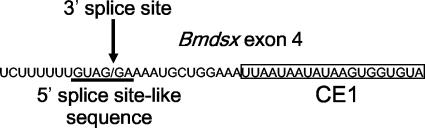Abstract
The Bombyx mori homolog of doublesex, Bmdsx, plays an essential role in silkworm sexual development. Exons 3 and 4 of Bmdsx pre-mRNA are specifically excluded in males. To explore how this occurs, we developed a novel in vivo sex-specific splicing assay system using sexually differentiated cultured cells. A series of mutation analyses using a Bmdsx minigene with this in vivo splicing assay system identified three distinct sequences (CE1, CE2, and CE3) positioned in exon 4 as exonic splicing silencers responsible for male-specific splicing. Gel shift analysis showed that CE1 binds to a nuclear protein from male cells but not that from female cells. Mutation of UAA repeats within CE1 inhibited the binding of the nuclear protein to the RNA and caused female-specific splicing in male cells. We have identified BmPSI, a Bombyx homolog of P-element somatic inhibitor (PSI), as the nuclear factor that specifically binds CE1. Down-regulation of endogenous BmPSI by RNA interference significantly increased female-specific splicing in male cells. This is the first report of a PSI homolog implicated in the regulated sex-specific splicing of dsx pre-mRNA.
Sexual reproduction is the primary means to maintain variation for evolutionary survival. Sex determination cascades that drive the differentiation of dimorphic gonads, however, are some of the most rapidly evolved developmental events (28). Genetic and environmental cues direct the generation of two distinct sexual phenotypes via several key molecular signals. In Drosophila melanogaster and Caenorhabditis elegans, the ratio of X chromosomes to autosomes (X/A) is the primary signal for sex determination (19, 23). In mammals, however, maleness is determined by the Y-linked gene SRY (17, 41). Nevertheless, recent studies indicate some downstream sex determination genes are functionally similar in diverse species. Of these, DM domain (Doublesex/Mab-3 DNA-binding motif) genes have been shown in the past several years to regulate sexual development in multiple metazoan phyla, including arthropods, nematodes, and vertebrates, and thus this gene family may represent the first example of widespread conservation of sexual regulatory genes (20, 46). The DM domain is a cysteine-rich DNA-binding motif first recognized in proteins encoded by the Drosophila sex determination gene doublesex (10, 48). As the name doublesex (dsx) suggests, this gene functions in both sexes: the primary transcript of dsx undergoes sex-specific alternative splicing, producing either a male-specific isoform, DSXM, or a female-specific isoform, DSXF (3, 6). Throughout most of the somatic tissues of the fruit fly, DSXM directs male development and DSXF directs female development.
The mechanism of sex-specific dsx splicing has been well studied. Female-specific splicing of dsx requires TRA, TRA-2, and an exonic splicing enhancer (ESE) element located within the untranslated region of the fourth exon (29, 34, 35). Both TRA and TRA-2 contain Arg/Ser-rich (RS) domains, protein interaction domains characteristic of the Ser/Arg (SR) family of essential splicing factors. SR proteins are thought to function by binding to ESEs and by recruiting the splicing machinery to adjacent introns via protein interactions involving the RS domain (4, 13). The dsx ESE, designated dsxRE, serves as a binding site for the cooperative assembly of the multicomponent splicing enhancer complex containing TRA, TRA-2, and one or more SR proteins (26, 27, 45). This complex functions to activate the upstream female-specific 3′ splice site, most likely by facilitating interactions of U2AF (11, 12, 49) and/or other general splicing factors (21, 25) with the RNA.
In the silkworm, Bombyx mori, the chromosomal sex determination mechanism is distinct from that of D. melanogaster, with female (ZW) being the heterogametic sex and male (ZZ) the homogametic sex. It has been shown genetically that female sex in B. mori is determined by the presence of a dominant feminizing factor on the W chromosome (16). Despite this difference, we have recently identified a dsx homolog in B. mori (36). Like dsx, the primary transcript of the Bmdsx gene is alternatively spliced in males and females to yield sex-specific mRNAs that encode male-specific (BmDSXM) and female-specific (BmDSXF) polypeptides (36, 42). Transgenic analysis of Bmdsx revealed that Bmdsx functions as a double-switch gene at the final step in the B. mori sex determination cascade (43, 44). Despite these similarities between dsx and Bmdsx, the underlying mechanism for sex-specific splicing is clearly different. We have demonstrated that female splicing of Bmdsx pre-mRNA represents the default mode when tested in HeLa nuclear extracts and also that the female exon is devoid of putative TRA/TRA-2-binding sites (42). These findings indicate that the female exon is selectively repressed in male silk moths by a yet-unknown mechanism.
To understand the molecular basis underlying sex differentiation, we reasoned that utilizing a splicing assay system that exploits cultured cells that differ only in sex would provide a powerful tool for analyzing the regulatory mechanisms of sex-specific alternative splicing. Because there were no reports of sexually differentiated cell lines in B. mori, or in other animals, we established the first sexually differentiated cell lines and used these novel cell lines to develop an in vivo splicing assay system that led to the identification of cis-acting elements and trans-acting cellular factors that negatively regulate splicing of Bmdsx female exons.
MATERIALS AND METHODS
Establishment of NIAS-Bm-F1 and NIAS-Bm-M1 cell lines.
Fertilized eggs of the silkworm mutant strain named Sex-limited black egg, which has a male-specific defect in egg pigmentation, were used for establishing the primary culture. In this mutant, female eggs display the same mauve coloration as normal eggs, while male eggs fail to develop egg pigmentation and thus display primrose yellow coloration (see Fig. 1A, below). Therefore, sexing of embryos can be done according to egg color. Female eggs and male eggs were separately cut and opened in saline solution using a razor blade. The excised embryos were cut into small pieces approximately 1 mm in length in culture medium and placed in culture dishes. Embryos from approximately 100 eggs were pooled to set up a primary culture in a 25-ml culture flask. The medium used for the primary culture was MGM-464 medium supplemented with 30% fetal bovine serum (FBS) and antibiotics (antibiotic-antimycotic; Gibco, Auckland, New Zealand). The culture was incubated at 25°C, and every 14 days half of the medium was replaced with fresh medium. Primary cultures were prepared on 13 November 2000. In the primary culture, a number of cells migrated from the adhered explants and formed cell sheets or networks around the explants. The primary cultured cells derived from female embryos were subcultured first on 30 December 2001, while the primary cultured cells derived from male embryos were subcultured first on 8 March 2002. The medium was gradually changed to IPL-41 medium from MGM-464 medium, and also the FBS volume was decreased from 20% to 10%. Finally, on 9 April 2002, the cells could be cultured with IPL-41 medium containing a 10% volume of FBS. The intervals of subculturing gradually shortened, and at the 15th passage, subculturing was at weekly intervals. At this point, each subculture was considered to have entered the continuously growing population phase. The cell line originated from female embryos was designated NIAS-Bm-F1, and the cell line from male embryos was designated NIAS-Bm-M1. The doubling time of both cell lines is approximately 3 days.
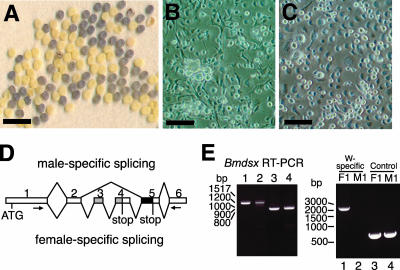
FIG. 1.
Characterization of NIAS-Bm-M1 and NIAS-Bm-F1 cell lines. (A) Photograph of fertilized eggs, laid by Sex-limited black egg strain females, that show a male-specific defect in egg pigmentation. Primrose yellow eggs are males, and mauve eggs are females. Bar, 5 mm. (B and C) NIAS-Bm-M1 (B) and NIAS-Bm-F1 (C) cells were photographed using phase-contrast microscopy. Bars, 100 μm. (D) Schematic diagram of alternative splicing in Bmdsx pre-mRNA. Boxes represent exons. The gray region indicates the female-specific open reading frame (ORF). The black region indicates the male-specific ORF. The numbers above the diagram represent exon labels. V-shaped lines above (skipping of alternative exons) and below (inclusion of alternative exons) the diagram represent the endogenous Bmdsx splice variants observed in males and females. Stop codons are indicated. The arrows indicate the approximate location of the primers that were used for the RT-PCR in panel E. Skipping of exons 3 and 4 results in an 880-bp PCR product, while inclusion results in a 1,134-bp product. (E) The left panel shows RT-PCR analysis of endogenous Bmdsx transcripts in adult females (lane 1), NIAS-Bm-F1 cells (lane 2), adult males (lane 3), and NIAS-Bm-M1 cells (lane 4) using primers as indicated in panel D. The right panel shows molecular sexing of NIAS-Bm-F1 cells (lane 1) and NIAS-Bm-M1 cells (lane 2) by PCR using oligonucleotide primers derived from the W chromosome-specific retrotransposon. As a control reaction, a genomic fragment of the Bmdsx gene (mapped on chromosome 25) was amplified from NIAS-Bm-F1 cells (lane 3) and NIAS-Bm-M1 cells (lane 4). PCR products were separated on 2% agarose gels and stained with ethidium bromide. Molecular sizes in base pairs are indicated to the left of each panel.
