Abstract
Escherichia coli WG5, the strain recommended by the International Organization for Standardization (ISO) to detect somatic coliphages, was transformed to F+ by introducing the plasmid Famp, which rendered it capable of simultaneously detecting both somatic and F-specific coliphages. Indeed, this strain, CB390, proved as effective in detecting similar numbers of phages as the sum of somatic and F-specific bacteriophages detected by the host strains recommended by both the ISO and the U.S. Environmental Protection Agency standardized methods.
The suitability of coliphages as indicators of virus and fecal contamination of water and food has long been studied, and there is abundant scientific literature addressing this topic compiled in several reviews (6, 8, 9, 12, 17). The recently legislated Ground Water Rule (22), as well as the regulation on the quality of drinking water in the Canadian State of Québec (2), proposes that coliphages be used as fecal indicators.
Somatic coliphages and F-specific coliphages are being used for these purposes, with available standardized methods for both the former (1, 11, 20, 21) and the latter (10, 20, 21). On some occasions, it might prove advantageous to have the added numbers of both phage groups. Examples can be found in groundwater monitoring procedures (22) or in the evaluation of disinfection of secondary wastewater effluents with UV irradiation (16). In this disinfection, whereas somatic coliphages predominate in secondary effluents, RNA F-specific phages are the most abundant after UV irradiation (16). This can be achieved by adding the values obtained by the standardized methods or by using a host strain capable of measuring both. Ideally, such a host strain should detect numbers of both groups of phages identical to those detected by the standardized methods. E. coli C-3000 (ATCC 15597) is one such host strain that has been used for this purpose. In a study comparing all of the host strains also tested here (except E. coli WG5), strain C-3000 was reported to recover lower amounts of phages than the sum of somatic and F-specific coliphages counted by the standardized methods (19).
The aim of the research reported here was to introduce the F-plasmid into Escherichia coli WG5, which is the strain recommended by the International Organization for Standardization (ISO) to detect somatic coliphages and, by doing this, to enable this strain to simultaneously detect somatic and F-specific coliphages. Also, the adequacy of the strain was determined by comparing its efficiency in detecting coliphages of the two groups in natural samples to those detected by the host strains recommended in standardized methods and the C-3000 strain.
The Famp plasmid (3) was used to transform the E. coli WG5 to an F+ strain. The plasmid was purified by using the Qiagen plasmid mini purification kit (Qiagen, Inc., Valencia, CA). Electroporation-competent cells were prepared from 10- to 50-ml cultures in Super Optimal Broth medium (7). Cells were then transformed by electroporation according to the method of Sambrook and Russell (18). Transformants were selected by plating on Super Optimal Catabolite medium (7) containing 100 μg of ampicillin ml−1. Thirty ampicillin-resistant transformants were isolated after the transformation experiments. These transformants were then tested for their efficiency in counting phages from suspensions of φX174 and MS2 and from naturally occurring suspensions of phages prepared as reference material, as previously described (14). φX174 and MS2 are the reference phages used in the standardized methods for enumerating somatic and F-specific coliphages (10, 11, 20, 21). All of the transformants proved sensitive to MS2, which indicates that they had become F+ and had retained sensitivity to φX174. Five of the isolates that gave the highest recoveries for each of the phages and the naturally occurring reference material were further twice tested for their efficiency in recovering somatic and F-specific RNA phages using the procedures and media recommended in the ISO methods (10, 11). After these verification steps, strain CB390 was selected for further study.
All of the media recommended in the standardized ISO methods were tested for enumerating φX174 (11) and MS2 (10) on CB390 strain. In addition, the following combination was tested. CB390 was grown in MSB broth (11) with 100 μg per ml of ampicillin, and TYG agar and TGY semisolid agar (10) supplemented with ampicillin (100 μg ml−1), Ca2+, and Mg2+ as in the ISO method (11) were used for the lower and upper layers in the double-agar-layer test. There were not significant differences (analysis of variance [ANOVA], P > 0.05) between the media, but numbers detected by the later combination proved slightly higher, so this is the combination of media used in the assays to enumerate phages with strain CB390 described above. All of the statistical analyses were carried out by using the Statgraphics statistical analysis software package (Statgraphics Plus 5.1; StatPoint, Inc.).
