Abstract
Genomic imprinting governs allele-specific gene expression in an epigenetically heritable manner. The characterization of histone modifications at imprinted gene loci is incomplete, and whether specific histone marks determine transcription or are dependent on it is not understood. Using chromatin immunoprecipitations, we examined in multiple cell types and in an allele-specific manner the active and repressive histone marks of several imprinted loci, including H19, KvDMR1, Snrpn promoter/exon 1, and IG-DMR imprinting control regions. Expressed alleles are enriched for specific actively modified histones, including H3 di- and trimethylated at Lys4 and acetylated histones H3 and H4, while their silent counterparts are associated with repressive marks such as H3 trimethylated at Lys9 alone or in combination with H3 trimethylated at Lys27 and H4/H2A symmetrically dimethylated at Arg3. At H19, allele-specific histone modifications occur throughout the entire locus, including nontranscribed regions such as the differentially methylated domain (DMD) as well as sequences in the H19 gene body that are not differentially methylated. Significantly, the presence of active marks at H19 depends on transcriptional activity and occurs even in the absence of the DMD. These findings suggest that histone modifications are dependent on the transcriptional status of imprinted alleles and illuminate epigenetic mechanisms of genomic imprinting.
Genomic imprinting is a mechanism of transcriptional regulation through which expression of a subset of mammalian genes occurs exclusively from the maternal or paternal allele (35, 51). Notably, imprinted genes are found in large clusters throughout the genome, and the imprinted regulation of genes in the cluster is typically mediated through a short DNA sequence called the imprinting control region (ICR). The monoallelic expression of imprinted genes likely results from epigenetic modifications that differentially mark the parental alleles at the ICR during gametogenesis. Imprinted genes often exhibit allelic differences in DNA methylation and chromatin structure at the ICR and elsewhere, as well as in asynchronous replication, each of which may represent, or contribute to, the marking of the parental chromosomes. While the exact nature of the epigenetic mark remains to be established fully and may be gene or cluster specific, one step towards this goal is understanding the specific DNA methylation patterns and chromatin modifications that are important for imprinted expression.
The H19 gene, which encodes a noncoding RNA (ncRNA) and is located within a large imprinted domain at the distal end of mouse chromosome 7, is expressed exclusively from the maternal allele (4). Imprinting of H19 is controlled by a 2-kb ICR, also known as the differentially methylated domain (DMD) (Fig. 1A) (44). The DMD exhibits paternal-allele-specific DNA methylation and has been proposed to harbor the imprinting mark that distinguishes the parental alleles of H19 and the linked and oppositely expressed Igf2 gene (12, 29). The differential DNA methylation of the DMD is present in the early embryo, is resistant to the genome-wide demethylation that occurs during preimplantation, and persists throughout subsequent development (47, 48). The H19 paternal allele is also hypermethylated at the promoter, consistent with its silent transcriptional status. However, DNA methylation in the promoter region occurs after implantation and may result from spreading of DNA methylation from the DMD sequence.
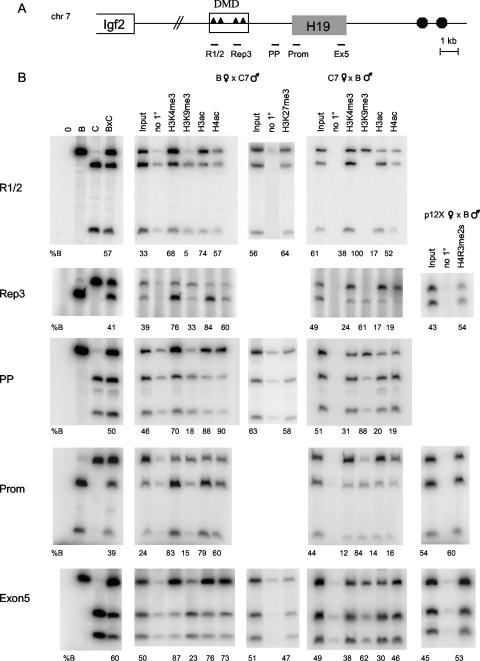
FIG. 1.
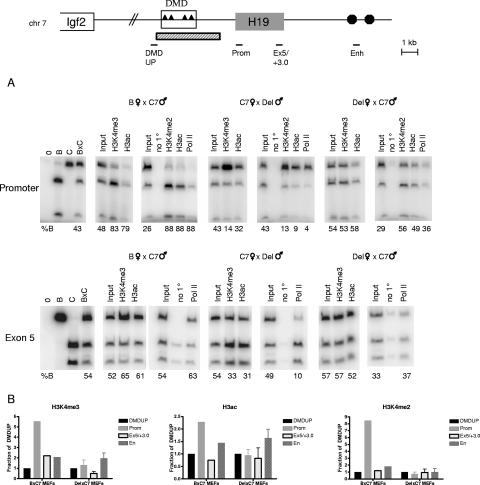
Allelic chromatin modifications across the H19 locus in MEFs. (A) Schematic of the H19/Igf2 locus. Triangles represent the four CTCF-binding sites within the 2-kb DMD located upstream of the H19 gene, and circles indicate the endodermal enhancers. The H19 regions analyzed in panel B are indicated below the diagram. (B) Allele-specific ChIPs were carried out with antibodies against H3K4me3, H3K9me3, H3ac, H4ac, H3K27me3, H4R3me2s, or no primary antibody (no 1°). DNAs from control (B, C, and B × C genomic DNAs; left panels), input, and precipitated samples were subjected to PCR for the indicated region of H19 and then digested with a restriction enzyme that is polymorphic between B and C. The numbers underneath each panel indicate the percent B allele relative to total. The genotype of the MEFs used (B × C7, C7 × B, or p12X × B) and the parental origin of each allele are indicated at the top. Note that ChIP assays were performed multiple times and the allele-specific assays were conducted on each ChIP sample, but only one representative experiment is shown for each antibody and region.
The DMD is critical for the regulation of H19 imprinted expression, as deletion or mutation of DMD sequence leads to loss of allele-specific expression and differential DNA methylation at this locus (14, 15, 27, 39, 43, 44). Four repeats within the DMD that represent binding sites for the CTCF protein play a critical role in mediating the various functions of the DMD, including the insulator or enhancer blocker activity that prevents Igf2 from accessing shared enhancers on the maternal allele (5, 25, 27, 28, 42). Additionally, the DMD and associated CTCF-binding sites are important for transcriptional activation of the H19 maternal allele, as deletion of the CTCF-binding sites within the DMD causes a delay in H19 gene activation in mouse embryos (14, 45).
The paternal-allele-specific DNA methylation of the DMD is established during male gametogenesis, after erasure of DNA methylation at this locus in primordial germ cells (PGCs) (10, 24, 49). Curiously, the two parental alleles are DNA methylated at different times during male germ cell development (11). These data suggest that the parental chromosomes are differentiated through an epigenetic signal other than DNA methylation during this time. A strong candidate for this signal is chromatin structure. In support of this hypothesis, specific active but not repressive modifications were observed during spermatogenesis at H19 (13), although the allelic pattern of the marks was not determined and the analysis was conducted in more differentiated cell types. Ideally, chromatin analysis would be performed in the PGCs, but the small number of cells makes such an analysis technically infeasible at this time. Nevertheless, the characterization of the chromatin profile across the H19 locus during development would allow a better understanding of the role that chromatin plays in the discrimination of parental alleles at this gene. In fact, a recent comprehensive investigation of the chromatin profile of the 250-kb Igf2r imprinted locus has provided important new mechanistic information regarding the control of imprinting across a domain that is regulated by the transcription of a large ncRNA (38).
