Abstract
Bacteria employ quorum sensing, a form of cell-cell communication, to sense changes in population density and regulate gene expression accordingly. This work investigated the rewiring of one quorum-sensing module, the lux circuit from the marine bacterium Vibrio fischeri. Steady-state experiments demonstrate that rewiring the network architecture of this module can yield graded, threshold, and bistable gene expression as predicted by a mathematical model. The experiments also show that the native lux operon is most consistent with a threshold, as opposed to a bistable, response. Each of the rewired networks yielded functional population sensors at biologically relevant conditions, suggesting that this operon is particularly robust. These findings (i) permit prediction of the behaviors of quorum-sensing operons in bacterial pathogens and (ii) facilitate forward engineering of synthetic gene circuits.
In bacteria, broadcasting of metabolite or small peptide signaling molecules enables sensing of changes in population density (18, 37, 57). This mechanism, known as quorum sensing, has been implicated in regulating the virulence factors in a number of bacterial pathogens, such as Vibrio cholerae (61), the bacterium responsible for the severe diarrheal disease cholera; Pseudomonas aeruginosa (30), the opportunistic pathogen responsible for death in cystic fibrosis patients and high mortality rates in immunocompromised individuals; and Staphylococcus aureus (32), a major culprit of infections in surgical wounds. Consequently, improved understanding of quorum-sensing regulation should provide more insight into combating these pathogens by using either traditional chemotherapy or emerging technologies such as quorum-sensing inhibitors (40) or regulated degradation of quorum-sensing signals (13, 14).
The lux module of the marine bacterium Vibrio fischeri, a facultative symbiont of luminescent fish or squid, serves as one model system for understanding quorum sensing. V. fischeri employs the lux module to regulate gene expression as a function of the population density. The key element of this system is a regulatory cassette consisting of the genes encoding LuxI and LuxR. LuxI is an acylhomoserine lactone (acyl-HSL) synthase; LuxR is a transcriptional regulator activated by the acyl-HSL. The acyl-HSL signaling molecule is produced inside the cell but can freely diffuse across the cell membrane into the environment. Therefore, the acyl-HSL concentration is low at low cell density. As the cell density increases, the signal accumulates in the environment and inside the cell. The signal can then bind LuxR to stabilize the transcription factor, which then activates gene expression. This process, in which activation of gene expression occurs only after attaining a critical cell density, is known as autoinduction.
In this work, we sought to computationally and experimentally address the fundamental question of how bacteria regulate cell-cell communication. Quorum sensing is often described in terms of a discrete switch; at low cell densities, the signal molecule is present at low concentrations, while at high cell densities, the signaling molecule accumulates to a concentration sufficient to activate gene expression (19). Meanwhile, the emergence of the synthetic gene circuit discipline has reemphasized the importance of accounting for the continuous nature of gene regulation (24). In this spirit, we sought to ascertain how quorum-sensing regulation works as a function of the population density in both a qualitative and a quantitative fashion. This goal differs from many of the recent characterizations of the lux system (15, 44, 51) because we examined the behavior of the entire regulatory module, not just a subset of it. Several synthetic-biology works have employed the entire lux regulatory module (3, 5, 6, 28, 59), but the focus of these works has been to achieve a desired engineering goal, not to characterize how the lux module functions.
One of the original published studies of quorum sensing in V. fischeri elegantly illustrated the autoinduction of bioluminescence as a function of cell density (43), and computational modeling hypothesized that this autoinduction resulted from bifurcation due to positive feedback on the lux regulatory elements (27). Yet, this characterization is far from complete. The simple lux module consists of three regulatory components, luxR, luxI, and the lux promoter; the native lux operon reflects merely one possible combination of these components. Combinatorial reshuffling of regulatory components has been shown to yield diverse logical outputs (23), and applying the same strategy to the lux system should elucidate possible responses of other quorum-sensing systems. Additionally, it is unclear how simple rearrangements in network architecture combined with system-specific parameters (e.g., transcriptional regulation and translational efficiency) will affect circuit performance. Specifically, do these rearrangements render functional circuits over biologically relevant conditions? If so, then the responses of these rearrangements can be used to predict the responses of other naturally occurring quorum-sensing operons. Finally, the computational prediction of bifurcation in the native lux operon has not been experimentally demonstrated. In this paper, we seek to address each of these issues.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strain used in this study was Escherichia coli DH5α [F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1]. E. coli was cultured at 37°C in Luria-Bertani (LB) medium or on LB agar plates. The acyl-HSL used in this study was 3-oxohexanoyl-dl-homoserine lactone (3OC6HSL; Sigma Aldrich). Antibiotics were added at concentrations of 50 μg/ml kanamycin and 100 μg/ml chloramphenicol to maintain the plasmids.
Construction of the quorum-sensing plasmids used in this work.
