Abstract
Transcription can enhance recombination; this is a ubiquitous phenomenon from prokaryotes to higher eukaryotes. However, the mechanism of transcription-associated recombination in mammalian cells is poorly understood. Here we have developed a construct with a recombination substrate in which levels of recombination can be studied in the presence or absence of transcription. We observed a direct enhancement in recombination when transcription levels through the substrate were increased. This increase in homologous recombination following transcription is locus specific, since homologous recombination at the unrelated hprt gene is unaffected. In addition, we have shown that transcription-associated recombination involves both short-tract and long-tract gene conversions in mammalian cells, which are different from double-strand-break-induced recombination events caused by endonucleases. Transcription fails to enhance recombination in cells that are not in the S phase of the cell cycle. Furthermore, inhibition of transcription suppresses induction of recombination at stalled replication forks, suggesting that recombination may be involved in bypassing transcription during replication.
High-fidelity pathways that repair damaged DNA are essential to maintain genomic integrity. The repair of DNA double strand breaks (DSBs) is crucial for the maintenance of genome stability, and defects in the cellular response to DSBs have been linked to a number of inherited human cancer-prone syndromes (7). Homologous recombination is a major mechanism involved in the repair of DNA breaks generated spontaneously during replication (3, 32) or by DNA damaging agents in mitotically dividing cells (see reference 42 for a review). Recombination is stimulated by UV damage or chemical damage (15) and also by transcription (2).
A link between transcription and homologous recombination has long been established, though the mechanism has begun to be deciphered only recently. Transcription enhances homologous recombination in all organisms from prokaryotes to higher eukaryotes (reviewed in reference 2), a phenomenon referred to as transcription-associated recombination (TAR).
Escherichia coli provided the first evidence of TAR in prokaryotes (11, 18, 44). Recombination enhanced by RNA polymerase I (RNAPI)-driven transcription was first shown in Saccharomyces cerevisiae in transcriptionally active DNA in the recombination hot spot HOT1 (38, 45). Later, RNAPII-dependent transcription was also shown to enhance recombination in S. cerevisiae (43) and in Schizosaccharomyces pombe (14).
In mammalian cells, transcription of heteroallelic neomycin genes stimulated recombination between plasmids transfected into Chinese hamster ovary cells by approximately sixfold (27), and transcription resulted in a two- to sevenfold increase in intrachromosomal recombination between duplicated neomycin genes stably integrated into a Chinese hamster ovary cell line (26).
Many theories have been postulated to explain the mechanism of TAR (see reference 2 for a review). The collision between transcription and replication machineries could result in a possible recombination intermediate for TAR. Such a collision may result in replication fork stalling, and recombination is the major mechanism involved in resolving the stalled forks (24). Recent studies with S. cerevisiae have provided supporting evidence. In direct repeat constructs under the control of regulated promoters in S. cerevisiae, TAR required head-on collisions with replication machinery (28). TAR occurred at high levels when replication fork progression was opposed to transcription and was far less pronounced when both transcription and replication machineries traveled in the same direction (28). This is also true in ribosomal DNA repeats of S. cerevisiae, where RNAPI-dependent transcription results in a collision with the replication machinery, leading to fork blockage and recombination (41).
TAR could also be a result of defects in transcription elongation (29). The THO-TREX complex in S. cerevisiae acts at the interface between transcription and mRNA export. Mutation of any of the proteins in this complex leads to a hyperrecombination phenotype associated with impaired transcription elongation (8). The THO mutants showing a hyperrecombination phenotype also showed impaired replication fork progression due to RNA-DNA hybrids blocking the replication machinery and leading to replication fork stalling (47). Furthermore, mutants of the TREX complex with aberrant mRNA processing also showed elevated levels of recombination that was dependent on transcription (23). Interestingly, an hpr1 (gene encoding one of the proteins of the THO complex) point mutant derived by random mutagenesis showed a strong defect in transcription and a general defect in mRNA export but no hyperrecombination phenotype (17). This was due to the absence of replication fork blockage in this mutant, suggesting that stalled replication is a prerequisite for hyperrecombination.
Thus, the data for yeast suggest that stalled replication forks either formed by collision between transcription and replication machineries or due to RNA-DNA hybrids formed during transcription elongation in S phase are the recombination intermediates for TAR. Here, in order to extend our understanding of the underlying mechanisms controlling TAR in mammalian cells, we designed a recombination reporter substrate containing two nonfunctional copies of the neomycin phosphotransferase (neo) gene. This reporter substrate offers the possibility of studying the influence of transcription on levels of homologous recombination in a highly controlled fashion. Using this substrate, we examined the possible link between TAR and transcription elongation and whether TAR in mammalian cells arises as a consequence of stalled forks occurring due to the collision between transcription and replication machineries. In conclusion, our results are in agreement with the hypothesis that TAR is a likely result of collision between DNA replication machinery and RNA polymerase in mammalian cells.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
The pBI-LMscIneo (TARneo) recombination construct was made by subcloning parts in three steps into the pBluescript II SK(+) vector (Stratagene). The truncated 3′ neomycin gene was extracted from the pMC1neopolyA plasmid (Stratagene) by cleavage with XhoI/NcoI and subsequently blunt end subcloned into the EcoRV site of the pBluescript II SK(+) vector. Secondly, a hygromycin resistance cassette (hygR) was blunt end subcloned into the SmaI site of the resulting pBluescript II SK(+) (3′ neo) vector by extracting hygR after cleavage of the pREP4 plasmid (Invitrogen) with NruI. To receive a nonfunctional neomycin gene, a linker containing an I-SceI restriction site was subcloned into the MscI site in a pMC1neopolyA plasmid (Stratagene), thus introducing a stop codon in the reading frame of the neomycin gene. To check the correct orientation and sequence of the subcloned linker, the resulting pMC1neopolyA plasmid was subjected to DNA sequencing. Subsequently, the nonfunctional neomycin gene was PCR amplified with primers including XbaI sites and the product was subcloned into the XbaI site of pBluescript II SK(+) (hygR, 3′ neo). Finally, the pMscIneo recombination substrate was released by digestion with SacI/KpnI and made blunt ended with a Klenow fragment. This approximately 4-kb sequence was subcloned into the EcoRV site of the pBI-L vector (Clontech) to receive the pBI-LMscIneo recombination substrate. The oligonucleotide constituting the I-SceI (underlined) linker was as follows: CCACGTAGGGATAACAGGGTAAT. The following primers were used for the PCR amplification of the nonfunctional neomycin gene: forward, 5′GCTCTAGAATGGGATCGGCCATTGAACAAGAT3′; reverse, 5′GCTCTAGAAGCTATGACCATGATTACGCCAAGC3′.