Previous analyses have reported differential allelic modifications for various imprinted genes, including H19, but at discrete locations and in limited cell types (8, 13, 19, 22, 31, 37, 50). Initial studies examining the H19 chromatin structure in fibroblasts showed that the parental alleles exhibit differential histone acetylation in exon 1 (37), as well as at the H19 promoter and promoter-proximal (PP) region (22). A more recent study reported differential chromatin modifications at the DMD in embryonic stem (ES) cells and neonatal liver (13). Specifically, the maternal DMD was enriched for acetylated histone H3 (H3ac) and for histone H3 dimethylated at Lys4 (H3K4me2). In contrast, the paternal allele was preferentially bound by H3 trimethylated at Lys9 (H3K9me3) and histone H4 trimethylated at Lys20 (H4K20me3) (13). Together with data from other imprinted loci, these analyses suggest associations of active histone modifications with the expressed allele and of repressive histone modifications with the silenced allele. However, it is unclear whether the histone modifications at ICRs are responsible for allelic identity or merely reflect the transcriptional activity of the allele. Finally, to understand the mechanism of imprinting at various imprinted clusters, it would be desirable to conduct comparative chromatin profiles at distinct ICRs. Although this has been performed to a limited extent at a number of loci by multiple laboratories employing different antibodies, no comprehensive analysis has been performed in a single study in various cell types.
The goal of this study was to characterize the allele-specific chromatin modifications at several imprinting domains, including multiple regions across the H19 locus and the KvDMR1, Snurf/Snrpn (abbreviated as Snrpn), and IG-DMR ICRs. We also tested whether histone modifications are dependent on transcriptional activity at H19. These experiments were carried out in multiple cell lines that reflected distinct developmental time points, including a pluripotent lineage such as ES cells as well as more differentiated cell types such as mouse embryonic fibroblasts (MEFs) and neonatal liver. Our data show that the H19 parental chromosomes exhibit differential chromatin modifications throughout the entire locus, including at the DMD, promoter, and transcription unit. Histone H3 trimethylated at Lys4 (H3K4me3), H3K4me2, H3ac, and acetylated histone H4 (H4ac) are preferentially bound on the maternal allele, while H3K9me3 occurs almost exclusively on the paternal chromosome. In contrast to previous reports (26), we did not observe preferential association of another repressive modification, histone H4/H2A symmetrically dimethylated at Arg3 (H4R3me2s), with one parental allele. Furthermore, whereas the highest level of the histones associated with active chromatin occurs at the H19 promoter, the repressive H3K9me3 modification is distributed more evenly across the entire locus. These differential modifications, including H3K27me3, are also present at KvDMR1, IG-DMR, and the Snrpn ICR, although we fail to observe differential H3K27me3 at H19. Finally, we analyzed the chromatin properties of the H19 locus in DMD-deleted MEFs, which do not express H19, and in neonatal liver harboring the same DMD deletion, where H19 is expressed. These data clearly show that transcriptional activity at H19 is critical for establishing the chromatin modifications at this locus, even in the absence of the DMD.
MATERIALS AND METHODS
Isolation of MEFs and neonatal liver.
MEFs were isolated from individual day 13.5 embryos generated from reciprocal crosses between C57BL/6 (B) mice and mice with Mus musculus castaneus chromosome 7 (C7) (34) or chromosomes 7, 12, and X (p12X) (J. Mager, C. Krapp, M. Mann, and M. S. Bartolomei, unpublished) in a B background. The head and internal organs were removed, and the embryos were minced and incubated in trypsin for 30 min at 37°C. The cells were then resuspended in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (HyClone), 2 mM l-glutamine (Invitrogen), and 50 μg/ml gentamicin (Invitrogen). For the Δ3.8 kb-5′H19 deletion samples, MEFs and neonatal livers were isolated from reciprocal crosses between Δ3.8 kb-5′H19 mice (46) and C7 mice. The ES cells were derived from blastocysts isolated from matings between B females and p12X males. Differentiation into embryoid bodies was carried out by culturing for 7 days in medium without leukemia inhibitory factor.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were carried out using the ChIP assay kit (Upstate) according to the manufacturer's instructions, with a few modifications. For neonatal tissues, nuclei were isolated as described previously (3) and frozen in digestion buffer and 50% glycerol. Prior to carrying out the ChIP analysis, the samples were thawed, washed three times in digestion buffer, and then resuspended in phosphate-buffered saline supplemented with protease inhibitors (Sigma). Cross-linking for all cells was performed with 1% formaldehyde (Sigma) for 15 min at room temperature, and then the reaction was quenched with 0.125 M glycine for 5 min at room temperature. Following lysis, sonication was carried out with a Branson Sonifier 250 at 30% output for four pulses of 10 s each. The sonicated cell supernatant was diluted 10-fold in ChIP dilution buffer (Upstate), and 1% of the material was removed prior to the addition of antibodies (input). Eighty to 100 μg of chromatin was used for each IP reaction with antibodies against H3K4me3 (Abcam, ab8580; lot no. 83188, 94105, 77499, and 197627), H3K4me2 (Upstate, 07-030; lot no. 26335 and 29698), H3K9me3 (Upstate, 07-442; lot no. 32493 and 33453), H3ac (Upstate, 06-599; lot no. 25233 and 31994), H4ac (Upstate, 06-866; lot no. 31992), H4R3me2s (Abcam, ab5823; lot no. 97452), H3K27me3 (Upstate, 07-449; lot no. 24440, and Abcam ab6002; lot no. 142927 and 134747), and RBP1 (RNA polymerase II [PolII]) (Neoclone; W0011). For the final step, samples were resuspended in 30 μl of Tris-EDTA and 1 μl was used for each PCR. Input DNAs were diluted 10-fold.
Allele-specific ChIP PCRs.
All PCRs except those for KvDMR1 were carried out with Ready-To-Go PCR beads (Amersham) using 0.3 μM of each primer (Table 1) and 0.1 μCi of [32P]dCTP. For KvDMR1, Herculase DNA polymerase (Stratagene) was used for the PCR, with 1× Herculase buffer, 0.3 μM of each primer, 0.8 mM deoxynucleoside triphosphates, 4% dimethyl sulfoxide, and 0.1 μCi of [32P]dCTP. Products were resolved on 7% polyacrylamide gels, and the relative band intensities were quantified using ImageQuant (Molecular Dynamics).
TABLE 1.
