Abstract
Myeloid leukemia factor 1 (MLF1) stabilizes the activity of the tumor suppressor p53 by suppressing its E3 ubiquitin ligase, COP1, through a third component of the COP9 signalosome (CSN3). However, little is known about how MLF1 functions upstream of the CSN3-COP1-p53 pathway and how its deregulation by the formation of the fusion protein nucleophosmin (NPM)-MLF1, generated by t(3;5)(q25.1;q34) chromosomal translocation, leads to leukemogenesis. Here we show that MLF1 is a cytoplasmic-nuclear-shuttling protein and that its nucleolar localization on fusing with NPM prevents the full induction of p53 by both genotoxic and oncogenic cellular stress. The majority of MLF1 was located in the cytoplasm, but the treatment of cells with leptomycin B rapidly induced a nuclear accumulation of MLF1. A mutation of the nuclear export signal (NES) motif identified in the MLF1 sequence enhanced the antiproliferative activity of MLF1. The fusion of MLF1 with NPM translocated MLF1 to the nucleolus and abolished the growth-suppressing activity. The introduction of NPM-MLF1 into early-passage murine embryonic fibroblasts allowed the cells to escape from cellular senescence at a markedly earlier stage and induced neoplastic transformation in collaboration with the oncogenic form of Ras. Interestingly, disruption of the MLF1-derived NES sequence completely abolished the growth-promoting activity of NPM-MLF1 in murine fibroblasts and hematopoietic cells. Thus, our results provide important evidence that the shuttling of MLF1 is critical for the regulation of cell proliferation and a disturbance in the shuttling balance increases the cell's susceptibility to oncogenic transformation.
Myeloid leukemia factor 1 (MLF1) was first identified as the carboxyl-terminal component of the leukemic fusion protein nucleophosmin (NPM)-MLF1, generated by t(3;5)(q25.1;q34) chromosomal translocation (46), which is associated with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (33). NPM is a ubiquitously expressed nucleolar phosphoprotein which has multiple functions including shuttling from the nucleolus to the cytoplasm (4), ribosomal biogenesis (49), centrosome duplication (31), and stabilization of the Arf-Mdm2-p53 tumor suppressor pathway (2, 7, 20, 21). However, the biochemical activity of MLF1 has not been well characterized, although we and other groups reported several proteins that interact with MLF1 such as CSN3, 14-3-3ζ, MADM, and MLF1IP/KLIP1/CENP-U(50) (17, 25, 47).
In biological studies, there have been several observations suggesting that MLF1 is physiologically involved in a tumor suppressor pathway. MLF1 has been found to be overexpressed in more than 25% of MDS-associated cases of AML, in the malignant transformation phase of MDS, and in lung squamous cell carcinoma (27, 38). The aberrant overexpression is usually related to mutations and to inactivation of p53 in various cell lines (47). We recently reported that MLF1 is a negative regulator of cell cycle progression that functions upstream of the tumor suppressor p53 and its novel E3 ubiquitin ligase COP1 (11, 47). MLF1 suppresses the activity of COP1 through physical interaction with CSN3, the third subunit of the COP9 signalosome complex (CSN), and consequently induces the arrest of cell growth due to an accumulation of p53. Although the regulatory mechanism of this novel CSN-COP1-p53 pathway located downstream of MLF1 remains to be elucidated, in plants, CSN and COP1 function together as a repressor of photomorphogenesis, i.e., light-mediated development (36). A knockdown of CSN3 in mammalian cells results in a reduced amount of CSN complex and in a failure to suppress COP1-mediated degradation of p53 after exposure to the MLF1 signal and genotoxic stress (47), which implies that CSN is required for the proper functioning of COP1 with regard to its mammalian substrates.
The fusion protein NPM-MLF1 consists of more than half of the amino terminus of NPM and almost the entire MLF1 sequence (46). In leukemia, NPM can fuse with two additional partners, creating NPM-ALK and NPM-RARα, which are associated with anaplastic large-cell lymphomas and acute promyelocytic leukemia, respectively (28, 35). These three translocations exhibit totally different clinical properties, implying that the deregulation of the carboxyl-terminal proteins determines the phenotype of malignancy. It is believed that the functional importance of the NPM region is mainly for dimerization or heteromerization, leading to the constitutive activation of signaling pathways or the sequestration of normal counterparts (3, 34). NPM-MLF1 appears to be unique in its oncogenic properties. In a normal setting, both NPM and MLF1 are independently involved in distinct pathways that are essential for p53 accumulation, leading to cell growth arrest or apoptosis. Under oncogenic stress conditions, NPM stabilizes the Arf-Mdm2-p53 pathway by binding directly to Arf and by recruiting it to the nucleoli, which leads to the inactivation of Mdm2 and the accumulation of p53. Recently, cytoplasmic NPM mutants, designated NPMc+, have been identified in approximately 50 to 60% of patients with normal karyotypic AML (13), suggesting that a disturbance in the shuttling balance of NPM is a leukemogenic event. Similarly, MLF1 stabilizes p53 by suppressing COP1 activity through CSN3 and MLF1 relocates from the cytoplasm to the nucleolus by forming a leukemic fusion protein with NPM (NPM-MLF1). However, the oncogenic properties of NPM-MLF1 and the role of MLF1 in the leukemic protein have not been characterized.
In this study, we addressed two important questions about the properties of MLF1: how does cytoplasmic MLF1 interact with the CSN3-COP1-p53 tumor suppressor pathway, despite the fact that these downstream factors are mainly located in the nucleus; and what are the transforming function of NPM-MLF1 and the contribution of the MLF1 sequence in NPM-MLF1 in terms of cell transformation? We show here that MLF1 contains the nuclear export signal (NES) sequence and the putative nuclear localization signal (NLS) sequences and shuttles between the cytoplasm and the nucleus. MLF1 containing a mutant NES accumulated in the nucleus and enhanced p53-mediated growth arrest. (During the preparation of our manuscript, Winteringham et al. reported the presence of a NES sequence in murine MLF1 [45]. The region of the murine motif is identical to that in human MLF1.) Second, we found that NPM-MLF1 antagonizes the activation of p53 and facilitates escape from senescence. Disruption of the MLF1-derived NES motif in NPM-MLF1 abrogated the cellular transformation activity mediated by this fusion protein, suggesting that a shuttling imbalance of MLF1 due to aberrant genetic alterations increases susceptibility to oncogenic transformation.
MATERIALS AND METHODS
Plasmid construction and retroviral production.
We constructed a green fluorescent protein (GFP) fusion protein expression vector by modifying the retroviral vector pMSCV-IRES-puro (Clontech) (47). cDNA fragments of the wild-type MLF1 (MLF1/WT), the mutant MLF1, the NPM-MLF1 fusion protein, the mutant NPM-MLF1 fusion protein, and oncogenic Ras (Ha-RasV12) were subcloned into the retroviral vector, in frame with GFP. For viral production, the plasmid was cotransfected into 293T cells (provided by David Baltimore) together with a plasmid encoding an ecotropic helper virus containing a defective virion-packaging (ψ2) sequence. Culture supernatants containing retroviruses harvested 48 to 72 h after transfection were used to infect proliferating murine embryonic fibroblasts (MEFs) and primary murine bone marrow (BM) cells.
Cell culture, transfection, and assays.
293T human kidney cells, NIH 3T3 (Arf-null, p53 WT) mouse fibroblasts (provided by C. J. Sherr and M. F. Roussel), and Arf-inducible NIH 3T3 metallothionein (MT)-Arf cells (22) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin (GIBCO/BRL) and transfected with expression vectors via the calcium phosphate-DNA precipitation method (6, 42). For subcellular localization analysis, 293T cells were photographed under (GFP) fluorescence microscopy (Olympus, Japan) at 48 to 72 h after transfection with and without treatment with leptomycin B (LMB; 10 ng/ml; provided by M.Yoshida) (48). For the colony formation assay, NIH 3T3 cells transfected in 35-mm-diameter dishes were replated at 1:10 and 1:100 dilutions onto duplicate 100-mm-diameter dishes, selected in the presence of puromycin (5 μg/ml; Clontech) for 5 days, and allowed to form colonies for an additional 9 days in the absence of the drug. Only GFP-positive colonies were enumerated under fluorescence microscopy. For the Arf induction assay, GFP-positive mixed populations of MT-Arf cells were selected in culture with puromycin (5 μg/ml), followed by cell sorting (FACS Vantage; Becton Dickinson), and expanded in 60-mm-diameter culture dishes for further analysis. For flow cytometry analysis of DNA content, cells were suspended in a 1-ml solution of 0.1% sodium citrate and 0.1% Triton X-100 containing 50 μg/ml of propidium iodide and treated with 1 μg/ml of RNase for 30 min at room temperature. Fluorescence from the propidium iodide-DNA complex was measured with a FACScan flow cytometer (Becton Dickinson), and the percentages of cells in phases G1, S, and G2/M of the cell cycle were determined with Modifit cell cycle software. Bromodeoxyuridine (BrdU) incorporation was determined using a BrdU labeling and detection kit (Roche) according to the manufacturer's instructions. Briefly, cells were incubated in 10 μM bromodeoxyuridine for 15 min, treated with 70% ethanol-glycine (50 mM, pH 2.0), and stained with anti-BrdU mouse monoclonal antibody and fluorescein isothiocyanate-linked anti-mouse immunoglobulin G.
