Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) activity is regulated through association with ligands that include the thiazolidinedione class of antidiabetic drugs, as well as derivatives of polyunsaturated fatty acids. Induction of PPARγ target gene expression involves ligand-dependent reconfiguration of the ligand-binding domain (LBD), followed by recruitment of specific transcriptional coactivators. In this study, we have identified an amino acid (F372) within helix 7 of the LBD that is required for the response of PPARγ to endogenous ligands. Additionally, the data show that this amino acid is also required for expression of a novel subset of adipocyte genes (group 2), including fibroblast growth factor 21 (FGF21), and that the FGF21 gene is a direct target of PPARγ. Expression of the group 2 genes is selectively repressed by the NAD-dependent deacetylase SIRT1 in mature 3T3-L1 adipocytes, since knockdown of SIRT1 through the constitutive expression of a corresponding RNA interference enhances their expression without affecting the expression of classic adipogenic genes, such as adiponectin and FABP4/aP2. It appears that many of the group 2 genes repressed by SIRT1 in mature adipocytes correspond to the same set of genes that are selectively activated by treatment of fat cells with the PPARγ ligand, troglitazone. These data support a role for helix 7 of the LBD of PPARγ in regulating adipocyte function and suggest that inhibition of SIRT1 in adipocytes induces the same insulin-sensitizing action as PPARγ ligands.
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor expressed in many tissues but is most abundantly produced in adipose tissue, where it acts as the master regulator of adipogenesis, as well as a regulator of the multiple functions of mature adipocytes (6-8, 21, 32, 37). The transcriptional activity of PPARγ is regulated in part by association with lipophilic ligands that include derivatives of polyunsaturated fatty acids, such as eicosinoids, as well as the thiazolidinedione (TZD) class of synthetic insulin sensitizers (19, 20). PPARγ consists primarily of three regulatory domains comprised of a ligand-independent transactivation domain at the N terminus, a central DNA-binding domain, and a C-terminal ligand-binding domain that facilitates ligand-dependent transactivation and heterodimerization with the retinoic acid X receptor (RXR) (18). Heterodimers of PPARγ and RXR bind to DNA consensus sites within the promoters/enhancers of target genes, which consist of direct repeats of the nuclear receptor half-site spaced by a single base pair (DR-1). Activation of transcription at these target genes involves a complex process in which the docked PPARγ/RXR heterodimers, following association with ligands, recruit a series of coactivators, including the p160/SRC family members, that initiate formation of the RNA polymerase II/transcriptional complex involving components of the Mediator complex (11, 24, 29).
Our understanding of the mechanisms by which PPARγ activates transcription has been derived from studies employing synthetic ligands, such as TZDs. It is generally accepted that in the unliganded state, PPARγ associates with the corepressor NCoR or SMRT to repress target gene expression. Entry of the TZD into the large ligand-binding pocket stabilizes helix 12 of transactivation domain 2 (AF2), which dislodges the corepressors and forms a binding site for the p160 family of coactivators that facilitates the pharmacological activation of PPARγ target gene expression (24, 27). Recent studies investigating the role of PPARγ in regulating inflammatory genes in macrophages presented an additional model by which TZDs might organize the recruitment of various nuclear coregulators. In this model, TZDs induce the SUMOylation of PPARγ on K365 within helix 7 of the ligand-binding domain, which targets PPARγ to NCoR/HDAC3 complexes on inflammatory gene promoters (26). These observations suggest that helix 7, in addition to helix 12, might participate in mechanisms by which ligands regulate the association of PPARγ with specific coactivators or corepressors. In support of this, our recent studies have identified helix 7 as a component of the functional interaction between β-catenin, the coactivator of the canonical Wnt signaling pathway, and PPARγ (22). Since the endogenous ligand for PPARγ has not been identified, the physiological mechanisms by which PPARγ regulates target gene expression in various cell types are not known.
The differentiation of preadipocytes into adipocytes depends on stimulation of PPARγ activity that is facilitated by a C/EBPβ-associated induction of PPARγ2 gene expression, as well as production of endogenous ligands (8). As mentioned above, the physiological ligand for PPARγ has not been identified, but recent studies suggest that signaling pathways involving cyclic AMP, C/EBPβ, and xanthine oxidoreductase activate a transient increase in ligand production during the initial 2 to 4 days of adipogenesis in 3T3-L1 preadipocytes (4, 14, 23, 35). The levels/activities of these ligands subside dramatically during terminal differentiation to the extent that mature adipocytes express low levels of activity. Despite this apparent decrease in endogenous ligand activity, PPARγ is capable of maintaining expression of most of its target genes in mature adipocytes. In this regard, it is interesting that some target genes are expressed at low levels in adipocytes but are responsive to activation of PPARγ by TZDs. Specifically, genes coding for glycerol kinase (GyK) and the oxidized LDL receptor (OLR-1) are PPARγ target genes that are normally expressed at low abundance in white adipose tissue and mature adipocytes in culture. Exposure of 3T3-L1 adipocytes to TZDs induces transcription of mRNAs for GyK and OLR-1 (5, 13). Additional studies by Guan and coworkers (12) have shown that PPARγ is bound to PPAR response elements (PPREs) in the promoter of the transcriptionally inactive GyK gene in mature adipocytes, in addition to being bound to the enhancer of the transcriptionally active aP2 gene. The data suggest that endogenous ligands are unable to dislodge corepressors from PPARγ on the GyK gene but do facilitate this process, along with recruitment of p160 coactivators to PPARγ, on the aP2 gene. Moreover, it appears that exposure of mature adipocytes to TZDs can activate GyK expression by regulating the switch in corepressor/coactivator recruitment to the PPARγ bound to the corresponding promoter.
In the present study, we investigated mechanisms by which PPARγ might induce expression of selected genes in response to different effectors. The data show that PPARγ regulates at least two programs of gene expression during adipogenesis in 3T3-L1 preadipocytes. One program (group 1) consists of classic adipogenic genes, including those for FABP4/aP2, adiponectin, and perilipin, and following its induction, this program continues to be expressed throughout terminal adipogenesis and in mature adipocytes. The other program (group 2) consists of a diverse array of genes, some of which appear to be involved in glucose homeostasis and insulin action, including those for fibroblast growth factor 21 (FGF21) and the oxidoreductase Ero1-Lα. Expression of these group 2 genes can be selectively activated in mature adipocytes by synthetic PPARγ ligands or suppression of SIRT1 activity. Our studies also show that helix 7 within the ligand-binding domain plays a critical role in the response of PPARγ to endogenous ligands. Mutation of select amino acids within helix 7, specifically F372, renders PPARγ completely incapable of activating adipogenic gene expression in response to endogenous ligand activity. Exposure of cells expressing the mutant F-PPARγ to TZDs induces expression of the adipogenic program containing adiponectin and aP2 but is incapable of inducing the program containing FGF21 and Ero1-Lα.
MATERIALS AND METHODS
Materials.
Dexamethasone (DEX), 3-isobutyl-1-methylxanthine (MIX), and insulin were purchased from Sigma (St. Louis, MO). Leupeptin, aprotinin, and puromycin were purchased from American Bioanalytical (Natick, MA), while Dulbecco's modified Eagle's medium (DMEM) was purchased from Mediatech, Inc. (Herndon, VA), and calf serum and Trizol were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was obtained from Gemini Bio-Products, and troglitazone was obtained from Biomol International.
Antibodies.
Monoclonal anti-PPARγ antibody and polyclonal C/EBPα were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and polyclonal aP2 serum was kindly provided by D. Bernlohr (University of Minnesota), while antiperilipin antibody was kindly provided by A. Greenberg (Tufts University, Boston, MA) and polyclonal anti-ACRP30 (adiponectin) was obtained from Affinity BioReagents (Golden, CO). Anti-Ero1 polyclonal antibody was purchased from Abnova Co. (Taiwan, China).
Plasmids and cell lines.
Replacement of phenylalanine 372 of PPARγ1 with an alanine was achieved by performing site-directed mutagenesis of the pBabe-WT-PPARγ plasmid using a QuikChange II XL kit (Stratagene) following the manufacturer's instructions. The pSUPER-SIRT1 small interfering RNA (siRNA) plasmid was generously provided by Jim Xiao of Boston University School of Medicine and consisted of the vector recently described (28). Generation of appropriate retrovirus particles was performed as follows. HEK-293T cells were grown to 70% confluence in 100-mm-diameter dishes, at which stage they were transfected with a DNA-Fugene cocktail consisting of 36 μl Fugene 6 transfection reagent, 6 μg retrovirus plasmid, 6 μg pVPack-VSV-G vector, 6 μg pVPack-GAG-POL vector, and 164 μl DMEM without FBS. Twenty-four hours later, the medium was replaced with 6 ml fresh DMEM containing 10% FBS. One day after that, the culture medium, containing high-titer retrovirus, was harvested and filtered through a 0.45-μm-pore-size filter. The viral filtrate was used to infect both 3T3-L1 preadipocytes (control and SIRT1 siRNA cells) and Swiss 3T3 fibroblasts (wild-type [WT], E-, EF-, F-, and DD-PPARγ cell lines).
Cell culture.
The Swiss fibroblast cell lines expressing WT and mutant forms of PPARγ (E, EF, F, and DD) generated as previously described (22) and murine 3T3-L1 preadipocytes were grown in DMEM containing 10% FBS (fibroblasts) or 10% calf serum (preadipocytes) until they were confluent and were then maintained in the same medium for an additional 2 days. Differentiation was induced at 2 days postconfluence (day zero) by adding fresh DMEM containing 10% FBS, 0.5 mM MIX, 1 μM DEX, and 1.67 μM insulin with or without 5 μM troglitazone. The immortilized primary brown preadipocytes (a gift of C. R. Kahn, Joslin Diabetes Center, Boston, MA) (9, 10) were grown to confluence in differentiation medium composed of DMEM containing 10% FBS supplemented with 20 nM insulin and 1 nM 3,3′,5-triiodo-l-thyronine). Two days postconfluence, the cells were induced to differentiate by exposure to DEX, MIX, insulin, 0.125 mM indomethacin, and 10% FBS. The cells were refed every 2 days.