Plasmid constructions.
An 823-bp Bombyx mori cytoplasmic actin gene BmA3 promoter fragment was synthesized by PCR amplification using primers A3F4 (5′-GGTTGCTGAGCTTGATACTGAGTCAGCCCGCGATTGGTG-3′) and A3DSXR (5′-GACACCATTGCCCGTACGAGTCCTTCT-3′). A fragment containing Bmdsx exon 2 along with part of the downstream intron (143 bp for exon 2 and 39 bp for intron 2) was amplified with primers MINIF1 (5′-CCGTCCCCTCGGAGACGCTTGTGGAG-3′) and MINIR1 (5′-AAGGTTCCTTTGCGTTATTCTAATCAACGCTACACTCG-3′). A fragment containing Bmdsx exon 3 along with part of the flanking introns (160 bp for intron 2, 82 bp for exon 3, and 57 bp for intron 3) was amplified with primers MINIF2 (5′-TGTAGCGTTGATTAGAATAACGCAAAGGAACCTTTGCC-3′) and MINIR2 (5′-AAATCACGACTGGTTAGATGTATCTGTCGCGTGTGTTG-3′). A fragment containing nucleotide −155 upstream of the Bmdsx exon 4 to +44 downstream of the Bmdsx exon 4 was amplified by PCR using primers MINIF3 (5′-ACACGCGACAGATACATCTAACCAGTCGTGATTTCCAC-3′) and MINIR3 (5′-AATTATCCCAAATGGGGGTTTCACTAATGTGAACAGAG-3′). A fragment comprising Bmdsx exon 5 along with part of the upstream intron (169 bp for intron 4 and 218 bp for exon 5) was amplified with primers MINIF4 (5′-GTTCACATTAGTGAAACCCCCATTTGGGATAATTTAGC-3′) and MINIR4 (5′-CGCGGATCCCTGTATCGGCGCGCAGTGTCGTCGC-3′). A 1,129-bp fragment containing the simian virus 40 polyadenylation sequence was amplified using primers MAcPAF2 (5′-TAGCGACGACGCCTCGAGAAGTCTTACG-3′) and polyAR3 (5′-CGTTGCTTCCACTATGAGCAGCCTCCTCACTACTTCTGG-3′). Primers A3DSXR, MINIF1, MINIR1, MINIF2, MINIR2, MINIF3, MINIR3, MINIF4, MINIR4, and FAcPAF were bridging primers. To link the DNA fragments produced by the above PCRs at the same time, PCR amplification was performed with primers 2-44F (5′-GGTTGCTGAGCTTGATAC-3′) and 2-22R (5′-GGTTGCTGAGCTTGATAC-3′) using a mixture of all the above PCR fragments. The resulting product was cloned into the pGEM-T Easy vector (Promega, Madison, WI) to generate the Bmdsx minigene construct pWTE2.5. Bmdsx exon 2, exon 3, exon 4, and exon 5 of pWTE2.5 were replaced with silk protein P25 exon 1, exon 2, exon 3, and exon 4 to generate pMutE2, pMutE3, pMutE4, and pMutE5, respectively (see Fig. 2A, below). These substitutions were performed by PCR using bridging primers. The four 5′-most and the three 3′-most base pairs of each exon were maintained in these substituted exons to leave the splice sites unchanged. Primer sequences and further details are available on request. To generate linker scan mutants, pls1 to pls16, 10 bp of the 167-bp Bmdsx exon 4 were replaced consecutively with a 10-bp NotI linker (AGCGGCCGCA) by PCR utilizing the GeneTailor site-directed mutagenesis system (Invitrogen, Carlsbad, CA). Sixteen PCR primer pairs were used that deleted 10 bp each throughout Bmdsx exon 4 (except for the four 5′-most and the three 3′-most nucleotides). At their 5′ ends, the primers contained the 10-bp NotI linker sequence. Primer sequences and further details are available on request. pCE1Mut1 and pCE1Mut2 were generated by PCR using the GeneTailor site-directed mutagenesis system as recommended by the manufacturer. The primers used were CE1M1F (5′-TGCTGGAAATTAATAATATAGATGGTGTACTG-3′)/CE1M1R (5′-TATATTATTAATTTCCAGCATTTTCCTACA-3′) and CE1M2F (5′-CTGGAAATTAATAATATAAGCAGTGTACTGTC-3′)/CE1M2R (5′-CTTATATTATTAATTTCCAGCATTTTCCTA-3′), respectively. In all cases the template was the plasmid pWTE2.5. The nucleotide sequences of the resulting constructs were confirmed by DNA sequencing.
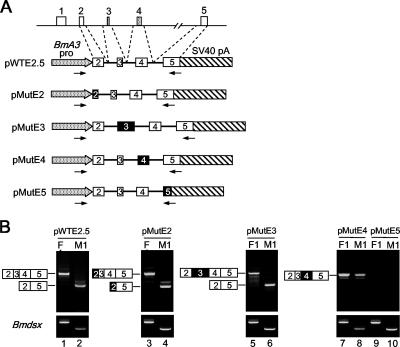
FIG. 2.
Sequences within Bmdsx exon 4 determine the inclusion of exons 3 and 4. (A) The genomic sequences in the Bmdsx gene included in the minigene construct are shown at the top. Schematic diagrams of the wild-type (pWTE2.5) and mutant (pMutE2, pMutE3, pMutE4, and pMutE5) Bmdsx minigene constructs used in panel B are also indicated. The open boxes represent the Bmdsx exons, whereas the exons replaced with the Bombyx silk protein P25 exons are indicated by filled boxes (see Materials and Methods). The numbers within each box represents exon labels. The arrows indicate the approximate locations of the primers that were used for RT-PCR in panel B. BmA3 pro, Bombyx actin 3 promoter sequence; SV40 pA, simian virus 40 polyadenylation sequence; lines, intron sequences. (B) The upper panel indicates RT-PCR analysis results upon transient transfection of the minigene constructs shown in panel A into NIAS-Bm-F1 cells (lanes F1) and NIAS-Bm-M1 cells (lanes M1). To identify the transcript splicing pattern from each minigene construct, PCR products were cloned and sequenced. The lower panel shows the RT-PCR analysis of endogenous Bmdsx transcripts using primers as indicated in Fig. 1D. PCR products were separated on 2% agarose gels and stained with ethidium bromide. The structure of the RT-PCR products is indicated schematically to the left of each panel.
DNA transfection of cells.
Transfections were performed on 60-mm by 15-mm dishes. Cells were plated 1 to 2 days before each transfection at a density of 5 × 105 cells/dish, such that on the day of the transfection they would be 50 to 70% confluent. Each plasmid containing either the wild-type or a mutant Bmdsx minigene (3 μg/dish) was transfected into the cells using Cellfectin (Invitrogen, Carlsbad, CA) at a Cellfectin/DNA (wt/wt) ratio of 8:1. The transfection mixture was replaced with serum-containing medium 5 h after transfection. Poly(A)+ RNA was isolated 48 h after transfection using a Micro-FastTrack 2.0 mRNA isolation kit (Invitrogen, Carlsbad, CA) according to the protocol provided by the manufacturer.
RT-PCR analysis.