GST pull-down assay.
cDNA fragments containing the MLF1/WT (residues 1 to 268) and the MLF1 mutant (residues 1 to 130 and 126 to 268) coding sequences were inserted into the pGEX vector (Pharmacia) in frame with glutathione S-transferase (GST). The nucleotides corresponding to the putative NES sequence of MLF1 (residues 88 to 100) and the mutant NES (leucine-to-alanine substitution at residue 89) sequence were synthesized and inserted into the pGEX vector in frame with GST. GST fusion proteins were expressed in bacteria and were purified as described previously (41). Crude cell extracts containing endogenous CRM1 proteins were isolated from proliferating NIH 3T3 cells in an EBC buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, and 1 mM EDTA) supplemented with 5 mg/ml of aprotinin, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM NaF, 0.1 mM NaVO4, and 1 mM dithiothreitol. Binding was performed by incubating GST fusion proteins immobilized on the beads with cell lysates in the EBC buffer supplemented with 0.5% bovine serum albumin at 37°C for 1 h, and the protein complexes were washed in the same buffer. Bound CRM1 proteins were detected by immunoblotting using a CRM1-specific antibody. The same amounts of beads used for the binding assay were subjected to gel electrophoresis and stained with Coomassie blue to quantitatively examine the GST proteins.
MEF analyses.
Primary MEFs (passaged 1 to 3 times after they were prepared from 13.5-day-old embryos) were cultured in DMEM supplemented with 10% FBS, 2 mM glutamine, 100U/ml of penicillin, 100 μg/ml of streptomycin, and 0.1 mM nonessential amino acids and infected twice with pMSCV-GFP-NPM-MLF1, pMSCV-GFP-NPM-MLF1/L89A and empty pMSCV-GFP retroviral supernatants in the presence of Polybrene (4 μg/ml). GFP-positive cells with puromycin resistance (5 μg/ml) were propagated and maintained according to a 3T6 protocol based on a schedule described by Todaro and Green (40). In brief, after cells were selected in puromycin, they were seeded at 6 × 105 cells per 60-mm dish and set in culture for 3 days, which is referred to as passage 1 in this protocol. The cells were then replated at approximately 6 × 105 to 7 × 105 cells per 60-mm-diameter dish every 3 days, and the total number of cells proliferating from passage 1 was counted. For the colony formation assay at low density, MEFs were seeded at 6 × 105 cells per 60-mm dish and infected twice with pMSCV-GFP-NPM-MLF1, pMSCV-GFP-NPM-MLF1/L89A, and empty pMSCV-GFP retroviral supernatants with and without pMSCV-GFP-RasV12 retroviral supernatants in the presence of Polybrene (4 μg/ml). At 17 days after retroviral infection, foci were fixed in 4% paraformaldehyde and stained in 0.4% crystal violet. For colony formation in soft agar, cells cloned from individual foci were resuspended in complete medium containing 0.3% low-melting-point agarose and seeded at 1 × 104 cells in 1 ml of soft agar per 35-mm dish. The formation of spherical colonies was evaluated after 2 weeks by microscopy.
Protein analyses.
Cell lysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were performed as described previously (22, 47). In brief, to prepare lysates directly for immunoblotting, cell lysates in an EBC buffer were mixed with the same amount of 2× SDS sample buffer (80 mM Tris-HCl, pH 6.8, 0.2 M dithiothreitol, 2% SDS, 20% glycerol, and 0.1% bromophenol blue) and boiled for 4 min. Protein samples were separated on SDS-polyacrylamide gels under reducing conditions, transferred to a polyvinylidene difluoride membrane (Millipore), and immunoblotted with the antibodies indicated. Proteins were detected with an ECL blotting system (Amersham) according to the manufacturer's instructions. MLF1 and NPM-MLF1 proteins were detected using a mouse monoclonal antibody to MLF1 (3E9) generated with bacterially produced human MLF1 polypeptides (27). Rabbit polyclonal antibodies to CRM1 (H-300), p53 (FL-393), and CDK2 (M2), and a goat polyclonal antibody to p21 (C-19) were purchased from Santa Cruz Biotechnology. A mouse monoclonal antibody to γ-tubulin (GTU-88) and a rabbit polyclonal antibody to p19Arf (Ab80) were obtained from Sigma and Abcam, respectively. Rabbit polyclonal antibodies to COP1 and p21 were generated using bacterially produced polypeptides in our laboratory. A mouse monoclonal antibody to MDM2 was provided by Arnold J. Levine.
Retroviral infection and transformation assays with primary BM cells.
BM cells were aseptically isolated from the femurs and tibias of 6- to 8-week-old C57BL/6 mice (CLEA Japan, Inc.) and incubated overnight in BM medium (DMEM supplemented with 15% heat-inactivated FBS, 5% WEHI3B-conditioned medium, murine interleukin-3 [mIL-3; 6 ng/ml], human IL-6 [hIL-6; 10 ng/ml], murine stem cell factor [mSCF; 50 ng/ml], 2 mM glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin) (recombinant cytokines were from Genzyme). Nucleated BM cells were plated at approximately 107 cells per well in 6-well plates and infected twice with 5 ml of culture supernatants containing retroviruses encoding GFP, GFP-NPM-MLF1, and GFP-NPM-MLF1/L89A, according to the spin infection procedure, in the presence of Polybrene (4 μg/ml). For the liquid culture, infected BM cells were selected in puromycin (5 μg/ml) for 5 days and split and maintained every 4 days in BM medium.
For the colony formation assay, BM cells were cultured in BM medium for 72 h after retroviral infection, and GFP-positive populations of BM cells were isolated by cell sorting with FACS Vantage. Cells were plated at 1,000 cells per 35-mm dish onto methylcellulose-based medium containing mIL-3 (10 ng/ml), hIL-6 (10 ng/ml), mSCF (50 ng/ml), and human erythropoietin (3 U/ml) (MethoCult GF M3434; StemCell Technologies, Inc.). After 3 weeks, the colonies containing more than 100 cells were enumerated. For the second plating, the cells were collected from the methylcellulose-based medium, counted, and replated at 1,000 cells per 35-mm dish into fresh medium. Remaining cells were cultured in BM medium. Some colonies and liquid cultures were cytocentrifuged onto glass slides, stained with a May-Grunwald Giemsa solution (Merck), and viewed by phase-contrast microscopy in order to evaluate the lineage.
RESULTS
The cytoplasmic localization of MLF1 is regulated by a CRM1-dependent nuclear export system.
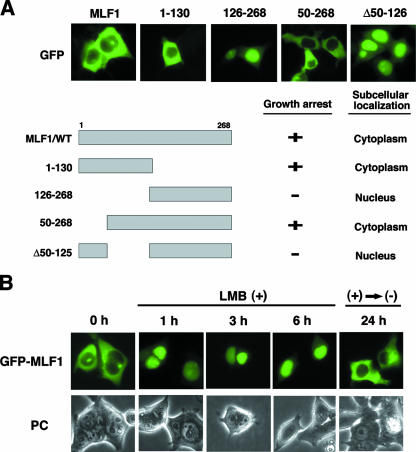
We found that some MLF1 mutants are detected exclusively in the nucleus, while the MLF1/WT is found largely in the cytoplasm, with a small fraction in the nucleus (47). To narrow down the region of MLF1 required for the cytoplasmic distribution, we introduced expression vectors encoding a series of MLF1 deletion mutants fused with GFP into 293T cells. An MLF1 mutant lacking the N-terminal 49-amino-acid sequence (MLF1/50-268), but not one lacking 125 amino acids (MLF1/126-268), was detected in the cytoplasm. Furthermore, an MLF1 mutant lacking residues 50 to 125 (MLF1/Δ50-125) was found in the nucleus, indicating that the region (amino acids 50 to 125) contains a sequence required for the cytoplasmic localization (Fig. 1A).