Microarray gene chips.
Swiss WT PPARγ and EF-PPARγ cells were differentiated in the presence or absence of troglitazone for 5 days as described above. Additionally, control and SIRT1 knockdown 3T3-L1 preadipocytes were differentiated as described above for 0, 4, and 10 days (see Tables S1A and S1B in the supplemental material). Total RNA was isolated from all cells using Trizol reagent (Invitrogen), and microarray analysis was performed by the Microarray Resource (Boston University School of Medicine). Briefly, double-stranded cDNA was synthesized from 10 μg of RNA using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) and purified using a Phase-Lock gel (PLG Heavy; Brinkmann Instruments, Westbury, NY). Biotin-labeled cRNA was then generated using an RNA transcript-labeling kit (Enzo Diagnostics, Farmingdale, NY) and purified using RNeasy affinity columns (Qiagen). After treatment at 94°C for 35 min in 40 mM Tris-acetate, pH 8.1, 100 mM potassium acetate, and 30 mM magnesium acetate, 15 μg of fragmented cRNA was hybridized to the Affymetrix Gene Chip mouse expression set MOE430A2.0 array at 45°C for 16 h at 60 rpm using controls supplied by the manufacturer (Affymetrix). The arrays were then washed and stained according to the standard protocol for antibody amplification for eukaryotic targets (Affymetrix). The stained Gene Chip arrays were scanned at 488 nm using an Affymetrix Gene Chip Scanner 3000 (Affymetrix). The scanned images were then quantified and scaled using Microarray Suite 5.0 software (Affymetrix).
Oil Red O staining.
The cells were seeded in 35-mm plates, and at the specified stage of differentiation, they were rinsed with phosphate-buffered saline (PBS) and fixed with 10% formalin in PBS for 15 min. After two washes in PBS, the cells were stained for at least 1 h in freshly diluted Oil Red O solution (six parts Oil Red O stock solution and four parts H2O; Oil Red O stock solution is 0.5% Oil Red O in isopropyl alcohol). The stain was then removed, and the cells were washed twice with water and then photographed.
Western blot analysis of proteins.
Equal amounts of protein extracted from the total cell layer were fractionated on 8% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Perkin-Elmer Life Sciences). Following transfer, the membranes were blocked with 10% nonfat dry milk in phosphate-buffered saline-0.1% Tween 20 and probed with the antibodies corresponding to the various target proteins indicated in each figure. Horseradish peroxidase-conjugated secondary antibodies (Sigma) and an enhanced-chemiluminescence substrate kit (Perkin-Elmer Life Sciences) were used for detection of specific proteins.
Analysis of RNA.
Total cellular RNA was prepared using TRIzol reagent (InVitrogen) according to the manufacturer's instructions. cDNAs were made from equivalent amounts of total RNA by using a Reverse Transcription System (Promega) as described previously (33). Primer sequences used for amplification were synthesized (Integrated DNA Technologies Inc., Coralville, IA) as follows: PPARγ forward primer, GAGCATGGTGCCTTCGCTGAT, and PPARγ reverse primer, CAACCATTGGGTCAGCTCTTG; C/EBPα forward primer, AAGGTGCTGGAGTTGACCAGT, and C/EBPα reverse primer, TAGAGATCCAGCGACCCGAAA; PGC-1α forward primer, GTCAACAGCAAAAGCCACAA, and PGC-1α reverse primer, TCTGGGGTCAGAGGAAGAGA; pex11a forward primer, CCGACTTTTCAGAGCCACTC, and pex11a reverse primer, CGGTTAGGTTGGCTAATGT; ERO1 forward primer, GAAGGATCCATGGGCCGCGCCTGGG, and ERO1 reverse primer, CGCCGTCGACGGCACATTCCAACCG; OLR1 forward primer, GTCATCCTCTGCCTGGTGTT, and OLR1 reverse primer, TTCTTCCGATGCAATCCAAT; ELOVL3 forward primer, TCGTCTGCAAAATCGAAATG, and ELOVL3 reverse primer, GGGAAACCATACAGGGAGGT; FGF21 forward primer, CTGGGGGTCTACCAAGCATA, and FGF21 reverse primer, AAGGCTCTACCATGCTCAGG; mGST1 forward primer, ATGAGGTGTTGATGGCCTTT, and mGST1 reverse primer, GGTTTCCCATAGGTGTGTGC; Nr1h3 (LXRα) forward primer, CCTGATTCTGCAACGGAGTT, and Nr1h3 (LXRα) reverse primer, GGCTCACCAGCTTCATTAGC; Ephx2 forward primer, CCTGGCACTGCCTAGAGACT, and Ephx2 reverse primer, GCTTTATGCACAGCGATGAA; Scd3 forwad primer, TGCTGCAAGAAGAGATGACG, and Scd3 reverse primer, CTCTTGTGACTCCCGTCTCC; adipoq forward primer, GTTGCAAGCTCTCCTGTTCC, and adipoq reverse primer, CAGACTTGGTCTCCCACCTC; GyK forward primer, CCGAAGGAAATTCTGCAGTC, and GyK reverse primer, CGCCTAGTGCAGTTGTTTCA; aP2 forward primer, AATGTGTGATGCCTTTGTGG, and aP2 reverse primer, AATTTCCATCCAGGCCTCTT; Ndrg1forward primer, CGAGAGCTACATGACGTGGA, and Ndrg1reverse primer, CTGGCAGAAGGCATGTATCC; Egln3 forward primer, GAGATGCCTCTGGGACACAT, and Egln3 reverse primer, TTCTGCCCTTTCTTCAGCAT; Egln1 forward primer, AGCCATGGTTGCTTGTTACC, and Egln1 reverse primer, TGTCCACGAGTCCACAGAAG; Cbr1 forward primer, GACCGGTGCTAACAAAGGAA, and Cbr1 reverse primer, GCTCCTTCTTCTGGGCTTTT; Mrap forward primer, CTTCGTGGTGCTCCTCTTTC, and Mrap reverse primer, GACCTGGTCCTAGGGGAGAG; Trib3 forward primer, GATGCCAAGTGTCCAGTCCT, and Trib3 reverse primer, TCTCCCTTCGGTCAGACTGT; Ndg2 forward primer, CGGGTCTTATGGCTCAGATG, and Ndg2 reverse primer, GAAGCCCTCACACAGGGTTA; Cox7a1 forward primer, GCTCTGGTCCGGTCTTTTAG, and Cox7a1 reverse primer, CCAGCCCAAGCAGTATAAGC; Adipsin forward primer, TGATGTGCAGAGTGTAGTGCCTCA, and Adipsin reverse primer, ACGTAACCACACCTTCGACTGCAT; Hsd11b forward primer, CAGAAATGCTCCAGGGAAAG, and Hsd11b reverse primer, GATCTTCCTTCCTGGGTTCC; UCP-1 forward primer, TCTTCTCAGCCGGAGTTTCAGCTT, and UCP-1 reverse primer, TGTTGACAAGCTTTCTGTGGTGGC; and glyceraldehyde-3-phosphate dehydrogenase forward primer, ATCACCATCTTCCAGGAGCGA, and glyceraldehyde-3-phosphate dehydrogenase reverse primer, GTTGTCATGGATGACCTTGGCC.
Plasmid constructs and luciferase reporter gene assays.
The mouse FGF21 promoter constructs −1537/+54, −1299/+54, and −553/+54 were generated by PCR using C57BL/6NCrl mouse genomic DNA and the following oligonucleotides: −1537 forward, 5′-AAGCCTCACCTTGACACC-3′; −1299 forward, 5′-CAGGAAACAACCCAGCTC-3′; −553 forward, 5′-AGTGCAGACAAGTCCCCT-3′; and +54 reverse, 5′-GGCAGCTGGAATTGTGTT-3′. The PCR-amplified fragments were cloned into KpnI and BglII sites of the luciferase reporter plasmid pGL3. For transfection assays, the Swiss fibroblasts (control expressing a pBabe-puro empty vector) or cells expressing a WT PPARγ were seeded in 24-well plates in triplicate for 24 h, at which time 500 ng of the FGF21 promoter plasmids or pBabe-PPARγ plasmid plus 20 ng of Renilla luciferase plasmid were transfected into each well using Fugene 6 (DNA/Fugene 6 ratio, 1:6). Twenty-four hours later, when appropriate, the cells were treated for 48 h with 1 μM GW1929 and were then washed twice with phosphate-buffered saline and lysed with 100 μl of passive lysis buffer. Luciferase/Renilla assays were performed using the Dual-Luciferase Reporter Assay System kit (Promega, Madison, WI) and a Luminoskan Ascent luminometer (Thermo Labsystems, Franklin, MA). The average ratio (from three wells) of luciferase activity (relative light units) to Renilla activity was calculated. The same experiment was repeated at least three times. The final value with standard deviation was calculated based on all repeats.
