Abstract
It is thought that degradation of nuclear proteins by the ubiquitylation system requires nuclear-cytoplasmic trafficking of E3 ubiquitin ligases. The von Hippel-Lindau (VHL) tumor suppressor protein is the substrate recognition component of a Cullin-2-containing E3 ubiquitin ligase that recruits hypoxia-inducible factor (HIF) for oxygen-dependent degradation. We demonstrated that VHL engages in nuclear-cytoplasmic trafficking that requires ongoing transcription to promote efficient HIF degradation. Here, we report the identification of a discreet motif, DXGX2DX2L, that directs transcription-dependent nuclear export of VHL and which is targeted by naturally occurring mutations associated with renal carcinoma and polycythemia in humans. The DXGX2DX2L motif is also found in other proteins, including poly(A)-binding protein 1, to direct its transcription-dependent nuclear export. We define DXGX2DX2L as TD-NEM (transcription-dependent nuclear export motif), since inhibition of transcription by actinomycin D or 5,6-dichlorobenzimidazole abrogates its nuclear export activity. Disease-causing mutations of key residues of TD-NEM restrain the ability of VHL to efficiently mediate oxygen-dependent degradation of HIF by altering its nuclear export dynamics without affecting interaction with its substrate. These results identify a novel nuclear export motif, further highlight the role of nuclear-cytoplasmic shuttling of E3 ligases in degradation of nuclear substrates, and provide evidence that disease-causing mutations can target subcellular trafficking.
Ubiquitylation is a multiprotein pathway that destines marked proteins for degradation by the 26S proteasome (22, 59). The conjugation of ubiquitin to proteins requires the action of three different enzymes: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. The process of ubiquitylation begins with the loading of a ubiquitin molecule onto the E1 ubiquitin-activating enzyme. This is followed by the transfer of ubiquitin from the E1 to the E2 ubiquitin-conjugating enzyme. Finally, transfer of ubiquitin from the E2 to the lysine residue of a target substrate is catalyzed by the E3 ubiquitin ligase. Selectivity of this pathway relies heavily on E3 ubiquitin ligases, which ultimately dictate substrate specificity. E3 ubiquitin ligases can act individually or form a multisubunit complex that may include a member of the Cullin family of proteins to covalently modify a vast array of cellular proteins. In view of the essential role of E3 ubiquitin ligases in regulation of many aspects of cellular functions and biological processes, there is mounting evidence that loss of function or deregulation of E3 ligases contributes to the development of disease.
Degradation of nuclear substrates by the ubiquitylation system often requires nuclear-cytoplasmic trafficking of both the E3 ubiquitin ligase and the substrate protein (2, 54). One example is the ubiquitin-mediated degradation of the p53 tumor suppressor protein by the Mdm2 (murine double minute 2) E3 ubiquitin ligase (45, 47). Mdm2 shuttles continuously between the nucleus and the cytoplasm in order to efficiently degrade nuclear p53 (12, 53). Cancer-causing point mutations that disrupt nuclear export of Mdm2 are impaired in mediating proteasomal degradation of p53 (37). Nuclear export of the ROC1-SCFFbw1a E3 ubiquitin ligase is also required for the proteasomal degradation of the Smad3 transcription factor (13). Another example is the cyclin-dependent kinase inhibitor p27Kip1, which requires nuclear export by Jab1 for proteasome-mediated degradation. A mutant form of p27Kip1 that fails to assemble with Jab1 cannot be exported from the nucleus and is not degraded by the proteasome (54, 58).