FIG. 1.
The cytoplasmic localization of MLF1 is disrupted by LMB. (A) Subcellular localization and schematic representation of MLF1 deletion mutants. 293T cells were transfected with the GFP-MLF1/WT and GFP-MLF1 mutant expression vectors and viewed by using fluorescence (GFP) microscopy (upper panels). Results of growth inhibition and subcellular localization are summarized on the right (lower panel). (B) Effects of LMB treatment on the subcellular distribution of MLF1. 293T cells transfected with GFP-MLF1 were treated with 10 ng/ml of LMB and photographed at 1, 3, and 6 h by fluorescence (GFP) microscopy. Cells were also photographed at 24 h after the removal of LMB from the culture. PC, phase contrast.
To test the possibility that the subcellular distribution of MLF1 is regulated by an NES-mediated system, we used LMB, which specifically inhibits NES-dependent transport by competing for interaction with an NES receptor, CRM1 (14, 16, 48). GFP-fused MLF1 was detected largely in the cytoplasm, with a small fraction found in the nucleus (Fig. 1B, left panel), but upon treatment with LMB, GFP-MLF1 rapidly moved into the nucleus within 1 h (Fig. 1B, middle three panels). Removal of LMB allowed MLF1 to relocate to the cytoplasm (Fig. 1B, right panel). These results indicate that MLF1 continuously shuttles between the nucleus and the cytoplasm in proliferating mammalian cells.
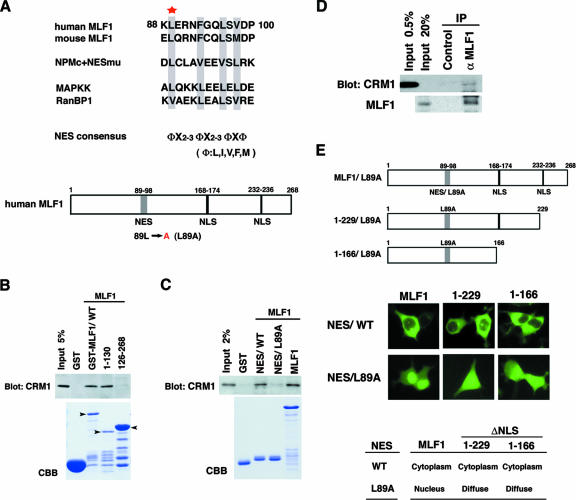
After a detailed examination of the amino acid sequence 50 to 125, we identified a putative NES consensus sequence at residues 89 to 96. This sequence was conserved in human and mouse MLF1 and contained four regularly spaced hydrophobic amino acids, which is characteristic of a typical NES motif (Fig. 2A) (12, 23). To determine whether MLF1 can interact directly with CRM1, we generated GST-tagged recombinant proteins and performed an in vitro binding assay using a lysate prepared from murine NIH 3T3 fibroblasts. Full-length MLF1/WT and the N-terminal-half mutant MLF1/1-130 efficiently bound to CRM1 in vitro, whereas the C-terminal-half mutant MLF1/126-268 did not (Fig. 2B), consistent with the idea that the putative NES sequence found at amino acids 89 to 96 is the CRM1-interaction site. To directly test whether the sequence 89 to 96 functions as the CRM1-binding site, we performed an in vitro binding assay and found that CRM1 did bind to the GST recombinant protein containing the minimum region of this putative NES sequence, whereas conversion of the conserved leucine-to-alanine substitution at residue 89 (NES/L89A) abolished the interaction (Fig. 2C). Furthermore, we tested whether MLF1 binds to CRM1 in living cells. In human leukemia K562 cells, which stably express endogenous MLF1 and CRM1, CRM1 protein was detected in an anti-MLF1 immunoprecipitate (Fig. 2D, upper panel), indicating that MLF1 interacts with CRM1 in vivo. We also identified a putative consensus sequence of NLSs at residues 168 to 174 and 232 to 236 (Fig. 2A). To determine whether these two regions are required for the nuclear import of MLF1, we introduced GFP-tagged expression vectors carrying MLF1 deletion mutations lacking putative NLS sequences of the C terminus (MLF1/1-229 and MLF1/1-166) with and without a leucine-to-alanine point mutation (MLF1/L89A) in the NES sequence into 293T cells (Fig. 2E, upper panel). Wild-type MLF1 was detected largely in the cytoplasm, with a small fraction in the nucleus, while MLF1/L89A was found exclusively in the nucleus (Fig. 2E, left panel). MLF1 deletion mutants (MLF1/1-229 and MLF1/1-116) with intact NES sequences were detected in the cytoplasm, whereas GFP signals were distributed diffusely in both the cytoplasm and the nucleus in the cells expressing the identical deletion mutants harboring a leucine-to-alanine mutation in the NES sequence (1-229/L89A and 1-166/L89A) (Fig. 2E, middle and right panels). These results show that MLF1 contains an NES motif in the N terminus and NLS motifs in the C terminus and shuttles between the nucleus and the cytoplasm by interacting directly with CRM1.
FIG. 2.
A putative NES in MLF1 and binding to CRM1. (A) Putative NES sequences of human and mouse MLF1 are aligned with known NES sequences identified in the NPMc+ mutant, mitogen-activated protein kinase kinase (MAPKK), and RanBP1. Well-conserved hydrophobic residues identical to a consensus sequence are boxed. The star indicates the location of residue 89, where we generated a lencine-to-alanine point mutant. Schematic representations of the primary structure of human MLF1 and its leucine-to-alanine mutant (L89A) are shown at the bottom. (B) MLF1 interacts with CRM1 in vitro. GST-control cell fusion or GST-MLF1 mutant fusion proteins shown at the top of the panel were incubated with NIH 3T3 cell lysates containing endogenous CRM1 proteins. Bound proteins were detected by immunoblotting with antibody to CRM1. The amounts of GST proteins absorbed on the beads were evaluated by Coomassie brilliant blue staining (CBB). The positions of the GST-MLF1/WT and the GST-MLF1 mutant fusion proteins are indicated with arrowheads. (C) The NES core sequence of MLF1 specifically binds to CRM1 in vitro. GST fusion proteins with an intact NES core sequence (NES/WT) corresponding to residues 88 to 100 of MLF1 or the leucine-to-alanine NES mutant (NES/L89A) were incubated with NIH 3T3 cell lysates. Bound proteins were detected by immunoblotting, and the amounts of absorbed GST proteins were evaluated as described in the legend to panel B. (D) Specific interaction between endogenous MLF1 and CRM1 proteins. Endogenous MLF1 protein was immunoprecipitated from the K562 cell lysate shown at the top and analyzed by immunoblotting with antibodies to CRM1 (upper panel) and MLF1 (lower panel). (E) Schematic representations of MLF1 deletion mutants of putative NLS sequences with a NES/L89A mutation and subcellular localization. 293T cells were transfected with the GFP-MLF1/WT and the GFP-MLF1/L89A NLS region deletion mutant (MLF1/1-229, MLF1/1-166, MLF1/1-229/L89A, and MLF1/1-166/L89A) expression vectors (upper panel) and viewed using fluorescence (GFP) microscopy (middle panel). The results of subcellular localization are summarized in the lower panel.
Functional significance of MLF1-derived NES for both MLF1 and NPM-MLF1 activities.