Five hundred micrograms of DNase-treated poly(A)+ RNA was denatured at 70°C for 10 min and immediately chilled on ice. First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the protocol provided by the manufacturer, with random hexamers as primers. A 1/20 portion of the resulting reaction mixture was used for PCR in a 50-μl total volume. To analyze splicing patterns of Bmdsx mRNAs transcribed from pWTE2.5, pMutE2, pMutE3, pMutE4, pCE1Mut1, and pCE1Mut2, PCR was performed using primer pairs A3F3 (5′-AATGGCTCCGGTATGTGCAAG-3′) and FR4 (5′-GCGCAGTGTCGTCGCTACAAGG-3′). To analyze splicing patterns of Bmdsx mRNAs transcribed from pMutE5, primer P25E4R4 (5′-GAGTCGCGGCCGCCATTAGGGTTAATGTGGAAG-3′) was used instead of primer FR4. To amplify the endogenous Bmdsx mRNAs, primers FF6 (5′-GTATACGGCCGACGAGGACCCAGCAATGG-3′) and FR4 were used. All PCRs were performed using LA Taq (Takara Bio Inc., Tokyo, Japan) according to the protocol provided by the manufacturer. PCR conditions were as follows: 94°C for 3 min followed by 31 cycles of 94°C for 15 seconds, 57°C for 30 seconds, and 72°C for 1 min. PCRs were in the linear phase (not in the plateau phase) under these conditions, as verified by using different amounts of cDNA. PCR products were analyzed on a 2% agarose gel and visualized with ethidium bromide. Product DNAs were quantified using a LAS3000 Bioimage analyzer (Fuji Photo Film, Tokyo, Japan). The identity of the PCR products was verified by sequence analyses.
Nuclear extract preparation.
NIAS-Bm-M1 and NIAS-Bm-F1 cells were maintained in IPL-41 with 10% FBS under a humidifying atmosphere at 27°C. About 2 × 107 cells were harvested, and nuclear extracts were prepared as described by Mine et al. (31) with a slight modification. Briefly, the cells were suspended in five packed cell volumes of buffer C (31) and allowed to swell on ice for 3 min. Nuclei were pelleted and resuspended in 2.2 packed cell volumes of buffer B′ (31). For lysis, the nuclei were drawn up in a syringe and rapidly pushed through a 26-gauge needle. This was repeated approximately 10 times. Extraction of nuclei was carried out on ice for 30 min with constant mixing. After extraction, nuclei were pelleted and the supernatant was dialyzed against buffer DB (31). The dialyzed nuclear protein extracts were then frozen in liquid N2 and stored at −80°C.
Synthesis of labeled RNA.
A 130-bp fragment of Bmdsx exon 4 was amplified by PCR using primers EMSAF1 (5′-CCGGATCCTAATACGACTCACTATAGGGCGTTAATAATATAAGTGGTGTA-3′) and EMSAR1 (5′-AAATATTTCCGTTGAAATTC-3′). EMSAF1 contains the T7 promoter site (underlined). This PCR product was transcribed using a RiboScribe T7 probe synthesis kit (Epicentre, Madison, WI) in the presence of [α-32P]UTP to generate a radiolabeled E4 RNA. A 140-bp fragment containing the T7 promoter site was amplified from pBluescript SK(+) using M13 to -20 and T3 primers. The resulting product was transcribed using a RiboScribe T7 probe synthesis kit to synthesize an [α-32P]UTP-labeled NC RNA. The CE1, CE2, CE3a, CE3b, CE3c, CE1mut1, CE1mut2, and CE1s DNAs were synthesized as two complementary oligonucleotides containing the T7 promoter. cDNA fragments were combined at equal molar amounts, heated at 95°C for 5 min, and then slowly cooled to anneal. The resulting products were transcribed using a RiboScribe T7 probe synthesis kit in the presence of [α-32P]ATP to generate radiolabeled CE1, CE2, CE3a, CE3b, CE3c, CE1mut1, CE1mut2, and CE1s RNAs. Cold RNA was produced using AmpliScribe T7 transcription kit (Epicentre, Madison, WI) according to the protocol provided by the manufacturer.
RNA mobility shift assays.
RNA mobility shift assays were performed essentially according to the methods of Chabot et al. (7). Binding reactions were performed in a 25-μl reaction mixture under splicing conditions (7). After a 15-min incubation on ice, the sample was adjusted to 1 mg/ml with heparin and incubated on ice for 2 min. For competition experiments, the indicated amount of cold competitor was incubated on ice for 8 min before addition of the probe. The reactions were run on a 4.5% native acrylamide gel (29:1 acrylamide-bisacrylamide, 5% glycerol, 50 mM Tris [pH 8.8], 50 mM glycine) in Tris-glycine running buffer (50 mM Tris [pH 8.8], 50 mM glycine). The gels were dried and visualized using a BAS2500 Bioimage analyzer (Fuji Photo Film, Tokyo, Japan).
UV cross-linking assays.
Five μg of nuclear extract or the indicated amount of recombinant protein was incubated with radiolabeled RNA in a final volume of 10 μl in 1× binding buffer (50 mM Tris-HCl [pH 7.5], 40 mM KCl, 15 mM NaCl, 0.5 mM dithiothreitol, 50 mg/ml bovine serum albumin) at room temperature for 20 min. Heparin was added to 1 mg/ml, incubated for 2 min, and UV cross-linked on ice at 254 nm for 10 min in a Bio-Rad GS UV cross-linker, followed by digestion with RNase A at 37°C for 30 min.
RNA affinity chromatography.
Twenty nanomoles of RNA oligonucleotide was coupled to adipic acid hydrazide agarose (Sigma) as described previously (5). The beads with bound RNA were then washed with 2 M NaCl and equilibrated with buffer DB. They were incubated with 3 mg of NIAS-Bm-M1 cell nuclear extract for 20 min at room temperature in 1,250 μl (final volume), pelleted by centrifugation at 3,000 rpm for 3 min, and washed five times with 5 ml of buffer DB containing 4 mM MgCl2. After the final centrifugation, 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added to the beads and heated for 5 min at 95°C before loading on a 10% SDS-PAGE gel. The Coomassie-stained band was excised and treated with 0.1 mg of Achromobacter protease I (a gift from Takeharu Masaki, Ibaraki University) at 37°C for 12 h in 0.1 M Tris-HCl (pH 9.0) containing 0.1% SDS. Peptides generated were extracted from the gel and separated on columns of DEAE-5PW (1 by 20 mm; Tosoh, Tokyo, Japan) and Inertsil ODS-3 (1 by 100 mm; GL Sciences Inc., Tokyo, Japan) connected in series with a model 1100 liquid chromatography system (Agilent Technologies, Waldbronn, Germany). Peptides were eluted at a flow rate of 0.02 ml/min using a linear gradient of 0 to 60% solvent B, where solvents A and B were 0.09% (vol/vol) aqueous trifluoroacetic acid and 0.075% (vol/vol) trifluoroacetic acid in 80% (vol/vol) acetonitrile, respectively. Selected peptides were subjected to Edman degradation using a Procise cLC protein sequencing system (Applied Biosystems) and to matrix-assisted laser desorption ionization time of flight mass spectrometry on an Ultraflex apparatus (Bruker-Franzen Analytik, Bremen, Germany) in reflector mode using alpha-cyano-4-hydroxycinnamic acid as a matrix.
Recombinant protein purification.
The BmPSI gene was amplified by PCR using a 5′ primer containing a SalI site (GTCGACATGAGTGATTATTCTTCTATGG) and a 3′ primer containing a NotI site (GCGGCCGCTCACTGCTGGTGGTCGGAGCC). The amplified product was cloned into the pGEX-6P-3 vector using the SalI and NotI sites. The fusion protein was prepared and purified as described previously (24). The fusion protein was subsequently cut with Precision protease (Amersham Pharmacia Biotech) and repurified with glutathione-Sepharose 4B to remove the glutathione-S-transferase protein. The purified protein was extensively dialyzed against buffer DB. The concentration of the recombinant protein was measured by the Bradford method using a protein assay from Bio-Rad.
RNA interference (RNAi) procedure.
cRNA strands were transcribed using the AmpliScribe T7 transcription kit, mixed, and annealed by heating to 95°C for 1 min and then allowing to cool to room temperature. For each gene, the following sequences were used to generate double-stranded RNA (dsRNA) (based on the start codon of the coding sequence): Bmsqd2, nucleotides (nt) 1 to 493; BmELAV, nt 1 to 616; BmPSI, nt 37 to 298 (dsRNA c), nt 1041 to 1209 (dsRNA b), and nt 1225 to 1467 (dsRNA a). Six μg of dsRNA was transfected into 5 × 105 NIAS-Bm-M1 cells using 24 μl of Cellfectin in IPL-41 medium without FBS. After 5 h, 2 ml of IPL-41 medium with 20% FBS was added to bring the final FBS concentration to 10%.
