Abstract
The Hsp90 molecular chaperone is a highly abundant eukaryotic molecular chaperone. While it is understood that Hsp90 modulates a significant number of proteins, the mechanistic contributions made by Hsp90 to a client protein typically are not well understood. Here we investigate the yeast Hsp90 regulatory roles with telomerase. Telomerase lengthens chromosome termini by specifically associating with single-stranded telomeric DNA and appending nucleotides by reverse transcription. We have found that the yeast Hsp90 homolog Hsp82p promotes both telomerase DNA binding and nucleotide addition properties. By isolating telomerase from different allelic backgrounds we observed distinct defects. For example, in an hsp82 T101I strain telomerase displayed decreased nucleotide processivity, whereas both DNA binding and extension activities were lowered in a G170D background. The decline in telomerase DNA binding correlated with a loss of Hsp82p association. No matter the defect, telomerase activity was recovered upon Hsp82p addition. Importantly, telomere length and telomerase telomere occupancy was yeast Hsp90 dependent. Taken together, our results indicate that Hsp82p promotes telomerase DNA association and facilitates DNA extension once telomerase is engaged with the DNA.
The Hsp90 molecular chaperone is a highly conserved and abundant protein that has evolved into an essential eukaryotic protein (4, 13, 41). Given recent proteomic and genetic screens, Hsp90 has a role in a multitude of normal cellular functions and is involved in a number of diseases ranging from conformational protein folding problems to cancer (31). Accelerating the interest in Hsp90 is its developing use as a therapeutic target for diverse diseases (27, 42). Presumably, the broad therapeutic spectrum results from Hsp90's role in maintaining the conformation, stability, and activity of many key cellular proteins that includes intracellular hormone receptors, cRaf, Her2, Akt, Cdk4, p53, and telomerase. Despite its apparent central role in the eukaryotic molecular chaperone system and disease relevance, its mechanistic role in client protein regulation is not well understood. Here we investigate the yeast Hsp90 contributions to telomerase activities.
Telomerase maintains genomic integrity, in part, by preserving chromosome length after DNA replication (5, 34). Since conventional DNA polymerases require priming events to initiate synthesis, the extreme terminus of each lagging strand cannot be completed—commonly referred to as the end replication problem (39). In the absence of a compensatory process, this limitation would lead to chromosome erosion with each round of replication. Almost all eukaryotes circumvent this problem by adding a tandem array of simple sequence repeats to each terminus that buffers against the loss. Depending upon the organism, telomerase increases chromosome ends between a few hundred to a few thousand nucleotides to create the telomeric DNA end (34). Perhaps unexpectedly, it was realized that the length of each telomere is not added at once but rather telomerase typically appends six to eight nucleotides per binding event (29). While it had been argued that telomerase might not need to be processive to maintain telomere length (22), recent studies in yeast indicate that telomeres can be extended over 100 nucleotides per cell cycle (23, 36). The mechanism(s) that governs the necessary repetitive telomerase-DNA binding and extension events is poorly understood.
The Hsp90 and p23 molecular chaperones were the first two telomerase cofactors found to alter DNA extension activity in vitro (15). At the time it was suggested that these two chaperones serve to assemble the telomerase catalytic protein with its RNA template—telomerase is a specialized reverse transcriptase that uses an RNA moiety to specifically extend the 3′ DNA end (34). However, recent reports indicate that Hsp90 and p23 can function after assembly. DNA extension by the mature telomerase enzyme is reduced upon Hsp90 inhibition (20). However, if a telomeric DNA substrate is added prior to the Hsp90 inhibitor, the effect is alleviated, which suggests Hsp90 supports telomerase DNA binding. The yeast p23 homolog Sba1p, on the other hand, was shown to promote telomerase dissociation from DNA in vitro and affect telomere maintenance and telomerase telomere occupancy in vivo (37). Unfortunately, correlative in vivo roles for human Hsp90 have not been reported.
In contrast to the mammalian system, Hsp90-dependent changes to yeast telomerase activity in vitro have not been shown, and the reported effects on telomere length have been conflicting. In one report, telomere length was unaltered upon disruption of either one of the two yeast Hsp90 genes HSP82 or HSC82; in addition, no telomere changes were observed in yeast expressing only low Hsp82p levels (∼10% of normal) (14). However, in a systematic evaluation of ∼4,800 haploid gene deletion strains, the hsc82-null mutation was found to have shortened telomeres (2). In an attempt to better understand the role(s) yeast Hsp90 proteins might have with telomerase we investigated the following: (i) telomere lengths in strains expressing various yeast Hsp90 alleles, (ii) Hsp82p-mediated effects on telomerase DNA binding and extension activities in vitro, (iii) Hsp82p telomerase interactions in vitro, and (iv) yeast Hsp90-dependent effects on telomerase telomere occupancy in vivo.
MATERIALS AND METHODS
Yeast strains.
The parent Saccharomyces cerevisiae strain used in the present study was W303 (MATa). The Hsp82-expressing strains were kindly provided by S. Lindquist (Whitehead Institute), the Hsc82-expressing strains were kindly provided by J. L. Johnson (University of Idaho), and wild-type 13 × Myc-EST2 yeast were kindly provided by E. Blackburn (University of California, San Francisco).
Telomere length analysis.
Genomic DNA was prepared from the indicated strains, it was digested with XhoI, resolved, and transferred to Immobilon-Ny+ membrane (Millipore, Inc.). The telomeric DNA was identified after hybridization in Ekono buffer (ISC BioExpress, Inc.) supplemented with a radiolabeled telomeric oligonucleotide [(TGTGGGT)4]. As a loading and migration control we used an established probe that recognizes a 1,621-bp fragment of chromosome IV (11). After appropriate washes using SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-sodium dodecyl sulfate (SDS) buffers, the telomeric and control DNA was visualized by using a PhosphorImager (Molecular Dynamics, Inc.).
Telomerase extract preparation.
The DEAE and MonoQ telomerase extracts were prepared by using established protocols (29, 37). In brief, 6 liters of the indicated yeast were grown to an optical density at 595 nm of 1.0, and cells were harvested by centrifugation and washed once with cold lysis buffer (20 mM Tris [pH 8.0], 1.1 mM MgCl2, 0.1 mM EDTA, 500 mM sodium acetate [pH 5.0], 1.5 mM dithiothreitol [DTT], 10% glycerol, 0.1% Triton X-100, 0.2% NP-40, 1.0 mM phenylmethylsulfonyl fluoride, 1.0 μg of pepstatin ml−1, 1.0 μg of leupeptin ml−1, and 0.25 mM aprotinin); the cell pellet was then frozen in liquid nitrogen. Whole-cell extracts were prepared by mechanical disruption by pulverizing the frozen pellet in a coffee grinder (Mr. Coffee, Inc.) for ∼1 min in the presence of an equivalent volume of dry ice; the resulting cell-dry ice powder was transferred to ice-cold lysis buffer and allowed to thaw at 4°C while nutating. The lysate was clarified by centrifugation at 20,000 × g for 20 min, the supernatant was collected and applied to DEAE FF Sepharose (8 ml; GE, Inc.), and protein was eluted with a sodium acetate (pH 5.0) gradient of 500 to 1,000 mM in TMG buffer (20 mM Tris [pH 8.0], 1.1 mM MgCl2, 0.1 mM EDTA, 1.5 mM DTT, 10% glycerol, 0.1% Triton X-100). For the DEAE extracts, the protein that eluted between 600 and 750 mM sodium acetate was pooled and concentrated to a volume of ∼100 μl by using a microconcentrator device (Ultrafree 4, 10-kDa cutoff; Millipore, Inc.), 2 ml of TMG30 (sodium acetate [pH 7.0]) was added to adjust the pH and salt concentration, the volume was reduced to ∼100 μl, 100 μl of cold 80% glycerol was added, and the mixed samples were stored at −20°C. For the MonoQ extracts, the pooled volume was diluted 1:2 (sample:TMG30), the diluted sample was applied to MonoQ resin (1.7 ml; GE, Inc.), the telomerase fraction was eluted with TMG500, the sample was concentrated to ∼100 μl, 2 ml of TMG30 (pH 7.0) was added to adjust the pH and salt concentration, the volume was reduced to ∼100 μl, 100 μl of cold 80% glycerol was added, and the mixed samples (MonoQ fraction) were stored at −20°C prior to quantification of protein and TLC1 RNA levels.