Plasmids pGLRKM-101, pPSSUB-101, and pPSSUB-102 were the kind gifts of Ron Weiss (Princeton University). Plasmids pPSSUB-101 and pLuxRI were described previously by Basu et al. (6) and You et al. (59), respectively. Both pPSSUB-101 and pPSSUB-102 contain a destabilized version of green fluorescent protein, GFPLVA [originating from pGFP(LVA); BD Biosciences Clontech], which in E. coli has a reported half-life of 40 min (2) and a rapid maturation time (detectable after only 10 min under fed-batch conditions [41]). pluxG-102 was constructed by using overlap extension PCR (25) to fuse the luxR promoter from pGLRKM-101 (3-G2-lux and 5-G-NotI) with luxR and its ribosome binding site (RBS) from pLuxRI (5-G2-lux and 3-XhoI-LuxR). The primers used to amplify the respective fragments are in parentheses; for the sequences of these primers, see Table S1 in the supplemental material. The resulting fragment was cloned into pPROLar.A122 (BD Biosciences Clontech) between the XhoI and NotI sites. pluxG-102C is identical to pluxG-102, except for the RBS. PCR was used to introduce a new luxR RBS (5-G-NotI and 3-KpnI-GC). The resulting fragment was cloned into pluxG-102 between the NotI and KpnI sites. pluxG-103 was constructed by amplifying the luxI gene from pLuxRI (5-Rseq and 5-AatII-LuxI). The resulting fragment was cloned into pluxG-102 between the AatII and BamHI sites. pluxRp-101 was constructed by using overlap extension PCR to fuse the luxI promoter from pPSSUB-101 (5-SalI-plux and 3-pluxC) with the luxR gene from pluxG-102 (5-LuxRC and 5-seq). The resulting fragment was cloned into pPROLar.A122 between the SalI and BamHI sites. pluxRp-103 is identical to pluxRp-101, except that a KpnI site was introduced upstream of the luxR gene. Overlap extension PCR was used to introduce this site (5-Rp-SalI and 3-Rp-3 and 5-Rp-3 and 3-HindIII-LuxRa). The resulting fragment was cloned into pluxRp-101 between the SalI and HindIII sites. pluxRp-103E is identical to pluxRp-103, except for the RBS. PCR was used to introduce a new luxR RBS (5-Rp-SalI and 3-KpnI-RpE). The resulting fragment was cloned into pluxRp-103 between the SalI and KpnI sites. All constructs were verified by DNA sequencing. For plasmid maps of these constructs, see Fig. S1 in the supplemental material.
Periodic-dilution experiments.
Steady states of quorum-sensing constructs were assayed by using a periodic-dilution experiment to maintain the cultures in the growth phase; a similar experiment outlined by Chen and Weiss (9) served as the template for the following procedure. One-milliliter cultures in LB medium with the appropriate antibiotics were started from plates and grown for roughly 8 h at 37°C. A portion of the 1-ml culture was diluted into 50- or 100-ml cultures of LB medium containing 0.05 M morpholinepropanesulfonic acid (MOPS; pH 7.0) and the appropriate antibiotics (hereafter called LBM) and then grown at 37°C with shaking for 15 h to appropriate densities. When noted, these overnight cultures were induced with 3OC6HSL at 11 h into the growth step. The overnight cultures were transferred to 50-ml centrifuge tubes, pelleted, washed, repelleted, and then resuspended to the desired absorbance (A600; all absorbance readings in this report were performed at 600 nm) in LBM. The resuspensions were diluted with LBM in 48-well blocks to various absorbances at a final volume of 2.2 ml. The absorbance and fluorescence (excitation at 470 nm, emission at 509 nm, gain of 69) of a 200-μl volume were assayed in a 96-well microplate with a clear bottom (Corning Costar) by using a microtiter plate reader (Infinite M200; Tecan); this reading corresponds to the t = 0 h measurement. The blocks were then covered with Aeroseal (Daigger) and shaken at 37°C. At the end of each hour, the absorbance and fluorescence of 200 μl were assayed; note that the t = 0 h measurement corresponds to the reading at the beginning of the hour as opposed to the end. A portion of each culture was then removed, and fresh LBM was added to reset the initial absorbance to a predetermined constant value while maintaining the culture volume at 2 ml. For the absorbance-CFU/ml calibration curve used to calculated the volume removed, see Fig. S2 in the supplemental material. This curve fits the data using the Hill function
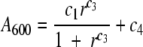 |
(1) |
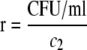 |
(2) |
in which c1 = 2.93, c2 = 3.77 ×108 CFU/ml, c3 = 1.02, and c4 = 0.0445. This process was repeated seven or eight times until both the absorbance and fluorescence values equilibrated. At this time, the cultures were pelleted, decanted, washed in 1 ml phosphate-buffered saline (PBS; pH 7.0), pelleted, decanted, and resuspended in 1.8 ml PBS. The absorbance and fluorescence of 200 μl were again assayed. Absorbance and fluorescence readings were corrected by subtracting the respective readings of 200 μl PBS. If the corrected fluorescence readings were nonpositive, then the cultures were resuspended to an A600 of ∼0.3 and assayed again. All steady-state measurements correspond to the corrected absorbance reading in LBM medium and the fluorescence or absorbance reading in PBS at the final time point. Unless otherwise noted, the results from at least three different experiments (each experiment being started from a different colony) were used to calculate the means (points) and standard deviations (error bars) shown in each of the steady-state figures. Also, samples were drawn at the next to last time point to (i) measure the number of viable cells by serial dilution and plating and (ii) confirm the accuracy of the calibration curve.
Modeling of the lux system.
