Abstract
PHAX (phosphorylated adaptor for RNA export) is the key regulator of U snRNA nuclear export in metazoa. Our previous work revealed that PHAX is phosphorylated in the nucleus and is exported as a component of the U snRNA export complex to the cytoplasm, where it is dephosphorylated (M. Ohno, A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj, Cell 101:187-198, 2000). PHAX phosphorylation is essential for export complex assembly, whereas its dephosphorylation causes export complex disassembly. Thus, PHAX is subject to a compartmentalized phosphorylation/dephosphorylation cycle that contributes to transport directionality. However, neither essential PHAX phosphorylation sites nor the modifying enzymes that contribute to the compartmentalized system have been identified. Here, we identify PHAX phosphorylation sites that are necessary and sufficient for U snRNA export. Mutation of the phosphorylation sites inhibited U snRNA export in a dominant-negative way. We also show, by both biochemical and RNA interference knockdown experiments, that the nuclear kinase and the cytoplasmic phosphatase for PHAX are CK2 kinase and protein phosphatase 2A, respectively. Our results reveal the composition of the compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export. This finding was surprising in that such a specific system for U snRNA export regulation is composed of two such universal regulators, suggesting that this compartmentalized system is used more broadly for gene expression regulation.
The presence of the nuclear envelope (NE) in the eukaryotic cell requires an efficient mechanism for macromolecular exchange across the NE. Exchange is achieved through the nuclear pore complexes that are embedded in the NE. The identification of importin-β family members as transport mediators has greatly increased our knowledge of transport between the nucleus and the cytoplasm and has made it possible to develop a simple model of import and export (reviewed in references 2, 9, 11, and 27). In the model, an import receptor binds to a cargo and carries it into the nucleus. RanGTP (the nuclear form of Ran) then binds to the receptor, leading to the release of the cargo. Similarly, an export receptor binds to a cargo in the nucleus together with RanGTP, forming a trimeric export complex. The complex translocates to the cytoplasm and disassembles due to GTP hydrolysis triggered by activating factors for Ran's GTPase in the cytoplasm. Thus, the asymmetric distribution of RanGTP between the nucleus and the cytoplasm regulates interactions between transport receptors and their cargos, thereby playing a major role in maintaining transport directionality.
RNA transport usually requires more complex mechanisms, and one example is U snRNA export. Major spliceosomal U snRNAs such as U1, U2, U4, and U5 are transcribed in the nucleus by RNA polymerase II and acquire a monomethylated cap structure. In metazoa, U snRNAs initially are exported to the cytoplasm, where they are assembled into complexes with a group of Sm proteins. This Sm-core assembly process is assisted by a protein complex, the SMN complex (24, 30). Subsequently, the cap structure is hypermethylated, and the snRNPs are imported back to the nucleus (20, 22).
U snRNA export requires a monomethyl cap structure on the RNA and the leucine-rich nuclear export signal (NES) receptor CRM1 (8, 10, 12). The interaction between CRM1 and U snRNA is mediated by two adaptors. The first adaptor is the nuclear cap binding complex (CBC), a heterodimeric protein complex (13, 17, 28). CBC binds specifically to the monomethyl cap structure of nascent RNA polymerase II transcripts (34) and promotes U snRNA export as well as pre-mRNA processing (7, 12, 13).
The other adaptor required for U snRNA export is PHAX (phosphorylated adaptor for RNA export) (29). PHAX binds to both CBC and U snRNA, forming a trimeric complex (the precomplex). The precomplex can efficiently interact with CRM1 in a RanGTP-dependent manner, forming a higher-order complex (the U snRNA export complex) (29). Although the NES of PHAX is essential for the precomplex to interact with CRM1, this binding is not constitutive, unlike regular NES-CRM1 interactions. Phosphorylation of PHAX is essential for the formation of the export complex but not of the precomplex (29). After translocating to the cytoplasm through the nuclear pore complex, the U snRNA export complex disassembles in a manner that involves both GTP hydrolysis by Ran and dephosphorylation of PHAX (29). After disassembly, all of the protein components of the export complex, including PHAX, must recycle back to the nucleus, where PHAX gets rephosphorylated for the next round of U snRNA export. Thus, in the case of U snRNA export, the asymmetric distribution of active (phosphorylated) PHAX between the nucleus and the cytoplasm contributes to the directionality of transport, reminiscent of the effect of the asymmetric distribution of active (GTP-bound) Ran in transport mediated by the importin-β family. Although there already are many examples of the regulation of nuclear transport by means of the phosphorylation of transport cargos (reviewed in reference 18), this PHAX system is unique in that the activity of the transport factor, rather than that of the transport cargos, is regulated by phosphorylation.
Interestingly, PHAX also was shown to be involved in the intranuclear transport of a subset of small nucleolar RNAs (snoRNAs), including U3, U8, and U13 (1, 35). Before being transported to the nucleoli, these snoRNAs transit to the Cajal bodies (CBs), where they are modified and assembled into RNPs (26, 33). PHAX appears to be required for this transport (1). Thus, PHAX is not only an export factor for U snRNAs but also is an intranuclear transport factor for snoRNAs. It is not known whether regulation of PHAX activity by phosphorylation/dephosphorylation also is involved in the intranuclear RNA transport.
In spite of the importance of the regulated phosphorylation of PHAX, neither the essential phosphorylation sites nor the enzymes that contribute to PHAX phosphorylation/dephosphorylation have been identified. Here, we identify the PHAX phosphorylation sites that are essential for U snRNA export. We also identify, by both in vitro and in vivo experiments, that the kinase and the phosphatase for PHAX are CK2 kinase and protein phosphatase 2A (PP2A), respectively. We further show that CK2 is mainly nuclear, whereas PP2A is mainly or exclusively cytoplasmic in mammalian cells and Xenopus oocytes. Our results clarify the composition of the novel compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export. These results are unexpected and surprising, since two such universal regulators, CK2 and PP2A, constitute such a specific system for U snRNA export regulation. We therefore suggest that this mechanism is more broadly used during gene expression.
MATERIALS AND METHODS
Cell culture.
Human cervical carcinoma HeLa cells and mouse embryonic fibroblast NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin in an atmosphere containing 5% CO2.
DNA constructs.
The human PHAX, CK2α, CK2α′, and PP2A-Bα cDNAs were PCR amplified from a Marathon Ready HeLa cDNA library (BD Biosciences/Clontech) and cloned into the BamHI and XhoI sites of a pcDNA3 vector containing Flag or Myc tag sequence. To construct short interfering RNA (siRNA)-resistant CK2α and CK2α′ cDNAs, the nucleotide sequence in each cDNA was mutated without amino-acid-coding alterations to avoid repression by the siRNAs. The expression vector Flag-mPHAX ΔNLS+NES was constructed by adding two tandem copies of viral NS2 NES sequences (MTKKFGTLTI) (4) to the C terminus of FLAG-mPHAX ΔNLS, in which two major nuclear localization signals (NLSs) of PHAX were mutated (31). Other mouse PHAX (mPHAX) mutants were constructed by PCR-based mutagenesis with KOD-Plus (Toyobo).
Recombinant proteins and band shift assay.
Recombinant PHAX, CRM1, RanQ69L, CBP80, and CBP20 proteins were expressed and purified as described previously (29). Band shift assays were performed as described previously (29).
