Abstract
The proteins ERK1 and ERK2 are highly similar, are ubiquitously expressed, and share activators and substrates; however, erk2 gene invalidation is lethal in mice, while erk1 inactivation is not. We ablated ERK1 and/or ERK2 by RNA interference and explored their relative roles in cell proliferation and immediate-early gene (IEG) expression. Reducing expression of either ERK1 or ERK2 lowered IEG induction by serum; however, silencing of only ERK2 slowed down cell proliferation. When both isoforms were silenced simultaneously, compensating activation of the residual pool of ERK1/2 masked a more deleterious effect on cell proliferation. It was only when ERK2 activation was clamped at a limiting level that we demonstrated the positive contribution of ERK1 to cell proliferation. We then established that ERK isoforms are activated indiscriminately and that their expression ratio correlated exactly with their activation ratio. Furthermore, we determined for the first time that ERK1 and ERK2 kinase activities are indistinguishable in vitro and that erk gene dosage is essential for survival of mice. We propose that the expression levels of ERK1 and ERK2 drive their apparent biological differences. Indeed, ERK1 is dispensable in some vertebrates, since it is absent from chicken and frog genomes despite being present in all mammals and fishes sequenced so far.
Numerous cell surface agonists activate the signaling cascade Ras/Raf/MEK/ERK. In contrast to the upstream activation steps of the cascade, which have few known substrates, ERK phosphorylates hundreds of substrates on serine and threonine residues (52). ERK substrates are localized in all cell compartments: at the membrane, (e.g., epidermal growth factor receptor), in the cytosol (e.g., DUSP6-MKP-3), on the cytoskeleton (e.g., cortactin), and in the nucleus (e.g., Elk-1). The serine/threonine protein kinase ERKs play key roles in cell proliferation, cell differentiation, and cell death. The spatio-temporal regulation of ERK activation dictates the biological outcome (34). Therefore, for all these reasons, ERK activation is a key signaling step for the regulation of many biological responses.
Two isoforms convey ERK activity in many vertebrates; these are ERK1 and ERK2, which are 84% identical at the amino acid level (8). Among the mitogen-activated protein kinases, ERK5 (55) is the closest kinase to ERK1/2, with all three kinases being activated on the threonine and tyrosine residues of the TEY sequence. However, ERK5 cannot be considered an ERK1/2 isoform, since ERK5 and ERK1/2 are activated by different kinase modules. ERK5 is activated by the kinase MEK5 (55), unlike ERK1 and -2, which are activated by MEK1/2 (23).
Many observations indicate that ERK1 and ERK2 are very similar. ERK1 and ERK2 are ubiquitously expressed; however, their relative distributions across tissues differ (8). ERK1 and -2 display the same subcellular localization; both isoforms translocate from the cytosol to the nucleus upon stimulation of resting cells (25). ERK1 and -2 are serine/threonine protein kinases that phosphorylate substrates on the consensus PXS/TP sites. Specificity is provided not by a domain near the catalytic site but by a common docking domain and a docking groove that are located on the back of the kinase with respect to the catalytic site (42, 43). Interestingly, both the common docking domain and groove are nearly identical in ERK1 and ERK2. Indeed, evidence indicates that ERK1 and ERK2 seem to have the same substrate specificities (52). Furthermore, ERK1 and -2 are activated by the same upstream kinase module and display identical kinetics of activation. No specific agonist able to activate only one of the two kinases has been discovered so far.
In contrast to the similarities between ERK1 and ERK2 presented above, their invalidation in mice indicates clear differences between ERK1 and ERK2. Invalidation of ERK2 leads to early embryonic death around embryonic day 6.5 as a consequence of placental defects (18, 36, 51). In contrast, mice lacking ERK1 live and reproduce normally (30, 32). Minor defects in erk1−/− mice have been reported, such as impaired terminal differentiation of T lymphocytes (32), decreased adiposity with fewer adipocytes (7), and facilitated learning and memory (28). However, Selcher et al. (40) have demonstrated that mice lacking ERK1 are unimpaired in memory formation probed by the same test of passive avoidance. Surprisingly, in a recent study Vantaggiato et al. (47) proposed that ERK1 and ERK2 have opposite roles in Ras-mediated signaling, with ERK1 antagonizing the positive signaling provided by ERK2. Similarly, in the JNK signaling pathway, earlier studies on Jnk1−/− and Jnk2−/− mice suggested that the JNK1 and JNK2 isoforms had opposite effects on cJun expression and on proliferation (37, 45). However, recent studies, using a chemical genetic approach, demonstrate unequivocally that both JNK1 and JNK2 are positive regulators of these processes (21). In fact, the opposite phenotypes caused by JNK1 and JNK2 deficiency could be due to competition between JNK1 and JNK2 for partners.
As demonstrated for JNK, assessing the functions of two kinase isoforms is challenging. The discrepancy between the different studies on ERK1 and ERK2 led us to design experiments to scrutinize the specific role of ERK1 and ERK2 in two ERK-dependent cellular processes: exponential growth and immediate-early gene (IEG) transcription. These two processes were chosen because they require distinct types of ERK activation, either persistent (over several days) or rapid (for less than 1 hour).
Our results indicate that both ERK1 and ERK2 are positive regulators of cell proliferation and IEG transcription. The low threshold of ERK activity required for exponential proliferation of fibroblasts led us initially to think that only ERK2 was necessary for proliferation to proceed. However, the positive contribution of ERK1 to cell proliferation was uncovered when the ERK2 level was markedly reduced and became limiting. When ERK1 expression alone was abrogated, total ERK activation was minimally affected because of compensating activation of ERK2. Careful measurement of isoform stoichiometry indicates that ERK1 is four times less abundant than ERK2 in NIH 3T3 cells. Interestingly, activated ERK1 is also four times less abundant than activated ERK2 in stimulated NIH 3T3 cells. This correlation between the quantities of ERK isoforms and their activation ratio is reported here for the first time and suggests that both isoforms are activated equally by the upstream kinase module. We propose that this correlation is a general phenomenon, since we observed it across a wide variation of ERK1/ERK2 ratios displayed in mouse brain structures. In vitro the specific activities of ERK1 and ERK2 are indistinguishable, reinforcing the notion that ERK isoforms contribute to ERK signaling according to the ratio of their expression levels.
Transcription of IEGs requires a very robust ERK activity; in this case removal of either ERK1 or ERK2 markedly reduced transcription of ERK-responsive genes. Overall we observed that the biological outcome is tightly linked to the total ERK activity, irrespective of isoform specificity. We propose that the threshold of total ERK activity required for a given response and the relative ratio between the isoforms expressed dictate the direct effect of ERK1 or ERK2 ablation.
Finally, genetic data on mouse survival are presented to strengthen the conclusion that ERK dosage is crucial.
MATERIALS AND METHODS
Cell culture and reagents.
NIH 3T3 cells were obtained from the American Type Culture Collection (Manassas, VA). NIH 3T3 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% bovine serum (Gibco no. 16170078), penicillin, and streptomycin and incubated at 37°C in a 5% CO2 incubator. U0126 was purchased from Promega, and PD184352 was a generous gift of Michiaki Kohno and Sir Philip Cohen. The plasmid pSUPER expressing short hairpin RNAs (shRNAs) upon H1 promoter-driven transcription was a generous gift from Reuven Agami (10). The plasmid pBabepuro was described elsewhere (29).
Transfection procedure.
NIH 3T3 cells were plated at a density of 1.8 × 106 cells per 10-cm dish and transfected with 30 to 100 μg of plasmid DNA (containing 3 μg of pBabepuro plasmid) by calcium phosphate precipitation overnight. At 36 h posttransfection, puromycin (9 μM) was added and left for 24 h to kill nontransfected cells. Stringency and speed of selection were increased by cotreatment with cyclosporine (5 μM) as described previously (54). For the rest of the experiment, no selection was applied. Cells were then trypsinized, allowed to attach to new plates for 3 h, and retrypsinized to count only healthy cells. For exponential growth, cells were plated at densities of 20, 30, and 40 cells per mm2 in 12-well plates or in 10-cm dishes for immunoblotting of cells that were strictly in the exponential phase. For serum removal and restimulation studies, cells were plated at a density of 2.2 × 106 per 10-cm dish for 12 h prior to serum removal for 24 h and stimulation as described in the figure legends. Cell proliferation was measured by fixing plates daily with 5% ice-cold trichloroacetic acid, neutralization with phosphate-buffered saline, and automated counting of DAPI (4′,6′-diamidino-2-phenylindole)-stained nuclei with ImageJ software. The average number of nuclei was calculated from counting 10 fields of 2.41 mm2 (50× magnification on a Zeiss Axiovert microscope). The initial cell number influenced the rate of cell proliferation, and therefore great care was taken to compare only plates of transfected cells that were seeded at the same densities.
Immunoblotting.
Cells were lysed in Laemmli sample buffer, separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (10% acrylamide-bisacrylamide [29:1] gels), and transferred onto polyvinylidene difluoride (Immobilon-P from Millipore). Phosphorylated ERK1/2 was detected with the monoclonal anti-phospho-ERK1/2 antibody M8159 from Sigma. Total ERK was detected with mixes of different antibodies: rabbit serum E1B (1/3,000) (epitope against the last 16 amino acids of Chinese hamster ERK1) plus rabbit anti-rat ERK1 no. 61 from Zymed (no. 61-7400) (1/4,000) or mouse monoclonal ERK2 from BD Pharmingen (no. 610103) plus mouse monoclonal ERK1 from BD Pharmingen (no. 554100). Monoclonal mix 1 was made by diluting BD-ERK1 (1/5,000) plus BD-ERK2 (1/1,000), and monoclonal mix 2 was made by diluting BD-ERK1 (1/2,000) plus BD-ERK2 (1/1,000). The antihemagglutinin (anti-HA) antibody was purchased from Covance (no. MMS-101R), and vesicular stomatitis glycoprotein (VSVG) was a generous gift of B. Goud. The anti-ARD1 was described previously (6). Secondary antibodies coupled to horseradish peroxidase were purchased from Promega, and chemiluminescence was used to detect immunoreative bands on autoradiography film or with the Gnome detector from Syngene (United Kingdom).
Nude mice were killed in accordance with the European Community council directive of 24 November 1986. Mouse brain was isolated, and the apparent anatomical structure was sliced out and frozen instantly in liquid nitrogen. The name of the structure was given according to the virtual mouse necropsy web site: (http://www.niaid.nih.gov/dir/services/animalcare/MouseNecropsy/necropsy.html). Frozen samples were then resuspended in 1.5× Laemmli sample buffer and sonicated immediately. The protein concentration was measured by the bicinchoninic acid method (Pierce). The levels of ERK and phosphorylated ERK were determined by immunoblotting of a gel loaded with 30 μg and 15 μg of protein, respectively. Normalization was performed as indicated in the figure legends on the same blots after stripping off the anti-ERK antibodies.
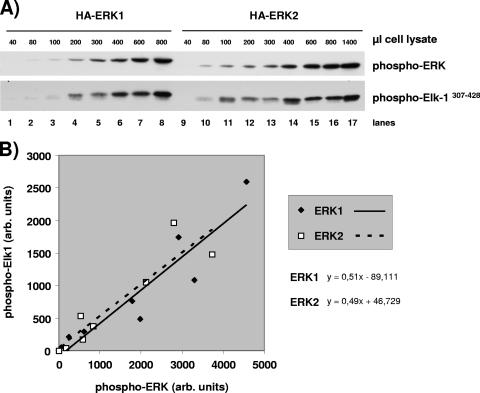
Specific kinase activities of HA-ERK1 and HA-ERK2 determined using a nonradioactive immunoprecipitation kinase assay.
CCl39 cells that express stably mouse HA-ERK1 or mouse HA-ERK2 were plated at a density of 14 × 106 cells per 10-cm dish for 24 h prior to serum removal for 24 h and stimulation with 10% fetal calf serum (FCS) for 5 min. The cells were then lysed with 1.4 ml of lysis buffer containing 50 mM Tris-HCl (pH 7.6), 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 40 mM beta-glycerol phosphate, 1% Triton X-100, 0.1% bovine serum albumin, 0.1 mg/ml soybean tryspsin inhibitor (Sigma), 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 40 mM para-nitrophenyl phosphate, 1 mM benzamidine, 1 mM sodium orthovanadate, 2 mM dithiothreitol, and protease inhibitor cocktail (Roche). Prior to immunoprecipitation, Sepharose beads coupled to protein G (Amersham Biosciences) were washed twice and incubated with the lysis buffer for 1 h. The cell extracts were centrifuged at 20 × 103 × g. Increasing quantities of supernatant were incubated with 3 μl of anti-HA antibody together with 20 μl of beads for 1 h at 4°C. For each point, the final immunoprecipitation volume was adjusted to 1.4 ml with the lysis buffer. The beads were pelleted and washed twice with lysis buffer and then once with kinase buffer containing 20 mM HEPES (pH 7.6), 10 mM MgCl2, 0.5 mM MnCl2, 40 mM para-nitrophenyl phosphate, 1 mM dithiothreitol, and 80 mM NaCl.