The von Hippel-Lindau tumor suppressor protein (VHL) is a vital component of the VBC-Cul2 E3 ubiquitin ligase complex, as it acts as the substrate recognition protein to provide specificity to the degradation process (25, 27, 30, 38, 39, 50). VHL promotes the recruitment, ubiquitylation, and subsequent proteasomal degradation of the alpha subunit of hypoxia-inducible factor (HIF) in an oxygen-dependent manner (26, 41). Under conditions of normal oxygen tension (normoxia), HIFα is hydroxylated at key prolyl residues within the oxygen-dependent degradation domain by prolyl hydroxylases (5, 8, 24, 26). This posttranslational modification promotes the interaction between HIFα and VHL and leads to ubiquitin-mediated degradation of HIFα (62). Under conditions of low oxygen tension (hypoxia), prolyl hydroxylation does not occur, leading to the stabilization of HIFα, since it fails to assemble with VHL (26, 46, 62). Stabilization of HIFα results in increased transcription of an array of hypoxia-inducible genes, including vascular endothelial growth factor, glucose transporter 1, and transforming growth factor α, among others, that modulate angiogenesis, glycolysis, and growth (11, 18, 20, 55, 56). Numerous inactivating mutations of the VHL gene lead to the stabilization of HIFα and are associated with the VHL cancer syndrome, in which afflicted individuals develop different tumors, such as renal clear cell carcinoma (RCC), retinal angioma, nervous system hemangioblastoma, and pheochromocytoma (6, 23, 33, 40). Inactivating mutations of VHL often prevent assembly with its substrate, HIF, or core components of the E3 ubiquitin ligase, elongins B and C, and Cullin 2, resulting in constitutive activation of HIFα targets (7, 24, 28, 35, 39, 46).
Nuclear-cytoplasmic trafficking is essential for the E3 ubiquitin ligase function of VHL and oxygen-dependent degradation of HIFα (17, 34). Failure of VHL to continuously shuttle between the nuclear and cytoplasmic compartments leads to the stabilization of HIFα (17, 34, 42-44). VHL engages in nuclear-cytoplasmic shuttling dynamics independently of the classical, leucine-rich, nuclear export sequence (NES) (9, 60) but accumulates in the nucleus upon addition of inhibitors of RNA polymerase II (Pol II) activity (17, 34). Interestingly, the general RNA metabolism and translation initiation factor poly(A)-binding protein 1 (PABP1) exhibits similar transcription-dependent trafficking dynamics as VHL, since it also accumulates in the nucleus upon addition of inhibitors of RNA Pol II activity (1). These results suggest the existence of a transcription-dependent nuclear export pathway that is employed by VHL, PABP1, and perhaps other proteins and that operates independently of the classical NES/CRM1 system.
Here we report the identification of a novel and discreet nuclear export motif, DXGX2DX2L. We define this motif as TD-NEM (transcription-dependent nuclear export motif), since it mediates nuclear export of proteins in a manner that requires ongoing RNA Pol II-dependent transcription but operates independently of the classical NES pathway. Disease mutations of TD-NEM of VHL alter its ability to be exported from the nucleus and to mediate oxygen-dependent degradation of HIFα without affecting interaction with its substrate. These results highlight the requirement of nuclear-cytoplasmic trafficking of E3 ubiquitin ligases for degradation of their nuclear substrates, provide evidence that mutations targeting subcellular trafficking can lead to disease, and identify a novel motif that mediates efficient nuclear export of proteins.
MATERIALS AND METHODS
Cell culture, transfections, drug treatments, and hypoxia treatment.
786-0 (VHL-negative) renal carcinoma cells, MCF-7 cells, and NIH 3T3 cells were obtained from the American Type Culture Collection. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum and 1% penicillin-streptomycin in a 37°C and 5% CO2 environment. Transient transfections in MCF-7 cells were conducted with Effective transfection reagent (Qiagen). Transfected cells were incubated for 24 h before any manipulations or drug treatment. Stable cell lines were generated by stably transfecting 786-0 cells with the following Flag-tagged constructs: F-VHL-GFP, F-VHL(D121G)-GFP, F-VHL(D126Y)-GFP, and F-VHL(G123A)-GFP, followed by G418 selection. Where indicated, cells were treated at 37°C with a final concentration of 2 μM actinomycin D (ActD), 10 μM leptomycin B (LMB), or 25 μg/ml 5,6-dichlorobenzimidazole (DRB) for 1 h prior to photobleaching experiments and 10 μM ActD or 10 μM LMB for 3 h prior to live cell fluorescence imaging. Proteasome inhibitor treatment was performed with MG132 (Calbiochem) at a final concentration of 10 μM for 2 h before the cells were harvested. Hypoxia treatment was performed in a hypoxic chamber at 37°C under a 1% O2, 5% CO2, and N2-balanced atmosphere.