RNA microinjection into Xenopus oocytes.
RNA microinjection into Xenopus oocytes was performed as described previously (15).
Preparation of subcellular fractions.
HeLa or NIH 3T3 cells were harvested and washed twice with ice-cold phosphate-buffered saline (PBS). Subcellular fractionation was performed essentially as described previously (3). The total cell lysate was prepared by briefly sonicating the cells in RSB-150 (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 2.5 mM MgCl2) containing 0.5% Triton X-100, 0.5 mM dithiothreitol (DTT), and protease inhibitors. The lysate was centrifuged in a microcentrifuge for 10 min, and the supernatant was recovered.
Purification of PHAX phosphatase.
The substrate for the phosphatase assay was prepared by in vitro phosphorylation of recombinant PHAX Δ3-8 mutant protein with recombinant CK2 kinase (Boehringer Mannheim) and [γ-32P]ATP (Amersham). Phosphatase fractions were mixed with the γ-32P-labeled PHAX Δ3-8 protein in a volume of 30 μl in 20 mM morpholinepropanesulfonic acid (pH 7.0), 10 mM MgCl2, 1 mM DTT, 1 mg/ml bovine serum albumin (BSA). After incubation for 10 min at 30°C, 1 ml of stop solution (7% charcoal, 10% ethanol, 0.1 M HCl, 10 mM KH2PO4) was added to the reaction mixture. The precipitated protein was removed by centrifugation, and the radioactivity in an aliquot of the supernatant was measured using a scintillation counter.
All of the procedures for the phosphatase purification were performed at 4°C. The cytoplasmic S100 extract from HeLa cells (3) was fractionated by ammonium sulfate precipitation, and the pellet from 30 to 50% saturation was collected. The pellet was dissolved in buffer A (20 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 5% glycerol, 1 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride), and the ammonium sulfate concentration of the solution was brought up to 0.5 M. The solution was applied to a phenyl-Sepharose column equilibrated with buffer A containing 0.5 M ammonium sulfate. The bound material was step eluted with 0.5, 0.25, and 0 M ammonium sulfate in buffer A. The active 0 M eluate was applied to a MonoQ (fast protein liquid chromatography; Pharmacia) column, and the bound protein was eluted with a linear gradient of 0.2 to 0.5 M NaCl in buffer A. The active fractions were collected, desalted by a G-25 column, concentrated by a MonoQ (SMART system; Pharmacia) column, and then fractionated by a Superose 6 (SMART system; Pharmacia) column. Two peaks of active fractions, A (∼250 kDa) and B (>440 kDa), were obtained, and each peak fraction was further purified separately by a MonoQ SMART system using an NaCl gradient of 0.2 to 0.45 M in buffer B (the same as buffer A, except pH 8.0). The proteins in the active fractions were fractionated by 4 to 20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were visualized by Coomassie staining.
Mass spectrometry.
Each protein band was excised from the polyacrylamide gel, reduced, alkylated, and digested overnight with trypsin (32). Proteins subsequently were identified by high-mass-accuracy matrix-assisted laser desorption ionization (MALDI) peptide mapping. To this end, a 0.5-μl aliquot of each digestion supernatant was analyzed by MALDI mass spectrometry on a Reflex time-of-flight mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with delayed extraction and detector bias gating (16). The resulting peptide masses then were searched against a comprehensive nonredundant protein database using the program PeptideSearch without taxonomy or molecular weight restrictions (21).
siRNA-mediated knockdown.
All siRNAs were obtained from Invitrogen (Stealth siRNAs of the 25-mer duplex). The target sequences were the following: 5′-GGAAAUCAAGAUGACUACCAGCUGG-3′ for human CK2α, 5′-CCUGACAGACUUUGAUAUCCGGUUU-3′ for human CK2α′, 5′-CCAACCAGAGUGACCUGAUUGAGCA-3′ for human CK2β, 5′-GUGCAAGUGGCAAGCGAAAGAAAGA-3′ for human PP2A-Bα, 5′-UCAGUUGGUGAGGAUAGCCAAGGUU-3′ for mouse CK2α, 5′-GCACAGGGAUGUGAAACCUCACAAU-3′ for mouse CK2α′, 5′-GGAGAAUAUUUGCCAACGCCCACAC-3′ for mouse PP2A-Bα, and 5′-CGAAUCCUACAAAGCGCGCACGUGA-3′ for the control siRNA.
Transient transfection of siRNAs was carried out using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol. At 48 h after the first transfection, the cells were diluted 1/4 or 1/5, and a second transfection of siRNAs was performed if necessary. At 24 h after the second transfection (i.e., 72 h after the first transfection), the cells were transfected with the Flag-mPHAX plasmid. The cells were harvested 24 h later directly in SDS sample buffer and were analyzed by Western blotting. The rescue expression plasmids were transfected with Lipofectamine 2000 reagent (Invitrogen) simultaneously with the Flag-mPHAX plasmid.
Western blot analysis.
To improve the separation of the phosphorylated and nonphosphorylated forms of PHAX, 6 M urea was included in the separating gel of SDS polyacrylamide gel. Western blot signals were visualized using enhanced chemiluminescence (Amersham) and quantified using an LAS-3000 luminoimage analyzer (Fujifilm). The following antibodies were used for Western blot analysis: anti-CK2α/α′ (BD Biosciences), anti-CK2β 6D5 (Calbiochem), anti-PP2A-A 4G7 (Upstate Biotechnology Inc.), anti-PP2A-B 2G9 (Upstate Biotechnology Inc.), PP2A-C 7A6 (Upstate Biotechnology Inc.), anti-nucleolin 4E2 (MBL), anti-hnRNP-C1/C2 4F4 (Sigma), anti-Flag M2 (Sigma), anti-Myc MC045 (Nacalai Tesque), anti-GAPDH 6C5 (Ambion), and anti-PHAX polyclonal antibody (29).
Immunofluorescence.
For immunofluorescence, cells were grown on coverslips to 50 to 70% confluence, fixed in 4% formaldehyde in PBS at room temperature or in methanol at −20°C for 15 min, and washed with PBS. Cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min, washed twice with PBS, and blocked with 3% BSA in PBS at 4°C. Cells were incubated with primary antibodies at appropriate dilutions for 1 h at room temperature in a humidified chamber. Cells were washed three times with TBS (40 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 0.1% Tween-20. The antibodies used for immunofluorescence were anti-CK2α (Upstate Biotechnology Inc.), anti-PP2A-B 2G9 (Upstate Biotechnology Inc.), anti-Flag M2 (Sigma), and anti-PHAX antibody (29). Cells then were incubated with Alexa 568-conjugated secondary antibodies and 0.1 μg/ml Hoechst 33342 and were washed. The coverslips were mounted and examined by fluorescence microscopy (Nikon TE300) or by confocal laser-scanning microscopy (Leica TCS-SP2). Fluorescence intensities were quantified with KyPlot 5.0 (KyensLab Inc.).
RESULTS
Identification of PHAX phosphorylation sites essential for U snRNA export.
