | + | + | + | + | Hexose |
| Arbutin | + | + | + | − | Aryl-monosaccharide |
| Esculin | − | − | − | − | Aryl-monosaccharide |
| Salicin | + | + | + | − | Aryl-monosaccharide |
| Amygdalin | + | + | +/− | − | Aryl-disaccharide |
| Cellobiose | + | + | + | − | Disaccharide |
| Maltose | + | + | + | − | Disaccharide |
| Lactose | + | + | +/− | + | Disaccharide |
| Sucrose | + | + | − | − | Disaccharide |
| Trehalose | + | + | + | − | Disaccharide |
| Gentiobiose | + | + | + | + | Disaccharide |
| Melibiose | + | − | − | − | Disaccharide |
| Raffinose | + | − | − | − | Trisaccharide |
| Gluconate | +/− | + | − | − | Sugar acid |
| Glucuronate | + | + | − | − | Sugar acid |
| Galacturonate | + | − | − | − | Sugar acid |
| Polysaccharides | |||||
| Galacto-oligosaccharides (Vivinal, n > 1, no lactose) | + | +/− | − | − | Di- plus higher galacto-oligosaccharides |
| Galacto-oligosaccharides (Vivinal, n > 2, no lactose) | − | − | − | − | Tri- plus higher galacto-oligosaccharides |
| Fructan (garlic) | − | + | − | +/− | Branched inulin (β-2,1-fructan with β-2,6 branching) |
| Fructan (blue agave) | +/− | + | − | − | Branched inulin (β-2,1-fructan with β-2,6 branching) |
| Fructan (chicory, Frutafit IQ) | +/− | + | − | − | Linear inulin (β-2,1-fructan; avg DP, ca. 9) |
| Fructan (chicory, Frutafit TEX) | − | +/− | − | − | Linear inulin (β-2,1-fructan; avg DP, >23) |
| Levan (Bacillus subtilis) | − | +/− | − | − | β-2,6-Fructan with β-2,1 branching |
| Pectin (apple) | − | − | − | − | |
| Arabinogalactan | − | − | − | − | |
| β-Cyclodextran | − | − | − | − | |
| Inulin | − | − | − | − | |
| Starch | + | + | + | − | |
| Glycogen | − | − | − | − | |
Partially based on API test described in reference 50. Symbols: −, no growth (API score = 0 to 1); +/−, limited growth (API score = 2 to 3); +, good growth (API score = 4 to 6). In addition, all strains do not grow on glycerol, erythritol, adonitol, dulcitol, inositol, sorbitol, xylitol, d-arabitol, l-arabitol, α-methyl-d-mannoside, α-methyl-d-glucoside, β-methyl-d-xyloside, melezitose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, sorbose, 2-ketogluconate, and 5-ketogluconate.
DP, degree of polymerization.
Very few phenotypic differences were found between the four L. lactis strains in their resistance/sensitivity to various antibiotics (chloramphenicol, erythromycin, tetracycline, vancomycin, ciprofloxacin, neomycin, penicillin G, and trimethoprim) or metal ions (arsenate, arsenite, Cd, Hg, Cr, Cu, Mn, and Pb) (see Table S3 in the supplemental material). Strain SK11 was generally most sensitive to antibiotics, whereas strain KF282 was significantly most resistant to arsenate/arsenite. The latter resistance can be explained by putative plasmid-located genes for arsenate/arsenite resistance (arsR-arsD-arsA) in strain KF282 (see below). Therefore, no further search was done for differences in genotype relating to such resistances, with the exception of resistance to nisin, which is described separately below.
Sequencing and comparative analysis of L. lactis genomes.
A low-coverage, diagnostic sequencing of the genomes of L. lactis strains KF147 and KF282 was performed (Table 1). The assembled contigs suggest that the sizes and G+C contents of the two genomes are similar (∼2.5 to 2.6 Mb and 34.9%, respectively) and comparable to those of L. lactis strains IL-1403 (2.365 Mb and 35.4%, respectively) (7), SK11 (2.438 Mb and 35.8%, respectively) (39), and MG1363 (2.529 Mb and 35.8%, respectively) (71). The initial BLAST analysis showed that the large majority of contigs of both L. lactis plant isolates were highly similar to one another (>95% nucleotide sequence identity) and to the IL-1403 sequence (>95% identity) and that the genes showed conserved gene order. Therefore, contigs larger than 1,000 nt were first mapped onto the IL-1403 template genome using Projector 2.0 (for examples, see Fig. S1A and S1B for KF147 and KF282, respectively, in the supplemental material). This mapping provided a fast first estimate of genome coverage, contig order, gap sizes, and possible positions of large insertions and deletions relative to IL-1403.
Closer inspection showed that many unmatched contigs actually contained additional large segments that were highly matched (>90% identity) to the IL-1403 genome. These matches were not found by Projector because the contigs either (i) contained repeats (these are filtered out before mapping), (ii) contained a large deletion but were otherwise >90% identical to IL-1403 (exceeding maximum allowed length deviation in Projector), or (iii) contained a large insertion relative to IL-1403. These partially matching contigs provided further clues for positions of insertions and deletions. Together, at least 1.8 Mb (>75%) of the genome of each plant isolate was found to match the IL-1403 genome, and the majority of these matches were found in both plant isolates.
To characterize genotypic differences between plant and dairy strains in more detail, we subsequently focused on large regions/clusters that were absent on the contigs from strain KF147 or KF282 compared to strain IL-1403 and on functions encoded on contigs that did not map to IL-1403 (Table 3). Since these regions were highly similar in strains KF147 and KF282, we performed high-quality resequencing only of the relevant regions of the KF147 genome.
TABLE 3.
Main differences in predicted functionalities encoded in gene clusters of L. lactis genomes
| Function | Result for strainh:
|
|||
|---|---|---|---|---|
| KF147 | KF282 | IL-1403 | SK11 | |
| Citrate/malate utilization | NO | NO | YESa | NO |
| Lactose/d-galactose utilization | yes | yes | yes | yesb |
| Sucrose utilization | YES | YES | NO | NO |
| Arabinose utilizationg | YES | YES | NO | NO |
| Xylose metabolism | YES | YES | YESe | NO |
| α-Galactoside utilizationg | YES | NO | NO | NO |
| β-Glucoside utilizationg | extrac | extrac | yes | yes |
| Galacturonate utilizationg | YES | NO | NO | NO |
| Xylan breakdowng | yes | yes | no | yes |
| Glucan breakdowng | yes | no | no | no |
| Arabinan breakdown | yes | no | no | no |
| Fructan/mannan breakdowng | YES | YES | NO | NO |
| Iron-siderophore transporterg | yes | yes | no | no |
| Polyamine ABC transporter | extrac | extrac | yes | yes |
| Potassium-transporting ATPaseg | yes | yes | no | no |
| Nisin biosynthesis | YESd | YES | NO | NO |
| Nisin resistance | YES | YES | NO | NO |
| NRPS | yes | yes | no | no |
| EPS/TA biosynthesisg | differentf | differentf | differentf | differentf |
| Prophages | differentf | differentf | differentf | differentf |
| Plasmids | differentf | differentf | no | differentf |
IL-1403 does not grow on citrate since the plasmid containing the citrate permease gene citP has been cured.
Plasmid encoded.
Extra genes with functions similar to those of genes in strain IL-1403.
Nisin biosynthesis genes present but nonfunctional due to internal deletion.