RESULTS
Our previous investigations demonstrated that four amino acids, E367, F372, D378, and D379, within helix 7 of the ligand-binding domain of PPARγ facilitate a functional interaction between PPARγ and β-catenin (22). To gain insight into the potential involvement of helix 7 in regulating the transcriptional activity of PPARγ during adipogenesis, we expressed a series of mutant PPARγ proteins in Swiss 3T3 fibroblasts in which E367, F372, D378, or D379 was modified to alanine and assessed their abilities to induce adipogenic gene expression. First, we observed that ectopic expression of the WT PPARγ was capable of inducing the conversion of these fibroblasts into adipocytes simply by exposure to DEX, MIX, and insulin without the need for an exogenous ligand, such as troglitazone (Fig. 1A). These data suggest that Swiss fibroblasts produce endogenous ligands that can activate the ectopic PPARγ following exposure to the normal cocktail of adipogenic inducers. In fact, exposure to troglitazone appeared to have no additional effect on the morphological features of these Swiss adipocytes (Fig. 1A). Additionally, the Western blot in Fig. 1B, lanes 1 and 5, shows abundant expression of the adipogenic proteins, C/EBPα, perilipin, and aP2, and a low level of β-catenin production in the Swiss WT PPARγ cells induced to differentiate in the presence or absence of troglitazone. The data also show that the relative abundance of transcriptionally active WT PPARγ was very low due to its rapid turnover. The mutant PPARγ corresponding to E367A (E-PPARγ) retained the ability to induce adipogenesis in the presence or absence of troglitazone (Fig. 1A), which included degradation of β-catenin (Fig. 1B). It is interesting, however, that this alteration appeared to stabilize PPARγ in the absence of ligand, while exposure to troglitazone resulted in a significant decrease in its abundance (Fig. 1B, compare lanes 2 and 6). The most interesting data came from analyzing the expression of the mutant PPARγ corresponding to E367A and F372A (EF-PPARγ). Figure 1 demonstrates that mutation of F372 to alanine, in addition to E367A, completely destroyed the ability of PPARγ to respond to an endogenous ligand, since Swiss cells expressing EF-PPARγ remained as fibroblasts (Fig. 1A) and did not express adipogenic genes or down-regulate β-catenin (Fig. 1B). Additionally, this mutant PPARγ appeared to be quite stable. More importantly, exposure of the Swiss EF-PPARγ cells to troglitazone induced their conversion into adipocytic cells (Fig. 1A) and expression of C/EBPα, perilipin, and aP2 (Fig. 1B). These data are consistent with the notion that F372 and E367 within helix 7 participate in the response of PPARγ to endogenous ligands, whereas responses to exogenous synthetic ligands, such as troglitazone, are less dependent on these amino acids. Mutation of both D378 and D379 to alanine completely destroyed the ability of PPARγ to respond to both endogenous and exogenous ligands, since the corresponding mutant PPARγ was incapable of inducing either morphological conversion or adipogenic gene expression (Fig. 1A and B, lanes 4 and 8).
FIG. 1.
EF-PPARγ does not respond to endogenous ligands but is activated by troglitazone. Swiss cells expressing various forms of PPARγ (WT, E, EF, and DD) were cultured until they were confluent; after 2 days, they were exposed to DEX, MIX, and insulin, with or without 5 μM troglitazone (TROG). (A) Day 5 cells were fixed, stained with Oil Red O, and photographed. (B) Total cellular proteins were collected at day 5 and subjected to Western blot analysis with the indicated antibodies. (C) Profile of ∼1,700 mRNAs expressed in Swiss fibroblasts in response to WT or EF-PPARγ. Total RNAs of WT-PPARγ and EF-PPARγ cells at day 5 (with or without troglitazone) were isolated using Trizol Reagent (Invitrogen), and microarray analysis was performed as described in Materials and Methods. The colors correspond to signal intensities.
The data presented in Fig. 1B suggested to us that analysis of mRNA expression in Swiss WT PPARγ versus Swiss EF-PPARγ cells might permit the identification of PPARγ target gene programs responding to endogenous versus exogenous ligands. Consequently, total RNA was harvested from Swiss fibroblasts expressing either WT or EF-PPARγ proteins 5 days following exposure to the adipogenic inducers in the presence or absence of troglitazone and was subjected to oligonucleotide microarray analysis employing Affymetrix chips. The data revealed that the abundances of 1,767 genes of the ∼22,690 represented on the array differed at least twofold between the highly differentiated Swiss WT PPARγ cells and the undifferentiated Swiss EF-PPARγ cells (minus troglitazone). A cluster analysis of these genes is shown in Fig. 1C, in which the genes that are highly expressed in WT PPARγ cells (minus troglitazone) relative to their expression in EF-PPARγ cells (minus troglitazone) are arranged in descending order of their relative abundances. Genes that are highly expressed in adipocytes (WT PPARγ) compared to fibroblasts (EF-PPARγ minus troglitazone) cluster together and include those coding for adipogenic, lipogenic, and mitochondrial proteins. In contrast, many genes are expressed at much lower abundance in the adipocytes (WT PPARγ cells) compared to the fibroblasts (EF-PPARγ minus troglitazone) and include components of the Wnt signaling pathway, as well as inflammatory proteins, several of which have previously been reported to be down-regulated during adipogenesis. Figure S1 in the supplemental material represents the relative abundances of selected genes present in each of these clusters and reveals that many of the genes displayed significantly more than a fivefold difference in abundance between the two cell types. In fact, some mRNAs, such as adiponectin and Fsp27, were expressed at least 104-fold more abundantly in the adipocytes (WT PPARγ) than in the fibroblasts (EF-PPARγ minus troglitazone). Figure 1C also illustrates that treatment of the WT PPARγ cells with troglitazone did not significantly alter the overall pattern of gene expression but appeared to enhance the level of adipogenic gene expression while suppressing even further the fibroblastic mRNAs (Fig. 1C, compare lane 2 with lane 1). More importantly, the EF-PPARγ cells that were completely unresponsive to endogenous ligands (minus troglitazone) were extensively induced to express multiple adipogenic, lipogenic, and mitochondrial genes following their exposure to troglitazone (Fig. 1C, compare lane 3 with lane 4). These EF-PPARγ cells also down-regulated expression of the fibroblastic genes in response to troglitazone, consistent with their attaining an adipocyte-like morphology (Fig. 1A).
Identification of a subset of PPARγ-responsive genes.
To gain more insight into the gene programs regulated by PPARγ in response to endogenous versus exogenous ligands, a more detailed analysis of individual genes was performed, as shown in Tables S1A and S1B in the supplemental material. We also analyzed the profiles of mRNAs expressed during the differentiation of 3T3-L1 preadipocytes for comparison with the mRNAs expressed in Swiss PPARγ cells by performing additional Affymetrix array analysis of mRNAs isolated from the preadipocytes at 0, 4, and 10 days of differentiation. Table S1A in the supplemental material lists a selection of classic adipogenic genes that were induced to various extents during adipogenesis in 3T3-L1 preadipocytes (columns 5, 6, and 7), which include genes coding for proteins involved in lipid storage/metabolism (i.e., FABP4), as well as endocrine functions (i.e., adiponectin). All of these mRNAs were expressed much more abundantly in Swiss fibroblasts expressing WT PPARγ than in cells expressing EF-PPARγ. In fact, the difference in the levels of expression of these mRNAs in EF-PPARγ cells versus WT PPARγ cells was comparable to the difference in their expression levels in preadipocytes versus mature adipocytes (see Table S1A in the supplemental material; compare columns 1 and 3 with columns 5 and 7). It is also relevant to point out that the expression of at least three genes, Resistin (Retn), Hsd11β1, and Orosomucoid (Orm1) genes, was enhanced by WT PPARγ in Swiss cells in response to endogenous ligands (minus troglitazone) and during adipogenesis in 3T3-L1 preadipocytes. Interestingly, troglitazone significantly attenuated the expression of these genes in WT PPARγ (see Table S1A in the supplemental material, Retn, Hsd11b1, and Orm1; compare column 2 with column 1), consistent with reports that TZDs selectively repress the expression of these genes following their dramatic induction during adipogenesis in 3T3-L1 cells (2, 3, 34). Taken together, the data in Table S1A in the supplemental material are consistent with the notion that PPARγ can induce expression of the majority of the classic adipogenic genes in Swiss fibroblasts in response to an endogenous ligand to the same extent as that occurring during normal adipogenesis in 3T3-L1 preadipocytes. Furthermore, mutation of critical amino acids within helix 7 (EF-PPARγ) prevents PPARγ from responding to endogenous ligand activity (Fig. 1C; see Table S1A, column 3, in the supplemental material). As shown in Fig. 1C, however, exposure of EF-PPARγ to troglitazone can induce expression of most of these classic adipogenic genes to levels attained in 3T3 adipocytes (see Table S1A in the supplemental material; compare column 4 with column 7).
Following a more extensive analysis of the array data, it was observed that not all genes that are highly expressed in 3T3-L1 or WT PPARγ adipocytes were induced in the EF-PPARγ cells by troglitazone. In fact, it appeared that activation of the mutant EF-PPARγ by the exogenous ligand, while capable of converting these fibroblasts into adipocytic cells that contained small lipid droplets and expressed many of the markers of mature adipocytes (Fig. 1), was incapable of inducing the entire adipogenic program (see Table S1B in the supplemental material). More specifically, the data shown in Table S1B in the supplemental material suggest that WT PPARγ induces the expression of a group of responsive genes (columns 1 and 2) that are significantly less responsive to stimulation of EF-PPARγ by troglitazone (column 4). This subset of PPARγ target genes (referred to here as group 2) includes proteins that have not previously been shown to be associated with PPARγ activity, such as the endoplasmic reticulum oxidoreductase Ero1-Lα, FGF21 and genes coding for components of the glycolytic pathway. Table S1B in the supplemental material also shows that some of these genes, including those for Mrap, KLF15, Klb (βKlotho), and Pdxp, are induced severalfold during adipogenesis but are unresponsive to troglitazone activation of EF-PPARγ. Other genes are moderately responsive to adipogenic signals in 3T3-L1 preadipocytes (i.e., the glycolytic genes), but almost all of these genes are induced in response to troglitazone activation of WT PPARγ, but not of EF-PPARγ.
