Abstract
Activation of eukaryotic gene transcription involves the recruitment by DNA-binding activators of multiprotein histone acetyltransferase (HAT) and Mediator complexes. How these coactivator complexes functionally cooperate and the roles of the different subunits/modules remain unclear. Here we report physical interactions between the human HAT complex STAGA (SPT3-TAF9-GCN5-acetylase) and a “core” form of the Mediator complex during transcription activation by the MYC oncoprotein. Knockdown of the STAF65γ component of STAGA in human cells prevents the stable association of TRRAP and GCN5 with the SPT3 and TAF9 subunits; impairs transcription of MYC-dependent genes, including MYC transactivation of the telomerase reverse transcriptase (TERT) promoter; and inhibits proliferation of MYC-dependent cells. STAF65γ is required for SPT3/STAGA interaction with core Mediator and for MYC recruitment of SPT3, TAF9, and core Mediator components to the TERT promoter but is dispensable for MYC recruitment of TRRAP, GCN5, and p300 and for acetylation of nucleosomes and loading of TFIID and RNA polymerase II on the promoter. These results suggest a novel STAF65γ-dependent function of STAGA-type complexes in cell proliferation and transcription activation by MYC postloading of TFIID and RNA polymerase II that involves direct recruitment of core Mediator.
Regulation of eukaryotic gene transcription involves the orchestrated recruitment by sequence-specific DNA-binding regulators of two broad classes of transcription cofactors—i.e., chromatin-modifying cofactors and general cofactors—that function in concert on target promoters. Chromatin-modifying cofactors include a variety of histone-modifying and ATP-dependent nucleosome-remodeling enzymes that, respectively, add (or remove) various covalent modifications on histones and alter the structure and positioning of nucleosomes (reviewed in reference 42). General cofactors such as the Mediator complex and the TATA-binding protein (TBP)-associated factors (TAFs) of the TFIID complex directly interact with the basal/general transcription initiation factors and/or RNA polymerase II (Pol II) on most promoters and stimulate the assembly and function of transcription initiation complexes (reviewed in references 35, 65, and 71).
The prototypical histone acetyltransferase (HAT) GCN5 is part of different chromatin-modifying multiprotein complexes, which have been identified and characterized to various extents in different organisms and include large SAGA/STAGA-type complexes from yeasts to humans, and the smaller yeast ADA and Drosophila ATAC complexes (reviewed in reference 39). Yeast SAGA-type complexes include SAGA (28) and its derivative SLIK/SALSA/SAGA(alt) complex (4, 62, 69, 77). Mammalian STAGA-type complexes include the STAGA (50), TFTC (10), and PCAF (57) complexes, which are highly similar but contain either GCN5 (in STAGA/TFTC) or its paralog, PCAF (p300/CBP-associated factor). In contrast to GCN5, PCAF is dispensable for mouse development and is expressed in a more tissue-restricted manner (57, 80, 81). Yeast and human SAGA/STAGA-type complexes are structurally related and composed of similar subunits including the following (known human subunits in parentheses): Spt3 (SPT3), Spt7 (STAF65γ), Spt8, Spt20/Ada5, Taf5 (PAF65β/TAF5L), Taf6 (PAF65α/TAF6L), Taf9 (TAF9), Taf10 (TAF10), Taf12 (TAF12), Ada1 (STAF42), Ada2 (ADA2B), Ada3 (ADA3), Gcn5/Ada4 (GCN5 or PCAF), Tra1 (PAF400/TRRAP), Sgf73 (ataxin-7), Sgf29 (STAF36), Sgf11, Ubp8, Sus1, and Chd1. Human complexes also associate with SAP130, a spliceosome-associated protein (see references 39, 49, 61, and 72 and references therein). Notably, all eukaryotic SAGA/STAGA-type complexes differ from other GCN5 complexes (i.e., ADA type and ATAC type), other Tra1/TRRAP-containing complexes (e.g., NuA4/TIP60 type), and from TFIID by the presence of several SAGA/STAGA-specific subunits, which include the “Spt group” of components (described above and see references 13 and 39).
Transcription profiling and genome-wide location analyses in yeast suggest that Gcn5 complexes may regulate most protein-coding genes; however, only a minority (5 to 10%) of yeast genes appear to be absolutely dependent on Gcn5 or on the integrity of the SAGA complex (34, 64). Genetic and biochemical studies in yeast indicated that Spt7, Spt20, and Ada1 subunits are important for the structural integrity of the SAGA complex (28, 68, 77). Spt7 interacts with Taf10 (a Taf shared by SAGA-type and TFIID complexes) via complementary histone fold domains (26) and is essential for the association of Spt3, Spt8, Taf6, Taf5, Ada1, and Tra1 with an independent Gcn5/Ada module containing Gcn5, Ada2, Ada3, and Spt20/Ada5 (69). Conversely, partial SAGA subcomplexes containing Spt7 can assemble in the absence of both Spt20/Ada5 and Ada1 (77). Intriguingly, a truncated Spt7 protein is specifically associated with the SAGA-type complex SLIK/SALSA/SAGA(alt), which lacks Spt8 and is more prevalent under amino acid starvation. SLIK/SALSA/SAGA(alt) has altered TBP-binding and transcription regulatory activities that are only partially understood (4, 62, 69, 77). These observations have suggested a modular and dynamic composition of SAGA-type complexes and a pivotal role for Spt7 in controlling SAGA organization and function in yeast. The proposed ortholog of yeast Spt7 in mammalian cells is STAF65γ (51), a protein component specific of STAGA-type complexes (18, 51). STAF65γ stably heterodimerizes through its histone fold domain with TAF10 (as yeast Spt7) and may also interact with TAF8 (see reference 18 and references therein), an alternative histone fold partner of TAF10 that is specific to TFIID complexes (18, 30). The role of the STAF65γ-TAF8 interaction is, however, unknown. More generally, the functions of STAF65γ and other Spt-like subunits of STAGA-type complexes remain to be determined in higher eukaryotes.
In Saccharomyces cerevisiae, SAGA is recruited to promoters by activators and binds to acetylated histones H3 and H4 via the bromodomain of Gcn5 and may also recognize methylated lysine 4 of histone H3 via a chromodomain in Chd1. SAGA recruitment leads to Gcn5-dependent histone hyperacetylation and facilitation of nucleosome remodeling, preinitiation complex assembly, and transcription elongation (reviewed in reference 42). SAGA can also more directly stimulate the recruitment of TBP and Pol II via a Gcn5-independent pathway that requires Spt3 (5, 19, 37) and involves the direct physical interaction of Spt3 and/or Spt8 with TBP (21, 66, 75). Several reports have also indicated a dependency on SAGA for Mediator recruitment by Gal4 at GAL1 and by Gcn4 at ARG4 and SNZ1 promoters (38, 40, 41, 63). However, the mechanisms have remained elusive and the dependency on SAGA, which varies greatly in different studies, is also controversial in certain cases (6, 11, 14). In addition, SAGA deubiquitylates histone H2B via its Ubp8 component, a step that controls histone H3 methylation and activation of selected genes, probably at the level of transcription elongation (33, 78).
Mammalian STAGA-type complexes have been shown to stimulate transcription activation by the model Gal4-VP16 activator on chromatin templates in vitro in a HAT activity-dependent manner (31, 51, 53). In addition, numerous studies have indicated regulatory functions for mammalian GCN5 and PCAF HATs via acetylation of various transcription factors and suggested coactivator roles in vivo for individual subunits of GCN5/PCAF complexes, including GCN5, PCAF, TRRAP, and specific ADA proteins (reviewed in references 70 and 83). However, since GCN5/PCAF HATs have altered activities and specificities within larger multiprotein assemblies and some of the above proteins are shared with several HAT complexes (described above and see references 13, 39, and 83), it is generally unclear whether the functions observed reflect the natural activities of specifically STAGA-type complexes. Several studies, however, have indicated concerted functions of GCN5 with other STAGA subunits, including (i) TRRAP during transactivation by MYC (44, 52), E2F4 (36), nuclear hormone receptors (82), and BRCA1 (58); (ii) ADA2B for transactivation by the B-cell-specific activator Pax5/BSAP (3); and (iii) ataxin-7 for transactivation by the photoreceptor-specific activator CRX and regulation of photoreceptor-specific genes in the retina (32, 59).
Here, we have further analyzed the functions of STAGA-type complexes in the context of transcription activation by the MYC oncoprotein in human cells. MYC binds E-box DNA elements as a heterodimer with MAX and activates transcription of target genes via mechanisms that are not yet fully understood but appear to involve either modification of chromatin structure through (in part) recruitment of TRRAP-containing HAT complexes or HAT-independent stimulation of a transcription step(s) postrecruitment of Pol II (reviewed in reference 16). In this study, we have used RNA interference (RNAi) to knock down STAF65γ, the Spt7-like subunit specific of STAGA-type complexes, in human cancer cell lines. A novel STAF65γ-dependent function of STAGA in cell proliferation and transcription activation by MYC is presented that is distinct from the classical HAT and TBP/Pol II-recruiting activities of SAGA-type complexes and involves physical recruitment of a “core” Mediator complex (i.e., lacking MED12 and MED13) to MYC and the telomerase reverse transcriptase (TERT) promoter in vivo. Our data suggest that mammalian STAGA complexes contribute to both functions of MYC in histone acetylation and activation postrecruitment of Pol II via, respectively, their TRRAP-GCN5 and SPT-TAF components.
MATERIALS AND METHODS
DNA plasmids.