Xylose utilization gene cluster is present but inactive due to deletion.
Functionality is present, but the genes and gene clusters are quite different from each other.
The gene clusters identified in strain KF147 are shown in Fig. 1 to 4, and their sequences have been submitted to GenBank.
Italicized uppercase shows agreement with phenotype (see Table S2 in the supplemental material); roman uppercase shows absence of agreement with phenotype; roman lowercase indicates that the phenotype was not tested.
Genes/gene clusters absent or different in plant L. lactis strains.
The number of genes and functions of strain IL-1403 that appear to be absent in the genomes of strains KF147 and KF282 is quite limited; the main differences, which are due to the absence of large continuous regions, are described below and summarized in Table 3.
(i) Citrate/malate utilization.
Both plant L. lactis isolates (and strain SK11) lack a segment of about 21 kb corresponding to genes mae to ymcE of strain IL-1403 encompassing the complete citrate/malate utilization gene cluster mae-maeP- citRCDEFXG, followed by genes encoding many hypothetical proteins and transposases. This suggests that strains KF147, KF282, and SK11 cannot cometabolize citrate/malate, a trait that is typical only of dairy Lactococcus lactis subsp. diacetylactis strains. Indeed, these strains were unable to ferment citrate (Table 2).
(ii) TA biosynthesis.
Extracellular polysaccharides (EPS) and (lipo)teichoic acids (TA) of bacteria could play a role in attachment to host surfaces and interactions with other bacteria in biofilms. Both plant L. lactis isolates (and strain SK11) lack a segment of about 20 kb corresponding to genes yjdE to tagF of IL-1403, encompassing a large gene cluster for TA biosynthesis (tagRLHGZYXDF). However, different TA biosynthesis clusters appear to be inserted in the equivalent positions in the genomes of the L. lactis plant isolates (see below).
(iii) Transposable elements.
Most prophages of strain IL-1403 (i.e., pi1, pi2, pi3, ps1, ps2, and ps3) are found to be missing in the genomes of both L. lactis KF147 and L. lactis KF282. In some cases other prophages may be inserted in equivalent positions (see below). Numerous IS elements of strain IL-1403 were not found in the plant L. lactis isolates, as flanking genes separated by these IS elements in IL-1403 were often directly adjacent in strains KF147 and KF282. Also absent in these plant-derived strains is a fragment of about 17 kb, encoding an integrase (XerD), a type I restriction-modification system (HsdRMS), transposases, and many hypotheticals, located in strain IL-1403 between the eno and ygfA genes (which are adjacent in strain KF147).
Novel genes/gene clusters in plant L. lactis strains: complex sugar metabolism.
Plant polysaccharides can be divided into structural carbohydrates (cellulose, xylan, pectin, and arabinan) and storage carbohydrates (sucrose, starch, and fructan). A variety of genes and gene clusters were identified in the genomes of the L. lactis plant isolates that encode proteins assumed to be involved in utilization of plant polysaccharides for growth. The main examples are described below and summarized in Table 3. In addition, a comparison was made of predicted carbohydrate-active enzymes (glycoside hydrolases and esterases) in the genomes of L. lactis strains KF147, KF282, IL-1403, and SK11 based on the CAZy database families (11), as summarized in Table 4.
TABLE 4.
Putative numbers of encoded glycoside hydrolases and carbohydrate esterases in L. lactis genomes
| Subfamilya | No. of encoded enzymes in strain genome
|
Annotation | EC no. | |||
|---|---|---|---|---|---|---|
| KF147 | KF282 | IL-1403 | SK11 | |||
| Glycoside hydrolaseb | ||||||
| 1 | 9 | 10 | 6 | 8 | Beta-glucosidase/phospho-beta-glucosidase | EC 3.2.1.21/3.2.1.86 |
| 2 | 1 | 1 | 1 | Beta-galactosidase | EC 3.2.1.23 | |
| 3 | 1 | 1 | 1 | 1 | Glycosidase | EC 3.2.1.-c |
| 5 | 1 | Endoglucanase | EC 3.2.1.- | |||
| 8 | 2 | 1 | 1 | Endoglucanase/endoxylanase | EC 3.2.1.- | |
| 11 | 1 | Xylanase | EC 3.2.1.8 | |||
| 13 | 1 | 1 | 1 | Alpha-1-6-glucosidase | EC 3.2.1.70 | |
| 13 | 1 | 1 | 1 | 1 | Amylopullulanase | EC 3.2.1.41 |
| 13 | 1 | 1 | 1 | 1 | Exo-alpha-1,4-glucosidase | EC 3.2.1.20 |
| 13 | 2 | 2 | 2 | 1 | Alpha-amylase | EC 3.2.1.1 |
| 13 | 1 | 1 | 1 | 1 | Oligo-1,6-glucosidase | EC 3.2.1.10 |
| 13 | 1 | 1 | 1 | 1 | Neopullulanase | EC 3.2.1.135 |
| 13 | 1 | Sucrose phosphorylase | EC 2.4.1.7 | |||
| 13 | 1 | 1 | 1 | 1 | 1,4-Alpha-glucan branching enzyme | EC 2.4.1.18 |
| 18 | 1 | 1 | 1 | 1 | Chitinase | EC 3.2.1.14 |
| 20 | 1 | 1 | 1 | Lacto-N-biosidase | EC 3.2.1.52 | |
| 23 | 1 | 1 | 1 | 1 | Peptidoglycan lytic transglycosylase | EC 3.2.1.- |
| 25 | 4 | 4 | 1 | 4 | N-Acetylmuramoyl-l-alanine amidase/lysozyme | EC 3.5.1.28/3.2.1.17 |
| 31 | 1 | 1 | Alpha-glucosidase | EC 3.2.1.20 | ||
| 32 | 1 | Sucrose-6-phosphate hydrolase | EC 3.2.1.26 | |||
| 36 | 1 | Alpha-galactosidase | EC 3.2.1.22 | |||
| 38 | 2 | 2 | 1 | Alpha mannosidase/alpha-fructosidase | EC 3.2.1.24? | |
| 43 | 2 | 1 | 1 | 1 | Beta-xylosidase/alpha-l-arabinofuranosidase | EC 3.2.1.37/3.2.1.55 |
| 43 | 1 | 1 | Ribulokinase? | EC 2.7.1.16 | ||
| 65 | 1 | 1 | 1 | 1 | Trehalose 6-phosphate phosphorylase | EC 2.4.1.216 |
| 65 | 1 | 1 | 1 | 1 | Maltose phosphorylase | EC 2.4.1.8 |
| 67 | 1 | 1 | 1 | Alpha-glucuronidase | EC 3.2.1.139 | |
| 73 | 4 | 4 | 4 | 4 | N-Acetylmuramidase | EC 3.5.1.28 |
| 77 | 1 | 1 | 1 | 1 | 4-Alpha-glucanotransferase | EC 2.4.1.25 |
| 85 | 1 | 1 | 1 | Endo-beta-N-acetylglucosaminidase | EC 3.2.1.96 | |
| 92 | 1 | 1 | 1 | Alpha-1,2-mannosidase | EC 3.2.1.- | |
| Carbohydrate esterase | ||||||
| 1 | 1 | 1 | 1 | 1 | Acetyl esterase | EC 3.1.1.- |
| 4 | 3 | 4 | 3 | 4 | Polysaccharide deacetylase | EC 3.1.1.-/3.5.1.- |
| 7 | 1 | 1 | 1 | Acetyl xylan esterase | EC 3.1.1.- | |
| 9 | 1 | 1 | 1 | 1 | N-Acetylglucosamine-6-phosphate deacetylase | EC 3.5.1.25 |
| 10 | 1 | 1 | 1 | 1 | Acetyl esterase | EC 3.1.1.- |
| ? | 1 | 1 | Sialic acid-specific 9-O-acetylesterase | EC 3.1.1.53 | ||
Subfamilies were derived from the CAZy database.