Expression vectors.
Human full-length VHL and deletion, truncation, or point mutants of VHL were cloned into pcDNA3.1 between an NH2-terminal Flag tag and a COOH-terminal green fluorescent protein (GFP) tag, as previously described (4, 17, 34). F-GFP and F-GFP-NES were previously described by Groulx et al. (16) and Lee et al. (34). The PK-GFP-NLS construct used for the in vivo fluorescence loss in photobleaching (FLIP) nuclear export assay consisted of pyruvate kinase (PK), which does not encode any localization determinant, GFP, and a nuclear localization signal (NLS) derived from the simian virus 40 large T antigen as previously described (16, 29). Human full-length VHL and deletion mutants and the strong NES from the human immunodeficiency virus Rev were inserted into a F-PK-GFP-NLS construct that was previously described by Groulx et al. (16), between the Flag tag and PK using the Apa1 and Xho1 restriction sites. cDNAs corresponding to VHL residues 114 to 138 [VHL(114-138)], PABP1(296-317), and cyclin C(158-179), which encode TD-NEM sequences and the full-length cyclin C, were inserted into PK-GFP-NLS using Apa1 and Xho1 restriction sites. The human full-length PABP1 and the deletion mutant Δ296-317 were fused to GFP-F to produce the GFP-F-PABP1 and GFP-F-PABP1(Δ296-317) fusion proteins.
Live cell fluorescence imaging.
Images of living cells transiently expressing GFP from experiments where photobleaching was not utilized were imaged with an Axiovert S100TV microscope (Carl Zeiss MicroImaging, Inc.) equipped with a 40×, 1.2 C-Apochromat water immersion objective using a digital charged-coupled-device camera (Empix). Cell nuclei were stained with Hoechst 33342 (Sigma). Images were captured using the Northern Eclipse software package (Empix).
Photobleaching and microscopy.
Cells were cultured and transfected directly onto 35-mm dishes with coverslip bottoms (MatTek). Photobleaching and live cell microscopy were performed using a confocal microscope (LSM5 Pa laser scanning microscope; Carl Zeiss Canada). In all experiments cells were maintained at 37°C in an environmental chamber. A 63× plan Apo oil immersion lens with a 1.4 numerical aperture was used for bleaching and imaging. Indicated areas were exposed to three rapid pulses of a 488-nm argon laser at 100%, and image acquisition was at 1% of full laser power. For cytoplasmic FLIP experiments of cells expressing a GFP-tagged fusion protein, a large cytoplasmic region was initially bleached with three rapid pulses to eliminate the dominant cytoplasmic signal. This was followed by repetitive bleaching in a small region of the cytoplasm and imaged at 30-second intervals. For cytoplasmic FLIP experiments of cells expressing a PK-GFP-NLS-tagged fusion protein, cells were repeatedly bleached in a small cytoplasmic region and imaged at 30-second intervals. Small bleached areas for cytoplasmic FLIPs were kept consistent in terms of size and distance from the nucleus. Fluorescence loss in the unbleached areas was quantified as previously described (43, 51) using the following equation: Irel = (It/I0)·(N0/Nt), where It is the average intensity of the unbleached nucleus or cell at time point t, I0 is the average prebleached intensity of the nucleus or cell of interest, and N0 and Nt are the average nuclear or cellular fluorescence intensity of a neighboring cell in the same field of vision prebleach or at time t, respectively. This calculation accounts for any losses in fluorescence by normalizing the fluorescence of the cell of interest to that of a neighboring cell of approximately equal size and fluorescent intensity. Nuclear FLIPs were performed by repetitive bleaching of a small nuclear area and imaging at 15-second intervals. Loss of nuclear fluorescence was quantified using the above equation; however, the value for It in this case was that of the bleached nucleus. For fluorescence recovery after