Primers and conditions for allele-specific PCR analysis
| Region | Primers | Restriction enzyme | Productsa |
|---|---|---|---|
| H19 DMD (R1/2) | 4017F (5′-CAGGACTCAAAGGAACATGCTAC), 3594R (5′-GCAATCCGTTTTAGGACTGCG) | DpnII | B 398; C 300, 98 |
| H19 Rep3 | Rep3F2 (5′-CAGTTGTGTTTCTGGAGGG), Rep3R2 (5′-TAGGAGTATGCTGCCACC) | Tsp45I | B 108, 28; C 137 |
| H19 PP | 205 (5′-GACTGGTCAGCCCTTGAGTCC), 501i (5′-GTCAGGACTCTCTTGCGATG) | AciI | B 315; C 112, 203 |
| H19 Prom | H19GB374F (5′-TGGGCAGTGAGTCTCCTTCT), H19GB675R (5′-GCCACTGTCTCCAAGGACTC) | HphI | B 215, 86; C 301 |
| H19 exon 5 | RT1 (5′-GCACTAAGTCGATTGCACTGG), HE5 (5′-AACACTTTATGATGGAACTGC) | BglI | B 212; C 124, 88 |
| Snrpn UP | SnUPCHIPsF (5′-AATCTGTGTGATGCTTGCAATCACTTGG), SnUPCHIPsR (5′-ATAGGATGCACTTTCACTACTAGAATCC) | SmlI | B 278, 142; C 420 |
| IG-DMR | IG-DMR207 (5′-TACGGAGATGTGCTGTGGAC), IG-DMR442 (5′-CTCGCTAGTTCACGGAGGTC) | NcoI | B 104, 94; C 198 |
| KvDMR1 | KvDMRfor (5′-GCGGGTTTCTTCTCTGAGTC), KvDMRrev (5′-TGTCCTAGGCCACTCACCTT) | BmgBI | B 254, 98; C 352 |
The expected sizes of the C57BL/6 (B) and M. musculus castaneus (C) alleles after digestion of the PCR products with the indicated restriction enzymes are shown.
Real-time ChIP PCRs.
Real-time PCR analysis was carried out with the LightCycler real-time PCR system (Roche). Briefly, reactions were set up in triplicate using the Ready-To-Go PCR beads (Amersham), with a 5-min initial incubation with the TaqStart antibody (Clontech), followed by addition of 0.3 μM primers, 1× SYBR green (Roche), and 1.5 to 3.0 mM MgCl2. The following primer pairs were used: for H19 DMDUP, 5488F (5′-CCCATAGTCCTTCCTGGGTA-3′) and 5357R (5′-TGATGTGCCACCTGGATAGA-3′); for H19 Rep3, Rep3F2 (5′-CAGTTGTGTTTCTGGAGGG-3′) and Rep3R2 (5′-TAGGAGTATGCTGCCACC-3′); for H19 Prom, H19GB374F (5′-TGGGCAGTGAGTCTCCTTCT-3′) and H19GB675R (5′-GCCACTGTCTCCAAGGACTC-3′); for H19 Exon 5, RT1 (5′-GCACTAAGTCGATTGCACTGG-3′) and HE5 (5′-AACACTTTATGATGGAACTGC-3′); for H19 (+3,000), H19-2819 (5′-ATGGGATGGCACACAGCGAAAG-3′) and H19-3106 (5′-CTCGGGAGTTGGGATTAGTGTG-3′); and for H19 enh, H19-7079 (5′-AGATTCCTGGAGGGACCATG-3′) and H19-7243 (5′-CCCGTCCCTCACAGCACTACC-3′). Data analysis was performed using the LightCycler 4.0 software, using relative quantification (Monocolor) to determine the ratio of each IP sample relative to input.
Expression analysis.
RNA was isolated using Dynal beads (Invitrogen) and reverse transcribed (16). For H19, allele-specific expression was determined using the LightCycler real-time PCR system (Roche) (46). Crossing points are provided by the LightCycler data analysis software as a measure of the total expression level for each sample. Melting peak areas were calculated after background subtraction, and the percent expression of each allele was calculated relative to the total expression level of both alleles (34). For Igf2, the allele-specific PCR conditions have been described previously (45). As an internal control, ARPPO (acidic phosphoprotein P0 subunit) expression was determined using the LightCycler system with Ready-To-Go PCR beads (Amersham), with an initial incubation with the TaqStart antibody (Clontech), followed by addition of 0.3 μM of each primer (Arbp0 no. 72L, TCCCACTTACTGAAAAGGTCAAG; Arbp0 no. 72R, TCCGACTCTTCCTTTGCTTC), 3.0 mM MgCl2, and 1× SYBR green (Molecular Probes). Total MEF RNA was isolated using TRIzol (Invitrogen), and Northern blotting was performed as described for H19 (14) and for Igf2, using a rat cDNA Igf2 probe.
RESULTS
Differential chromatin modifications at H19 in MEFs.
To study the role of histone modifications in regulating imprinting of the H19 gene, we first characterized the allelic pattern of chromatin modifications across the locus and then determined the relative level of these histones at each region. Since H19 is a relatively compact gene, we were able to analyze the chromatin structure across over 12 kb of sequence spanning most of the locus. Finally, the allelic analysis of chromatin marks enabled us to compare the active and silent alleles in the same reaction.
For our allele-specific assays, we focused on five H19 regions (Fig. 1A). First, we chose two elements (R1/2 and Rep3) within the DMD. Importantly, significant levels of transcription were not evident in the DMD, which is 2 to 4 kb 5′ to the promoter. Second, we selected two regions around the H19 promoter: one located 0.8 kb upstream of the transcription start site (the PP region) and another one 400 bp downstream (Prom). These sequences acquire paternal-allele-specific methylation after fertilization. Finally, we chose a region located close to the 3′ end of the H19 gene (Ex5). This sequence is transcribed on the maternal allele but harbors no differential methylation (3, 17).
For our analysis, we generated hybrid MEFs derived from reciprocal intercrosses between B mice and C7 or p12X mice in a B background. Due to polymorphisms between the two strains, we were able to distinguish the parental alleles at H19 and other genes on chromosomes 7 and 12 (see below). H19 is expressed and correctly imprinted in MEFs, making these cells a suitable model for the study of H19 regulation (Fig. 2). We also employed ES cells that were F1 hybrid between B and p12X to determine the pattern of chromatin marks in a pluripotent cell lineage.
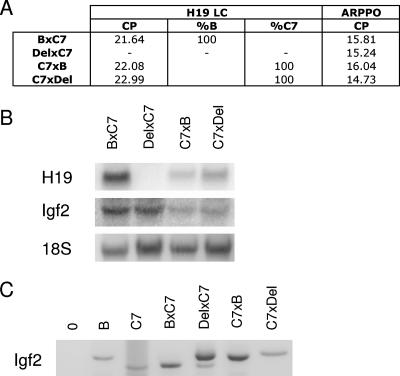
FIG. 2.
H19 and Igf2 expression in wild-type and reciprocal deletion MEFs. (A) H19 allele-specific expression was assayed using real-time PCR. The crossing points (CP) of the samples are provided as a measurement of their respective expression levels, and the percent expression of each allele as determined by melting curve analysis is indicated. Expression data for the ARPPO gene obtained using real-time PCR are included as a control. (B) Northern blotting analysis for H19 and Igf2. (C) Allele-specific expression for Igf2 was determined using reverse transcription-PCR followed by digestion with a polymorphic restriction enzyme that differentially cuts the B and C alleles. In summary, the Del × C7 MEFs do not express H19, while Igf2 is biallelically expressed in these cells. In contrast, the C7 × Del MEFs express H19 from the wild-type C7 allele and Igf2 from the Del paternal allele.