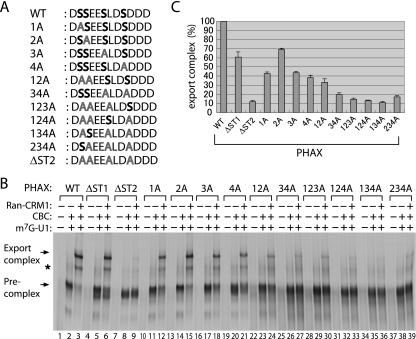
We first examined which phosphorylation sites of mPHAX are important for U snRNA export. Since our previous study showed that PHAX can be phosphorylated and activated by recombinant CK2 kinase in vitro (29), CK2 was a good candidate for the PHAX kinase. A computer search of the PHAX protein sequence revealed eight clusters of serines and threonines that matched the consensus phosphorylation site sequence of CK2 kinase (ST1 to ST8) (Fig. 1A) (19, 23). Accordingly, we constructed six PHAX phosphorylation site mutants. In five of them, all of the serine and threonine residues in each cluster were replaced by alanines (ΔST1 to ΔST5) (Fig. 1B). In the sixth mutant, ST6 to ST8 were simultaneously removed by a small C-terminal deletion (ΔST6-8) (Fig. 1). We also employed the previously described ΔNES mutant that has alanine substitutions in the NES sequence and therefore is defective in the formation of the export complex but not the precomplex (29). The activity of the mutant proteins in U snRNA export complex assembly was examined by an in vitro assay (29). In brief, 35S-labeled PHAX mutant proteins were produced by in vitro translation in rabbit reticulocyte lysate and were incubated with in vitro-transcribed m7G-capped U1 RNA and various recombinant components, including CBC, CRM1, and RanQ69L-GTP. Complex formation was examined by gel mobility shifts, followed by autoradiography (Fig. 1B). The wild-type (WT) protein efficiently formed both the precomplex (containing U1 RNA, CBC, and PHAX) and the export complex (containing U1 RNA, CBC, PHAX, CRM1, and RanGTP) in the presence of corresponding components (lanes 3 and 4), whereas the ΔNES mutant protein was defective in the formation of the export complex but not the precomplex, as expected (lanes 7 and 8). All of the phosphorylation mutants could form the precomplex, which was consistent with the fact that PHAX phosphorylation is not required for precomplex formation (29). All of the mutants except ΔST1 and ΔST2 also could form the export complex with efficiency comparable to that of the WT protein. However, the ΔST2 mutant was severely defective in export complex formation, whereas the ΔST1 mutant was slightly less efficient in export complex formation than the WT (Fig. 1B; also see Fig. 3C for quantification). These results suggested that phosphorylation of the ST2 cluster is most important for PHAX activity and that phosphorylation of the ST1 cluster also contributes, although to a lesser extent.
FIG. 1.
PHAX phosphorylation sites required for export complex formation. (A) The putative CK2 phosphorylation site clusters (ST1 to ST8), as well as a previously identified NES found in the mPHAX, are schematically shown. Amino acid sequences in a single-letter code of each ST region are shown below the diagram. The serine (S) and threonine (T) residues mutated to alanines in the experiments depicted in panel B are in boldface. The consensus sequence for CK2 phosphorylation also is indicated, in which the target S and T residues are in boldface and X, pS, and pY represent any amino acid, phosphorylated serine, and phosphorylated tyrosine, respectively (19). (B) Six putative PHAX phosphorylation site mutants were constructed. In five of them, all of the serine and threonine residues in each cluster were mutated to alanines (ΔST1 to ΔST5). In the ΔST6-8 mutant, ST6 to ST8 were simultaneously removed by a C-terminal deletion. A ΔNES mutant that has alanine substitutions in the NES sequence (29) also was employed. 35S-labeled PHAX WT or mutant proteins were produced by in vitro translation in rabbit reticulocyte lysate and were incubated with in vitro-transcribed m7G-capped U1 RNA (m7G U1 RNA; 1 μM) and various recombinant components, including CBC (1 μM) and a mixture of CRM1 and RanQ69L-GTP (CRM1+Ran; 0.3 μM each). Complex formation was examined by the gel mobility shift assay, followed by autoradiography. Ex and Pre indicate export complex and precomplex, respectively.
FIG. 3.
Identification of essential serine residues in ST2 for export complex formation. (A) The amino acid sequences of the PHAX ST2 site of the WT and alanine substitution mutants used in the experiments. (B) WT and mutant PHAX proteins were assayed for export complex formation as described in the legend to Fig. 1B. The asterisk shows the nonspecific band shift sometimes seen. (C) Relative efficiency of export complex formation from two independent experiments, as described for panel B.
To confirm that phosphorylation of the ST2 cluster is important for U snRNA nuclear export, we employed the well-established Xenopus oocyte microinjection system (Fig. 2). When a mixture of 32P-labeled RNAs was injected into the nucleus, either alone or together with recombinant WT PHAX, export of both U1 and U5 snRNAs was stimulated, whereas neither tRNA nor dihydrofolate reductase (DHFR) mRNA export was affected (Fig. 2, compare lanes 3 and 4 to 5 and 6). Nonexported U6 RNA control remained in the nucleus. When recombinant PHAX ΔNES was injected with the same mixture of labeled RNAs, U snRNA export was greatly reduced, whereas that of DHFR and tRNA was hardly affected (Fig. 2, lanes 9 and 10). Importantly, recombinant PHAX ΔST2 produced a specific dominant-negative effect on U snRNA export similar to that of PHAX ΔNES (lanes 7 and 8), although somewhat weaker. These results strongly support the idea that phosphorylation of the ST2 cluster is important for U snRNA export in vivo.
FIG. 2.
PHAX phosphorylation site cluster ST2 is required for U snRNA export. A mixture of 32P-labeled DHFR mRNA, m7G-capped U1ΔSm, m7G-capped U5ΔSm, m7G-capped U6Δss, and initiator methionyl tRNA was injected into the nucleus of Xenopus oocytes either alone (lanes 1 to 4) or together with 330 fmol/oocyte of recombinant WT PHAX (WT; lanes 5 and 6), ΔST2 mutant (lanes 7 and 8), or ΔNES mutant (lanes 9 and 10). RNA was extracted from nuclear (N) and cytoplasmic (C) fractions immediately (lanes 1 and 2) or 1 h (lanes 3 to 10) after injection and were analyzed by denaturing PAGE, followed by autoradiography.
There are four serine residues in the ST2 cluster. To obtain information regarding which of the four serine residues is important, we constructed mutants in which one or more serine residues in ST2 were changed to an alanine(s) in multiple combinations (Fig. 3A), and the activity of the mutant proteins in U snRNA export complex assembly was assayed in the same manner as that described for Fig. 1B (Fig. 3B and C). The presence of all four serine residues was required for the full activity. Alanine substitution for any one of the four serines significantly reduced PHAX activity. Double, triple, and quadruple mutations generally showed further reductions of the activity (Fig. 3C). These results strongly suggested that the phosphorylation of all four residues contributed to the full activity in U snRNA export complex assembly. We do not know whether the phosphorylation of the intact serine residues in a given mutant is affected by the mutation.
PHAX is phosphorylated by CK2 in vivo.