The pellets were resuspended in 20 μl of kinase buffer supplemented with 50 μM ATP and 1.5 μg glutathione S-transferase (GST)-Elk-1307-428 fusion protein and were incubated for 26 min at 37°C. The precipitates were dissolved by adding 20 μl 3× Laemmli buffer. The beads were pelleted, and the supernatant was boiled at 95°C for 5 min after adding dithiothreitol to a final concentration of 1 mM. The supernatants were analyzed by immunoblotting probed with anti phospho-Elk-1 antibody (Santa Cruz no. sc-8406) and monoclonal phospho-ERK1/2 antibody (Sigma). The TrueBlot Ultra horseradish peroxidase-conjugated anti-mouse antibody (eBioscience) was used as the second antibody. The chemiluminescence was detected by autoradiography and measured directly with the Gnome detector from Syngene (United Kingdom).
Plasmids.
The plasmid coding for GST-Elk-1 (positions 307 to 428) was provided by Robert Hipskind, and plasmid pBA4, containing mouse ERK2, was a generous gift of Michael Webber (19). A VSVG tag was added at the C terminus by site-directed mutagenesis of the stop codon as described previously (25) (sequence, ERK2-KGEGPPGPYTDIEMNRLGK). An HA tag was added at the N terminus by site-directed mutagenesis of the initiating methionine (sequence, MYDVPDYASLPGNW-ERK2). Mouse ERK1 was cloned by nested reverse transcription-PCR (RT-PCR) with the following primers: N terminal, GAGGCGGGAGGAGTGGAGATGGC; C terminal external, CTAACAATCTGCAGAGAAGG; and C terminal internal, GTTCTTGTTAGGGGCCCTCTGGC. RT-PCR was performed with the Phusion enzyme as described by the manufacturer (Finnzymes). A VSVG tag was added at the C terminus of mouse ERK1 by mutating the stop codon, and an HA tag was added at the N terminus by mutating the initiating methionine, exactly as was done for mouse ERK2.
shRNA sequences.
The control plasmid was either empty pSUPER plasmid or pSUPER-SIMA, which expresses an shRNA that targets Drosophila HIF1-alpha as described previously (5). pSUPER-ERK2 expressed the targeting sequence GCGCTTCAGACATGAGAAC, which is conserved between mouse and human ERK2, and pSUPER-ERK2 bis expressed the targeting sequence GGGTTCTTGACAGAGTACGTAG. pSUPER-ERK1 expressed the targeting sequence CATGAAGGCCCGAAACTAC. Sequence integrity was verified by sequencing the clones from both directions.
Quantitative RT-PCR.
Transfected cells were selected as described above, plated at a density of 2.2 × 106 cells per 10-cm dish, grown for 12 h prior to serum deprivation for 24 h, and stimulated with 10% FCS as described in the figure legends. mRNA was isolated by lysing the cells with TRIzol (Invitrogen), and residual genomic DNA was digested with RNase-free DNase (QIAGEN) and purified on RNeasy columns (QIAGEN). RT-PCR was initiated by oligo(dT) priming, and reverse transcription was performed at 37°C with the Omnicript enzyme (QIAGEN) according to the manufacture's instructions. Quantitative RT-PCR was performed on an Applied Biosystems 7300 cycler with Mastermix RT-SN2X-03+ for SYBR green dye (Eurogentec, Belgium). The PCR primers were chosen on the PrimerBank website (49) (PrimerBank IDs junB-6680512a1, egr1-6681285a3, zfp36-6756059a1, id3-6680341a1, and fosl1-6753896a3). Specific down-regulation of ERK1 or ERK2 mRNA was evaluated with the primers erk1-21489933a3 and erk2-6754632a2.
Genomic sequence analysis.
Predicted protein sequences of ERKs were obtained from the Ensembl web server. Alignment of the predicted ERKs was performed with the Multalin sequence alignment algorithm on the web server (12). For sequences that were not properly predicted in spring of 2007, such as that for the ERK1 protein from the fish Oryzias latipes, exons homologous to mouse ERK1 were searched in the translation of the entire genome of Oryzias latipes by the BLAST search algorithm of Ensembl. In August 2007, our predicted sequence of ERK1 from Oryzias latipes was annotated in the 46th release of the Ensembl database; it is available with the Ensembl ID ENSORLG00000011993. For the Gallus gallus (chicken) and Xenopus tropicalis (frog) genomes, ERK1 was not annotated in summer 2007. Furthermore, searching all exons homologous to mouse ERK1 in the translation of the entire genome of G. gallus (genome sequence version 2.1) and X. tropicalis (genome sequence version 4.1) did not allow retrieval of any ERK1 sequence; however, ERK2 and ERK5 exons were readily identified due to their homology to mouse ERK1 exons. Similarly, for X. tropicalis, searching the entire translated genomic DNA sequence with peptides for all the conserved exons of mouse MEK2 did not reveal any MEK2 exons. However, X. tropicalis MEK1 and MEK5 were readily identified by this search approach. Confirmation of the lack of ERK1 in X. tropicalis and G. gallus was done by searching expressed sequenced tag (ESTs) for ERKs. ESTs for X. tropicalis, G. gallus, and Danio rerio (zebra fish) were searched with the BLASTN algorithm of the NCBI website by retrieving sequences similar to a fragment of the mouse ERK1 cDNA (parameters, expect 1 and filter default). The fragment of the mouse ERK1 cDNA was constituted by joining exons 3, 4, and 5 (ENSMUSE00000489861, ENSMUSE00000477246, and ENSMUSE00000130333). These exons were chosen for their high conservation between mouse ERK1 and mouse ERK2. In spring 2007, in the zebra fish EST database there were 53 ERK1 ESTs and 30 ERK2 ESTs, in the frog EST database there were 38 ERK2 ESTs and no ERK1 ESTs, and in the chicken EST database there were 9 ERK2 ESTs and no ERK1 ESTs. Since ERK1 ESTs were readily found in zebra fish, which is evolutionarily more distant from mouse than chicken or frog, ERK1 ESTs should have been found in chicken and frog if ERK1 were present in their genomes. For all three animals, more ESTs than ERKs were retrieved and corresponded mainly to other kinases (e.g., cyclin-dependent kinases and stress mitogen-activated protein kinases). Hence, the lack of ERK1 ESTs in the frog and chicken databases was not due to a narrow search strategy. Similarly, retrieving sequences similar to that of mouse MEK2 in the frog EST database of GenBank revealed four ESTs for MEK1 and none for MEK2 and then ESTs for MEK6, MEK5, MEK3, confirming the absence of MEK2 in frog.
RESULTS
ERK activity is required for NIH 3T3 cell proliferation.
It was first established for fibroblasts that ERK activation is a requisite for growth factor-induced cell growth (33). However, this is not the case for all cell types; for example, embryonic stem cells grow independently of ERK activation (11). Similarly, several tumor cell lines proliferate with low levels of ERK activation (20). Our shRNAs were designed against mouse sequences, and thus we searched a mouse cell line whose proliferation was ERK dependent.
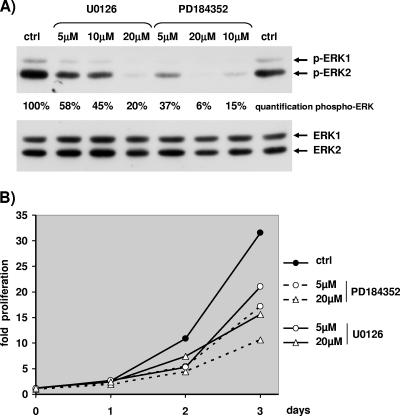
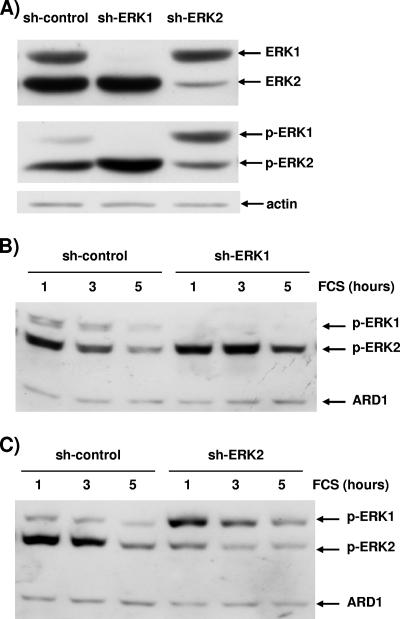
Hence, we tested whether exponential growth of NIH 3T3 cells was ERK dependent. The results of a typical experiment with exponentially growing NIH 3T3 cells treated with MEK inhibitors U0126 and PD184352 are depicted in Fig. 1. Great care was taken in all experiments, including this one, to measure phosphorylated ERK in cells grown in the same conditions as the ones plated to measure proliferation (12-well plates for counting and 10-cm plates for protein extracts). Addition of MEK inhibitors was sufficient to lower the levels of phospho-ERK1 and phospho-ERK2 after two days of treatment (Fig. 1A, upper blot). For a given concentration, PD184352 was more potent than U0126 to reduce ERK activation. For both inhibitors, a gradual increase of the concentration led to a gradual reduction of ERK activation. Interestingly, in NIH 3T3 cells the level of phospho-ERK2 was more elevated than the level of phospho-ERK1; hence, at high concentrations of inhibitors only the remaining level of phospho-ERK2 was detectable, being higher than threshold of detection. We tried unsuccessfully to lower significantly further the phospho-ERK level by changing the medium with fresh U0126 daily (data not shown).
FIG. 1.
Proliferation of NIH 3T3 cells requires MEK/ERK activation. Exponentially growing NIH 3T3 cells were treated with dimethyl sulfoxide (0.2%), with increasing concentrations of U0126 (5 μM, 10 μM, and 20 μM), or with increasing concentrations of PD184352 (5 μM, 10 μM, and 20 μM) on day 0. (A) Cells were lysed at day 2, and Western blotting was used to visualize expression of ERKs (antibodies E1B and ERK1 no. 61) or phosphorylated ERKs. Quantification of phospho-ERK1/2 was performed with light capture. (B) NIH 3T3 cells were plated and treated at day 0 (8 h after the seeding of the cells) as for panel A, and nuclei were counted daily as explained in Materials and Methods. Fold proliferation was calculated by dividing the number of cells counted each day by the number of cells counted at 8 h postplating (day 0).
As indicated in Fig. 1B, at 3 days after addition of MEK inhibitors, cell proliferation was markedly reduced, by about 50% with 20 μM U0126 and 66% with 20 μM PD184352. Lower doses of inhibitors were still effective to slow cell growth (about 33% reduction with 5 μM U0126 and 46% reduction with 5 μM PD184352). Globally the rate of cell proliferation correlated with the level of ERK activation. However, even when high levels of inhibitors were used, a significant proliferation of the cells was observed. For example, 20 μM PD184352 diminished ERK activation by about 94% and slowed cell proliferation by about 66%. Changing the growth medium every day in the presence of high concentrations of inhibitor led to the same slow growth (data not shown). Hence, a weak level of ERK activity was sufficient to allow NIH 3T3 cells to proliferate slowly; however, the rate of growth was gradually diminished when ERK activation was gradually decreased, and thus NIH 3T3 cells are a good model to test our hypothesis.
Ablation of ERK1 has no impact on cell proliferation.
NIH 3T3 cells were transfected with the plasmid pSUPER-shERK1 as described in Materials and Methods. Only highly transfected cells were selected owing to a 10-fold excess of shRNA-expressing plasmid above the pBabePuro selection plasmid. After selection of transfected cells with puromycin, puromycin was omitted for the rest of the experiment.
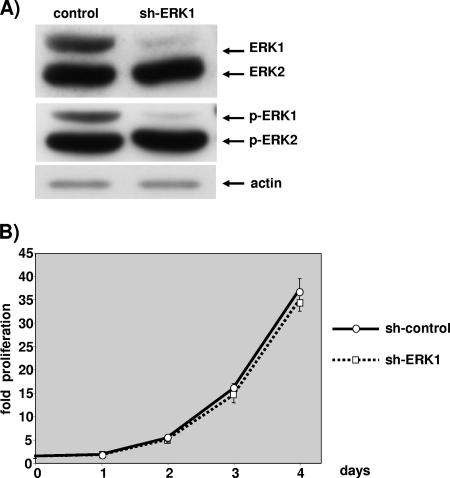
As shown in Fig. 2A, cells expressing the shERK1 plasmid did not display any ERK1 protein. Quantification performed by comparing serial dilutions of control transfected extracts to reach the low level of shERK1-transfected cells indicated about a 95% diminution in ERK1 protein expression in cells expressing shERK1 (data not shown). As expected, pSUPER-shERK1 targeted ERK1 very specifically, since the level of ERK2 was absolutely normal (Fig. 2A). The cell extract was obtained after 2.5 days of exponential growth; however, the silencing of ERK1 was still maximal at 4 to 5 days postseeding, ERK1 expression started to rebound at between 5 and 6 days postselection (data not shown).