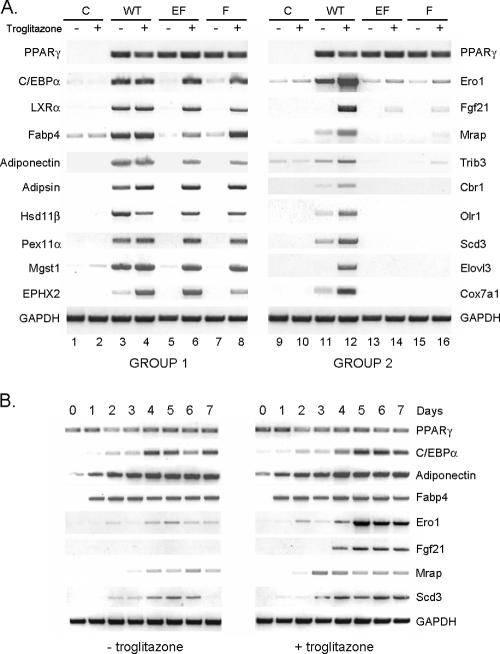
To confirm the oligonucleotide microarray data, a series of reverse transcription (RT)-PCR analyses were performed in which the relative abundances of select mRNAs expressed in the Swiss cell lines were measured. Since the data presented in Fig. 1 suggest that F372 is the amino acid that appears to influence the transcriptional activity of PPARγ, we generated an additional cell line corresponding to Swiss fibroblasts expressing PPARγ in which only F372 was changed to alanine (F-PPARγ cells). We also analyzed Swiss fibroblasts that do not contain an ectopic PPARγ and therefore are completely incapable of adipogenesis even in the presence of troglitazone (control cells). Figure 2A shows the constitutive expression of the corresponding PPARγ mRNAs in each of the Swiss PPARγ cell lines and the absence of any PPARγ mRNA in the control cells. The panel on the left demonstrates expression of select target genes (from Table S1A in the supplemental material) that are induced in the WT PPARγ cells exposed to endogenous (Fig. 2A, lane 3), as well as exogenous (Fig. 2A, lane 4), PPARγ ligands. The expression of most of these genes is unaffected by troglitazone, with the exception of EPHX2, which appears to be enhanced even further by the exogenous ligand. This set of classic adipogenic genes, as expected, is not expressed in the EF- or F-PPARγ cells in response to the endogenous ligands (Fig. 2A, lanes 5 and 7). Furthermore, activation of these mutant cell lines (EF- and F-PPARγ) by exposure to troglitazone significantly induces expression of all of these genes (Fig. 2A, lanes 6 and 8), as shown in Table S1A in the supplemental material. In contrast, the subset of genes (group 2) presented in the panel on the right (selected from Table S1B in the supplemental material) responds quite differently to the action of the mutant PPARγ molecules. These genes are induced to various extents by endogenous ligands in cells expressing WT PPARγ and are enhanced manyfold by exposure to troglitazone (Fig. 2A, compare lanes 11 and 12 with 9 and 10). More importantly, this subset of genes is unresponsive to activation of EF- or F-PPARγ by troglitazone, as well as the endogenous ligands (Fig. 2A, compare lanes 13, 14, 15, and 16 with lanes 11 and 12). We also performed Western blot analysis of C/EBPα, FABP4/aP2, adiponectin (group 1, EF-PPARγ-responsive genes), and Ero1-Lα (a group 2, non-EF-PPARγ-responsive gene) to confirm the RT-PCR data. Figure S2 in the supplemental material shows that troglitazone stimulation of all forms of PPARγ, including WT PPARγ, E-PPARγ, EF-PPARγ, and F-PPARγ, leads to abundant expression of C/EBPα, FABP4/aP2, and adiponectin. In contrast, expression of Ero1-Lα is completely unresponsive to troglitazone stimulation of F-PPARγ or EF-PPARγ but responds to WT and E-PPARγ activities. It is interesting that analysis of the proteins in the culture media showed that adiponectin is secreted from cells expressing WT and E-PPARγ but is absent from the media of F-PPARγ and EF-PPARγ cells. These data suggest that some group 2 proteins likely participate in processes responsible for secretion of adiponectin. In fact, recent studies by others and by us have demonstrated a role for Ero1-Lα in regulating the secretion of adiponectin from adipocytes (30, 36).
FIG. 2.
(A) Identification of two groups of PPARγ-responsive genes: group 1 (including adiponectin) is responsive, whereas group 2 (including Ero1 and FGF21 genes) is completely unresponsive, to troglitazone activation of EF-PPARγ or F-PPARγ. Swiss fibroblasts (lane C) and Swiss-PPARγ (lanes WT, EF, and F) cells were cultured until they were confluent; after 2 days, they were exposed to DEX, MIX, and insulin, with or without 5 μM troglitazone, for 5 days. Total RNAs of the cells were isolated using Trizol reagent (Invitrogen) and subjected to RT-PCR analysis as described in Materials and Methods. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Troglitazone selectively enhances expression of the group 2 PPARγ-responsive genes during the differentiation of Swiss fibroblasts expressing WT PPARγ. Swiss WT PPARγ cells were cultured in 10% FBS until they reached confluence. Two days postconfluence, the cells were induced to differentiate by exposure to DEX, MIX, insulin, and 10% FBS with or without troglitazone. At days 0, 1, 2, 3, 4, 5, 6, and 7 of differentiation, cells were harvested for RT-PCR analysis as described in Materials and Methods.
The group 2 subset of genes can be selectively activated in response to troglitazone during the differentiation of Swiss 3T3 fibroblasts into adipocytes.
The data in Fig. 2A and Table S1B in the supplemental material show that many of the group 2 genes are constitutively expressed at a low level during normal adipogenesis in response to endogenous ligands but appear to be responsive to potent exogenous ligands. To gain greater insight into the mechanisms regulating these two programs of gene expression, Swiss WT PPARγ cells were induced to differentiate in the absence or presence of troglitazone, and the expression of selected genes was analyzed each day using RT-PCR technology. Figure 2B demonstrates the constant and abundant expression of the WT PPARγ throughout 7 days of differentiation, which resulted in a robust and sustained induction of the group 1 genes, such as those for adiponectin and C/EBPα, in response to endogenous (minus troglitazone), as well as exogenous (plus troglitazone), ligands. Interestingly, the group 2 genes, including those for Ero1-Lα, Scd3, and FGF21, are transiently expressed at a very low level during the initial 2 to 4 days of adipogenesis and are then down-regulated as differentiation proceeds in the absence of troglitazone. Differentiation of these WT PPARγ cells in the presence of troglitazone has a minimal effect on expression of adiponectin and C/EBPα mRNAs but enhances, as well as maintains, expression of Ero1-Lα, Scd3, and FGF21 throughout the 7-day culture period.
Selected group 2 genes are transiently expressed during the differentiation of brown and white preadipocytes.
The data presented in Fig. 2A and B were derived from nonadipogenic fibroblasts forced to differentiate into adipocytes by overexpression of PPARγ. We considered it important, therefore, to determine whether this interesting pattern of PPARγ target gene expression occurs in preadipocytes undergoing differentiation into brown, as well as white, adipocytes in response to activation of endogenous adipogenic transcription factors. To this end, we analyzed the expression of genes during the differentiation of 3T3-L1 white preadipocytes and immortalized primary brown preadipocytes. Figure 3 shows the expected induction of PPARγ, C/EBPα, LXRα, and adiponectin mRNAs 2 days after exposure of the preadipocytes to DEX, MIX, insulin, and 10% FBS. Furthermore, expression of these adipogenic genes remained at a high level throughout the differentiation of both brown and white preadipocytes. To confirm that the immortalized primary brown preadipocytes underwent differentiation into brown adipocytes, we also analyzed the expression of PGC-1α and UCP-1, and the data show that the expression of these mRNAs was initiated at 2 days and was maintained throughout brown adipogenesis (Fig. 3B). In contrast, the group 2 genes that responded poorly to expression of F-PPARγ in the Swiss cells were induced in response to activation of endogenous PPARγ in both the brown and white preadipocytes;, however, the level of expression of the corresponding mRNAs dropped significantly during terminal adipogenesis, as observed in the Swiss WT PPARγ cells differentiated in the absence of troglitazone (Fig. 2B).
FIG. 3.
Selected group 2 PPARγ-responsive genes are transiently induced during the initial phase of adipogenesis in white 3T3-L1 preadipocytes (A) and immortalized primary brown preadipocytes (B). (A) 3T3-L1 white preadipocytes were cultured in 10% calf serum until they reached confluence. Two days postconfluence, the cells were induced to differentiate by exposure to DEX, MIX, insulin, and 10% FBS. At the indicated days of differentiation, cells were harvested for RT-PCR analysis as described in Materials and Methods. (B) Immortalized brown preadipocytes were grown to confluence in differentiation medium composed of DMEM containing 10% FBS supplemented with 20 nM insulin and 1 nM 3,3′,5-triiodo-l-thyronine. At 2 days postconfluence, the cells were induced to differentiate by exposure to DEX, MIX, insulin, 0.125 mM indomethacin, and 10% FBS. On the indicated days of differentiation, cells were harvested for RT-PCR analysis as described in Materials and Methods.
Differential responses of group 1 and group 2 genes to PPARγ agonists and antagonists.