ChIPs were carried out on fixed chromatin isolated from B × C7, C7 × B, B × p12X, and p12X × B cells. We used antibodies against histones that associate with active chromatin (H3K4me2, H3K4me3, H3ac, and H4ac), as well as antibodies against histones found in repressive chromatin (H3K9me3, H3K27me3, and H4R3me2s). The immunoprecipitated chromatin was eluted, purified, and subjected to semiquantitative allele-specific PCR analysis for the indicated regions (Fig. 1A). For each PCR, parental alleles were distinguished by digesting the product with polymorphic restriction enzymes that cut differentially between B and C alleles. In the input lanes, the alleles were represented at roughly equal frequencies for all regions tested.
We found significant differences in the chromatin present at the DMD on the parental alleles. For both R1/2 and Rep3 regions, the maternal allele was preferentially associated with active chromatin modifications. H3K4me3, H3K4me2, and H3ac were significantly enriched on the maternal H19 DMD. A similar trend was also seen with H4ac, although to a lesser extent (Fig. 1B and data not shown). In contrast, the silent paternal allele was bound almost exclusively by H3K9me3. For example, at Rep 1/2, 95% of the chromatin precipitated by the antibody against H3K9me3 derived from the paternal DMD. Interestingly, no differential H4R3me2s or H3K27me3 was evident at the DMD or any of the other H19 regions tested, as we consistently observed an equal distribution of the parental alleles (Fig. 1B). Using reciprocal B × C7 and C7 × B MEFs, we confirmed that our results were due to specific allelic preferences of the chromatin rather than simply an artifact of the ChIPs or PCR assays.
We also observed strong differences in the chromatin modifications associated with the parental alleles at the PP region, which resides in a nontranscribed portion of the locus, and at the Prom and Ex5 sequences, which represent actively transcribed regions. As seen with the DMD, the PP, Prom, and Ex5 maternal alleles were enriched for H3K4me3, H3K4me2, H3ac, and H4ac (Fig. 1B and data not shown). The degrees of enrichment for the maternal copy were comparable at these different regions and for the different chromatin modifications. The paternal alleles were preferentially bound by H3K9me3. However, H4R3me2s and H3K27me3 did not preferentially immunoprecipitate with either parental allele. These results are consistent with data showing H3K4me3, H3K4me2, and H3ac in the vicinity of promoters (7). We observed similar chromatin modifications in undifferentiated ES cells at all regions tested, except for Ex5, where only H3K4me3 was preferentially bound (data not shown).
Differential chromatin modifications at other ICRs.
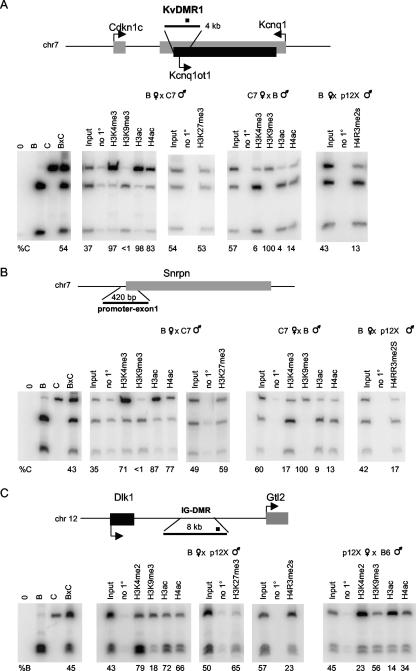
We next analyzed the ICRs for other imprinted genes. In addition to comparing the chromatin modifications at the various ICRs, these experiments enabled us to determine whether our ChIP assays were working as expected, especially at KvDMR1, which has been studied extensively (30, 31, 50). KvDMR1, the ICR for a cluster of imprinted genes at the distal end of mouse chromosome 7, is maternally methylated and harbors the promoter for the paternally expressed Kcnq1ot1 transcript (18, 33). We analyzed the chromatin modifications at KvDMR1 in reciprocal B × C7 and C7 × B MEFs and in B × p12X MEFs, since Kcnq1ot1 is expressed and imprinted in these cells (data not shown). We observed a very strong association of active chromatin modifications (H3K4me3, H3K4me2, H3ac, and H4ac) with the transcribed paternal allele (Fig. 3A and data not shown). In contrast, the repressive chromatin modifications H3K9me3 and H4R3me2s, but not H3K27me3, were found on the silent maternal allele. With ES cells we obtained results similar to those for MEFs, except for H3K27me3, which was detected preferentially on the maternal allele (data not shown). These data are consistent with a previous report describing H3K4me2 and H3K27me3 on the paternal and maternal KvDMR1 alleles, respectively, in ES cells (50), and they suggest that different ICRs carry both common and distinct histone marks.
FIG. 3.
Allelic chromatin modifications at the KvDMR1, Snrpn, and IG-DMR ICRs in MEFs. Allele-specific ChIP assays at KvDMR1 (A), Snrpn promoter-exon 1 (B), and IG-DMR (C) were performed. A diagram of the region analyzed is included for each panel. For KvDMR1 (A), the location of the 300-bp region amplified (black square) is indicated within the 4-kb ICR. For Snprn (B), the primers amplified a 420-bp region within the Snprn promoter/exon1. For IG-DMR, we analyzed a 198-bp region located towards the 3′ end of the 8-kb DMR (black square). ChIP assays were carried out with antibodies detecting the modified histones H3K4me3, H3K4me2, H3K9me3, H3ac, H4ac, H3K27me3, and H4R3me2s, as indicated. Paternally and maternally expressed genes in panels A and C are designated with dark and gray boxes, respectively. See the Fig. 1 legend for details.
We also assayed the promoter/exon 1 of the Snrpn gene, which is part of a maternally methylated ICR that regulates the paternal-allele-specific expression of a cluster of genes on the central region of mouse chromosome 7. In MEFs and ES cells, two cell types where Snprn expression is exclusively paternal (data not shown), the expressed paternal allele is preferentially immunoprecipitated with antibodies against H3K4me3, H3K4me2, H3ac, and H4ac. In contrast, the silent maternal allele is preferentially bound by the H3K9me3 and H4R3me2s in MEFs and by H3K9me3, H4R3me2s, and H3K27me3 in ES cells (Fig. 3B and data not shown). Previous experiments with a tissue composed of differentiated cells, brain, showed a subset of these modifications at the Snprn locus, with H3K4me2, H3ac, and H4ac enriched on the expressed paternal allele and histone H3 dimethylated on Lys9 on the maternal allele (19).
Finally, we extended our analysis to characterize the chromatin modifications of IG-DMR, the paternally methylated ICR in the Dlk1-Gtl2 imprinted domain that is located 10 kb upstream of the Gtl2 promoter and regulates imprinting of Gtl2 and other maternally expressed genes on mouse chromosome 12. In MEFs, where Gtl2 is expressed and correctly imprinted (data not shown), the maternal IG-DMR allele was enriched for H3K4me3 and H3K4me2 (Fig. 3C and data not shown). We also observed a slight enrichment of H3ac and H4ac for the maternal allele. The paternal allele was preferentially bound by H3K9me3 and by H4R3me2s. Analogous to what we observed for H19, H3K27me3 was not enriched on the silent paternal allele. A similar pattern of chromatin modifications was detected at IG-DMR in ES cells, as reported previously (13), with the exception of H3K9me3, for which we observed an enrichment on the paternal allele (data not shown).