The levels of TLC1 RNA were determined for the telomerase extracts by using reverse transcription quantitative PCR and Northern blot analysis. Prior to quantification the fractions were subjected to DNase treatment using a TurboDNase kit (Ambion, Inc.) according to the manufacturer's instructions. For reverse transcription quantitative PCR, the TurboDNase-treated samples were used for reverse transcription reactions as recommended by the manufacturer (Invitrogen, Inc.) using an oligonucleotide specific to the substrate region of the TLC1 RNA (TGTGTGGGTGTGGTGATGGTAGG). After reverse transcription, an aliquot (2.0 μl) of each reaction was used for quantitative PCR according to the manufacturer's instructions (Bio-Rad, Inc.) using oligonucleotides specific for TLC1 (GATGGTGAAGAGATAGTGTCGGATTCG and TGTGTGGGTGTGGTGATGGTAGG). The copy number of TLC1 was determined by using a dilution series of known amounts of isolated in vitro-transcribed TLC1 RNA to generate a standard curve. For Northern analysis TurboDNase samples were resolved by denaturing agarose electrophoresis and then transferred to Zeta Probe membrane (Bio-Rad, Inc.). Hybridization was performed in Ekono hybridization buffer (ISC BioExpress, Inc.) with a radiolabeled TLC1 probe. Sequential washes were performed with 40 mM NaHPO4-1.0 mM EDTA-5% SDS and 40 mM NaHPO4-1.0 mM EDTA-1% SDS. The blot was visualized by using a PhosphorImager. The copy number of the TLC1 RNA was determined by blotting a dilution series of known amounts of isolated in vitro transcribed TLC1 RNA to generate a standard curve; in general, the extract preparations contain equivalent TLC1 RNA levels (∼1 ng of TLC1 RNA μl of extract−1).
Telomerase DNA extension assays.
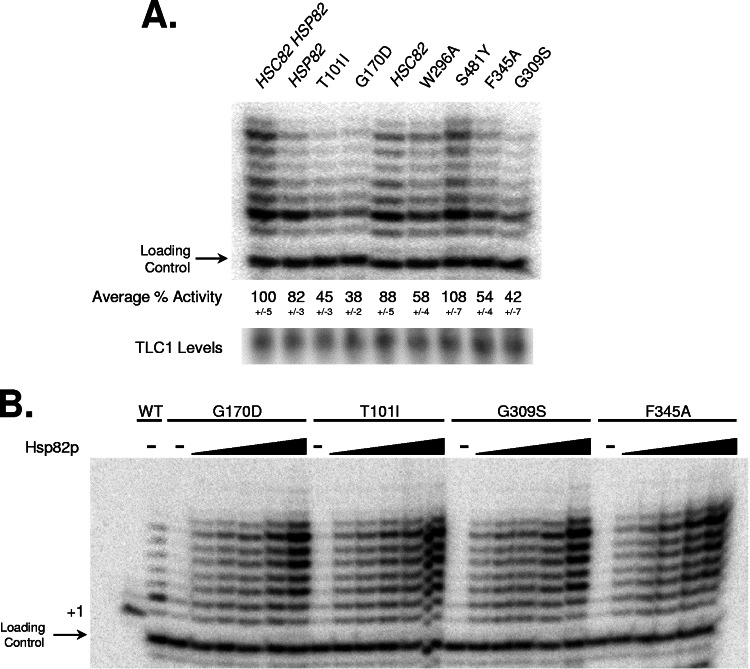
Immobilized DNA extension reactions used DEAE fractions with equivalent amounts of TLC1 RNA in extension buffer (50 mM Tris [pH 8.0], 1.0 mM MgCl2, 1.0 mM spermidine, 1.0 mM DTT, 0.5% glycerol, 50 μM dTTP, 10 μCi of [α-32P]dGTP [3,000 Ci mmol−1; Amersham, Inc.]) supplemented with 2.0 pmol of telomeric seven-base 3′ overhang DNA substrates immobilized on paramagnetic beads (Promega, Inc.). The reactions were incubated for 30 min at 30°C, and the DNA-bound beads were collected by magnetic separation, washed twice with 200 μl of 1× EcoRI buffer (100 mM Tris [pH 7.5], 50 mM NaCl, 10 mM MgCl2, and 0.025% Triton X-100; New England Biolabs, Inc.), and resuspended in 50 μl of 1× EcoRI restriction buffer supplemented with 100 μg of bovine serum albumin ml−1 and 10 U of EcoRI and incubated 1 h at 37°C. The beads were magnetically separated from the cleaved DNA fragments, a PNK end-labeled loading control primer was added, the DNAs were subjected to ethanol precipitation, and the products were reconstituted in formamide-NaOH loading buffer, subjected to denaturing gel electrophoresis (14% acrylamide), and visualized by using a PhosphorImager. For the DNA extension assays shown in Fig. 2A, the percent activities were calculated from six independent experiments by normalizing the intensities of the extension products by the intensities of the loading control in each lane; the band intensities were determined by using the ImageQuant program (Molecular Dynamics, Inc.). The normalized values were averaged, and the percent activities for each extract were determined relative to the extract prepared from the parental strain (HSC82 HSP82).
FIG. 2.
Telomerase DNA extension activity is Hsp82p dependent in vitro. Telomerase-mediated extension of an immobilized seven-base single-stranded 3′ overhang G-rich DNA substrate was examined using conventional DEAE telomerase extracts prepared from yeast expressing the indicated hsc82 or hsp82 alleles. (A) The abilities of the indicated DEAE telomerase extracts to extend a telomeric DNA substrate in the presence of [α-32P]dGTP and dTTP was determined. The extract prepared from the parental strain is HSC82 HSP82, and those from the hsp82 strains are clustered on the left (HSP82, T101I, and G170D), while extracts made from the hsc82 strains are shown on the right (HSC82, W296A, S481Y, F345A, and G309S). The normalized percent DNA extension activities derived from six independent experiments for each extract is shown below each lane. The Northern blot analysis showing the relative TLC1 RNA levels in each extract is shown below the DNA extension data; TLC1 levels were also checked by reverse transcription real-time PCR analysis (data not shown). (B) Purified Hsp82p was sufficient to restore telomerase DNA extension activity in vitro. Recombinant, purified Hsp82p was titrated (0.2, 0.5, 1.0, 2.0, and 4.0 μM) into the indicated telomerase extracts; endogenous cellular Hsp82p levels in yeast have been calculated to be ∼17 μM (26). The extracts were prepared from the parental HSC82 HSP82 (WT), hsp82 allele (G170D or T101I), or hsc82 allele (G309S or F345A) strains. All extension reactions contained equivalent levels of TLC1 RNA and a polynucleotide kinase end-labeled 27-base oligonucleotide was added prior to the precipitation of all of the DNA extension products to serve as a loading control. The resolved extension products were visualized by using a PhosphorImager.