Members of subfamilies 1, 25, and 43 are not always orthologs.
In an EC number, the hyphen indicates that the full number has not been assigned yet.
(i) Xylan/xylose degradation.
Xylan is the main component of hemicellulose and the second most abundant component of plant materials. Xylans are complex structures requiring many enzymes acting synergistically for breakdown (4, 10, 16, 17, 37). Xylan is composed of β-1,4-linked d-xylose units which can be substituted with different side groups such as l-arabinose, d-galactose, acetyl, feruloyl, p-coumaroyl, or glucuronic acid. Xyloglucans are dominant hemicellulosic polysaccharides, consisting of a d-glucose backbone substituted by d-xylose. In gram-positive bacteria, xylan/xylose utilization genes have been described for several bacilli (4, 10, 37, 57). The key enzyme in the degradation of xylan is an extracellular endo-1,4-β-xylanase (xynA). This enzyme releases short xylose oligomers (xylobiose, xylotriose, and xylotetraose) which can be substituted with various side chains such as l-arabinose, d-glucuronic acid, or its 4-O-methyl ether. Such substituted xylose oligomers are then cleaved by α-glucuronidase and α-l-arabinofuranosidase to yield xylose oligomers, d-glucuronic acid, and arabinose, while the xylo-oligomers are hydrolyzed to xylose by β-xylosidase. Xylose is converted into xylulose-5-phosphate, which can enter the pentose phosphate cycle.
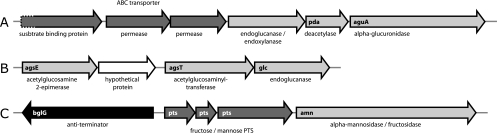
Both plant L. lactis strains have (i) a cluster of six genes encoding endoxylanase, polysaccharide deacetylase, α-glucuronidase (aguA), and a sugar ABC transporter (Fig. 1A); (ii) a separate gene for acetyl-xylan esterase (Table 4); and (iii) a cluster of three genes encoding a putative ABC transporter for (substituted) xylo-oligosaccharides (see Tables S1 and S2 in the supplemental material). The last transporter is very similar to a Bacillus clausii gene cluster that is directly downstream of xylan utilization genes, encoding xylan α-1,2-glucuronidase and β-xylosidase (Y. Takaki et al., unpublished data; accession code NC_006582). Both plant L. lactis strains also have uptake and utilization gene clusters for the breakdown products xylose and d-glucuronate (see below).
FIG. 1.
Novel gene clusters for polysaccharide breakdown identified in L. lactis KF147. (A) Xylan breakdown; (B) glucan breakdown; (C) fructan/mannan breakdown. Color coding: light gray, enzyme; dark gray, transport; black, regulation; white, hypothetical/other. For detailed annotation see Table S2 in the supplemental material. Contigs are 1963_z898 (A), y771 (B), and x474 (C).
(ii) Starch breakdown.
Starch consists of amylose and amylopectin. Both contain glucose residues that are mainly linked to each other via α(1>4) bonds. Amylose is linear with few branches, while amylopectin has many branches. All four L. lactis strains contain the same large cluster (mapA-agl-amyY-maa-dexA-dexC-malEFG) for starch breakdown (by α-glucosidases and α-amylases) to dextrin and maltose and subsequent uptake (maltose transporter) and conversion. Only SK11 does not grow on starch, but it is not clear why, since there is no apparent gene inactivation.
(iii) Glucan breakdown (e.g., cellulose and xyloglucan).
Strain KF147 has a unique cluster of four genes encoding putative a UDP-N-acetyl-glucosamine-2-epimerase, a hypothetical membrane protein, an N-acetyl-glucosaminyl-transferase, and an endo-1,4-beta-glucanase/endocellulase of glycoside hydrolase family 8 (Fig. 1B). The best homologs (up to 66% identity) are encoded in similar conserved clusters in Streptococcus mutans and in Lactobacillus acidophilus group genomes. The putative function of this cluster could be breakdown of complex glucans by removal of N-Ac-glucosamine side chains and cleavage of the main glucan backbone.
(iv) Fructan/mannan breakdown.
The phenotypic assays show that some fructans can be degraded by one or both plant L. lactis strains (Table 2). Fructans are the dominant carbohydrate reserve in many plants. Fructans such as inulin, levan, and graminan are polymers of fructose. Some plants cell walls contain galactomannans and glucomannans (including mung bean [18]), which are polymers of primarily mannose (α-d-mannosides). Degradation of mannans to d-mannose can be followed by conversion to d-fructose by mannose isomerase (EC 5.3.1.7; no known sequences). Both KF147 and KF282 strains have a gene cluster encoding either a mannose or a fructose phosphotransferase system (PTS; three genes), a transcriptional antiterminator (BglG family), and a putative α-mannosidase/α-fructosidase of glycosyl hydrolase family 38 (Fig. 1C). This cluster is not present in other L. lactis genomes, but it is highly similar (40 to 60% amino acid identity) to gene clusters in Clostridium difficile and Listeria genomes. These proteins could contribute to degradation of fructans and/or mannans and uptake of monosaccharides for growth.
(v) Side chain removal.
Strain KF282 and strain SK11 encode a putative sialic acid-specific 9-O-acetylesterase (EC 3.1.1.53), which is not present in the genome of strain KF147 or IL-1403. The putative function of this enzyme is to cleave acetyl groups off sialic acid (=N-Ac-neuraminic acid) side chains of glycans. The best homologs are found in Bacteroides, Bifidobacterium, Caldicellulosiruptor, Pseudoalteromonas, and Flavobacterium species; in other genomes, including L. lactis SK11, this gene is typically located in a gene cluster encoding glucan/xylan-degrading enzymes.
Novel genes/gene clusters in plant L. lactis strains: simple sugar metabolism. (i) Arabinose metabolism.
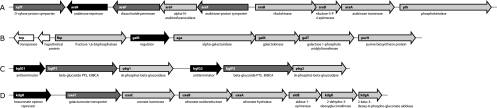
Arabinose is a major sugar moiety in various hemicellulosic and pectic plant polysaccharides, such as arabinan (homoglycan), arabinoglycan, and arabinoxylan. Both plant L. lactis strains, KF147 and KF282, grow on l-arabinose, in contrast to L. lactis strains IL-1403 and SK11 (Table 2). A novel arabinose transport and metabolism gene cluster, araRPFTBDA, was found in both plant strains but not in IL-1403 and SK11 (Fig. 2A). These genes encode proteins for uptake of arabinose (and possibly oligomers) and conversion to d-xylulose-5P, which can enter the pentose phosphate pathway. The cluster also encodes a glycosyl hydrolase family 43 protein; this family contains members that are arabinanases or xylanases (http://www.cazy.org). Arabinanases hydrolyze the α-1,5-linked l-arabinofuranoside backbone of plant cell wall arabinans. Compared to strain IL-1403, this gene cluster is found to be inserted between the xylT and ptk genes (Fig. 2A), which are adjacent in IL-1403. Arabinose operons have not been described before in L. lactis, but they do occur in lactobacilli and bacilli (54). L. lactis strain KF147 also encodes an extra glycosyl hydrolase family 43 protein (Table 4). The best homologs (30 to 40% sequence identity) of this KF147 glycosyl hydrolase are found in Deinococcus, Myxococcus, and Bacteroides species, in which they are generally encoded together with other enzymes for xylan or arabinan degradation.