Urbanowski et al. (53) determined the binding characteristics of the LuxR protein to the lux promoter. Gel shift experiments and DNA protection assays demonstrated that stimulation of LuxR-DNA complex formation by varying either the 3OC6HSL or the LuxR concentration is noncooperative (Hill coefficients of 0.9 and 0.85, respectively). One kinetic mechanism that captures both of these features is the following model:
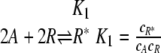 |
(3) |
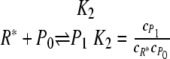 |
(4) |
in which A is the 3OC6HSL signaling molecule, R is the LuxR protein, R* is the activated transcription factor, P0 is the unbound DNA, P1 is the LuxR-DNA complex, and cj refers to the concentration of species j. For this model,
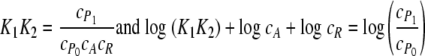 |
(5) |
Plotting the right-hand side of equation 5 as a function of the logarithm of either A or R renders the Hill plot. It is clear that such a plot would give a Hill coefficient of 1.0 for varying the concentration of either A or R while holding the other concentration constant. Hence, this mechanism captures the essential features of the gel shift experiments and DNA protection assays.
Models were analyzed by using the continuation package CL_MATCONT, a variant of the MATCONT package (11), to solve for the steady states as a function of the population density. The modeling results presented in this paper assume that the system is in a continuous-flow setting; see section S1 in the supplemental material for a more detailed account of this modeling. Modeling the system discretely to describe the periodic-dilution experiment did not significantly alter the qualitative or quantitative results because the growth time of 1 h between dilutions was relatively short (data not shown).
RESULTS
The mathematical model predicts that shuffling of the network architecture can result in graded, threshold, or bistable gene expression.
Given the reporter gene gfplva and the luxI and luxR components, there are three possible ways of regulating GFPLVA expression as a function of population density. These shuffled network architectures are illustrated in Fig. 1. Here, p(luxI) and p(luxR) refer to the luxI and luxR promoters in the native lux operon. In architecture a, luxI and luxR are constitutively expressed from the p(luxR) promoter and gfplva is expressed from the p(luxI) promoter. In architecture b, luxR is constitutively expressed from the p(luxR) promoter and luxI and gfplva are expressed from the p(luxI) promoter. Modeling suggests that exchanging the placement of luxI and luxR in this architecture yields behavior similar to that which results from nominal architecture b since LuxI and LuxR demonstrate comparable cooperativities, although the precise behavior of the circuit depends on the parameter values. Finally, in architecture c, luxI, luxR, and gfplva are all expressed from the p(luxI) promoter. The network architectures presented in Fig. 1 correspond experimentally to the plasmid combinations outlined in Table 1.
FIG. 1.
Possible architectures for quorum-sensing gene circuits. Architecture a: constitutive expression of luxI and luxR, p(luxI)-regulated expression of gfplva. Architecture b: constitutive expression of luxR, p(luxI)-regulated expression of luxI and gfplva. Architecture c: p(luxI)-regulated expression of luxI, luxR, and gfplva.
TABLE 1.
Plasmids used in this studya
| Architecture | Product(s) | Plasmid(s) |
|---|---|---|
| a | LuxR + LuxI | pluxG-103 |
| GFPLVA | pPSSUB-101 | |
| b | LuxR | pluxG-102, pluxG-102C |
| GFPLVA + LuxI | pPSSUB-102 | |
| c | LuxR | pluxRp-101, pluxRp-103E |
| GFPLVA + LuxI | pPSSUB-102 |
Constructs in the same row differ only in the promoter region (−20 to −1 positions upstream of the first gene), which effectively alters the RBS.
To explore the possible ranges of system behavior, we constructed mass action kinetic models for all three of the architectures in Fig. 1; Fig. 2 presents these models. All reactions are elementary as written except for cell growth, which is assumed to follow a logistic growth expression, and the transcription factor binding events, whose expressions are given by equations 3 and 4. These models represent a highly simplified version of the actual system; transcription and translation are lumped into first-order processes limited by a DNA catalyst, a simple expression approximates cell growth, and the transcription factor binding reactions are assumed to be at equilibrium. We also assume that the signaling molecule freely diffuses into and out of the cell membrane, whereas all of the other intracellular species cannot freely diffuse. Finally, we assume that the system is well mixed and deterministic.
FIG. 2.
Kinetic mechanisms used to model the behaviors of the three design architectures. G, C, and Q0 refer to GFPLVA, bacterial cells, and the DNA encoding constitutive expression, respectively.
Figure 3 plots the steady-state response of each network architecture for GFPLVA fluorescence per cell as a function of cell density. Network architecture a leads to a graded response; fluorescence per cell increases linearly with cell density on a log-log scale until the fluorescence saturates. In contrast, network architecture b results in a threshold response characteristic of a soft switch; below (above) a certain density, the fluorescence per cell is minimal (maximal). This threshold-like behavior stems directly from the positive feedback on the signaling molecule obtained by placing the luxI gene under the control of the p(luxI) promoter. Finally, network architecture c exhibits bistability over a range of densities. Note that the middle loci of steady states are unstable. Here, fluorescence per cell is hysteretic in that either maximal or minimal fluorescence results, depending upon the initial condition of the system.
FIG. 3.