Typically, the extension assay DNA substrate had a seven-base, 3′ overhang sequence of GGGTGTG, except for the dTTP titration experiments in which the DNA substrate terminated with GGTGTGG. Optimal alignment of these DNA substrates using strictly base-pairing preferences would allow only short extensions (e.g., three nucleotides to be added to the GTG substrate); however, a prior study (7a) indicates that the 3′ section of the telomerase template is preferentially used for DNA extensions, which would permit longer extensions on these substrates. For the dTTP titration experiment, the Km for dTTP was determined by calculating the ratio of the band intensities between the +1 and +2 products; the level of each product was determined by using ImageQuant. The Km for nucleotide was determined by plotting the ratio of the +2/+1 intensities to the concentration of dTTP, and a curve was fitted to the data using the equation v = {Vmax ([dTTP])}/(Km + [dTTP]), where v equals +2/+1 and Vmax equals 0.84, as determined at saturating levels of dTTP (50 μM).
Protein purification.
Amino-terminal polyhistidine Hsp82 fusion proteins (WT, G170D, or T101I) were expressed in Rosetta (Novagen, Inc.) bacteria using pET expression constructs, and the proteins were purified as previously described (37). In brief, the Hsp82 protein was isolated on cobalt resin in accordance with the manufacturer's instructions (Clontech, Inc.), followed by further resolution over DEAE FF resin and a Superdex-200 size exclusion column (GE, Inc.).
Fluorescence anisotropy assay.
The anisotropy reactions were performed in TMG30 (20 mM Tris [pH 8.0], 1.1 mM MgCl2, 0.1 mM EDTA, 1.5 mM DTT, 10% glycerol, 0.1% Triton X-100, 30 mM sodium acetate [pH 7.0]) supplemented with a fluorescein-labeled telomere oligonucleotide (12.5 nM; fl-GTGTGGTGTGTGGG). The reactions were incubated 5 min at 25°C before determining the anisotropy values using an Ultra Evolution plate reader (Tecan, Inc.). The apparent dissociation constants (Kd) were determined by fitting a curve as follows: [m1 + m2 + m0 − 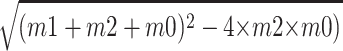 /(2 × m2)]m3, where m0 is the total TLC1 RNA, m1 is the Kd, m2 is the total oligonucleotide, and m3 is the maximal change in anisotropy, which is set for each experiment (37).
/(2 × m2)]m3, where m0 is the total TLC1 RNA, m1 is the Kd, m2 is the total oligonucleotide, and m3 is the maximal change in anisotropy, which is set for each experiment (37).
ChIP assay.
Chromatin immunoprecipitation (ChIP) analysis was performed essentially as described previously (37). To determine the relative telomere occupancy by telomerase 13 × Myc-EST2 yeast were grown to an optical density at 595 nm of 0.5, radicicol (100 μM) or an equivalent volume of dimethyl sulfoxide (DMSO) was added, and the cells were incubated 2 h at 30°C prior to formaldehyde addition. Target DNA levels were determined by quantitative PCR in accordance with the manufacturer's instructions (Bio-Rad, Inc.) using primers specific for a Y′ element found within 11 subtelomeric regions, oligonucleotides that amplify a single subtelomeric region on chromosome XV, or a primer set specific for an internal control, nontelomeric DNA region (YJL052W). The enrichments were determined by normalizing the ratio of the specific to nonspecific telomeric values (e.g., α-Myc ChIP cycle threshold [CT] values/normal mouse immunoglobulin G ChIP CT value) at the telomeric DNA targets to the ratio of the specific to the nonspecific YJL052W signals.
ATPase and in vitro chaperone assays.
The ATPase activity of Hsp82p was measured by using an EnzChek phosphate assay kit (Molecular Probes, Inc.), and the abilities of the Hsp82 derivatives to maintain denatured luciferase in a refoldable state over an extended period was determined as previously described (9, 37). In brief for the ATPase assays, the indicated Hsp82 protein (1 μM per monomer) was incubated at either 30 or 37°C with 1 mM ATP in ATPase buffer (40 mM HEPES [pH 7.5], 150 mM KCl, 5 mM MgCl2, 2 mM DTT). The reactions were incubated for 90 min and then diluted 1:5 into a 100-μl phosphate assay reaction as outlined by the manufacturer's instructions. A standard curve was generated using the inorganic phosphate solution provided by the manufacturer (Molecular Probes, Inc.).
For the in vitro chaperone assays, firefly luciferase was denatured by incubating the protein at 30°C for 30 min in 6 M guanidinium hydrochloride diluted 125-fold (final concentration, 3.4 nM) into refolding buffer (25 mM HEPES [pH 7.5], 50 mM KCl, 5.0 mM MgCl2, 10 mM DTT, 1.0 mM ATP) and supplemented with either 3.2 μM bovine serum albumin (BSA) or the indicated Hsp82 protein. The reactions were incubated 1 h at 37°C, human Hsp70 (1.6 μM) and Hdj1 (3.2 μM) were added, and the reactions were incubated an additional 2 h. Aliquots were removed after 2 h, and the recovered luciferase activity was determined. As a positive folding control, the denatured luciferase was diluted directly into refolding buffer supplemented with Hsp70 and Hdj1. The percent recovery of luciferase activity was determined by using native luciferase diluted to the same extent in refolding buffer supplemented with BSA (3.2 μM).
DNA pull-down assay.
To investigate the lifetime of the wild-type Hsp82p interaction with DNA bound telomerase, W303 DEAE extract (20 ng of TLC1 RNA/sample) was incubated with immobilized seven-base 3′ overhang DNA (20 pmol/sample) for the indicated time points at 30°C, the beads were collected by magnetic separation and washed once with TMG30 buffer, and the bound proteins were eluted by boiling in SDS sample buffer. To determine the abilities of the various Hsp82 proteins to associate with DNA bound telomerase, the indicated DEAE extracts with equivalent levels of TLC1 RNA were incubated for 2 min at room temperature prior to washing and elution. In either case, the associated Hsp82p or Sba1p were detected and visualized by Western blot analysis.
RESULTS
Yeast telomere length is Hsp90 dependent.