The hTERT(−211/+40) luciferase reporter vector (p2xEB) was a gift from J. Carl Barrett, pCbS-Flag-Myc wt and Δ1-110 were a kind gift from Michael D. Cole, the glutathione S-transferase (GST)-Myc expression vectors were from R. Bernards, and the pRSETA-His6-GCN5-S expression vector was from Richard A. Currie (22). The expression vector for Flag- and hemagglutinin (HA)-tagged human STAF65γ (pIRESneo-fh:STAF65γ) was kindly provided by Robert G. Roeder. The pIRESneo-fh:STAF65γ “recoded” vector was derived from pIRESneo-fh:STAF65γ by site-directed mutagenesis of the sequence GGAGGAACCTGTGAGCGAC to GGAGGAACCTGTCTCCGAC (substituted nucleotides underlined), using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. The pSUPER-STAF65γ828 vector encodes a specific shRNA that targets the sequence above. The shRNA expression vectors pSUPER-MYC827 and pSUPER-GL2 were described previously (22).
Cell culture, transfection, reporter gene, colony formation assays, RNAi, and RT-PCR.
HeLa, HeLa S3, U2OS, and HEK 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C with 5% CO2. HL-60 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C with 5% CO2. To induce HL-60 cell differentiation, cells at a density of 4 × 105 cells/ml were treated with 1.25% (vol/vol) dimethyl sulfoxide (Merck) for 48 h (79). HeLa S3 suspension cells were adapted from monolayer and maintained at 37°C in minimum essential medium (S-MEM; Gibco-Invitrogen) supplemented with 5% bovine calf serum. The fh:GCN5 cell line expressing N-terminal Flag- and HA-tagged human GCN5L was obtained by stable transfection of HeLa S3 cells with pFH:hGCN5L-IRESneo vector (44) and G418 selection as described previously for fh:SPT3 cells (51). Transfection conditions and luciferase and β-galactosidase assays were essentially as previously described (22). Results are means ± standard deviations from three independent experiments performed in duplicate. For the colony formation assay, HeLa or U2OS cells at ∼90% confluence were transfected with the indicated pSUPER-shRNA vectors using Lipofectamine 2000. After 24 h, equal numbers of cells were replated onto 10-cm dishes. Twenty-four hours after replating, the cells were selected in medium containing 1.5 μg/ml puromycin. The selection medium was changed every 3 days. After approximately 3 to 4 weeks, the colonies were stained with crystal violet. For transient knockdown, HeLa cells were transfected in 35-mm plates with Lipofectamine 2000 (Invitrogen) and 2 μg of pSUPER-shRNA vector and once again 24 h later. Six hours after the second transfection, the medium was replaced with puromycin-containing medium (0.5 μg/ml), and after 48 to 72 h, total RNA and protein were extracted for reverse transcription-PCR (RT-PCR) and Western blot analyses. STAF65γ-knockdown cell lines were obtained by stable transfection of HeLa, HeLa S3, fh:SPT3, and fh:GCN5 cell lines with pSUPER-STAF65γ828 as described previously for MYC-knockdown cells (22). All experiments were performed with cells at low-passage number as both STAF65γ- and MYC-knockdown cells eventually stop dividing after 1 to 2 months in culture. RT-PCR analyses were performed as previously described (22), and PCR products from serial dilutions of cDNA, and within the linear range of amplification, were quantified by using the NIH Image software. The following primers were used for RT-PCR: TERT, TCTGGATTTGCAGGTGAACAGCC and GGGTGGCCATCAGTCCAGGATGG; CDK4, GTGGACATGTGGAGTGTTGG (HCD1) and GCCCTCTCAGTGTCCAGAAG (HCD2); HSP60, ACAAGTGATGTTGAAGTGAATG (HHSP1) and ATTGCTGGAATTTTGAGTGTTC (HHSP2); GAPDH (glyceraldehyde-3-phosphate dehydrogenase), CTCAGACACCATGGGGAAGGTGA (HG1) and ATGATCTTGAGGCTGTTGTCATA (HG2); and CAD, GGTGGATCTGGAGCATGAGTGGA (CAD1) and AGATGGAAGCGGCCATCAGGAAG (CAD2) (56). β-Actin primers were described previously (22).
Antibodies, Western blotting, and ChIP.
A rabbit polyclonal antibody for STAF65γ was custom-made against the peptide GIPQSPDDSDSSYGS (Invitrogen). The following antibodies were kindly provided by Robert G. Roeder: TBP, TAF8 (TAF43), TAF9 (TAF31), TAF12 (TAF20/15), MED17 (TRAP80), MED16 (TRAP95), and an independent STAF65γ antibody. The TAF10 (TAF30) monoclonal antibody 1H8 and the TAF5L (PAF65β) antibody were kind gifts from Laszlo Tora and Yoshihiro Nakatani, respectively. Other antibodies were as follows: TRRAP (T-17; sc-5405; and H-300; sc-11411), GCN5 (N-18; sc-6303), TAF9/TAFIIp32 (N-16; sc-1247), p300 (N-15; sc-584), Sp1 (H-225; sc-14027), MYC (N-262; sc-764), USF (C-20; sc-229), MAX (C-17; sc-197), MED1/TRAP220 (C-19; sc-5334), MED14/CRSP150 (C-17; sc-9420), MED12/TRAP230 (A-18; sc-5374), MED13/TRAP240 (E-20; sc-12013), CDK8 (C-19; sc-1521), TERT (H-231; sc-7212), and Pol II large subunit (N-20; sc-899), TAFIIp250 (6B3; sc-735), all from Santa Cruz Biotech.; anti-acetyl-histone H3 (06-599) and anti-acetyl-histone H4 (06-866) from Upstate; and anti-Flag M2 monoclonal antibody (F3165) and anti-Flag M2 agarose (A2220) (Sigma). Secondary antibodies for Western blotting were anti-rabbit (88-1688) and anti-mouse (88-7788) TrueBlot (eBioscience). Western blots and chromatin immunoprecipitation (ChIP) assays were performed as described previously (22).
Protein immunoprecipitation and GST pull-down.
Whole-cell extracts and coimmunoprecipitation of endogenous or ectopic Flag-tagged proteins and associated factors were performed as described previously by using immunoprecipitation (IP) buffer containing 180 mM NaCl (22, 44). Endogenous TFIID was immunoprecipitated from whole-cell extracts prepared in lysis buffer (22) containing 400 mM NaCl, and immunoprecipitations and washes were performed in the same buffer; these conditions were also used for immunoprecipitation of endogenous SPT3 (STAGA) in Fig. 1C. For immunoprecipitation of endogenous MED1 and associated factors (see Fig. 6A), 600 μg HeLa nuclear extracts (precleared with 20 μl protein G-Sepharose [Amersham] for 30 min at 4°C) was incubated with 2 μg MED1 antibody overnight at 4°C and immunoprecipitated and washed in IP buffer containing 180 mM NaCl as described above (44). For immunoprecipitation of Flag-SPT3, Flag-STAF65γ, and Flag-GCN5 (Fig. 6B and C), 600 μg of nuclear extracts prepared from the fh:SPT3, fh:STAF65γ, and fh:GCN5 cell lines, respectively, was incubated with anti-Flag M2-agarose in IP buffer containing 150 mM KCl and washed three times with 1 ml IP buffer containing 180 mM NaCl, and bound proteins were eluted with FLAG peptide (0.2 mg/ml), as indicated in Fig. 6B, or with sodium dodecyl sulfate (SDS) loading buffer (Fig. 6C). Highly purified STAGA and Mediator complexes were prepared from nuclear extracts of the fh:SPT3 cell line (51) and the Flag-Nut2/MED10 cell line (48), respectively, by anti-Flag M2 affinity and extensive washes in 330 mM KCl-containing buffers, as described previously (29, 51). For Fig. 6E, 20 μl of affinity-purified STAGA and Mediator complexes was precleared with protein G-Sepharose for 30 min at 4°C and then incubated with 2 μg of MED1 antibody for 2 h at 4°C, precipitated with 10 μl protein G-Sepharose, washed twice with 1 ml BC150 (20 mM Tris-HCl [pH 7.9], 150 mM KCl, 20% glycerol, 0.05% IGEPAL CA-630, 0.2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 10 mM 2-mercaptoethanol), and bound proteins were eluted with SDS loading buffer. GST-MYC and GST-VP16 pull-down assays were performed essentially as previously described (44, 51).
FIG. 1.
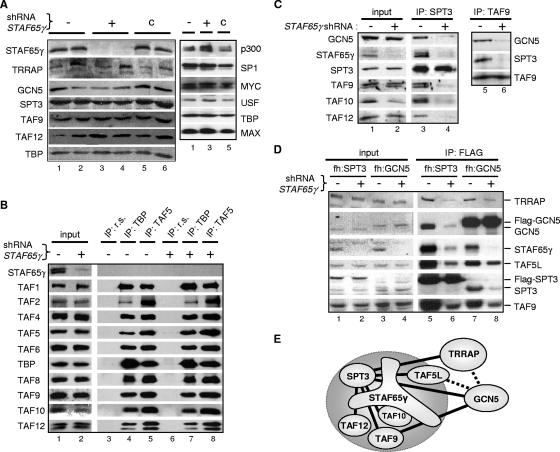
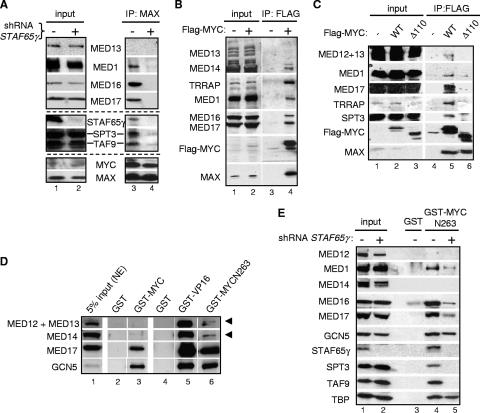
Knockdown of STAF65γ affects the integrity of STAGA but not TFIID complexes in human cells. (A) STAF65γ-knockdown HeLa S3 cell lines. Western blot analyses were performed with whole-cell extracts from parental HeLa S3 cells (−) (lanes 1 and 2); two STAF65γ-knockdown HeLa S3 cell lines stably transfected with the STAF65γ shRNA vector pSUPER-STAF65γ828 (+) (clone 1 in lanes 3 and clone 4 in lane 4); and two control HeLa S3 cell lines (lanes 5 and 6) stably transfected with a different shRNA vector, pSUPER-STAF65γ218 (c), which retained normal STAF65γ protein levels. The indicated antibodies were used to probe different stripes of the blots (panels) shown. (B and C) Whole-cell extracts (input) of STAF65γ-knockdown cells (+) (clone 1) and control HeLa S3 cells stably transfected with the empty pSUPER vector (−) were immunoprecipitated with either preimmune rabbit serum (IP: r.s.) or TBP (IP: TBP), TAF5 (IP: TAF5), SPT3 (IP: SPT3), or TAF9 (IP: TAF9) antisera, and complexes were analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with specific antibodies to the indicated proteins. (D) Whole-cell extracts (input) of parental (−) and STAF65γ-knockdown (+) fh:SPT3 and fh:GCN5 cell lines were immunoprecipitated with the Flag antibody (IP: FLAG) and analyzed by Western blotting with antibodies to the indicated proteins. (E) Diagram summarizing the interaction results of panels C and D. Binary associations are shown as straight lines between two subunits (except for the direct interaction of STAF65γ with TAF10). Solid lines interrupted by STAF65γ indicate a strong dependency on STAF65γ for the corresponding binary association. Dashed lines indicate a partial dependency on STAF65γ. The large circle highlights SPT-TAF subunits.