FIG. 2.
ERK1 ablation has no impact on cell proliferation. NIH 3T3 cells were transfected with 3 μg of pBabePuro plasmid and 27 μg of control plasmid or 27 μg pSUPER-ERK1 (sh-ERK1). After selection, cells were plated under conditions of exponential growth. (A) The levels of ERKs and phosphorylated ERKs were evaluated by immunoblotting at 2.5 days postplating as for Fig. 1. The level of actin analyzed by immunoblotting is given for loading normalization. (B) Cells plated on 12-well plates were fixed at 6 h postplating (day 0) and daily up to 4 days postplating. Proliferation was calculated as described for Fig. 1. Data are representative of at least seven independent experiments. Values are the averages and standard deviations of cell counts obtained when cells were plated at 20, 30, and 40 cells per mm2.
The remaining level of ERK1 was sufficiently low that the level of phospho-ERK1 was nearly abrogated (Fig. 2A, lower blot). This was not the case with another plasmid that did not reduce sufficiently the level of the ERK1 protein (data not shown). We took great care in measuring the level of phospho-ERKs in exponentially growth conditions; thus, 10-cm diameter plates were seeded at the density of the 12-well plates used for measuring cell proliferation, and lysis was performed at 2.5 days postseeding. This is a critical step, since confluence markedly affects ERK activation measured by the phospho-ERK level (data not shown).
Cells lacking ERK1 grew at the rate of control transfected cells for at least 4 days postseeding (Fig. 2B). The results are representative of seven independent experiments which all indicated that removal of ERK1 had no impact on cell proliferation. For each experiment, puromycin-resistant cells were seeded at three cell densities, and great care was taken to compare only plates that displayed exactly the same number of cells at 6 h postseeding. No difference in cell attachment was noted (data not shown). Hence, silencing of ERK1 alone has no impact on NIH 3T3 cell proliferation.
Reducing ERK2 markedly slows cell proliferation.
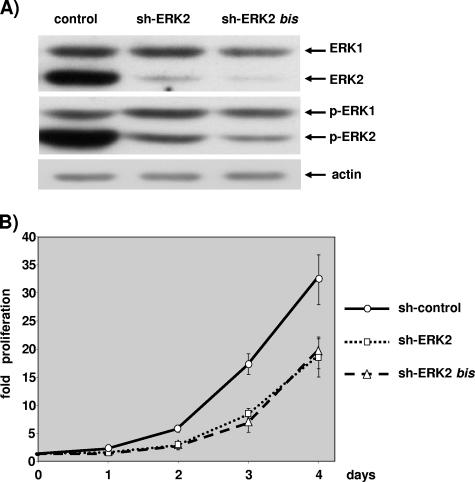
Plasmids expressing shRNA targeting ERK2 were transfected and selected as described above and in Materials and Methods. Two plasmids expressing shRNA sequences unrelated to ERK2 very efficiently reduced the level of the ERK2 protein (Fig. 3A). Quantification performed by comparing serial dilutions of control transfected cell extracts with shRNA-expressing extracts indicated that the ERK2 level was reduced by about 90% to 95% in transfected cells (data not shown). Selectivity towards ERK2 is very high, since the level of ERK1 was not affected upon transfection with either plasmid targeting ERK2 (Fig. 3A).
FIG. 3.
ERK2 ablation slows cell proliferation. NIH 3T3 cells were transfected with 3 μg of pBabePuro plasmid and 27 μg of control plasmid, 27 μg pSUPER-ERK2 (sh-ERK2), or 27 μg pSUPER-ERK2bis plasmids (sh-ERK2bis). After selection, cells were plated under conditions of exponential growth. (A) The levels of ERKs and phosphorylated ERKs were evaluated by immunoblotting at 2.5 days postplating as for Fig. 2. The level of actin analyzed by immunoblotting is given for loading normalization. (B) Cells plated on 12-well plates were fixed and nuclei counted as described for Fig. 2. These data are representative of at least four independent experiments. Values are as described for Fig. 2.
The level of phospho-ERK2 was markedly reduced in cells that were transfected with one or the other plasmid targeting ERK2 (50% reduction [data not shown]). Reducing the level of phospho-ERK2 in exponentially growing cells was sufficient to reduce markedly the rate of cell proliferation (Fig. 3B). The two sequences were equally effective in reducing cell proliferation; however, a third sequence that did not reduce the level of phospho-ERK2 as much had limited impact on cell proliferation (data not shown).
The results in Fig. 3 are representative of four independent experiments with one or the other plasmid that targets ERK2, and they indicate that ERK2 is required for NIH 3T3 cell proliferation to proceed normally.
Removal of both ERK1 and ERK2 seems equally effective as removal of only ERK2 in slowing cell proliferation.
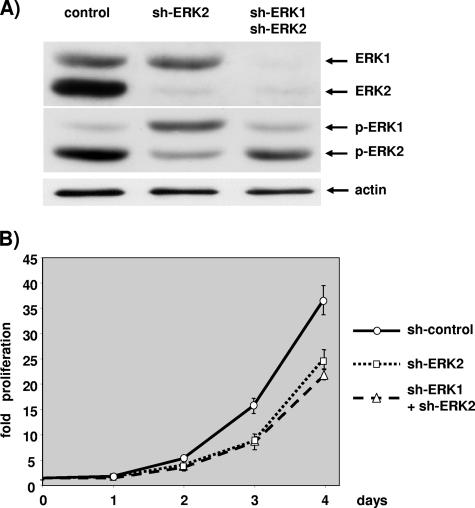
A lack of an effect of ERK1 silencing could be explained by the weak contribution of ERK1 to the total ERK activity, as indicated by the low ratio of phospho-ERK1 to phospho-ERK2 in the control transfected cells in all the experiments presented above. Hence, we decided to lower the ERK1 expression in limiting conditions of ERK2 activation. Initially we tried to silence ERK1 completely in a background of mild ERK2 silencing, and we observed no increase of the effect on cell proliferation for ERK1 and ERK2 silencing over the sole silencing of ERK2 (data not shown). Figure 4 depicts one experiment where ERK1 was expressed or not in conditions where ERK2 was maximally silenced.
FIG. 4.
Double silencing of ERK1 and ERK2 is equally effective as silencing ERK2 alone in slowing cell proliferation. Cells were transfected with 3 μg of pBabePuro plasmid and 27 μg of control plasmid, 13 μg pSUPER-ERK2 plus 13 μg of control plasmid (sh-ERK2), or 13 μg pSUPER-ERK2 plus 13 μg pSUPER-ERK1 plasmids (sh-ERK1+sh-ERK2). After selection, cells were plated under conditions of exponential growth. (A) The levels of ERKs and phosphorylated ERKs were evaluated by immunoblotting at 2.5 days postplating as described for Fig. 2. The level of actin was determined as a control for loading. (B) Cells plated on 12-well plates were fixed and counted as described for Fig. 2. Data are representative of at least four independent experiments. Values are as described for Fig. 2.
Cells transfected with the plasmid expressing shERK2 displayed a very low level of ERK2 expression, while ERK1 expression was equal to that of control transfected cells (Fig. 4A). In cells expressing both shERK1 and shERK2, expression of both ERK isoforms was barely detectable compared to that in control transfected cells. Importantly, the very low level of ERK2 expression was barely detectable for single and double silencing.
Interestingly, silencing of ERK2 alone strongly reduced ERK2 phosphorylation while concomitantly increasing ERK1 phosphorylation compared to that in control transfected cells (Fig. 4A, middle blot). Furthermore, when both ERK1 and ERK2 were silenced simultaneously, although the levels of ERK1 and ERK2 were barely detectable, the phosphorylation level of ERK1 and ERK2 was not as diminished as expected: ERK1 phosphorylation was similar to that in control transfected cells, and ERK2 phosphorylation was reduced compared to that in control transfected cells but was markedly higher than that in cells where ERK2 alone was silenced. Interestingly, the ratio of phospho-ERK1 to phospho-ERK2 is inverse for single ERK2 silencing and double ERK1 and ERK2 silencing.
Removal of ERK2 alone or simultaneous removal of the two ERKs lowered the proliferation of NIH 3T3 cells compared to control transfected cells (Fig. 4B). However, cells grew at the same rate whether ERK2 expression was abrogated alone or expression of ERK1 and ERK2 was abrogated simultaneously. At first glance, this experiment confirmed that removal of ERK1 had no impact on cell proliferation. However, clearly there was no correlation between the level of ERK2 activation and the rate of cell proliferation: double-silenced and single-ERK2-silenced cells grew at the same rate despite different levels of phospho-ERK2.
Both isoforms compete for upstream activating signals.
The dramatic inversion of the ratio between the phospho-ERK isoforms in ERK2-silenced cells versus ERK1- and ERK2-silenced cells (Fig. 4A) prompted us to evaluate more closely this shift of activation among isoforms.
A typical experiment with exponentially growing cells in which either ERK1 or ERK2 expression has been reduced is presented in Fig. 5A. The immunoblot that displays ERK levels confirms that the vectors ablate expression of their target very effectively and highly specifically, since expression of the targeted isoform is nearly abolished while expression of the other ERK isoform is unaltered. When assessing the level of phospho-ERK in these cells, one must note first that removal of ERK1 increased activation of ERK2, as was reported for mouse embryo fibroblasts derived from knockout animals (32). Furthermore, this experiment indicated unambiguously that removal of ERK2 led to increased activation of ERK1 compared to that in control transfected cells. Can this phenomenon be observed only in exponentially growing cells? To answer this question, cells transfected in this experiment were also plated at high density to reach confluence at 1.5 days postseeding, serum deprived overnight, and stimulated with 10% FCS. Other experiments indicate that confluence did not affect the level of silencing (data not shown); hence, at the time of stimulation, silencing of ERK1 or ERK2 was identical as shown by immunoblotting for total ERK (Fig. 5A). Removal of ERK1 also increased phospho-ERK2 levels during acute stimulation (Fig. 5B). The overactivation of ERK2 was more obvious after 3 and 5 h of FCS stimulation.
FIG. 5.
The remaining ERK isoform is overactivated. Cells were transfected with 3 μg of pBabePuro plasmid and 27 μg of control plasmid, 27 μg pSUPER-ERK1 (sh-ERK1), or 27 μg pSUPER-ERK2 (sh-ERK2). After selection, cells were plated at low density for exponential growth (A) or at high densities with serum removal at 24 h prior to stimulation (B and C). The data were generated from one transfection, and hence the levels of ERK1 and ERK2 shown in panels A, B, and C are identical. The invariant level of actin and ARD1 (31 kDa) was used as a loading control in panels B and C. (A) Cells either transfected with control plasmid (sh-control) or lacking ERK1 (sh-ERK1) or ERK2 (sh-ERK2) were lysed at 2.5 days postplating during exponential growth. (B) Control transfected cells (sh-control) and cells lacking ERK1 (sh-ERK1) were stimulated for 1, 3, and 5 h with 10% FCS. (C) Control transfected cells (sh-control) and cells lacking ERK2 (sh-ERK2) were stimulated for 1, 3, and 5 h with 10% FCS. Data are representative of three similar experiments.
The phospho-ERK1 level was much higher in cells lacking ERK2 than in control cells at all time points of stimulation (Fig. 5C). The phospho-ERK2 level was still detectable owing to a residual pool of ERK2 (Fig. 5A). After 5 h of serum stimulation, removal of ERK2 led to inversion of the activation ratio between the isoforms compared to that in control transfected cells. The results indicate that ERK activation was transferred to the residual pool of ERK molecules, in exponentially growing cells as well as during acute stimulation (Fig. 5).
ERK1 becomes a positive activator of cell proliferation when ERK2 activation is severely limiting.