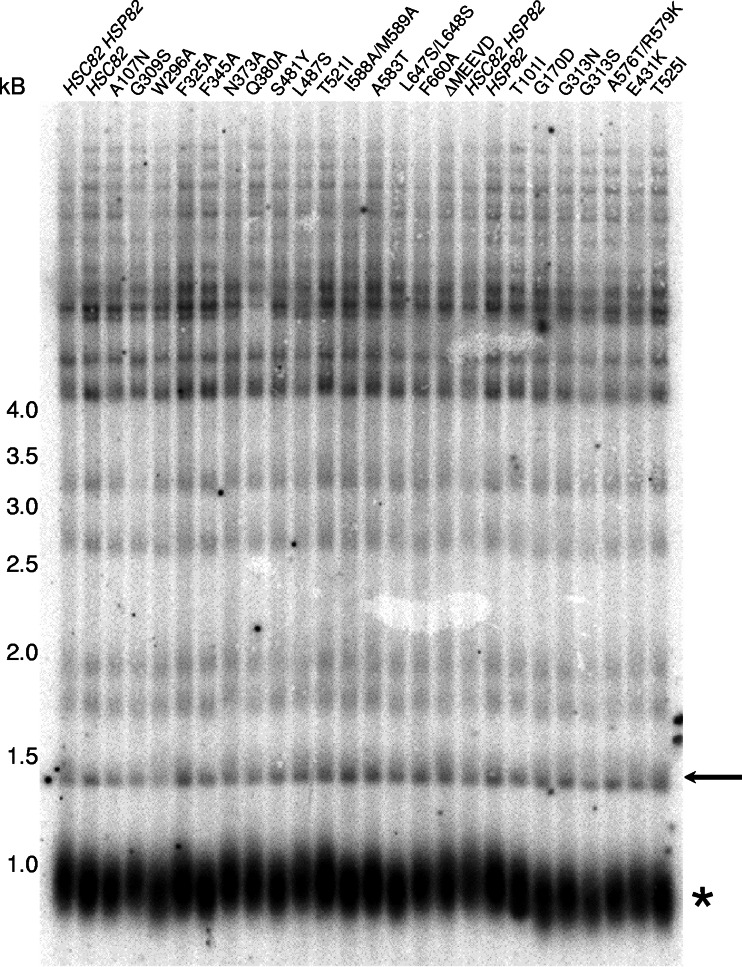
We addressed whether proper telomere maintenance is yeast Hsp90 dependent by Southern blot analysis. While a telomere length defect might be more striking if the cellular yeast Hsp90 (i.e., Hsp82p and Hsc82p) activities could be completely eliminated, the essential nature of this protein family prohibits this test (4). Since a prior study indicates that even low Hsp82p levels are sufficient to support telomere length (14), we used an alternate tactic in which telomere length was assessed in the absence of any wild-type Hsp82 or Hsc82 protein. To achieve this end point, we used established engineered yeast strains in which the endogenous HSP82 and HSC82 genes have been disrupted and viability is maintained by plasmid-born expression of either a wild-type (HSC82 or HSP82) or a mutant allele (Fig. 1) (19, 25). The various alleles were previously identified through either genetic screens or site-directed engineering, and the plasmid-borne expression results in protein accumulation levels that are comparable to the endogenous levels (19, 25). While changes to cochaperone interactions or effects on nonendogenous target proteins (e.g., glucocorticoid receptor or v-Src kinase) have been reported, the impact of these mutations on an endogenous client protein is not well understood. Hence, we attempted to use the hsp82 or hsc82 allele collection to identify mutations displaying telomere length phenotypes.
FIG. 1.
Yeast telomere DNA length is yeast Hsp90 dependent. Genomic DNA was isolated from the parental (HSC82 HSP82) or yeast expressing a single hsp82 or hsc82 allele, as indicated. The hsc82 alleles are loaded on the left side of the Southern blot and are listed from wild-type HSC82 to the ΔMEEVD strain, and the hsp82 yeast are loaded on the right side of the Southern blot and are listed from the wild-type HSP82 to the T525I allele. The subtelomeric DNA was visualized via Southern blot analysis with a radiolabeled telomere probe (TGTGGGT)4. The asterisks indicate the approximate position of the Y′ subtelomeric fragments. As a migration control, the position of a nontelomeric DNA (arrow) was visualized using a probe specific to an internal fragment of chromosome IV. The approximate positions of coresolved DNA molecular weight markers are indicated.
In general, we found that either wild-type Hsp82p or Hsc82p expression was sufficient for telomere DNA length maintenance. We did observe an intermittent mild decrease in telomeric DNA (<50 bp) in the individual gene knockouts (Fig. 1, HSC82 lane; data not shown); the inconsistent, weak phenotype might explain the disparity in the reported telomere effects for single hsc82 or hsp82 gene nulls (2, 14). When both wild-type proteins were absent, we observed a wide telomere DNA phenotype distribution in the 24 different alleles examined. The hsp82 G170D allele displayed the most significant decline in telomere length (∼50%), the other hsp82 alleles showed more mild shortening, along with the hsc82 W296A, F345A and S481Y mutants, whereas the other hsc82 alleles had little to no apparent effect (Fig. 1). Importantly, regardless of the telomere shortening extent the effects were reproducible for a given allele, and the declines in telomeric DNA length were specific since no significant change in mobility was found for a nontelomeric, internal DNA region (Fig. 1, arrow). Hence, the hsp82/hsc82 allele-specific phenotypes indicate that a wild-type yeast Hsp90 protein is required for proper telomere DNA length maintenance.
Telomerase DNA extension activity is yeast Hsp90 dependent.
To address whether the observed telomere shortening might arise from a telomerase activity defect, we prepared standard telomerase DEAE extracts from various allelic backgrounds (29). In general, we found a reduced telomerase DNA extension activity in the mutant backgrounds (Fig. 2A); the relative activity of each extract compared to the parental strain preparation is provided. Since the extracts all contained equivalent telomerase levels (i.e., TLC1 amounts) (Fig. 2A), the changes in the DNA extension activity likely result from the mutations in either Hsp82p or Hsc82p. With the exception of the S481Y and G309S backgrounds, we observed a good correlation between the relative DNA extension levels in vitro and general telomere lengths in vivo (Fig. 1 and 2). The S481Y background had slightly shortened telomeres and yet displayed a DNA extension activity comparable to the wild type, and the G309S yeast had no apparent decline in telomere length and yet displayed reduced extension activity in vitro. The S481Y result might indicate that Hsc82p has a telomere role independent of telomerase, whereas the G309S data suggest that some mutations can be compensated for in vivo. However, given the more general correlation between the in vitro and in vivo defects, we suggest that the primary supporting role for Hsp82p and Hsc82p at the telomere is with telomerase. Importantly, titration of recombinant Hsp82p into the various extracts was sufficient to recover extension activity (Fig. 2B); supplementation of a control protein (e.g., BSA) had no apparent effect (data not shown). In addition to the general increase in DNA extension levels, Hsp82p supplementation promoted longer telomerase products as the pronounced pausing at the +2 position in the unsupplemented reactions was relieved (Fig. 2B). These data provide the first evidence that Hsp82p might modulate nucleotide processivity by telomerase.
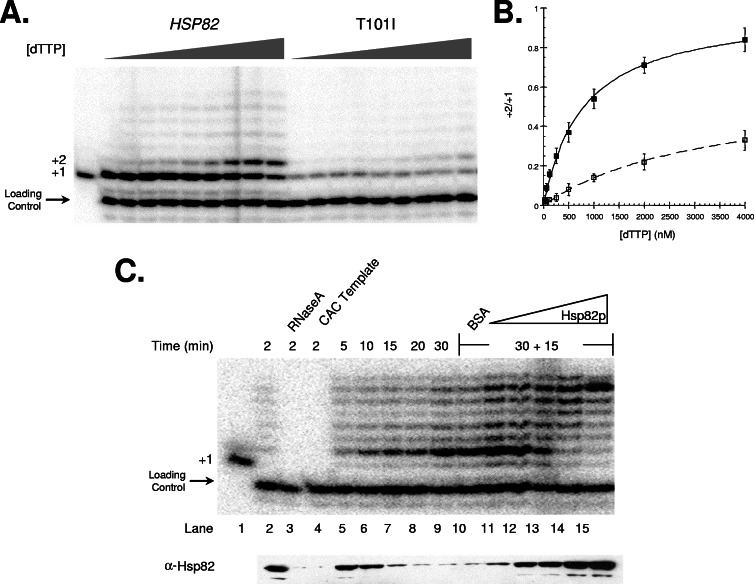
To more directly assess whether Hsp82p has a role in telomerase nucleotide addition, we performed DNA extension assays under limiting concentrations of nucleotide. We reasoned that if Hsp82p promotes telomerase nucleotide addition, then the enzyme might display an increased reliance on nucleotide availability in the absence of a wild-type Hsp90 chaperone. In our assays, we exploited a telomeric DNA substrate terminating with TGG at the 3′ end. Alignment of the TLC1 template and this substrate would permit incorporation of a single G nucleotide and then, depending upon the availability of dTTP the complex, would pause (29). As expected, we observe a single extension product at +1 in the presence of only [α-32P]dGTP using extracts prepared from either wild-type or T101I yeast (Fig. 3A, lanes 2 and 11, respectively). However, as dTTP is titrated into the reactions longer extensions are observed (Fig. 3A). While the intensity of the +1 products varied between the wild-type and T101I extracts, we found that an ∼4-fold-higher level of dTTP was required for telomerase to add a T at the +2 position in the T101I extract relative to WT (Fig. 3B); the Km for nucleotide in the wild-type extract was found to be 797 ± 6 nM, whereas it increased to 3,314 ± 47 nM in the T101I background. Thus, telomerase nucleotide affinity is altered in the absence of wild-type Hsp82p.