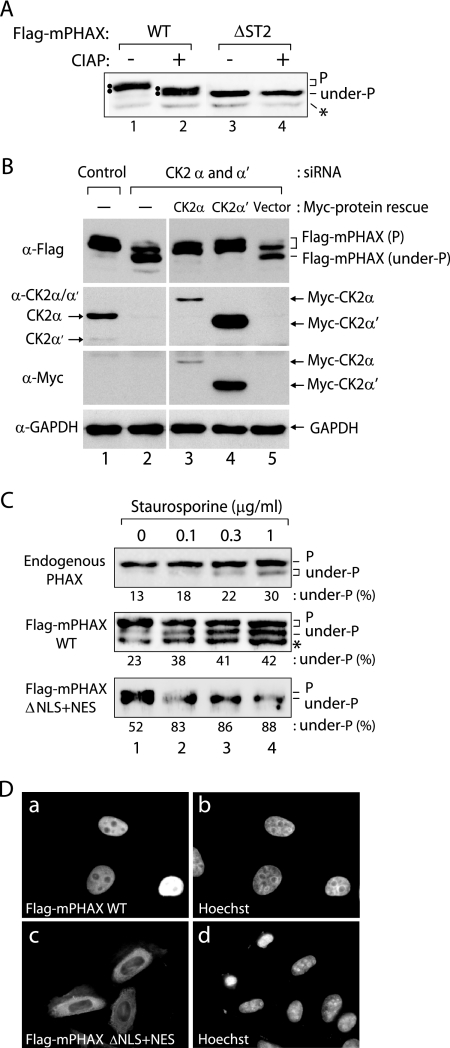
The results described so far strongly suggested that CK2 is the kinase responsible for PHAX activation. To test this hypothesis in vivo, we next examined whether PHAX is converted to the unphosphorylated form when CK2 kinase in HeLa cells is depleted by siRNA-mediated knockdown (Fig. 4). To distinguish between phosphorylated and unphosphorylated forms of PHAX, we took advantage of the previous observation that phosphorylated PHAX showed reduced mobility in SDS-PAGE compared to that of the unphosphorylated form (29). We found that the inclusion of 6 M urea in the separating gel could further enhance the mobility shift. When HeLa cells were transfected with a plasmid expressing Flag-tagged mPHAX (Flag-mPHAX), the expressed PHAX protein was detected clearly as closely migrating double bands in a Western blot analysis with anti-Flag antibody (Fig. 4A, lane 1). When the lysate of the transfected cells was treated with calf intestine alkaline phosphatase (CIAP) prior to the Western blot analysis, the mobility of PHAX protein increased, forming two bands, the upper of which comigrated with the lower band shown in lane 1 (Fig. 4A, lane 2). When more extensive CIAP treatment was performed, PHAX migrated as one band comigrating with the lower band of lane 2 (data not shown). When the ΔST2 mutant protein was expressed, it migrated as one band comigrating with the lower band of lane 2, regardless of CIAP treatment (Fig. 4A, lanes 3 and 4). Although the lower band likely is an unphosphorylated form, since we do not know for sure if the lower band contains no phosphoresidues we refer to this lower band as underphosphorylated. These results indicated that the phosphorylated and underphosphorylated forms of PHAX can be distinguished by the mobility shift in Western blot analysis, and that the majority of the mobility shift is due to the phosphorylation of ST2, the most important phosphorylation site cluster for the PHAX activity.
FIG. 4.
PHAX is phosphorylated by CK2 in vivo. (A) HeLa cells were transfected with the plasmid expressing either Flag-tagged WT mPHAX (Flag-mPHAX WT) or the ΔST2 mutant (ΔST2). After 24 h, the lysate of the transfected cells either was left untreated (−) or was treated with CIAP (+), and Western blot analysis was performed with anti-Flag antibody. Phosphorylated (P) and underphosphorylated (under-P) forms of Flag-mPHAX are indicated. Double bands are indicated by two dots. A degradation product of PHAX is indicated by the asterisk. (B) HeLa cells were treated with siRNAs against CK2α and CK2α′ (lanes 2 to 5) or the control siRNA (Control; lane 1) for 72 h and subsequently were transfected with the plasmid expressing Flag-mPHAX WT. For lanes 3 to 5, either the vector (lane 5) or the plasmids expressing Myc-tagged CK2α (lane 3) or CK2α′ (lane 4) that contained silent mutations conferring resistance to the siRNAs also were transfected with the Flag-mPHAX plasmid. After 24 h, the total cell lysate was prepared from the transfected cells and was analyzed by Western blotting using antibodies denoted on the left side of the figure. The identities of detected bands are indicated on the left side (for endogenous CK2α and CK2α′) or on the right side (for all other proteins). (C) HeLa cells either left untransfected (top row), transfected with Flag-mPHAX WT (middle row), or transfected with Flag-mPHAX ΔNLS+NES (bottom row) were treated with various concentrations of staurosporine (Sigma) for 3 h. The treated cells were analyzed by Western blotting with anti-PHAX antibody (top row) or anti-Flag antibody (middle and bottom rows). P and under-P represent phosphorylated and underphosphorylated forms of PHAX, respectively. The percentage of the underphosphorylated form is shown below each panel. The asterisk indicates a degradation product of PHAX. (D) HeLa cells transfected with Flag-mPHAX WT (images a and b) or Flag-mPHAX ΔNLS+NES mutant (images c and d) were analyzed by indirect immunofluorescence using an anti-Flag antibody (images a and c). Images b and d show DNA staining by Hoechst 33342 of images a and c, respectively.
CK2 kinase is a heterotetrameric enzyme, composed of two catalytic subunits (α and α′) and two regulatory β subunits (reviewed in references 5 and 19). When both CK2α and CK2α′ subunits were knocked down with siRNAs (>95% reduction) (Fig. 4B, lane 2), the Flag-mPHAX protein transiently expressed in HeLa cells was largely converted to the underphosphorylated form (Fig. 4B, lane 2). When either Myc-tagged CK2α or CK2α′ was transiently expressed from siRNA-resistant plasmids, the Flag-mPHAX protein was completely converted back to the phosphorylated forms, even under the CK2 knockdown condition (Fig. 4B, lanes 3 and 4). When the β subunit was knocked down, similar results were obtained (see Fig. 6A). Furthermore, similar results also were obtained when mouse NIH 3T3 cells were employed (see Fig. S1 in the supplemental material). These results indicated that CK2 is the kinase that phosphorylates PHAX in vivo.
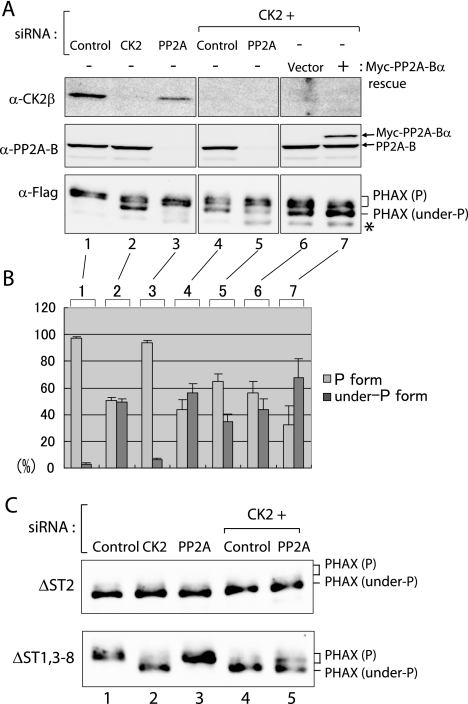
FIG. 6.