FIG. 6.
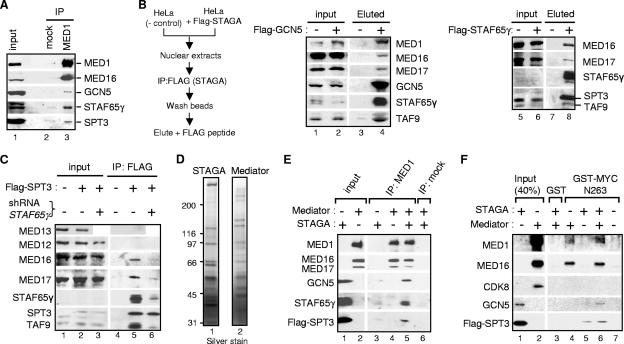
Physical interaction of STAGA and Mediator in human cells and in vitro. (A) HeLa S3 nuclear proteins were immunoprecipitated (IP) with a MED1-specific antibody (lane 3) or protein G-agarose (mock, lane 2) and analyzed by Western blotting with specific antibodies to the indicated proteins. (B) Nuclear extracts (input) of HeLa S3 (−) and derivative cell lines expressing (+) Flag-GCN5 or Flag-STAF65γ were immunoprecipitated with the FLAG antibody M2 resin, and complexes were eluted with excess Flag peptide (Eluted), as indicated in the scheme to the left. Proteins were analyzed by Western blotting. (C) Nuclear extracts of HeLa S3 cells (lanes 1 and 4), fh:SPT3 cells (lanes 2 and 5), and fh:SPT3 cells expressing the STAF65γ shRNA (lanes 3 and 6) were immunoprecipitated with the Flag antibody resin (IP:FLAG; lanes 4 to 6) and analyzed by Western blotting. The weak (or undetectable) signals for STAF65γ in input lanes (B and C) are due to the use of a short autoradiographic exposure (same gel/exposure for input and IP lanes) to prevent saturation of signals in the IP lanes. STAF65γ levels in fh:SPT3 cells ± shRNA vector are shown in Fig. 1D (lanes 1 and 2). (D) Immunoaffinity-purified STAGA (Flag-SPT3; lane 1) and Mediator (Flag-Nut2; lane 2) complexes resolved by SDS-polyacrylamide gel electrophoresis and silver stained. Positions of size markers in kDa are shown. (E) Direct interaction of purified human STAGA and Mediator complexes in vitro. STAGA and Mediator complexes (shown in panel D) either alone or combined (as indicated by + and − signs) were immunoprecipitated with a MED1-specific antibody (IP: MED1) or with protein G agarose (IP: mock) and analyzed by Western blotting with the indicated antibodies. Elution buffer containing the Flag peptide was used instead of the corresponding complex (−) in lanes 3 and 4. (F) In vitro pull-down assays were performed with immobilized GST and GST-MYC(1-263) and affinity-purified Flag-SPT3 (STAGA) and/or Flag-Nut2 (Mediator) complexes, as indicated (− and + signs). Forty percent of each input complex (lanes 1 and 2) and GST/GST-MYC-bound proteins (lanes 3 to 7) was analyzed by Western blotting with antibodies to the indicated subunits.
RESULTS
STAF65γ is required for the integrity of the STAGA complex.
Human STAF65γ is highly similar to yeast Spt7 but lacks the N-terminal Spt7 bromodomain (51). Mass spectrometry analyses of the proteins stably associated with Flag-tagged STAF65γ in human cells have only identified STAGA components (A. Gamper and R. G. Roeder, unpublished data). Thus, STAF65γ is an SPT7-like subunit specific of STAGA-type complexes (see also below). To analyze the role of STAF65γ in human cells, RNAi with shRNA vectors was used to stably knock down its expression in HeLa S3 cells. As shown in Fig. 1A, Western blot analyses of whole-cell extracts indicated that STAF65γ expression was specifically suppressed in cell lines transfected with a specific STAF65γ shRNA (lanes 3 and 4). The steady-state protein levels of other STAGA subunits, including TRRAP, GCN5, SPT3, TAF9, and TAF12, and other transcription factors (e.g., p300, SP1, MYC, MAX, USF, and TBP) were not significantly changed compared to those of parental HeLa S3 cells (lanes 1 and 2) or to control HeLa S3 cell lines stably transfected with a different shRNA vector that retained normal STAF65γ levels (lanes 5 and 6). Interestingly, the cellular levels of TAF10 were reduced in the STAF65γ-depleted cells, while all other TAFs analyzed were unaffected (Fig. 1B, compare lanes 1 and 2). Significantly, immunoprecipitation of TFIID with TBP and TAF5 antibodies and analysis of associated proteins by Western blotting indicated that STAF65γ depletion did not affect the association of TBP and TAF5 with each other and with TAF1, TAF2, TAF4, TAF6, TAF8, TAF9, TAF10, and TAF12 (compare lanes 4 and 5 with, respectively, lanes 7 and 8). Thus, while STAF65γ depletion reduces the steady-state levels of TAF10, it does not affect the cellular levels of TAF8 (a histone fold partner of TAF10 in TFIID) nor the incorporation within TFIID of normal levels of TAF10 and all other TAFs tested. This result suggests that TFIID complexes are not significantly altered in STAF65γ-knockdown cells.
To determine whether STAF65γ depletion alters the STAGA complex, endogenous SPT3 was immunoprecipitated from extracts of the STAF65γ-knockdown HeLa cells characterized above and from control HeLa cells (i.e., stably transfected with an empty pSUPER vector) and associated STAGA subunits were analyzed by Western blotting. As expected, SPT3 was associated with GCN5, STAF65γ, TAF9, TAF10, and TAF12 in control HeLa cells (Fig. 1C, lane 3). In STAF65γ-knockdown cells, however, SPT3 did not interact efficiently with GCN5, TAF9, TAF10, or TAF12 (lane 4). Similarly, immunoprecipitation of endogenous TAF9 indicated its reduced association with GCN5 and SPT3 in STAF65γ-knockdown cells (lane 6). Thus, in contrast to TFIID, the incorporation of TAF9, TAF10, and TAF12 within STAGA requires STAF65γ. To analyze additional STAGA components and avoid possible disruption of complexes by antibodies recognizing endogenous epitopes, STAF65γ was also knocked down in HeLa S3 cell lines that express Flag-tagged SPT3 (fh:SPT3 cells) or GCN5 (fh:GCN5 cells) and immunoprecipitation was performed with the Flag antibody (Fig. 1D). Immunoprecipitation of Flag-SPT3 from cells expressing the STAF65γ shRNA resulted in the expected reduced coprecipitation of STAF65γ but also in a proportionally reduced association of all the other STAGA subunits tested, including TRRAP, GCN5, TAF5L, and TAF9 (lane 5 versus lane 6). Similarly, immunoprecipitation of Flag-GCN5 from STAF65γ-depleted cells resulted in reduced coprecipitation of SPT3, TAF9, and, to a lesser extent, TRRAP and TAF5L (lane 7 versus lane 8). These results demonstrate that STAF65γ is important for the structural integrity of the STAGA complex in human cells and specifically indicate that STAF65γ is essential for the association of TAF9, SPT3, and GCN5 with each other and of SPT3 with TAF10, TAF12, TAF5L, and TRRAP but appears to be only partially required for GCN5 association with TRRAP and TAF5L. These binary interactions and their dependency on STAF65γ are summarized schematically in Fig. 1E. In addition, these results suggest that STAF65γ knockdown may destabilize its histone fold partner TAF10 without affecting the fraction of cellular TAF10 associated with TAF8/TFIID. Thus, we conclude that SPT-TAF (STAGA type) but not TBP-TAF (TFIID) complexes are selectively disrupted in STAF65γ-knockdown cells.
Function of STAF65γ in cell proliferation and MYC-activated transcription.