FIG. 3.
Hsp82p activates the DNA extension activity of stalled, DNA-bound telomerase. (A) The dTTP concentration needed for telomerase nucleotide addition in the presence or absence of wild-type Hsp82p was determined by using a dTTP titration. Telomerase extracts prepared from either yeast expressing wild-type HSP82 or the T101I allele were incubated with [α-32P]dGTP, seven-base 3′-overhang DNA substrate and various amounts of dTTP (0, 10, 25, 50, 100, 200, 400, 800, or 1,600 nM). (B) The data from three independent dTTP titration experiments were quantified and plotted as indicated. (C) DNA extension and Hsp82p association using wild-type telomerase extract was monitored at the indicated time points. To control for telomerase dependence, RNase A was added to the reaction prior to incubation with the DNA substrate or, alternatively, a substrate terminating in CAC was used. After 30 min at 30°C (to allow DNA extension and telomerase stalling), the reactions were supplemented with BSA (4.0 μM) or Hsp82p (0.1, 0.2, 0.5, 1.0, 2.0, and 4.0 μM) and incubated for an additional 15 min. As controls, the position of +1 is indicated, and an end-labeled 27-base oligonucleotide was added prior to the precipitation of all of the telomerase reactions. The resolved extension products were visualized by using a PhosphorImager, and the relative levels of DNA-bound, telomerase-associated Hsp82p were determined by Western blot analysis.
It is well established that yeast telomerase has poor DNA extension properties compared to human telomerase, since the yeast enzyme does not add multiple DNA repeats, nor does it typically append all of the bases for a single telomeric repeat (29, 34). Based on these reports and our findings, we investigated whether Hsp82p is limiting for nucleotide addition in extracts prepared from wild-type yeast. In addition, we checked whether Hsp82p is associated with DNA-bound telomerase during nucleotide addition. To accomplish these goals, we exploited an immobilized telomeric DNA substrate as a means to rapidly pull down telomerase and its associated cofactors. To control for telomerase specific cofactors, the pull-down was accomplished using G-rich telomeric DNA in the absence or in the presence of RNase A or with a DNA substrate that terminated with CAC; C-terminal primers do not support telomerase binding (21, 37). Importantly, we found that Hsp82p was associated with the DNA substrate in a telomerase-dependent manner (Fig. 3C, lanes 2 to 4).
The duration of the Hsp82p interaction, however, was relatively short and decreased within minutes after assembly of the DNA-bound complex; we suspect telomerase undergoes a conformational change upon DNA binding that decreases the affinity of interaction with Hsp82p. In contrast to Hsp82p, telomerase forms long-lived complexes with the DNA substrate (29, 37). Perhaps notably, the amount of DNA-associated Hsp82p correlated with the telomerase DNA extension activity—as extension product levels neared saturation (i.e., telomerase was no longer actively adding nucleotides), the relative amount of bound Hsp82p declined (Fig. 3C, lanes 1 and 5 to 9).
To address whether Hsp82p dissociates because telomerase can no longer add nucleotides or whether telomerase can no longer add nucleotides because it is not associated with Hsp82p, we supplemented stalled telomerase complexes with purified Hsp82p. Since it is well established that yeast telomerase remains bound after extending a DNA substrate (29, 37), any effects that might be observed likely involve the modulation of DNA-bound telomerase. Significantly, Hsp82p addition both enhances the total amount of extended DNA and results in a predominance of longer products (Fig. 3C, lanes 11 to 15); as a control, we added BSA and observed no effect on DNA extension activity or Hsp82p association (Fig. 3C, lane 10). In addition, the association between Hsp82p and telomerase was reestablished, which suggests that DNA-bound telomerase is able to interact with Hsp82p, albeit at a reduced affinity relative to free telomerase. Taken together, the presented data indicate that Hsp82p interacts with DNA-bound telomerase to facilitate nucleotide processivity.
Telomerase DNA binding is compromised in the G170D background.
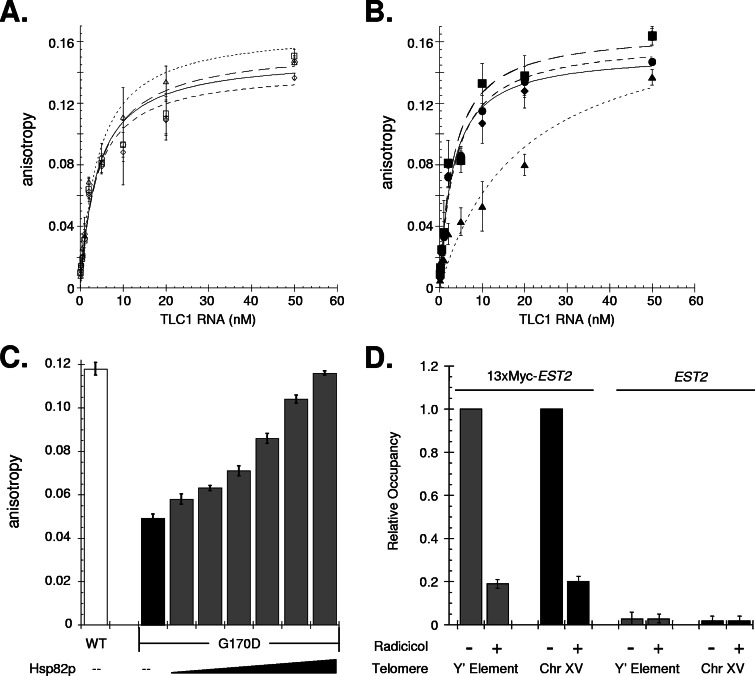
In addition to investigating telomerase nucleotide addition activity, we also examined whether Hsp82p has a role in assembling the telomerase DNA complex. A recent report has shown that human Hsp90 is required for human telomerase to effectively bind a telomeric DNA substrate (20). To monitor telomerase DNA binding, we prepared MonoQ telomerase extracts, which exhibit RNA-dependent telomerase DNA-binding activity (37). The MonoQ extracts were then titrated into a set level of fluorescein-labeled, single-stranded, telomeric primer, and the change in anisotropy was monitored; an increase in the anisotropy value is indicative of DNA binding. In addition to preparing telomerase extracts from yeast maintained at 30°C, we also made MonoQ extracts from yeast incubated at 37°C for 30 min prior to harvesting and extract production. By heat treating the cells prior to collection, we are exploiting the rapid inactivation kinetics of the G170D protein and preparing telomerase extracts lacking functional Hsp82 chaperone protein (25). Using this strategy, we found that only the 37°C G170D extract displayed a decreased DNA-binding activity (Fig. 4B); the telomerase DNA-binding affinity was reduced fivefold in the G170D background for a Kd of ∼1 to ∼5 nM. For all extracts the change in anisotropy was RNA dependent (i.e., telomerase dependent), since the addition of RNase A abolished the binding activity (data not shown). Importantly, telomerase DNA binding in the G170D 37°C extract was recovered upon the addition of purified Hsp82p (Fig. 4C). The positive effect of Hsp82p supplementation was specific to the G170D 37°C extract since no apparent change in anisotropy was observed upon Hsp82p addition into the other extracts (data not shown). Hence, Hsp82p can directly modulate telomerase DNA binding in vitro.