RESULTS
Establishment of sexually different cell lines and application to in vivo analyses for Bmdsx alternative splicing using a minigene.
In a previous study, we showed that exons 3 and 4 of Bmdsx pre-mRNA are specifically excluded in males (42). To explore how these exons are selected sex specifically and to identify sequence elements targeted by trans-acting factors, we developed a novel in vivo sex-specific splicing assay system using cultured cells that differ in sex. Anecdotal evidence suggested that B. mori cultured cell lines were easily established from embryos. Hence, we used silkworm embryos for establishing the male and female specific cell lines (for details, see Materials and Methods). To discriminate male embryos from female ones, fertilized eggs from the silkworm strain named Sex-limited black egg were used for establishing the primary culture. In this strain, female eggs display the same mauve coloration as normal eggs, whereas male eggs display a primrose yellow coloration (Fig. 1A). As shown in Fig. 1B and C, we successfully established a male cell line, NIAS-Bm-M1, derived from male embryos at the blastokinesis stage, as well as a female cell line, NIAS-Bm-F1, from female embryos. Molecular sexing by PCR using oligonucleotide primers derived from the W chromosome-specific retrotransposon confirmed that NIAS-Bm-M1 cells were all chromosomally male and NIAS-Bm-F1 cells were chromosomally female (Fig. 1E, right panel). To clarify the expression pattern of Bmdsx, RT-PCR analysis was performed using the primer sets illustrated in Fig. 1D to amplify male-type Bmdsx mRNA from NIAS-Bm-M1 cells and female-type Bmdsx mRNA from NIAS-Bm-F1 cells (Fig. 1E, left panel). These results further demonstrated that NIAS-Bm-M1 cells are male while NIAS-Bm-F1 cells are female.
We used a minigene construct (pWTE2.5) transiently transfected into NIAS-Bm-M1 and NIAS-Bm-F1 cells to test whether the minigene Bmdsx exons 3 and 4 underwent sex-specific splicing similar to the endogenous Bmdsx gene (Fig. 2A). Since the intronic sequences of the Bmdsx gene are very long (>40 kbp), the intronic sequences in the minigene were largely deleted. As shown in Fig. 2B, male-specific exclusion of exons 3 and 4 in both the endogenous Bmdsx pre-mRNA and the exogenous pWTE2.5 wild-type minigene pre-mRNA was observed in NIAS-Bm-M1 cells. In contrast, no exon exclusion was observed in the NIAS-Bm-F1 cell line, as all exons from both the endogenous Bmdsx pre-mRNA and the exogenous pWTE2.5 wild-type minigene pre-mRNA were present. Thus, in the minigene pre-mRNA, exons 3 and 4 were skipped or included in a sex-specific manner similar to exons 3 and 4 of the endogenous Bmdsx pre-mRNA. These results indicated that the pWTE2.5 wild-type minigene contained all the cis-acting regulatory elements involved in sex-specific alternative splicing and that the NIAS-Bm-M1 and NIAS-Bm-F1 cells are valid for splicing assay experiments.
Bmdsx exon 4 contains cis-acting elements that regulate the sex-specific splicing.
We first tested the possibility that sequences within the exons governed sex-specific splicing. With this reasoning, the Bmdsx exon sequences were replaced with unrelated sequences of approximately the same length from the Bombyx silk protein P25 exon (Fig. 2A). As shown in Fig. 2B, exclusion of exons 3 and 4 was observed in a male-specific manner when the male and female culture cells were transfected with either pMutE2 or pMutE3. These results indicated that substitution of exons 2 and 3 had no effect on sex-specific splicing regulation. In contrast, substitution of exon 4 resulted in nearly complete inclusion of exons 3 and 4 in male culture cells (Fig. 2B, lane 8). Interestingly, this substitution resulted in inclusion of both exons 3 and 4 at the same time, suggesting that the Bmdsx exon 4 contains one or more sequences that repress inclusion of not only exon 4 itself but also exon 3 in male cells. No amplified products were observed in either male or female culture cells transfected with the mutant exon 5 construct pMutE5 (Fig. 2B, lanes 9 and 10). These results are not a PCR artifact, since the same reactions using pMutE5 as a template successfully amplified the appropriate size of DNA fragment (data not shown). This result therefore indicated that the pMutE5 transcripts were degraded in both male and female culture cells. At present we do not know exactly the reason for this degradation but surmise that substitution of exon 5 inhibited splicing of the pMutE5 minigene pre-mRNA, with the resultant unspliced pre-mRNA degraded in the nuclei of the male and female culture cells. These results indicated that one or more sequences necessary for male-specific exon skipping are present in the Bmdsx exon 4. The same results were obtained when the Bmdsx exon sequences were replaced with unrelated sequences of approximately the same length from the bacterial ampicillin resistance gene (data not shown).
Linker scan mutations define distinct regulatory elements in Bmdsx exon 4.
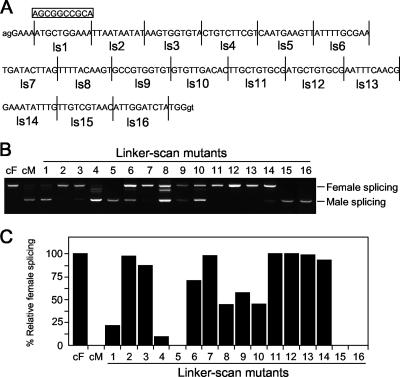
To identify the sequences within Bmdsx exon 4 that govern male-specific alternative splicing, we generated linker scan (ls) mutants by consecutively replacing 10-bp segments within the 167-bp Bmdsx of exon 4 with a 10-bp linker sequence (Fig. 3A). After transient transfection into NIAS-Bm-F1 cells, none of the mutations impaired female splicing (data not shown). On the other hand, a number of ls mutations had significant effects on male-specific splicing: 85 to 100% of the transcripts in male culture cells transfected with mutants ls2, ls3, ls7, ls11, ls12, ls13, and ls14 contained exons 3 and 4 (Fig. 3B and C). These effects were similar to those observed following substitution of exon 4 (compare Fig. 2B, lane 8,with 3B, ls2, ls3, ls7, ls11, ls12, ls13, and ls14). Thus, we concluded that the regions of exon 4 encompassing nucleotides 15 to 34, 65 to 74, and 105 to 144 most likely represent strong ESSs that repress the inclusion of exons 3 and 4. As a matter of convenience, we have denoted these elements as CE1, CE2, and CE3, respectively. About 50% of the transcripts from mutants ls8, ls9, and ls10 represented female splicing (Fig. 3C, ls8, ls9, and ls10), with exclusion of exon 3 occasionally observed (Fig. 3B; band below the band representing female-splicing in lane ls8). These results indicated that the region encompassing nucleotides 75 to 104 might function as a weak and incomplete ESS.
FIG. 3.
Identification of negatively acting regulatory sequences within Bmdsx exon 4 by linker scan mutagenesis. (A) Sequence of Bmdsx exon 4, indicating the positions of the introduced linker scan mutants (ls1 to ls16). Segments flanked by vertical lines were replaced consecutively by a 10-bp NotI linker sequence (boxed). Uppercase lettering, exon sequence; lowercase lettering, intron sequences. (B) RT-PCR analysis of NIAS-Bm-M1 cells transiently transfected with the ls mutants using primers as indicated in Fig. 2A. As a control, RT-PCR amplification was also performed from cytoplasmic RNA of NIAS-Bm-F1 cells (lane cF) and NIAS-Bm-M1 cells (lane cM) transiently transfected with the wild-type minigene construct pWTE2.5. Labels to the left of the gel refer to female splicing and male splicing. (C) Quantification of the RT-PCR analysis shown in panel B. Results are expressed as the percentage of RT-PCR products representing female splicing relative to total RT-PCR products.
A NIAS-Bm-M1 nuclear factor specifically binds to CE1.