The fact that mutations within helix 7 rendered PPARγ unresponsive to endogenous ligands and responsive to troglitazone, at least for the group 1 genes, encouraged us to determine the effects of other ligands that had previously been shown to possess a range of activities. Consequently, WT PPARγ and EF-PPARγ cells were induced to differentiate in the presence or absence of 15δ-PGJ2, 9-fluorenylmethoxy carbonyl (FMOC)-leu, troglitazone, rosiglitazone, or GW1929 for 5 days, and the expression of selected genes was analyzed by RT-PCR. Figure 4A shows that all of the exogenous ligands had little or no additional effect on the expression of selected group 1 genes in WT PPARγ cells, since their levels of expression were already at a maximum, due presumably to the stimulation of the ectopic PPARγ by endogenous ligands (compare lanes 2 to 6 with lane 1). In contrast, expression of the group 2 genes was enhanced to various extents by exposure of the WT PPARγ cells to the exogenous ligands. In the case of the EF-PPARγ cells, exposure to the different ligands resulted in a significantly more varied response than that observed in the WT PPARγ cells. Specifically, FMOC-leu was incapable of stimulating the expression of any of the selected group 1 or group 2 genes, and 15δ-PGJ2 activated only FABP4 expression. The TZDs, troglitazone and rosiglitazone, induced the expression of selected group 1 genes, including those for C/EBPα, adiponectin, and FABP4, but had a negligible effect on the expression of the group 2 genes, such as those for Ero1-Lα and Mrap. Interestingly, GW1929, an extremely potent synthetic PPARγ ligand in which N-tyrosine moieties have been substituted for the TZD head group, is capable of inducing expression of the group 2 genes, as well as group 1 genes, in the EF-PPARγ cells. These data clearly show differential responses of the two groups of genes to different ligands; we asked, therefore, whether the genes also showed similar differential responses to a PPARγ antagonist. To address this question, WT PPARγ cells were induced to differentiate in the presence or absence of T0070907 or GW9662 (two PPARγ antagonists) with or without troglitazone, and the corresponding cellular RNAs were analyzed by RT-PCR. Figure 4B demonstrates that T0070907 and GW9662 moderately attenuated the ability of WT PPARγ to induce expression of C/EBPα and adiponectin in response to endogenous ligands. The presence of troglitazone overcame the inhibitory effect of the antagonists (Fig. 4B, compare lanes 5 and 6 with lanes 3 and 4). In contrast, the antagonists almost completely blocked expression of the group 2 genes, including those for Ero1-Lα, Mrap, Elovl3, Egln1, SCD3, and OLR-1, in response to stimulation of WT PPARγ by endogenous ligands, with T0070907 being the most potent. Again, this effect was overcome somewhat by troglitazone. Taken together, the data in Fig. 4A and B show that activation of the group 2 genes by PPARγ requires more potent ligands and is significantly more sensitive to antagonists than that of the group 1 genes. We also considered it important to determine whether mutation of F372 had simply dampened the ligand-binding affinity of PPARγ and, consequently, had significantly shifted the dose response to troglitazone to higher concentrations. To investigate this possibility, WT PPARγ and EF-PPARγ cells were induced to differentiate for 5 days in the presence of increasing concentrations of troglitazone. At this stage, cells were harvested for analysis of selected genes by RT-PCR. Figure 4C shows abundant expression of selected group 1 genes, including those for C/EBPα, adiponectin, and FABP4/aP2, in WT PPARγ cells due to endogenous ligands (lane 1), with no significant change in expression in response to increasing doses of troglitazone (lanes 2 to 8). As expected, the group 2 genes, encoding FGF21 and OLR-1, are not expressed in WT PPARγ without an exogenous ligand (lane 1) but can be induced by troglitazone in a dose-dependent manner (lanes 2 to 8). Analysis of gene expression in the EF-PPARγ cells showed that the group 1 genes, for C/EBPα, adiponectin, and FABP4/aP2, are not expressed in the absence of exogenous ligand (Fig. 4C, lane 9) but, as expected, are induced in response to doses of troglitazone (250 to 500 nM) previously shown to be specific for PPARγ (Fig. 4C, lanes 10 to 16). Of importance is the observation that expression of FGF21 and OLR1 cannot be activated in the EF-PPARγ cells by doses of troglitazone (10 μM) that far exceed the dose that is specific for PPARγ. These data demonstrate clearly that mutation of F372 within helix 7 prevented PPARγ from responding to endogenous ligands and, additionally, prevented PPARγ from inducing expression of the group 2 genes in response to the TZD troglitazone.
FIG. 4.
The differential responses of WT PPARγ versus EF-PPARγ to selected PPARγ ligands and antagonists. (A) Swiss-PPARγ (WT and EF) cells were cultured until they were confluent; after 2 days, they were exposed to DEX, MIX, and insulin, with or without the following PPARγ ligands: FMOC-leu (15 μM), 15δ-PGD2 (7 μM), troglitazone (5 μM), rosiglitazone (10 μM), and GW1929 (10 μM). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) WT PPARγ cells were differentiated as in panel A by exposure to DEX, MIX, and insulin in the presence or absence of troglitazone with or without either T0070907 (10 μM) or GW9662 (10 μM) (PPARγ antagonists). (C) WT PPARγ and EF-PPARγ cells were induced to differentiate with DEX, MIX, insulin, and the indicated doses of troglitazone (Trog). In panels A, B, and C, total RNA of the cells was isolated at day 5 using Trizol reagent (Invitrogen) and subjected to RT-PCR analysis of the indicated group 1 and group 2 genes as described in Materials and Methods.
PPARγ directly regulates expression of FGF21.
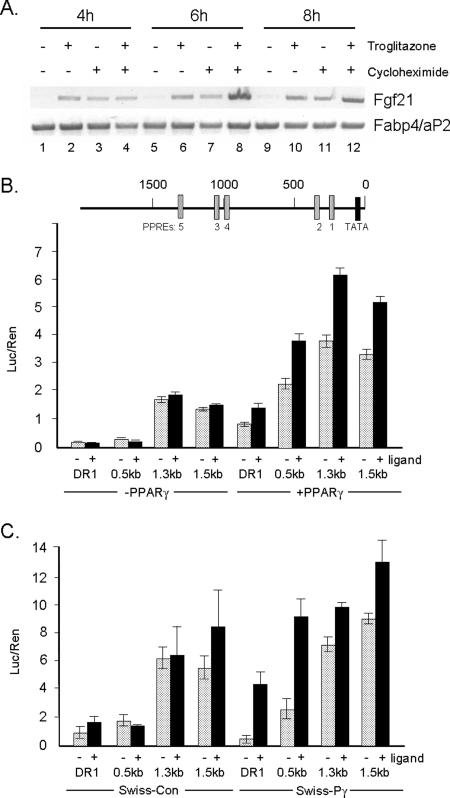
It is conceivable that the inability of the mutant PPARγ (EF or F) to induce expression FGF21 and OLR1 by troglitazone, as well as exogenous ligands, is due to the fact that the corresponding genes might not be direct targets of PPARγ. Other studies, however, have shown direct induction of the OLR1 gene promoter by PPARγ (5). We considered it important and of significant interest to determine whether the FGF21 gene is also a direct target of PPARγ. To this end, we performed two sets of experiments. First, we determined whether the induction of FGF21 gene expression occurred in the absence of ongoing protein synthesis. To do this, WT PPARγ cells were induced to differentiate for 5 days without a synthetic ligand, at which stage troglitazone (5 μM) or cycloheximide (5 μg/ml) was added alone or together for 4, 6, or 8 h, and at each time, RNA was analyzed by RT-PCR. Figure 5A shows significant expression of FABP4/aP2a mRNA at all three times due to its activation by endogenous ligand activity during the 5 days of differentiation of the WT PPARγ cells. In contrast, there are virtually undetectable levels of FGF21 mRNA expression in the absence of an exogenous ligand (Fig. 5A, lanes 1, 5, and 9). Interestingly, exposure to troglitazone rapidly induces FGF21 mRNA expression during the 8-h exposure time (Fig. 5A, lanes 2, 6, and 10), and this event occurs in the presence of cycloheximide (Fig. 5A, lanes 4, 8, and 12), showing that the FGF21 gene is a direct target of PPARγ. Also of interest is the observation that FGF21 mRNA expression is induced simply due to exposure to cycloheximide (Fig. 5A, lanes 3, 7, and 11). This is usually indicative of the existence of a repressor that is removed due to its rapid turnover in the absence of ongoing protein synthesis. To demonstrate further that PPARγ directly activates FGF21 gene expression, we performed a series of FGF21 gene promoter/luciferase reporter assays. To this end, fragments (−500, −1300 and −1500) of the upstream region of the FGF21 gene were cloned into the pGL3 luciferase reporter plasmid as shown in Fig. 5B. Analysis of the sequence of the proximal 1,500 bp of the gene showed the presence of at least five DR-1 response elements, which are highly homologous to a consensus PPRE and therefore have the potential to associate with PPARγ/RXRα heterodimers. Figure 5B shows that transfection of the bp −500 fragment plasmid, which contains two PPREs, into control Swiss fibroblasts in the presence or absence of a potent PPARγ ligand, GW1929, expressed a low basal level of luciferase activity equivalent to a control DR-1/luciferase reporter composed of consensus PPREs. Interestingly, the 1,300-bp and 1,500-bp fragments expressed higher levels of luciferase activity, but the presence of GW1929 had no affect on this activity. Transfection of the reporter plasmids, along with a PPARγ expression plasmid, however, resulted in a significant increase in the activities of all three FGF21 gene fragments, which was enhanced even further in the presence of GW1929. We also analyzed FGF21 promoter activity in control (pBabe-puro) and Swiss WT PPARγ cells by transfecting each of the luciferase reporter plasmids in the presence or absence of GW1929. The results in Fig. 5C are consistent with those in Fig. 5B, showing that the transcriptional activity of the 500-bp fragment of the FGF21 gene was significantly higher in the Swiss cells expressing PPARγ than in control Swiss cells and that this activity was enhanced manyfold by GW1929. The activity of the 1,300- and 1,500-bp fragments in WT PPARγ cells in the presence of GW1929 was only slightly higher than that of the 500-bp region, suggesting that elements within this proximal promoter are likely responsible for the observed PPARγ-dependent activities of all three fragments.