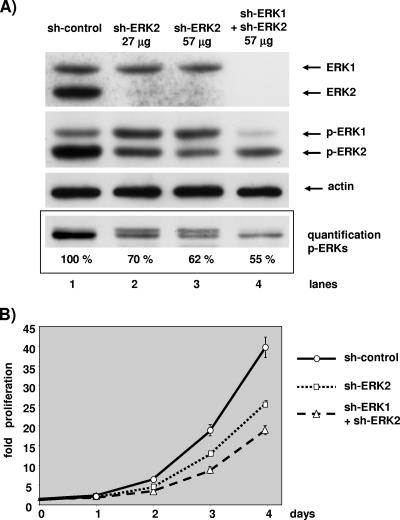
It may be concluded from the results shown in Fig. 5 that MEK activates the remnant pool of ERKs present in the cell, independently of isoforms. Hence, evaluating the role of ERK1 at limiting levels of ERK2 activation is difficult, since the silencing of ERK1 can “reactivate” the remnant pool of ERK2. We decided to lower further the level of the ERK2 protein in the doubly transfected cells in order to compare the same level of phospho-ERK2 in cells lacking ERK2 alone or lacking both ERK1 and ERK2. Figure 6 displays the results of one such experiment. ERK2 was nearly undetectable in cells transfected with 27 μg or 57 μg of shERK2 plasmid and in cells transfected with 40 μg of shERK1 plasmid in the presence of 57 μg of shERK2 plasmid (Fig. 6A, upper blot). Only cells expressing the shERK1 plasmid showed a drop in the ERK1 level compared to control transfected cells.
FIG. 6.
At a constant level of ERK2 activity, ERK1 silencing further slows cell proliferation. NIH 3T3 cells in a 10-cm dish were transfected with a total of 100 μg of plasmid containing 3 μg of pBabePuro plasmid for selection and 97 μg control plasmid (lane 1), 27 μg of pSUPER-ERK2 plasmid and 70 μg of control plasmid (lane 2), 57 μg of pSUPER-ERK2 plasmid and 40 μg of control plasmid (lane 3), or 57 μg of pSUPER-ERK2 plasmid and 40 μg of pSUPER-ERK1 plasmid (lane 4). After selection, cells were plated under conditions of exponential growth. (A) The levels of ERKs and phosphorylated ERKs were evaluated by immunoblotting at 2.5 days postplating as described for Fig. 2. The level of actin was determined for loading normalization. Results of a blot of a 14% SDS-polyacrylamide gel, which minimizes size differences between ERK1 and ERK2, for quantification by light capture are shown. The level of actin is given for loading normalization. (B) The cell proliferation assay and data presentation are as described for Fig. 2. These data are representative of three similar experiments.
When ERK2 alone was lowered, the phospho-ERK1 level was higher than that in control transfected cells; in contrast, in cells lacking ERK1 and ERK2, the phospho-ERK1 level was much lower than that in control transfected cells (Fig. 6A). The phospho-ERK2 level was lowered in cells transfected with 27 μg of shERK2 plasmid compared to control transfected cells; it became even lower when cells were transfected with 57 μg of shERK2 plasmid alone. In cells lacking ERK1 and ERK2, the phospho-ERK2 level was not lowered as much as in cells transfected with 57 μg of shERK2 alone but was nearly as low as in cells transfected with 27 μg of shERK2. Hence, we decided to compare the proliferation of cells transfected with 27 μg of shERK2 alone with that of cells doubly transfected. In these two populations of cells the level of phospho-ERK2 was nearly identical; however, in the doubly transfected cells phospho-ERK1 was markedly reduced. Thus, we were then able to assess the role of ERK1 when ERK2 activation was low and comparable in two populations of cells (Fig. 6A, lanes 2 and 4).
Removal of ERK2 diminished the rate of cell proliferation compared to control transfected cells (Fig. 6B). Interestingly, the removal of ERK1 and ERK2 reduced cell proliferation more than the sole removal of ERK2, despite an equivalent level of active ERK2 (Fig. 6A). Hence, for the first time we demonstrate that ERK1 plays a positive role in the control of cell proliferation. This role of ERK1 was unmasked in this experiment designed to avoid “reactivation” of ERK2 in doubly transfected cells.
Cells grew at the same rate with different levels of phospho-ERK2 (Fig. 4) but also grew at different rates with nearly equivalent levels of phospho-ERK2 (Fig. 6). These two observations indicate that the rate of cell proliferation was not correlated with the level of ERK2 activation. To determine whether the rate of proliferation was correlated with the level of total ERK activation, we decided to evaluate the total level of phospho-ERK in Fig. 6A. To avoid problems of threshold detection of immunoblots, we intended to combine the phospho-ERK1 and phospho-ERK2 signals as one band for quantification of the chemiluminescence by camera capture. In Fig. 6A, one gel is presented and three independent gels were used for quantification. Silencing of ERK2 with 27 μg of shERK2 reduced the total ERK phosphorylation by 30% over that in control transfected cells, whereas silencing of ERK2 and ERK1 reduced total ERK phosphorylation by about 45%. Hence, we observe a correlation between the rate of cell proliferation and the level of total ERK phosphorylation.
Removal of ERK1 alone or ERK2 alone is sufficient to abrogate IEG transcription.
To confirm that ERK1 and ERK2 are both positive contributors of ERK signaling, we decided to study the effect of removal of the ERK isoforms on another ERK-dependent biological response. We chose to study IEG transcription since it is well documented that transcription of many IEGs is ERK dependent and quantitative measurement is easily performed. shRNA-expressing plasmids were transfected, and cells were selected and plated at high density for 1 day prior to serum depletion overnight. Following 45 min of serum stimulation, the mRNA levels of several genes were measured by quantitative RT-PCR.
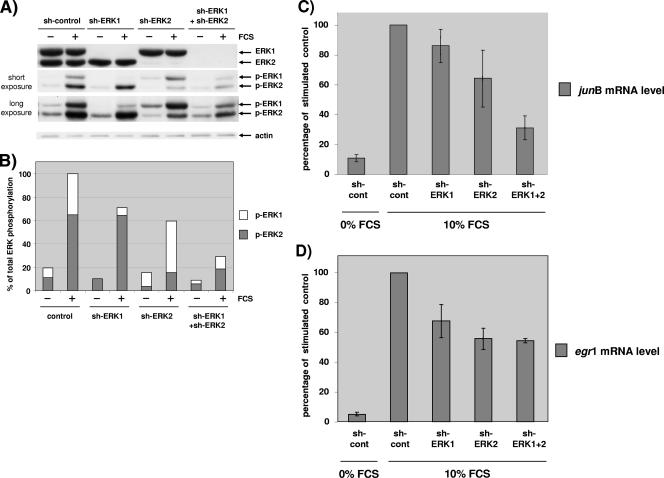
Transfection with shRNA efficiently silenced specifically ERK1 or ERK2, since nearly all of the targeted isoform was removed while the other was expressed at normal levels (Fig. 7A). In this experiment, we sought to silence both isoforms at the same time to compare the biological impact of this double silencing with the impact of single silencings.
FIG. 7.
Both ERK1 and ERK2 contribute to stimulate IEG transcription. NIH 3T3 cells in a 10-cm dish were transfected with a total of 100 μg of plasmid containing 3 μg of pBabePuro plasmid for selection. Cells were transfected with 97 μg of control plasmid (sh-control), 27 μg of pSUPER-ERK1 plasmid and 70 μg of control plasmid (sh-ERK1), 27 μg of pSUPER-ERK2 plasmid and 70 μg of control plasmid (sh-ERK2), or 40 μg of pSUPER-ERK1 plasmid and 57 μg of pSUPER-ERK2 plasmid (sh-ERK1+ shERK2). After selection, cells were plated under conditions of high density and arrested 24 h prior to stimulation for 45 min with 10% FCS. (A) The levels of ERK1/2 and phosphorylated ERK1/2 were evaluated by immunoblotting analysis as described for Fig. 2. For the phospho-ERK1/2 immunoblot, short and long exposures are shown. The level of actin is given for loading normalization. (B) Quantification of phosphorylated ERK1 and ERK2 from the blot in panel A after acquisition of the emitted light. For each bar, the levels of phospho-ERK1 and phospho-ERK2 relative to the total phospho-ERK level of stimulated control cells are shown. (C) Quantitative RT-PCR of junB in cells transfected and stimulated as described above (same transfection). The level was measured in triplicate and expressed as a percentage of junB expressed after 45 min in serum-stimulated control-transfected cells. (D) Quantitative RT-PCR of egr1 in the same extracts as for panel C for junB. The median and range from four experiments performed identically are represented in panels C and D. Results in all conditions were statistically lower than the control (P < 0.05 as determined by the Friedman test). Similar results were obtained for the single ablation of ERK1 or ERK2 when NIH 3T3 cells were transfected with 30 μg of plasmid (five additional independent experiments).
Hence, we transfected higher levels of shRNAs-expressing plasmids against ERK1 and ERK2 in the double silencing than in the single silencing, to counteract compensation on the remaining ERKs as shown in Fig. 5. The middle immunoblot of Fig. 7A represents the level of phospho-ERKs in transfected cells stimulated or not for 45 min with FCS. ERK1 silencing decreased markedly the level of phospho-ERK1. As expected ERK2 silencing decreased markedly the level of phospho-ERK2 and concomitantly increased the level of phospho-ERK1. In doubly silenced cells, both phospho-ERK1 and phospho-ERK2 were lower than in control transfected cells. Furthermore, the levels of phospho-ERK2 were comparable in cells where only ERK2 was silenced and in cells where both ERK1 and ERK2 were silenced, due to attempts to avoid compensation by transfecting larger amounts of plasmids in the double silencing. Unfortunately, in this experiment we were unable to reduce ERK1 phosphorylation in the double silencing as effectively as in the single ERK1 silencing despite a larger amount of sh-ERK1 plasmid transfected in doubly transfected cells. This observation stresses once more the difficulty of avoiding compensatory activation of the remaining pool of ERKs.
Figure 7B represents the quantification of the respective phospho-ERK levels in Fig. 7A. Interestingly, when observing the phospho-ERK levels in stimulated cells, a gradation in the reduction of phospho-ERKs was observed. Removal of ERK1 decreased phospho-ERKs by about 25%, removal of ERK2 decreased phospho-ERKs by about 40%, and removal of both ERK1 and ERK2 decreased both phospho-ERKs by about 70% (Fig. 7A and B). The total active ERK level is obtained by adding the residual activated pool of the two ERKs. Interestingly, the contribution of ERK1 or ERK2 to the total phospho-ERK level appears to be reversed during ERK1 silencing versus ERK2 silencing. In addition, when both ERK1 and ERK2 are silenced, the ratio between active ERK1 and active ERK2 returns to the ratio in control transfected cells, although the total level of active ERKs is much reduced.
Stimulation of serum-deprived cells for 45 min led to a massive induction of the levels of junB (Fig. 7C) and egr1 (Fig. 7D) mRNAs (11.4- and 13.8-fold, respectively). In preliminary experiments we demonstrated that this induction was ERK dependent, since it was markedly reduced on MEK inhibition with 50 μM U0126 or 2 μM PD184352 (data not shown).
Silencing of either ERK1 or ERK2 alone was sufficient to lower the mRNA level of junB compared to that in control transfected cells, while silencing of both ERK1 and ERK2 lowered further the junB mRNA level, to below that of ERK2 alone (Fig. 7C).
Since ERK2 phosphorylation was nearly the same in the single silencing with sh-ERK2 as in the double silencing, there is no correlation between the level of junB mRNA expression and ERK2 activation. In contrast, in the double silencing there was a decrease in total ERK phosphorylation compared to single silencing, and hence this experiment confirms that total ERK activation correlates with junB mRNA induction.
Regulation of the egr1 mRNA level was somewhat different when the same extracts were examined. First, silencing of ERK1 or silencing of ERK2 was nearly equally effective in reducing the egr1 mRNA level (Fig. 7D, bars 3 and 4). Second, induction of the egr1 mRNA level was reduced equally when ERK2 was silenced alone or in conjunction with ERK1 (Fig. 7D, last bar). This experiment confirms that ERK1 plays a positive role in increasing the mRNA levels of ERK-responsive genes. However, there is not always a straightforward correlation between the level of total ERK activity and induction of the mRNA level (as shown for egr1 mRNA induction). Experimental variation certainly plays a role, and it is also obvious that ERK-independent signaling pathways contribute to induce the mRNA levels of many IEGs; hence, full mRNA induction may require full ERK activation, but a threshold ERK activity may be sufficient for robust induction to occur.
Stoichiometric ratio of isoforms in NIH 3T3 cells.
The results presented above (Fig. 6 and 7) indicate that both ERK isoforms are positive activators of cell proliferation and IEG transcription; furthermore, the level of phosphorylated ERK1 is consistently lower than the level of phospho-ERK2 in NIH 3T3 cells (Fig. 1A). Hence, when ERK activation is not limiting, removal of ERK1 may have no more effect than removal of 20 to 30% of ERK2. Is the low level of phospho-ERK1 relative to the level of phospho-ERK2 due to inefficient activation? To understand better the consequences of the lack of ERK1 or of ERK2, we sought to determine precisely the stoichiometric ratio between ERK1 and ERK2 in our model system and to compare it to the precise ratio between phospho-ERK1 and phospho-ERK2.