photobleaching (FRAP) experiments, a large nuclear region was photobleached once with three rapid pulses and images were collected every 5 seconds. Recovery of the fluorescent signal within the bleached region was calculated as described by Phair and Misteli (51) and the following equation: Irel = (It/I0)·(T0/Tt), where Tt is the total cellular intensity at time t, T0 is the total cellular intensity before bleaching, I0 is the intensity in the bleached area before bleaching, and It is the intensity of the previously bleached region at time t. Pseudocolor images were generated to highlight differences in GFP fluorescence, with red representing high fluorescent intensity and light blue representing low fluorescent intensity. The quantification graphic was generated by using FLIP/FRAP software. For all bleaching experiments, 10 data sets were analyzed for each result. Pseudocoloring for bleaching experiments was achieved by applying the gradient map function of Photoshop (Adobe) to a montage of picture frames prepared with ImageJ software (National Institutes of Health, Bethesda, MD). The Northern Eclipse (Empix), Excel (Microsoft), and FreeHand (Macromedia) software packages were also used to capture images, analyze the data, and generate graphs.
Polykaryon assay.
MCF-7 cells were transfected to express fluorescently labeled proteins and incubated under standard conditions for 24 h. Usually, 40 to 60% of cells presented strong fluorescence as observed by 488-nm fluorescence microscopy. Cells were trypsinized 24 h after transfection and mixed with untransfected NIH 3T3 cells in a ratio of 1 to 10. The cell mixture was plated in 35-mm dishes with coverslip bottoms and incubated overnight under standard cell culture conditions. The confluent cell layer was visually inspected for even distribution of fluorescent cells among untransfected cells. Cells were washed twice with prewarmed phosphate-buffered saline (PBS) and fused for 2 min by addition of a prewarmed 50% solution of polyethylene glycol (PEG) in PBS (Sigma-Aldrich). PEG was removed thoroughly by four washes with prewarmed PBS, and cells were then replenished with warmed standard cell culture medium. Hoechst staining of DNA was used to identify donor and acceptor cells. Cells were observed under phase-contrast microscopy for fusion events and were monitored for the redistribution of nuclear expression of PK-GFP-NLS-tagged proteins.
In vitro nuclear export assay.
The in vitro export assay was performed as described by Groulx et al. (16). Briefly, cells were plated and grown on a 35-mm coverslip plate. Cells were washed with transport buffer (TB) containing 20 mM HEPES pH 7.3, 110 mM KO-acetate (KOAc), 5 mM NaOAc, 2 mM Mg(OAc)2 and permeabilized at 4°C for 5 min with TB containing 50 μg/ml digitonin and a protease inhibitor mixture (Hoechst stain 33258 [Sigma] was used to monitor the permeabilization). After several washes with TB at 4°C, cells were incubated for 30 to 45 min at 20°C in the presence of a standard mixture that included TB, 2 mM ATP, 2 mM GTP, and an ATP-regenerating system (5 mM creatine phosphate and 20 units/ml creatine phosphokinase). Where indicated, MCF-7 total cell lysate was added to the standard mixture.
Immunofluorescence.
Cells were seeded onto coverslips and fixed with prechilled methanol for 10 min at −20°C followed by prechilled acetone for 1 min at −20°C. Anti-PABP1 monoclonal antibody was used (Upstate). Cells were incubated for 1 h with a primary antibody solution containing 10% (vol/vol) fetal bovine serum and 1% (vol/vol) Triton X-100 at room temperature in a humidified chamber. Cells were then washed several times in PBS before a 1-h incubation with a secondary Texas Red-labeled antibody (Jackson ImmunoResearch) at room temperature in a dark humidified chamber. Hoechst stain 33342 (Sigma) was added to visualize nuclei, and coverslips were mounted using Fluoromount G (EMS).