We have shown previously that STAGA interacts with the transactivation domain (TAD)/transformation domain of the MYC oncoprotein in cells and in vitro and that TRRAP and GCN5 cooperatively stimulate the transcription-activating function of MYC TAD fused to the Gal4 DNA-binding domain (44). To further address the role of the cellular STAGA complex in native MYC functions in human cells, we tested whether STAF65γ depletion affects cell proliferation and transcription of natural MYC-dependent genes. As shown in the colony formation assays of Fig. 2A, transfection of the STAF65γ shRNA vector (but not a control GL2 shRNA vector targeting firefly luciferase) specifically reduced the growth of HeLa cell colonies but had no effect on U2OS osteosarcoma cells. Similarly, the knockdown of MYC with a specific shRNA vector did not affect U2OS cell growth (data not shown). Note that the shRNA vector efficiently knocked down STAF65γ in U2OS cells (data not shown). Cell count/growth curves verified the reduced proliferation rate of STAF65γ-knockdown cell lines as compared to parental HeLa cells, with no apparent reduction in cell viability (data not shown). Importantly, the inhibition of HeLa cell proliferation was not due to a nonspecific “off-target” effect of the shRNA used. Indeed, the STAF65γ shRNA did not inhibit the growth of HeLa cells that express an ectopic Flag-STAF65γ “recoded” gene/cDNA (fh:STAF65γ “recoded”) carrying substitutions within the shRNA target sequence (Fig, 2B, left panel, compare two bottom plates). These substitutions effectively prevented the knockdown of the “recoded” Flag-STAF65γ protein (Fig. 2B, right panels, Western blots). In contrast, the STAF65γ shRNA inhibited proliferation of HeLa cells expressing an ectopic wild-type fh:STAF65γ gene (Fig. 2B, left panel, compare top two plates). The above results suggested that human STAF65γ (and thus the integrity of STAGA) is not generally required for growth and viability of all cell types but may have cell/context-dependent and gene-specific functions in cell proliferation, perhaps by functioning (in part) as a transcription cofactor for MYC and/or other regulators of cell proliferation.
FIG. 2.
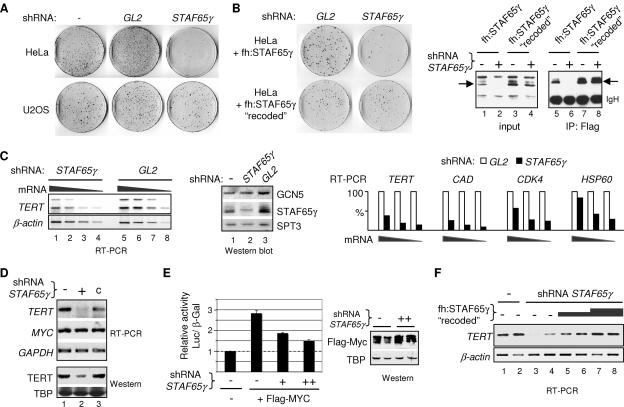
STAF65γ is required for proliferation of HeLa cells and expression of MYC target genes and mediates MYC activation of TERT transcription. (A) Colony formation assays with HeLa and U2OS cells transfected with the empty pSUPER vector (−) or the shRNA-expressing vectors pSUPER-GL2 (GL2) and pSUPER-STAF65γ828 (STAF65γ). A representative result of three experiments done in duplicate is shown. (B) Colony formation assay as in panel A but with two HeLa cell lines stably expressing ectopic Flag-tagged STAF65γ (top) or STAF65γ “recoded” (bottom), as indicated. Right panels are Western blots probed with the Flag antibody of total extracts (input, lanes 1 to 4) and anti-Flag immunoprecipitated proteins (IP: Flag, lanes 5 to 8) from these two cell lines after transient transfection with pSUPER (−) or pSUPER-STAF65γ828 (+). Arrows point to the Flag-STAF65γ proteins. IgH, immunoglobulin heavy chain. (C) Knockdown of STAF65γ in HeLa cells specifically reduces TERT, CAD CDK4, and HSP60 mRNA levels. HeLa cells were transiently transfected with pSUPER-STAF65γ828 (STAF65γ) or pSUPER-GL2 (GL2). TERT (two alternative splicing variants) and β-actin transcripts were analyzed by RT-PCR of serial dilutions of total cDNA (mRNA); a negative image of an ethidium bromide-stained agarose gel is shown (left panel; RT-PCR). Whole-cell extracts from the above transfected cells (shRNA: STAF65γ and GL2) and from untransfected cells (−) were also analyzed by Western blotting with the indicated antibodies (middle panel, Western blot, lanes 1 to 3). The bar graph shows the quantitation of TERT, CAD, CDK4, and HSP60 transcripts in transfected cells by RT-PCR and serial dilution of total cDNA (mRNA). RT-PCR signals in STAF65γ shRNA-expressing cells (black bars) and in GL2 control-transfected cells (white bars) were normalized to β-actin signals at each dilution, and the normalized values for GL2 cells were arbitrarily set to 100% (white bars). (D) RT-PCR and Western blot analyses of whole-cell extracts from parental HeLa S3 (−), STAF65γ-knockdown HeLa S3 (+; clone 1), and stably transfected control HeLa S3 (c) cell lines shown in Fig. 1A. (E) Role of endogenous STAF65γ in TERT promoter activation by MYC. HeLa cells were transiently transfected with a TERT(−211/+41)-luciferase reporter gene construct and the pCMV-β-galactosidase reference gene and with expression vectors for Flag-MYC and the STAF65γ shRNA (+ and ++) or with corresponding empty vectors (−), as indicated. Relative luciferase activities (normalized to β-galactosidase) are the means ± standard deviation from three independent experiments performed in duplicate. Corresponding whole-cell extracts of transfected cells (duplicates) were analyzed by Western blotting (right panel; Western) with anti-Flag (Flag-MYC) and anti-TBP antibodies. (F) A “recoded” Flag-STAF65γ restores endogenous TERT transcription in STAF65γ-knockdown cells. Adherent HeLa cell lines stably transfected with the empty pSUPER vector (lanes 1 and 2) or with the pSUPER-STAF65γ828 (shRNA STAF65γ) vector (clone 7; lanes 3 to 8) were transiently transfected with the empty pIRESneo vector (lanes 1 to 4) or increasing amounts of pIRESneo-fh:STAF65γ “recoded”: 2 μg (lanes 5 and 6) and 4 μg (lanes 7 and 8). TERT and β-actin transcripts were analyzed by RT-PCR.
The possible requirement of STAF65γ for MYC-dependent gene activation was tested by RT-PCR in HeLa cells transiently transfected with the STAF65γ shRNA vector (or the GL2 shRNA vector, as control). STAF65γ knockdown decreased the levels of specific transcripts from several genes that are directly activated by MYC, including TERT (6- to 8-fold reduction), CAD (7- to 12-fold reduction), CDK4 (∼4-fold reduction), and HSP60 (∼3- to 4-fold reduction) but did not affect β-actin transcripts (Fig. 2C). Similarly, both TERT mRNA and protein levels were reduced in stable STAF65γ-knockdown HeLa cells, although the reduction in total TERT protein was less pronounced (Fig. 2D). Furthermore, in transient cotransfection assays the STAF65γ shRNA vector specifically reduced MYC activation of a minimal TERT promoter (−211 to +40) containing the two essential MYC/E-box elements linked to the luciferase reporter gene, but did not affect the activity of the cotransfected CMV-Flag-MYC and CMV-β-Gal genes (Fig. 2E). Similar results were obtained in the stable STAF65γ-knockdown HeLa cell lines (data not shown). Moreover, transcription of the endogenous TERT gene in STAF65γ-knockdown HeLa cells was stimulated in a dose-dependent manner by transfection of the “recoded” fh:STAF65γ expression vector (Fig. 2F). Altogether these results demonstrate a specific requirement of STAF65γ for transcription of several MYC-dependent genes, including MYC-dependent transcription activation of TERT in HeLa cells.
MYC recruits STAGA to the endogenous TERT promoter in human cells.
To test whether STAGA binds to the native TERT promoter in a MYC-dependent manner, we performed ChIP experiments with MYC-knockdown HeLa cells stably transfected with a pSUPER-MYC-shRNA vector (22). As shown previously (22), MYC-knockdown cells have reduced MYC expression and TERT transcription (Fig. 3A, RT-PCR and Western panels). In control HeLa cells (stably transfected with the empty pSUPER vector), MYC, SPT3, TAF9, and GCN5 bound specifically to the TERT promoter, but not to a β-globin locus used as a background reference (Fig. 3A, middle panel, ChIP, lanes 5, 7, 9, and 11). This demonstrates that the native human TERT promoter is indeed a direct target of STAGA in these cells. In contrast, the association of MYC, SPT3, TAF9, and GCN5 with the TERT promoter was drastically reduced in MYC-knockdown cells (lanes 6, 8, 10, and 12). This defective STAGA recruitment in MYC-knockdown cells was specific since several other factors, including components of the basal transcription machinery bound to the TERT and GAPDH promoters (see below) and transcription of the β-actin and GAPDH genes was unaffected (Fig. 3A, left panel) (22). To further verify the essential role of MYC in STAGA recruitment, Flag-MYC was transiently overexpressed in U2OS cells, which have their endogenous TERT gene repressed by MAD1, a member of the MAD family of E-box-binding MYC antagonists (43). MYC overexpression stimulated TERT transcription (Fig. 3B, RT-PCR panel, lane 2) and induced binding of MYC, SPT3, and TAF9 to the endogenous TERT promoter, as determined by ChIP (right panel, lanes 4, 6, and 8).
FIG. 3.