We normalized the immunoblot signals for ERK1 and ERK2 by comparing NIH 3T3 cell extracts with standard quantities of exogenously expressed ERK1 and ERK2. We used epitope-tagged ERK1 and ERK2 expressed in mammalian cells as standards to ensure proper posttranslational modification. Since most ERK antibodies are directed towards the N- or C-terminal end of the molecule, two sets of standards were used to avoid bias. Hence, the full-length mouse ERK1 and ERK2 were epitope tagged either at the N terminus with the HA epitope (9 amino acids) or at the C terminus with the VSVG epitope (11 amino acids).
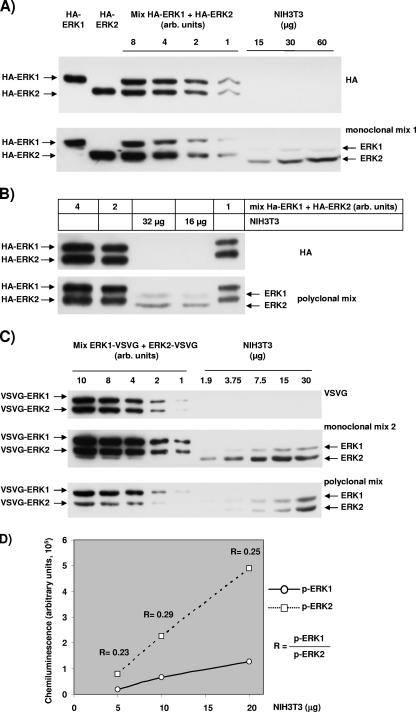
Figure 8 shows representative immunoblots that allowed us to quantify the ratio between ERK1 and ERK2 in NIH 3T3 cells. The chemiluminescence was acquired with radiography film (higher resolution) and with light acquisition equipment for precise quantification and calculation. For Fig. 8A, B, and C, the different gels were loaded with identical extracts, either from HEK293 cells transfected with epitope-tagged ERKs or from NIH 3T3 cells.
FIG. 8.
Quantification of the relative levels of ERK1 and ERK2 and the relative levels of activated ERK1 and activated ERK2. HEK293 cells were transfected with plasmids expressing mouse ERK1 or mouse ERK2 that was tagged at the N terminus with the HA epitope or at the C terminus with the VSVG epitope. Extracts from these cells were mixed to load equal quantities of the two isoforms tagged with the same epitope in order to normalize the signal obtained with antibodies that recognize endogenous mouse ERKs in NIH 3T3 cell extracts. (A) Two gels were loaded identically, and the upper blot was revealed with an anti-HA antibody and the lower blot with ERK monoclonal antibody mix 1 as described in Materials and Methods. The first six lanes were loaded with decreasing amount of HEK transfected cell lysate and the last three with increasing quantities of NIH 3T3 cell extracts. (B) As in panel A, two gels were loaded identically, and the upper blot was revealed with an anti-HA antibody and the lower with the ERK polyclonal mix of antibodies (E1B and ERK1 no. 61). Lanes 1, 2, and 5 were loaded with decreasing amounts of HA-ERKs for normalization, and lanes 3 and 4 were loaded with NIH 3T3 cell extracts. (C) Three gels were equally loaded and revealed with either a VSVG antibody (upper blot), anti-ERK monoclonal antibody mix 2 (middle blot), or a polyclonal mix of antibodies (E1B and ERK1 no. 61). The first five lanes were loaded with decreasing amounts of HEK293 cells transfected with ERK-VSVG, whereas the last five lanes were loaded with increasing quantities of NIH 3T3 cell extracts. (D) Quantification of phospho-ERK1 and phospho-ERK2 in increasing amounts of NIH 3T3 extracts of cells stimulated for 45 min with 10% FCS. The ratio of phospho-ERK1 to phospho-ERK2 is indicated for each quantity of NIH 3T3 extract loaded.
For Fig. 8A and B, normalization was performed with exogenously expressed ERK1 and ERK2 that were epitope tagged at the N terminus. Gels were loaded with extracts from HEK293 cells transfected with mouse HA-ERK1 or mouse HA-ERK2 or with nontransfected NIH 3T3 cell extracts. In the linear range of the HA signal, HA-ERK2 was 10% more abundant than HA-ERK1 in the mix (Fig. 8A, upper blot). The monoclonal antibody mix 1 recognized preferentially HA-ERK2 (HA-ERK1 = 0.486 × ERK2) (Fig. 8A, lower blot). For the NIH 3T3 extracts, only the loading of 15 μg and 30 μg gave linear results (data not shown), with a much stronger signal for ERK2 than for ERK1 (ERK1apparent = 0.132 × ERK2apparent). After correction for the loading of the HA isoforms (10% difference) and for the affinity of the antibodies, the ratio was determined to be ERK1normalized = × 0.3 ERK2normalized.
The signal detected with the HA antibody in Fig. 8B indicated that less HA-ERK1 than HA-ERK2 was loaded on the gel (HA-ERK1 = 0.81 × HA-ERK2). However, the signal with a polyclonal mix of anti-ERKs antibodies (E1B and ERK1 no. 61), was stronger for HA-ERK1 than for HA-ERK2 (HA-ERK1 = 1.36 × HA-ERK2). This ratio indicated that on an immunoblot where there was more ERK2 than ERK1, this mix of antibodies recognized more ERK1. Despite this bias, in extracts of NIH 3T3 cells ERK2 was recognized to a greater extent than ERK1 (ERK1apparent = 0.44 × ERK2apparent). After correction for the loading and the isoform bias of the mix of antibodies, this normalization in NIH 3T3 cells corresponded to ERK1normalized = 0.26 × ERK2normalized.
In Fig. 8C, normalization was performed with exogenously expressed ERK1 and ERK2 that were epitope tagged at the C terminus. Gels were loaded with extracts from HEK293 cells transfected with mouse ERK1-VSVG or mouse ERK2-VSVG or with nontransfected NIH 3T3 cell extracts. Equal loading of the two tagged isoforms was obtained, as shown by equal signals detected with the VSVG antibody (Fig. 8C, upper blot). Parallel revelation with the second mix of monoclonal ERK antibodies on a gel loaded equally to the first showed a ratio between ERK1 and ERK2 that differed slightly from that obtained with the mix 1 used for Fig. 8A, middle panel. Indeed, the signal of ERK1-VSVG was stronger than that of ERK2-VSVG when revealed with monoclonal mix 2, indicating that the mix had more affinity for ERK1. In contrast, the signal obtained for NIH 3T3 cells extracts was much stronger for ERK2 than ERK1 (Fig. 8C, middle panel). This indicates that there was more ERK2 than ERK1 in NIH 3T3 cells (ERK1normalized = 0.22 × ERK2normalized). The lower blot in Fig. 8C shows the ERK-specific signals obtained on a gel loaded equally incubated with a mix of polyclonal antibodies. This polyclonal mix recognized even more ERK1, since the signal was stronger for ERK1-VSVG than for ERK2-VSVG although equal quantities were loaded on the gel. As shown with 30 μg of NIH 3T3 cell extracts and with a longer exposure captured on a charge-coupled device camera rather than on film, this mix of antibodies recognized slightly more ERK2 than ERK1. Again, this result indicated that there was more ERK2 than ERK1 in these cells (ERK1normalized = 0.18 × ERK2normalized).
Overall, the average calculation with the four different approaches in NIH 3T3 cells gave ERK1normalized = 0.245 × ERK2normalized. The results of quantification were nearly identical when exogenous ERK standards were tagged at the N terminus or C terminus. If total ERK is adjusted to 100%, ERK1 = 20% and ERK2 = 80% in NIH 3T3 cells.
We then sought to measure as precisely as possible the ratio of activated ERK isoforms in NIH 3T3. Note that the phospho epitope is identical in ERK1 and ERK2, and hence no normalization is necessary for SDS-polyacrylamide gel electrophoresis with proteins of nearly the same molecular weight. We performed immunoblotting experiments with anti-phospho-ERK antibodies in extracts from exponentially growing and 24-hour-arrested cells stimulated for either 7 min or 45 min with 10% FCS. Figure 8D shows the chemiluminescent signal measured with the anti-phospho-ERK antibody in diluted extracts of NIH 3T3 stimulated for 45 min with 10% FCS. The average ratio is phospho-ERK1 = 0.25 × phospho-ERK2. For cells stimulated for 7 min the ratio was 0.27, and for exponentially growing cells the ratio was 0.27 (data not shown). If total phospho-ERK is adjusted to 100%, then phospho-ERK1 = 20% and phospho-ERK2 = 80% in NIH 3T3 cells.
In conclusion, we demonstrate for the first time that the ratio between the activated ERK isoforms correlated exactly with the ratio of their relative expression. In addition, we never observed a preferential activation of one isoform when stimulating arrested NIH 3T3 with various agonists (fibroblast growth factor, platelet-derived growth factor, thrombin, and insulin, alone or in combination) (data not shown). Therefore, we propose that ERK1 and ERK2 compete equally for phosphorylation by MEK1/2 and that the ratio of phospho-ERK1 to ERK2 reflects the mass ratio of ERK1 to ERK2.
Comparison of the in vitro specific activities of HA-ERK1 and HA-ERK2.
After determining that ERK1 and ERK2 are activated relative to each other with respect to their expression ratio, we sought to compare their specific activities in vitro. For this, CCL39 hamster fibroblasts were transfected with mouse HA-ERK1 or mouse HA-ERK2 and stable clones obtained. Cells were serum starved for 24 h and then stimulated for 5 min with 10% FCS to maximally activate ERKs (endogenous ERKs and HA-ERKs). First we determined that GST-Elk307-428 phosphorylation increased linearly for at least 30 min when the immunoprecipitated HA-ERKs were incubated in kinase buffer with an excess of GST-Elk307-428 (evaluation of excess by amido black staining [data not shown]). Then, increasing amounts of immunoprecipitated HA-ERKs were incubated with GST-Elk307-428. The expression levels of HA-ERK1 and HA-ERK2 were distinct in the two chosen stable clones, and hence we decided to measure simultaneously in each kinase assay lysate the amount of active ERK immunoprecipitated and the amount of phosphorylated GST-Elk307-428. The phospho-ERK epitope is identical for ERK1 and ERK2, and therefore the signal is directly indicative of the amount of active ERK. Phosphorylation of the substrate GST-Elk307-428 was determined by immunoblotting with the anti-phospho-Elk-Ser383 antibody. To minimize bias induced by variation in protein transfer for different gels, the levels of HA-ERKs was determined for a single SDS-polyacrylamide gel, and similar levels of phospho-Elk307-428 were detected in a single SDS-polyacrylamide gel. An augmentation of phospho-HA-ERKs was observed in kinase assays performed with increasing quantities of cell extracts (Fig. 9A, upper blot). Phospho-Elk1307-428 levels were determined in the same kinase assay (Fig. 9A, lower blot). We observed that comparable levels of phospho-HA-ERK1 and phospho-HA-ERK2 induced comparable phosphorylation of the substrate GST-Elk307-428 (compare lane 5 with lane 13). Figure 9B shows the linear relationship between the level of phospho-ERKs and the phosphorylation of GST-Elk1307-428, each measured in the same kinase assay lysate. The slope of the linear regression is virtually identical for phospho-HA-ERK1 and phospho-HA-ERK2 to phosphorylate GST-Elk1307-428. This result demonstrates for the first time that the in vitro kinase activities of HA-ERK1 and HA-ERK2 are almost identical. Considering that it is impossible to obtain cells lacking ERK2 from erk2−/− mouse embryos (36) or to purify to homogeneity ERK1 separate from ERK2, our comparison of the specific activities of transfected HA-ERK1 and HA-ERK2 seems to be at present the most precise way to compare the intrinsic kinase activities of ERK1 and ERK2.
FIG. 9.
Determination of HA-ERK1- and HA-ERK2-specific kinase activities. HA-ERK1 or HA-ERK2 was immunoprecipitated from increasing volumes of cell lysate obtained from CCL39 cells that stably express HA-ERK1 or HA-ERK2. An in vitro kinase assay using the GST-Elk-1307-428 fusion protein as the substrate was performed on each immunoprecipitate. (A) In the lysates of the kinase assays, the activated forms of HA-ERKs and the phosphorylation of the GST-Elk-1307-428 fusion protein were determined by immunoblotting using the anti-phospho-ERK1/2 and anti-phospho-Elk-1-ser383 antibodies, respectively. (B) The chemiluminescence was measured directly with a Gnome detector from Syngene (United Kingdom). The chemiluminescence corresponding to the phosphorylation of the GST-Elk-1307-428 fusion protein is plotted as a function of the chemiluminescence corresponding to the activated form of HA-ERK1 or HA-ERK2.
Close correlation between expression of the ERK isoforms and their relative activation in brain structures.