Strain CB390 was sensitive to bacteriophages SOM1, SOM2, SOM4, SOM28, and SCH10 (15) and P1 and φX174 (4), which represent the main somatic coliphages families and groups, as well as, to phages MS2, GA, Qβ, and FI, representing the four serotypes of F-specific RNA phages (5).
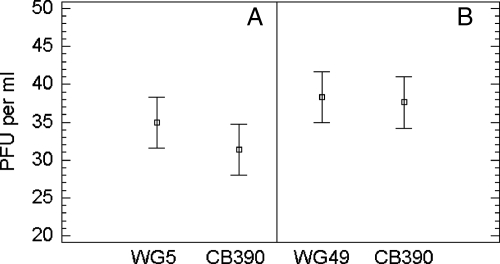
Moreover, strain CB390 detected numbers of φX174 similar (ANOVA, P > 0.05) to those detected by E. coli WG5 and numbers of MS2 similar (ANOVA, P > 0.05) to those detected by Salmonella enterica serovar Typhimurium WG49, as shown in Fig. 1.
FIG. 1.
Mean values (bars show the 95% confidence intervals) of PFU of phage φX174 on host strains WG5 and CB390 (A) and phage MS2 on host strains WG49 and CB390 (B). The host strains used to count the bacteriophages are listed on the x axis. Values are the means of 10 independent assays.
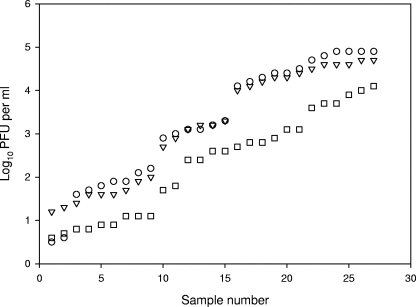
Also, when very different natural matrices, including raw municipal sewage, slaughterhouse wastewater, tertiary effluent (UV irradiated and chlorinated), river water, and primary and dehydrated sludge were tested, strain CB390 detected slightly lower numbers of phages but not significantly (Fisher least significant differences, P > 0.05) than the sum detected by the WG5 and WG49 strains. The values are plotted in Fig. 2 (SigmaPlot 2000, v6.10; SPSS Science, Inc., Chicago, IL). Other than UV-irradiated samples in which F-specific phages were more abundant, and wherein, consequently, WG49 and CB390 detected more phages than did WG5, both WG5 and CB390 detected similar numbers of phages (Fisher least significant differences, P > 0.05) and higher numbers than those detected by WG49 in all of the other samples. More than 150 plaques formed on CB390 were replicated over WG5 and WG49, and the ratios between the numbers of those replicated in each strain maintained the ratios found between the numbers of somatic coliphages and F-specific phages detected by the ISO methods for the same samples.
FIG. 2.
Counts of bacteriophages enumerated on CB390 (▿), WG5 (○), and WG49 (□). The three values following the vertical line belong to the same sample. Values correspond to 27 independent assays.