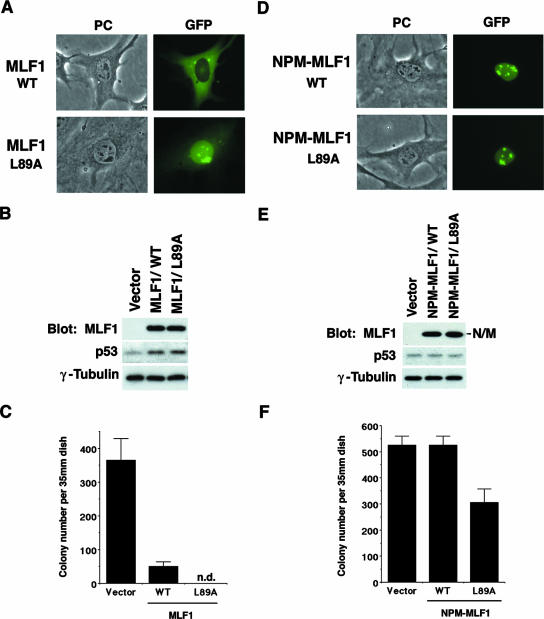
To assess the physiological significance of the subcellular distribution of MLF1 protein, we ectopically expressed an MLF1 mutant (MLF1/L89A) harboring a leucine-to-alanine mutation in the NES sequence. NIH 3T3 mouse fibroblasts were transfected with expression vectors, which encode the MLF1/WT and MLF1/L89A mutant proteins fused with GFP and contain a puromycin resistance gene as a selection marker. The intracellular localization of the MLF1/WT and the MLF1/L89A mutant was basically the same as that shown in Fig. 2E and 3A. The transfected cells expressed equivalent amounts of the MLF1/WT and MLF1/L89A proteins at the time of selection in puromycin (Fig. 3B) and contained upregulated levels of p53 (the level was slightly [ca. 5%] higher in cells transfected with MLF1/L89A than in the MLF1/WT transfectants). Consistent with our previous findings, the ectopic expression of MLF1 induced p53-dependent cell cycle arrest in mouse fibroblasts (47). After cultures were incubated for 14 days in puromycin, the number of GFP-positive colonies derived from MLF1-transfected cells was markedly decreased (ca. 13.7%) compared with that derived from GFP-transfected control cells. Interestingly, very few GFP-positive colonies expressing the MLF1/L89A mutant proteins were detected after selection (Fig. 3C). Cells expressing MLF1/L89A seemed to stop proliferating after several rounds of cell division, and we detected only a group of scattered GFP-positive cells. Thus, the exclusive expression of MLF1 in the nucleus enhanced the antiproliferative activity of MLF1.
FIG. 3.
Disruption of NES enhances MLF1-induced growth arrest. NIH 3T3 cells (Arf null, p53 WT) were transfected with GFP alone, the GFP-fused WT (MLF1 or NPM-MLF1), and the NES mutant (MLF1/L89A or NPM-MLF1/L89A [leucine 89 of MLF1 sequence is replaced by alanine]) expression vectors and selected in puromycin for 5 days. (A) Subcellular localization of MLF1/L89A. Before puromycin selection, GFP-positive cells were viewed using fluorescence microscopy and photographed. PC, phase contrast. (B) Expression levels of GFP-MLF1, GFP-MLF1/L89A, and endogenous p53 proteins at an early phase are shown by Western blotting with antibodies to MLF1, p53, and γ-tubulin. (C) At 14 days posttransfection, GFP-positive colonies were enumerated. Data are the averages of three independent experiments shown as means ± standard deviations (SD). (D) Subcellular localization of NPM-MLF1 and NPM-MLF1/L89A. Before puromycin selection, GFP-positive cells were viewed using fluorescence microscopy and photographed. (E) Expression levels of the GFP-NPM-MLF1, GFP-NPM-MLF1/L89A, and p53 proteins at an early phase are shown by Western blotting with antibodies to MLF1, p53, and γ-tubulin. (F) At 14 days posttransfection, GFP-positive colonies were enumerated. Data are averages of three independent experiments shown as means ± SD. Results shown in panels B and E are representative of three independent experiments.
In contrast to MLF1, the leukemic NPM-MLF1 fusion protein was located predominantly in the nucleolus and lost antiproliferative activity (Fig. 3D, E, and F). NPM-MLF1 contains 175 amino acids of N-terminal NPM and almost the entire MLF1 sequence, lacking only the last 16 N-terminal amino acids (46). Since our previous results showed that the MLF1 deletion mutant lacking 22 N-terminal amino acids (MLF1/23-268) retains both growth inhibitory activity and cytoplasmic localization (47), it seems likely that the relocation to the nucleolus is responsible for the loss of growth-suppressing activity. To determine whether the MLF1-derived NES sequence plays a role in NPM-MLF1, we mutated the conserved leucine residue located in the MLF1-derived NES sequence to alanine (NPM-MLF1/L89A) and expressed the mutant in NIH 3T3 cells. Because our NIH 3T3 cells harbor a global deletion at the ARF locus but retain an intact p53 allele, we can exclude any possible effect of the NPM domain on the Arf-Mdm2-p53 growth-suppressing pathway. Transfection with expression vectors containing GFP alone and GFP-fused NPM-MLF1 produced the equivalent number of colonies after selection in the presence of puromycin, whereas cells expressing GFP-fused NPM-MLF1/L89A formed significantly fewer (ca. 70%) colonies (Fig. 3F). NPM-MLF1/L89A was detected largely in the nucleolus (Fig. 3D). These results suggest that the MLF1-derived NES sequence is functional in NPM-MLF1 and that the disruption of this motif impaired the cell proliferation in an Arf-independent manner. Presumably, in addition to their relocating to the nucleolus, shuttling between the cytoplasm, the nucleoplasm, and the nucleolus is essential for the function of MLF1 and NPM-MLF1.
NPM-MLF1 actively induces cell immortalization and oncogenic transformation.
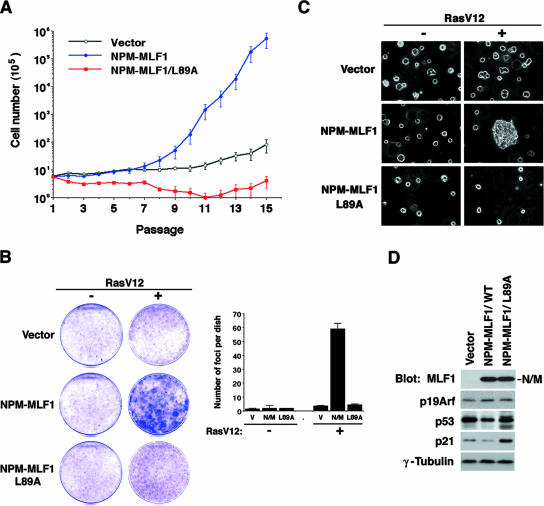
We examined further the effects of ectopic expression of the leukemic NPM-MLF1 fusion protein on the growth of primary MEFs. We infected MEFs with retroviruses that expressed GFP alone, GFP-fused NPM-MLF1, and NPM-MLF1/L89A mutant proteins and selected them in puromycin. GFP-positive MEF cultures were propagated according to the “3T6” protocol (40), and the first culture plated after the selection was referred to as passage 1 (Fig. 4A). All MEF cultures infected with retroviral vectors ceased to proliferate and entered senescence at passages 2 to ∼4. The population of MEFs infected with the GFP control did not increase for more than 10 passages, but some cells regained growth capability at passages 15 to 17. In contrast, MEFs expressing NPM-MLF1 reproducibly recovered from senescence much earlier than the GFP control MEFs. At passages 7 to ∼8, NPM-MLF1-infected MEFs started to proliferate significantly faster and reached higher cell densities than control cells, after which they continued to proliferate exponentially without recurrently entering into senescence (Fig. 4A). Reverse transcription (RT)-PCR and DNA sequencing revealed that, at passage 16, NPM-MLF1-expressing MEFs contained a missense mutation in one allele, but not in both alleles, of the p53 gene locus, suggesting that NPM-MLF1 can weaken the p53-associated growth-suppressing activity but presumably do not completely inactivate the p53 pathway. The growth rate of MEFs expressing NPM-MLF1/L89A was much lower than that of MEFs expressing NPM-MLF1 or GFP alone (Fig. 4A). Consistent with the results obtained for NIH 3T3 cells, the MLF1-derived NES sequence plays an important role in NPM-MLF1-associated activity. We observed little enhancement of apoptosis for the wild-type MEFs and little effect on the proliferation of p53-null MEFs after the infection of retroviruses encoding wild-type and mutant forms of NPM-MLF1 and MLF1 (see Fig. S1 in the supplemental material), indicating that the defect in proliferation is due to the p53-dependent proliferation and not to apoptosis. Therefore, the ectopic expression of NPM-MLF1 in MEFs facilitated escape from senescence much earlier and the MLF1-derived NES sequence is essential for this function.
FIG. 4.