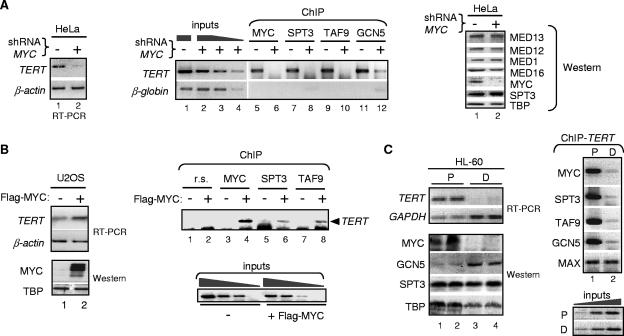
MYC recruits STAGA to the endogenous TERT promoter in human cells. (A) MYC knockdown decreases TERT transcription and recruitment of STAGA to the TERT promoter. MYC-knockdown (+ shRNA MYC) and control (−) HeLa cells were analyzed by RT-PCR (left panel) and by ChIP with MYC, SPT3, TAF9, and GCN5 antibodies, as indicated (middle panel). Lanes 1 to 4 show PCRs with serial dilutions of input chromatin (inputs). The TERT promoter region between −226 and +120 (containing the two E-boxes at −187 and +28) was amplified with specific primers (22). A region of the β-globin gene third intron was amplified as a nonspecific/background reference. Negative images of ethidium bromide-stained agarose gels are shown. A Western blot of whole-cell extracts from parental (−) and MYC-knockdown (+) HeLa S3 cells was probed with specific antibodies to the indicated proteins (right panel; Western). (B) Overexpression of Flag-MYC induces TERT transcription and STAGA binding to the endogenousTERT promoter in U2OS cells. Cells were transiently transfected with empty (−) or Flag-MYC (+) expression vectors and analyzed by RT-PCR and Western blotting (with MYC and TBP antibodies), and by ChIP, as described above. Preimmune rabbit serum (r.s.) was used in the control ChIP assays (lanes 1 and 2). (C) Proliferating (P) HL-60 cells were induced to differentiate (D), transcripts for TERT and GAPDH were analyzed by RT-PCR (top panel), and whole-cell extracts were analyzed by Western blotting with the indicated antibodies (bottom panel). Right panels show ChIP analyses of the TERT promoter with chromatin from P and D cells using antibodies to MYC, SPT3, TAF9, GCN5, and MAX.
To test whether STAGA binding to the TERT promoter is associated with MYC-dependent TERT transcription and cell proliferation in a different human cell line, we performed ChIP experiments in HL-60 cells, a MYC-dependent acute promyelocytic leukemia cell line, both under proliferating conditions and after induction of differentiation along the granulocytic pathway (79). The endogenous TERT gene was transcribed in proliferating HL-60 cells before induction (Fig. 3C, RT-PCR panel, lanes 1 and 2), and its promoter was bound by the MYC/MAX complex (as expected) but also by the STAGA components SPT3, TAF9, and GCN5 (ChIP panel, lane 1). After induced differentiation, MYC protein and TERT mRNA levels decreased (RT-PCR and Western blot panels, lanes 3 and 4) and this correlated with a reduced binding of MYC, SPT3, TAF9, and GCN5 to the promoter (ChIP panel, lane 2). Note that MAX binding was unaffected, as expected from the replacement of MYC/MAX by MAD/MAX complexes on the TERT promoter during HL-60 cell differentiation (79). Altogether, the above results demonstrate a MYC-dependent recruitment of STAGA to the endogenous TERT promoter in human cells and are consistent with the requirement of STAF65γ for HeLa cell proliferation and MYC-dependent activation of TERT transcription and with the previously described direct interaction of STAGA with MYC TAD (44).
Selective role of STAF65γ in MYC-dependent recruitment of SPT3, TAF9, and “core” Mediator components to the TERT promoter in human cells.
The dependency on STAF65γ for TERT transcription and the MYC-directed binding of STAGA components to the TERT promoter suggested a critical STAF65γ-dependent function of the STAGA complex on this promoter. To further address this, we analyzed by ChIP the recruitment of STAGA subunits and components of the transcription machinery and the acetylation state of histones on the native TERT promoter in control and STAF65γ-knockdown HeLa cell lines. As shown in Fig. 4A, the knockdown of STAF65γ did not affect the binding of MYC (lanes 9 and 10) or its associated cofactors GCN5 (lanes 15 and 16), TRRAP (lanes 17 and 18), and p300 (lanes 19 and 20), which are all recruited by MYC to the TERT promoter in vivo (Fig. 3A) (22, 56). Consistent with this, the levels of acetylated histones H3 and H4 were also unaltered (lanes 21 to 24). In stark contrast, no detectable binding of TAF9 (lane 12) or SPT3 (lane 14) was observed in STAF65γ-knockdown cells, despite normal TAF9 and SPT3 expression in these cells (see Fig. 1A). The selective association of both TRRAP and GCN5, but not SPT3 or TAF9, with the TERT promoter in STAF65γ-depleted cells is consistent with the direct interaction of MYC with both TRRAP and GCN5 in vitro (60; N. Zhang, S. Lo, and E. Martinez, unpublished data). Note that this selective subunit association could reflect the binding of TRRAP and GCN5 in complexes unrelated to STAGA and/or could indicate the association of TRRAP and GCN5 as part of a partial STAGA complex that lacks the STAF65γ-dependent group of “SPT-TAF” components (i.e., STAF65γ, SPT3, TAF9, and associated histone fold partners). These two scenarios are not necessarily mutually exclusive, and the latter possibility would be in accord with the requirement of STAF65γ for the association of TRRAP and GCN5 with SPT3 and TAF9 (Fig. 1E). In any case, these results indicate that STAF65γ is required for the normal recruitment of the STAGA complex—or minimally its STAF65γ-dependent SPT-TAF subunits—to the native TERT promoter in human cells and suggest a STAF65γ-dependent transactivating function of STAGA distinct from stimulation of histone acetylation.
FIG. 4.
STAF65γ is required for MYC-dependent recruitment of selected STAGA and core Mediator components to the endogenous TERT promoter in human cells. (A to D) Equivalent amounts of chromatin from control pSUPER-transfected (−), STAF65γ-knockdown (shRNA STAF65γ + in panels A to C) or MYC-knockdown (shRNA MYC + in panel D) HeLa S3 cell lines were used in ChIP experiments with specific antibodies or preimmune rabbit serum (r.s.), as indicated. AcH3 and AcH4 are anti-acetylated histone H3 and H4 antibodies. PCR analyses were performed with primers specific for the TERT and GAPDH promoters and for a region in the third intron of the β-globin gene, as in Fig. 3.
Since SAGA in yeast directs the recruitment of the general transcription machinery to specific promoters via both GCN5-dependent and GCN5-independent pathways and yeast SPT3 plays an essential role in the binding of TBP to GCN5-independent promoters (see introduction), we analyzed the effect of STAF65γ knockdown on TFIID and Pol II recruitment to the TERT promoter. Unexpectedly, and in contrast to the strong inhibitory effect of STAF65γ knockdown on both TERT transcription and MYC-dependent recruitment of SPT3 and TAF9, the binding of the TFIID-specific subunits TAF1 and TAF5 was not affected and only a marginal reduction in TBP and Pol II binding was observed (Fig. 4B, lanes 15 to 22). Thus, these results indicate that STAGA, or minimally its STAF65γ-dependent group of SPT-TAF components, is not essential for the loading of TFIID and Pol II on the native human TERT promoter.
We analyzed similarly the recruitment of subunits of the Mediator complex. Unexpectedly, while a significant binding of MED1, MED17, MED12, and MED13 was observed on the MYC-independent GAPDH promoter, a preferential binding of MED1 and MED17 (relative to MED12 and MED13) was observed on the TERT promoter (Fig. 4C, lanes 11, 13, 15, and 17 [see also below]). Interestingly, the association of MED1 and MED17 with the TERT promoter was strongly reduced in STAF65γ-knockdown cells (Fig. 4C, lanes 12 and 14), while the weak binding of MED12 and MED13 was not affected (lanes 15 to 18). Note that the expression of MED subunits was unaltered in STAF65γ-knockdown cells (e.g., Fig. 5A). In contrast, all MED subunits analyzed bound to the GAPDH promoter similarly in control and STAF65γ-knockdown cells (Fig. 4C, lanes 11 to 18). In accord with the above results, the knockdown of MYC inhibited the binding of MED1, MED16, and MED17 to the TERT promoter but not to the GAPDH promoter (Fig. 4D, lanes 12, 14, and 16), while only a modest reduction in the binding of TBP and Pol II and in the level of histone H3 acetylation was observed (lanes 22, 24, and 28). Note that the expression of these MED subunits was also unaffected in MYC-knockdown cells (see Fig. 3A, Western panel, lane 2). Surprisingly, acetylation of histone H4 was not significantly altered in MYC-knockdown cells (Fig. 4D, lane 26) despite the fact that MYC recruits p300, GCN5, and other TRRAP-associated HATs such as Tip60 (Fig. 3) (9, 22-24, 56); however, the potential association of Tip60 with the TERT promoter remains to be shown. The above results indicate that MYC is required for recruitment of both STAGA and core Mediator subunits (MED1, MED16, and MED17) to the TERT promoter but not the variably associated MED12 and MED13 components specific to large Mediator complexes. Interestingly, the association of MED12 and MED13 relative to the other MED subunits differed on the TERT and GAPDH promoters. Furthermore, these data demonstrate that STAF65γ is essential for MYC-directed recruitment of SPT-TAF and core Mediator components but not for histone acetylation or binding of TFIID and Pol II on the TERT promoter in HeLa cells.
FIG. 5.