Qualitative steady-state behaviors of (a) design architecture a, constitutive expression of both luxR and luxI; (b) architecture b, constitutive expression of luxR and p(luxI)-mediated expression of luxI; and (c) architecture c, p(luxI)-mediated expression of both luxI and luxR (the dashed line denotes the unstable loci of steady states). Plot d presents a phase plot for a simplified model of the system illustrating the dependence of the steady state on the initial condition (both cell density and 3OC6HSL concentration) for a steady-state cell density of 109 CFU/ml. Solid lines denote the null clines (where the derivative of either the cell density or the 3OC6HSL concentration is zero). Dashed lines denote representative phase traces for this system that were obtained by solving for time courses of the dynamic model with various initial conditions and projecting the solutions onto the cell density-3OC6HSL axes. Arrows denote the directionality of these traces, filled points denote stable attractors, and the unfilled point denotes the unstable attractor. Fluor., fluorescence; A.U., arbitrary units.
In continuous-growth cultures, the presence of bistability can be detected by varying the initial concentration of the signaling molecule, 3OC6HSL. Because bistability results in two stable steady states (or attractors) for a given density, the initial 3OC6HSL concentration can determine to which steady state the system evolves. Figure 3d presents a phase portrait in two dimensions (3OC6HSL concentration and population density) for a simplified model of the system. Here, starting the experiment from the same population density but different initial concentrations of 3OC6HSL leads to different steady-state attractors.
Previous work indicates that LuxR is capable of positively stimulating transcription from the p(luxR) promoter to a small degree (46, 48). In contrast, we have assumed that the p(luxR) promoter is constitutive. This discrepancy does not significantly affect the model predictions because the observed positive feedback from the p(luxR) promoter is significantly smaller than that observed from the p(luxI) promoter (48; also see section S2 in the supplemental material).
Shuffling the architecture of the lux operon yields graded, threshold, and bistable responses as predicted by the mathematical model.
We constructed each of the three different network architectures shown in Fig. 1. Constitutive expression and lux-mediated expression were obtained by expressing genes from the p(luxR) and p(luxI) promoters, respectively. We then performed periodic-dilution experiments to determine the steady-state behavior of each architecture. The results are presented in Fig. 4. The qualitative features of the shuffled architectures are consistent with the model predictions. Namely, shuffling the network architecture of the lux operon yields graded, threshold, and bistable responses, as predicted by the mathematical model and shown in Fig. 4.
FIG. 4.
Experimental steady-state results for architectures a, b, and c. In the bistable region, architecture c exhibits memory in that the initial condition can drive the system to different steady-state attractors, as demonstrated in plot b. Here, initial cultures were grown to a low absorbance (A600 of <0.2), grown to a high absorbance (A600 of >0.7), or induced with 100 nM 3OC6HSL. The negative (neg.) control results (pPROLar.A122 and pPSSUB-102) are plotted for comparison. Fluor., fluorescence; A.U., arbitrary units.
The dynamic evolution of all of the architectures and the negative control is presented in Fig. 5 to illustrate both the dynamics of the periodic-dilution experiment and the notion of bistability. This figure demonstrates equilibration first of the absorbances in approximately 2 to 3 h and then the fluorescences in the last 5 to 8 h. Note that the z axis is a plot of the corrected fluorescence values (i) in LBM as opposed to PBS and (ii) not normalized by the absorbance reading. Also, the two highest absorbances in Fig. 5c did not equilibrate until later in the experiment since the experiment was started from a low-absorbance culture.
FIG. 5.
Dynamic periodic-dilution results for the shuffled network architectures. Corrected absorbance and fluorescence dynamics over the duration of the periodic-dilution experiment are shown for (a) architecture a, (b) architecture b, (c) architecture c low (A600o of <0.2), (d) architecture c high (A600o = 0.799), (e) architecture c induced with 100 nM 3OC6HSL, and (f) the negative control. Architecture a is pluxG-103 and pPSSUB-101, architecture b is pluxG-102 and pPSSUB-102, architecture c is pluxRp-103E and pPSSUB-102, and the negative control is pPROLar.A122 and pPSSUB-102. A600o is the absorbance of the overnight culture used to start the periodic-dilution experiment. Each plot uses the same shading to highlight differences in the measured fluorescence values. Corr., corrected; Fluor., fluorescence; A.U., arbitrary units.
In the bistable region, the system exhibits memory in that the initial condition can drive the system to different steady-state attractors. Figure 5c to e illustrates the dynamics of this process for architecture c. Initiating the periodic-dilution experiment from a culture grown to a low initial absorbance (A600o of <0.2) maintains the system in a low-fluorescence state, whereas initiating the experiment from a culture induced with 100 nM 3OC6HSL results in high fluorescences for all of the densities tested. Initiating the experiment from a culture grown to a high initial absorbance (A600o of >0.7) results in density-dependent fluorescence. We also note that in each experiment, the same 1-ml inoculating culture was used to start a low culture and either a high or a 3OC6HSL culture. Thus, low fluorescence readings most likely resulted from the network architecture, as opposed to disabling of the quorum-sensing operon by, for example, mutation.
Shuffling the architecture of the lux operon yields functional population sensors.