FIG. 2.
Novel gene clusters for sugar metabolism identified in L. lactis KF147. (A) Arabinose metabolism; (B) α-galactoside metabolism; (C) β-glucoside metabolism; (D) galacturonate metabolism. Color coding: light gray, enzyme; dark gray, transport; black, regulation; white, hypothetical/other. For detailed annotation see Table S2 in the supplemental material. Contigs are z738_873 (A), z563tr (B), z439 (C), and x474 (D).
(ii) Xylose metabolism.
Both plant strains grow on d-xylose, in contrast to strains IL-1403 and SK11 (Table 2). Both plant L. lactis strains and IL-1403 have a d-xylose proton symporter, encoded by xylT, but the xylT of IL-1403 could be nonfunctional since it is C-terminally truncated. Perhaps the arabinose transporters AraT and AraP of L. lactis strains KF147 and KF282 also contribute to xylose uptake, as both pentoses are structurally very similar, but as yet there is no experimental evidence for this.
(iii) α-Galactoside metabolism.
α-Galactosides such as stachyose, raffinose (=melitose), and melibiose are typical plant oligosaccharides. α-Galactosidase (encoded by gene aga) is required to cleave off terminal d-galactose moieties. Only strain KF147 was found to grow on α-galactosides (raffinose and melibiose were tested), in contrast to the other L. lactis strains (Table 2). In agreement with this phenotype, only strain KF147 was found to have a gene cluster for α-galactoside breakdown and subsequent d-galactose conversion by the Leloir pathway (Fig. 2B). This α-galactoside gene cluster, fbp-galR-aga-galK-galT, is highly similar (90 to 94% nucleotide identity) to that of Lactococcus raffinolactis ATCC 43920 (GenBank accession code AY164273.1) (8). The fbp, galK, and galT genes each have a paralog in strain KF147, each of which is also present in other L. lactis strains.
(iv) β-Glucoside metabolism.
β-Glucosides like cellulose, cellobiose, β-1,3-1,4-glucans (lichenin), and aryl-β-glucosides (e.g., arbutin, salicin, and amygdalin) are important components of plant cell walls. The L. lactis plant isolates KF147 and KF282 grow well on several of these β-glucosides (Table 2), and both genomes encode more β-glucosidases than strains IL-1403 and SK11 do (Table 4). In particular, an additional cluster of six genes is found encoding two sets of (6-P)-β-glucosidase, β-glucoside PTS, and a transcriptional antiterminator (Fig. 2C), all of them most similar to Listeria and Clostridium genes.
(v) Uronic acid metabolism.
Some strains of Bacillus subtilis can use glucuronate and galacturonate as primary carbon sources (42). All four L. lactis strains have a gene cluster, kdgR-uxuB-uxuA-uxuT-hypAE-uxaC-kdgK-kdgA, for uptake and degradation of d-glucuronate (building block of xylan), yet only the two plant isolates were observed to grow on glucuronate (Table 2).
Strain KF147 is found to have an additional gene cluster, exuR-uxaT-uxaC-uxaB-uxaA-hypAE-kdgK2-kdgA2, for uptake and degradation of d-galacturonate, the building block of pectin (Fig. 2D). Pectins are a family of complex polysaccharides containing 1,4-linked galacturonate moieties, with possible variations in the backbone (rhamnose units) or side chain substitutions (53, 72). Pectins can be degraded by extracellular pectate lyases and pectinases (polygalacturonase and pectin esterase), converting pectin into oligogalacturonate and subsequently into d-galacturonate (17). These pectinolytic enzymes were not found in the genomes of strains KF147 and KF282 or other lactococci. This galacturonate utilization cluster has not been described before in lactic acid bacteria and is most similar to gene clusters from Clostridium acetobutylicum, Flavobacterium johnsoniae, and Bacillus licheniformis; in the genomes of B. licheniformis and F. johnsoniae this cluster is linked to a gene for polygalacturonase (EC 3.2.1.15) for pectate degradation. Only strain KF147 grows well on d-galacturonate, in agreement with the genotype, suggesting that it can grow on pectin breakdown products generated by other bacteria or fungi on the plant surface.
(vi) Gluconate metabolism.
Both plant L. lactis strains, KF147 and KF282, grow on gluconate, in contrast to strains IL-1403 and SK11 (Table 2). The plant strains both have a gene cluster encoding a gluconate transporter (gntP), gluconate kinase (gntK) (EC 2.7.1.12), and 6-phosphogluconate dehydrogenase (EC 1.1.1.44). The same genes are found in the genomes of L. lactis IL-1403 and SK11 but may be nonfunctional, since the gntP gene of strain IL-1403 appears to be interrupted by a frameshift and/or transposase and the SK11 GntP transporter appears to be truncated at its C terminus.
(vii) Sucrose metabolism.
Both plant L. lactis isolates grow on sucrose, in contrast to strains IL-1403 and SK11. Only the KF147 and KF282 strains are found to have the sacBK and sacAR operons encoding a sucrose PTS, fructokinase, sucrose-6-phosphate hydrolase (EC 3.2.1.26), and a sucrose regulator, with >95% identity to the proteins encoded by the nisin-sucrose conjugative transposon of L. lactis NZ9800 (38) (data not shown). In addition, only strain KF147 encodes a putative sucrose phosphorylase (EC 2.4.1.7; GtfA), not found before in L. lactis strains, with the best homologs (65 to 70% identity) in Leuconostoc species, Listeria species, lactobacilli, and streptococci. The presence of this sucrose phosphorylase suggests that there should also be a permease for uptake of sucrose.
Nisin production/resistance.
Both plant strains are much more resistant than strains IL-1403 and SK11 to the lantibiotic peptide nisin; strains KF147 and KF282 survived >2,000 ng/ml nisin, while IL-1403 and SK11 survived at maximally 125 and 25 ng/ml, respectively. In a nisin biosynthesis assay, only strain KF282 showed a high growth inhibition of the indicator strain. The gene cluster nisZABTCIPRKFEG for biosynthesis of nisin Z and immunity to nisin Z (19, 28, 36) is found to be present in both KF147 and KF282, in contrast to strains IL-1403 and SK11. However, strain KF147 clearly has an internal deletion in parts of the nisB and nisC genes, which should lead to inactivation of the encoded nisin biosynthetic enzymes. Hence, the genotype predicts that both plant strains should be resistant to nisin (presence of immunity genes) but that only strain KF282 should be a nisin producer. This genotype agrees completely with the phenotypes found by us and Kelly et al. (31).
Survival/stress response. (i) NRPS.
Several contigs were identified in both L. lactis strain KF147 (total of ∼40 kb; data not shown) and L. lactis strain KF282 encoding enzymes which are characteristic of nonribosomal peptide or polyketide biosynthesis, such as phospho-pantetheine protein transferase, thioesterase, and multimodular peptide synthetases with adenylation, condensation, and acyl carrier protein domains (20, 70). Such systems are commonly found in environmental bacteria such as Bacillus, Pseudomonas, and Streptomyces spp., where they can play a role in survival, defense, signaling, or adhesion (20). This is the first identification of such a system in Lactococcus lactis. Studies are under way to sequence the complete gene clusters and identify their nonribosomal peptide synthesis (NRPS)/polyketide synthesis products.