FIG. 5.
PPARγ directly activates the FGF21 gene. (A) Troglitazone activates FGF21 gene expression in the absence of ongoing protein synthesis. WT PPARγ cells were induced to differentiate with DEX, MIX, and insulin for 5 days, at which time troglitazone (5 μM) was added in the presence or absence of cycloheximide (5 μg/ml) for the indicated times. Cells were then harvested for extraction of RNA, followed by RT-PCR analysis of FGF21 and FABP4/aP2 mRNAs as described in Materials and Methods. (B) Reporter assays were performed in control Swiss fibroblasts following transfection of individual FGF21 luciferase (Luc) plasmids (bp −1500, −1300, and −500 fragments), along with a PPARγ (pBabe-PPARγ) or control (pBabe-Puro) expression plasmid and a Renilla (Ren)-based pGL3 reporter as a control in the presence or absence of the potent PPARγ ligand GW1929. The scheme at the top shows the presence of putative PPREs in the upstream region of the gene that have the following DR-1 sequences: 1, AGACCAAGGAGCA; 2, AGACCCAAGGCCC; 3, TGGCCTGTGGCCA; 4, TGAGCACAAGGCCC; and 5, AGTTCCAGGGCCA. (C) Reporter assays were also performed in Swiss fibroblasts stably expressing a WT PPARγ or a pBabe-puro empty vector (control cells) following transfection of the FGF21 promoter reporter plasmids, along with the Renilla control vector, in the presence or absence of GW1929. In both assays B and C, a set of cells were also transfected with a reporter plasmid consisting of the PPRE from the aP2 gene upstream of luciferase within pGL3 (DR-1). The transcriptional activity of each of the fragments of the FGF21 gene promoter is shown as the ratio of luciferase activity to Renilla activity (Luc/Ren) as described in Materials and Methods.
Expression of many of the group 2 genes is actively repressed by SIRT1 during terminal adipogenesis.
A recent report demonstrated that activation of SIRT1 in adipocytes triggers lipolysis and loss of fat by mechanisms involving repression of PPARγ activity (28). We asked, therefore, whether there is a role for SIRT1 in facilitating the differential expression of the group 1 and group 2 genes as preadipocytes become mature fat cells. Consequently, we analyzed the expression of adipogenic genes during differentiation of 3T3-L1 preadipocytes, in which SIRT1 expression is suppressed due to constitutive production of a corresponding SIRT1 RNA interference. The Western blot in Fig. 6A shows the extensive reduction in SIRT1 expression in the RNA interference cells compared to the abundant production in a control line of 3T3-L1 cells expressing the vector alone. It is also relevant that expression of SIRT1 increased severalfold during the early phase of adipogenesis in the control cells but then subsided to preadipocyte levels during the terminal phase. It is also important to point out that there was no significant effect of knockdown of SIRT1 on the production of adiponectin. Total RNA was harvested from SIRT1 knockdown, as well as control, cells at selected times throughout differentiation. RNAs from preadipocytes (day 0) and cells at a middle (day 4) and late (day 10) phase of adipogenesis were subjected to oligonucleotide microarray analysis employing Affymetrix chips, as discussed above. The relative expression levels of selected mRNAs corresponding to both group 1 and group 2 genes were analyzed, as shown in Tables S1A and S1B in the supplemental material, respectively. As discussed above, Table S1A in the supplemental material is a list of classic adipogenic genes (group 1) that are induced during adipogenesis in 3T3-L1 preadipocytes and are differentially responsive to WT PPARγ versus EF-PPARγ. Suppression of SIRT1 activity causes a transient increase (∼50%) in the expression levels of most of these genes at day 4 of differentiation in 3T3-L1 cells compared to their levels of expression at this stage of differentiation in control cells (see Table S1A in the supplemental material; compare column 9 with column 6). This difference in expression correlates with a significant increase in SIRT1 expression during early adipogenesis in control 3T3-L1 preadipocytes (Fig. 6A). Interestingly, these genes appeared to reach a maximum level of expression by day 4 in the knockdown cells, whereas it required 10 days for them to reach this maximum in the control cells (see Table S1A in the supplemental material; compare columns 7 and 10). The data are consistent with the notion that the preadipocytes lacking SIRT1 activity differentiate much faster than control cells, reaching terminal adipogenesis within 4 days compared to 10 days in the control cells. Table S1B in the supplemental material shows the expression profiles of the group 2 mRNAs in control and SIRT1 knockdown 3T3-L1 cells and Swiss Pγ cells. As observed for the group 1 genes in Table S1A in the supplemental material, expression of the group 2 genes also increased at day 4 of differentiation in the knockdown cells, even though most of the genes did not normally show enhanced expression at this stage of differentiation in control cells (see Table S1B in the supplemental material; compare column 9 with column 6). More importantly, most of the group 2 genes were expressed at significantly higher levels at day 10 in the SIRT1 knockdown cells than in control cells (see Table S1B in the supplemental material; compare column 10 with column 7). In fact, some genes, most notably Ero1-Lα (487%), Hig1 (262%), and Trib3 (290%), were enhanced manyfold in response to reduction in SIRT1 abundance. Additionally, all the genes coding for glycolytic enzymes, as well as glucose transporter 1, were also induced in the SIRT1 knockdown cells. It is also worth mentioning that the extent of induction of each of these group 2 genes appeared to correlate with the level of induction by troglitazone in WT PPARγ cells (see Table S1B in the supplemental material; compare columns 1 and 2 with 7 and 10). To confirm the data presented in Tables S1A and S1B in the supplemental material, Fig. 6B shows an RT-PCR analysis of RNAs harvested from control and SIRT1 knockdown 3T3-L1 cells at times throughout differentiation. It is quite apparent that the knockdown of SIRT1 had a selective effect on the group 2 genes compared to the group 1 genes. Specifically, expression of C/EBPα and adiponectin mRNAs (group 1) showed a modest increase at the early stage of adipogenesis in the SIRT1 knockdown cells (Fig. 6B), as presented in Table S1A in the supplemental material, but no significant increase as these cells matured into adipocytes. In contrast, expression of group 2 genes, including Ero1-Lα, Scd3, FGF21, and Elovl3, was dramatically enhanced in the SIRT1 knockdown cells (Fig. 6B).
FIG. 6.

Suppression of SIRT1 by expression of a corresponding SIRT1 siRNA selectively enhances the expression of group 2 PPARγ-responsive genes during the differentiation of 3T3-L1 preadipocytes. Control and SIRT1 siRNA cells were cultured in 10% calf serum until they reached confluence. At 2 days postconfluence, the cells were induced to differentiate by exposure to DEX, MIX, insulin, and 10% FBS. On the indicated days of differentiation, cells were harvested for Western blot analysis (A) and RT-PCR analysis (B) of the indicated gene products as described in Materials and Methods. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Group 2 genes are selectively induced in mature adipocytes by exposure to PPARγ ligands.
The data presented in Fig. 6B and Table S1B in the supplemental material suggested that several of the group 2 genes are actively repressed in mature adipocytes by mechanisms involving SIRT1. We asked, therefore, whether exposure of such cells to a synthetic PPARγ ligand could overcome the repression and stimulate their expression. To test this notion, normal 3T3-L1 preadipocytes were induced to differentiate by following standard procedures, and at days 2, 4, and 6, the differentiating cells were exposed to troglitazone for 2 days, at which time total RNA was harvested for analysis by RT-PCR. Figure 7A demonstrates extensive induction of selected group 2 genes at different stages of the differentiation process. Specifically, ELOVL3 was induced by exposure of the 3T3-L1 cells to troglitazone as early as day 4 of adipogenesis, and the corresponding mRNA levels remained elevated throughout differentiation. FGF21 and Ero1-Lα gene expression was also enhanced severalfold but occurred only in more mature adipocytes. Expression of the selected members of the group 1 genes (C/EBPα, adiponectin, and FABP4 genes), however, was essentially unresponsive to the exogenous ligand, since the level of expression was already at a maximum due to their induction by endogenous PPARγ ligands. These data are consistent with the hypothesis that a subset of the group 2 genes, including FGF21 and Ero1-Lα genes, are actively repressed by SIRT1 in mature adipocytes and that this repression can be overcome by exposure to troglitazone. The fact that attenuation of SIRT1 (Fig. 6B) or exposure to the PPARγ ligand (Fig. 7A) did not induce expression of the classic adipogenic genes in group 1 suggests that they are distinct from the group 2 genes, since they are presumably in a constant state of optimum transcriptional activity. These data suggest that SIRT1 and PPARγ ligands reciprocally regulate PPARγ activity on group 2 genes; we asked, therefore, whether SIRT1 might attenuate the response of PPARγ to its ligands. To test this notion, we determined the dose of troglitazone required to induce expression of select group 2 genes in control versus SIRT1 knockdown 3T3-L1 preadipocytes at 4 days of differentiation. Specifically, differentiating cells were exposed to increasing doses of troglitazone for 2 days, at which stage RNA was analyzed for expression of selected genes by RT-PCR. Figure 7B shows that expression of FGF21 and Egln1 genes (group 2 genes) was induced in control cells following exposure to doses of troglitazone in the range of 1 to 5 μM; in contrast, induction of these genes in SIRT1 knockdown cells required a significantly lower dose of troglitazone (250 nM). These data suggest that SIRT1 attenuates the response of PPARγ to an exogenous ligand. Additionally, Fig. 7B also confirms that there was a negligible effect of either knockdown of SIRT1 or ligands on the expression of the group 1 genes for adiponectin or FABP4.