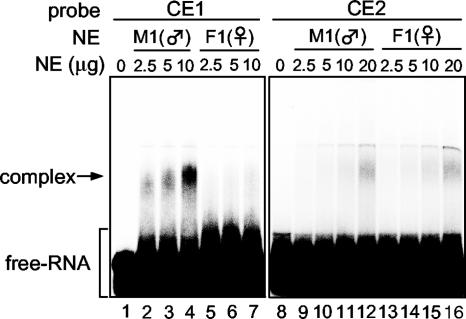
To test whether some protein factors bind to CE1, CE2, and CE3, we first carried out a gel shift analysis using nuclear factors extracted from male culture cells (Fig. 4). When a 32P-labeled RNA probe E4 (Fig. 4A), including the CE1, CE2, and CE3 regions, was incubated with nuclear factors from NIAS-Bm-M1 cells, two bands with different mobilities from that of the probe RNA were detected (Fig. 4B). To determine which band reflected specific interaction of the E4 RNA with nuclear factors, a 32P-labeled control RNA carrying only plasmid sequence was subjected to the same reaction. Comparing the gel shift patterns of the control RNA with the pattern of the E4 RNA revealed that the upper band was specific for E4 RNA-protein interaction. We further carried out a series of competition experiments using cold competitor RNAs covering the CE1, CE2, or CE3 regions (Fig. 4A). As shown in Fig. 4C, CE3b did not compete for binding of the complex even when present at 100-fold excess (Fig. 4C, lane 13). CE3a and CE3c also had little to no effect on complex formation (Fig. 4C, lanes 8 to 10 and lanes 14 to 16). The upper shifted band was weakly affected when CE2 was used as a competitor (Fig. 4C, lanes 5 to 7). On the other hand, CE1 had a strong inhibitory effect on the RNA-protein interaction. Addition of unlabeled CE1 resulted in a dose-dependent reduction in the formation of the complex (Fig. 4C, lanes 1 to 3). These experiments therefore suggested that most of the nuclear factors interact with the CE1 region. We next performed the same gel shift analysis using CE1, CE2, CE3a, and CE3c as 32P-labeled probes. Consistent with the previous results, a band with retarded mobility was detected when 32P-labeled CE1 was incubated with increasing amounts of nuclear extract (Fig. 4D, lanes 1 to 4). In contrast, 32P-labeled CE2, CE3a, and CE3c did not form any complex (Fig. 4D, lanes 5 to 8, 9 to 12, and 13 to 16). Overexposure of the gel shift autoradiograph, however, revealed that a very faint band was observed when the 32P-labeled CE2 was incubated with the highest concentrations of the nuclear extract (data not shown). These results demonstrated that the NIAS-Bm-M1 cell nuclear extract contains factors that bind to CE1 and CE2 RNAs.
FIG. 4.
Complex formation between NIAS-Bm-M1 cell nuclear proteins and RNA sequences in exon 4 that are essential for male-specific splicing. (A) Schematics of the 32P-labeled probes and competitor RNAs. (B) Radiolabeled transcripts were incubated for 15 min on ice in the presence (+) or absence (-) of a NIAS-Bm-M1 cell nuclear extract (M1 cell NE). Complexes were separated on a nondenaturing 4.5% acrylamide gel. The positions of the free RNA and complex are indicated on the left. Asterisks denote an unspecific complex. NC, control RNA containing 140 nt from pBluescript. (C) The RNA-protein interaction between a labeled E4 probe and the NIAS-Bm-M1 cell nuclear extract in the presence of increasing amounts of CE1, CE2, CE3a, CE3b, and CE3c unlabeled competitors (80, 800, and 2,400 ng). Complexes were separated on a nondenaturing 4.5% acrylamide gel. (D) Radiolabeled probes were incubated for 15 min on ice in the presence of increasing amounts of a NIAS-Bm-M1 cell nuclear extract (2.5, 5, and 10 μg). Other experimental procedures were the same as those for panels B and C.
We then performed the gel shift analysis using nuclear proteins extracted from female culture cells and compared the gel shift patterns of the female cell nuclear extract with that of the male cell nuclear extract (Fig. 5). When 32P-labeled CE1 was incubated with increasing amounts of the male cell nuclear extract, the same shifted band as observed in Fig. 4D was detected (Fig. 5A, lanes 2 to 4). This band shift was not observed following incubation with the female cell nuclear extract (Fig. 5A, lanes 5 to 7). A very faint band was also observed when 32P-labeled CE2 was incubated with 20 μg of male and female nuclear proteins; however, the intensity of the shifted band was almost equal in male and female cells. These results clearly indicated that a trans-acting factor specifically recognizes the CE1 region in a sequence-specific manner and that it exists only in the nuclei of male cells.
FIG. 5.
Nuclear factors interacting with CE1 and CE2. Radiolabeled CE1 and CE2 RNAs were incubated with the indicated amounts of NIAS-Bm-M1 (M1) and NIAS-Bm-F1 (F1) cell nuclear extracts. Complexes were separated on a nondenaturing 4.5% acrylamide gel. The position of the free RNA and complex are indicated on the left. NE, nuclear extract.
A CE1-nuclear protein interaction is required for male-specific splicing.
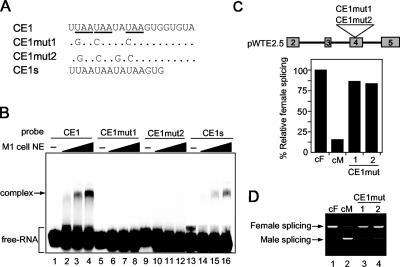
CE1 consists of a 20-nt UA-rich sequence containing three copies of a UAA repeat motif. A database search for exonic splicing signals within CE1 revealed that the first 8-nt sequence (UUAAUAAU) completely matches functional silencers in the PESS (http://cubweb.biology.columbia.edu/pesx) ESS database. To identify the sequence within the CE1 region essential for the protein interaction, we introduced a series of point mutations into the CE1 region. Mutation of the UAA repeats to GAA or CAA within CE1 (Fig. 6A, CE1mut1) completely disrupted the RNA-protein interaction (Fig. 6B, lanes 6 to 8). Similar results were obtained when CE1mut2 was used as a probe (Fig. 6B, lanes 10 to 12). In addition, the first 15 nt of CE1 were able to form the RNA-protein complex (Fig. 6B, lanes 14 to 16).
FIG. 6.
Binding of the nuclear factor to CE1 correlates with repression activity. (A) Sequence of the CE1 probe and the mutated sequences of two types of CE1 mutants (CE1mut1 and CE1mut2). Dots represent identical nucleotides in CE1 and the mutated CE1 sequences. UAA repeats are underlined. CE1s consists of the first 15 nt of CE1. (B) Radiolabeled probes were incubated for 15 min on ice in the presence of increasing amounts of a NIAS-Bm-M1 cell nuclear extract (2.5, 5, and 10 μg). Complexes were separated on a nondenaturing 4.5% acrylamide gel. The positions of the free RNA and complex are indicated on the left. (C) The upper panel shows a schematic diagram of the pWTE2.5 derivative mutant minigenes in which the CE1 sequence was replaced with the CE1mut1 or CE1mut2 mutant sequence. The lower panel indicates the quantification of the RT-PCR analysis shown in panel D. Results are expressed as the percentage of RT-PCR products representing female splicing relative to total RT-PCR products. Data represent the means of four independent experiments. (D) RT-PCR analysis of NIAS-Bm-M1 cells transiently transfected with the mutant minigenes shown in panel C, using primers as indicated in Fig. 2A. As a control, RT-PCR amplification was also performed from cytoplasmic RNA of NIAS-Bm-F1 cells (lane cF) and NIAS-Bm-M1 cells (lane cM) transiently transfected with the wild-type minigene construct pWTE2.5. Labels to the left of the gel refer to female splicing and male splicing.
To test whether the male-specific splicing is affected by the mutations that eliminate the CE1-protein interaction, we constructed mutant minigenes by introducing either the CE1mut1 or the CE1mut2 substitution into pWTE2.5 (Fig. 6C, upper panel). After transient transfection into NIAS-Bm-M1 cells, all mutations had a significant effect on male-specific splicing: more than 85% of the transcripts in male culture cells transfected with the mutant minigenes contained exons 3 and 4 (Fig. 6B and C). In contrast, these mutations had no effect on female splicing (see Fig. S1 in the supplemental material). These results suggest that the CE1-nuclear protein interaction is essential for male-specific splicing and that this interaction is required for repressing the inclusion of exons 3 and 4 in male culture cells.
CE1 RNA beads specifically retain an 80-kDa protein, BmPSI.