Accelerated immortalization and transformation of MEFs expressing NPM-MLF1. (A) Growth curves of MEFs according to the 3T6 protocol. Primary MEFs were infected with retroviruses that expressed GFP alone or GFP-fused NPM-MLF1 or NPM-MLF1/L89A protein and then selected in puromycin. At 3-day intervals, cells were harvested, counted, and passaged by replating at 6 × 105 cells per 60-mm-diameter dish. Data are averages of three independent experiments shown as means ± standard deviations (SD). (B) Colony formation assay using primary MEFs infected with empty, NPM-MLF1, or NPM-MLF1/L89A expression vectors with or without oncogenic Ras. Foci of transformed cells were stained with crystal violet and enumerated. Data are averages of two independent experiments shown as means ± SD. (C) Colony formation in soft agar. Primary MEFs were infected with empty, NPM-MLF1 or NPM-MLF1/L89A expression vectors with or without oncogenic Ras. (D) MEFs pooled from the focus formation assay shown in panel B were lysed and analyzed by immunoblotting with antibodies to MLF1, p19Arf, p53, p21, and γ-tubulin. Results shown in panels B (left), C, and D are representative of two independent experiments.
To further examine the activity associated with NPM-MLF1, MEFs were infected with NPM-MLF1 and NPM-MLF1/L89A together with the activated form of Ras (Ha-RasV12), and the transforming activity was evaluated by colony formation assays (37). Infection with NPM-MLF1or Ha-RasV12 alone resulted in few foci, but we observed a significant increase in the number of foci in those cultures that were coinfected with NPM-MLF1 and Ha-RasV12 (Fig. 4B). These cells also acquired the ability to proliferate in soft agar (Fig. 4C). However, coinfection with NPM-MLF1/L89A and Ha-RasV12 neither increased the number of foci nor enabled the cells to proliferate in soft agar (Fig. 4B and 4C). The expression of p19Arf was induced, but the activation of p53 was blunted in MEFs expressing NPM-MLF1 but not NPM-MLF1/L89A (Fig. 4D). Thus, the expression of NPM-MLF1 in MEFs facilitated an early escape from senescence and induced oncogenic transformation in collaboration with Ras. The MLF1-derived NES sequence is also crucial for these functions.
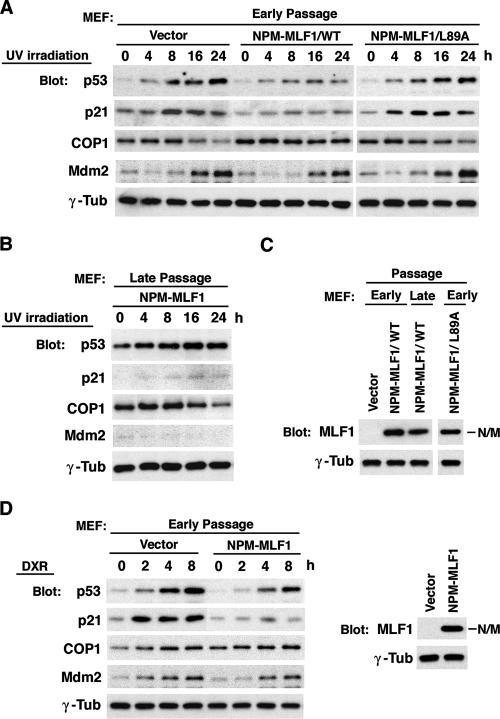
NPM-MLF1 impairs p53 activation induced by genotoxic stress and oncogenic cellular stress.
NPM and MLF1 wild types are both involved in the stabilization of p53 protein through the Arf-Mdm2-p53 and CSN3-COP1-p53 pathways, respectively. Therefore, it is likely that the leukemic fusion protein formed by these two interferes with both the Mdm2 and the COP1 pathways and, thereby, counteracts p53 accumulation in cells exposed to genotoxic stress. To explore this possibility, we examined the effect of NPM-MLF1 expression on genotoxic stress-induced activation of p53. We infected MEFs at passage 2 with retroviruses expressing GFP alone or GFP-fused NPM-MLF1 or NPM-MLF1/L89A protein. After selection in puromycin, these MEFs were treated with UV light (25 J/m2), and the kinetics of endogenous p53, p21, COP1, and Mdm2 protein expression were analyzed (Fig. 5A). UV irradiation induced a distinct accumulation of p53, p21, and Mdm2 and a concomitant decrease of COP1 in GFP control cells but failed to induce full upregulation of p53 and p21 expression in cells infected with the NPM-MLF1 retrovirus. The level of COP1 seemed to decline less in cells expressing NPM-MLF1 than in control cells. NPM-MLF1/L89A expression had little effect on suppressing p53 stabilization following UV irradiation. MEFs expressing NPM-MLF1 were immortalized at a late passage and harbored a mutant p53 gene at one allele, with the other locus intact, expressing a higher basal level of p53. Upon stimulation of these cells with UV, the level of p53 increased further, but little expression of p21 and MDM2 occurred. Interestingly, a marked downregulation of COP1 expression was observed in these cells in response to UV irradiation (Fig. 5B), presumably because these cells tolerate high levels of mutant p53. Importantly, the level of NPM-MLF1 protein remained unchanged between the early and late passages (Fig. 5C). To determine whether NPM-MLF1 impairs p53 activation in response to other genotoxic stress signals, we continuously treated MEFs with doxorubicin (DXR; 0.5 μg/ml) and analyzed the kinetics of the p53, p21, COP1, and Mdm2 proteins (Fig. 5D). MEFs expressing NPM-MLF1 were defective in the full activation of p53 and p21 in response to DXR compared with that of cells infected with the GFP control. These results indicate that the expression of NPM-MLF1 affects the normal stabilization of p53 in response to genotoxic stress, in which the MLF1-derived NES sequence plays an essential role.
FIG. 5.
NPM-MLF1 fusion protein prevents full induction of p53 by genotoxic stress. Primary MEFs were infected with retroviruses expressing GFP alone or GFP-fused NPM-MLF1 or NPM-MLF1/L89A protein and then selected in puromycin. NPM-MLF1 cells were propagated to the late passage (p16), and the p53 status was determined by RT-PCR and DNA sequencing. (A and B) Cells were treated with 25 J/m2 of UV irradiation, and the expression levels of p53, p21, COP1, Mdm2, and γ-tubulin were measured by immunoblotting at the indicated times after exposure. (C) Expression levels of NPM-MLF1 proteins at early and late passages are shown by immunoblotting with an antibody to MLF1. (D) Cells were treated in culture with 0.5 μg/ml of DXR, and p53, p21, COP1, Mdm2, and γ-tubulin were measured by immunoblotting at the indicated times after coculture with DXR (left panel). The expression level of NPM-MLF1 protein is shown by immunoblotting with an antibody to MLF1 (right panel). All the data shown in panels A, B, C, and D are representative of two independent experiments.