FIG. 4.
Telomerase DNA binding is Hsp82p dependent in vitro. Fluorescence anisotropy was used to detect telomerase binding to a fluorescein-labeled telomeric oligonucleotide. MonoQ telomerase extracts were prepared from the parental yeast (WT; □) or cells only expressing single hsp82 alleles (HSP82, ○; T101I, ⧫; G170D, ▴) grown at 30°C and incubated at 30° (A) or 37°C (B), as indicated, for 30 min prior to cell collection and extract preparation. To measure the telomerase DNA-binding affinity, the MonoQ fractions were titrated into a reaction containing a set level of oligonucleotide (12.5 nM). (C) Recombinant Hsp82p is sufficient to modulate telomerase DNA-binding activity. Purified recombinant Hsp82p was titrated (0.1, 0.2, 0.5, 1.0, 2.0, and 4.0 μM) into anisotropy reactions containing G170D 37°C MonoQ extract. For comparison, the binding activities of unsupplemented telomerase fractions with equivalent TLC1 and protein levels from WT 37°C (□) and G170D 37°C (▪) yeast are shown. (D) Telomere association by telomerase was monitored by using the ChIP assay. The relative levels of telomerase-telomere binding were determined in a 13 × Myc-EST2 strain exposed to either carrier (DMSO) or radicicol (100 μM) for 2 h at 30°C. Telomerase residency at a population of telomeres was determined using PCR and oligonucleotides select for a subtelomeric Y′ element found at 11 chromosomal termini (Y′ elements) ( ) or at a single telomere using primers specific for a subtelomeric region of chromosome XV (Chr XV, ▪); all values were normalized to the signal from an internal nontelomeric DNA. As a control, ChIP trials were carried out with yeast expressing nontagged Est2 (EST2). All data represent average values (mean ± the standard deviation) from three independent assays.
) or at a single telomere using primers specific for a subtelomeric region of chromosome XV (Chr XV, ▪); all values were normalized to the signal from an internal nontelomeric DNA. As a control, ChIP trials were carried out with yeast expressing nontagged Est2 (EST2). All data represent average values (mean ± the standard deviation) from three independent assays.
To address whether yeast Hsp90 can effect telomerase association with telomeres in vivo, we utilized the ChIP assay. To determine the influence of Hsp82p and Hsc82p, we exploited the Hsp90 protein inhibitor radicicol, which permits a rapid means to inactivate Hsp82p and Hsc82p in vivo. We performed ChIP trials using yeast expressing Myc-tagged Est2p treated either with carrier (DMSO) or radicicol for 2 h at 30°C. We found that in the presence of radicicol telomerase telomere binding decreased ∼80% at either a single telomere (Chr XV) or at a telomere population (Y′ elements) (Fig. 4D). Importantly, the ChIP signal was dependent upon the presence of epitope-tagged Est2p, since no significant signal was apparent using wild-type yeast expressing an untagged Est2 protein. Although it is possible that the radicicol effect involves other cellular proteins since type II DNA topoisomerases are inhibited by radicicol (12), we found a comparable decline in Myc-Est2p telomere occupancy when the Hsp90 inhibitor geldanamycin was used, which does not affect type II DNA topoisomerases (12). Hence, yeast Hsp90 activity is likely required for proper telomerase telomere association in vivo.
The Hsp82p mutants have altered chaperone and ATPase activities.
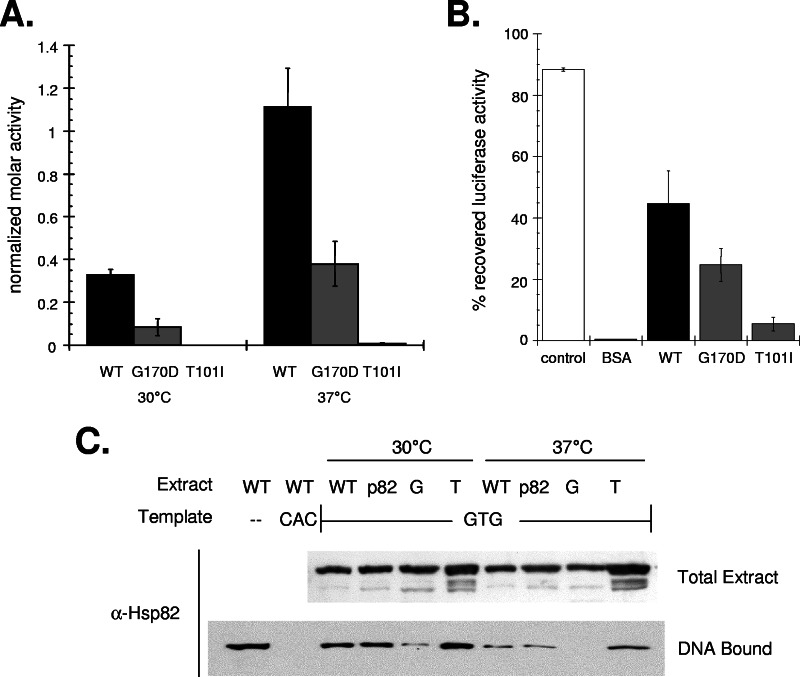
In an attempt to understand why telomerase activities decline in the mutant hsp82 backgrounds, we purified and characterized recombinant WT, G170D, and T101I Hsp82 proteins. We examined the two known intrinsic functions for Hsp90 family proteins: ATP hydrolysis and molecular chaperone activity. A prior report on the ATPase rate for these proteins had shown that T101I has a substantially lower rate relative to WT or G170D (30). Likewise, we found that T101I displayed the lowest ATPase rate not only at 30° but also at 37°C (Fig. 5A). Thus, reduced Hsp82p ATP hydrolysis might contribute to the decreased nucleotide processivity in the mutant hsp82 backgrounds. However, it likely does not account for the reduced telomerase DNA-binding activity in G170D extract since the rate does not correlate with a change in DNA-binding activity (i.e., the WT rate is higher and T101I is lower, and yet only G170D displays a DNA-binding defect).
FIG. 5.
Hsp82p dissociation correlates with a decreased telomerase DNA-binding activity. The ATPase rate (A) and in vitro chaperone activities (B) of WT, T101I, and G170D Hsp82 proteins were determined by using standard assays. The data represent average values (mean ± the standard deviation) from three independent assays. (C) The relative associated levels of the Hsp82p derivatives in extracts prepared from the parental HSC82 HSP82 yeast (WT), strains expressing HSP82 (p82), or the mutant alleles G170D (G) or T101I (T) with DNA-bound telomerase were determined by using immobilized telomeric DNA and Western blot analysis with an antibody that specifically recognizes Hsp82p (DNA bound). Each reaction contained comparable TLC1 RNA amounts (data not shown) and Hsp82 protein levels prior to DNA binding (Total Extract).