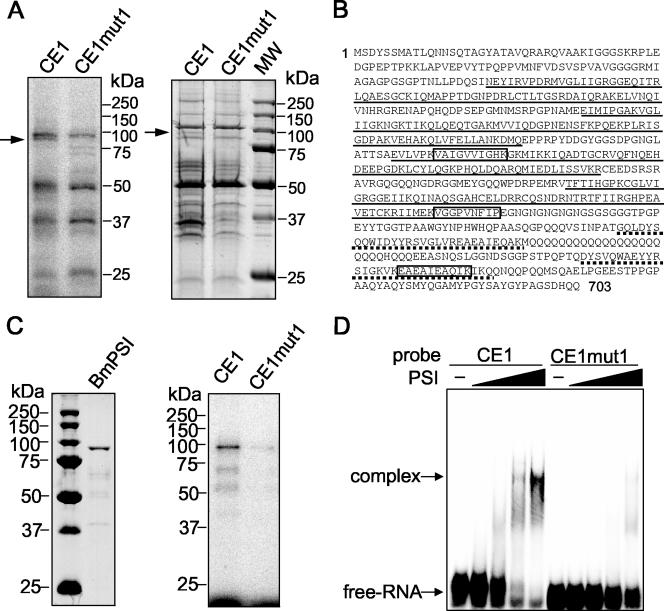
UV cross-linking was used to identify proteins in a NIAS-Bm-M1 cell nuclear extract that bind to CE1. Following incubation with a NIAS-Bm-M1 cell nuclear extract, 32P-labeled RNA probes were digested with RNase A and proteins were resolved by SDS-polyacrylamide gel electrophoresis. Comparing the cross-linking patterns of the mutant RNA (CEmut1) with the pattern of CE1 RNA revealed a specific band migrating at approximately 80 kDa (Fig. 7A, left panel).
FIG. 7.
Identification of BmPSI as a nuclear factor binding specifically to CE1. (A) The left panel shows UV cross-linking of NIAS-Bm-M1 cell nuclear extract to radiolabeled CE1 and CE1mut1 RNAs. Proteins cross-linked to RNase A-digested samples were electrophoresed on an SDS-10% polyacrylamide gel. The arrow indicates an approximately 80-kDa protein band that is present in the CE1 lane as opposed to the CE1mut1 lane. Approximate locations of molecular mass markers are shown on the right. The right panel shows the result of affinity chromatography using adipic acid dehydrazide beads derivatized with CE1 and CE1mut1 RNAs following incubation with a NIAS-Bm-M1 cell nuclear extract. The arrow indicates the protein band with a molecular mass corresponding to the molecular mass of the protein band at 80 kDa that was UV cross-linked to the labeled CE1 RNA. Lane MW, molecular size markers. (B) The full amino acid sequence of BmPSI, with the open boxes corresponding to the sequenced peptides from the excised 80-kDa band. Underlined sequences highlight the four KH-domains. Sequences underlined with dots indicate A and B motifs. (C) The purified recombinant protein (left panel) and its reactivity with labeled CE1 and CE1mut1 RNAs in a UV cross-linking assay (right panel). (D) Radiolabeled CE1 and CE1mut1 RNAs were incubated in the presence of 1.25, 2.5, 5, and 10 pmol of recombinant BmPSI. Complexes were separated on a nondenaturing 4.5% acrylamide gel.
To identify the 80-kDa protein, we performed RNA affinity chromatography using CE1 RNA covalently linked to adipic acid dehydrazide agarose beads. After incubating the RNA beads with a NIAS-Bm-M1 cell nuclear extract, the bound proteins were analyzed on an SDS-PAGE gel and stained with Coomassie blue. A comparison of the proteins retained by the mutant RNA (CEmut1) beads and by the CE1 RNA beads again revealed a protein of approximately 80 kDa that was specifically retained by the CE1 RNA (Fig. 7A, right panel). Internal sequence analysis of the excised 80-kDa band resulted in three peptides, and a search in KAIKObase (http://sgp.dna.affrc.go.jp/KAIKO/index.html) revealed that those sequences were 100% identical to residues 306 to 315, 474 to 482, and 637 to 646 of a predicted protein encoded by EST clone fdpeP24-F18 (Fig. 7B). This protein is similar to the P-element somatic inhibitor (PSI)-like protein of Apis mellifera (identity, 46%; similarity, 58%). PSI was originally identified in D. melanogaster as a splicing inhibitor that represses splicing of the P-element third intron by binding to a pseudo-splice site using four KH-type RNA-binding motifs (1, 39). The overall domain structure of BmPSI is similar to that of PSI, with four KH domains and a carboxyl terminus containing two tandem repeats of a 30-amino-acid motif homologous to the PSI-AB motif (Fig. 7B). As a matter of convenience, we have denoted the 80-kDa protein as BmPSI.
In addition to BmPSI, CE1 RNA also specifically pulled down 35-kDa and 42-kDa proteins. Internal sequencing analysis of these bands revealed that the 35-kDa protein is Bmsqd2 protein (a member of hnRNP A/B) and the 42-kDa protein corresponds to Bombyx ELAV homologous protein (BmELAV). However, these proteins may reflect experimental artifacts of the affinity chromatography, since prominent bands corresponding to these proteins were not observed in the UV cross-linking experiment (Fig. 7A, left panel).
Recombinant BmPSI binds specifically to CE1.
To confirm that BmPSI can interact specifically with CE1, recombinant BmPSI protein was prepared using an Escherichia coli expression system (for details, see Materials and Methods). Figure 7C, left panel, shows the purified recombinant BmPSI protein with the expected 80-kDa molecular mass. Figure 7C, right panel, shows that the recombinant BmPSI can specifically cross-link with the 32P-labeled CE1 RNA. The specificity of the interaction of recombinant BmPSI with CE1 was also assessed with the mobility shift assay. 32P-labeled CE1 RNA was incubated in the presence of increasing amounts of recombinant BmPSI. As shown in Fig. 7D, complex formation occurred with CE1 RNA but not with CE1mut1 RNA. It is reasonable to suppose that BmPSI binds with high affinity to CE1, since the amount of the recombinant BmPSI used in the mobility shift assay illustrated in Fig. 7D was very low (1.25 to ∼10 pmol).
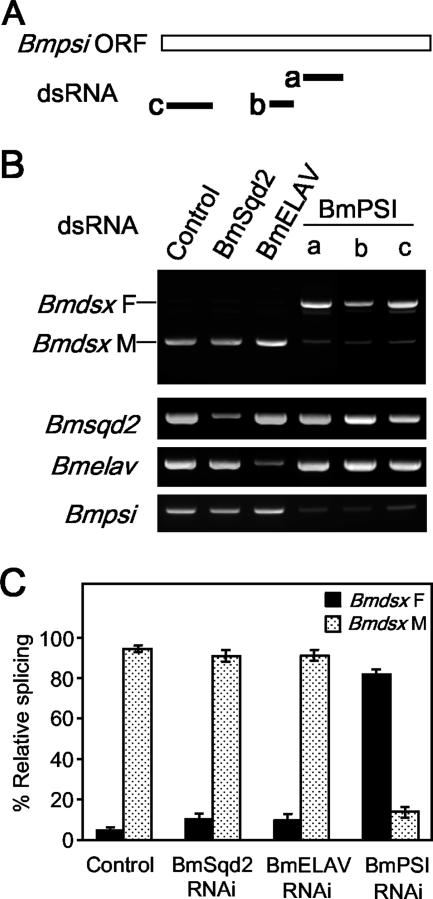
Down-regulation of BmPSI by RNA interference increases female-specific splicing.
To further test whether BmPSI plays a role in regulating male-specific splicing of Bmdsx pre-mRNA, we tried an RNAi approach to reduce BmPSI expression in male cells. To prevent the identification of false positives through “off-target effects,” several nonoverlapping dsRNAs were designed to different regions of the Bmpsi sequence (Fig. 8A) and then transfected into NIAS-Bm-M1 cells. Although we were not able to completely block Bmpsi expression, all three dsRNAs caused a consistent and significant increase in female-specific splicing (Fig. 8B and C). No variation in sex-specific splicing pattern and/or BmPSI endogenous level was observed when the cells were transfected with dsRNAs targeting Bmsqd2 or Bmelav. Therefore, we decided to rule out these proteins as candidates for the cellular factor implicated in Bmdsx sex-specific splicing. The treatment by the above dsRNAs targeting Bmpsi did not affect alternative splicing in other genes examined, such as BmSxl (see Fig. S2 in the supplemental material), suggesting a relative specificity of BmPSI in regulation of Bmdsx gene sex-specific splicing.