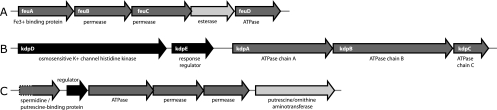
A preliminary analysis shows that many predicted open reading frames had the best sequence identity (up to 40%) to Bacillus biosynthesis systems for bacitracin, bacillibactin, or bacillomycin (data not shown). The nonribosomally synthesized peptide of L. lactis could be involved in plant interactions (defense or adhesion) or could be a siderophore for binding of Fe ions (12). The production of low-molecular-weight Fe(III) chelators (siderophores) enables microorganisms to efficiently scavenge iron even in aerobic environments where iron exists primarily as insoluble hydroxides (47). Both plant L. lactis strains also have a unique cluster of five genes (Fig. 3A) encoding a FeuABC-like transporter, most similar (33 to 53% identity) to the B. subtilis uptake system for the catecholate siderophores enterobactin and bacillibactin (41, 44), supporting the hypothesis that the product of the L. lactis NRPS system could be a siderophore. Alternatively, the NRPS product could be a lipopeptide surfactant that promotes biofilm formation, like surfactin produced by Bacillus (34, 73).
FIG. 3.
Novel gene clusters for transporters identified in L. lactis KF147. (A) ABC-type iron-siderophore transporter; (B) high-affinity potassium-transporting ATPase; (C) spermidine/putrescine ABC transporter. Color coding: light gray, enzyme; dark gray, transport; black, regulation. For detailed annotation see Table S2 in the supplemental material. Contigs are x180tr (A), x2018tr (B), and x159 (C).
(ii) Cation transporters.
Both plant L. lactis strains have a kdpDEABC gene cluster encoding the high-affinity ATP-driven K+ transport system Kdp (potassium-transporting ATPase) and its two-component regulator KdpDE (Fig. 3B). This system has not been found before in L. lactis and has best hits to Listeria innocua (52 to 74% identity). In Escherichia coli, KdpDE regulates expression of the kdpFABC operon, which encodes the high-affinity K+ transport system KdpFABC. At low potassium concentrations or by an increase of the osmolality of the medium, the sensor kinase KdpD autophosphorylates itself and transfers the phosphoryl group to the cytoplasmic response regulator KdpE (23, 24), which in its phosphorylated form induces the expression of the kdpFABC operon (29). Both plant L. lactis strains also appear to have an NhaP-type Na+/H+ and K+/H+ antiporter, most similar (∼50% identity) to those in lactobacilli but not found previously in L. lactis. This antiporter could play a role in pH regulation and osmoregulation.
(iii) Polyamine ABC transporter.
Polyamines (putrescine, spermidine, and spermine) are necessary for cell growth (9, 27, 61). They are among the major polycations in cells, together with Ca2+ and Mg2+. Polyamines and Mg2+ can bind to intracellular polyanions such as nucleic acids and ATP to modulate their function. A PotABCD spermidine/putrescine uptake system is present in L. lactis IL-1403 and SK11 and in both plant L. lactis genomes, with 99% identity. A second spermidine/putrescine ABC transporter together with a putrescine/ornithine aminotransferase is found only in both plant L. lactis strains, with >80% sequence identity (Fig. 3C). This system is most similar (44 to 55% identity) to PotABCD of Clostridium beijerincki and various bacilli.
Cell envelope.
The physicochemical environment in the natural habitat of L. lactis plant isolates is very different from that of dairy isolates. Plant-associated bacteria are often found in biofilms, using EPS and large proteins to adhere to plants (14). As a consequence, major adaptations in gene sets for biosynthesis of components involved in the physical interaction with the extracellular space are expected, such as in cell-bound polysaccharides, TA, and proteins.
(i) EPS/TA biosynthesis.
Both plant L. lactis strains have several large contigs (and scaffolds) with putative EPS/TA biosynthesis genes, in particular strain KF147, which has three large clusters totaling over 42 kb (see Tables S1 and S2 in the supplemental material). The genes in these clusters show highest sequence similarity to L. lactis, Streptococcus thermophilus, Streptococcus pneumoniae, and Enterococcus faecalis EPS/TA clusters, but gene order in these clusters is generally very different. The only exception appears to be the epsXABCD gene cluster, which is highly similar (>90% identity at amino acid sequence level) to that of L. lactis plasmid pNZ4000 (69). These epsXABCD genes in L. lactis KF147 are part of a larger EPS biosynthesis gene cluster of ∼13 kb (Fig. 4) that is flanked by two small integrase gene fragments and appears to be inserted in the chromosome between the ybeF and tgt genes compared to L. lactis IL-1403 (see Table S2 in the supplemental material). A TA biosynthesis gene cluster of ∼22 kb is present in strain KF147 in the same position as the TA cluster in the IL-1403 genome (between genes yjdF and deoB), but the composition and sequence similarity of these clusters differ considerably (see Fig. S2 and Table S2 in the supplemental material).
FIG. 4.
Novel gene cluster for EPS biosynthesis identified in L. lactis KF147. Color coding: light gray, enzyme; dark gray, transport; black, regulation. Polysaccharide biosynthesis proteins are presumed to be enzymes. For detailed annotation see Table S2 in the supplemental material. The contig is x896.
(ii) Extracellular proteins.
Several differences were found in the presence of encoded extracellular proteins of the L. lactis strains. Extracellular proteins of gram-positive bacteria are often large multidomain proteins, with a C-terminal, sortase-dependent LPxTG-type peptidoglycan anchor (6). The intercellular adhesion gene cluster icaABC of strain IL-1403, encoding a putative N-acetyl-glucosaminyltransferase, a polysaccharide deacetylase, and a collagen adhesion protein, respectively, is absent in strain KF147 but present in KF282. IcaC of strain KF282 is an extracellular peptidoglycan-bound protein since it has a signal peptide and an LPxTG-type anchor. Analogously, the gene cluster yoiABC, possibly of similar function since it also encodes a glycosyl transferase and a large LPxTG-anchored extracellular protein (YoiC, 1,441 residues), is also shared by IL-1403 and KF282 but absent in KF147. Another large LPxTG-type extracellular protein, YihD (1,063 residues), with multiple serine-rich repeats is also absent only in strain KF147.
In contrast, the genomes of both L. lactis plant isolates are found to encode two novel putative extracellular proteins with an LPxTG anchor and a so-called collagen-binding domain, which we call LpxA (822 residues) and LpxB (790 residues; two paralogs found in strain KF282). Recently an LpxA ortholog of 815 residues (83% identity) has been identified in L. lactis MG1363 (71), and part of the lpxA gene has also been found in L. lactis MG1614 (51), but no homolog of LpxB has been identified in other bacteria.
These differences in extracellular proteins may relate to differences in niche interactions, e.g., biofilm formation and binding to plant hosts.
Transposable elements. (i) Phages.