Together our experiments show enrichment of H3K4me2, H3K4me3, H3ac, and H4ac on expressed alleles and of H3K9me3 at the repressed alleles at all ICRs studied. In contrast, the repressive modifications H3K27me3 and H4R3me2s are variable and could represent a property of the specific ICR. Consistent with this idea, H3K27me3 is associated only with the two ICRs that overlap with the promoter regions of imprinted genes, namely, Snrpn and KvDMR1.
Chromatin modifications across the H19 locus in MEFs.
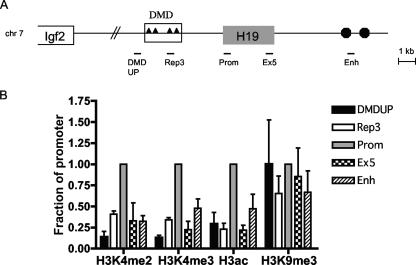
Although we observed a clear association of active and repressive chromatin modifications with opposite parental alleles at H19, it remained possible that specific histones were not distributed uniformly throughout the locus. Thus, we next determined the relative amount of precipitated chromatin for a given histone modification across 12 kb of H19 sequence. This analysis did not distinguish alleles, but instead ascertained the level of a modified histone at the two alleles. We developed real-time PCR assays at five regions throughout the locus: DMDUP, located upstream of the DMD at −5.5 kb relative to the start of H19 transcription; Rep3, within the DMD; Prom (+400 bp); Ex5/+3.0, at the end of the H19 gene body; and En, a sequence within the endodermal enhancers located at + 7.0 kb (Fig. 4A). Samples were precipitated with antibodies reflective of either active (H3K4me3, H3K4me2, and H3ac) or repressive (H3K9me3 and H4R3me2s) chromatin modifications and analyzed by real-time PCR relative to the input DNA. To make comparisons among the independent experiments, the fraction of precipitated DNA relative to input was normalized to the value obtained at the promoter. For the active chromatin modifications, we typically observed the highest level of precipitated chromatin at the promoter region (Fig. 4B). The DMDUP region showed the lowest level (10 to 30% of promoter values), whereas Rep3 and Ex5 exhibited consistently higher levels of actively modified histones (20 to 40%). Interestingly, the enhancer region displayed significant levels of precipitated chromatin (30 to 50%).
FIG. 4.
Profile of chromatin modifications across the H19 locus in MEFs. (A) Schematic of the H19 and Igf2 loci, with the H19 regions analyzed in the ChIP assays indicated. (B) Pattern of chromatin modifications across the H19 locus. Samples were precipitated with H3K4me2, H3K4me3, H3ac, or H3K9me3 and analyzed by real-time PCR relative to the input DNA. To make comparisons among the independent experiments, the fraction of precipitated DNA relative to input was normalized to the value obtained at the promoter region, which typically had the highest level. The graphs represent averages and standard deviations from at least three independent experiments. The material used to perform the real-time PCR assays is the same as that analyzed allele specifically in Fig. 1.
When the profile of the repressive H3K9me3 modification across the H19 locus was assayed, two striking differences from the results with the active chromatin marks were noted. First, the fraction of precipitated chromatin appeared to be relatively constant throughout the locus (Fig. 4B). Second, reduced levels of precipitated chromatin for H3K9me3 occurred at all regions tested (data not shown). This result is consistent with the prevalence of H3K9me3 in pericentric heterochromatin. Nevertheless, for the regions where we were able to test the allelic pattern of H3K9me3 (Rep3, Prom, and Ex5), this modification was represented almost exclusively on the silent paternal allele (Fig. 1B). The pattern of H4R3me2s resembled more closely that for H3K4me3, H3K4me2, and H3ac, with the highest level of precipitated chromatin at the promoter (data not shown). However, the significance of this result is unclear, as we did not observe an allelic preference for H4R3me2s at any H19 region tested.
Active chromatin modifications at H19 in DMD deletion MEFs and neonatal liver.
Since we typically observed the highest level of H3K4me3, H3K4me2, and H3ac precipitated chromatin at the H19 promoter, we hypothesized that this effect was related to the transcriptional activity at the promoter. To determine whether transcription is required for the observed profile of active chromatin at H19, we took advantage of a mouse strain that harbored either a maternally or paternally inherited 3.8-kb deletion that spans the entire DMD as well as sequence between the DMD and the promoter (Δ3.8 kb-5′H19, or abbreviated as Del here [46]). MEFs with a maternally inherited deletion exhibit no H19 mRNA (Fig. 2A and B). In contrast, H19 is expressed from the maternally inherited deleted allele in neonatal liver (46). We first performed allele-specific ChIPs in the reciprocal deletion MEFs and in B × C7 controls using antibodies against H3K4me3, H3K4me2, and H3ac, as well as PolII. These modifications and PolII were preferentially bound to the maternal promoter and exon 5 in B × C7 and C7 × Del MEFs, which both express H19 from this allele (Fig. 5A and data not shown). Strikingly, in the Del × C7 cells, in which H19 is not expressed, no enrichment of H3K4me3, H3K4me2, H3ac, or PolII at the maternal H19 promoter was detected. These data strongly suggested that transcriptional activity on one allele influences the preferential association of chromatin modifications with that region. We also examined the distribution of the active chromatin marks across the H19 locus in wild-type and DMD deletion MEFs using real-time PCR. Consistent with our previous data (Fig. 4), we observed a significant increase in the amount of precipitated chromatin at the promoter for H3K4me3, H3K4me2, and H3ac for the B × C7 cells (Fig. 5B). In contrast, the Del × C7 MEFs did not exhibit this effect, and only low levels of these modifications occurred at the promoter. Interestingly, for the Del × C7 MEFs, the enhancers harbored the highest levels of precipitated chromatin for both H3K4me3 and H3ac (Fig. 5B). This effect is likely due to the transcription of Igf2 from both the normally expressed paternal allele (C7) and the deleted maternal chromosome (Fig. 2C).
FIG. 5.
Chromatin modifications at H19 in MEFs harboring a DMD deletion. Schematic of the H19/Igf2 locus is shown at the top, with the regions analyzed for both the allelic and real-time PCR marked below the diagram and the extent of the Δ3.8kb-5′H19 deletion (Del) (46) indicated by the hatched rectangle. (A) Allelic chromatin modifications and PolII at the H19 promoter and exon 5 (see the Fig. 1 legend for details) in wild-type (B × C7), Del × C7, and C7 × Del MEFs. (B) Profile of H3K4me3, H3K4me2, and H3ac across the H19 locus in wild-type B × C7 and Del × C7 MEFs. ChIP samples were analyzed by real-time PCR, and the fraction of precipitated chromatin relative to input was normalized to the value obtained at the DMDUP region. Results from a representative experiment are shown for wild-type MEFs. For the deletion MEFs, the graphs represent averages and standard deviations from three independent experiments.