FIG. 8.
Down-regulation of BmPSI by RNAi increases female-specific splicing. (A) Schematic diagram of three dsRNAs (a, b, and c) specific for BmPSI. (B) NIAS-Bm-M1 cells were transfected with dsRNAs specific for DsRed (control), BmSqd2, BmELAV, or BmPSI. The upper panel shows the Bmdsx splicing pattern as detected by RT-PCR. Labels to the left of the gel refer to female splicing (Bmdsx F) and male splicing (Bmdsx M). The lower panel shows the specific down-regulation of BmSqd2, BmELAV, and BmPSI expression in dsRNA-treated cells. The RNA level of each gene was detected by RT-PCR using primers specific for each gene. (C) Quantification of the results derived from three independent experiments. Results are expressed as the percentage of RT-PCR products representing female or male splicing relative to total RT-PCR products.
DISCUSSION
In the present study, we successfully established both male- and female-specific cell lines (NIAS-Bm-M1 and NIAS-Bm-F1, respectively) derived from silkworm embryos. Our data demonstrate that NIAS-Bm-M1 and NIAS-Bm-F1 cells maintain several aspects of in vivo sexual dimorphism (Fig. 1). As far as we know, this study is the first report of the establishment of stable cell lines that only differ in sex. We expect that our cell lines will be a helpful tool for understanding the molecular mechanism governing sex determination and sexual differentiation.
The substitution experiments using a novel in vivo sex-specific splicing assay system with the two sexually different cell lines revealed that Bmdsx exon 4 contains male-specific negative regulatory elements referred to as CE1, CE2, and CE3 (Fig. 3). It is clear from our studies that a CE1-nuclear protein interaction is essential for male-specific splicing and that this interaction is required for repressing female-specific splicing (i.e., inclusion of exons 3 and 4) in male cells (Fig. 6). Furthermore, the fact that the trans-acting factor interacting with CE1 exists in male cells but not in female cells (Fig. 5) is consistent with our finding that CE1 is essential for male-specific splicing. We have identified BmPSI, a Bombyx homolog of PSI, as the factor that binds specifically to CE1. PSI is a KH-domain RNA-binding protein that is responsible for alternative splicing of P-element transposase pre-mRNA in Drosophila somatic tissues (1, 39). In vitro and in vivo assays support the role of BmPSI in CE1 activity. First, recombinant BmPSI specifically interacts with CE1 in UV cross-linking and gel mobility shift assays. Second, down-regulation of endogenous BmPSI expression in male culture cells by RNAi leads to an increase in female-specific splicing. Collectively, these results indicate that BmPSI mediates the repressing activity of CE1. Based on the data reported here, we propose a possible model for the regulatory mechanism underlying sex-specific alternative splicing of Bmdsx. In male cells, an interaction between BmPSI and CE1 prevents the use of exons 3 and 4, leading to an enhancement in the production of male-type Bmdsx. However, in female cells, exons 3 and 4 are selected to generate female-type Bmdsx mRNA because of either insufficient levels of BmPSI or the presence of a trans-acting factor(s) that counteracts the activity of BmPSI. Northern blot analysis showed that no differences in expression patterns of BmPSI mRNAs were seen between male and female cells (see Fig. S3 in the supplemental material), suggesting the possibility that the activity of BmPSI is different between males and females. For several KH-domain RNA-binding proteins, their ability to bind RNA is negatively regulated by tyrosine phosphorylation (14, 30, 47). The KH-domain protein Sam68 is acetylated in vivo, and its acetylation correlates with enhanced RNA-binding activity (2). Likewise, the RNA-binding activity of BmPSI may be sex specifically regulated by some posttranslational modifications.
How does BmPSI binding mediate inhibition of female-specific splicing? D. melanogaster PSI specifically binds to a cis-acting negative regulatory element within the exon upstream of IVS3, which contains pseudo-5′-splice sites. PSI recruits U1 snRNP to the pseudo-splice site, thereby preventing U1 snRNP from binding to the accurate IVS3 5′ splice site (40). The functional interaction between PSI and U1 snRNP is mediated by the C-terminal domain of the U1 snRNP-specific 70K (U1-70K) protein and the AB motif of PSI (24). Interestingly, a sequence showing weak homology to the 5′ splice site consensus sequence lies 13 nt upstream of the CE1 cis-acting element (Fig. 9). Like PSI, BmPSI may recruit U1-snRNP to the 5′ splice site-like sequence upstream of CE1, thereby preventing U1snRNP from binding to the accurate 5′ splice site. As shown in Fig. 9, the 5′ splice site-like sequence lies within the 3′ splice site immediately upstream of exon 4. Therefore, recruitment of U1-snRNP to this 5′ splice site-like sequence may preclude binding of the splicing factor(s) assembling on the 3′ splice site, such as U2AF, leading to exon 4 skipping. However, the binding site of BmPSI is 141 nt upstream of the 5′ splice site, whereas the binding site of PSI is so close to the 5′ splice site (14 nt upstream of the 5′ splice site) that it has been proposed to interfere with spliceosome assembly at the 5′ splice site (40). Thus, it seems unlikely that BmPSI can inhibit the use of the 5′ splice site of Bmdsx exon 4, or other components may be required to act together with BmPSI to block the 5′ splice site. At present, it remains unknown how BmPSI induces exon 3 skipping. Studies are under way to clarify this point.
FIG. 9.
5′ splice site-like sequence upstream of CE1. The sequence around the 3′ splice site of Bmdsx exon 4 is shown. The 5′ splice site-like sequence similar to the 5′ splice site consensus sequence (GURAGU) is underlined. The 3′ splice site of Bmdsx exon 4 is indicated by an arrow. The CE1 sequence is boxed.
The gel shift analysis showed that CE2 interacts with a nuclear factor in both male and female cells; however, the intensity of the shifted band representing this interaction was very weak (Fig. 5). It seems likely that this nuclear factor exists in both male and female cells and functions as a splicing cofactor that facilitates male-specific splicing, such that BmPSI and the CE2-binding protein are needed to inhibit the use of exons 3 and 4 in male cells. The same gel shift analysis showed that CE3 does not interact with any nuclear proteins, even though CE3 is essential for male-specific splicing (Fig. 4). This discrepancy may be due to an experimental artifact of the linker scanning/minigene assay. Alternatively, CE3 functions via its RNA secondary structure in male-specific alternative splicing. Indeed, several studies have shown an effect of RNA secondary structure on alternative splicing in cellular pre-mRNAs and some viral mRNAs. In some cases, RNA secondary structure acts as a splicing enhancer by decreasing the physical distances between splice sites (8, 9, 32) or by maintaining one or more cis-acting regions of pre-mRNA in a favorable configuration for interaction with trans-acting factors (15, 33, 38). In this way, it is possible to envision that the CE3 element in the Bmdsx exon 4 functions to maintain the correct display of the CE1 (or CE2) element rather than in binding specific splicing regulatory proteins.
In our previous study, an in vitro splicing reaction utilizing HeLa cell nuclear extracts showed that female-type Bmdsx mRNA represented default splicing (42). In contrast, default processing in Drosophila yields male-type dsx mRNA with exon 4 excluded. In females, TRA/TRA-2 promote the incorporation of exon 4. This pattern of sex-specific splicing is also observed in the Queensland fruit fly, Bactrocera tryoni, the Phorid fly, Megaselia scalari, and the housefly, Musca domestica, where it occurs at equivalent positions in the corresponding dsx genes (18, 22, 37). The presence of putative TRA/TRA-2-binding sites in the female-specific exon of dsx in Bactrocera, Megaselia, and Musca gives further support to the notion that female exon selection by activation is common in dipteran insects. In contrast, TRA/TRA-2-binding motif-related sequences are not present in the Bmdsx genomic sequence (42). Our data presented here provide evidence for BmPSI-mediated regulation of Bmdsx pre-mRNA sex-specific alternative splicing. Since BmPSI does not exhibit any sequence relationship to known SR proteins, such as TRA and TRA-2, the regulatory mechanism of sex-specific alternative splicing of Bmdsx pre-mRNA is distinct from that of dsx. Furthermore, to the best of our knowledge there are no reports describing PSI involvement in regulating sex-specific splicing of dsx pre-mRNA or the sex determination cascade. Further work is needed to elucidate how BmPSI represses inclusion of female-specific exons in a male-specific manner.