Efficient interaction of MYC with core Mediator requires STAF65γ and an intact TAD. (A) STAF65γ-dependent association of STAGA and core Mediator subunits with endogenous MYC/MAX in HeLa cells. MAX was immunoprecipitated with a specific antibody (IP: MAX) from whole-cell extracts (input) of control pSUPER-transfected (−) and STAF65γ-knockdown (+) HeLa S3 cell lines, and associated proteins were analyzed by Western blotting with specific antibodies to the indicated proteins. (B) HEK293 cells were transfected with an empty pCbS vector (−) or pCbS-Flag-MYC (+), and whole-cell extracts (input) were immunoprecipitated with the Flag antibody (IP: FLAG) and analyzed by Western blotting. (C) Western blot as in panel B, but with HEK293 cells transfected with pCbS-Flag-MYC (WT) or Δ1-110 (Δ110) together with pCbS-HA-MAX. The top stripe (MED12 + 13) was probed with both MED12 and MED13 antibodies. The lack of MAX signal in “input” (lanes 1 to 3) is due to the short exposure used to prevent saturation of the signals in IP: FLAG lanes 5 and 6. (D and E) GST-pulldown experiments were performed with equivalent amounts of GST, GST-VP16, GST-MYC, or GST-MYC(1-263) resins (as indicated) and with nuclear extracts (NE) from parental HeLa S3 cells (D) and pSUPER-transfected (−) or STAF65γ-knockdown (+) HeLa S3 cell lines (E). Input nuclear extracts (input) and proteins bound to GST, GST-VP16, and GST-MYC were analyzed by Western blotting.
STAGA is required for the interaction of selected Mediator subunits with MYC in human cells and in vitro.
Since both MYC and STAF65γ were required for core Mediator recruitment to the TERT promoter, we analyzed the interaction of MYC with Mediator components in human cells and the possible role of STAF65γ by coimmunoprecipitation. To avoid any possible displacement of MYC-associated factors by MYC-binding antibodies, a MAX-specific antibody was used to immunoprecipitate endogenous MYC/MAX heterodimers from control and STAF65γ-knockdown HeLa cells. As shown in Fig. 5A, MYC/MAX interacted with various STAGA subunits, as previously reported (44), and with MED1, MED16, and MED17, but not with MED13 in control HeLa cells (lane 3). Interestingly, MYC/MAX interaction with both STAGA and Mediator was significantly impaired in STAF65γ-knockdown cells, while the association of MYC with MAX was unaffected (lane 4). To verify the interaction of Mediator with MYC, Flag-MYC was expressed in HEK293 cells and immunoprecipitated with the Flag antibody. Consistent with the above results, we observed a Flag-MYC interaction with MED1, MED16, and MED17 and more weakly with MED14 but not with MED13 (Fig. 5B). Note that a very weak interaction of MED12 and MED13 was occasionally observed after longer exposure of the Western blots (e.g., Fig. 5C, lane 5, top stripe). To further test the requirement of the N-terminal MYC TAD region for Mediator binding, a Flag-MYCΔN110 deletion mutant that lacks residues 1 to 110 important for STAGA binding (44)—and mimics the shorter natural “MYC-S” variants (67)—was cotransfected with MAX and analyzed as described above. Flag-MYCΔN110 had a reduced ability to interact with Mediator components compared to full-length Flag-MYC (Fig. 5C, lane 6). Interestingly, a residual but significant interaction of Mediator subunits with Flag-MYCΔN110 was reproducibly observed, indicating that Mediator can also bind to MYC independently of TAD residues 1 to 110. We found that the region of TAD residues 1 to 110 of MYC plays a more important role for STAGA and Mediator binding when MAX is cotransfected with MYC (data not shown), suggesting that this TAD region contributes to cofactor recruitment in the context of physiological MYC/MAX heterodimers within cells.
To determine whether the N-terminal TAD of MYC is sufficient for recruitment of Mediator and to analyze the role of STAF65γ, in vitro pull-down assays were performed with GST-MYC fusion proteins and nuclear extracts of wild-type and STAF65γ-knockdown cells. The full-length MYC protein fused to GST (GST-MYC), but not GST alone, interacted with STAGA (as reported previously [44]) and with the Mediator component MED17, but not significantly with MED12, MED13, and MED14 in wild-type HeLa nuclear extracts (Fig. 5D, lane 3). Similarly, a GST-MYCN263 fusion protein that contains the N-terminal 1 to 263 TAD residues of MYC, but lacks the DNA-binding and MAX-interacting “basic helix-loop-helix” domain, was sufficient to recruit STAGA (GCN5, STAF65γ, SPT3, and TAF9) and the core Mediator subunits MED1, MED16, and MED17 but did not significantly interact with MED12, MED13, and MED14 (Fig. 5D, lane 6, and Fig. 5E, lane 4). This differential binding of Mediator components was activation domain specific, as all MED subunits tested, including MED12, MED13, and MED14, interacted efficiently with the VP16 activation domain (GST-VP16; Fig. 5D, lane 5). In contrast, in nuclear extracts of STAF65γ-knockdown cells, GST-MYCN263 did not detectably bind SPT3 or TAF9 and the interaction with MED1, MED16, and MED17 was significantly reduced, although not completely abolished (Fig. 5E, lane 5). Notably, the binding of GCN5 and TBP to GST-MYCN263 was independent of STAF65γ (lane 4 versus 5). Thus, the MYC TAD is sufficient to recruit selected MED components in nuclear extracts (i.e., MED1, MED16, and MED17 but not, or less stably, MED12, MED13, and MED14), and this depends on STAF65γ but does not require MYC association with MAX or binding to DNA/chromatin. Altogether, these results show that MYC interacts with STAGA and specific Mediator core components in human cells and in vitro in a manner that is dependent on STAF65γ and the MYC TAD 1-110 region.
STAGA and Mediator complexes physically interact in human cells and in vitro.
To address the mechanisms by which STAF65γ facilitates Mediator recruitment by MYC, we analyzed the possible physical interaction of STAGA and Mediator complexes in HeLa cells. Mediator complexes immunoprecipitated with a specific MED1 antibody were found associated with the STAGA subunits GCN5, STAF65γ, and SPT3 (Fig. 6A, lane 3). Conversely, immunoprecpitation of STAGA complexes with SPT3 and TAF9 antibodies indicated their association with MED1, MED16, and MED17 (data not shown). The association of STAGA with Mediator was not due to formation of insoluble aggregates as soluble Flag-GCN5 and Flag-STAF65γ complexes eluted from the anti-FLAG M2 resin under nondenaturing conditions (i.e., by competition with excess FLAG peptide) also contained MED1, MED16, and MED17 (Fig. 6B). It is worth noting that these STAGA-Mediator interactions were observed for complexes washed in 180 mM NaCl but generally not for highly purified complexes isolated and washed at 300 mM or higher salt concentrations (29, 51); nonetheless, small amounts of MED17 were occasionally detected by Western blotting in highly purified STAGA complexes, consistent with the above results (E. Martinez, unpublished observations). To further test the role of STAF65γ on the intracellular interaction between STAGA and Mediator, Flag-SPT3 was immunoprecipitated from fh:SPT3 cells and cell derivatives that express the STAF65γ shRNA (described in Fig. 1C). Consistent with the above results, Flag-SPT3/STAGA was associated with MED16 and MED17 but not with MED12 or MED13 (Fig. 6C, lane 5). In STAF65γ-knockdown cells, however, the interaction of Flag-SPT3 with MED16 and MED17 was significantly reduced (Fig. 6C, lane 6). These results demonstrate an interaction of STAGA and Mediator complexes in human cells that is dependent on STAF65γ.
To determine whether STAGA and Mediator complexes directly interact, highly purified STAGA (Flag-SPT3) and Mediator (Flag-Nut2) complexes (Fig. 6D) were combined in vitro and immunoprecipitated with a specific MED1 antibody. As shown in Fig. 6E, STAGA components were specifically coimmunoprecipitated with MED1-containing Mediator complexes (lane 5). Consistent with this, GST pull-down experiments with purified Mediator and STAGA indicated an increased binding of MED1, GCN5, and SPT3 to GST-MYCN263 when both complexes were present and also the ability of specific Mediator components (i.e., MED16 and, weakly, MED1 but not CDK8) to interact directly with MYC TAD residues 1 to 263 (Fig. 6F). Collectively, these results demonstrate a direct interaction of STAGA with a MED1-containing Mediator complex and suggest that both complexes interact with MYC TAD in a partially interdependent manner.
DISCUSSION
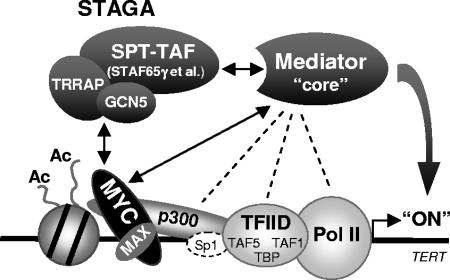
In this study, we have uncovered a novel function of the human STAGA coactivator complex in physical recruitment of Mediator and stimulation of MYC-dependent transcription and cell proliferation, which requires the integrity of the complex or minimally its STAF65γ-dependent SPT-TAF module (which includes SPT3, TAF9, TAF10, TAF12, and associated histone fold partners; Fig. 1E). We have shown that STAF65γ is required for activation of TERT transcription by MYC and for the association of both SPT3 (STAGA) and MYC with Mediator in human cells. Consistent with this, we found that STAGA and Mediator complexes directly interact and bind in a partially interdependent manner to the TAD of MYC in vitro. Collectively, our results suggest a model in which MYC interacts with the STAGA complex via its TRRAP and/or GCN5 subunits, while the STAF65γ-dependent SPT-TAF components physically recruit a core Mediator complex (i.e., devoid of MED12 and MED13) to the TERT promoter and direct transcription activation at a step(s) mostly distinct from, and likely subsequent to, the loading of TFIID and Pol II (Fig. 7).
FIG. 7.
Model for STAGA-dependent transcription activation of TERT by MYC involving recruitment and/or stabilization of a core Mediator complex at the promoter. Double arrows indicate direct interactions between MYC, STAGA, and Mediator described here and previously (44). Dashed lines indicate previously reported interactions of Mediator with p300, TFIID, and Pol II (see references 7 and 47 and references therein), which could also take place on the TERT promoter but are not sufficient for the stable recruitment of core Mediator in the absence of either MYC or the STAF65γ-dependent SPT-TAF subunits of STAGA. The binding of Sp1/3 to TERT GC-boxes (dashed oval Sp1) is expected but was not tested here. “(Ac)” indicates acetylation of nucleosomal histones H3 and H4, which is independent of the STAF65γ/SPT-TAF components of STAGA (see also Discussion).