Bioinformatic analysis.
Proteins containing a TD-NEM were identified using the emotif and My Genomics Resource Center software.
Immunoprecipitation and immunoblotting.
Cells were lysed in lysis buffer containing 0.5% Igepal CA630, 100 mM NaCl, 20 mM Tris-HCl (pH 7.6), 5 mM MgCl2, and 1 mM sodium orthovanadate with 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 1 μg/ml pepstatin. Cell lysates were incubated with anti-Flag M2 beads (Sigma) overnight while tumbling at 4°C. Beads were washed several times and eluted with Flag peptides (Sigma). For total cell lysates, cells were washed several times in PBS, lysed with 4% sodium dodecyl sulfate (SDS) in PBS, and boiled for 5 min, and the DNA was sheared by passage through a 19-gauge needle. The protein concentration was quantified using the bicinchoninic acid method (Pierce). Samples were separated on denaturing polyacrylamide gels. Western blots gels were transferred onto polyvinylidene difluoride membranes and blocked in skimmed milk powder in PBS containing 0.2% Tween 20 before incubation with Flag-M2 (Sigma), HIF2α (Novus Biologicals), actin (Sigma), and GFP (AVES Lab Inc.) antibodies. Membranes were washed with 0.2% Tween-PBS and blotted with a secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories) and detected by using Western Lightning chemiluminescence reagent plus (Perkin-Elmer).
RESULTS
Nuclear export of VHL is mediated by a discreet transcription-dependent motif.
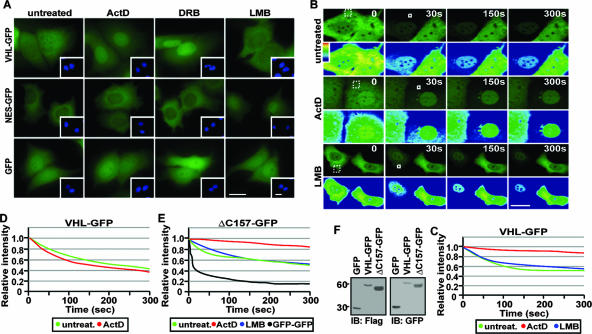
VHL engages in a nuclear export pathway that requires ongoing RNA Pol II activity (17, 34). As previously reported, addition of the RNA Pol II inhibitors ActD or DRB resulted in nuclear accumulation of the highly cytoplasmic VHL-GFP but had no effect on GFP alone or when fused to a strong, classical, leucine-rich nuclear export signal (see Materials and Methods) (Fig. 1A) (17, 34). In contrast, LMB, a compound that abolishes CRM1-dependent nuclear export of NES-containing proteins (10, 14, 31, 57), had no effect on the steady-state localization of VHL-GFP but caused nuclear accumulation of NES-GFP (Fig. 1A). Nuclear accumulation by ActD was due to a considerable decrease in the rate of nuclear export of VHL-GFP, as demonstrated by cytoplasmic FLIP experiments (Fig. 1B; see also the quantification in panel C) and by cellular fusion experiments (data not shown) (34). In the cytoplasmic FLIP experiments, a small region of the cytoplasm is repetitively bleached (Fig. 1B), and a decrease in nuclear GFP fluorescence indicates that the GFP-tagged fusion protein has been exported from the nucleus to the cytoplasm. Furthermore, nuclear FLIP revealed similar intranuclear dynamics between ActD-treated and untreated cells, demonstrating that ActD (or DRB) did not cause nuclear retention of VHL-GFP (Fig. 1D; see also Fig. S1 in the supplemental material). Loss of GFP fluorescence in the FLIP experiments was not due to fragmentation of the GFP-fusion proteins, as demonstrated by immunoblot analysis (Fig. 1F). Next, it was important to determine whether the signal mediating nuclear export was encoded by VHL or was present within the E3 ubiquitin ligase complex. We also tested a C-terminal deletion of the α-domain of VHL (ΔC157), which is required for assembly with core E3 ubiquitin ligase components elongins B and C and Cullin 2 (4, 46). ΔC157 retained the ability to engage in transcription-dependent nuclear export, since it accumulated in the nucleus upon addition of ActD or DRB (data not shown) and displayed reduced nuclear export dynamics in the presence of RNA Pol II inhibitors as demonstrated by cytoplasmic FLIP analysis (Fig. 1E). These data indicate the presence of a transcription-dependent nuclear export signal between residues 1 and 157 of VHL and suggest that formation of the E3 ubiquitin ligase complex is not required for export of VHL.