To build a TetOff plasmid encoding zeocin resistance, pCDNA3.1Cat/Zeo (Invitrogen) was digested with DrdI to extract the zeocin resistance cassette. Simultaneously, the pTetOffneo plasmid (Clontech) was digested with XhoI to release the neomycin resistance cassette. The resulting vector and the zeocin resistance cassette were made blunt ended with a Klenow fragment (New England Biolabs) and ligated to receive pTetOffzeo.
Cell culture.
Cells were cultured in Dulbecco's minimum essential medium (DMEM) supplemented with 10% fetal calf serum, 1× nonessential amino acids, and 90 U/ml of penicillin-streptomycin at 37°C and 5% CO2. S8TofZM3 and SPD8 cells were maintained in 6-thioguanine to prevent the spontaneous reversion of the hprt gene prior to the treatment. The S8TofZM3 cells were also maintained in 1 μg/ml doxycycline to keep the transcription from the TetOff promoter turned off and 0.25 mg/ml of zeocin and 120 U/ml hygromycin to maintain the pTetoffZeo and pBI-LMscI vectors, respectively.
Establishment of stable cell lines.
Stable S8TofZM clones were obtained by electroporating 5 × 106 SPD8 cells with pTetoffZeo and then pBI-LMscI (250 V, 1,500-μF capacitance) in a Equibio EasyjectT plus machine (Geneflow). Following the first transfection and selection in 0.5 mg/ml zeocin, the resulting SPD8-TetOff clones were screened for low background expression combined with efficient reporter gene induction by removing doxycycline and assaying for luciferase activity. Subsequently, the selected SPD8-TetOff clones were transfected with pBI-LMscI. The double transfectants were selected in DMEM supplemented with 360 U/ml of hygromycin and 0.25 mg/ml zeocin.
Luciferase activity.
A Promega firefly luciferase assay system was used for assaying the luciferase activity, following the manufacturer's instructions. A Lucy 2 luminometer (Anthos Labtec Instruments, Salzburg, Austria) was used to measure relative light units, which give a measure of the amount of luciferase expressed.
Southern analysis.
Southern blotting was carried out as previously described (35) prior to assays with S8TofZM3 cells in order to check that the cells contained an intact copy of the TARneo substrate. The pBI-LMscI vector was digested with XhoI and MluI restriction endonucleases, and the 900-bp fragment containing a part of the 5′ neo gene was used as the probe. EcoRV restriction endonuclease was used for the digestion of genomic DNA.
Recombination and fluctuation assays.
Unless otherwise specified, all treatments in the recombination assays were performed for 24 h. S8TofZM3 cells (1.5 ×106) were inoculated on 10-cm dishes in 10 ml DMEM without 6-thioguanine and incubated overnight prior to treatment. The cells were allowed to recover for 48 h after treatments and cloning and selection were performed. For selection, 3 × 105 cells per dish were plated in the presence of 1× HAsT (50 μM hypoxanthine, 10 μM l-azaserine, 5 μM thymidine), and hypoxanthine phosphoribosyltransferase (HPRT)-positive revertants were selected for 10 days. To determine cloning efficiency, 500 cells were plated per dish without a selective agent. The number of colonies on cloning plates gives a measure of the clonogenic survival. The cloning was performed in duplicate and selection in triplicate.
In the case of the TARneo construct, 1.0 × 106 cells were inoculated onto 10-cm dishes in DMEM overnight. The cells were treated for 24 h in the presence or absence of 1 μg/ml doxycycline (without or with transcription, respectively). The cells were allowed to recover for 48 h after the treatment before subsequent cloning and selection as described above. For the fluctuation assay, 1,000 cells were plated in 24-well plates with or without 1 μg/ml of doxycycline for 10 days. The total number of cells in each well was then determined, and cells were replated on 10-cm dishes for selection. Ten milliliters of DMEM with 1 mg/ml of G-418 was added per selection plate with or without 1 μg/ml of doxycycline to select for either long-tract gene conversion (LTGC)/sister chromatid exchange (SCE) or both LTGC/SCE and short-tract gene conversion (STGC), respectively.
After 10 days, the colonies were fixed using methylene blue in methanol (4 g/liter) and counted, and the recombination levels were calculated using the following formulae: recombination frequency = number of colonies on selection plates/(number of cells plated on selection × cloning efficiency), where cloning efficiency is the number of colonies on cloning plates/500; and recombination rate (25) = ln(number of selection plates with no colonies/total number of plates)/number of cell generations per culture.
PCR analysis.
Genomic DNA was extracted from each recombinant using the FlexiGene DNA kit (QIAGEN), and PCR was performed using Go-Taq master mix (Promega) according to the manufacturer's instructions with an annealing temperature of 59°C and an extension time of 30 s. The primers used were as follows: forward primer (Pf), 5′GCCGCATCGATAAGCTTGTC3′; reverse primer 1 (Pr1), 5′CGTCGTGGATTACCCTGTTA3′; and reverse primer 2 (Pr2), 5′GTCGTGGCCAGCCACGATA3′.
Propidium iodide staining and cell cycle analysis.
Cells (1.5 × 106) were plated on 10-cm plates overnight prior to treatment. Cells were then trypsinized, washed in phosphate-buffered saline, and fixed in 70% (vol/vol) ethanol at −20°C overnight. The cells were then washed in phosphate-buffered saline twice and resuspended in 300 μl (50 μg/ml) of propidium iodide stain and 50 μl of (5 mg/ml) RNase A and incubated at 4°C for at least 1.5 h before being acquired on a Becton Dickinson FACSort machine and analyzed using Cell Quest software.
RESULTS
Establishment of cell lines to study the effect of transcription on recombination.