While several phages of L. lactis IL-1403 are not found in the plant L. lactis strains (see above), numerous other putative phage-encoding genes were identified in both plant strains, with the best homologs either in other L. lactis strains or in other microorganisms (data not shown). Particularly striking is the presence in L. lactis KF282 of some new (pro)phages resembling known phages of Streptococcus pyogenes and Lactobacillus johnsonii. A gene cluster equivalent to prophage Lj965 of L. johnsonii NCC533 genes LJ0307 to LJ0324 (21.5 kb) was identified with conserved gene order and 24 to 54% amino acid sequence identity, as well as a gene cluster equivalent to phage SSI-1 of S. pyogenes serotype M3 genes SPs1133 to SPs1144 (8.2 kb) with conserved gene order and 26 to 59% amino acid sequence identity (data not shown).
(ii) Plasmids/transposons.
Both L. lactis plant isolates have putative plasmids or transposons, based on the presence of characteristic functions encoded on several contigs, e.g., replication, partitioning (ParA-ParB), conjugation protein, recombinase, excisionase, transposon protein, transfer protein, transposase, and IS elements (data not shown). The total size of the putative plasmids is estimated to be between 50 and 70 kb in each strain, with the largest continuous assembled fragment of 25 kb found in strain KF147; sodium dodecyl sulfate-polyacrylamide gels show at least one dominant band of >20 kb for strain KF147 (data not shown). Several of these putative plasmid fragments of the two strains are highly similar to each other, suggesting that the two strains have similar plasmids. Other putative plasmid-encoded functions on these contigs are, e.g., arsenate/arsenite resistance (arsR-arsD-arsA in strain KF282), abortive infection (abiN), metal-transporting ATPase, SOS response (umuC), cell division (ftsK/spoIIIE), and cell surface proteins.
Different parts of the nisin conjugative transposon were identified, in both strain KF147 and strain KF282 (see above), but these regions were not yet sequenced in detail to assess the completeness of the transposon.
Evolutionary aspects.
One of the questions that now arises is whether these unique genes in L. lactis plant isolates were acquired by recent horizontal gene transfer or whether they are more ancient, having been lost more recently in dairy isolates as an adaptation to the nutrient-rich milk environment, such as found for the yoghurt bacteria Streptococcus thermophilus (7) and Lactobacillus bulgaricus (66). To address this question, we calculated the G+C content of the unique genes and gene clusters of L. lactis KF147 and compared them to their best BLAST hits, as summarized in Table 5 (more details of individual genes can be found in Table S2 in the supplemental material). Strikingly, nearly all the unique gene clusters have a G+C content close to or slightly lower than the average G+C content of 35 to 36% for L. lactis genomes. In contrast, their best BLAST hits generally have a higher G+C content, with the exception of the clostridial best hits, which have a lower average G+C content, in all cases closer to the average G+C content for their own genomes (Table 5 footnote). This strongly suggests that most of these “unique” gene clusters of plant isolates are actually more ancient and were lost in many dairy isolates. This hypothesis is supported both by the fact that most of the best BLAST hits are from phylogenetically related Lactobacillales (see, for instance, Fig. 5 of reference 40), indicative of presence in a common ancestor, and by the fact that some gene clusters are still found in L. lactis subsp. cremoris strain SK11 or MG1363, but not in L. lactis subsp. lactis IL-1403 (Table 5).
TABLE 5.
Evolutionary relatedness of extra regions in L. lactis KF147a
| Contig | Gene number(s) | % GC (avg) | Main function(s) | Best BLAST hit(s)b | Hit GC% (avg) | % Amino acid identity |
|---|---|---|---|---|---|---|
| z738_873 | 8 to 14 | 36.3 | Arabinose metabolism | Enterococcus faecium, Lactobacillus brevis, Lactobacillus sakei, Bacillus coagulans, Oceanobacillus | 42.2 | 58-79 |
| z563tr | 3 to 7 | 43.5 | α-Galactoside metabolism | L. raffinolactis | 42.2 | 94-97 |
| 2 | 45.0 | Purine biosynthesis protein PurH | S. pneumoniae | 47.1 | 82 | |
| 1 | 35.3 | Msm operon regulatory protein | S. mutans | 33.6 | 39 | |
| z439 | 6 to 11 | 34.0 | β-Glucoside metabolism | C. difficile, C. beijerincki, Streptococcus suis, S. mutans, Listeria monocytogenes, Erwinia carotovora | 37.7 | 37-67 |
| 2 to 5 | 30.2 | Various | C. acetobutylicum, Oenococcus oeni, Lactobacillus salivarius | 35.5 | 35-55 | |
| x474 | 3 to 10 | 36.3 | Galacturonate metabolism | L. brevis, Leuconostoc mesenteroides, E. faecium, L. lactis MG1363/SK11/IL-1403, B. coagulans | 40.8 | 45-79 |
| 11 to 16 | 33.0 | Fructan/mannan breakdown | C. difficile | 29.6 | 27-58 | |
| 1963_z898 | 1 to 8 | 35.6 | Xylan breakdown, transport | L. lactis subsp. cremoris MG1363/SK11 | 37.4 | 74-89 |
| x245 | 2 | 36.3 | Acetylxylan esterase | Bacillus stearothermophilus | 42.9 | 49 |
| 1, 3 | 38.1 | Hypothetical | L. lactis subsp. cremoris SK11 | 38.4 | 89-98 | |
| x2117 | 2, 5 to 9 | 34.7 | Xylo-oligosaccharide transport | L. lactis subsp. cremoris MG1363/SK11 | 36.0 | 86-99 |
| 4 | 30.7 | 6-Aminohexanoate dimer hydrolase | Bacillus thuringiensis | 33.2 | 39 | |
| y771 | 1 to 4 | 31.5 | Glucan breakdown | L. brevis, Lactobacillus reuteri, Streptococcus sanguinis | 41.6 | 27-66 |
| 7 to 11 | 30.0 | Various | E. faecalis, L. lactis subsp. lactis IL-1403 | 34.3 | 44-66 | |
| x896 | 23 to 27 | 33.2 | EPS biosynthesis cluster I | L. lactis subsp. cremoris | 33.7 | 91-98 |
| 28 to 33 | 27.8 | EPS biosynthesis cluster I | S. thermophilus, Lactobacillus rhamnosus, O. oeni, Nitrobacter spp., Aneurinibacillus spp. | 38.5 | 30-44 | |
| 34 and 35 | 33.2 | EPS biosynthesis cluster I | S. thermophilus | 33.9 | 97-98 | |
| x726 | 3 to 10 | 31.5 | EPS biosynthesis cluster II | L. lactis subsp. cremoris MG1363/SK11 | 32.6 | 75-92 |
| z1990 | 2 to 21 | 32.4 | TA biosynthesis cluster | L. lactis subsp. cremoris MG1363/SK11/IL-1403 | 34.1 | 49-99 |
| x180tr | 3 to 8 | 31.0 | Iron-siderophore transporter | B. clausii, Bacillus weihenstephanensis, Pseudomonas syringae, Clostridium spp. | 44.3 | 33-49 |
| x159 | 1 to 8 | 36.1 | Polyamine ABC transporter | C. beijerincki | 32.3 | 39-72 |
| x2018tr | 3 to 9 | 33.2 | Potassium ATPase transporter | L. innocua, C. acetobutylicum | 35.1 | 52-74 |
| n346 | 10 to 12 | 32.1 | ABC transporter | E. faecalis | 35.9 | 63-69 |
| 2, 6 to 9 | 32.7 | Hypothetical | Lactobacillus casei, L. mesenteroides, L. lactis subsp. cremoris SK11, Rhodobacter sphaeroides | 43.7 | 36-74 | |
| n760 | 3 to 8 | 33.3 | Restriction-modification system | B. subtilis, S. suis | 36.9 | 36-63 |
| 425 | 5 to 8 | 27.8 | ABC transporter | Desulfitobacterium hafniense | 48.3 | 38-57 |
| 10 to 18 | 29.6 | Various | Burkholderia, Oceanobacillus, Nocardioides, Pyrococcus, Symbiobacterium, Anaplasma, Clostridium spp. | 51.1 | 26-49 |
Details for each gene can be found in Table S2 in the supplemental material.