FIG. 7.
(A) Troglitazone selectively activates expression of group 2 genes in 3T3-L1 adipocytes. 3T3-L1 preadipocytes were differentiated using standard conditions, and at day 2, day 4, and day 6, the differentiating cells were exposed to 5 μM troglitazone for 2 days. Untreated and treated cells were harvested for analysis of select mRNAs using RT-PCR as described in Materials and Methods. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Knockdown of SIRT1 in 3T3-L1 preadipocytes enhances the expression of FGF21 in response to exposure to troglitazone. Control and SIRT1 knockdown 3T3-L1 preadipocytes were differentiated for 4 days, at which time the cells were exposed to the indicated doses of troglitazone for 2 days. Total RNA of the cells was isolated at day 6 and subjected to RT-PCR for analysis of the indicated group 1 and group 2 genes as described in Materials and Methods. (C) Model proposing interplay between PPARγ and SIRT1 in controlling adipocyte function. PPARγ functions to regulate adipocyte formation and function. Endogenous ligands activate PPARγ, requiring participation of both helices 7 and 12 to orchestrate adipogenesis. In mature adipocytes, SIRT1 mediates hormonal and nutrient control of select PPARγ target genes that are involved in controlling metabolism by suppressing the actions of endogenous ligands. The TZD family of synthetic PPARγ ligands can overcome the suppressive effects of SIRT1 acting through helix 7, as well as helix 12, to induce the metabolic genes.
DISCUSSION
Our recent studies have demonstrated that helix 7 within the ligand-binding domain of PPARγ facilitates a functional interaction between β-catenin and PPARγ (22). Additionally, other investigations by Pascual and coworkers (26) identified K365 in helix 7 of PPARγ1 as a target for ligand-dependent SUMOylation, which regulates the repression of inflammatory genes by PPARγ in macrophages. These observations suggested to us that helix 7 might also participate in ligand-dependent control of PPARγ target gene expression during adipogenesis. In the present studies, we show that ectopic expression of a WT PPARγ in Swiss mouse fibroblasts induces expression of the adipogenic program in response to the normal cocktail of adipogenic inducers, including DEX, MIX, insulin, and FBS, without the need for additional stimulation with an exogenous synthetic PPARγ ligand, such as troglitazone. In fact, treatment with troglitazone appeared to have only a minimal effect on the already robust expression of the adipogenic genes (Fig. 1C; see Table S1A in the supplemental material). In contrast, expression of mutant PPARγ, in which F372 within helix 7 of the ligand-binding domain has been changed to alanine, completely destroyed the ability of PPARγ to induce adipogenesis in response to endogenous ligands (minus troglitazone). Interestingly, F372A-PPARγ can respond to troglitazone, and in doing so, it activates the expression of many of the genes induced by the WT PPARγ, although a subset of these genes are unresponsive to troglitazone-activated F372A-PPARγ (Fig. 2A and Table S1B in the supplemental material, group 2 genes). This subset consists of a diverse group of genes encoding a novel set of adipocyte proteins, such as Ero1-Lα and FGF21, as well as components of the glycolytic pathway and regulators of glucose uptake. Many of these group 2 genes are constitutively produced at a low level during adipogenesis, but their expression can be activated in mature adipocytes by exposure to potent PPARγ ligands or suppression of SIRT1 activity (Fig. 6 and 7). The studies also show that PPARγ directly regulates expression of the FGF21 gene through elements located within the 500-bp upstream region of the gene (Fig. 5). Taken together, these data are consistent with the notion that PPARγ can differentially regulate multiple programs of gene expression in response to ligands activating different regions of the ligand-binding domain. Moreover, activation of the group 2 genes by PPARγ requires its association with a potent ligand to overcome the selective suppressive effects of SIRT1.
The molecular mechanisms responsible for distinguishing one set of target genes from another likely involves recruitment of different coregulators to PPARγ docked on the promoters/enhancers of the genes. In fact, Chui et al. and Guan et al. have recently shown that GyK and OLR1 genes are actively repressed in mature adipocytes by recruitment of NCoR/HDAC3 complexes to PPARγ docked on PPRE in the promoters of the corresponding genes (5, 12). Interestingly, exposure of adipocytes to TZDs dislodges the repressor complexes from these sites by mechanisms involving PPARγ coactivator 1α (PGC-1α), leading to expression of the genes. These authors also demonstrated that other adipogenic genes, such as the FABP4/aP2 genes, that are abundantly expressed in mature adipocytes in response to endogenous ligand activity have PPARγ docked on their enhancers in association with the SRC/p160 family of coactivators. The data presented here are consistent with these observations, but they also suggest that a subset of group 2 genes, including Ero1-Lα and FGF21 genes, are selectively repressed by mechanisms dependent on SIRT1 activity. A mechanism under consideration involves a regulated SUMOylation of K365 within helix 7 of PPARγ that is docked on the specific set of target genes (group 2) destined for suppression by SIRT1 in mature adipocytes. SUMOylated PPARγ then recruits select corepressors, such as NCoR/HDAC3, as well as SIRT1, to the target genes that are then subsequently repressed. PPARγ, which is docked on the genes (group 1) that remain active during this process, likely escapes SUMOylation. Formulation of this model is based on the recent findings of Pascual and coworkers, which demonstrated that ligand-dependent SUMOylation of PPARγ on K365 of helix 7 induces the PPARγ-associated repression of inflammatory genes in macrophages (26). K365 could also be a target of acetylation, in which case acetylated K365 would prevent SUMOylation and consequently maintain PPARγ in an active state. It follows, therefore, that deacetylation of K365 by SIRT1 should facilitate SUMOylation, resulting in repression of PPARγ on selected target genes. An important question in considering this model is by what means the SUMOylation process selects PPARγ molecules that are docked on the targets that will be repressed during terminal adipogenesis. One possibility is that the environment surrounding the PPREs within these genes facilitates SUMOylation. For instance, the mechanism could involve docking of other nuclear factors that are induced during adipogenesis to sites flanking the PPREs. These factors could then participate in recruitment of the SUMOylation machinery to PPARγ and the resulting repression of these genes.
It is also possible that SIRT1 regulates the expression/activities of coregulators whose association with PPARγ is dependent on helix 7 of the ligand-binding domain. In fact, it is reasonable to suggest a role for PGC-1α, since Guan and coworkers have previously shown induction of this coactivator in white adipocytes in response to TZDs (12). Furthermore, these investigators demonstrated the involvement of PGC-1α in the selective activation of the GyK gene by TZDs. Additionally, other studies have shown that SIRT1 deacetylates PGC-1α and, in doing so, regulates its ability to modulate the activities of different transcription factors (31). Consequently, it is conceivable that the selective expression of the group 2 genes, which include GyK and OLR1 genes, in mature adipocytes involves induction of PGC-1α by TZDs and its activation through the suppression of SIRT1 activity.
Another important component of the model explaining how PPARγ activates different programs of gene expression at precise times during adipogenesis is the roles played by specific ligands. As stated earlier, the endogenous ligands responsible for stimulating PPARγ activity during adipogenesis have not yet been identified. Several studies, however, have identified signaling pathways and transcription factors that appear to regulate ligand production (4, 14, 17, 23, 35). The combined data are consistent with a regulated process induced during the initial days of adipogenesis involving cyclic AMP signaling and enzymes that convert polyunsaturated fatty acids into eicosinoids. It is interesting that ligand activity peaks after 2 to 4 days of adipogenesis in 3T3-L1 preadipocytes but then rapidly subsides during terminal adipogenesis. Several questions result from these observations; most notably, how does PPARγ continue to maintain the expression of most target genes, such as the FABP4/aP2 genes, in the presence of lower concentrations of these ligands? There are many explanations for this apparent conundrum, including changes in expression and activities of coregulators during adipogenesis that require lower levels of PPARγ activity to facilitate the associated recruitment/dislodgment process.
The subset of adipogenic genes (group 2) that have been identified in this report contains several members that have not previously been shown to be regulated during adipogenesis or responsive to the activity of PPARγ. In the case of the endoplasmic reticulum oxidoreductase Ero1-Lα, studies have recently shown that this protein is involved in regulating the secretion of adiponectin from mature adipocytes (30, 36). Furthermore, expression of Ero1-Lα mediates the nutrient control of adiponectin secretion by responding to the activity of the NAD-dependent deacetylase SIRT1 (30); these data are consistent with the observations presented here showing that the group 2 set of adipogenic genes is regulated by SIRT1. It is also noteworthy that we have identified FGF21 as a direct target of PPARγ, since it has recently been shown to be a hormone produced in the liver in response to activation of PPARα and acts as a component of the body's adaptation to fasting (1, 15). Other studies have also shown that it is a potent regulator of glucose uptake in 3T3-L1 adipocytes and primary human adipocytes (16). Our data show that FGF21 is not only produced in hepatocytes, but can also be induced in 3T3-L1 adipocytes by exposure to potent PPARγ ligands or suppression of SIRT1 activity, suggesting that this secreted factor might act in an autocrine, as well as a paracrine, fashion to regulate insulin-responsive glucose uptake in adipocytes. With regard to FGF21 signaling, the data in Table S1B in the supplemental material show that a gene (Klb-βKlotho) coding for an important component of FGF21 receptor (FGFR1 and -4) complex (25) is also responsive to both PPARγ and SIRT1 activities, categorizing it as a member of the group 2 gene family. Additionally, components of the glycolytic pathway and regulators of glucose uptake are also members of this novel group 2 set of adipocyte genes. In fact, studies by others (31) have shown an increase in the expression of liver pyruvate kinase and glucokinase in response to knockdown of SIRT1 in hepatocytes. We propose (Fig. 7C), therefore, that SIRT1 can control metabolic homeostasis by regulating expression of the group 2 subset of adipocyte genes in response to metabolic effectors, resulting in production of multiple proteins, including insulin sensitizers, such as adiponectin (through Ero1-Lα), as well as intracellular regulators of glucose uptake/metabolism (i.e., FGF21 and βKlotho). It appears that synthetic PPARγ ligands also target this gene program in adipocytes by selectively overcoming the suppressive effects of SIRT1 on these genes.