In order to study the mechanism of TAR in mammalian cells, we designed a recombination substrate containing two inactive repeats of the neomycin phosphotransferase gene (neo), in which transcription can be regulated using tetracycline (Fig. 1A), and subcloned it into a commercial vector. This vector is labeled pBI-LMscI and the construct TARneo. Stable transfectants of the Chinese hamster cell line SPD8 (10, 15) were established by transfection of the pTetOff plasmid, which contains the tetracycline-dependent transactivator. The selected clones were then transiently transfected with pBI-L vector, which has the luciferase reporter gene, and individual clones were isolated based on their luciferase activities (data not shown). Subsequently, we established double-stable transfectants of the selected SPD8-TetOff clone by transfection with pBI-LMscI. We selected two clones, S8TofZM3 and S8TofZM24, based on a low background of recombinants combined with efficient induction of luciferase expression (Fig. 2A). Clones containing an intact TARneo substrate were verified by Southern blotting (data not shown).
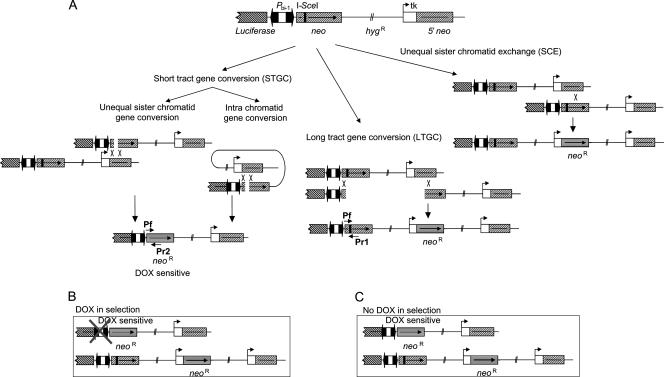
FIG. 1.
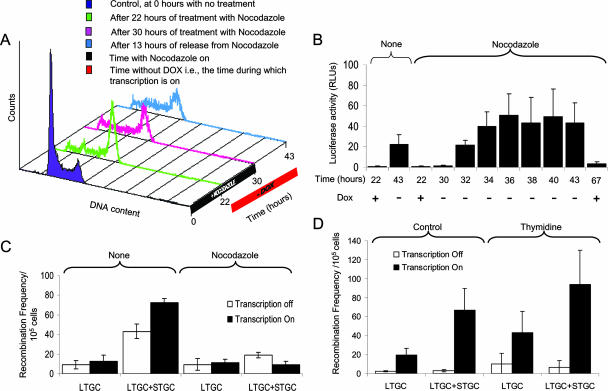
Schematic illustration of the TARneo substrate. (A) The substrate contains two inactive neo repeats. The downstream neo gene is controlled by a constitutive thymidine kinase promoter and has a 3′ deletion, resulting in a nonfunctional gene product. The upstream neo gene includes the full coding sequence but has an I-SceI restriction site introduced that creates a stop codon. The two neo genes are in direct orientation, separated by a functional hygromycin resistance cassette. The full recombination substrate was subcloned into a commercial vector containing a bidirectional promoter (Pbi-1) based on a tetracycline-controlled system (TetOff) and allowed inducible transcription over the upstream neomycin gene. The TetOff promoter codirectionally controls the transcription of the construct and the reporter luciferase gene, enabling the quantification of transcription through the substrate (4). The Pbi-1 promoter is negatively controlled by the presence of the tetracycline derivative doxycycline (DOX). Transcription is turned off in the presence of doxycycline and turned on in its absence. The construct can revert back to its active form by recombination, and the recombinants can be selected using G-418. STGC gives rise to a recombination product in which the upstream neo gene controlled by the inducible promoter is reverted. The second recombination product is a result of either LTGC or SCE, in which the downstream neo gene is reverted. The primers used to distinguish between the products of STGC (Pf and Pr2) and LTGC/SCE (Pf and Pr1) are also depicted. (B) Presence of doxycycline during selection with G-418 turns off Pbi-1, leading to the selection of LTGC/SCE alone since the product of STGC is not expressed. (C) When there is no doxycycline during selection, both the products of STGC and LTGC/SCE are selected with G-418.
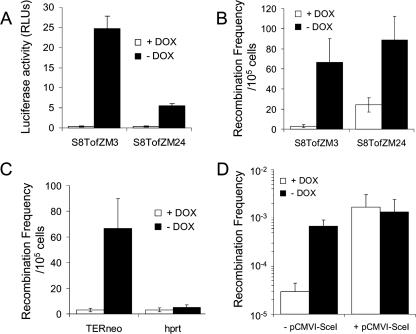
FIG. 2.
Locus-specific homologous recombination is enhanced by transcription, and transcription does not further enhance DSB-induced recombination. (A) Luciferase assay on the substrate in S8TofZM3 and S8TofZM24 cells confirms that 1 μg/ml of doxycycline effectively inhibits transcription in the TARneo substrate. (B) Recombination frequencies in the TARneo substrate in the S8TofZM3 and S8TofZM24 cells in the presence (transcription off) or absence (transcription on) of 1 μg/ml doxycycline. (C) Recombination frequencies in the TARneo and hprt substrates of S8TofZM3 cells in the presence or absence of 1 μg/ml doxycycline (transcription off or on, respectively). (D) Recombination frequencies in S8TofZM3 cells with or without pCMVI-SceI (with or without DSB) in the presence (transcription off) or absence (transcription on) of 1 μg/ml doxycycline. Results presented represent averages from at least three independent experiments, and error bars denote standard deviations. DOX, doxycycline; RLUs, relative light units.
Homologous recombination is efficiently enhanced by transcription.
In both S8TofZM3 and S8TofZM24 cells, doxycycline effectively controlled transcription through the substrate as indicated by the luciferase activity (Fig. 2A). We observed 64-fold and 14-fold increases in transcription in the S8TofZM3 and S8TofZM24 cells, respectively, in the absence of doxycycline compared to levels in its presence. This increase in transcription led to a 22-fold and 4-fold enhancement in recombination in the TARneo substrate in S8TofZM3 and S8TofZM24 cells, respectively (Fig. 2B). Together, these results suggest a potent ability for transcription to induce homologous recombination in the TARneo substrate.
The increase in recombination we see above might be due to the global effect of long-term doxycycline treatment on rates of homologous recombination. To test this, we examined the rate of revertants in the unrelated hprt gene in the S8TofZM3 cells after treatment with doxycycline. This cell line carries a spontaneous partial duplication of exon 7 in the hprt gene, rendering the gene nonfunctional, and a functional HPRT can be regained by a homologous recombination event (15). We did not observe any significant increase in the rate of homologous recombination in the hprt gene (Fig. 2C), indicating that the increased rate of TAR we demonstrate using the TARneo construct is a locus-specific event.