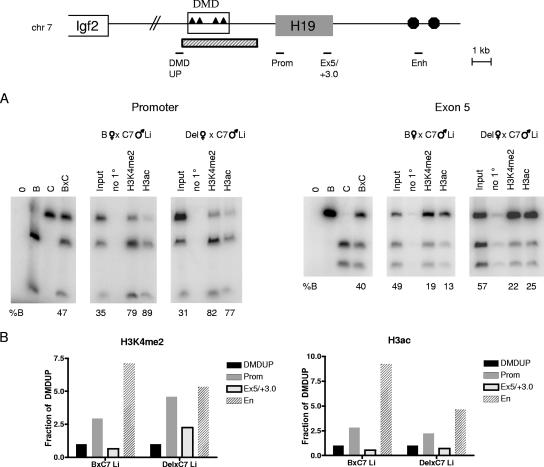
The absence of an allelic preference for active chromatin in Del × C7 MEFs, as well as the low level of these marks at the promoter, could be explained by the removal of specific DMD sequence from the maternal chromosome if elements within this region were important for recruiting actively modified histones. Alternatively, these results could be a direct consequence of the lack of transcriptional activity at H19 from the deleted allele. To distinguish between these possibilities, we extended our ChIP analysis to neonatal liver samples harboring the same DMD deletion. As stated above, H19 is expressed from the maternal DMD deletion allele in neonatal liver (46), thus allowing us to test specifically whether transcription or the DMD is required for the preferential association of active chromatin marks with the maternal allele throughout the locus and for the enrichment of these modifications at the promoter. We first assayed two histone modifications representative of active chromatin, H3K4me2 and H3ac, at the H19 promoter and in exon 5 (Fig. 6A). As in the wild-type MEFs, H3K4me2 and H3ac were preferentially associated with the maternal allele in B × C7 neonatal liver. Strikingly, the Del × C7 liver sample harbored a chromatin profile similar to that of B × C7 liver at both regions tested. Thus, in contrast to the Del × C7 MEFs, which exhibit no H19 expression and no allelic preference for active chromatin modifications, in the Del × C7 liver, where H19 is expressed, actively modified histones were enriched on the maternal allele. Consistent with this allelic chromatin pattern in the Del × C7 neonatal liver, we also found a strong enrichment in the total level of chromatin precipitated by H3K4me2 and H3ac at the promoter and enhancer (Fig. 6B). These data support the model that transcriptional activity at H19, even in the absence of the DMD, is critical for establishing the chromatin configuration at this locus.
FIG. 6.
Chromatin modifications at H19 in DMD-deleted neonatal liver nuclei. (A) Allelic chromatin modifications at the H19 promoter and exon 5 in wild-type (B × C7) and Del × C7 neonatal liver. (B) Profile of H3K4me2 and H3ac across the H19 locus in the neonatal liver samples used for panel A. See the Fig. 5 legend for details. Results of representative experiments are shown.
DISCUSSION
In this study, we conducted an extensive chromatin analysis of H19 and three additional ICRs to compare modification of maternally and paternally methylated ICRs, as well as to determine the effect of transcription on imprinted gene histone marks. Our approach offers the advantage of a direct comparison of both active and repressive histone modifications at multiple ICRs in a variety of cell types in which the imprinted expression pattern is known. We were also able to use cell lines and tissues with a deletion of the H19 DMD that differ in their H19 expression status to show that transcriptional activity is critical for establishing the chromatin modifications at this locus, even in the absence of the DMD.
Whereas previous studies have examined the chromatin structure at defined regions of H19 (13, 22, 37), this is the first report that offers a comprehensive analysis of both the allelic pattern and the relative levels of specific chromatin modifications across the H19 locus in multiple cell types. Our analysis clearly showed that the allelic preference for the active or repressive chromatin marks occurs throughout the entire H19 locus, including the upstream DMD, the promoter, and the transcription unit, thus defining a locus-wide domain of active or silent chromatin on each parental allele. While H3ac, H4ac, H3K4me2, and H3K4me3 were preferentially associated with the maternal chromosome at all regions tested, the highest level of these chromatin modifications was associated with the H19 promoter and enhancers, suggesting a close relationship between chromatin state and transcriptional status. We did not observe any differences between the patterns of H3K4me2 and H3K4me3 across the H19 locus, and the high levels of these modifications at the H19 promoter are consistent with the enrichment of both methylated states in the vicinity of the 5′ ends of genes (2, 7). Although H3K4me3, H3K4me2, and H3ac are normally associated with promoters (7), at the maternal H19 DMD these marks occur in the absence of transcription or associated binding of PolII (data not shown). As the DMD contains binding sites for CTCF, the presence of active marks in this region may be related to its function as an insulator/enhancer blocker. In fact, a recent report documents the colocalization of both H3K4me2 and H3K4me3, as well as the histone variant H2A.Z, at presumed CTCF insulator sites throughout the genome (2). Interestingly, these same modifications, as well as histone acetylation, can also mark active enhancers (2, 7). Consistent with these data, we observed high levels of these modifications at the H19 enhancers. A recent study reported that a significant proportion of inactive protein-coding genes contain active modifications and PolII at their promoters in human ES cells (23), perhaps allowing for easier reprogramming of gene expression during differentiation. Since our results showed a significant enrichment but, in most cases, not an exclusive association of actively modified histones with expressed alleles, we cannot rule out that the repressed alleles may retain low levels of active chromatin. However, it is also possible that the silent imprinted alleles belong to the small class of repressed genes that lack active modifications at their promoters due to stricter mechanisms that prevent transcriptional initiation (23).
Our results further demonstrate that the pattern of active histone modifications on the H19 maternal allele is strictly dependent upon its transcriptional activity. We utilized MEFs and liver samples that harbor a 3.8-kb DMD deletion and which differ in their H19 expression status, such that transcriptional activity occurs only in neonatal liver. This represented an ideal system to test whether specific sequences such as the DMD or transcriptional activity at the locus is important for establishment of the active chromatin marks. In contrast to their wild-type counterparts, MEFs with a maternal DMD deletion in which H19 is not expressed did not exhibit preferential association of active chromatin modifications with the maternal allele and failed to show peak binding of these histones at the promoter and enhancers. Interestingly, in neonatal liver, where H19 is transcribed from the DMD deletion allele, the allelic enrichment and binding pattern of actively modified histones throughout the locus were restored. Thus, these data showed very clearly that transcription is a prerequisite for the association of specific active chromatin marks with the maternal H19 allele. At this time, however, the available H19 alleles do not allow us to determine whether transcriptional activation is also required for the establishment of these marks in the DMD. Analysis of chromatin modifications under conditions when H19 is not expressed may offer some clues about how the DMD modifications are set up.
Nevertheless, because we observed differential chromatin structure at H19 in the absence of the DMD, it is likely that this region is dispensable for the establishment of active chromatin marks at the maternal promoter and gene body. How are the parental alleles distinguished in the absence of the DMD? Upon its insertion at a heterologous locus, the DMD did not acquire DNA methylation in the male germ line (36), suggesting that it is not sufficient for this process. In addition, we have shown that the DMD is not absolutely necessary for differential DNA methylation, as the 3.8-kb deletion allele exhibited some allele-specific methylation (45). Given these results, we postulate that sequences outside the DMD may serve this function in its absence. These elements may play a role in setting up differential chromatin marks at H19 by mediating transcriptional initiation and the subsequent recruitment of active histone modifications specifically on the maternal allele.