PHAX is dephosphorylated by PP2A in vivo. (A) HeLa cells were transfected with siRNAs against CK2β (CK2), PP2A-Bα (PP2A), the control siRNA, or in combination. Transiently expressed Flag-mPHAX protein was examined by Western blot analysis for its phosphorylation status, as described in the legend to Fig. 4. For lanes 6 and 7, Myc-tagged PP2A-Bα (lane 7) or the vector (lane 6) was transiently expressed on top of CK2 knockdown. The asterisk indicates a degradation product of PHAX. (B) Quantitation of the ratio (in percentages) of phosphorylated (P form; light gray bars) and underphosphorylated (under-P form; dark gray bars) forms of Flag-mPHAX from four (lanes 1 to 5) or two (lanes 6 and 7) independent experiments, as described for panel A. Averages and standard errors are shown. (C) Experiments similar to those described for panel A were performed, except that Flag-ΔST2 (upper) and Flag-ΔST1,3-8 (lower) were employed instead of Flag-mPHAX WT.
The transiently expressed PHAX protein was quite efficiently converted to the underphosphorylated form upon CK2 depletion by siRNAs. However, examination of the phosphorylation state of the endogenous PHAX protein by Western blot analysis with α-PHAX antibody showed that the CK2 knockdown induced only a slight conversion of endogenous PHAX to the underphosphorylated form (data not shown). To examine why the endogenous PHAX protein was largely unaffected, HeLa cells were treated with increasing concentrations of a potent kinase inhibitor, staurosporine, which previously was shown to inhibit PHAX phosphorylation in vitro (29). Even at the highest dose, only a small fraction of endogenous PHAX was converted to the underphosphorylated form (Fig. 4C, first row). When HeLa cells were transfected with the plasmid expressing Flag-mPHAX and 24 h later were treated with the same drug, the conversion of Flag-mPHAX to the underphosphorylated form also was inefficient (Fig. 4C, second row). Based on these results, we hypothesized that a large fraction of endogenous PHAX is not engaged in constant shuttling in HeLa cells. The majority of endogenous PHAX does not appear in the cytoplasm and therefore does not have a chance to be dephosphorylated by the putative cytoplasmic phosphatase. Assuming that this hypothesis is correct, PHAX would be efficiently converted to the underphosphorylated form if the protein was forced to localize to the cytoplasm. To test this possibility, we attempted to construct an artificial PHAX protein that would be cytoplasmic by mutating two of the four NLSs (31) and simultaneously introducing two copies of the constitutive minute virus of mice NES to the C terminus (4). This artificial PHAX protein (Flag-mPHAX ΔNLS+NES) was largely localized to the cytoplasm at steady state (Fig. 4D, image c). Approximately half of this mutant protein was in the underphosphorylated form even in the absence of Staurosporine (Fig. 4C, lane 1). This pattern was reminiscent of the behavior of PHAX in the Xenopus oocyte cytoplasm, in which PHAX is subject to rapid cycles of dephosphorylation and rephosphorylation (29). Importantly, the Flag-mPHAX ΔNLS+NES protein was efficiently converted to the underphosphorylated form upon staurosporine treatment (Fig. 4C, bottom row). These results are consistent with the idea that a large fraction of endogenous PHAX is not constantly shuttling in HeLa cells under normal conditions. This small fraction of endogenous PHAX presumably is sufficient for exporting the necessary quantity of U snRNAs. These results also suggested that the conversion of Flag-mPHAX to the underphosphorylated form in the CK2 knockdown cells mainly was due to the lack of the first phosphorylation event of the newly synthesized PHAX protein.
Biochemical purification of PHAX phosphatase from HeLa cells.
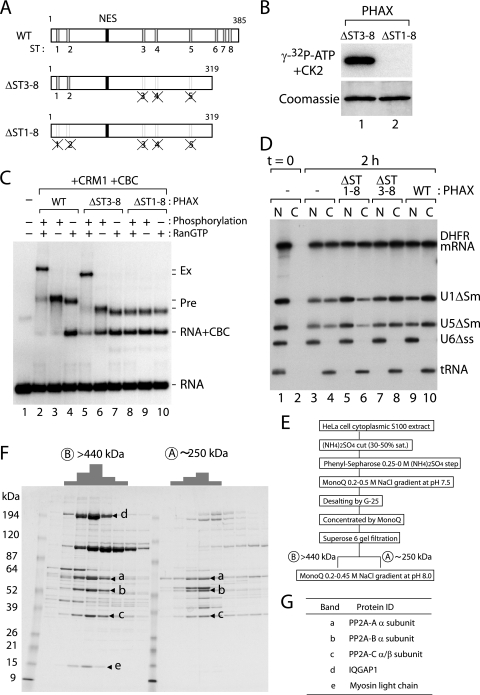
Unlike the case for the PHAX kinase, we did not have any information on the identity of the PHAX phosphatase. Experiments using a series of phosphatase inhibitors with different inhibitory spectra did not yield convincing results (data not shown). We therefore decided to biochemically purify PHAX phosphatase activity from HeLa cells. To obtain an ideal substrate for purifying the putative PHAX phosphatase, we constructed a PHAX mutant in which all of the ST clusters except ST1 and ST2 were mutated by means of combining the preexisting ΔST mutants (ΔST3-8) (Fig. 5A). We also made a control substrate in which all of the ST clusters were mutated (ΔST1-8) (Fig. 5A). When recombinant PHAXΔST1-8 and ΔST3-8 proteins were subjected to in vitro phosphorylation by recombinant CK2 and [γ-32P]ATP, the ΔST1-8 protein was not labeled at all, whereas the ΔST3-8 protein was efficiently labeled (Fig. 5B), indicating that there are no CK2 phosphorylation sites outside of ST1 to ST8. Moreover, the ΔST3-8 protein exerted strong phosphorylation-dependent activity comparable to that of the WT protein in the U snRNA export complex assembly, whereas the ΔST1-8 protein was inactive, although it could efficiently form the precomplex (Fig. 5C). Furthermore, the ΔST3-8 protein specifically stimulated U snRNA export as efficiently as the WT protein, whereas the ΔST1-8 protein specifically inhibited U snRNA export in Xenopus oocyte microinjection experiments (Fig. 5D). These results, taken together, indicated that the phosphorylation of ST1 and ST2 clusters is both necessary and sufficient for U snRNA export complex assembly and that phosphorylated ΔST3-8 protein could serve as a substrate for purifying the critical PHAX phosphatase activity.
FIG. 5.