Average GC% of hit genomes: Anaplasma marginale, 49.8; Bacillus clausii, 44.8; Bacillus coagulans, 46.2; Bacillus subtilis, 43.5; Bacillus stearothermophilus, 52.7; Bacillus thuringiensis, 35.4; Bacillus weihenstephanensis, 35.4; Burkholderia spp., ∼68.0; Clostridium acetobutylicum, 30.9; Clostridium difficile, 29.1; Clostridium beijerincki, 29.8; Desulfitobacterium hafniense, 48.0; Enterococcus faecalis, 37.5; Enterococcus faecium, 37.6; Lactobacillus brevis, 46.1; Lactobacillus casei, 46.7; Lactobacillus sakei, 41.3; Lactobacillus reuteri, 38.9; Lactobacillus rhamnosus, not available; Leuconostoc mesenteroides, 37.7; Lactobacillus raffinolactis, not available; Lactobacillus salivarius, 33.0; L. lactis subsp. lactis IL-1403, 35.3; L. lactis subsp. cremoris SK11, 35.8; L. lactis subsp. cremoris MG1363, 35.8; Listeria innocua, 37.4; Listeria monocytogenes, 38.0; Nocardioides spp., 71.4; Oceanobacillus iheyensis, 35.7; Oenococcus oeni, 37.9; Pseudomonas syringae, 57.9; Pyrococcus horikoshii, 41.9; Rhodobacter sphaeroides, 69.0; Streptococcus mutans, 36.8; Streptococcus pneumoniae, 39.7; Streptococcus sanguinis, not available; Streptococcus suis, 41.1; Streptococcus thermophilus, 39.1; Symbiobacterium thermophilum, 68.7.
However, one clear candidate of horizontal gene transfer is the gene cluster for α-galactoside metabolism (contig z563tr), which is highly similar to that in Lactococcus raffinolactis (8), but in both bacteria the G+C content is much higher than expected for Lactococcus strains.
Conclusions and outlook.
The L. lactis strains KF147 and KF282 isolated from mung bean sprouts and mustard and cress, respectively, are found to have many adaptations to the plant environment, particularly for growth on plant carbohydrates. Mung bean (Phaseolus vulgaris) cell walls consist mainly of arabinose (most dominant), uronic acids, galactose, xylose, mannose, and glucose (25, 56). White mustard (Sinapsis alba) cell walls (mucilage) consist mainly of glucose (most dominant), galactose, mannose, rhamnose, arabinose, galacturonic acid, and xylose (13, 26, 52); their polysaccharides are mainly 1,4-linked β-d-glucan (branched cellulose), complex pectin, and xyloglucan. The adaptation to growth on substrates derived from these plant cell walls is evident from the presence of gene sets for the degradation of complex plant polymers such as xylan, arabinan, glucans, and fructans but also for the uptake and conversion of typical plant cell wall degradation products such as α-galactosides, β-glucosides, arabinose, xylose, galacturonate, glucuronate, and gluconate. Lactococci growing on plants generally live in synergy with other microbes in biofilms (14), including various bacteria and fungi, which could have similar and complementary enzymes (e.g., pectinases), allowing lactococci to grow on the plant cell wall breakdown products generated by other microbes.
Other plant niche adaptations include genes for defense (such as nisin biosynthesis and immunity) and stress response. The latter involves several extra putative transport systems for uptake of iron (possibly involving a siderophore), potassium, and polyamines. In most cases these genotypes agree well with the observed phenotypes (Table 3). Many of these genes and gene clusters have been identified for the first time in L. lactis. For instance, in addition to the new genes for plant sugar metabolism, the nonribosomal peptide biosynthesis gene cluster is new for lactococci, and the only other lactic acid bacterium with a known but unrelated NRPS cluster is Lactobacillus plantarum WCFS1 (32).
Our approach of low-cost diagnostic sequencing provides a first quick view of the gene pool present within lactococci from plant environments and yields insight into their molecular basis of adaptation. It also indicates that, to achieve insight into the pangenome of Lactococcus lactis, or any other microbe for that matter, it is essential to isolate and sequence strains from a wide variety of environments to allow for inclusion of all possible adaptation mechanisms. Based on this newly identified repertoire of genes in L. lactis, we have constructed a first-generation L. lactis pangenome microarray, using ultra-high-density DNA arrays (Nimblegen technology) for the assessment of genomic diversity within a large collection of Lactococcus lactis strains from dairy and nondairy origins, with the aim of correlating genomic makeup with phenotypic traits and better defining evolution of the Lactococcus lactis branch (G. Felis et al., unpublished results).
Supplementary Material
Acknowledgments
We thank Bill Kelly for the L. lactis strains, Iris van Swam for DNA isolation, Sacha van Hijum for assistance with Projector 2 (67), Bernhard Henrissat for CAZy analysis (11), and Ronald de Vries and Christof Francke for stimulating discussions.
This research was partly funded by the Kluyver Centre for Genomics of Industrial Fermentation, a Centre of Excellence of The Netherlands Genomics Initiative.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayad, E. H., A. Verheul, C. De Jong, J. T. Wouters, and G. Smit. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9:725-735. [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beg, Q. K., M. Kapoor, L. Mahajan, and G. S. Hoondal. 2001. Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 56:326-338. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Boekhorst, J., M. W. de Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 187:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher, I., C. Vadeboncoeur, and S. Moineau. 2003. Characterization of genes involved in the metabolism of alpha-galactosides by Lactococcus raffinolactis. Appl. Environ. Microbiol. 69:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, S. 1997. A guide to the polyamines. Oxford University Press, Oxford, United Kingdom.