Our data also raise some interesting questions about the complex interplay between chromatin modifications and promoter activity. Transcriptional activation at H19 appears to be necessary for the association of H3K4me2, H3K4me3, and H3ac with the maternal promoter and downstream regions. Whether other actively modified histones and/or additional chromatin-remodeling factors are required to facilitate the opening of chromatin for transcriptional initiation remains to be determined. The onset of transcription is followed by binding of hypermethylated H3 at Lys4 and hyperacetylated H3 and H4. In support of our model at H19, the establishment of H3K4me3 has been shown to depend on prior transcriptional activation as well as H2B monoubiquitination (reviewed in reference 40). Once bound to the maternal H19 allele, 2- and 3-K4 methylations likely promote transcription through the recruitment of nucleosome-remodeling factors such as CHD1 or NURF or other histone-modifying enzymes (6, 40).
Our analysis of four different ICRs in multiple cells types allows us to draw several conclusions about the nature of the chromatin modifications at imprinted genes. Overall, active chromatin modifications are enriched on the expressed alleles, while repressive histone marks reside on the silent copies, highlighting important similarities in the mechanisms that regulate imprinted and nonimprinted genes. For the active chromatin modifications, we observe significant enrichment of H3K4me2, H3K4me3, H3ac, and H4ac on the ICRs associated with the normally active alleles, which also lack DNA methylation. The enrichment of active chromatin in these regions may protect the ICRs from acquiring DNA methylation, thus playing a critical role in the epigenetic marking of the parental chromosomes. A possible explanation of this is found during germ cell development, when ICRs are demethylated in migrating germ cells and are poised to assume sex-specific patterns. Maternal and paternal alleles subsequently acquire methylation with different kinetics at both the H19 and Snprn loci (11, 32). Although it has been questioned how the alleles are distinguished, it may be more appropriate to ask why the alleles are distinguished. Possibly, actively modified histones on the formerly expressed allele need to be removed before the DNA can be methylated. In support of this, low levels of H3K4me2 and somewhat higher levels of H4ac were observed during spermatogenesis at H19 (13). Determining both the allelic chromatin pattern and the transcriptional activity of H19 in both wild-type and DMD-deleted male PGCs will undoubtedly provide critical clues about the nature of the epigenetic mark at this locus.
For the heterochromatic modifications, H3K9me3 associated preferentially with the H19 DMD, KvDMR1, Snrpn promoter/exon 1 and IG-DMR on the normally silent alleles. Interestingly, although for H19 the amount of precipitated H3K9me3 was smaller than what was observed for the active histones, the preference of this modification for the paternal allele was very significant. These results underscore the necessity of analyzing the allelic pattern of a given histone modification, in addition to its relative levels. Furthermore, whereas at the Igf2r imprinted cluster the localization of H3K9me3 was limited to the silent Air and Igf2r promoters and coincided with regions of DNA methylation (38), at H19 H3K9me3 was present on the paternal allele throughout the entire domain, including sequences that were not DNA methylated. This difference may reflect distinct mechanisms of imprinting control at these clusters (insulator/enhancer blocker at H19 versus ncRNA at Igf2r), or it may simply be due to the fact that the H19 locus is more compact. In contrast to the strong allelic preference for H3K9me3 throughout the H19 locus, preferential binding of H3K27me3 was not observed at the H19 DMD (Fig. 1B and data not shown). This result is consistent with other reports describing nonoverlapping regions of H3K9me3 and H3K27me3, suggesting that these modifications reflect different types of heterochromatin (9, 38). H3K27me3, which is conferred by the Polycomb repressive complex 2, is currently the only histone modification that is convincingly epigenetically transmitted (1). This modification exhibited a strong association with the repressed alleles at the KvDMR1 and Snrpn ICRs, the two ICRs analyzed that overlap with promoters. In contrast to our findings at H19, at KvDMR1 and Snrpn, H3K9me3 and H3K27me3 colocalized in ES cells on the silent allele, likely reflecting distinct mechanisms that regulate these different imprinting clusters. In fact, H3K27me3 is proposed to mediate the silencing of the genes controlled by KvDMR1 in the placenta (31, 50).
In addition to the modifications observed in our study, H4K20me3 was also reported at the paternal H19 and Rasgrf1 ICRs and maternal KvDMR1 in ES cells and adult liver (13). The presence of histone methylation at the ICRs that also harbor DNA methylation is consistent with a link between the two states. H3K9me3 histone methyltransferases may directly bind sites of DNA methylation through methyl-CpG-binding domains (21). Conversely, the HP1 adaptor protein, DNMT1, and the G9a histone methyltransferase colocalized at several endogenous promoters, and binding of methylated K9 by HP1 lead to the recruitment of DNMT1 and stimulated DNA methylation (41). Furthermore, loss of both H4K20me3 and DNA methylation was observed at pericentric chromatin regions in cancer cells (20). Thus, although histone and DNA methylation can mutually reinforce each other, the precise sequence of events important for imprinted gene expression remains to be determined. Another repressive histone modification, H4R3me2s, was recently proposed to play a role in the methylation of the H19 DMD in the male germ line (26), although this mark had not previously been examined at the other imprinted ICRs. H4R3me2s was not enriched on the paternal H19 DMD in MEFs, perhaps reflecting a developmental and cell-type-specific difference, but allelic bias of this modification occurred at KvDMR1, Snprn, and IG-DMR ICRs, suggesting that it may also play a role at other imprinting genes. Characterization of the germ line allelic distribution pattern of H4R3me2s, as well as those of H3K9me3 or H4K20me3, is essential for understanding how these marks may contribute to the regulation of imprinted expression. Although no significant levels of H3K9me3 and H4K20me3 were detected at H19 at a time when the DMD was methylated in late spermatogenesis (13), these modifications may be targeted to the parental alleles differentially earlier in PGC development.
In conclusion, our comparison of active and repressive histone modifications at multiple imprinted ICRs revealed that these regions are marked by common as well as locus-specific chromatin modifications. Thus, a single histone code may not mark all the active and silent alleles of imprinted genes, and the identity of the specific chromatin modifications at a given locus may depend on the presence of other epigenetic features such as DNA methylation. For example, for genes whose imprinting requires DNA methylation, such as H19, fewer repressive modifications may be sufficient to mediate silencing in concert with DNA methylation. In contrast, other imprinted loci that are not as dependent on DNA methylation, such as genes in the KvDMR1 cluster in placenta, must employ additional repressive modifications (i.e., H3K27me3 and H4R3me2s in addition to H3K9me3) for transcriptional repression (references 31 and 50 and this study). Finally, our data from the H19 locus highlight the need to expand the current dogma of imprinting control to include allelic chromatin modifications and promoter activity along with DNA methylation.
Acknowledgments
We thank G. Blobel and S. Blythe for advice on the ChIP assays and G. Blobel for critical comments on the manuscript.
This work was supported by U.S. Public Health Service grant GM51279. R. I. Verona and K. J. Reese were supported by the Lalor Foundation.
Footnotes
Published ahead of print on 29 October 2007.