Similar to the nonessential gene-specific function of Spt7 (SAGA) in yeast, we have shown that STAF65γ plays a structural role within the human STAGA complex and is not essential for viability of U2OS and HeLa cells or for normal growth of U2OS cells. However, STAF65γ is important for proliferation of HeLa cells, which depend on MYC for growth, suggesting the intriguing possibility that STAF65γ (or an associated SPT-TAF component) may have MYC- and/or cell type-specific functions. The exact nature of this cell-type specificity, however, remains to be elucidated, and it will be important to determine whether STAF65γ is required specifically for MYC-induced cell proliferation and transformation. Our results demonstrate that STAF65γ is required for MYC-dependent activation of TERT transcription (as well as for transcription of several other MYC target genes) and for the association of SPT3, TAF9, and probably other components of the SPT-TAF group with the TRRAP and GCN5 subunits. Thus, TRRAP and GCN5 may belong to a separate module(s) in the complex (e.g., a “TRRAP/domain I” module and a “GCN5/ADA2/ADA3” module) (49, 72). Consistent with this, MYC directly interacts with both TRRAP (60) and GCN5 (N. Zhang, S. Lo and E. Martinez, unpublished results) and recruits TRRAP to various promoters (23, 24) and GCN5 (as shown here) to the TERT promoter independently of STAF65γ. It is possible that, in addition to MYC, other promoter-associated factors, perhaps including acetylated histones, might contribute to the stable binding of TRRAP and/or GCN5 to the TERT promoter. However, MYC is essential for the stable recruitment of all STAGA components analyzed and interacts directly with a purified STAGA complex in vitro (44). Our results do not exclude the possibility that the STAF65γ-independent binding of TRRAP and GCN5 to the TERT promoter might instead (or in addition) reflect the recruitment by MYC of other complexes, i.e., distinct from STAGA-type complexes.
In contrast to the STAF65γ-independent recruitment of TRRAP and GCN5, the MYC-directed recruitment of SPT3 and TAF9 to the TERT promoter is strictly dependent on STAF65γ. While STAF65γ and SPT3 are specific to STAGA-type complexes, TAF9 is shared by STAGA-type and TFIID complexes: note that the polyclonal TAF9 antibodies used also recognize the highly similar TAF9b (formerly TAF9L) paralog that coexists with TAF9 in both TFIID and STAGA/TFTC (25). Since we have shown that STAF65γ depletion selectively affects the association of TAF9 with STAGA but not with TFIID (Fig. 1), these results suggest the intriguing possibility that TAF9 is recruited to the TERT promoter as part of the SPT-TAF module of STAGA. This would be consistent with increasing evidence indicating the existence of structurally and functionally distinct TFIID complexes, including TFIID complexes that lack TAF9 (reviewed in reference 55). Note, however, that we cannot formally exclude the possibility that TAF9 is associated with the TERT promoter (as part of TFIID) in STAF65γ-knockdown cells but cannot be detected by ChIP.
In addition to SPT3 and TAF9 components of STAGA, STAF65γ is also strictly required for MYC-dependent recruitment of core Mediator subunits (MED1 and MED17) to the TERT promoter. This supports observations in yeast indicating at least a partial dependency on SAGA for Mediator recruitment to certain yeast promoters (6, 38, 40, 41, 63). However, neither MYC nor STAF65γ is essential for TFIID or Pol II binding to the human TERT promoter in HeLa cells (although we note a moderate decrease in the association of TBP and Pol II). Moreover, STAF65γ is also dispensable for histone H3 and H4 acetylation. Altogether, these results suggest a role of the STAF65γ-dependent SPT-TAF components of STAGA in MYC-dependent TERT transcription by (at least in part) enhancing core Mediator association with the promoter and facilitating a transcription step(s) postrecruitment of TFIID and Pol II. This model is in accord with previous reports indicating that MYC activates transcription of TERT and other target genes after Pol II engagement, possibly by stimulating Pol II C-terminal domain phosphorylation and/or promoter clearance or elongation (8, 17, 20) and with recent studies indicating a role of yeast SAGA in transcription elongation (12, 27, 76, 78). Similarly, Mediator has also been shown to play a role postrecruitment of Pol II during the initiation phase of transcription in mouse cells (74), stimulates TFIIB recruitment and initiation/reinitiation in vitro (2, 45, 85), and physically and/or functionally associates with various elongation factors, including TFIIS (76), Brd4/P-TEFb (84), and DSIF (46), and with coding regions of yeast genes (1, 87). Clearly, it will be important to identify the steps downstream of TFIID/Pol II loading that are controlled by STAGA and core Mediator on the TERT promoter and the detailed activation mechanisms involved. The results presented here nonetheless provide the first evidence for a concerted function of STAGA and core Mediator in MYC-dependent transcription activation in human cells at a step mostly distinct from histone acetylation and binding of TFIID and Pol II to the promoter. Importantly, our results also indicate a partial dependency on MYC (but not STAF65γ) for histone H3 acetylation on the endogenous TERT promoter in HeLa cells, which is consistent with previous findings in different cells/systems (8, 56). These results suggest that MYC-dependent histone H3 acetylation does not require the STAF65γ-dependent SPT-TAF components of STAGA (or Mediator) but most likely depends on GCN5, p300/CBP, and/or other recruited HATs (8, 22, 23, 44, 52, 56). Thus, we propose that the STAGA complex may contribute, perhaps in a cell context-dependent manner, to the two distinct transactivating functions of MYC (i) at the level of chromatin acetylation and (ii) at subsequent steps after Pol II engagement, via separate TRRAP-GCN5 and SPT-TAF components, respectively.
We have demonstrated that MYC and STAF65γ (STAGA) are required for the recruitment of core Mediator components (i.e., MED1, MED16, and MED17) but not MED12 or MED13 to the endogenous TERT promoter in HeLa cells. This is consistent with our in vitro interaction assays indicating that, in contrast to core MED subunits, MED12 and MED13 in nuclear extracts and CDK8 in our affinity-purified Flag-Nut2 Mediator complexes interact poorly with MYC TAD. In addition, the interaction in vitro of MYC TAD with MED1 but not MED16, in purified Flag-Nut2/Mediator complexes, is enhanced by STAGA. This supports the reported heterogeneity of purified Flag-Nut2 MED10-containing Mediator complexes (86) and suggests that STAGA may facilitate the interaction of MYC TAD with selectively MED1-containing core complexes—e.g., those lacking the CDK8 module (which comprises MED12, MED13, cyclin C, and CDK8). Consistent with this, the preferential interaction of MYC TAD with core MED subunits in nuclear extracts was both activation domain specific (i.e., not observed with GST-VP16) and at least partially STAF65γ dependent. Note that, in contrast to MED12 and MED13, CDK8 in nuclear extracts interacts efficiently with MYC TAD, but this association is independent of STAF65γ (our data not shown) and of cyclin C (20) and thus might not reflect an interaction with a bona fide Mediator complex.
Our ChIP analyses have further uncovered a differential association of MED12 and MED13 (relative to core MED components) on the TERT and GAPDH promoters. Note that this differential association does not indicate the levels of the different MED subunits on either promoter. This contrasts with the reported uniform binding of Mediator components including subunits of the CDK8 module in genomewide location analyses in yeast (1, 87). However, this differential interaction of MED subunits is in agreement with two previous reports describing the recruitment to native human promoters of CDK8-containing and CDK8-lacking Mediator complexes by, respectively, repressive and active forms of C/EBPβ (54) and MED1-containing and MED1-deficient Mediator complexes by the estrogen receptor and p53, respectively (86). Thus, at least in human cells, the association of Mediator components with promoters appears to vary in a promoter-specific and regulated manner, consistent with the original biochemical characterization of different forms of human Mediator (see references 15, 47, 65, and 71 and references therein).
We have shown that purified STAGA and Mediator complexes directly interact and bind to MYC TAD both independently and, at least for specific components (i.e., MED1, GCN5, and SPT3), also in an interdependent manner. Thus, it is possible that MYC might also recruit specific Mediator components to promoters in vivo independently of STAGA and Mediator might help STAGA recruitment to certain human promoters. The latter possibility would be consistent with the observed dependency on Mediator for SAGA recruitment to certain promoters in yeast (63). Moreover, the interdependent recruitment of SAGA and Mediator to specific yeast promoters (6, 38, 40, 41, 63) strongly suggests analogous physical interactions of SAGA and Mediator complexes in yeast and highly conserved mechanisms for SAGA/STAGA-dependent transcription activation from yeasts to humans.
In addition to STAGA and core Mediator, MYC also recruits p300 to the TERT promoter (22) and this is independent of STAF65γ/STAGA and core Mediator (as shown here). How MYC coordinates the recruitment of p300, STAGA, and core Mediator remains to be determined. Beyond cooperative interactions, coactivator exchange mechanisms may be required as proposed for the concerted interactions of p300 and Mediator with PGC1-α and Gal4-VP16 (7, 73). In summary, our results support and extend the notion that at least certain HATs such as p300 (7) and HAT complexes such as STAGA (this report) can directly communicate with Mediator via physical interactions and suggest that eukaryotic SAGA/STAGA-type complexes may coordinate different steps in transcription activation: from chromatin modification and facilitated recruitment of TBP/TFIID and Pol II to subsequent steps in transcription that require components of the SAGA/STAGA-specific SPT-TAF module and interactions with a specific core Mediator complex.
Acknowledgments
We thank J. C. Barrett, R. Bernards, M. D. Cole, R. A. Currie, S. Malik, Y. Nakatani, R. G. Roeder, and L. Tora for generous gifts of reagents. We also thank R. G. Roeder and A. Gamper for communication of unpublished results and R. G. Roeder and S. Malik for stimulating discussions and comments on the manuscript. We also thank L. Tora for critical comments on the manuscript.