- 10.Collins, T., C. Gerday, and G. Feller. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3-23. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 12.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, S. W., M. A. Eskin, Y. Wu, and S. Ding. 2006. Synergisms between yellow mustard mucilage and galactomannans and applications in food products—a mini review. Adv. Colloid Interface Sci. 128-130:249-256. [DOI] [PubMed] [Google Scholar]
- 14.Danhorn, T., and C. Fuqua. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61:401-422. [DOI] [PubMed] [Google Scholar]
- 15.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries, R. P., H. C. Kester, C. H. Poulsen, J. A. Benen, and J. Visser. 2000. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr. Res. 327:401-410. [DOI] [PubMed] [Google Scholar]
- 17.de Vries, R. P., and J. Visser. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65:497-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbein, A. D. 1969. Biosynthesis of a cell wall glucomannan in mung bean seedlings. J. Biol. Chem. 244:1608-1616. [PubMed] [Google Scholar]
- 19.Entian, K. D., and W. M. de Vos. 1996. Genetics of subtilin and nisin biosyntheses: biosynthesis of lantibiotics. Antonie Leeuwenhoek 69:109-117. [DOI] [PubMed] [Google Scholar]
- 20.Finking, R., and M. A. Marahiel. 2004. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58:453-488. [DOI] [PubMed] [Google Scholar]
- 21.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galesloot, T. E., I. Hassing, and J. Stadhouders. 1961. Agar media for the isolation and enumeration of aroma bacteria in starters. Netherlands Milk Dairy J. 15:145-150. [Google Scholar]
- 23.Gassel, M., and K. Altendorf. 2001. Analysis of KdpC of the K+-transporting KdpFABC complex of Escherichia coli. Eur. J. Biochem. 268:1772-1781. [PubMed] [Google Scholar]
- 24.Gassel, M., T. Mollenkamp, W. Puppe, and K. Altendorf. 1999. The KdpF subunit is part of the K+-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 274:37901-37907. [DOI] [PubMed] [Google Scholar]
- 25.Gooneratne, J., P. W. Needs, P. Ryden, and R. R. Selvendran. 1994. Structural features of cell wall polysaccharides from the cotyledons of mung bean Vigna radiata. Carbohydr. Res. 265:61-77. [Google Scholar]
- 26.Gould, S. E., D. A. Rees, and N. J. Wight. 1971. Polysaccharides in germination. Xyloglucans (′amyloids') from the cotyledons of white mustard. Biochem. J. 124:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi, K., and K. Kashiwagi. 1999. Polyamine transport in bacteria and yeast. Biochem. J. 344:633-642. [PMC free article] [PubMed] [Google Scholar]
- 28.Immonen, T., and P. E. Saris. 1998. Characterization of the nisFEG operon of the nisin Z producing Lactococcus lactis subsp. lactis N8 strain. DNA Seq. 9:263-274. [DOI] [PubMed] [Google Scholar]
- 29.Jung, K., and K. Altendorf. 2002. Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:223-228. [PubMed] [Google Scholar]
- 30.Kelly, W., and L. Ward. 2002. Genotypic vs. phenotypic biodiversity in Lactococcus lactis. Microbiology 148:3332-3333. [DOI] [PubMed] [Google Scholar]
- 31.Kelly, W. J., G. P. Davey, and L. J. Ward. 1998. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int. J. Food Microbiol. 45:85-92. [DOI] [PubMed] [Google Scholar]
- 32.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kok, J., G. Buist, A. L. Zomer, S. A. van Hijum, and O. P. Kuipers. 2005. Comparative and functional genomics of lactococci. FEMS Microbiol. Rev. 29:411-433. [DOI] [PubMed] [Google Scholar]
- 34.Koumoutsi, A., X. H. Chen, A. Henne, H. Liesegang, G. Hitzeroth, P. Franke, J. Vater, and R. Borriss. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 36.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 37.Kulkarni, N., A. Shendye, and M. Rao. 1999. Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 23:411-456. [DOI] [PubMed] [Google Scholar]
- 38.Luesink, E. J., J. D. Marugg, O. P. Kuipers, and W. M. de Vos. 1999. Characterization of the divergent sacBK and sacAR operons, involved in sucrose utilization by Lactococcus lactis. J. Bacteriol. 181:1924-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarova, K. S., and E. V. Koonin. 2007. Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 42.Mekjian, K. R., E. M. Bryan, B. W. Beall, and C. P. Moran, Jr. 1999. Regulation of hexuronate utilization in Bacillus subtilis. J. Bacteriol. 181:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 44.Miethke, M., O. Klotz, U. Linne, J. J. May, C. L. Beckering, and M. A. Marahiel. 2006. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 61:1413-1427. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 46.Nomura, M., M. Kobayashi, T. Narita, H. Kimoto-Nira, and T. Okamoto. 2006. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J. Appl. Microbiol. 101:396-405. [DOI] [PubMed] [Google Scholar]
- 47.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quevillon, E., V. Silventoinen, S. Pillai, N. Harte, N. Mulder, R. Apweiler, and R. Lopez. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116-W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rademaker, J., H. Herbet, M. Starrenburg, S. Naser, D. Gevers, W. Kelly, J. Hugenholtz, J. Swings, and J. van Hylckama Vlieg. 2007. Diversity analysis of dairy and nondairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 73:7128-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravn, P., J. Arnau, S. M. Madsen, A. Vrang, and H. Israelsen. 2000. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene 242:347-356. [DOI] [PubMed] [Google Scholar]
- 52.Rees, D. A., and N. J. Wight. 1969. Molecular cohesion in plant cell walls. Methylation analysis of pectic polysaccharides from the cotyledons of white mustard. Biochem. J. 115:431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridley, B. L., M. A. O'Neill, and D. Mohnen. 2001. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929-967. [DOI] [PubMed] [Google Scholar]
- 54.Sa-Nogueira, I., T. V. Nogueira, S. Soares, and H. de Lencastre. 1997. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology 143:957-969. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 56.Shiga, T. M., F. M. Lajolo, and T. M. C. C. Filisetti. 2003. Cell wall polysaccharides of common beans (Phaseolus vulgaris L.). Cienc. Tecnol. Aliment. Campinas 23:141-148. [Google Scholar]
- 57.Shulami, S., O. Gat, A. L. Sonenshein, and Y. Shoham. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smit, B. A., W. J. Engels, J. T. Wouters, and G. Smit. 2004. Diversity of L-leucine catabolism in various microorganisms involved in dairy fermentations, and identification of the rate-controlling step in the formation of the potent flavour component 3-methylbutanal. Appl. Microbiol. Biotechnol. 64:396-402. [DOI] [PubMed] [Google Scholar]
- 59.Smit, B. A., J. E. van Hylckama Vlieg, W. J. Engels, L. Meijer, J. T. Wouters, and G. Smit. 2005. Identification, cloning, and characterization of a Lactococcus lactis branched-chain α-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 71:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smit, G., B. A. Smit, and W. J. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 61.Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53:749-790. [DOI] [PubMed] [Google Scholar]
- 62.Tanous, C., A. Kieronczyk, S. Helinck, E. Chambellon, and M. Yvon. 2002. Glutamate dehydrogenase activity: a major criterion for the selection of flavour-producing lactic acid bacteria strains. Antonie Leeuwenhoek 82:271-278. [PubMed] [Google Scholar]
- 63.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tramer, J., and G. G. Fowler. 1964. Estimation of nisin in foods. J. Sci. Food Agric. 15:522-528. [Google Scholar]
- 66.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Hijum, S. A., A. L. Zomer, O. P. Kuipers, and J. Kok. 2005. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res. 33:W560-W566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Hylckama Vlieg, J. E., J. L. Rademaker, H. Bachmann, D. Molenaar, W. J. Kelly, and R. J. Siezen. 2006. Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 17:183-190. [DOI] [PubMed] [Google Scholar]
- 69.van Kranenburg, R., M. Kleerebezem, and W. M. de Vos. 2000. Nucleotide sequence analysis of the lactococcal EPS plasmid pNZ4000. Plasmid 43:130-136. [DOI] [PubMed] [Google Scholar]
- 70.Weber, T., and M. A. Marahiel. 2001. Exploring the domain structure of modular nonribosomal peptide synthetases. Structure 9:R3-R9. [DOI] [PubMed] [Google Scholar]
- 71.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willats, W. G., L. McCartney, W. Mackie, and J. P. Knox. 2001. Pectin: cell biology and prospects for functional analysis. Plant Mol. Biol. 47:9-27. [PubMed] [Google Scholar]
- 73.Yao, S., X. Gao, N. Fuchsbauer, W. Hillen, J. Vater, and J. Wang. 2003. Cloning, sequencing, and characterization of the genetic region relevant to biosynthesis of the lipopeptides iturin A and surfactin in Bacillus subtilis. Curr. Microbiol. 47:272-277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.