Since DSBs are strong inducers of recombination (16), we next investigated the effect of transcription on DSB-induced recombination by inducing a DSB at the I-SceI site (using the pCMVI-SceI vector) on the upstream neo gene in the TARneo construct in S8TofZM3 cells and measuring recombination frequencies in the presence or absence of transcription (without or with doxycycline, respectively). Consistent with previous reports (9, 22), recombination was stimulated by DSBs created in vivo at the I-SceI site. However, transcription (no doxycycline) did not further stimulate DSB-induced recombination (Fig. 2D), suggesting that transcription has no further effect on recombination once a DSB is induced, as shown in previous reports (40, 48).
Transcription-associated recombination events involve gene conversion.
The design of the TARneo construct (Fig. 1A) allows us to separate the recombination products rising from STGC or LTGC. Inclusion of doxycycline during selection favors the products of LTGC/SCE alone (Fig. 1B), while exclusion of doxycycline selects the products of both LTGC/SCE and STGC (Fig. 1C). Furthermore, single-strand annealing, which occurs independently of RAD51-induced strand invasion, is not selected for in this substrate since it would generate a nonfunctional neo gene product.
There was an eightfold increase in recombination when only LTGC/SCE was selected for (presence of doxycycline) (Fig. 3A). In contrast, on excluding doxycycline during selection (selecting LTGC/SCE plus STGC), there was a 22-fold increase in the recombination frequency. Hence, the 2.7-fold further increase in LTGC/SCE plus STGC over LTGC/SCE alone reflects STGC. We also measured the recombination rate using fluctuation assays (Fig. 3B). While the recombination assay gives the frequency of recombination, i.e., the number of recombinants in a pool of cells, the fluctuation assay measures the recombination rate, i.e., the frequency at which recombination occurs per cell division. In line with the increase in the recombination frequency, there was a 9.8-fold increase in the recombination rate when doxycycline was included during selection (LTGC/SCE), while there was 22-fold increase when doxycycline was excluded (LTGC/SCE and STGC). Hence, the 2.2-fold further increase reflects STGC. This suggests that TAR induces both LTGC/SCE and STGC.
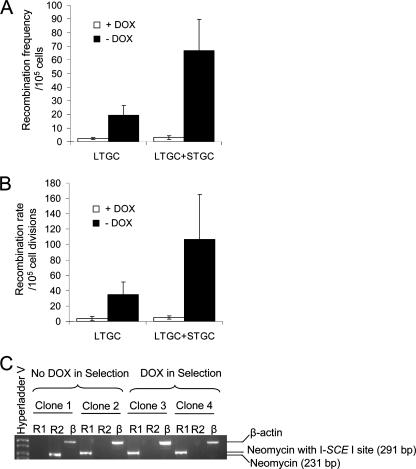
FIG. 3.
Transcription enhances both short-tract and long-tract gene conversion events in mammalian cells. Recombination frequency (A) and Recombination rate (B) in the TARneo substrate of S8TofZM3 cells in the presence or absence of 1 μg/ml doxycycline to turn off or turn on transcription, respectively. Products of LTGC/SCE alone or both LTGC/SCE and STGC are distinguished by selection in the presence or absence of 1 μg/ml doxycycline, respectively. Results presented represent averages from at least three independent experiments, and error bars denote standard deviations. (C) PCR analysis of two colonies each from selection plates with or without doxycycline, expanded from the recombination assay (see panel A) in order to distinguish between the products of STGC (PCR product corresponds to the recombined upstream neomycin without the I-SceI site) and LTGC/SCE (PCR product corresponds to the upstream neomycin with the I-SceI site). The different types of recombination events detected genetically correspond to the expected molecular structure. The primers used are depicted in Fig. 1. R1, Pf plus Pr1; R2, Pf plus Pr2; β, β-actin primers; DOX, doxycycline.
In order to further verify this, we isolated individual colonies from the selection plates of the fluctuation assays not treated with doxycycline and investigated the recombination pathway by which they reverted. These are the products of either LTGC/SCE or STGC. Those colonies that initially arose by STGC will not be able to survive the subsequent G-418 treatment in the presence of doxycycline, but this treatment will make no difference to the survival of those which arose by LTGC/SCE. Out of 30 colonies isolated from plates initially selected with G-418 alone, nearly half the colonies (14/30) survived well with doxycycline during subsequent selection with G-418. This suggests that the colonies that could not survive arose by STGC initially. This establishes that STGC and LTGC/SCE are equally enhanced by transcription in mammalian cells.
The different types of recombination events detected genetically were analyzed using PCR to verify that they correspond to the expected molecular structure. In order to achieve this, two reverse primers were designed (Fig. 1) in such a way that reverse primer 1 (Pr1) gives a 291-bp product only with the products of LTGC/SCE, corresponding to the upstream neomycin gene with the I-SceI site, and the reverse primer 2 (Pr2) gives a 231-bp product only with the products of STGC, corresponding to the reverted upstream neomycin gene without the I-SceI site. As the forward primer is placed in the multiple cloning site of the vector, only the upstream neomycin gene is amplified and not the downstream repeat (due to the large hygromycin cassette between them). Using these primers, PCR analysis was performed on the genomic DNA extracted from 25 different colonies isolated from the selection plates of the recombination assays with or without doxycycline in selection. Representative PCR products from four different colonies, two each from selection plates with or without doxycycline, are shown in Fig. 3C. In agreement with the expected molecular structure, colonies 3 and 4 selected with G-418 and doxycycline (and which therefore must have been the products of LTGC/SCE) gave only a 291-bp product with Pr1 and no product with Pr2. This was also the case with colony 2 (selected without doxycycline), suggesting that it is a product of LTGC/SCE, whereas colony 1 (without doxycycline in selection) gave a 231-bp product with Pr2 and no product with Pr1, suggesting that it is a product of STGC.
Inhibition of transcription elongation has no major effect on transcription-associated recombination.