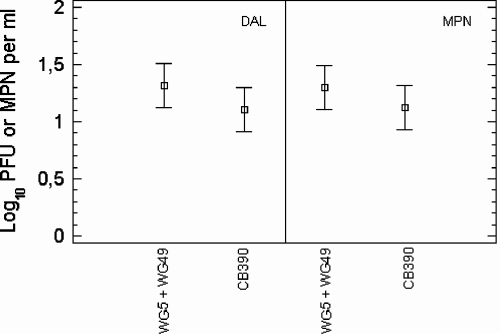
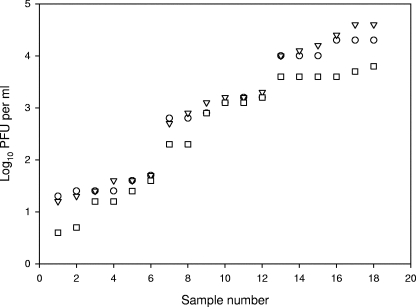
The next step was to determine the numbers of bacteriophages that CB390 strain detected in natural samples, as well as the sum of those detected by strains WG5 and WG49, using both the double-agar-layer method (10, 11) and the most-probable-number (MPN) method (10, 11). Knowing the MPN efficiency is essential for determining whether the presence or absence results are comparable. For this, naturally occurring phages in several sample types, including UV-irradiated secondary effluents (those containing more F-specific than somatic phages), were tested for PFU by both techniques. Test samples were diluted beforehand such that they contained between 1 and 100 phages per ml. The results are shown in Fig. 3. No significant differences (ANOVA, P > 0.05) were found either between the two procedures or between the phages counted by CB390 and the sum of those counted by WG5 and WG49. On the other hand, the number of phages detected by CB390, the sum of those detected by E. coli strains CN13 and HS (pFamp)R, which are the ones recommended by U.S. Environmental Protection Agency (20, 21), and those detected by strain C-3000 were determined by the double agar layer technique in a suite of natural samples similar to that previously described. The values are plotted in Fig. 4 (SigmaPlot 2000). Values detected by the CB390 strain, calculated as percentages of the sum of values detected by strains CN13 and HS, were significantly higher (ANOVA, P < 0.05) than those detected by the C-3000 strain and similar (ANOVA, P > 0.05) to the sum detected by CN13 and HS. One possible explanation for this is that the C-3000 strain has a narrower spectrum of infecting phages. In fact, upon challenge with the phages described above, C-3000 was not sensitive to φX174 and SOM2. Finally, 78 drinking water samples collected from two complex water treatment plants using either chlorine or chlorine plus ozone as disinfectants were tested for somatic coliphages on WG5, for F-specific phages on WG49, and for both simultaneously on CB390. In this assay, 1 liter of water was concentrated according to the method of Méndez et al. (13). Somatic coliphages were detected in two (2.3%) of the samples, phages were detected on strain CB390 in 1 (1.3%), and F-specific coliphages were detected in none of the samples. The maximum load detected by strain WG5 was 3 PFU per liter, and that detected by strain CB309 was 7 PFU per liter.
FIG. 3.
Mean values (bars show the 95% confidence intervals) of bacteriophages quantified on WG5 plus those enumerated on WG49 and those quantified on CB390 in different matrices using either the double-agar-layer assay (DAL) or the MPN method. The host strains used to count the bacteriophages are listed on the x axis. Values are the means of 12 independent assays.
FIG. 4.
Counts of bacteriophages enumerated on CB390 (▿), on CN13 plus HS (○), and on C-3000 (□). The three values following the vertical line belong to the same sample. The values correspond to 18 independent assays.
In conclusion, CB390 strain detects lower, but not statistically significant, values of somatic and F-specific phages than the sum of phages determined by the ISO methods. Moreover, it detects numbers similar to the sum of phages enumerated by the U.S. Environmental Protection Agency methods and significantly higher values than those quantified by C-3000 strain. Of course, the next step in the validation of the strain is a multilaboratory comparison of results using this new host compared to the conventional assays, but this exceeds the aims of the research described here.
Acknowledgments
This study was supported by grants from the Ministerio de Educación y Ciencia CTM 2005-02513, the CDTI (CENIT 2007-1039; SOSTAQUA), and the Generalitat de Catalunya (2005SGR00592). C.G. received a fellowship from the Spanish Agency for International Cooperation (MAE-AECI).
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.American Public Health Association. 2001. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 2.Anonymous. 2001. Loi sur la qualité de l'environnement: règlement sur la qualité de l'eau potable c. Q.-2, r. 18.1.1. Gazette Officielle du Québec 24, 3561. Government of Quebec, Montreal, Quebec, Canada.