A consequence of demonstrating a correlation between the relative expression of the ERK isoforms and their relative activation is the ability to perform phospho-ERK immunoblotting to study the relative levels of ERK isoforms. We tested whether this correlation holds true in mouse tissues. Since several tissue arrays indicate that the greatest disparities in ERK mRNA expression occur in brain structures, we measured the relative levels of ERK1 and ERK2 and their phosphorylation state in brain extracts.
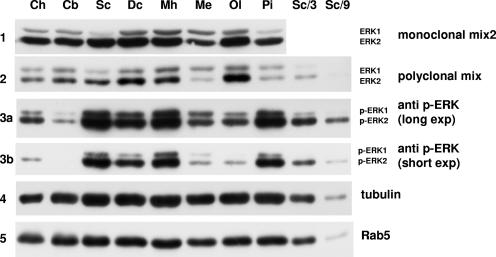
ERK1 and -2 levels in eight brain structures were detected with either the monoclonal ERK antibody mix 2 (Fig. 10, panel 1) or the polyclonal ERK antibody mix (Fig. 10, panel 2). At first sight, the relative abundances of ERK1 and ERK2 seemed to differ; however, as expected from the normalization determined for Fig. 8C, the polyclonal mix recognized ERK1 with greater affinity (Fig. 8C, lower blot). Direct comparison was thus impossible without normalization to adjust for the specific affinity for each isoform. After taking into account the higher affinity of the polyclonal mix of antibodies for ERK1, the calculated ratio observed between ERK1 and ERK2 (Fig. 10, panel 2) resembled very closely that obtained with the monoclonal mix 2 (Fig. 10, panel 1) (quantification not shown). In any case, both antibody mixes indicated that some brain structures, such as the superficial cortex, seemed devoid of ERK1 (13 times more ERK2 than ERK1 [calculation not shown]), whereas other areas, such as the medulla, contained more similar levels of the ERK isoforms (only twofold more ERK2 than ERK1). This result is in accordance with previous studies indicating that the brain stem, medulla, and sciatic nerve express relatively more ERK1 than ERK2 than most other tissues (31). Panels 3a and 3b in Fig. 10 show the relative phosphorylation states of ERK1 and ERK2 in the same brain extracts with long and short exposures of the blot, respectively. Although there were great disparities in the total ERK phosphorylation between samples, for all extracts the ratio between phospho-ERK1 and phospho-ERK2 very tightly correlated with the ratio between ERK1 and ERK2. For example, the medulla showed the highest level of ERK1 relative to ERK2 (blot 1) and also the highest level of phospho-ERK1 relative to phospho-ERK2 (blot 3a). In contrast, in the superficial cortex there was little ERK1 relative to ERK2 (blot 1) and there was little phospho-ERK1, as best seen with dilutions of the superficial cortex sample to avoid saturation of the signal (blot 3a) or with a shorter exposure of the same immunoblot (blot 3b).
FIG. 10.
Correlation between the levels of ERK isoforms in brain and their relative phosphorylation. Proteins from mouse brain were extracted in Laemmli sample buffer as described in Materials and Methods. Extracted brain structures: Ch, cerebellar hemisphere; Cb, cerebellum; Sc, superficial cerebrum; Dc, deep cerebrum; Mh, median eminence and hypophyse; Me, medulla; Ol, olfactory bulb; Pi, Pineal gland. The last two lanes are one-third and one-ninth dilutions of superficial cerebrum extract (third lane).
The results presented in Fig. 10 indicated that disparities between ERK1 and ERK2 expression in mouse tissues closely mimic the relative activation states of the two isoforms. In addition, these observations confirm that measuring phospho-ERKs allows prediction of the ratio between ERK1 and ERK2. This conclusion was drawn from experiments with mouse tissues, since the normalization of the antibodies was performed with mouse isoforms; however, the total conservation of the phospho epitope across species leads us to propose that measuring phospho-ERKs will predict the ERK1/ERK2 ratio at least in other mammals.
Normalization of protein loading was performed by measuring the abundance of tubulin and RabV proteins. Although there were minor loading differences, there were marked differences in total ERK expression. For example, in the pineal gland there was little ERK, whereas in the olfactory bulb ERK was very abundant. Interestingly, MEK1 seemed to be expressed at higher levels in tissues where ERK was abundant, and MEK2 was virtually not detected in brain extracts with our antibody (data not shown).
Genetic evidences for dosage requirement of total ERK activity.
Mice with erk1 gene invalidation are viable and reproduce normally, while invalidation of the erk2 gene leads to early embryonic lethality. ERK isoforms may have unique functions, explaining the contrast in phenotypes. Alternatively, low levels of ERK1 expression in some cells may explain the inability of ERK1 to compensate for a loss in ERK2, whereas ERK2 can compensate for ERK1 in nearly all cells. To test this hypothesis, we assessed whether it is detrimental or not to loose erk1 when only one allele of erk2 is expressed. Hence, we bred animals lacking one erk2 allele with animals lacking both erk1 alleles.
The erk1−/−; erk2+/+ animals and the erk1+/+; erk2+/− animals were backcrossed for nine and five generations, respectively, with C57BL/6 mice. When erk1−/−; erk2+/+ animals are crossed with erk1+/+; erk2+/− animals, 50% of the offspring ought to be double heterozygous animals (erk1+/−; erk2+/−). However, this breeding program led to the birth of 128 animals of which only 4 were double heterozygous; the remaining 124 animals were erk1+/−; erk2+/+. The frequency of birth of double heterozygous animals was thus only 3.4% instead of the 50% expected. This result indicated that it was very unfavorable to diminish the quantity of erk1 alleles when only one erk2 allele is present. This experiment was performed in the genetic background of C57BL/6 mice; however, the birth of double heterozygous animals was also observed in three other mouse genetic backgrounds (S. Meloche, personal communication).
Very interestingly, 59 pups were obtained by crossing the double heterozygous animals with each other (we were fortunate to obtain two females and two males in the F1 generation). Among the 59 animals that were born, 19 were double heterozygous (35%). As expected, no erk2−/− animals were born; furthermore, no animals were born with only one allele of erk2 and no allele of erk1. Hence, only 60% of the expected offspring could survive. Taking into account the survival rate, the frequency of double heterozygous pups should be 25% of the total expected offspring, i.e., 25 out of the 60 animals that could survive, which is 41%. Hence, the observed frequency of double erk heterozygotes born from double heterozygous parents is very close to the calculated frequency (35% versus 41%). Consequently, although obtaining double heterozygous animals was very detrimental in the F1 generation, transmission of double erk heterozygocity to the next generation became normal. In accordance with our observation in C57BL/6 mice, in three other independent genetic backgrounds no animals were ever born with only one allele of erk (S. Meloche, personal communication). In conclusion, erk gene dosage seems to be crucial for mouse survival; no animal can survive with one erk allele only, while animals survive with a minimum of only two erk2 alleles or with a minimum of one allele of erk1 and one allele of erk2.
DISCUSSION
ERK1 and ERK2 are 84% identical at the protein level (91% identical if the N-terminal stretch is not taken into account) and ubiquitously expressed, and no agonist is known to activate more specifically ERK1 over ERK2. Interestingly, the only known difference between isoforms is the formation of the complex P14/MP1 with MEK1, not with MEK2, in the signaling cascade (39). Although silencing or invalidation of either P14 (44, 50) or MP1 (35) reduces global ERK activation, it does not tip the activation towards ERK1 or ERK2. Similarly, silencing of MORG1, identified as a partner of MP1, reduces activation of both ERK1 and ERK2 upon serum stimulation (48). Therefore, the P14/MP1-specific action on MEK1 does not discriminate between specific functions for ERK1 and ERK2 (similarly for MP1/MORG1). Recently the crystal structure of ERK2 bound to a specific D-peptide that interacts with the only known ERK docking site was solved (CD domain and ED groove) (56). ERK activators, substrates, and phosphatases bind to ERK via this domain, which defines the specificity of interaction (42, 43). Crystallization of ERK2 bound to the D-peptide pinpointed all the amino acids of ERK2 that interact and hence the amino acids that confer specificity (56). Only one difference was observed between ERK1 and ERK2: an isoleucine in ERK1 versus a leucine in ERK2 (out of 11 amino acids of ERK that interact with the D-peptide). This substitution is unlikely to markedly affect interaction with partners, and indeed no specific substrates of ERK1 or ERK2 have yet been firmly established. The question as to why two so similar ERK isoforms exist in all mammals can be raised.
Many studies have started to address this question. When both isoforms are silenced in the same experimental design, data indicate usually a greater effect of the loss of ERK2 than of ERK1 (15, 26, 27, 41, 53), with the exception of the study of Vantaggiato et al., who indicated that ERK1 and ERK2 have opposite roles, with ERK1 counterbalancing the positive activation provided by ERK2 (47). However, this study has been seriously challenged by the recent publication of Sanjo et al. (38). Using B-cell-specific targeted mice, these authors demonstrate the absolute normal proliferation of B cells lacking ERK2 and propose that ERK1 (and traces of residual ERK2 caused by inefficient recombination in the B-cell population) compensate for the loss of ERK2 to result in normal proliferation (38).
Apart from the controversial study of Vantaggiato et al. (47), our results provide a rational explanation for the results observed in all the other experiments, and they stress the uppermost importance of assessing the phosphorylation/activation levels of the two isoforms in all studies to draw valid conclusions.
Indeed, we demonstrate that NIH 3T3 cell proliferation is not altered by silencing ERK1 alone, whereas it is markedly reduced upon silencing ERK2 alone. At first sight, silencing both isoforms seemed to be as effective as silencing ERK2 alone. However, the silencing of both ERK1 and ERK2 reestablishes the ratio in favor of ERK2 by increasing ERK2 activation; hence, the remaining ERK2 molecules were more activated than in the single ERK2 silencing, and the total ERK activity was no longer lowered in the double silencing. Clearly, there was no correlation between the rate of cell proliferation and the level of ERK2 activation. However, upon further lowering of ERK2 level in the “controlled” double silencing, we were able to achieve nearly the same phosphorylation of ERK2 in the double and single ERK2 silencing. Under these conditions, we clearly demonstrated that ERK1 activation played a positive role in driving cell proliferation.
Challenging cells with a very strong mitogenic agonist can lead to cell arrest; however, in NIH 3T3 cells silencing of ERK1 or ERK2 had no specific impact on the sub-G1 fraction under these conditions (data not shown). In contrast, serum removal or addition of staurosporine to the growth medium triggered apoptosis as evaluated by an increase in the sub-G1 fraction. In this case, ERK2 silencing potentiated more highly the increase in the sub-G1 fraction than ERK1 silencing (data not shown). Hence, both ERK1 and ERK2 provide prosurvival signals, with ERK2 being more effective than ERK1.
We sought to test whether the loss of ERK2 could be compensated for by exogenous expression of ERKs. First we tried to express full-length human cDNAs coding for ERK1 and ERK2 in NIH 3T3 cells lacking either ERK1 or ERK2. Although the small interfering RNA target sequences were silently mutated and the human ERKs were expressed without any epitope tag, we failed to reestablish consistently the activation level of endogenous mouse ERKs (data not shown). Interestingly, we were able to express more ERK1 than the endogenous level and about the same amount of ERK2 as endogenous ERK2, but for both kinases the level of activated human ERKs was much lower than that of endogenous mouse ERKs in control cells. Hence, expression of human ERKs did not restore either the loss of ERK1 or that of ERK2 in NIH 3T3 cells. After cloning mouse ERK1 for quantification studies, in cells lacking ERK2 we reexpressed mouse ERK1 or mouse ERK2 (full-length cDNAs without epitope tags whose small interfering RNA target sequences were silently mutated). For the first time we were able to reestablish to a large extent the level of active ERK2 in cells lacking ERK2 and to increase the level of active ERK1 in cells lacking ERK2 (see Fig. S1B at http://www.unice.fr/isbdc/pub/lefloch-suppl-MCB2008). Reexpression of ERK1 or ERK2 in cells lacking ERK2 proved to be sufficient to restore the induction of junB or egr1 mRNA to nearly normal levels (data not shown). This experiment indicated that ERK1 can substitute functionally for ERK2 in the induction of IEGs. Compensating for the decrease in cell proliferation induced by ERK2 silencing would be nearly impossible in our system, since the time courses of H1 promoter-driven silencing of ERK2 and Rous sarcoma virus promoter-driven ectopic ERK expression do not coincide (data not shown). Furthermore, the transfected mouse isoforms were not as efficiently activated as the endogenous ERKs (it took more expression of exogenous than endogenous ERKs to reach similar levels of activation [data not shown]).