We sought to determine if shuffling of network architecture would give rise to functional quorum-sensing behavior in the observable range of the periodic-dilution experiment. We define a functional population sensor as “on” at high absorbances (A600 of ∼1) and “off” at low absorbances (A600 of <10−2). In the previous section, we tuned the circuit behavior to fall within the observable range by manipulating the luxR RBS. Here, we maintained as much similarity in terms of promoter regions and RBSs as possible. According to our definition of a functional population sensor, Fig. 6a demonstrates that shuffling the network architecture yields functional sensors for architectures a and b. Architecture c, however, results in maximal GFPLVA expression, regardless of the initial condition. Prolonged maintenance of this high-expression state appears to be unfavorable; this particular combination (pluxRp-101 and pPSSUB-102) did not equilibrate in the time course of the experiment (see Fig. S3 and section S3 in the supplemental material). pluxRp-101 and pluxRp-103E differ by only the luxR RBS strength (pluxRp-101 being stronger than pluxRp-103E), and Fig. 4b demonstrates that the combination of pluxRp-103E and pPSSUB-102 exhibits bistability. Thus, we infer that the bistable region of pluxRp-101 and pPSSUB-102 occurs at densities lower than those observable from the periodic-dilution experiment.
FIG. 6.
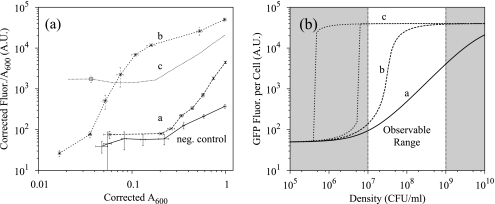
Comparison of the lux operon with shuffled network architectures. Panels: a, experimental results; b, model predictions. Architectures a and b yield a functional response in the observable range of the periodic-dilution experiment. Note, however, that network architecture c does not exhibit bistability within this range. The unshaded region in plot b corresponds to the observable range of the periodic-dilution experiment (A600 of 0.01 to 1). The negative (neg.) control results (pPROLar.A122 and pPSSUB-102) are presented in plot a for comparison. Fluor., fluorescence; A.U., arbitrary units.
Given the same set of parameter values for each reaction event, the model should predict the observed steady-state results by only shuffling the network architecture. By adjusting parameters that had no experimental basis for their values, we identified a set of parameters consistent with the observed data (see Table S3 in the supplemental material). The periodic-dilution experiment captures corrected absorbances within an A600 range of 0.01 to 1.0, corresponding to a range of roughly 107 to 109 CFU/ml. As shown in Fig. 6b, the model suggests that (i) the bistable region for architecture c occurs before the lowest density observable from the experiment and (ii) maximal induction of architecture a is achieved at densities higher than those observable from the experiment. Both predictions are consistent with the observed behavior.
The wild-type lux operon behavior is consistent with a threshold response.
In the bistable region (if there is one), different initial conditions can drive the system to different steady-state attractors. To test the native lux architecture, namely, network architecture b, for bistability, we varied the initial condition in three ways. Three cultures were respectively grown to an initial absorbance below the induction threshold, an initial absorbance above the induction threshold, and an initial absorbance above the induction threshold induced with 10 μM 3OC6HSL. Here we define the induction threshold as the absorbance at which p(luxI)-mediated gene expression is activated. The nominal luxR configuration for architecture b (plasmid pluxG-102) induces the threshold response at a low initial absorbance (A600 of ∼0.05; Fig. 6a). This low induction threshold made varying the initial condition difficult. Therefore, we altered the luxR RBS to increase the induction threshold (plasmid pluxG-102C), a manipulation suggested by the mathematical model as shown in Fig. 7a. Figure 7b presents the steady-state data confirming this model prediction.
FIG. 7.
Model prediction and experimental verification of the effect of altering luxR RBS strength. As shown in plot a, the model predicts that decreasing the luxR RBS strength increases the density of the induction threshold, that is, the minimum population density required for activation of the threshold response. Plot b verifies this prediction by comparing the responses for architecture b using plasmids pluxG-102 and pluxG-102C, which differ only by the luxR RBS. The arrows in plot b designate the induction threshold. The negative control results (pPROLar.A122 and pPSSUB-102) are presented in plot b for comparison. Fluor., fluorescence; A.U., arbitrary units.
Overnight cultures with different initial conditions were then used to start three respective periodic-dilution experiments using plasmids pluxG-102C and pPSSUB-102. For the evolution of the absorbances and fluorescences over time, see Fig. S4 in the supplemental material. After the system equilibrated (at t = 8 h), the system steady states were compared. Figure 8 demonstrates that, to the accuracy of the experiment, the steady states are virtually indistinguishable. Due to day-to-day variability in this experiment, the steady-state results for pluxG-102C and pPSSUB-102 presented in Fig. 8 were obtained from a single experiment. Comparing results from different days also demonstrates little difference in the steady states obtained from different initial conditions (see Fig. S5 in the supplemental material). These data do not completely rule out the possibility of bistability; for example, small positive feedback from the luxR promoter can lead to a small bistable region (see Fig. S7 in the supplemental material). Our data suggest that a bistable region, if present, is sufficiently small to render the behavior of the native lux architecture equivalent to a threshold-type response. Also, comparing Fig. 8 to Fig. 4b emphasizes that architecture c has a much larger region of bistability than architecture b.
FIG. 8.
Architecture b does not exhibit bistability under these circumstances. Here, initial cultures were grown to a low absorbance (A600o of <0.2), grown to a high absorbance (A600o of >0.7), or induced with 10 μM 3OC6HSL. For architecture b, consisting of pluxG-102C and pPSSUB-102, the same steady state is obtained, regardless of the initial conditions tested, to the accuracy of the experiment. The negative (neg.) control results (pPROLar.A122 and pPSSUB-102) are plotted for comparison. Fluor., fluorescence; A.U., arbitrary units.