REFERENCES
- 1.Bantignies, F., and G. Cavalli. 2006. Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 18275-283. [DOI] [PubMed] [Google Scholar]
- 2.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 71663-1673. [DOI] [PubMed] [Google Scholar]
- 4.Bartolomei, M. S., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351153-155. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 6.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120169-181. [DOI] [PubMed] [Google Scholar]
- 8.Carr, M. S., A. Yevtodiyenko, C. L. Schmidt, and J. V. Schmidt. 2007. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics 89280-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chadwick, B. P., and H. F. Willard. 2004. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. USA 10117450-17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, T. L., J. M. Trasler, S. B. Moss, G. J. Yang, and M. S. Bartolomei. 1999. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 5818-28. [DOI] [PubMed] [Google Scholar]
- 11.Davis, T. L., G. J. Yang, J. R. McCarrey, and M. S. Bartolomei. 2000. The H19 methylation imprint is erased and reestablished differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 92885-2894. [DOI] [PubMed] [Google Scholar]
- 12.DeChiara, T. M., E. J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64849-859. [DOI] [PubMed] [Google Scholar]
- 13.Delaval, K., J. Govin, F. Cerqueira, S. Rousseaux, S. Khochbin, and R. Feil. 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 26720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel, N., J. L. Thorvaldsen, and M. S. Bartolomei. 2006. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 152945-2954. [DOI] [PubMed] [Google Scholar]
- 15.Engel, N. I., A. E. West, G. Felsenfeld, and M. S. Bartolomei. 2004. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is revealed by CpG mutations. Nat. Genet. 36883-888. [DOI] [PubMed] [Google Scholar]
- 16.Fedoriw, A. M., P. Stein, P. Svoboda, R. M. Schultz, and M. S. Bartolomei. 2004. Transgenic RNAi reveals essentail function for CTCF in H19 gene imprinting. Science 303238-240. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson-Smith, A. S., H. Sasaki, B. M. Cattanach, and M. A. Surani. 1993. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362751-754. [DOI] [PubMed] [Google Scholar]
- 18.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deleion of KvDMR1. Nat. Genet. 32426-431. [DOI] [PubMed] [Google Scholar]
- 19.Fournier, C., Y. Goto, E. Ballestar, K. Delaval, A. M. Hever, M. Esteller, and R. Feil. 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 216560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraga, M. F., E. Ballestar, A. Villar-Garea, M. Boix-Chornet, J. Espada, G. Schotta, T. Bonaldi, C. Haydon, S. Ropero, K. Petrie, N. G. Iyer, A. Perez-Rosado, E. Calvo, J. A. Lopez, A. Cano, M. J. Calasanz, D. Colomer, M. A. Piris, N. Ahn, A. Imhof, C. Caldas, T. Jenuwein, and M. Esteller. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37391-400. [DOI] [PubMed] [Google Scholar]
- 21.Fuks, F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15490-495. [DOI] [PubMed] [Google Scholar]
- 22.Grandjean, V., L. O'Neill, T. Sado, B. Turner, and A. Ferguson-Smith. 2001. Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett. 488165-169. [DOI] [PubMed] [Google Scholar]
- 23.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajkova, P., S. Erhardt, N. Lane, T. Haaf, O. El-Maarri, W. Reik, J. Walter, and M. A. Surani. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 11715-23. [DOI] [PubMed] [Google Scholar]
- 25.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 26.Jelinic, P., J. C. Stehle, and P. Shaw. 2006. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 4e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaffer, C. R., M. Srivastava, K.-Y. Park, E. Ives, S. Hsieh, J. Batle, A. Grinberg, S.-P. Huang, and K. Pfeifer. 2000. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 141908-1919. [PMC free article] [PubMed] [Google Scholar]
- 28.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10853-856. [DOI] [PubMed] [Google Scholar]
- 29.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 37534-39. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, A., K. Green, C. Dawson, L. Redrup, K. D. Huynh, J. T. Lee, M. Hemberger, and W. Reik. 2006. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development 1334203-4210. [DOI] [PubMed] [Google Scholar]
- 31.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 361291-1295. [DOI] [PubMed] [Google Scholar]
- 32.Lucifero, D., M. R. Mann, M. S. Bartolomei, and J. M. Trasler. 2004. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum. Mol. Genet. 13839-849. [DOI] [PubMed] [Google Scholar]
- 33.Mancini-DiNardo, D., S. J. Steele, R. S. Ingram, and S. M. Tilghman. 2003. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum. Mol. Genet. 12283-294. [DOI] [PubMed] [Google Scholar]
- 34.Mann, M. R. W., Y. G. Chung, L. D. Nolen, R. I. Verona, K. E. Latham, and M. S. Bartolomei. 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69902-914. [DOI] [PubMed] [Google Scholar]
- 35.O'Neill, M. J. 2005. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum. Mol. Genet. 14(Spec. No. 1)R113-R120. [DOI] [PubMed] [Google Scholar]
- 36.Park, K. Y., E. A. Sellars, A. Grinberg, S. P. Huang, and K. Pfeifer. 2004. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol. Cell. Biol. 243588-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedone, P. V., M. J. Pikaart, F. Cerrato, M. Vernucci, P. Ungaro, C. B. Bruni, and A. Riccio. 1999. Role of histone acetylation and DNA methylation in the maintenance of the imprinted expression of the H19 and Igf2 genes. FEBS Lett. 45845-50. [DOI] [PubMed] [Google Scholar]
- 38.Regha, K., M. A. Sloane, R. Huang, F. M. Pauler, K. E. Warczok, B. Melikant, M. Radolf, J. H. A. Martens, G. Schotta, T. Jenuwein, and D. P. Barlow. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 3366-69. [DOI] [PubMed] [Google Scholar]
- 40.Sims, R. J., III, and D. Reinberg. 2006. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 202779-2786. [DOI] [PubMed] [Google Scholar]
- 41.Smallwood, A., P. O. Esteve, S. Pradhan, and M. Carey. 2007. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 211169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szabo, P. E., S.-H. Tang, A. Rentsendorj, G. P. Pfeifer, and J. R. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10607-610. [DOI] [PubMed] [Google Scholar]
- 43.Szabo, P. E., S. H. Tang, F. J. Silva, W. M. Tsark, and J. R. Mann. 2004. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell. Biol. 244791-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 123693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorvaldsen, J. L., A. M. Fedoriw, S. Nguyen, and M. S. Bartolomei. 2006. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol. Cell. Biol. 261245-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsen, J. L., M. R. Mann, O. Nwoko, K. L. Duran, and M. S. Bartolomei. 2002. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 222450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tremblay, K. D., K. L. Duran, and M. S. Bartolomei. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 174322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tremblay, K. D., J. R. Saam, R. S. Ingram, S. M. Tilghman, and M. S. Bartolomei. 1995. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9407-413. [DOI] [PubMed] [Google Scholar]
- 49.Ueda, T., K. Abe, A. Miura, M. Yuzuriha, M. Zubair, M. Noguchi, K. Niwa, Y. Kawase, T. Kona, Y. Matsuda, H. Fujimoto, H. Shibata, Y. Hayashizaki, and H. Sasaki. 2000. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5649-659. [DOI] [PubMed] [Google Scholar]
- 50.Umlauf, D., Y. Goto, R. Cao, F. Cerqueira, A. Wagschal, Y. Zhang, and R. Feil. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 361296-1300. [DOI] [PubMed] [Google Scholar]
- 51.Verona, R. I., M. R. Mann, and M. S. Bartolomei. 2003. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19237-259. [DOI] [PubMed] [Google Scholar]