- 3.Debartolomeis, J., and V. J. Cabelli. 1991. Evaluation of an Escherichia coli host strain for enumeration of F male-specific bacteriophages. Appl. Environ. Microbiol. 57:1301-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball. 2005. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, New York, NY.
- 5.Furuse, K. 1987. Distribution of coliphages in the environment: general considerations, p. 87-124. In S. M. Goyal, C. P. Gerba, and G. Bitton (ed.), Phage ecology. John Wiley & Sons, Inc., New York, NY.
- 6.Grabow, W. O. K. 2001. Bacteriophages: update on application as models for viruses in water. Water SA 27:251-268. [Google Scholar]
- 7.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 8.Hsu, F.-C., Y.-S. C. Shieh, and M. D. Sobsey. 2002. Enteric bacteriophages as potential fecal indicators in ground beef and poultry meat. J. Food Prot. 65:93-99. [DOI] [PubMed] [Google Scholar]
- 9.IAWPRC Study Group on Health-Related Water Microbiology. 1991. Bacteriophages as model viruses in water quality control. Water Res. 25:529-545. [Google Scholar]
- 10.International Organization for Standardization. 1995. Water quality: detection and enumeration of bacteriophages. 1. Enumeration of F-specific RNA bacteriophages. ISO 10705-1. International Organization for Standardization, Geneva, Switzerland.
- 11.International Organization for Standardization. 1995. Water quality: detection and enumeration of bacteriophages. 2. Enumeration of somatic coliphages. ISO 10705-2. International Organization for Standardization, Geneva, Switzerland.
- 12.Jofre, J. 2002. Bacteriophage as indicators, p. 354-363. In G. Bitton (ed.), Encyclopedia of environmental microbiology, vol. 1. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 13.Méndez, J., A. Audicana, A. Isern, J. Llaneza, M. L. Tarancón, B. Moreno, J. Jofre, and F. Lucena. 2004. Standardized evaluation of the performance of a simple membrane filtration-elution method to concentrate bacteriophages from drinking water. J. Virol. Methods 117:19-25. [DOI] [PubMed] [Google Scholar]
- 14.Méndez, J., J. Jofre, F. Lucena, N. Contreras, K. Mooijman, and R. Araujo. 2002. Conservation of phage reference materials and water samples containing bacteriophages of enteric bacteria. J. Virol. Methods 106:215-224. [DOI] [PubMed] [Google Scholar]
- 15.Muniesa, M., L. Mocé-Llivina, H. Katayama, and J. Jofre. 2003. Bacterial host strains that support replication of somatic coliphages. Antonie Leeuwenhoek 83:305-315. [DOI] [PubMed] [Google Scholar]
- 16.Payán, A. 2006. Bacteriófagos como modelo en el origen de la contaminación fecal y en aguas regeneradas. Ph.D. thesis. University of Barcelona, Barcelona, Spain.
- 17.Pillai, S. D. 2006. Bacteriophages as indicators, p. 205-222. In S. Goyal.(ed.), Food virology. Kluwer Academic Publishers, Amsterdam, The Netherlands.
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Sobsey, M. D., M. V. Yates, F.-C. Hsu, G. Lovelace, D. Battigelli, A. Margolin, S. D. Pillai, and N. Nwachuku. 2004. Development and evaluation of methods to detect coliphages in large volumes of water. Water Sci. Technol. 50:211-217. [PubMed] [Google Scholar]
- 20.U.S. Environmental Protection Agency. 2001. Method 1601: male-specific (F+) and somatic coliphage in water by two-step enrichment procedure. EPA document 821-R-01-030. Environmental Protection Agency, Washington, DC.
- 21.U.S. Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. EPA document 821-R-01-029. Environmental Protection Agency, Washington, DC.
- 22.U.S. Environmental Protection Agency. 2006. National primary drinking water regulations: ground water rule; final rule; 40 CFR parts 9, 141, and 142. Fed. Regist. 71:65574-65660. [Google Scholar]