TAR has previously been suggested to be coupled to transcription elongation (8, 29). Based on the data from S. cerevisiae, we hypothesized that inhibitors which block the movement of transcription elongation would enhance TAR in the TARneo construct, since they create intermediate structures similar to the ones seen in the THO mutants in S. cerevisiae. We therefore decided to examine the rates of recombination in S8TofZM3 cells after inhibition of transcription elongation with 2,3-dichlororibobenzimadazole (DRB) or actinomycin D. DRB inhibits CDK7 kinase associated with transcription factor II H, thereby preventing it from phosphorylating RNAPII and blocking it from proceeding in to the elongation phase (50). Actinomycin D inhibits elongation by binding DNA at the transcription initiation complex, thereby immobilizing it (36). Transcription as measured by luciferase assay is inhibited at 1 nM actinomycin D (Fig. 4A) and 75 μM DRB (Fig. 4B).
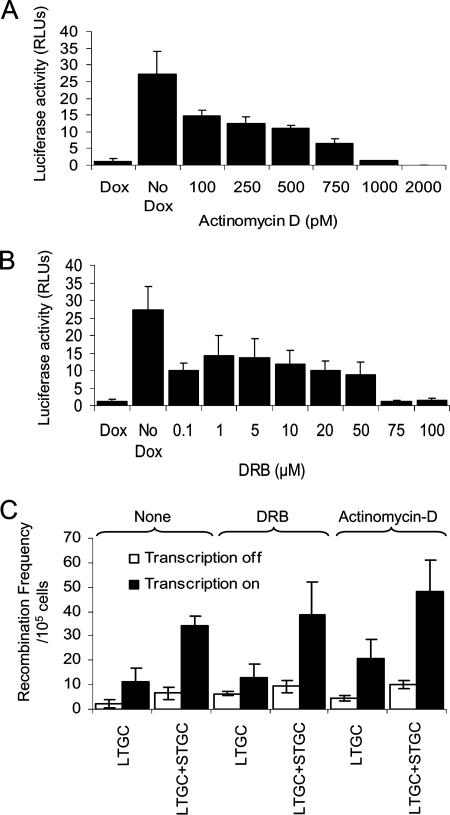
FIG. 4.
Inhibition of transcription elongation does not further affect transcription-associated recombination. Luciferase activity of the TARneo substrate of S8TofZM3 cells after treatment with increasing doses of actinomycin D (A) or DRB (B). (C) Recombination frequency in the TARneo substrate of S8TofZM3 cells after treatment with transcription elongation inhibitors 1 nM actinomycin D and 75 μM DRB in the presence (no transcription) or absence (transcription) of 1 μg/ml doxycycline. Results presented represent average from at least three independent experiments, and error bars denote standard deviations. Dox, doxycycline; RLUs, relative light units.
Treatment with 75 μM DRB or 1 nM actinomycin D resulted in a slight increase in the recombination frequency, though this was not statistically significant (Fig. 4C). A similar result was obtained after treatment with 500 μM novobiocin (see Fig. S1B in the supplemental material), which interferes with transcription by inhibiting DNA gyrase (46). The dosage was restricted to 500 μM due to the toxicity of the drug at higher doses (data not shown) and since there was a considerable reduction in the luciferase activity in the TARneo construct at this dose (see Fig. S1A in the supplemental material). These results suggest that inhibiting transcription elongation does not cause a significant increase in TAR in the TARneo construct.
Transcription-associated recombination is associated with cycling cells.
It has been shown in yeast that TAR is S phase associated and replication dependent (28, 47). Since homologous recombination is particularly important in late S/G2 phase (31, 34), we next investigated if TAR in mammalian cells is correlated to S phase. We hypothesized that if TAR is S phase associated, then a growth arrest in G2/M would decrease TAR. Transcription was turned on in the S8TofZM3 cells for 21 h, during which time cells were arrested in G2/M by treatment with 40 ng/ml nocodazole. Propidium iodide staining and fluorescence-activated cell sorting confirmed that nocodazole treatment for 22 h effectively arrested the cells in G2/M phase and that after 30 h cells were still arrested without displaying significant apoptosis. Thirteen hours after removal of nocodazole, most of the cells were still in G2/M phase, with only a very small percentage in S phase (Fig. 5A). Simultaneously, by assaying luciferase activity, it was determined that withdrawal of doxycycline from the medium for 21 h (taken away at 22 h and added back in after 13 h of release) was enough to sufficiently turn on transcription (Fig. 5B). Although nocodazole treatment inhibited transcription through the substrate (consistent with previous reports [19]), transcription was turned back on as soon as the cells were released from the treatment (Fig. 5B).
FIG. 5.
Transcription-associated recombination in the TARneo substrate is S phase dependent. (A) Fluorescence-activated cell sorting data representing the cell cycle profiles of S8TofZM3 cells after treatment with 40 ng/ml nocodazole and 1 μg/ml doxycycline. The results presented are a representative of at least three independent experiments. (B) Luciferase activity of the S8TofZM3 cells during treatment with 40 ng/ml nocodazole and 1 μg/ml doxycycline. (C) Recombination frequencies of the TARneo substrate in S8TofZM3 cells after treatment with 40 ng/ml nocodazole in the presence or absence of 1 μg/ml doxycycline. (D) Recombination frequencies of the TARneo substrate in S8TofZM3 cells after treatment with 10 mM thymidine in the presence or absence of 1 μg/ml doxycycline. Results presented represent averages from at least three independent experiments, and error bars denote standard deviations. DOX, doxycycline; RLUs, relative light units.
Based on these data, the experiment was designed such that transcription was switched on (no doxycycline) for 21 h, during which time cells were not in S phase (Fig. 5A), and its effect on TAR was measured. Arresting the cells in G2/M phase and thus hindering them from entering S phase completely abolished TAR (Fig. 5C), suggesting that TAR in S8TofZM3 cells is S phase associated.
Dependency on S phase suggested that collision between the machineries of transcription and replication could be responsible for TAR. If this is the case, then halting either replication or transcription elongation may decrease TAR. However, there was no significant difference in recombination levels in the cells treated with 10 mM thymidine relative to levels in the untreated cells (Fig. 5D). Likewise, treatment with more-stringent replication inhibitors, hydroxyurea and aphidicolin alone or together prior to the recombination assay, also showed no effect on recombination frequency (see Fig. S2 in the supplemental material), suggesting that inhibiting replication alone has no effect on TAR in S8TofZM3 cells.