We also examined the in vitro molecular chaperone activities of purified WT, G170D, and T101I Hsp82p. In short, we tested the abilities of the various derivatives to maintain a chemically denatured protein (firefly luciferase) in a folding competent state for an extended period (30 min) at 37°C; protein refolding (i.e., the recovery of enzymatic activity) was initiated upon Hsp70 and Hdj1 addition (9). Utilizing this assay we found that 45% of the total luciferase activity could be recovered in the presence of WT Hsp82p (Fig. 5B). While the G170D protein displayed a reduced capacity (∼50% of WT), the ability of T101I to maintain denatured luciferase in a refoldable state was more impaired (∼22% or WT) (Fig. 5B). In parallel to the ATPase data, the reduced chaperone activity might contribute to the decline in telomerase DNA extension, but no apparent connection exists between these two intrinsic properties of Hsp82p and the reduced telomerase DNA-binding activity in the G170D background.
Since no correlation existed between the decreased telomerase DNA-binding activity in the G170D extract and inherent Hsp82p functions, we evaluated the direct association between telomerase and the Hsp82p proteins both on and off the DNA. An initial examination of the Hsp82 protein levels in the various DEAE extracts did not reveal any substantial differences, suggesting that all of the derivatives coeluted with telomerase during the high-salt purification step; the telomerase fraction elutes from the DEAE anion-exchange resin with ∼650 mM salt (Fig. 5C). Next, we tested whether the Hsp82 derivatives remain telomerase associated upon DNA binding. Interestingly, we found that the various Hsp82 proteins remained telomerase associated even after DNA binding except in the case of the 37°C G170D extract, which showed no apparent interaction (Fig. 5C). Notably, these reactions were carried out under conditions in which telomerase DNA binding is saturated. Thus, the decline in telomerase DNA-binding activity in the G170D extract correlates with a reduced telomerase-Hsp82p interaction and not with diminished in vitro chaperone or ATPase activities. We suspect that the diminished interaction between telomerase and the G170D protein results from either a declined association half-life or a change in chaperone substrate specificity that occurs upon telomerase binding to the DNA. Regardless, the data demonstrate that an Hsp82p interaction is needed to support telomerase DNA-binding activity.
DISCUSSION
This study has important implications for our understanding of both Hsp90 and telomerase. In brief, yeast Hsp90 supports two telomerase functions (DNA binding and extension) that are required for proper telomere maintenance. Initial studies linking Hsp90 and telomerase advocated that the chaperone helped assemble the reverse transcriptase protein with the RNA template to form the core telomerase enzyme (15). A recent report extended human Hsp90's function by suggesting a postassembly role to foster telomerase DNA primer loading (20). Our work provides direct evidence that Hsp82p facilitates telomerase DNA binding. Together, the two studies indicate that this is a conserved Hsp90 task. Notably, the presented data uniquely demonstrate that Hsp82p operates with an enzyme after DNA binding. Perhaps the continued Hsp82p association is required to maintain an open telomerase conformation that favors acceptance of free nucleotide and fosters the DNA extension activity. The ability of Hsp82p to serve a DNA-bound enzyme might be a general function, since Hsp90 proteins have been found associated with additional DNA enzymes including other reverse transcriptases and the DNA helicase XPB, which is required for transcription initiation and DNA repair (6, 16, 17).
The short half-life observed for the Hsp82p-telomerase interaction in the DNA-bound structure (Fig. 3B) might help explain the high stalling rate for yeast telomerase DNA extension activity, which could account for the degenerate nature of yeast telomeric repeats (7, 40). In contrast to the yeast system, human Hsp90 displays a continued telomerase association through multiple DNA extension events, which might contribute to the ability of human telomerase to add multiple telomeric DNA repeats (8). Nevertheless, the data presented here indicate that yeast Hsp90 serves telomerase at a number of important steps, including DNA binding and extension, and demonstrates the multifaceted effects this molecular chaperone can have on a single client protein.
The multiple telomerase functions affected by Hsp82p parallels studies on the established Hsp90 client proteins steroid hormone receptors. Initially, Hsp90 was found as a component of the 9S untransformed steroid receptors for glucocorticoids or progesterones in which Hsp90 was required to maintain the high-affinity hormone binding state and prevent aggregation of the untransformed receptors (3, 28, 32). In addition, Hsp90 was suggested to prevent DNA binding by the receptor through retention in the cytoplasm (28). However, recent studies refute this last hypothesis and indicate that Hsp90 is found in the nucleus and facilitates rather than inhibits glucocorticoid receptor DNA binding (35). In general, Hsp90 appears to support the DNA-binding activities of heterologous transcription factors including p53, hypoxia-inducible factor 1, aryl hydrocarbon receptor, and myogenic determination protein (1, 18, 24, 33, 38, 43). Hence, Hsp90 maintains DNA-binding proteins in a state able to respond to activation signals (e.g., phosphorylation or hormone binding) and furthers the various pathways by promoting the DNA-bound complex.
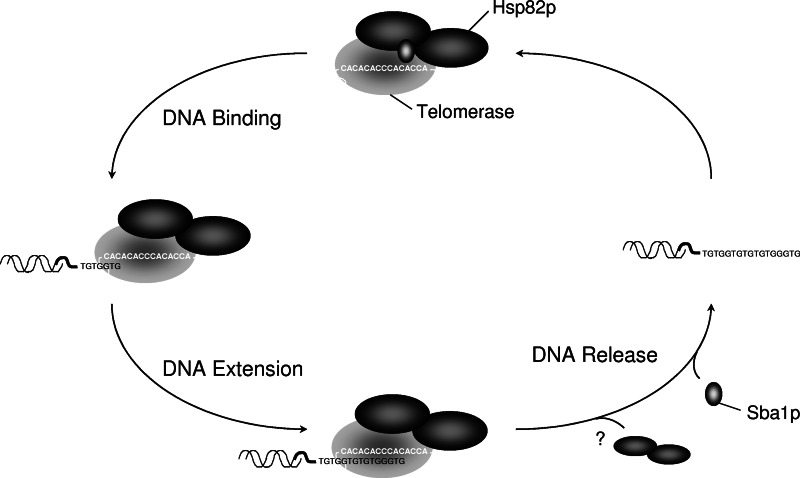
The presented data in conjunction with a prior study on the yeast p23 molecular chaperone Sba1p (37) suggest that these two chaperones jointly modulate telomerase DNA binding (Fig. 6). In our model, Hsp82p and Sba1p cooperate to promote a rapid telomerase DNA-binding cycle that would be highly beneficial for the addition of multiple telomeric DNA repeats during a single cell cycle: Hsp82p would promote assembly, while Sba1p fosters disassembly. The coordinated chaperone actions on telomerase also parallel an intracellular receptor paradigm (10, 35). Given the divergent nature of telomerase and intracellular hormone receptors, the common functional effects mediated by Hsp90 and p23 chaperones on these clients, and recent proteomic studies, we suggest that these two molecular chaperones serve to maintain a wide variety of proteins in a dynamic state with DNA (31). It will be interesting to discover which other DNA-associated proteins are clients for p23 and Hsp90 and the effect these chaperones have on the various DNA-related activities.
FIG. 6.
The Hsp82p and Sba1p molecular chaperones modulate telomerase DNA-associated activities. Hsp82p and Sba1p interact with telomerase prior to telomeric DNA binding. Hsp82p remains telomerase bound during DNA association to facilitate the assembly of the telomerase DNA complex and extension of the bound DNA substrate. The potential release of Hsp82p results in a stalled, nonproductive DNA-bound telomerase complex. Telomerase DNA dissociation is triggered by Sba1p to permit additional binding and extension events.