In conclusion, our studies have identified a novel regulatory region of the ligand-binding domain of PPARγ that facilitates the selective expression of different subgroups of adipocyte genes during the formation of mature fat cells. These findings should provide a greater understanding of the roles of PPARγ and its ligands in regulating physiological functions of adipocytes, most notably insulin responsiveness and energy balance. Furthermore, the identification of novel genes that respond to SIRT1, as well as PPARγ activity, such as Ero1-Lα and FGF21 genes, should provide additional targets for the development of effective therapeutics to combat obesity and its associated disorders.
Supplementary Material
Acknowledgments
We acknowledge the support of the Genome Center at Boston University School of Medicine and the valuable help of Marc Lenburg and Karen Schlauch in analyzing the microarray data. We are grateful to Kathryn Davis for critical reading of the manuscript and constructive comments. We thank L. Guarente for the pSUPER SIRT1 siRNA plasmid.
This work was supported by USPHS grants DK51586 and DK58825.
Footnotes
Published ahead of print on 22 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Badman, M. K., P. Pissios, A. R. Kennedy, G. Koukos, J. S. Flier, and E. Maratos-Flier. 2007. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 5426-437. [DOI] [PubMed] [Google Scholar]
- 2.Berger, J., M. Tanen, A. Elbrecht, A. Hermanowski-Vosatka, D. E. Moller, S. D. Wright, and R. Thieringer. 2001. Peroxisome proliferator-activated receptor-gamma ligands inhibit adipocyte 11β-hydroxysteroid dehydrogenase type 1 expression and activity. J. Biol. Chem. 27612629-12635. [DOI] [PubMed] [Google Scholar]
- 3.Castriota, G., G. M. Thompson, Y. Lin, P. E. Scherer, D. E. Moller, and J. P. Berger. 2006. Peroxisome proliferator-activated receptor gamma agonists inhibit adipocyte expression of α1-acid glycoprotein. Cell Biol. Int. 31586-591. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, K. J., I. Tzameli, P. Pissios, I. Rovira, O. Gavrilova, T. Ohtsubo, Z. Chen, T. Finkel, J. S. Flier, and J. M. Friedman. 2007. Xanthine oxidoreductase is a regulator of adipogenesis and PPARγ activity. Cell Metab. 5115-128. [DOI] [PubMed] [Google Scholar]
- 5.Chui, P. C., H. P. Guan, M. Lehrke, and M. A. Lazar. 2005. PPARγ regulates adipocyte cholesterol metabolism via oxidized LDL receptor 1. J. Clin. Investig. 1152244-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans, R. M., G. D. Barish, and Y. X. Wang. 2004. PPARs and the complex journey to obesity. Nat. Med. 10355-361. [DOI] [PubMed] [Google Scholar]
- 7.Fajas, L., M. B. Debril, and J. Auwerx. 2001. PPAR gamma: an essential role in metabolic control. Nutr. Metab. Cardiovasc. Dis. 1164-69. [PubMed] [Google Scholar]
- 8.Farmer, S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4263-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasshauer, M., J. Klein, K. M. Kriauciunas, K. Ueki, M. Benito, and C. R. Kahn. 2001. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol. Cell. Biol. 21319-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasshauer, M., J. Klein, K. Ueki, K. M. Kriauciunas, M. Benito, M. F. White, and C. R. Kahn. 2000. Essential role of insulin receptor substrate-2 in insulin stimulation of Glut4 translocation and glucose uptake in brown adipocytes. J. Biol. Chem. 27525494-25501. [DOI] [PubMed] [Google Scholar]
- 11.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417563-567. [DOI] [PubMed] [Google Scholar]
- 12.Guan, H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes Dev. 19453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan, H. P., Y. Li, M. V. Jensen, C. B. Newgard, C. M. Steppan, and M. A. Lazar. 2002. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat. Med. 81122-1128. [DOI] [PubMed] [Google Scholar]
- 14.Hamm, J. K., B. H. Park, and S. R. Farmer. 2001. A role for C/EBPβ in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J. Biol. Chem. 27618464-18471. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki, T., P. Dutchak, G. Zhao, X. Ding, L. Gautron, V. Parameswara, Y. Li, R. Goetz, M. Mohammadi, V. Esser, J. K. Elmquist, R. D. Gerard, S. C. Burgess, R. E. Hammer, D. J. Mangelsdorf, and S. A. Kliewer. 2007. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 5415-425. [DOI] [PubMed] [Google Scholar]
- 16.Kharitonenkov, A., T. L. Shiyanova, A. Koester, A. M. Ford, R. Micanovic, E. J. Galbreath, G. E. Sandusky, L. J. Hammond, J. S. Moyers, R. A. Owens, J. Gromada, J. T. Brozinick, E. D. Hawkins, V. J. Wroblewski, D. S. Li, F. Mehrbod, S. R. Jaskunas, and A. B. Shanafelt. 2005. FGF-21 as a novel metabolic regulator. J. Clin. Investig. 1151627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J. B., H. M. Wright, M. Wright, and B. M. Spiegelman. 1998. ADD1/SREBP1 activates PPAR gamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. USA 954333-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliewer, S. A., H. E. Xu, M. H. Lambert, and T. M. Willson. 2001. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog. Horm. Res. 56239-263. [DOI] [PubMed] [Google Scholar]
- 19.Krey, G., O. Braissant, F. L'Horset, E. Kalkhoven, M. Perroud, M. G. Parker, and W. Wahli. 1997. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 11779-791. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann, J. M., L. B. Moore, T. A. Smith-Oliver, W. O. Wilkison, T. M. Willson, and S. A. Kliewer. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J. Biol. Chem. 27012953-12956. [DOI] [PubMed] [Google Scholar]
- 21.Lehrke, M., and M. A. Lazar. 2005. The many faces of PPARγ. Cell 123993-999. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., H. Wang, Y. Zuo, and S. R. Farmer. 2006. Functional interaction between peroxisome proliferator-activated receptor gamma and beta-catenin. Mol. Cell. Biol. 265827-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madsen, L., R. K. Petersen, M. B. Sorensen, C. Jorgensen, P. Hallenborg, L. Pridal, J. Fleckner, E.-Z. Amri, P. Krieg, G. Furstenberger, R. K. Berge, and K. Kristiansen. 2003. Adipocyte differentiation of 3T3-L1 preadipocytes is dependent on lipoxygenase activity during the initial stages of the differentiation process. Biochem. J. 375539-549. [DOI] [PubMed] [Google Scholar]
- 24.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395137-143. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa, Y., H. Kurosu, M. Yamamoto, A. Nandi, K. P. Rosenblatt, R. Goetz, A. V. Eliseenkova, M. Mohammadi, and M. Kuro-o. 2007. βKlotho is required for metabolic activity of fibroblast growth factor 21. Proc. Natl. Acad. Sci. USA 1047432-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pascual, G., A. L. Fong, S. Ogawa, A. Gamliel, A. C. Li, V. Perissi, D. W. Rose, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 437759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116511-526. [DOI] [PubMed] [Google Scholar]
- 28.Picard, F., M. Kurtev, N. Chung, A. Topark-Ngarm, T. Senawong, R. Machado De Oliveira, M. Leid, M. W. McBurney, and L. Guarente. 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi, C., S. Surapureddi, Y. J. Zhu, S. Yu, P. Kashireddy, M. S. Rao, and J. K. Reddy. 2003. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARγ)-interacting protein, is required for PPARγ-mediated adipogenesis. J. Biol. Chem. 27825281-25284. [DOI] [PubMed] [Google Scholar]
- 30.Qiang, L., H. Wang, and S. R. Farmer. 2007. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-Lα. Mol. Cell. Biol. 274698-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodgers, J. T., C. Lerin, W. Haas, S. P. Gygi, B. M. Spiegelman, and P. Puigserver. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434113-118. [DOI] [PubMed] [Google Scholar]
- 32.Rosen, E. D., and B. M. Spiegelman. 2001. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 27637731-37734. [DOI] [PubMed] [Google Scholar]
- 33.Schadinger, S. E., N. L. Bucher, B. M. Schreiber, and S. R. Farmer. 2005. PPARγ2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am. J. Physiol. Endocrinol. Metab. 288E1195-E1205. [DOI] [PubMed] [Google Scholar]
- 34.Steppan, C. M., and M. A. Lazar. 2002. Resistin and obesity-associated insulin resistance. Trends Endocrinol. Metab. 1318-23. [DOI] [PubMed] [Google Scholar]
- 35.Tzameli, I., H. Fang, M. Ollero, H. Shi, J. K. Hamm, P. Kievit, A. N. Hollenberg, and J. S. Flier. 2004. Regulated production of a peroxisome proliferator-activated receptor-gamma ligand during an early phase of adipocyte differentiation in 3T3-L1 adipocytes. J. Biol. Chem. 27936093-36102. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z. V., T. D. Schraw, J. Y. Kim, T. Khan, M. W. Rajala, A. Follenzi, and P. E. Scherer. 2007. Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol. Cell. Biol. 273716-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willson, T. M., M. H. Lambert, and S. A. Kliewer. 2001. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu. Rev. Biochem. 70341-367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.