DISCUSSION
The modeling and experimental results presented in this paper illustrate how simple changes in network architecture can dramatically alter the steady-state behavior of a cell-cell signaling network. Increasing the number of lux regulatory elements under the control of the p(luxI) promoter respectively yields graded, threshold, and bistable responses. Previous implementations of synthetic bistable gene circuits (4, 7, 21, 26, 29) have all required some degree of cooperativity. Here, the modeling suggests that bistability can be engineered from noncooperative components by taking advantage of the fact that two components (3OC6HSL and LuxR) combine to form the activated transcription factor. Placing both luxR and luxI under the control of the p(luxI) promoter introduces a nonlinearity sufficient for bistability, as was demonstrated experimentally.
A previous modeling work suggested that the wild-type lux operon yields bistable behavior (27). The presence of bistability, however, has not been determined experimentally to the best of our knowledge. By manipulating the initial condition around the quorum-sensing induction density, we demonstrated that the response of the wild-type lux operon is more consistent with a threshold, as opposed to a bistable, response. We expect that understanding how different network architectures render different responses for the quorum-sensing system should provide valuable insight into designing new synthetic gene circuits. For example, several recent papers assume that formation of the activated lux transcription factor is either cooperative (49) or the result of elementary dimerization of two transcription factor-signaling molecule complexes (5, 36, 60). Coupled with positive feedback, both of these assumptions would lead to the erroneous prediction that architecture b (positive feedback on luxI) yields a significant bistable range. Using such assumptions in other modeling efforts will similarly yield incorrect predictions about the steady-state behavior of these engineered constructs.
The experimental results also raise questions about how the natural lux system evolved. Gene duplication, gene shuffling, and point mutations can each give rise to new genes and function (1). Recent theoretical and experimental results suggest that certain motifs are particularly “evolvable,” in that new functions readily arise from either gene shuffling of regulatory and output elements (23), point mutations to operons with dynamical plasticity (34, 54), a combination of the two (17, 55), or gene duplication and divergence (50). The work presented in this paper suggests that simple rearrangements of the lux regulatory module can result in qualitatively different steady-state population level responses. Additionally, simple shuffling of the architecture can give rise to population sensors that induce at closely clustered population densities (within 106 to 108 CFU/ml). Thus, shuffling from one architecture to another could maintain a quantitatively functional population sensor. Such robustness may explain the spate of recent synthetic-biology works that have successfully reengineered this operon to obtain effective quorum-sensing behavior by using a variety of different architectures (3, 5, 6, 28, 44, 59). Also, if quorum sensing indeed descended from a common ancestral operon, then these results suggest that any of these behaviors might be found in current quorum-sensing architectures.
Such a finding has particular import due to the fact that quorum sensing has been implicated as a global regulator of virulence in several human and agricultural pathogens (30, 31, 47, 61). The possibility of bistability arising in such pathogens could yield insight into understanding and controlling these pathogens. For example, recent modeling papers have predicted bistability in the regulation of the plant pathogen Agrobacterium tumefaciens (22) and the human pathogen P. aeruginosa (12, 16). If bistability is indeed present, then once virulence genes are turned on, the pathogen density must be reduced substantially below the induction density to turn off expression of the virulence factors. Elimination of the virulence factors themselves depends on the stability of these factors in the cellular milieu. Consequently, the virulence factors may not be readily down-regulated. Once quorum sensing is down-regulated, though, the bifurcation works in one's favor by making reinduction difficult. Unfortunately, the presence of bistability cannot be readily inferred by simply inspecting the DNA sequences of the quorum-sensing operons in pathogens. However, combining our findings with both existing knowledge of the operon structure and biochemical data on transcription factor cooperativity permits us to speculate as to the response of the quorum-sensing network. Namely, the modeling suggests that (i) a threshold response results from positive feedback on a single, noncooperatively bound regulatory element (either a LuxR or a LuxI homolog) and (ii) bistability results from either positive feedback on two noncooperatively bound regulatory elements or positive feedback on one or more cooperatively bound regulatory elements. Using these criteria, we predict, as shown in Table 2, that the LasR/LasI, EsaR/EsaI, and ExpR/ExpI systems should exhibit threshold responses while the TraR/TraI and CepR/CepI systems are capable of bistability. Bistability is likely a feature of gram-positive quorum-sensing pathogens as well. For example, S. aureus exhibits positive feedback on both the quorum-sensing signal and receptor (39). We stress that the presence of bistability should be rigorously examined in these systems by using either periodic-dilution experiments, as presented in this work, or by using other constant growth rate environments such as a chemostat or a turbidostat.
TABLE 2.
Predicted behavior of the quorum-sensing circuits of gram-negative plant and human pathogensa
| Pathogen | LuxI/LuxR homolog | Cooperative DNA binding | Feedbackb | Predicted response | Reference(s) |
|---|---|---|---|---|---|
| Agrobacterium tumefaciens | TraR/TraI | No | +, TraR/TraI | Bistable | 20, 62 |
| Burkholderia cepacia | CepR/CepI | Yes | +, CepI | Bistable | 58 |
| Erwinia chrysanthemi | ExpR/ExpI | No | +, ExpI | Threshold | 8, 42 |
| Pantoea stewartii | EsaR/EsaI | No | −, EsaR | Threshold | 38 |
| Pseudomonas aeruginosa | LasR/LasI | No | +, LasI | Threshold | 45 |
Based on the experimentally observed cooperativity of DNA binding and the feedback on quorum-sensing elements.