Acknowledgments
We dedicate this work to the cherished memory of Oyetunji A. Toogun, who was an excellent scientist, friend, and human being.
We are grateful to Peter L. Jones (UIUC) for useful comments on the manuscript and Zhi Li for technical support with the TLC1 Northern analysis. We thank Susan Lindquist (Whitehead Institute, Boston, MA) for the Hsp82p antibody, hsp82 alleles, and associated yeast strains; Jill Johnson (University of Idaho, Moscow) for the hsc82 alleles and associated yeast strains; Elizabeth Blackburn (University of California, San Francisco) for the 13 × Myc-EST2 yeast strain; and Johannes Buchner (Technische Universitat, Munich, Germany) for the pET28-HSP82 plasmid.
This study was supported by the Public Health Service grant DK074270.
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Antonsson, C., M. L. Whitelaw, J. McGuire, J. A. Gustafsson, and L. Poellinger. 1995. Distinct roles of the molecular chaperone hsp90 in modulating dioxin receptor function via the basic helix-loop-helix and PAS domains. Mol. Cell. Biol. 15:756-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askree, S. H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk, C. Coker, A. Krauskopf, M. Kupiec, and M. J. McEachern. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101:8658-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailly, A., J. F. Savouret, N. Sallas, and E. Milgrom. 1978. Factors modifying the equilibrium between activated and non-activated forms of steroid-receptor complexes. Eur. J. Biochem. 88:623-632. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech, T. R. 2004. Beginning to understand the end of the chromosome. Cell 116:273-279. [DOI] [PubMed] [Google Scholar]
- 6.Flom, G., J. Weekes, and J. L. Johnson. 2005. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr. Genet. 47:368-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forstemann, K., M. Hoss, and J. Lingner. 2000., Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 28:2690-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Forstemann, K., and J. Lingner. 2005. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMB. J. 6:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsythe, H. L., J. L. Jarvis, J. W. Turner, L. W. Elmore, and S. E. Holt. 2001. Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J. Biol. Chem. 276:15571-15574. [DOI] [PubMed] [Google Scholar]
- 9.Freeman, B. C., M. P. Myers, R. Schumacher, and R. I. Morimoto. 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding, and interaction with HDJ-1. EMBO J. 14:2281-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman, B. C., and K. R. Yamamoto. 2001. Continuous recycling: a mechanism for modulatory signal transduction. Trends Biochem. Sci. 26:285-290. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, K. L., and T. R. Cech. 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadelle, D., C. Bocs, M. Graille, and P. Forterre. 2005. Inhibition of archaeal growth and DNA topoisomerase VI activities by the Hsp90 inhibitor radicicol. Nucleic Acids Res. 33:2310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 14.Grandin, N., and M. Charbonneau. 2001. Hsp90 levels affect telomere length in yeast. Mol. Genet. Genomics 265:126-134. [DOI] [PubMed] [Google Scholar]
- 15.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. B. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., D. Flores, D. Toft, X. Wang, and D. Nguyen. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 78:13122-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hur, E., H. H. Kim, S. M. Choi, J. H. Kim, S. Yim, H. J. Kwon, Y. Choi, D. K. Kim, M. O. Lee, and H. Park. 2002. Reduction of hypoxia-induced transcription through the repression of hypoxia-inducible factor-1alpha/aryl hydrocarbon receptor nuclear translocator DNA binding by the 90-kDa heat-shock protein inhibitor radicicol. Mol. Pharmacol. 62:975-982. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. L., A. Halas, and G. Flom. 2007. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol. Cell. Biol. 27:768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keppler, B. R., A. T. Grady, and M. B. Jarstfer. 2006. The biochemical role of the heat shock protein 90 chaperone complex in establishing human telomerase activity. J. Biol. Chem. 281:19840-19848. [DOI] [PubMed] [Google Scholar]
- 21.Lue, N. F., and Y. Peng. 1998. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 26:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 23.Marcand, S., V. Brevet, and E. Gilson. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller, L., A. Schaupp, D. Walerych, H. Wegele, and J. Buchner. 2004. Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J. Biol. Chem. 279:48846-48854. [DOI] [PubMed] [Google Scholar]
- 25.Nathan, D. F., and S. Lindquist. 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard, D. 2006. Intracellular dynamics of the Hsp90 co-chaperone p23 is dictated by Hsp90. Exp. Cell Res. 312:198-204. [DOI] [PubMed] [Google Scholar]
- 27.Powers, M. V., and P. Workman. 2006. Targeting of multiple signalling pathways by heat shock protein 90 molecular chaperone inhibitors. Endocrinol. Relat. Cancer 13:S125-135. [DOI] [PubMed] [Google Scholar]
- 28.Pratt, W. B. 1993. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J. Biol. Chem. 268:21455-21458. [PubMed] [Google Scholar]
- 29.Prescott, J., and E. H. Blackburn. 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11:2790-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prodromou, C., B. Panaretou, S. Chohan, G. Siligardi, R. O'Brien, J. E. Ladbury, S. M. Roe, P. W. Piper, and L. H. Pearl. 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19:4383-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter, K., L. M. Hendershot, and B. C. Freeman. 2007. The cellular world according to Hsp90. Nat. Struct. Mol. Biol. 14:90-94. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez, E. R., S. Meshinchi, M. J. Schlesinger, and W. B. Pratt. 1987. Demonstration that the 90-kilodalton heat shock protein is bound to the glucocorticoid receptor in its 9S nondeoxynucleic acid binding form. Mol. Endocrinol. 1:908-912. [DOI] [PubMed] [Google Scholar]
- 33.Shaknovich, R., G. Shue, and D. S. Kohtz. 1992. Conformational activation of a basic helix-loop-helix protein (MyoD1) by the C-terminal region of murine HSP90 (HSP84). Mol. Cell. Biol. 12:5059-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smogorzewska, A., and de Lange, T. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 35.Stavreva, D. A., W. G. Muller, G. L. Hager, C. L. Smith, and J. G. McNally. 2004. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 24:2682-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixeira, M. T., M. Arneric, P. Sperisen, and J. Lingner. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117:323-335. [DOI] [PubMed] [Google Scholar]
- 37.Toogun, O. A., W. Zeiger, and B. C. Freeman. 2007. The p23 molecular chaperone promotes functional telomerase complexes through DNA dissociation. Proc. Natl. Acad. Sci. USA 104:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walerych, D., G. Kudla, M. Gutkowska, B. Wawrzynow, L. Muller, F. W. King, A. Helwak, J. Boros, A. Zylicz, and M. Zylicz. 2004. Hsp90 chaperones wild-type p53 tumor suppressor protein. J. Biol. Chem. 279:48836-48845. [DOI] [PubMed] [Google Scholar]
- 39.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239:197-201. [DOI] [PubMed] [Google Scholar]
- 40.Wang, S. S., and V. A. Zakian. 1990. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol. Cell. Biol. 10:4415-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wegele, H., L. Muller, and J. Buchner. 2004. Hsp70 and Hsp90-a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151:1-44. [DOI] [PubMed] [Google Scholar]
- 42.Whitesell, L., and S. L. Lindquist. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5:761-772. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelmsson, A., S. Cuthill, M. Denis, A. C. Wikstrom, J. A. Gustafsson, and L. Poellinger. 1990. The specific DNA binding activity of the dioxin receptor is modulated by the 90 kd heat shock protein. EMBO J. 9:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]