Careful measurement of ERK1 and ERK2 stoichiometric ratios indicates that ERK2 is four times more abundant than ERK1 in NIH 3T3 cells. Interestingly, at all the time points of stimulation studied, activated ERK2 was also four times more abundant than activated ERK1 (there are identical phospho-epitopes in the two isoforms). This exact correlation between the relative amounts of ERKs and their relative activation was observed in all mouse brain structures that we studied, despite marked differences in the ratio of ERKs in different brain structures. Incidentally, determining the ratio of activated ERKs can provide a good evaluation of the protein ratio of ERKs. The fact that the quantitative ratio of the isoforms correlates perfectly with their activation ratio indicates that MEKs indiscriminately activate ERKs. Indeed, we demonstrate for the first time that ERK2 activation is increased at all time points when ERK1 is silenced and confirm the reciprocal increased activation of ERK1 when ERK2 is silenced. In addition, we have shown that the in vitro specific activities of ERK1 and ERK2 were nearly identical, and hence the strength of signals conveyed by ERK isoforms seems to be directly proportional to their level of expression.
Our results also indicate that gene dosage correlates with biological outcome. Animals that expressed two erk2 alleles were viable (with or without erk1 alleles), animals that expressed one erk2 allele were viable only if one erk1 or two erk1 alleles were expressed, and no animals could survive without erk2. Interestingly, animals with only one erk1 allele and one erk2 allele (double heterozygotes) were born at a lower-than-expected frequency; however, these double heterozygous animals transmitted the double heterozygocity normally to their offspring. The normal transmission in F2 of double heterozygocity is unlikely to be due to a mixing of genetic backgrounds, since all the parental animals were backcrossed at least five generations in the C57BL/6 strain prior to crossing. The normal transmission in F2 of double heterozygocity reinforces the notion that the ERK signaling cascade is resilient: although the total level of ERK was probably limiting in double heterozygous animals, compensation in the F1 generation led to normal transmission in the F2 generation. We propose that there are critical steps during development that require a high level of ERK activity, and if the lack of ERK2 cannot be compensated for by ERK1 due to low expression of ERK1 in these critical cells, the erk2+/− and erk2−/− embryos die. If this critical step is bypassed, embryonic development can go on normally up to the next critical stage of development. Indeed, tetraploid rescue of erk2−/− embryos allows normal development of embryos at least up to day 14, whereas erk2−/− embryo are set to die at around embryonic day 6.5 (18). erk2−/− embryos die of placental defects (18, 36, 51), and tetraploid rescue allows expression of ERK2 in the placenta to avoid death while the embryos are still lacking any erk2 allele.
Why does the role of ERK2 seem preeminent over that of ERK1, at least in mammals? We formulate the hypothesis that this is due to a generally higher level of ERK2 expressed over ERK1. It seems that most if not all tissues express more ERK2 than ERK1 as evaluated by the ratio between phosphorylated forms or after normalization of total protein expressed (reference 31 and our observations). For example, we have shown that in brain structures, expression ranged from up to 13-fold more ERK2 than ERK1 in the superficial cortex to only twice as much ERK2 than ERK1 in the medulla. These measurements correlated well with previous quantitative Western blotting experiments (31) and mRNA expression measured by in situ hybridization (13, 24). In many experiments, silencing of ERK1 has no effects, probably because the abundant level of ERK2 can compensate for the loss of ERK1. Furthermore, Fujioka et al. have evaluated the percentage of ERK molecules activated following a 5-minute stimulation with epidermal growth factor to be 60% (16). Thus, there is a reserve of ERK to be activated. Only a great reduction of the total ERK level will have a biological consequence, except for biological responses that require full-blown ERK activation. Indeed, we observed that ERK1 silencing decreased the induction levels of several IEGs despite the fact that ERK1 represented 20% of ERKs in NIH 3T3 cells. Similarly, Liu et al. have shown in HeLa cells that ERK1 silencing slowed cell proliferation as effectively as ERK2 silencing (27). Unfortunately, those authors did not measure the level of activated ERKs, and hence we cannot conclude if the equal effectiveness of ERK1 and ERK2 single silencing was due to similar expression/activation of the two proteins.
The generally lower level of ERK1 means that silencing of ERK1 or erk1 gene invalidation has a minor impact on total ERK activity (14, 32). It does not mean that a lack of ERK1 is without consequence. Among binary responses of ERK activation, only those that require full-blown ERK activity will be affected directly by the lack of ERK1. However, processes that respond gradually to the strength of ERK activation may be affected partially by the lack of ERK1 or not affected if overactivation of ERK2 compensates for the lack of ERK1, similarly to the case for hypomorphic mutants of Drosophila. For example, our results indicate that removal of ERK1 in NIH 3T3 cells was sufficient to diminish the elevation of the mRNA levels of several IEGs. In addition, it was shown that animals lacking ERK1 display cutaneous lesions and hyperkeratosis and are resistant to development of induced skin papillomas (9). It was also shown that mice lacking ERK1 have lower adiposity and fewer adipocytes than wild-type animals. When these mice were challenged with a high-fat diet, they were resistant to obesity (7). It was also shown that mice lacking ERK1 display an enhancement of the striatum-dependent long-term memory. Another study indicated that these mice were more susceptible to experimental autoimmune encephalomyelitis than wild-type animals (3). Lack of thymocyte terminal differentiation in erk1−/− mice was observed initially (32) but was not confirmed in two more recent studies (14, 30). Unlike ERK1c, the splice variant ERK1b is present in rodents (2); however, this isoform ought to be detected by our antibodies against total ERK or active ERK, but no slower-migrating bands were detected. Even if ERK1b is expressed at very low levels, it is targeted as well as ERK1 with our shRNA, and hence ERK1b cannot explain the lack of effect of ERK1 silencing on cell proliferation.
An indication that ERK1 is not essential for vertebrates comes from the observation of the lack of the erk1 gene in at least two sequenced genomes, those of Gallus gallus and of Xenopus tropicalis (searches in whole genomes and in EST databases as described in Materials and Methods). Interestingly, the four fish genomes sequenced so far (Takifugu rubripes, Tetraodon nigroviridis, Oryzias latipes, and Danio rerio) contain erk1 and erk2, and therefore Gallus and Xenopus have lost the erk1 that was present in early vertebrates (the fish are closer to the common ancestor of all vertebrates). ERK isoforms arose in evolution at whole-genome duplication (WGD) that occurred when vertebrates appeared (22, 46). Indeed, only vertebrates have two erk genes. For example, the closest relatives of vertebrates for which the genomes have been sequenced, Ciona intestinalis and Ciona tropicalis, have only one isoform of ERK. Interestingly, it is impossible to trace back whether ERK1 or ERK2 is closer to Ciona's ERK by aligning the amino acids that are specific for each ERK isoform and conserved in all mammals and fishes sequenced (data not shown).
Why have mammals and nearly all vertebrates whose genomes have been sequenced kept two isoforms of ERK if these kinases fulfill highly similar functions? Recently, sequencing of the Paramecium genome revealed that WGDs were followed by a slow loss of redundant genes, keeping the few genes that evolved to raise new species. However, these authors demonstrated that the loss of redundant genes was slower if they were inserted into modules whose partners are linked by stoichiometric relationships (4). Since it is known that the proper subcellular localization of ERK requires a balanced amount of MEK (1, 17, 25), it can be assumed that these two genes are linked by a stoichiometric ratio, which could provide an explanation for keeping highly similar kinases in this module after a WGD. Indeed, it was shown recently that there is about twice as much MEK as ERK in HeLa and Cos-7 cells (16). Very interestingly, at least in Xenopus tropicalis, loss of ERK1 is also accompanied by loss of MEK2 (by genome analysis described in Materials and Methods); hence, at least in one vertebrate this essential signaling cascade is maintained with one isoform at each stage instead of two MEKs and two ERKs in all mammals and fishes sequenced so far. We cannot rule out completely specific functions for ERK1 or for its isoforms (ERK1b in rodents) in specialized cells; however, the lack of the erk1 gene in several vertebrates indicates that ERK1 and its isoforms can be fully replaced by ERK2 for all biological functions in these animals. In a whole organism, expressing two isoforms simultaneously at different levels may increase the range of possibilities to regulate finely the strength of ERK signaling in specialized cells. If this is true, the amounts of kinase isoforms expressed evolved to provide precisely the strength of signal needed in each cell; in fact, very different ERK1/ERK2 ratios can be observed in brain structures.
Acknowledgments
We appreciate the invaluable help of Danièle Roux with pSUPER-ERK2 cloning and of Christine Bourcier with mouse ERK1 cloning. We are grateful to Gilles Pagès and to Sylvain Méloche for sharing the results of mouse crosses. We are grateful to Elisabeth Goldsmith for helpful discussion concerning ERK2 versus ERK1 binding to the D-motif peptide and to Melanie Cobb for personal communication concerning the lack of ERK1 in the chicken genome. We thank Michiaki Kohno and Sir Philip Cohen for kindly providing us PD184352. We thank Krittalak Chakrabandhu for performing the RabVa immunoblotting, Richard Christen for elaborating genomic sequence analysis, and Robert Hipskind for kindly providing us the vector GST-Elk1307-428. We are grateful to Aurélie Rossin, Rosana Kral, and Sébastien Huault for helping with fluorescence-activated cell sorter analysis. We thank Emmanuel Chamorey for helping with statistical analysis and M. Christiane Brahimi-Horn for critical reading of the manuscript.
Financial support was provided by the Centre National de la Recherche Scientifique (CNRS), Centre A. Lacassagne, Ministère de l'Education de la Recherche et de la Technologie, Ligue Nationale Contre le Cancer (Equipe Labelisée), and Association pour la Recherche sur le Cancer (ARC contract 3338).
Footnotes
Published ahead of print on 29 October 2007.