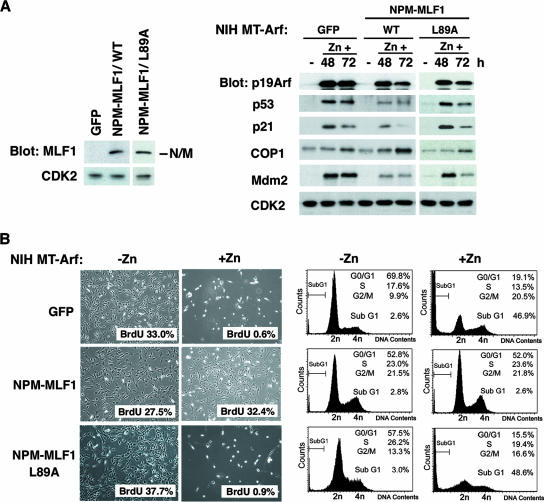
In addition to being activated by DNA damage, p53 is also activated by various oncogenic signals through p19Arf. The expression of Arf is triggered by the activation of E2F1, the overexpression of Myc, and the activating mutations of Ras (1, 32, 50). To investigate whether NPM-MLF1 expression affects the activation of p53 induced by oncogenic signals, we used a hemagglutinin (HA)-tagged Arf-inducible expression system based on NIH 3T3 murine fibroblasts, a cell type which lacks the endogenous Ink4a-Arf locus. In this NIH MT-Arf cell system, the induction of HA-tagged Arf is engineered under the control of the zinc-responsive MT promoter (Fig. 6) (22). MT-Arf cells were transfected with the GFP empty vector, GFP-fused NPM-MLF1, and NPM-MLF1/L89A, and the GFP-positive population of cells was isolated by cell sorting and seeded at a low density. Treatment of cells transfected with GFP with 100 μM Zn2+ induced an accumulation of HA-Arf, the stabilization of p53, and a marked increase in activation levels of p21 and Mdm2, whereas the level of p53 activation and the resultant induction of p21 and Mdm2 expression were much reduced in cells transfected with NPM-MLF1 (Fig. 6A, right panel). Interestingly, the expression of COP1 is upregulated in response to Arf, and the ectopic expression of NPM-MLF1 further enhanced the expression of COP1, but not that of Mdm2, compared with that of GFP control cells at 72 h. After they were treated with zinc for 96 h, the control cells underwent apoptosis and were gradually eliminated from the culture (Fig. 6B, upper panel), whereas cells expressing NPM-MLF1 were partially resistant to Arf-induced apoptosis and seemed to retain the ability to proliferate (Fig. 6B, middle panel). As expected, NPM-MLF1/L89A lost the ability to suppress the accumulation of p53 and the resultant apoptosis induced by Arf (Fig. 6A, right panel, and B, lower panel). To determine whether the cells expressing NPM-MLF1 were still proliferating, we examined BrdU incorporation and a cell cycle profile of these cells before and after the induction of Arf by Zn. More than 30% of cells expressing NPM-MLF1 incorporated BrdU even after the induction of Arf (Fig. 6B), indicating that the cells were in cycle and proliferating. In addition, cells expressing NPM-MLF1 showed little enhancement of apoptosis after the induction of HA-Arf (sub-G1 phase population, from 2.8% to 2.6%), whereas cells expressing GFP alone and NPM-MLF1/L89A exhibited a marked increase in the apoptotic population in the sub-G1 phase (from 2.6% to 46.9% and from 3.0% to 48.6%, respectively) (Fig. 6B, right panel). Thus, NPM-MLF1 can interfere with the activation of p53 and the accumulation of p21 and Mdm2 induced by Arf, which is consistent with the findings obtained from NPM-MLF1-expressing MEFs treated with UV irradiation and doxorubicin.
FIG. 6.
NPM-MLF1 fusion protein prevents full induction of p53 by oncogenic cellular stress. The inducible NIH MT-Arf cells were transfected with GFP control, GFP-fused NPM-MLF1, and NPM-MLF1/L89A expression vectors and then selected in puromycin, followed by cell sorting. (A) Expression levels of the NPM-MLF1 (WT and L89A) proteins are shown by immunoblotting with an antibody to MLF1 (left panel). Cells treated with zinc for the indicated times were harvested and analyzed by immunoblotting with antibodies to p19Arf, p53, p21, COP1, Mdm2, and CDK2 (right panel). (B) NIH MT-Arf cells treated with (+Zn) and without (-Zn) zinc for 96 h were photographed (left panel). At 72 h post-HA-Arf induction, cells were subjected to a BrdU incorporation assay (left panel) and a flow cytometric analysis of the DNA content (right panel) for cell cycle analysis. More than 500 BrdU-stained cells were enumerated. Percentages of BrdU uptake are shown at the bottom right of each photograph (left panel).
Murine hematopoietic progenitors expressing NPM-MLF1 acquire a growth advantage and p53 instability.
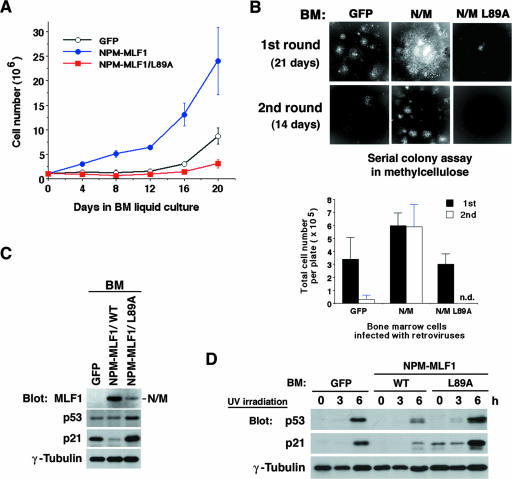
We next tested the effect of ectopic expression of the leukemic NPM-MLF1 fusion protein on the growth of mouse hematopoietic progenitor cells. The pMSCV retrovirus vector, which we used in this study, was derived from the embryonic murine stem cell virus (MSCV) and was designed to efficiently target murine hematopoietic stem cells and their progeny (18). We infected primary BM cells with retroviral supernatants containing MSCV-GFP alone, MSCV-GFP-fused NPM-MLF1, and NPM-MLF1/L89A. After cell selection in puromycin, viable GFP-positive cells were enumerated and split every 4 days. The levels of NPM-MLF1 and NPM-MLF1/L89A proteins in BM cells are shown in Fig. 7C. Growth curves show that BM cells expressing NPM-MLF1 grew markedly faster than the GFP-positive control cells, while cells expressing NPM-MLF1/L89A seemed to cease proliferation (Fig. 7A). To avoid clonal deviation of the infected cells in the liquid culture, we isolated the GFP-positive population of cells by cell sorting directly from the BM culture after 72 h postinfection and plated the cells in a methylcellulose-based medium in the presence of cytokines for hematopoietic colony formation assay. NPM-MLF1 expression did not induce any increase in the number of hematopoietic CFU (erythroid burst-forming units, granulocyte/macrophage, CFU, and CFU mixture) compared to that of GFP-positive control expression (data not shown), but we found that at 21 days after plating, colonies expressing NPM-MLF1 showed a marked increase in size and seemed to be still expanding, whereas control cells ceased to proliferate at a normal size (Fig. 7B, upper panel, “1st round”). In the secondary replating from the initial plates, the cells expressing NPM-MLF1 efficiently formed hematopoietic colonies, whereas the control cells barely retained the ability and formed only a small number and size of colonies (Fig. 7B, upper panel, “2nd round”). To evaluate the replicable capacity of the infected BM cells, we enumerated the total viable cell number per dish at 21 days after initial plating and at 14 days after secondary plating. The number of total cells per dish was much larger for NPM-MLF1-infected cells than for the GFP-positive control cells and those infected with NPM-MLF1/L89A (Fig. 7B, lower panel). These results indicated that NPM-MLF1 promotes cell proliferation in hematopoietic cells and that the MLF1-derived NES motif is required for the activity.
FIG. 7.
Growth advantage and p53 instability of hematopoietic cells expressing NPM-MLF1. Freshly isolated BM cells were infected with retroviruses expressing GFP alone or GFP-fused NPM-MLF1 or NPM-MLF1/L89A protein. (A) Growth curve for BM cells. Infected BM cells were selected in puromycin and maintained in BM medium. Cells were enumerated and split every 4 days. Data are averages of three independent experiments shown as means ± standard deviations. (B) GFP-positive cells in infected BM cells were isolated by cell sorting and plated in a methylcellulose-based medium. Colonies were viewed after 3 weeks (upper panels, “1st round”) (original magnification, ×16). Viable cells were harvested, counted (lower panel, 1st), and replated in a fresh medium. Colonies were viewed (upper panels), and viable cells were enumerated (lower panel) after 2 weeks (“2nd round”). (C) Infected BM cells, which were selected and maintained in BM medium as shown in panel A, were harvested after 3 weeks postinfection. Expression levels of NPM-MLF1, p53, p21 and γ-tubulin were shown by immunoblotting. (D) Cells, selected and maintained as described in the legend to panel C, were treated with 25 J/m2 of UV irradiation, and p53, p21 and γ-tubulin expression levels were measured by immunoblotting at the indicated times after exposure. Results shown in panels C and D are representative of two independent experiments.
To assess whether our findings obtained from the MEFs are common in the mechanism underlying the growth-promoting activity of NPM-MLF1 in hematopoietic cells, we examined the effect of the NPM-MLF1 fusion protein on the p53 tumor suppressor pathway in BM cells. We evaluated levels of endogenous p53 and p21 in infected BM cells from the liquid culture after selection in puromycin. In proliferating cells, the level of p21 was lower in NPM-MLF1-infected cells than in GFP-stained control cells (Fig. 7C). Treatment with UV irradiation (25 J/m2) induced a distinct accumulation of p53 and p21 in control cells but failed to induce full upregulation of p53 and p21 expression in cells infected with the NPM-MLF1 retrovirus (Fig. 7D). In contrast, NPM-MLF1/L89A seemed to enhance the activation of p53 and p21 after cultures were exposed to UV. Thus, the expression of NPM-MLF1 leads directly to the generation of hematopoietic populations with growth advantage and p53 instability because of the altered shuttling activity.