Supplementary Material
Acknowledgments
We thank Shinji Atsuzawa and Masaaki Kurihara for their excellent technical assistance.
This work was supported by the Bioarchitect Research Program and the Chemical Biology Research Project from RIKEN and by the Ministry of Education, Science, Sports and Culture, Grant-in-Aid for Young Scientists (B), 17780045, 2006.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, M. D., R. S. Tarng, and D. C. Rio. 1997. The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes Dev. 11129-138. [DOI] [PubMed] [Google Scholar]
- 2.Babic, I., A. Jakymiw, and D. J. Fujita. 2004. The RNA binding protein Sam68 is acetylated in tumor cell lines, and its acetylation correlates with enhanced RNA binding activity. Oncogene 233781-3789. [DOI] [PubMed] [Google Scholar]
- 3.Baker, B. S., and M. F. Wolfner. 1988. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 2477-489. [DOI] [PubMed] [Google Scholar]
- 4.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25106-110. [DOI] [PubMed] [Google Scholar]
- 5.Buratti, E., T. Dork, E. Zuccato, F. Pagani, M. Romano, and F. E. Baralle. 2001. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 201774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burtis, K. C., and B. S. Baker. 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56997-1010. [DOI] [PubMed] [Google Scholar]
- 7.Chabot, B., M. Blanchette, I. Lapierre, and H. La Branche. 1997. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 171776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier, B., and M. Rosbash. 1996. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA 2509-522. [PMC free article] [PubMed] [Google Scholar]
- 9.Deshler, J. O., and J. J. Rossi. 1991. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 51252-1263. [DOI] [PubMed] [Google Scholar]
- 10.Erdman, S. E., and K. C. Burtis. 1993. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1765-771. [DOI] [PubMed] [Google Scholar]
- 12.Graveley, B. R., K. J. Hertel, and T. Maniatis. 1999. SR proteins are “locators” of the RNA splicing machinery. Curr. Biol. 9R6-R7. [DOI] [PubMed] [Google Scholar]
- 13.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 61197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haegebarth. A., D. Heap, W. Bie, J. J. Derry, S. Richard, and A. L. Tyner. 2004. The nuclear tyrosine kinase BRK/Sik phosphorylates and inhibits the RNA-binding activities of the Sam68-like mammalian proteins SLM-1 and SLM-2. J. Biol. Chem. 27954398-54404. [DOI] [PubMed] [Google Scholar]
- 15.Handa, N., O. Nureki, K. Kurimoto, I. Kim, H. Sakamoto, Y. Shimura, Y. Muto, and S. Yokoyama. 1999. Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature 398579-585. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto, H. 1933. The role of the W chromosome for sex determination in the silkworm, Bombyx mori. Jap. J. Genet. 8245-258. [Google Scholar]
- 17.Hawkins, J. R. 1993. Mutational analysis of SRY in XY females. Hum. Mutat. 2347-350. [DOI] [PubMed] [Google Scholar]
- 18.Hediger, M., G. Burghardt, C. Siegenthaler, N. Buser, D. Hilfiker-Kleiner, A. Dubendorfer, and D. Bopp. 2004. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev. Genes Evol. 21429-42. [DOI] [PubMed] [Google Scholar]
- 19.Hodgkin, J. 1999. Sex, cell death, and the genome of C. elegans. Cell 98277-280. [DOI] [PubMed] [Google Scholar]
- 20.Hodgkin, J. 2002. The remarkable ubiquity of DM domain factors as regulators of sexual phenotype: ancestry or aptitude? Genes Dev. 162322-2326. [DOI] [PubMed] [Google Scholar]
- 21.Kan, J. L., and M. R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn, S., V. Sievert, and W. Traut. 2000. The sex-determining gene doublesex in the fly Megaselia scalaris: conserved structure and sex-specific splicing. Genome 431011-1020. [DOI] [PubMed] [Google Scholar]
- 23.Kuwabara, P. E. 1999. Developmental genetics of Caenorhabditis elegans sex determination. Curr. Top. Dev. Biol. 4199-132. [DOI] [PubMed] [Google Scholar]
- 24.Labourier, E., M. D. Adams, and D. C. Rio. 2001. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell 8363-373. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y., and B. J. Blencowe. 1999. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 27435074-35079. [DOI] [PubMed] [Google Scholar]
- 26.Lynch, K. W., and T. Maniatis. 1995. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 9284-293. [DOI] [PubMed] [Google Scholar]
- 27.Lynch, K. W., and T. Maniatis. 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 102089-2101. [DOI] [PubMed] [Google Scholar]
- 28.Marin, I., and B. S. Baker. 1998. The evolutionary dynamics of sex determination. Science 2811990-1994. [DOI] [PubMed] [Google Scholar]
- 29.McKeown, M., J. M. Belote, and R. T. Boggs. 1988. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell 53887-895. [DOI] [PubMed] [Google Scholar]
- 30.Messias, A. C., C. Harnisch, A. Ostareck-Lederer, M. Sattler, and D. H. Ostareck. 2006. The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation. J. Mol. Biol. 361470-481. [DOI] [PubMed] [Google Scholar]
- 31.Mine, E., H. Sakurai, S. Izumi, and S. Tomino. 1995. The fat body cell-free system for tissue-specific transcription of plasma protein gene of Bombyx mori. Nucleic Acids Res. 232648-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mougin, A., A. Gregoire, J. Banroques, V. Segault, R. Fournier, F. Brule, M. Chevrier-Miller, and C. Branlant. 1996. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA 21079-1093. [PMC free article] [PubMed] [Google Scholar]
- 33.Muro, A. F., M. Caputi, R. Pariyarath, F. Pagani, E. Buratti, and F. E. Baralle. 1999. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 192657-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagoshi, R. N., M. McKeown, K. C. Burtis, J. M. Belote, and B. S. Baker. 1988. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 53229-236. [DOI] [PubMed] [Google Scholar]
- 35.Nagoshi, R. N., and B. S. Baker. 1990. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 489-97. [DOI] [PubMed] [Google Scholar]
- 36.Ohbayashi, F., T. Shimada, T. Sugasaki, S. Kawai, K. Mita, T. Oshiki, and H. Abe. 1998. Molecular structure of the copia-like retrotransposable element Yokozuna on the W chromosome of the silkworm, Bombyx mori. Genes Genet. Systems 73345-352. [DOI] [PubMed] [Google Scholar]
- 37.Shearman, D. C. A., and M. Formmer. 1998. The Bactrocera tryoni homolog of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 71-12. [DOI] [PubMed] [Google Scholar]
- 38.Shi, H., B. E. Hoffman, and J. T. Lis. 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 172649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siebel, C. W., and D. C. Rio. 1990. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science 2481200-1208. [DOI] [PubMed] [Google Scholar]
- 40.Siebel, C. W., L. D. Fresco, and D. C. Rio. 1992. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 61386-1401. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths, M. J. Smith, J. W. Foster, A. M. Frischauf, R. Lovell-Badge, and P. N. Goodfellow. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346240-244. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, M. G., F. Ohbayashi, K. Mita, and T. Shimada. 2001. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem. Mol. Biol. 311201-1211. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, M. G., S. Funaguma, T. Kanda, T. Tamura, and T. Shimada. 2003. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev. Genes Evol. 213345-354. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, M. G., S. Funaguma, T. Kanda, T. Tamura, and T. Shimada. 2005. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol. Dev. 758-68. [DOI] [PubMed] [Google Scholar]
- 45.Tian, M., and T. Maniatis. 1994. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 81703-1712. [DOI] [PubMed] [Google Scholar]
- 46.Volff, J. N., D. Zarkower, V. J. Bardwell, and M. Schartl. 2003. Evolutionary dynamics of the DM domain gene family in metazoans. J. Mol. Evol. 57S241-S249. [DOI] [PubMed] [Google Scholar]
- 47.Wang, L. L., S. Richard, and A. S. Shaw. 1995. p62 association with RNA is regulated by tyrosine phosphorylation. J. Biol. Chem. 2702010-2013. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, L., J. Wilken, N. B. Phillips, U. Narendra, C. Chan, S. M. Stratton, S. B. Kent, and M. A. Weiss. 2000. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 141750-1764. [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo, P., and T. Maniatis. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 101356-1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.