Purification of PHAX phosphatase activity from HeLa cells. (A) Schematic representation of PHAX WT protein, the mutant in which all of the putative CK2 phosphorylation sites are removed (ΔST1-8), and the mutant in which only the important phosphorylation sites remain (ΔST3-8). (B) Recombinant proteins of PHAX ΔST3-8 and ΔST1-8 were purified from Escherichia coli, and an in vitro phosphorylation reaction with recombinant CK2 and [γ-32P]ATP was performed. The proteins were fractionated by SDS-PAGE, stained by Coomassie brilliant blue (lower), and autoradiographed (upper). (C) In vitro-transcribed 32P-labeled m7G-capped U1ΔSm RNA probe was incubated for 30 min at 25°C in the presence or absence of phosphorylated or unphosphorylated PHAX WT, ΔST3-8, or ΔST1-8 protein (0.5 μM); RanQ69L-GTP (RanGTP; 0.5 μM); CRM1 (0.5 μM); and CBC (0.5 μM), as indicated. The samples were fractionated by native 6% PAGE, followed by autoradiography. Free RNA probe and major complexes are indicated on the right. (D) A mixture of 32P-labeled RNAs as described in the legend to Fig. 2 was injected into the nucleus of Xenopus oocytes either alone (lanes 1 to 4) or together with 330 fmol/oocyte of recombinant ΔST1-8 mutant PHAX (ΔST1-8; lanes 5 and 6), ΔST3-8 mutant PHAX (ΔST3-8; lanes 7 and 8), or WT PHAX (WT; lanes 9 and 10). RNA was extracted from nuclear (N) and cytoplasmic (C) fractions immediately (lanes 1 and 2) or 2 h (lanes 3 to 10) after injection and were analyzed by denaturing PAGE, followed by autoradiography. (E) Purification procedure for PHAX phosphatase activity. See Materials and Methods for details. (F) The active fractions from the Superose 6 (A and B) gel filtration step were further fractionated by MonoQ, and the protein compositions of the MonoQ fractions active in PHAX dephosphorylation activity were analyzed by SDS-PAGE and Coomassie staining. The relative phosphatase activity of each fraction is schematically represented on top of the figure. The protein bands that correlated well with the phosphatase activity are indicated by arrow heads (a to e). (G) The identities of the protein bands (a to e in panel F) revealed by MALDI mass spectrometric analysis are shown.
We developed an assay to detect PHAX phosphatase activity. The recombinant ΔST3-8 protein, labeled with recombinant CK2 and [γ-32P]ATP, was incubated with various phosphatase fractions, and the phosphatase activity was measured by the release of the labeled phosphate from the ΔST3-8 protein (see Materials and Methods for details). When both the nuclear extract and the cytoplasmic S100 extract from HeLa cells were tested for PHAX phosphatase activity, the majority (>80%) of the activity was detected in the cytoplasmic S100 extract (data not shown), which is consistent with the previous finding that PHAX dephosphorylation occurs in the cytoplasm (29). We further purified PHAX phosphatase activity from the cytoplasmic S100 extract over several chromatographic steps, as summarized in Fig. 5E (see Materials and Methods for details). The phosphatase activity always fractionated as a single peak of activity during the purification procedure until the Superose 6 gel filtration step, at which point the activity clearly separated into two peaks, one of approximately 250 kDa and the other of more than 440 kDa. The phosphatase activity of each peak fraction was further purified by MonoQ. When the final purified fractions were subjected to SDS-PAGE and Coomassie staining, several protein bands that correlated well with the phosphatase activity were evident (Fig. 5F, bands a to d). Mass spectrometric analysis revealed that the three protein bands (a to c) that were found in both the 250- and the 440-kDa fractions were the three subunits (A, B, and C) of protein phosphatase 2A (PP2A) (Fig. 5G). The larger fraction (more than 440 kDa) contained two more proteins related to actin organization (band d, IQGAP1; and band e, myosin alkali light chain) in addition to the three subunits of PP2A (Fig. 5G). Careful comparison of the specific activities of the 250- and 440-kDa fractions revealed that they were very similar. Therefore, the significance of the two additional proteins in the larger fraction for PHAX phosphatase activity is not clear. In any case, these results revealed that the purified PHAX phosphatase activity was that of PP2A.
PHAX is dephosphorylated by PP2A in vivo.
The in vitro results suggested that PP2A is the PHAX phosphatase. To test whether this is the case in vivo, we knocked down PP2A in HeLa cells with siRNAs (Fig. 6). We chose to knock down the B subunit because the B subunit determines the substrate specificity (14, 25), and therefore the general toxicity to the cells due to the knockdown might be minimized. When PP2A-B (α isoform) was knocked down (>95% reduction) (Fig. 6A, lane 3), the phosphorylation state of the transfected Flag-mPHAX was unchanged, since it was already exclusively phosphorylated (Fig. 6A and B, lanes 1 and 3). When CK2β was knocked down (83% reduction), a fraction of Flag-mPHAX was converted to the underphosphorylated form, as was already described (Fig. 6A and B, lane 2). However, when both CK2β and PP2A-B were knocked down, the amount of the phosphorylated form was significantly increased compared to that after CK2β-only knockdown (Fig. 6A and B, compare lanes 4 and 5). When the Myc-tagged PP2A-B subunit was transiently expressed in the CK2 knockdown cells, the amount of PP2A-B protein increased only modestly (by ∼40%) (Fig. 6A, lane 7). Nevertheless, a significant increase of underphosphorylated PHAX was observed (Fig. 6A and B, compare lanes 6 and 7). Similar results were obtained with mouse NIH 3T3 cells (see Fig. S1 in the supplemental material). Furthermore, when ΔST2 protein was expressed, its mobility was unchanged under the various conditions (Fig. 6C, top row). However, ΔST1,3-8 protein, in which all of the ST clusters except ST2 were mutated, behaved in a fashion similar to that of WT Flag-mPHAX (Fig. 6C, bottom row), indicating that PP2A as well as CK2 acted on the ST2 cluster in vivo and caused the mobility shift. These results, taken together, strongly suggested that PP2A is the PHAX phosphatase in vivo as well as in vitro.
CK2 and PP2A constitute the compartmentalized phosphorylation/dephosphorylation system.
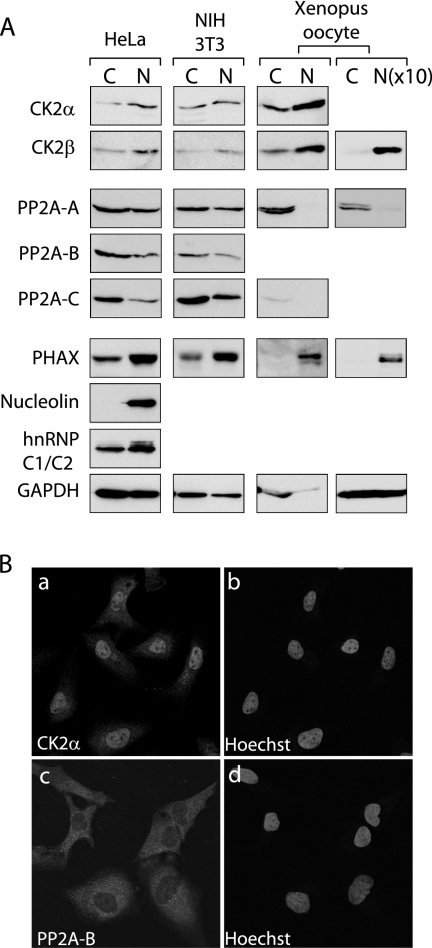
Our previous results indicated that net phosphorylation and dephosphorylation of PHAX occur in the nucleus and cytoplasm of Xenopus oocytes, respectively (29), suggesting that the putative PHAX kinase and phosphatase are mainly nuclear and cytoplasmic, respectively. We therefore next examined whether CK2 and PP2A follow this distribution. The cellular localization of CK2 and PP2A subunits was examined by Western blot analysis of the nuclear and the cytoplasmic fractions from HeLa cells, NIH 3T3 cells, and Xenopus oocytes (Fig. 7A), as well as by immunofluorescent cell staining of HeLa cells (Fig. 7B). As judged by the behavior of nuclear marker proteins (PHAX, nucleolin, and hnRNP C1/C2) and a cytoplasmic marker protein (glyceraldehyde-3-phosphate dehydrogenase), the subcellular fractionations shown in Fig. 7A appeared reasonable, although there is cross-contamination of the nuclear fraction by cytoplasmic material in HeLa and NIH 3T3 cells. Not surprisingly, the fractionation was much more precise in Xenopus oocytes (Fig. 7A). In this condition, both subunits of CK2 were found to be preferentially nuclear in all three cells. There seemed to be some CK2 present in the cytoplasm (Fig. 7A, first two rows). It was unlikely that this was due to the cross-contamination. Even in Xenopus oocytes, in which the nuclear and cytoplasmic fractionation was cleaner, some CK2 signal always was detected in the cytoplasmic fraction, consistent with our previous results that there is some PHAX phosphorylation activity in the cytoplasm of Xenopus oocytes (29). Consistently, the cell staining by α-CK2α antibody yielded mainly nuclear signal but significant cytoplasmic staining (Fig. 7B, image a). The relative CK2α concentrations of the nucleus and the cytoplasm were quantified to be 1 to 0.35 ± 0.02 (n = 45). In fact, it has been reported that the subcellular localization of CK2 subunits is even dynamic (6).