DISCUSSION
In this study, we demonstrated that MLF1 is a shuttling protein containing a functional NES sequence, which enables binding to CRM1. MLF1 is found predominantly in the cytoplasm, with a small fraction present in the nucleus, and ectopic expression of the protein induces growth arrest in a p53-dependent manner. Our findings provide a clue as to why cytoplasmic MLF1 is able to interact with its downstream elements CSN3, COP1, and p53, which are located mainly in the nucleus. A mutant MLF1 containing a disrupted NES sequence was relocated to the nucleus and exhibited a stronger induction of growth arrest than the WT protein did. In addition, our previous study showed that an MLF1 deletion mutant (designated MLF1/1-130), which lacked all of the potential NLS sequence, inhibited cell proliferation more efficiently than the WT protein (47). These results imply that in a normal setting, the proper shuttling of MLF1 is critical to controlling its activity. Furthermore, we showed here that the leukemic fusion protein NPM-MLF1, recruited to the nucleolus, has the ability to override cellular senescence and to facilitate oncogenic transformation. Importantly, the MLF1-derived NES sequence in NPM-MLF1 is functional and contributes to leukemogenesis because the mutant with a disrupted MLF1-derived NES sequence loses the transformation ability of NPM-MLF1. Thus, the mechanism of leukemogenesis initiated by the formation of NPM-MLF1 is associated strictly with an imbalance of MLF1 shuttling, not with a loss of the shuttling ability.
Although the regulatory mechanism of MLF1 shuttling is currently unknown, several possible explanations are emerging from the study of proteins that interact with MLF1. MLF1 contains the consensus sequence RSXSXP for binding to 14-3-3 proteins and interacts with 14-3-3ζ when a serine residue in this motif is phosphorylated (25, 30). 14-3-3 proteins function in part as a modulator of various signaling proteins including Raf-1, Bad, and Cdc25 (15). For instance, the binding of 14-3-3 to Raf-1, a Ser/Thr kinase downstream of Ras, keeps it in an inactive state by promoting a conformational change and prevents activation of the Ras signaling pathway (29). In response to DNA damage, the mitotic inducer Cdc25 is phosphorylated by Chk1 and sequestered to the cytoplasm by the 14-3-3 protein during the interphase of the cell cycle, which prevents activation of the cyclin B/Cdc2 kinase in the nucleus (5, 26). Likewise, it is likely that the binding of 14-3-3 to MLF1 in the cytoplasm is required for the inhibition of MLF1 activity by promoting a conformational change or by masking a certain motif leading to the regulatory segregation of MLF1 from the nucleus in proliferating cells. However, a kinase which phosphorylates MLF1 and thereby promotes its binding to 14-3-3 has yet to be identified, although kinase activity toward MLF1 has been detected (25).
We recently reported that CSN3, a subunit of the CSN, is a direct mediator of MLF1 signaling to the COP1-p53 pathway (47). CSN3 itself has no kinase activity, but the CSN complex associates with kinases including inositol 1,3,4-triphosphate 5/6-kinase (39), casein kinase II (CK2), and protein kinase D (43). The last two kinases phosphorylate p53 and c-Jun through CSN3. CK2 phosphorylates threonine 155 of p53, which promotes proteasome-mediated degradation (43). Interestingly, the 14-3-3-binding motif (residues 31 to 36) in MLF1 contains a putative CK2 phosphorylation site (residues 31 to 34). Therefore, it is possible that MLF1 is one of the substrates of CK2, although the CK2-interacting protein that determines the specificity of CK2 remains to be determined. A recent study showed that ATM kinase directly phosphorylates COP1, which in turn leads to the autodegradation of COP1 and consequent stabilization of p53 (10). Interestingly, CSN3 was also identified as a substrate of ATM in DNA damage-induced apoptosis, and its phosphorylation requires the interaction of ATM with another subunit of CSN (24). Since CSN and COP1 function cooperatively, these observations suggest that CSN integrates various signals triggered by the DNA damage into the p53 activation pathway through regulation of the COP1 E3 ligase activity. More studies will be required to clarify how the shuttling activity of MLF1 is involved in the function of the CSN-COP1 axis and their associated kinases.
We have reported, for the first time, that the leukemic fusion protein NPM-MLF1 exhibits transformation activity in collaboration with the active form of Ras, presumably by preventing full activation of the p53 pathway. Both NPM and MLF1 stabilize p53 protein through the Arf-Mdm2-p53 and CSN3-COP1-p53 pathways, respectively, whereas NPM-MLF1 impairs the expression of p53 in response to genotoxic or oncogenic stress. The region of NPM retained in NPM-MLF1 includes a NES sequence (residues 94 to 102) and a bipartite NLS sequence (residues 141 to 157) (19, 44), while the region of MLF1 contains a NES sequence and two putative NLS sequences (Fig. 2A, residues 168 to 174 and 232 to 236). The acquisition of additional NES and NLS sequences should confer more powerful shuttling activity on the leukemic fusion protein. This is reminiscent of the cytoplasmic NPM mutant (NPMc+) recently identified in characteristic karyotypic AML (13). In this mutant allele, an aberrant insertion at the 3′ region of the open reading frame in the NPM gene locus resulted in a frame shift mutation and the addition of a newly generated functional NES sequence to the C terminus. The NPMc+ mutant retains some shuttling ability because treatment with LMB relocated the protein to the nucleus. The NPMc+ mutant protein presumably interferes with the Arf-Mdm2-p53 pathway by relocating Arf from the nucleolus to the cytoplasm (8, 9). In the case of NPM-MLF1, disruption of the MLF1-derived NES sequence impaired the growth-promoting activity of the fusion protein, suggesting that accelerated shuttling, not a loss of shuttling, is critical for leukemogenesis. Because the t(3;5) chromosomal translocation, which produces NPM-MLF1, is associated with MDS and AML with multilineage dysplasia and is correlated with a poor prognosis (33, 46), a shuttling imbalance in MLF1 and NPM in normal CD34+ hematopoietic stem cells may perturb the regulation of p53 stability and contribute to the oncogenic conversion of normal stem cells to leukemic stem cells.
Supplementary Material
Acknowledgments
We thank C. J. Sherr and M. F. Roussel for the NIH 3T3 cell line and I. Nakamae for excellent technical assistance.