Plus and minus signs denote positive and negative feedback, respectively.
The difference in response for the LasR/LasI and CepR/CepI systems is particularly intriguing because (i) both systems are activated by the same acyl-HSL signal (3OC12HSL) and (ii) their respective hosts, P. aeruginosa and B. cepacia, are known to coinfect the lungs of cystic fibrosis patients, leading to impairment of lung function and eventually death (10). Interestingly, it appears that P. aeruginosa can induce the expression of quorum sensing-controlled virulence factors in B. cepacia under conditions in which B. cepacia cannot induce itself (35). These results suggest that (i) the population density of B. cepacia falls within its bistable region and (ii) the LasR/LasI system is inducing the CepR/CepI system. The coupling of these two species through a common signal renders a quorum-sensing response significantly different than the individual responses.
Understanding how cell-cell communication evolved in bacteria may provide insights into how such feedback evolved in higher organisms since combining such communication with positive feedback is a strategy employed by a wide variety of multi- and unicellular organisms to coordinate and regulate cellular decisions. In Drosophila, temporal gradients of regulatory proteins drive differential patterning of the embryo by using, for example, bistability generated by positive feedback on a signaling receptor (52, 56). In mammalian cells, interferon molecules secreted by virus-infected cells lead uninfected cells to up-regulate antiviral defenses via interferon-stimulated positive feedback (33). How might these complicated mechanisms have evolved? Our results suggest that the architecture of these systems might make them particularly evolvable. Namely, simple shuffling of finely tuned network architectures may render detuned yet functional operons with various degrees of positive feedback.
Supplementary Material
Acknowledgments
E.L.H. gratefully acknowledges support from the Caltech Center for Biological Circuit Design and the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 5F32CA120055. F.H.A. acknowledges support from the U.S. NSF and NIH.
We thank Ron Weiss, Subhayu Basu, and Ming-Tang Chen for providing genetic constructs and experimental advice. We also thank Cynthia Collins, Katie Brenner, Cara Tracewell, and Michael Dougherty for helpful discussions and experimental advice.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular biology of the cell, 4th ed. Garland Science, New York, NY.
- 2.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjørn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. C., E. J. Clarke, A. P. Arkin, and C. A. Voigt. 2006. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 355:619-627. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson, M., M. A. Savageau, J. T. Myers, and A. J. Ninfa. 2003. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113:597-607. [DOI] [PubMed] [Google Scholar]
- 5.Basu, S., Y. Gerchman, C. H. Collins, F. H. Arnold, and R. Weiss. 2005. A synthetic multicellular system for programmed pattern formation. Nature 434:1130-1134. [DOI] [PubMed] [Google Scholar]
- 6.Basu, S., R. Mehreja, S. Thiberge, M.-T. Chen, and R. Weiss. 2004. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA 101:6355-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becskei, A., B. Séraphin, and L. Serrano. 2001. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20:2528-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castang, S., S. Reverchon, P. Gouet, and W. Nasser. 2006. Direct evidence for the modulation of the activity of the Erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J. Biol. Chem. 281:29972-29987. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M., and R. Weiss. 2005. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 23:1551-1555. [DOI] [PubMed] [Google Scholar]
- 10.Chmiel, J. F., and P. B. Davis. 2003. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir. Res. 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhooge, A., W. Govaerts, and Y. A. Kuznetsov. 2003. MATCONT: a MATLAB package for numerical bifurcation analysis of ODEs. ACM Transact. Math. Software 29:141-164. [Google Scholar]
- 12.Dockery, J. D., and J. P. Keener. 2001. A mathematical model for quorum sensing in Pseudomonas aeruginosa. Bull. Math. Biol. 63:95-116. [DOI] [PubMed] [Google Scholar]
- 13.Dong, Y., L. Wang, J. Xu, H. Zhang, X. Zhang, and L. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 14.Dong, Y.-H., J.-L. Xu, X.-Z. Li, and L.-H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egland, K. A., and E. P. Greenberg. 2001. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J. Bacteriol. 183:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagerlind, M. G., S. A. Rice, P. Nilsson, M. Harlén, S. James, T. Charlton, and S. Kjelleberg. 2003. The role of regulators in the expression of quorum-sensing signals in Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 6:88-100. [DOI] [PubMed] [Google Scholar]
- 17.François, P., and V. Hakim. 2004. Design of genetic networks with specified functions by evolution in silico. Proc. Natl. Acad. Sci. USA 101:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 19.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner, T. S., C. R. Cantor, and J. J. Collins. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403:339-342. [DOI] [PubMed] [Google Scholar]
- 22.Goryachev, A. B., D. Jun Toh, K. B. Wee, T. Lee, H. Bao Zhang, and L. Hui Zhang. 2005. Transition to quorum sensing in an Agrobacterium population: a stochastic model. PLoS Comput. Biol. 1:0265-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guet, C. C., M. B. Elowitz, W. Hsing, and S. Leibler. 2002. Combinatorial synthesis of genetic networks. Science 296:1466-1470. [DOI] [PubMed] [Google Scholar]