FIG. 7.
Subcellular localization of CK2 and PP2A subunits. (A) Western blotting analysis of nuclear (N) and cytoplasmic (C) fractions from HeLa and NIH 3T3 cells and Xenopus oocytes with antibodies against various subunits of CK2 and PP2A. Antibodies against two nuclear proteins (nucleolin and hnRNP C1/C2) and a cytoplasmic protein (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were employed for checking the quality of the fractionations. The N and C fractions from equivalent cell numbers are compared for the left three columns, whereas a 10 times greater number of N fractions [N(×10)] were loaded for the right-most column. (B) Confocal laser-scanning microscopy of HeLa cells stained with anti-CK2α (a) and anti-PP2A-B (c) antibodies. Images b and d show DNA staining by Hoechst 33342 of images a and c, respectively.
By contrast, PP2A subunits were found to be mainly cytoplasmic in all three cell types. PP2A subunits, as judged by the localization of A and C subunits, were exclusively cytoplasmic in Xenopus oocytes. To directly compare relative concentrations of PP2A-A in the nucleus and cytoplasm, 10 times more nuclear fraction was loaded (Fig. 7A, right-most column). The signal intensities should directly show the relative concentrations of PP2A-A in the nucleus and cytoplasm, since the relative volume of the nucleus versus that in the cytoplasm of a Xenopus oocyte is approximately 1 to 10. Even in this condition, PP2A-A still was almost exclusively cytoplasmic (Fig. 7A, right-most column). In contrast, there seemed to be some nuclear fractionation of PP2A in HeLa cells and NIH 3T3 cells (Fig. 7A, PP2A rows). By immunofluorescence, there was some nuclear PP2A-B signal in HeLa cells (Fig. 7B, image b). The relative PP2A-B concentrations in the cytoplasm and nucleus were determined to be 1 to 0.47 ± 0.03 (n = 28). It can, however, be concluded that CK2 is more concentrated in the nucleus and PP2A is more concentrated in the cytoplasm in all three cell types. These distributions explain why net phosphorylation and net dephosphorylation of PHAX occur in the nucleus and the cytoplasm, respectively, and the two enzymes meet the criteria expected for the compartmentalized PHAX phosphorylation/dephosphorylation system.
DISCUSSION
In this study, the PHAX phosphorylation sites essential for U snRNA export were identified. In addition, the components of the compartmentalized PHAX phosphorylation/dephosphorylation system, the nuclear CK2 kinase and the cytoplasmic PP2A phosphatase, were identified.
CK2 as the PHAX kinase.
The PHAX kinase was shown to be CK2. CK2 is an essential, ubiquitously expressed serine/threonine kinase. CK2 is a heterotetrameric enzyme composed of two catalytic subunits (α and α′) and two regulatory β subunits. CK2 regulates a broad range of cellular functions and has more than 100 known cellular substrates (23). Phosphorylation by CK2 generally increases cell growth and division, and CK2 overexpression therefore is observed often in transformed cancer cells (reviewed in references 5 and 19). It is reasonable to think that CK2 phosphorylation of PHAX is regulated by the state of cell growth, and that an increase of phosphorylated PHAX may accompany the input of cell growth signals in order to increase the production of U snRNAs in response to the requirement for cell growth.
Under normal conditions in rapidly dividing cells, most PHAX is localized in the nucleus and therefore is fully phosphorylated. Our finding that endogenous PHAX is hardly converted to the unphosphorylated form in CK2 knockdown cells, even after 72 h of siRNA treatment, suggests that only a small fraction of cellular PHAX is engaged in U snRNA export, while the majority stays in the nucleus. Although both PHAX phosphorylation and the PHAX NES sequence are required for the U snRNA precomplex to interact with CRM1 and RanGTP, the interaction of phosphorylated PHAX alone with CRM1-RanGTP is unstable (29) but is stabilized by CBC and U snRNA. This cooperative assembly of the export complex should result in a reduction of the rate of export of free PHAX (and CBC) in the absence of RNA substrates, thus preventing futile shuttling of these components and simultaneously reducing the competition between PHAX and other cellular NES-containing proteins for CRM1-mediated export. Thus, it is reasonable that the majority of endogenous PHAX does not appear in the cytoplasm under the conditions of our experiments. A small fraction of endogenous PHAX may be enough to export sufficient U snRNAs for cell growth. At least a portion of nuclear PHAX has been shown to be engaged in the intranuclear movement of a subset of snoRNAs, an activity that does not involve transport to the cytoplasm.
The PHAX phosphorylation sites that are necessary and sufficient for U snRNA export complex assembly were determined to be the ST1 and ST2 serine clusters, both of which are relatively close to the PHAX N terminus. Of the two clusters, ST2 is much more important. The phosphorylation of all four serine residues in ST2 seems to be required for full PHAX activation, although we do not have direct evidence that all of the serine residues are phosphorylated. It is possible that the phosphorylation of only some serine residues actually is important for the export complex formation, while the phosphorylation of others is necessary only for the phosphorylation of the important residues. Also, we cannot exclude the possibility that the presence of some unphosphorylated serine residues is required for the PHAX activity. How does the phosphorylation of ST2 induce interaction with CRM1-RanGTP? One possibility is that the phosphorylation of ST2 changes the conformation of the PHAX protein and exposes the NES sequence. Similar types of regulation, involving exposure of transport signals, have been reported in other cases (reviewed in reference 18). An alternative possibility is that the NES sequence per se is not strong enough for CRM1 binding and that the phosphorylated ST2 region directly interacts with CRM1 to strengthen the interaction. Detailed structural analyses of the unphosphorylated and phosphorylated precomplexes would help answer this question.
PP2A as the PHAX phosphatase.