Transcription-associated recombination is likely a consequence of collision between transcription and replication machineries.
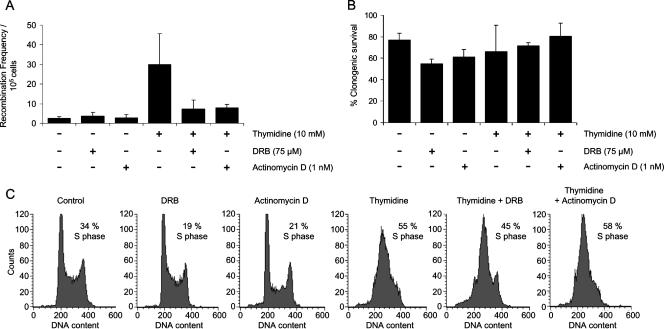
It has been shown for S. cerevisiae that head-on collision between transcription machinery and replication machinery results in TAR (28). Stalling replication forks causes homologous recombination (24). Therefore, replication forks stalled by the transcription machinery may explain enhanced TAR. It can be proposed that slowing replication forks with thymidine in normally cycling cells could enhance collisions. However, inhibiting transcription elongation at the same time might decrease the number of collisions, thus decreasing the number of recombination intermediates formed, in turn leading to a decrease in homologous recombination. In order to test this with mammalian cells, the SPD8 cells were treated with 10 mM thymidine alone and the transcription elongation inhibitor DRB (75 μM) or actinomycin D (1 nM), and the recombination frequency was then established using the hprt substrate (Fig. 6A). Consistent with previous results (24), there was an 11-fold increase in the frequency of HPRT-positive revertants following treatment with thymidine. Cotreatment with thymidine and an elongation inhibitor, however, decreased thymidine-induced recombination by threefold (statistically significant; P < 0.05). The clonogenic survival of the cells was similar after different treatments (Fig. 6B), suggesting that the decrease in recombination observed is not because of the toxicity of the drugs used.
FIG. 6.
Transcription-associated recombination is likely to be due to the collision between transcription and replication machineries. (A) Recombination frequency in the hprt substrate of SPD8 cells after treatment with 10 mM thymidine alone, the transcription elongation inhibitor DRB (75 μM) or actinomycin D (1 nM) alone, or both thymidine and transcription elongation inhibitors together. Results presented represent averages from at least three independent experiments, and error bars denote standard deviations. (B) Clonogenic survival of SPD8 cells after treatment with 10 mM thymidine, DRB (75 μM), or actinomycin D (1 nM) or both thymidine and transcription elongation inhibitors together. Results presented represent averages from at least three independent experiments, and error bars denote standard deviations. (C) Cell cycle profiles of SPD8 cells after treatment with 10 mM thymidine, DRB (75 μM), or actinomycin D (1 nM) or both thymidine and transcription elongation inhibitors together. The results presented here are representative of at least three independent experiments.
A similar decrease in recombination was also observed after combined treatment of SPD8 cells with thymidine and a more stringent transcription elongation inhibitor, α-amanitin. The dosage of α-amanitin was restricted to 5 μM due to its toxicity to the SPD8 cells (data not shown). Treatment at this dose resulted in a 1.7-fold reduction in thymidine-induced homologous recombination (see Fig. S3 in the supplemental material). Though α-amanitin is a more stringent transcription elongation inhibitor, the decrease is probably not as pronounced as in the case of treatment with DRB or actinomycin D due to the low dose of α-amanitin used. However, this decrease is still statistically significant (P < 0.05).
The decrease in homologous recombination after combined treatment with thymidine and transcription elongation inhibitors could be due to a decreased number of recombination intermediates formed due to fewer collisions between transcription and replication machineries, or it could be due to the effect of transcription elongation inhibitors on the cell cycle. The elongation inhibitors might arrest cells in the G1 phase of the cell cycle, which would prevent the action of thymidine during S phase. However, there were similar percentages of cells in S phase after treatment with thymidine alone or thymidine and a transcription elongation inhibitor together (Fig. 6C), ruling out the second possibility. This suggests that the decrease in homologous recombination after combined treatment is more likely due to the reduction in the number of collisions and thus the number of recombination intermediates formed, implying that TAR is a consequence of collision between transcription and replication machineries in mammalian cells.
Transcription-associated recombination is not influenced by inhibition of DNA damage signaling pathways.
We have shown here that TAR is S phase associated, is likely to be dependent on replication, and could be due to a collision between transcription and replication machineries, leading to replication fork stalling. Evidence suggests that the ATM and ATR kinases are activated in response to replication stress (1). Replication stress caused by agents such as thymidine leads to rapid activation of the ATM-mediated signaling cascade to initiate homologous recombination, and thymidine-induced replication fork stalling requires ATM and Chk1 for its homologous recombination repair (5, 37). Recently, Chk1 was proposed to be important during normal S phase to protect against DNA breakage (39). Furthermore, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and ATR were also shown to be required for the cellular response to replication stress and were suggested to play an important role in the repair of stalled replication forks (49). Our work suggests that replication stalling could be the cause of TAR, and it was previously shown that inhibiting ATM, Chk1, and ATR abrogates homologous recombination induced by the DNA stalling agents thymidine and hydroxyurea (5, 37, 49). We therefore investigated the role of the kinases ATM, DNA-PKcs, ATR, and Chk1 in TAR. Recombination frequencies were measured after each of the kinases were inhibited in S8TofZM3 cells by using specific inhibitors: ATM with 10 μM of KU55933 (6), DNA-PKcs with 10 μM of KU51777 (6) and Chk1 with 500 nM of CEP-3891 (37). Ten millimolar of caffeine (37), a nonspecific ATM and ATR inhibitor, was used to test the effect of combined inhibition of all phosphatidylinositol 3-kinase (PI 3-kinase)-related kinases on TAR. We observed that treatment with either caffeine or specific inhibitors of ATM, DNA-PKcs, or Chk1 had no significant effect on recombination frequencies (Fig. 7), suggesting that TAR at a single locus is independent of kinases within the DNA damage signaling pathways.
FIG. 7.