- 24.Haseltine, E. L., and F. H. Arnold. 2007. Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 36:1-19. [DOI] [PubMed] [Google Scholar]
- 25.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 26.Isaacs, F. J., J. Hasty, C. R. Cantor, and J. J. Collins. 2003. Prediction and measurement of an autoregulatory genetic module. Proc. Natl. Acad. Sci. USA 100:7714-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, S., P. Nilsson, G. James, S. Kjelleberg, and T. Fagerstrom. 2000. Luminescence control in the marine bacterium Vibrio fischeri: an analysis of the dynamics of lux regulation. J. Mol. Biol. 296:1127-1137. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi, H., M. Kaern, M. Araki, K. Chung, T. S. Gardner, C. R. Cantor, and J. J. Collins. 2004. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl. Acad. Sci. USA 101:8414-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer, B. P., A. U. Viretta, M. Daoud-El-Baba, D. Aubel, W. Weber, and M. Fussenegger. 2004. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 22:867-870. [DOI] [PubMed] [Google Scholar]
- 30.Lazdunski, A. M., I. Ventre, and J. N. Sturgis. 2004. Regulatory circuits and communication in gram-negative bacteria. Nat. Rev. Microbiol. 2:581-592. [DOI] [PubMed] [Google Scholar]
- 31.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 25:1389-1403. [DOI] [PubMed] [Google Scholar]
- 33.Marié, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayo, A. E., Y. Setty, S. Shavit, A. Zaslaver, and U. Alon. 2006. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 4:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenney, D., K. E. Brown, and D. G. Allison. 1995. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J. Bacteriol. 177:6989-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMillen, D., N. Kopell, J. Hasty, and J. J. Collins. 2002. Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl. Acad. Sci. USA 99:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 38.Minogue, T. D., M. Wehland-von Trebra, F. Bernhard, and S. B. von Bodman. 2002. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Mol. Microbiol. 44:1625-1635. [DOI] [PubMed] [Google Scholar]
- 39.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, S. Moghazeh, and R. P. Novick. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen, T. B., and M. Givskov. 2006. Quorum sensing inhibitors: a bargain of effects. Microbiology 152:895-904. [DOI] [PubMed] [Google Scholar]
- 41.Reischer, H., I. Schotola, G. Striedner, F. Pötschacher, and K. Bayer. 2004. Evaluation of the GFP signal and its aptitude for novel on-line monitoring strategies of recombinant fermentation processes. J. Biotechnol. 108:115-125. [DOI] [PubMed] [Google Scholar]
- 42.Reverchon, S., M. L. Bouillant, G. Salmond, and W. Nasser. 1998. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 29:1407-1418. [DOI] [PubMed] [Google Scholar]
- 43.Rosson, R. A., and K. H. Nealson. 1981. Autoinduction of bacterial bioluminescence in a carbon limited chemostat. Arch. Microbiol. 129:299-304. [Google Scholar]
- 44.Sayut, D. J., Y. Niu, and L. Sun. 2006. Construction and engineering of positive feedback loops. ACS Chem. Biol. 1:692-696. [DOI] [PubMed] [Google Scholar]
- 45.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shadel, G. S., and T. O. Baldwin. 1991. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J. Bacteriol. 173:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 48.Sitnikov, D. M., G. S. Shadel, and T. O. Baldwin. 1996. Autoinducer-independent mutants of the LuxR transcriptional activator exhibit differential effects on the two lux promoters of Vibrio fischeri. Mol. Gen. Genet. 252:622-625. [DOI] [PubMed] [Google Scholar]
- 49.Song, H., and L. You. 2006. Evolving sensitivity. ACS Chem. Biol. 1:681-682. [DOI] [PubMed] [Google Scholar]
- 50.Teichmann, S. A., and M. M. Babu. 2004. Gene regulatory network growth by duplication. Nat. Genet. 36:492-496. [DOI] [PubMed] [Google Scholar]
- 51.Trott, A. E., and A. M. Stevens. 2001. Amino acid residues in LuxR critical for its mechanism of transcriptional activation during quorum sensing in Vibrio fischeri. J. Bacteriol. 183:387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umulis, D. M., M. Serpe, M. B. O'Connor, and H. G. Othmer. 2006. Robust, bistable patterning of the dorsal surface of the Drosophila embryo. Proc. Natl. Acad. Sci. USA 103:11613-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voigt, C. A., D. M. Wolf, and A. P. Arkin. 2005. The Bacillus subtilis sin operon: an evolvable network motif. Genetics 169:1187-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner, A. 2005. Circuit topology and the evolution of robustness in two-gene circadian oscillators. Proc. Natl. Acad. Sci. USA 102:11775-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Y.-C., and E. L. Ferguson. 2005. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature 434:229-234. [DOI] [PubMed] [Google Scholar]
- 57.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 58.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C.-S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57:452-467. [DOI] [PubMed] [Google Scholar]
- 59.You, L., R. S. Cox III, R. Weiss, and F. H. Arnold. 2004. Programmed population control by cell-cell communication and regulated killing. Nature 428:868-871. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, T., L. Chen, and K. Aihara. 2005. Molecular communication through stochastic synchronization induced by extracellular fluctuations. Phys. Rev. Lett. 95:178103. doi: 10.1103/PhysRevLett.95.178103. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.