REFERENCES
- 1.Adachi, M., M. Fukuda, and E. Nishida. 2000. Nuclear export of MAP kinase (ERK) involves a MAP kinase kinase (MEK)-dependent active transport mechanism. J. Cell Biol. 148849-856. (Erratum, 149:754, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebersold, D. M., Y. D. Shaul, Y. Yung, N. Yarom, Z. Yao, T. Hanoch, and R. Seger. 2004. Extracellular signal-regulated kinase 1c (ERK1c), a novel 42-kilodalton ERK, demonstrates unique modes of regulation, localization, and function. Mol. Cell. Biol. 2410000-10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal, A., S. Dillon, T. L. Denning, and B. Pulendran. 2006. ERK1−/− mice exhibit Th1 cell polarization and increased susceptibility to experimental autoimmune encephalomyelitis. J. Immunol. 1765788-5796. [DOI] [PubMed] [Google Scholar]
- 4.Aury, J. M., O. Jaillon, L. Duret, B. Noel, C. Jubin, B. M. Porcel, B. Segurens, V. Daubin, V. Anthouard, N. Aiach, O. Arnaiz, A. Billaut, J. Beisson, I. Blanc, K. Bouhouche, F. Camara, S. Duharcourt, R. Guigo, D. Gogendeau, M. Katinka, A. M. Keller, R. Kissmehl, C. Klotz, F. Koll, A. Le Mouel, G. Lepere, S. Malinsky, M. Nowacki, J. K. Nowak, H. Plattner, J. Poulain, F. Ruiz, V. Serrano, M. Zagulski, P. Dessen, M. Betermier, J. Weissenbach, C. Scarpelli, V. Schachter, L. Sperling, E. Meyer, J. Cohen, and P. Wincker. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444171-178. [DOI] [PubMed] [Google Scholar]
- 5.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 224082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilton, R., N. Mazure, E. Trottier, M. Hattab, M. A. Dery, D. E. Richard, J. Pouyssegur, and M. C. Brahimi-Horn. 2005. Arrest-defective-1 protein, an acetyltransferase, does not alter stability of hypoxia-inducible factor (HIF)-1alpha and is not induced by hypoxia or HIF. J. Biol. Chem. 28031132-31140. [DOI] [PubMed] [Google Scholar]
- 7.Bost, F., M. Aouadi, L. Caron, P. Even, N. Belmonte, M. Prot, C. Dani, P. Hofman, G. Pages, J. Pouyssegur, Y. Le Marchand-Brustel, and B. Binetruy. 2005. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes 54402-411. [DOI] [PubMed] [Google Scholar]
- 8.Boulton, T. G., S. H. Nye, D. J. Robbins, N. Y. Ip, E. Radziejewska, S. D. Morgenbesser, R. A. DePinho, N. Panayotatos, M. H. Cobb, and G. D. Yancopoulos. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65663-675. [DOI] [PubMed] [Google Scholar]
- 9.Bourcier, C., A. Jacquel, J. Hess, I. Peyrottes, P. Angel, P. Hofman, P. Auberger, J. Pouyssegur, and G. Pages. 2006. p44 mitogen-activated protein kinase (extracellular signal-regulated kinase 1)-dependent signaling contributes to epithelial skin carcinogenesis. Cancer Res. 662700-2707. [DOI] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296550-553. [DOI] [PubMed] [Google Scholar]
- 11.Burdon, T., C. Stracey, I. Chambers, J. Nichols, and A. Smith. 1999. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 21030-43. [DOI] [PubMed] [Google Scholar]
- 12.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1610881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Benedetto, B., C. Hitz, S. M. Holter, R. Kuhn, D. M. Vogt Weisenhorn, and W. Wurst. 2007. Differential mRNA distribution of components of the ERK/MAPK signalling cascade in the adult mouse brain. J. Comp. Neurol. 500542-556. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, A. M., C. D. Katayama, G. Pages, J. Pouyssegur, and S. M. Hedrick. 2005. The role of erk1 and erk2 in multiple stages of T cell development. Immunity 23431-443. [DOI] [PubMed] [Google Scholar]
- 15.Fremin, C., F. Ezan, P. Boisselier, A. Bessard, G. Pages, J. Pouyssegur, and G. Baffet. 2007. ERK2 but not ERK1 plays a key role in hepatocyte replication: an RNAi-mediated ERK2 knockdown approach in wild-type and ERK1 null hepatocytes. Hepatology 451035-1045. [DOI] [PubMed] [Google Scholar]
- 16.Fujioka, A., K. Terai, R. E. Itoh, K. Aoki, T. Nakamura, S. Kuroda, E. Nishida, and M. Matsuda. 2006. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2818917-8926. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, M., Y. Gotoh, and E. Nishida. 1997. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 161901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatano, N., Y. Mori, M. Oh-hora, A. Kosugi, T. Fujikawa, N. Nakai, H. Niwa, J. Miyazaki, T. Hamaoka, and M. Ogata. 2003. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells 8847-856. [DOI] [PubMed] [Google Scholar]
- 19.Her, J. H., S. Lakhani, K. Zu, J. Vila, P. Dent, T. W. Sturgill, and M. J. Weber. 1993. Dual phosphorylation and autophosphorylation in mitogen-activated protein (MAP) kinase activation. Biochem. J. 29625-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshino, R., Y. Chatani, T. Yamori, T. Tsuruo, H. Oka, O. Yoshida, Y. Shimada, S. Ari-i, H. Wada, J. Fujimoto, and M. Kohno. 1999. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene 18813-822. [DOI] [PubMed] [Google Scholar]
- 21.Jaeschke, A., M. Karasarides, J. J. Ventura, A. Ehrhardt, C. Zhang, R. A. Flavell, K. M. Shokat, and R. J. Davis. 2006. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell 23899-911. [DOI] [PubMed] [Google Scholar]
- 22.Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann, E. Mauceli, L. Bouneau, C. Fischer, C. Ozouf-Costaz, A. Bernot, S. Nicaud, D. Jaffe, S. Fisher, G. Lutfalla, C. Dossat, B. Segurens, C. Dasilva, M. Salanoubat, M. Levy, N. Boudet, S. Castellano, V. Anthouard, C. Jubin, V. Castelli, M. Katinka, B. Vacherie, C. Biemont, Z. Skalli, L. Cattolico, J. Poulain, V. De Berardinis, C. Cruaud, S. Duprat, P. Brottier, J. P. Coutanceau, J. Gouzy, G. Parra, G. Lardier, C. Chapple, K. J. McKernan, P. McEwan, S. Bosak, M. Kellis, J. N. Volff, R. Guigo, M. C. Zody, J. Mesirov, K. Lindblad-Toh, B. Birren, C. Nusbaum, D. Kahn, M. Robinson-Rechavi, V. Laudet, V. Schachter, F. Quetier, W. Saurin, C. Scarpelli, P. Wincker, E. S. Lander, J. Weissenbach, and H. Roest Crollius. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431946-957. [DOI] [PubMed] [Google Scholar]
- 23.Kosako, H., E. Nishida, and Y. Gotoh. 1993. cDNA cloning of MAP kinase kinase reveals kinase cascade pathways in yeasts to vertebrates. EMBO J. 12787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lein, E. S., M. J. Hawrylycz, N. Ao, M. Ayres, A. Bensinger, A. Bernard, A. F. Boe, M. S. Boguski, K. S. Brockway, E. J. Byrnes, L. Chen, L. Chen, T. M. Chen, M. C. Chin, J. Chong, B. E. Crook, A. Czaplinska, C. N. Dang, S. Datta, N. R. Dee, A. L. Desaki, T. Desta, E. Diep, T. A. Dolbeare, M. J. Donelan, H. W. Dong, J. G. Dougherty, B. J. Duncan, A. J. Ebbert, G. Eichele, L. K. Estin, C. Faber, B. A. Facer, R. Fields, S. R. Fischer, T. P. Fliss, C. Frensley, S. N. Gates, K. J. Glattfelder, K. R. Halverson, M. R. Hart, J. G. Hohmann, M. P. Howell, D. P. Jeung, R. A. Johnson, P. T. Karr, R. Kawal, J. M. Kidney, R. H. Knapik, C. L. Kuan, J. H. Lake, A. R. Laramee, K. D. Larsen, C. Lau, T. A. Lemon, A. J. Liang, Y. Liu, L. T. Luong, J. Michaels, J. J. Morgan, R. J. Morgan, M. T. Mortrud, N. F. Mosqueda, L. L. Ng, R. Ng, G. J. Orta, C. C. Overly, T. H. Pak, S. E. Parry, S. D. Pathak, O. C. Pearson, R. B. Puchalski, Z. L. Riley, H. R. Rockett, S. A. Rowland, J. J. Royall, M. J. Ruiz, N. R. Sarno, K. Schaffnit, N. V. Shapovalova, T. Sivisay, C. R. Slaughterbeck, S. C. Smith, K. A. Smith, B. I. Smith, A. J. Sodt, N. N. Stewart, K. R. Stumpf, S. M. Sunkin, M. Sutram, A. Tam, C. D. Teemer, C. Thaller, C. L. Thompson, L. R. Varnam, A. Visel, R. M. Whitlock, P. E. Wohnoutka, C. K. Wolkey, V. Y. Wong, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445168-176. [DOI] [PubMed] [Google Scholar]
- 25.Lenormand, P., C. Sardet, G. Pagès, G. L'Allemain, A. Brunet, and J. Pouysségur. 1993. Growth factors induce nuclear translocation of MAP kinases (p42maPk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 1221079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, J., and S. E. Johnson. 2006. ERK2 is required for efficient terminal differentiation of skeletal myoblasts. Biochem. Biophys. Res. Commun. 3451425-1433. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., S. Yan, T. Zhou, Y. Terada, and R. L. Erikson. 2004. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene 23763-776. [DOI] [PubMed] [Google Scholar]
- 28.Mazzucchelli, C., C. Vantaggiato, A. Ciamei, S. Fasano, P. Pakhotin, W. Krezel, H. Welzl, D. P. Wolfer, G. Pages, O. Valverde, A. Marowsky, A. Porrazzo, P. C. Orban, R. Maldonado, M. U. Ehrengruber, V. Cestari, H. P. Lipp, P. F. Chapman, J. Pouyssegur, and R. Brambilla. 2002. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron 34807-820. [DOI] [PubMed] [Google Scholar]
- 29.Morgenstern, J. P., and H. Land. 1990. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 181068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nekrasova, T., C. Shive, Y. Gao, K. Kawamura, R. Guardia, G. Landreth, and T. G. Forsthuber. 2005. ERK1-deficient mice show normal T cell effector function and are highly susceptible to experimental autoimmune encephalomyelitis. J. Immunol. 1752374-2380. [DOI] [PubMed] [Google Scholar]
- 31.Ortiz, J., H. W. Harris, X. Guitart, R. Z. Terwilliger, J. W. Haycock, and E. J. Nestler. 1995. Extracellular signal-regulated protein kinases (ERKs) and ERK kinase (MEK) in brain: regional distribution and regulation by chronic morphine. J. Neurosci. 151285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pages, G., S. Guerin, D. Grall, F. Bonino, A. Smith, F. Anjuere, P. Auberger, and J. Pouyssegur. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science 2861374-1377. [DOI] [PubMed] [Google Scholar]
- 33.Pages, G., P. Lenormand, G. L'Allemain, J. C. Chambard, S. Meloche, and J. Pouyssegur. 1993. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc. Natl. Acad. Sci. USA 908319-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pouyssegur, J., V. Volmat, and P. Lenormand. 2002. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem. Pharmacol. 64755-763. [DOI] [PubMed] [Google Scholar]
- 35.Pullikuth, A., E. McKinnon, H. J. Schaeffer, and A. D. Catling. 2005. The MEK1 scaffolding protein MP1 regulates cell spreading by integrating PAK1 and Rho signals. Mol. Cell. Biol. 255119-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saba-El-Leil, M. K., F. D. Vella, B. Vernay, L. Voisin, L. Chen, N. Labrecque, S. L. Ang, and S. Meloche. 2003. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 4964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabapathy, K., K. Hochedlinger, S. Y. Nam, A. Bauer, M. Karin, and E. F. Wagner. 2004. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell 15713-725. [DOI] [PubMed] [Google Scholar]
- 38.Sanjo, H., M. Hikida, Y. Aiba, Y. Mori, N. Hatano, M. Ogata, and T. Kurosaki. 2007. Extracellular signal-regulated protein kinase 2 is required for efficient generation of B cells bearing antigen-specific immunoglobulin g. Mol. Cell. Biol. 271236-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaeffer, H. J., A. D. Catling, S. T. Eblen, L. S. Collier, A. Krauss, and M. J. Weber. 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 2811668-1671. [DOI] [PubMed] [Google Scholar]
- 40.Selcher, J. C., T. Nekrasova, R. Paylor, G. E. Landreth, and J. D. Sweatt. 2001. Mice lacking the ERK1 isoform of MAP kinase are unimpaired in emotional learning. Learn. Mem. 811-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinmetz, R., H. A. Wagoner, P. Zeng, J. R. Hammond, T. S. Hannon, J. L. Meyers, and O. H. Pescovitz. 2004. Mechanisms regulating the constitutive activation of the extracellular signal-regulated kinase (ERK) signaling pathway in ovarian cancer and the effect of ribonucleic acid interference for ERK1/2 on cancer cell proliferation. Mol. Endocrinol. 182570-2582. [DOI] [PubMed] [Google Scholar]
- 42.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2110-116. [DOI] [PubMed] [Google Scholar]
- 43.Tanoue, T., R. Maeda, M. Adachi, and E. Nishida. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20466-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teis, D., N. Taub, R. Kurzbauer, D. Hilber, M. E. de Araujo, M. Erlacher, M. Offterdinger, A. Villunger, S. Geley, G. Bohn, C. Klein, M. W. Hess, and L. A. Huber. 2006. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 175861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288870-874. [DOI] [PubMed] [Google Scholar]
- 46.Vandepoele, K., W. De Vos, J. S. Taylor, A. Meyer, and Y. Van de Peer. 2004. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. USA 1011638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vantaggiato, C., I. Formentini, A. Bondanza, C. Bonini, L. Naldini, and R. Brambilla. 2006. ERK1 and ERK2 mitogen-activated protein kinases affect Ras-dependent cell signaling differentially. J. Biol. 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vomastek, T., H. J. Schaeffer, A. Tarcsafalvi, M. E. Smolkin, E. A. Bissonette, and M. J. Weber. 2004. Modular construction of a signaling scaffold: MORG1 interacts with components of the ERK cascade and links ERK signaling to specific agonists. Proc. Natl. Acad. Sci. USA 1016981-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, X., and B. Seed. 2003. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wunderlich, W., I. Fialka, D. Teis, A. Alpi, A. Pfeifer, R. G. Parton, F. Lottspeich, and L. A. Huber. 2001. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 152765-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, Y., W. Li, J. Wu, U. A. Germann, M. S. Su, K. Kuida, and D. M. Boucher. 2003. Extracellular signal-regulated kinase 2 is necessary for mesoderm differentiation. Proc. Natl. Acad. Sci. USA 10012759-12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon, S., and R. Seger. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 2421-44. [DOI] [PubMed] [Google Scholar]
- 53.Zeng, P., H. A. Wagoner, O. H. Pescovitz, and R. Steinmetz. 2005. RNA interference (RNAi) for extracellular signal-regulated kinase 1 (ERK1) alone is sufficient to suppress cell viability in ovarian cancer cells. Cancer Biol. Ther. 4961-967. [DOI] [PubMed] [Google Scholar]
- 54.Zheng, G., and G. Lyons. 2002. Cyclosporin A improves the selection of cells transfected with the puromycin acetyltransferase gene. BioTechniques 3332-36. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, G., Z. Q. Bao, and J. E. Dixon. 1995. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 27012665-12669. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, T., L. Sun, J. Humphreys, and E. J. Goldsmith. 2006. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure 141011-1019. [DOI] [PubMed] [Google Scholar]