This work was supported by Grants-in-Aid for Scientific Research and for Cancer Research from the Ministry of Education, Science, and Culture of Japan.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395124-125. [DOI] [PubMed] [Google Scholar]
- 2.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischof, D., K. Pulford, D. Y. Mason, and S. W. Morris. 1997. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol. Cell. Biol. 172312-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56379-390. [DOI] [PubMed] [Google Scholar]
- 5.Chan, T. A., H. Hermeking, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 401616-620. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 72745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo, E., J. C. Marine, D. Danovi, B. Falini, and P. G. Pelicci. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4529-533. [DOI] [PubMed] [Google Scholar]
- 8.Colombo, E., P. Martinelli, R. Zamponi, D. C. Shing, P. Bonetti, L. Luzi, S. Volorio, L. Bernard, G. Pruneri, M. Alcalay, and P. G. Pelicci. 2006. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 663044-3050. [DOI] [PubMed] [Google Scholar]
- 9.den Besten, W., M. L. Kuo, R. T. Williams, and C. J. Sherr. 2005. Myeloid leukemia-associated nucleophosmin mutants perturb p53-dependent and independent activities of the Arf tumor suppressor protein. Cell Cycle 41593-1598. [DOI] [PubMed] [Google Scholar]
- 10.Dornan, D., H. Shimizu, A. Mah, T. Dudhela, M. Eby, K. O'Rourke, S. Seshagiri, and V. M. Dixit. 2006. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 3131122-1126. [DOI] [PubMed] [Google Scholar]
- 11.Dornan, D., I. Wertz, H. Shimizu, D. Arnott, G. D. Frantz, P. Dowd, K. O'Rourke, H. Koeppen, and V. M. Dixit. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 42986-92. [DOI] [PubMed] [Google Scholar]
- 12.Engelsma, D., R. Bernad, J. Calafat, and M. Fornerod. 2004. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 233643-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falini, B., C. Mecucci, E. Tiacci, M. Alcalay, R. Rosati, L. Pasqualucci, R. La Starza, D. Diverio, E. Colombo, A. Santucci, B. Bigerna, R. Pacini, A. Pucciarini, A. Liso, M. Vignetti, P. Fazi, N. Meani, V. Pettirossi, G. Saglio, F. Mandelli, F. Lo-Coco, P. G. Pelicci, and M. F. Martelli. 2005. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352254-266. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 15.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40617-647. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390308-311. [DOI] [PubMed] [Google Scholar]
- 17.Hanissian, S. H., U. Akbar, B. Teng, Z. Janjetovic, A. Hoffmann, J. K. Hitzler, N. Iscove, K. Hamre, X. Du, Y. Tong, S. Mukatira, J. H. Robertson, and S. W. Morris. 2004. cDNA cloning and characterization of a novel gene encoding the MLF1-interacting protein MLF1IP. Oncogene 233700-3707. [DOI] [PubMed] [Google Scholar]
- 18.Hawley, R. G., F. H. Lieu, A. Z. Fong, and T. S. Hawley. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1136-138. [PubMed] [Google Scholar]
- 19.Hingorani, K., A. Szebeni, and M. O. Olson. 2000. Mapping the functional domains of nucleolar protein B23. J. Biol. Chem. 27524451-24457. [DOI] [PubMed] [Google Scholar]
- 20.Itahana, K., K. P. Bhat, A. Jin, Y. Itahana, D. Hawke, R. Kobayashi, and Y. Zhang. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 121151-1164. [DOI] [PubMed] [Google Scholar]
- 21.Korgaonkar, C., J. Hagen, V. Tompkins, A. A. Frazier, C. Allamargot, F. W. Quelle, and D. E. Quelle. 2005. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 251258-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa, K., T. Tanaka, and J. Kato. 1999. p19ARF prevents G1 cyclin-dependent kinase activation by interacting with MDM2 and activating p53 in mouse fibroblasts. Oncogene 182718-2727. [DOI] [PubMed] [Google Scholar]
- 23.Kutay, U., and S. Guttinger. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15121-124. [DOI] [PubMed] [Google Scholar]
- 24.Lavin, M. F., D. Delia, and L. Chessa. 2006. ATM and the DNA damage response. Workshop on ataxia-telangiectasia and related syndromes. EMBO Rep. 7154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, R., L. N. Winteringham, J. H. Williams, R. K. McCulloch, E. Ingley, J. Y. Tiao, J. P. Lalonde, S. Tsai, P. A. Tilbrook, Y. Sun, X. Wu, S. W. Morris, and S. P. Klinken. 2002. MADM, a novel adaptor protein that mediates phosphorylation of the 14-3-3 binding site of myeloid leukemia factor 1. J. Biol. Chem. 27740997-41008. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397172-175. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, N., N. Yoneda-Kato, T. Iguchi, Y. Kishimoto, T. Kyo, H. Sawada, E. Tatsumi, and S. Fukuhara. 2000. Elevated MLF1 expression correlates with malignant progression from myelodysplastic syndrome. Leukemia 141757-1765. [DOI] [PubMed] [Google Scholar]
- 28.Morris, S. W., M. N. Kirstein, M. B. Valentine, K. G. Dittmer, D. N. Shapiro, D. L. Saltman, and A. T. Look. 1994. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 2631281-1284. [DOI] [PubMed] [Google Scholar]
- 29.Morrison, D. K., and R. E. Cutler. 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9174-179. [DOI] [PubMed] [Google Scholar]
- 30.Ohno, K., Y. Takahashi, F. Hirose, Y. H. Inoue, O. Taguchi, Y. Nishida, A. Matsukage, and M. Yamaguchi. 2000. Characterization of a Drosophila homologue of the human myelodysplasia/myeloid leukemia factor (MLF). Gene 260133-143. [DOI] [PubMed] [Google Scholar]
- 31.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103127-140. [DOI] [PubMed] [Google Scholar]
- 32.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395125-126. [DOI] [PubMed] [Google Scholar]
- 33.Raimondi, S. C., I. D. Dube, M. B. Valentine, J. Mirro, Jr., H. J. Watt, R. A. Larson, M. A. Bitter, M. M. Le Beau, and J. D. Rowley. 1989. Clinicopathologic manifestations and breakpoints of the t(3;5) in patients with acute nonlymphocytic leukemia. Leukemia 342-47. [PubMed] [Google Scholar]
- 34.Redner, R. L., J. D. Chen, E. A. Rush, H. Li, and S. L. Pollock. 2000. The t(5;17) acute promyelocytic leukemia fusion protein NPM-RAR interacts with co-repressor and co-activator proteins and exhibits both positive and negative transcriptional properties. Blood 952683-2690. [PubMed] [Google Scholar]
- 35.Redner, R. L., E. A. Rush, S. Faas, W. A. Rudert, and S. J. Corey. 1996. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood 87882-886. [PubMed] [Google Scholar]
- 36.Schwechheimer, C., and X. W. Deng. 2001. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol. 11420-426. [DOI] [PubMed] [Google Scholar]
- 37.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88593-602. [DOI] [PubMed] [Google Scholar]
- 38.Sun, W., K. Zhang, X. Zhang, W. Lei, T. Xiao, J. Ma, S. Guo, S. Shao, H. Zhang, Y. Liu, J. Yuan, Z. Hu, Y. Ma, X. Feng, S. Hu, J. Zhou, S. Cheng, and Y. Gao. 2004. Identification of differentially expressed genes in human lung squamous cell carcinoma using suppression subtractive hybridization. Cancer Lett. 21283-93. [DOI] [PubMed] [Google Scholar]
- 39.Sun, Y., M. P. Wilson, and P. W. Majerus. 2002. Inositol 1,3,4-trisphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J. Biol. Chem. 27745759-45764. [DOI] [PubMed] [Google Scholar]
- 40.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomoda, K., Y. Kubota, Y. Arata, S. Mori, M. Maeda, T. Tanaka, M. Yoshida, N. Yoneda-Kato, and J. Y. Kato. 2002. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J. Biol. Chem. 2772302-2310. [DOI] [PubMed] [Google Scholar]
- 42.Tomoda, K., Y. Kubota, and J. Kato. 1999. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398160-165. [DOI] [PubMed] [Google Scholar]
- 43.Uhle, S., O. Medalia, R. Waldron, R. Dumdey, P. Henklein, D. Bech-Otschir, X. Huang, M. Berse, J. Sperling, R. Schade, and W. Dubiel. 2003. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J. 221302-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, W., A. Budhu, M. Forgues, and X. W. Wang. 2005. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 7823-830. [DOI] [PubMed] [Google Scholar]
- 45.Winteringham, L. N., R. Endersby, S. Kolbeke, R. K. McCulloch, J. H. Williams, J. Stillitano, S. M. Cornwall, E. Ingley, and S. P. Klinken. 2006. Myeloid leukemia factor 1 associates with a novel hnRNP-U-like molecule. J. Biol. Chem. 28138791-38800. [DOI] [PubMed] [Google Scholar]
- 46.Yoneda-Kato, N., A. T. Look, M. N. Kirstein, M. B. Valentine, S. C. Raimondi, K. J. Cohen, A. J. Carroll, and S. W. Morris. 1996. The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene 12265-275. [PubMed] [Google Scholar]
- 47.Yoneda-Kato, N., K. Tomoda, M. Umehara, Y. Arata, and J. Y. Kato. 2005. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J. 241739-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida, M., and S. Horinouchi. 1999. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N. Y. Acad. Sci. 88623-36. [DOI] [PubMed] [Google Scholar]
- 49.Yung, B. Y., H. Busch, and P. K. Chan. 1985. Translocation of nucleolar phosphoprotein B23 (37 kDa/pI 5.1) induced by selective inhibitors of ribosome synthesis. Biochim. Biophys. Acta 826167-173. [DOI] [PubMed] [Google Scholar]
- 50.Zindy, F., R. T. Williams, T. A. Baudino, J. E. Rehg, S. X. Skapek, J. L. Cleveland, M. F. Roussel, and C. J. Sherr. 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc. Natl. Acad. Sci. USA 10015930-15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.