The PHAX phosphatase was shown to be PP2A. PP2A is an abundant serine/threonine phosphatase in eukaryotic cells that accounts for 0.05 to 0.1% of the total protein mass. PP2A regulates a broad range of cellular functions, and dephosphorylation by PP2A generally decreases cell growth and division, which is the opposite of the effect of phosphorylation by CK2 (14, 25). PP2A is a heterotrimeric complex in which a core dimmer of the catalytic C subunit and the structural A subunit recruits a third regulatory B subunit, and each subunit has isoforms. The B subunit is thought to determine the substrate specificity and subcellular localization and is the most variable. In combination, the number of the PP2A variants is more than 50 (14, 25). The combination of subunits that was identified as the in vitro PHAX phosphatase was Aα/Bα/Cα or β. We do not have any information on whether other PP2A variants also show PHAX phosphatase activity, but during the biochemical purification procedure, the activity was shown as a single peak until the gel filtration step, during which the activity separated into two peaks. However, both peaks contained the same type of PP2A. This may mean simply that this type of PP2A is the most abundant in HeLa cells. At the same time, it is unlikely that there is a second PHAX phosphatase that plays a major role in U snRNA export.
In the Superose 6 gel filtration step, the phosphatase activity clearly was separated into two peaks, one of approximately 250 kDa and the other of more than 440 kDa. The larger peak contained IQGAP1 and the myosin alkali light chain, as well as the three subunits of PP2A (Fig. 5F). As already mentioned, there seems to be no contribution of these two proteins to the PHAX phosphatase activity in vitro. Given the function of the two proteins, it is more likely that this particular PP2A complex is involved in the regulation of actin organization.
The compartmentalized phosphorylation/dephosphorylation system.
The two enzymes identified as the kinase and the phosphatase of PHAX constitute a compartmentalized phosphorylation/dephosphorylation system. This finding was unexpected and surprising, in that two such universal regulators, CK2 and PP2A, constitute such a specific system for U snRNA export regulation. It is therefore likely that this robust system is used more generally. It is interesting that CK2 and PP2A regulate gene expression to drive cell growth and division in opposite directions (reviewed in references 5, 14, 19, and 25). Phosphorylation by CK2 generally increases cell growth and division, and CK2 overexpression therefore is often observed in transformed cancer cells, whereas dephosphorylation by PP2A generally decreases cell growth and division. We speculate that the compartmentalized phosphorylation/dephosphorylation system identified in this study regulates the activities of a broad range of substrates related to the response to cell growth control signals.
Supplementary Material
Acknowledgments
We thank Odile Filhol for materials for the CK2 activity assay.
This work was supported by EMBL and CREST and by grants from the Ministry of Education, Science, Sports, and Culture of Japan.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Boulon, S., C. Verheggen, B. E. Jady, C. Girard, C. Pescia, C. Paul, J. K. Ospina, T. Kiss, A. G. Matera, R. Bordonne, and E. Bertrand. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell 16777-787. [DOI] [PubMed] [Google Scholar]
- 2.Cole, C. N., and C. M. Hammell. 1998. Nucleocytoplasmic transport: driving and directing transport. Curr. Biol. 8R368-R372. [DOI] [PubMed] [Google Scholar]
- 3.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelsma, D., R. Bernad, J. Calafat, and M. Fornerod. 2004. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 233643-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faust, M., and M. Montenarh. 2000. Subcellular localization of protein kinase CK2. A key to its function? Cell Tissue Res. 301329-340. [DOI] [PubMed] [Google Scholar]
- 6.Filhol, O., A. Nueda, V. Martel, D. Gerber-Scokaert, M. J. Benitez, C. Souchier, Y. Saoudi, and C. Cochet. 2003. Live-cell fluorescence imaging reveals the dynamics of protein kinase CK2 individual subunits. Mol. Cell. Biol. 23975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty, S. M., P. Fortes, E. Izaurralde, I. W. Mattaj, and G. M. Gilmartin. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 9411893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 9.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15607-660. [DOI] [PubMed] [Google Scholar]
- 10.Hamm, J., and I. W. Mattaj. 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63109-118. [DOI] [PubMed] [Google Scholar]
- 11.Hetzer, M. W., T. C. Walther, and I. W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21347-380. [DOI] [PubMed] [Google Scholar]
- 12.Izaurralde, E., J. Lewis, C. Gamberi, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376709-712. [DOI] [PubMed] [Google Scholar]
- 13.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78657-668. [DOI] [PubMed] [Google Scholar]
- 14.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353417-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarmolowski, A., W. C. Boelens, E. Izaurralde, and I. W. Mattaj. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, O. N., A. Podtelejnikov, and M. Mann. 1996. Delayed extraction improves specificity in database searches by matrix-assisted laser desorption/ionization peptide maps. Rapid Commun. Mass Spectrom. 101371-1378. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka, N., M. Ohno, I. Moda, and Y. Shimura. 1995. Identification of the factors that interact with NCBP, an 80 kDa nuclear cap binding protein. Nucleic Acids Res. 233638-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komeili, A., and E. K. O'Shea. 2001. New perspectives on nuclear transport. Annu. Rev. Genet. 35341-364. [DOI] [PubMed] [Google Scholar]
- 19.Litchfield, D. W. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 3691-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lührmann, R., B. Kastner, and M. Bach. 1990. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1087265-292. [DOI] [PubMed] [Google Scholar]
- 21.Mann, M., P. Hojrup, and P. Roepstorff. 1993. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol. Mass Spectrom. 22338-345. [DOI] [PubMed] [Google Scholar]
- 22.Mattaj, I. W. 1988. U snRNP assembly and transport, p. 100-114. In M. L. Birnstiel (ed.), Structure and function of major and minor small nuclear ribonucleoprotein particles. Springer-Verlag, New York, NY.
- 23.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17349-368. [DOI] [PubMed] [Google Scholar]
- 24.Meister, G., D. Buhler, R. Pillai, F. Lottspeich, and U. Fischer. 2001. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3945-949. [DOI] [PubMed] [Google Scholar]
- 25.Millward, T. A., S. Zolnierowicz, and B. A. Hemmings. 1999. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 24186-191. [DOI] [PubMed] [Google Scholar]
- 26.Narayanan, A., W. Speckmann, R. Terns, and M. P. Terns. 1999. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 102131-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno, M., M. Fornerod, and I. W. Mattaj. 1998. Nucleocytoplasmic transport: the last 200 nanometers. Cell 92327-336. [DOI] [PubMed] [Google Scholar]
- 28.Ohno, M., N. Kataoka, and Y. Shimura. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 186989-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno, M., A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101187-198. [DOI] [PubMed] [Google Scholar]
- 30.Pellizzoni, L., J. Yong, and G. Dreyfuss. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 2981775-1779. [DOI] [PubMed] [Google Scholar]
- 31.Segref, A., I. W. Mattaj, and M. Ohno. 2001. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 33.Verheggen, C., D. L. Lafontaine, D. Samarsky, J. Mouaikel, J. M. Blanchard, R. Bordonne, and E. Bertrand. 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 212736-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visa, N., E. Izaurralde, J. Ferreira, B. Daneholt, and I. W. Mattaj. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 1335-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins, N. J., I. Lemm, D. Ingelfinger, C. Schneider, M. Hossbach, H. Urlaub, and R. Luhrmann. 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16789-798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.