Transcription-associated recombination is independent of DNA damage signaling pathways. Recombination frequencies in the TARneo substrate of S8TofZM3 cells after treatment with the following PI 3-kinase-like kinase inhibitors: 10 μM of KU55933 (ATMi), 10 μM of KU51777 (DNA-PKi), 500 nM of CEP3891 (Chk1i), or 10 mM of caffeine (nonspecific inhibitor of PI 3-kinase-like kinases). Results presented represent averages from at least three independent experiments, and error bars denote standard deviations. DOX, doxycycline; RLUs, relative light units; ATMi, ATM inhibitor; DNA-PKi, DNA-PK inhibitor; Chk1i, Chk1 inhibitor.
DISCUSSION
The design of the substrate using two inactive neo repeats not only allows us to measure the amount of recombination through the substrate but also allows us to distinguish between the products of STGC and LTGC/SCE. Our work suggests that transcription induces gene conversion in the substrate, including both short tract and long tract. Previously we showed that homologous recombination repair of DSBs involves STGC in all the phases of the cell cycle (34). In contrast to this, transcription induces both STGC and LTGC, suggesting that TAR is not associated with a classical two-ended DSB. Our data further suggest that once recombination is initiated by an endonuclease-induced DSB, transcription does not have any further effect on recombination levels. Together with previous results obtained with yeast and mammalian cells, these results are consistent with the idea that transcription enhances spontaneous recombination by increasing the initiation events (13, 40, 48).
Recombination at the unrelated hprt locus is not affected by transcription in the TARneo substrate. This shows that we are looking at a locus-specific event in the TARneo construct due to the effect of transcription and not at a general effect of doxycycline. Recombination during mating-type switching in yeast, which is enhanced by transcription, is also shown to be site specific (20, 21). Taken together, these data suggest that locus specificity is a general feature of recombination associated with transcription.
S-phase association of TAR suggests that it is dependent on replication, since stalled replication forks are mainly repaired by homologous recombination. These stalled replication forks could be formed either by collision between replication and transcription machineries or by obstruction due to RNA-DNA hybrids that form during transcription. Yeast mutants that cannot process the nascent mRNA properly form RNA-DNA hybrids during transcription, resulting in hyperrecombination (23). Replication fork progression is impaired by transcription in these mutants, leading to the stalling of replication forks, a possible cause of hyperrecombination (47). In addition, the hyperrecombination in these mutants was higher in GC-rich regions, which are difficult to transcribe in yeast (29). However, since the SPD8 cells do not have any defect in mRNA processing, the collision between the machineries of transcription and replication, resulting in stalled replication forks, is probably the main cause of TAR in these cells. We saw that cells stalled in S phase with no transcription do not undergo as much homologous recombination as cells stalled to the same degree in S phase but with transcription, probably due to less-frequent collisions between transcription machinery and replication machinery. This suggests that TAR is due not just to fork stalling but to the collision between transcription and replication machineries in mammalian cells, as it is in yeast (28). Several recent publications on yeast have shown TAR as a consequence of stalled replication (17, 28, 47). Here, though we do not show any direct association between TAR and stalled replication forks in mammalian cells, we do show that TAR is S phase associated and that inhibiting transcription and replication at the same time abolishes thymidine-induced recombination, suggesting that stalled replication forks are involved in TAR in mammalian cells.
Since our data suggest that TAR is S phase associated, it is surprising that slowing down the replication forks with thymidine in the presence of transcription does not enhance TAR. In cells with direct repeat constructs under the control of the regulated promoters, a head-on collision between replication and transcription machinery was required for TAR (28). When both transcription and replication were in the same direction, the increase in recombination was not significant. This may explain the result with thymidine in S8TofZM3 cells. These cells have a stably transfected TARneo construct. It is possible that this construct integrated into the cells such that replication through this substrate is codirectional with transcription, which may explain why slowing down replication forks does not make a further difference to the TAR. Another possible explanation for this is that TAR requires only either transcription or replication machinery to be moving at any given time. When replication is stalled with thymidine, transcription through the substrate is still on, which may lead to collision between the two machineries. On the other hand, when transcription elongation was inhibited in these cells using chemical inhibitors, the cells were still actively dividing, resulting in the replication machinery colliding with the blocked RNA polymerase. This suggests that replication needs to stall for TAR to occur, and this can happen either when a moving transcription complex collides with replication fork or when the transcription complex itself is stalled, which will then obstruct the replication machinery.
Mutations in the THO complex in yeast indicated that TAR is associated with transcription elongation (2, 8, 29, 30). Both of the elongation inhibitors we used, DRB and actinomycin D, inhibit transcription elongation by blocking the movement of the RNA polymerase (36, 50). Theoretically, using these drugs to inhibit elongation creates the same scenario as in the THO mutants in yeast. We saw a very slight increase (not statistically significant) in recombination frequency on inhibition of transcription elongation, though not to the same extent as in yeast. This could also be due to the direction in which the construct integrated into the cells (codirectional), as explained above. Another possible explanation could be the time used for treatment. The effect of drugs is temporary (only 24 h), whereas in the THO mutants, there is a permanent defect in transcription.
Finally we have shown that TAR is not influenced by PI 3-kinase-related kinases or Chk1. ATM, ATR, DNA-PKcs, and a downstream target for ATR, Chk1, are known to be involved in DNA damage response in S phase (1, 12). They are also known to be involved in homologous recombination in response to DSBs and replication stress (5, 37). Hence, it is unexpected that TAR was found not be affected by the inhibition of any of these kinases independently or all together (using the nonspecific inhibitor caffeine). However, TAR might be independent of damage-signaling checkpoint kinases, since transcription occurs constitutively and PI 3-kinase-related kinases are activated only upon DNA stress, for example, during DNA damage. Furthermore, transcription stalling may need to occur within nucleoli in order to trigger a DNA damage response (33).
In conclusion, we have shown here that transcription-associated recombination is locus specific and involves gene conversion events. We also have shown that TAR takes place only in the cycling cells at the S phase of the cell cycle. Finally, our results suggest that recombination may be involved in bypassing transcription during replication.
Supplementary Material
Acknowledgments
We thank Helen Bryant for her scientific advice and discussions and Susan Newton for help with flow cytometry. We thank Graeme Smith and Stephen Trusko for materials.
The Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Swedish Research Council, the Swedish Pain Relief Foundation, the Medical Research Council, and the Biological & Biotechnological Sciences Research Council supported this work financially.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 152177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera, A. 2002. The connection between transcription and genomic instability. EMBO J. 21195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 3071235-1245. [DOI] [PubMed] [Google Scholar]
- 4.Baron, U., S. Freundlieb, M. Gossen, and H. Bujard. 1995. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 233605-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolderson, E., J. Scorah, T. Helleday, C. Smythe, and M. Meuth. 2004. ATM is required for the cellular response to thymidine induced replication fork stress. Hum. Mol. Genet. 132937-2945. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, H. E., and T. Helleday. 2006. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 341685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93477-486. [DOI] [PubMed] [Google Scholar]
- 8.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 113459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choulika, A., A. Perrin, B. Dujon, and J. F. Nicolas. 1995. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 151968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dare, E., L. H. Zhang, D. Jenssen, and V. Bianchi. 1995. Molecular analysis of mutations in the hprt gene of V79 hamster fibroblasts: effects of imbalances in the dCTP, dGTP and dTTP pools. J. Mol. Biol. 252514-521. [DOI] [PubMed] [Google Scholar]
- 11.Dul, J. L., and H. Drexler. 1988. Transcription stimulates recombination. II. Generalized transduction of Escherichia coli by phages T1 and T4. Virology 162471-477. [DOI] [PubMed] [Google Scholar]
- 12.Durocher, D., and S. P. Jackson. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13225-231. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Barrera, S., M. Garcia-Rubio, and A. Aguilera. 2002. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics 162603-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm, C., P. Schaer, P. Munz, and J. Kohli. 1991. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol. Cell. Biol. 11289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helleday, T., C. Arnaudeau, and D. Jenssen. 1998. A partial hprt gene duplication generated by non-homologous recombination in V79 Chinese hamster cells is eliminated by homologous recombination. J. Mol. Biol. 279687-694. [DOI] [PubMed] [Google Scholar]
- 16.Helleday, T., J. Lo, D. C. van Gent, and B. P. Engelward. 2007. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amsterdam) 6923-935. [DOI] [PubMed] [Google Scholar]
- 17.Huertas, P., M. L. Garcia-Rubio, R. E. Wellinger, R. Luna, and A. Aguilera. 2006. An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol. Cell. Biol. 267451-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda, H., and T. Matsumoto. 1979. Transcription promotes recA-independent recombination mediated by DNA-dependent RNA polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 764571-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanova, T. V., V. N. Ivanov, and E. S. Nadezhdina. 2001. Transcription factors NF-kappaB and AP-1/c-fos in cell response to nocodazole. Membr. Cell Biol. 14727-741. [PubMed] [Google Scholar]
- 20.Jung, S., K. Rajewsky, and A. Radbruch. 1993. Shutdown of class switch recombination by deletion of a switch region control element. Science 259984-987. [DOI] [PubMed] [Google Scholar]
- 21.Klar, A. J., J. N. Strathern, and J. B. Hicks. 1981. A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell 25517-524. [DOI] [PubMed] [Google Scholar]
- 22.Lukacsovich, T., D. Yang, and A. S. Waldman. 1994. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI. Nucleic Acids Res. 225649-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luna, R., S. Jimeno, M. Marin, P. Huertas, M. Garcia-Rubio, and A. Aguilera. 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell 18711-722. [DOI] [PubMed] [Google Scholar]
- 24.Lundin, C., K. Erixon, C. Arnaudeau, N. Schultz, D. Jenssen, M. Meuth, and T. Helleday. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 225869-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickoloff, J. A. 1992. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 125311-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickoloff, J. A., and R. J. Reynolds. 1990. Transcription stimulates homologous recombination in mammalian cells. Mol. Cell. Biol. 104837-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prado, F., and A. Aguilera. 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 241267-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prado, F., J. I. Piruat, and A. Aguilera. 1997. Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 162826-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rondon, A. G., S. Jimeno, M. Garcia-Rubio, and A. Aguilera. 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 27839037-39043. [DOI] [PubMed] [Google Scholar]
- 31.Rothkamm, K., I. Kruger, L. H. Thompson, and M. Lobrich. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 235706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 141-10. [PubMed] [Google Scholar]
- 33.Rubbi, C. P., and J. Milner. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 226068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh-Gohari, N., and T. Helleday. 2004. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 323683-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz, N., E. Lopez, N. Saleh-Gohari, and T. Helleday. 2003. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 314959-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobell, H. M. 1985. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 825328-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorensen, C. S., L. T. Hansen, J. Dziegielewski, R. G. Syljuasen, C. Lundin, J. Bartek, and T. Helleday. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7195-201. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, S. E., and G. S. Roeder. 1989. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 93464-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syljuasen, R. G., C. S. Sorensen, L. T. Hansen, K. Fugger, C. Lundin, F. Johansson, T. Helleday, M. Sehested, J. Lukas, and J. Bartek. 2005. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 253553-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taghian, D. G., and J. A. Nickoloff. 1997. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 176386-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi, Y., T. Horiuchi, and T. Kobayashi. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 171497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thacker, J. 1999. The role of homologous recombination processes in the repair of severe forms of DNA damage in mammalian cells. Biochimie 8177-85. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 44.Vilette, D., S. D. Ehrlich, and B. Michel. 1995. Transcription-induced deletions in Escherichia coli plasmids. Mol. Microbiol. 17493-504. [DOI] [PubMed] [Google Scholar]
- 45.Voelkel-Meiman, K., R. L. Keil, and G. S. Roeder. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 481071-1079. [DOI] [PubMed] [Google Scholar]
- 46.Webb, M. L., and S. T. Jacob. 1988. Inhibition of RNA polymerase I-directed transcription by novobiocin. Potential use of novobiocin as a general inhibitor of eukaryotic transcription initiation. J. Biol. Chem. 2634745-4748. [PubMed] [Google Scholar]
- 47.Wellinger, R. E., F. Prado, and A. Aguilera. 2006. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 263327-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng, Y. S., D. Xing, J. A. Clikeman, and J. A. Nickoloff. 2000. Transcriptional effects on double-strand break-induced gene conversion tracts. Mutat. Res. 461119-132. [DOI] [PubMed] [Google Scholar]
- 49.Yajima, H., K. J. Lee, and B. P. Chen. 2006. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 267520-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yankulov, K., K. Yamashita, R. Roy, J. M. Egly, and D. L. Bentley. 1995. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J. Biol. Chem. 27023922-23925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.