This work was supported by grant CA100464 from the National Institutes of Health.
Footnotes
Published ahead of print on 27 October 2007.
REFERENCES
- 1.Andrau, J. C., L. van de Pasch, P. Lijnzaad, T. Bijma, M. G. Koerkamp, J. van de Peppel, M. Werner, and F. C. Holstege. 2006. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22179-192. [DOI] [PubMed] [Google Scholar]
- 2.Baek, H. J., Y. K. Kang, and R. G. Roeder. 2006. Human Mediator enhances basal transcription by facilitating recruitment of TFIIB during preinitiation complex assembly. J. Biol. Chem. 28115172-15181. [DOI] [PubMed] [Google Scholar]
- 3.Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 236944-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 151935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, J. C., J. E. Choi, S. R. Lombardo, and M. Carey. 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23809-818. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard, C., J. Marquardt, A. Bras, R. H. Medema, and M. Eilers. 2004. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 232830-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchard, C., O. Dittrich, A. Kiermaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 152042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 27418285-18289. [DOI] [PubMed] [Google Scholar]
- 11.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 111301-1309. [DOI] [PubMed] [Google Scholar]
- 12.Carey, M., B. Li, and J. L. Workman. 2006. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19321-329. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, J. X., M. Gandolfi, and M. Ptashne. 2004. Activation of the Gal1 gene of yeast by pairs of ‘non-classical’ activators. Curr. Biol. 141675-1679. [DOI] [PubMed] [Google Scholar]
- 15.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30250-255. [DOI] [PubMed] [Google Scholar]
- 16.Cowling, V. H., and M. D. Cole. 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 16242-252. [DOI] [PubMed] [Google Scholar]
- 17.Cowling, V. H., and M. D. Cole. 2007. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 272059-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demeny, M. A., E. Soutoglou, Z. Nagy, E. Scheer, A. Janoshazi, M. Richardot, M. Argentini, P. Kessler, and L. Tora. 2007. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS ONE 2e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 132940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 27648562-48571. [DOI] [PubMed] [Google Scholar]
- 21.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 61319-1331. [DOI] [PubMed] [Google Scholar]
- 22.Faiola, F., X. Liu, S. Lo, S. Pan, K. Zhang, E. Lymar, A. Farina, and E. Martinez. 2005. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol. Cell. Biol. 2510220-10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 152069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frontini, M., E. Soutoglou, M. Argentini, C. Bole-Freysot, B. Jost, E. Scheer, and L. Tora. 2005. TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol. Cell. Biol. 254638-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gangloff, Y.-G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol. 211841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer, and A. G. Hinnebusch. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 2531-42. [DOI] [PubMed] [Google Scholar]
- 28.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 111640-1650. [DOI] [PubMed] [Google Scholar]
- 29.Gu, W., S. Malik, M. Ito, C.-X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 397-108. [DOI] [PubMed] [Google Scholar]
- 30.Guermah, M., K. Ge, C.-M. Chiang, and R. G. Roeder. 2003. The TBN protein, which is essential for early embryonic mouse development, is an inducible TAFII implicated in adipogenesis. Mol. Cell 12991-1001. [DOI] [PubMed] [Google Scholar]
- 31.Hardy, S., M. Brand, G. Mittler, J. Yanagisawa, S. Kato, M. Meisterernst, and L. Tora. 2002. TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J. Biol. Chem. 27732875-32882. [DOI] [PubMed] [Google Scholar]
- 32.Helmlinger, D., S. Hardy, G. Abou-Sleymane, A. Eberlin, A. B. Bowman, A. Gansmüller, S. Picaud, H. Y. Zoghbi, Y. Trottier, L. Tora, and D. Devys. 2006. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 4e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 172648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13573-585. [DOI] [PubMed] [Google Scholar]
- 35.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30235-239. [DOI] [PubMed] [Google Scholar]
- 36.Lang, S. E., S. B. McMahon, M. D. Cole, and P. Hearing. 2001. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 27632627-32634. [DOI] [PubMed] [Google Scholar]
- 37.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 151946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larschan, E., and F. Winston. 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, K. K., and J. L. Workman. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8284-295. [DOI] [PubMed] [Google Scholar]
- 40.Lemieux, K., and L. Gaudreau. 2004. Targeting of Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAFIIs, and RNA polymerase II. EMBO J. 234040-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leroy, C., L. Cormier, and L. Kuras. 2006. Independent recruitment of mediator and SAGA by the activator Met4. Mol. Cell. Biol. 263149-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 43.Lin, S. Y., and S. J. Elledge. 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113881-889. [DOI] [PubMed] [Google Scholar]
- 44.Liu, X., J. Tesfai, Y. A. Evrard, S. Y. Dent, and E. Martinez. 2003. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J. Biol. Chem. 27820405-20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik, S., H. J. Baek, W. Wu, and R. G. Roeder. 2005. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol. Cell. Biol. 252117-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik, S., M. J. Barrero, and T. Jones. 2007. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl. Acad. Sci. USA 1046182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30256-263. [DOI] [PubMed] [Google Scholar]
- 48.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5753-760. [DOI] [PubMed] [Google Scholar]
- 49.Martinez, E. 2002. Multi-protein complexes in eukaryotic gene transcription. Plant Mol. Biol. 50925-947. [DOI] [PubMed] [Google Scholar]
- 50.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 27323781-23785. [DOI] [PubMed] [Google Scholar]
- 51.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 216782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuguchi, G., A. Vassilev, T. Tsukiyama, Y. Nakatani, and C. Wu. 2001. ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J. Biol. Chem. 27614773-14783. [DOI] [PubMed] [Google Scholar]
- 54.Mo, X., E. Kowenz-Leutz, H. Xu, and A. Leutz. 2004. Ras induces mediator complex exchange on C/EBPβ. Mol. Cell 13241-250. [DOI] [PubMed] [Google Scholar]
- 55.Muller, F., M. A. Demeny, and L. Tora. 2007. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 28214685-14689. [DOI] [PubMed] [Google Scholar]
- 56.Nikiforov, M. A., S. Chandriani, J. Park, I. Kotenko, D. Matheos, A. Johnsson, S. B. McMahon, and M. D. Cole. 2002. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol. 225054-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 9435-44. [DOI] [PubMed] [Google Scholar]
- 58.Oishi, H., H. Kitagawa, O. Wada, S. Takezawa, L. Tora, M. Kouzu-Fujita, I. Takada, T. Yano, J. Yanagisawa, and S. Kato. 2006. An hGCN5/TRRAP histone acetyltransferase complex co-activates BRCA1 transactivation function through histone modification. J. Biol. Chem. 28120-26. [DOI] [PubMed] [Google Scholar]
- 59.Palhan, V. B., S. Chen, G. H. Peng, A. Tjernberg, A. M. Gamper, Y. Fan, B. T. Chait, A. R. La Spada, and R. G. Roeder. 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. USA 1028472-8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 151619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433434-438. [DOI] [PubMed] [Google Scholar]
- 62.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 228774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu, H., C. Hu, F. Zhang, G. J. Hwang, M. J. Swanson, C. Boonchird, and A. G. Hinnebusch. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 253461-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roeder, R. G. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579909-915. [DOI] [PubMed] [Google Scholar]
- 66.Sermwittayawong, D., and S. Tan. 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 253791-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spotts, G. D., S. V. Patel, Q. Xiao, and S. R. Hann. 1997. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 171459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1986-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sterner, D. E., R. Belotserkovskaya, and S. L. Berger. 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 9911622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taatjes, D. J., M. T. Marr, and R. Tjian. 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5403-410. [DOI] [PubMed] [Google Scholar]
- 72.Timmers, H. T., and L. Tora. 2005. SAGA unveiled. Trends Biochem. Sci. 307-10. [DOI] [PubMed] [Google Scholar]
- 73.Wallberg, A. E., S. Yamamura, S. Malik, B. M. Spiegelman, and R. G. Roeder. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol. Cell 121137-1149. [DOI] [PubMed] [Google Scholar]
- 74.Wang, G., M. A. Balamotis, J. L. Stevens, Y. Yamaguchi, H. Handa, and A. J. Berk. 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17683-694. [DOI] [PubMed] [Google Scholar]
- 75.Warfield, L., J. A. Ranish, and S. Hahn. 2004. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 181022-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wery, M., E. Shematorova, B. Van Driessche, J. Vandenhaute, P. Thuriaux, and V. Van Mullem. 2004. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 234232-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, P.-Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 225367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wyce, A., T. Xiao, K. A. Whelan, C. Kosman, W. Walter, D. Eick, T. R. Hughes, N. J. Krogan, B. D. Strahl, and S. L. Berger. 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell 27275-288. [DOI] [PubMed] [Google Scholar]
- 79.Xu, D., N. Popov, M. Hou, Q. Wang, M. Bjorkholm, A. Gruber, A. R. Menkel, and M. Henriksson. 2001. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 983826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26229-232. [DOI] [PubMed] [Google Scholar]
- 81.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 9711303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9553-562. [DOI] [PubMed] [Google Scholar]
- 83.Yang, X. J. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19535-545. [DOI] [PubMed] [Google Scholar]
- 85.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408225-229. [DOI] [PubMed] [Google Scholar]
- 86.Zhang, X., A. Krutchinsky, A. Fukuda, W. Chen, S. Yamamura, B. T. Chait, and R. G. Roeder. 2005. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol. Cell 1989-100. [DOI] [PubMed] [Google Scholar]
- 87.Zhu, X., M. Wiren, I. Sinha, N. N. Rasmussen, T. Linder, S. Holmberg, K. Ekwall, and C. M. Gustafsson. 2006. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell 22169-178. [DOI] [PubMed] [Google Scholar]