FIG. 1.
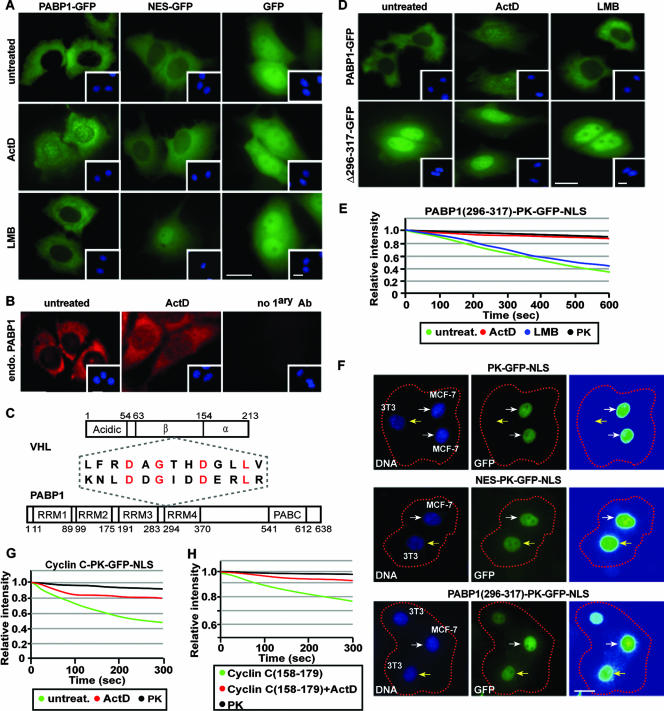
Transcription-dependent nuclear export of VHL. (A) ActD alters the steady-state localization of VHL. MCF-7 cells transiently expressing VHL-GFP, NES-GFP, or GFP were either untreated or treated with LMB (10 μM), ActD (8 μM), or DRB (25 μg/ml). Insets are the corresponding Hoechst staining of the cells. Bars, 10 μm. (B and C) Cytoplasmic FLIP reveals that ActD decreases nuclear export of VHL. MCF-7 cells transiently expressing VHL-GFP were treated with ActD (2 μM) or LMB (10 μM) for 1 h. Cells were initially bleached in a large cytoplasmic region (dashed squares) to reduce cytoplasmic signal and then submitted to repetitive bleaching in a small cytoplasmic region (white squares). Cells were imaged between pulses, and the corresponding kinetics for loss of nuclear fluorescence were calculated and plotted on a graph (C) (see Materials and Methods). Bar, 10 μm. (D) Nuclear FLIP analysis was performed on cells treated as for panel B by repetitive bleaching in a small area in the nucleus. The loss of nuclear fluorescence was graphed. (E) ΔC157 exports from the nucleus in a transcription-dependent manner. ΔC157-GFP-transiently expressing cells were treated and analyzed as described for panel B. (F) Western blot analysis using anti-Flag and anti-GFP antibodies verified that the Flag- and GFP-tagged VHL, ΔC157, and GFP fusion proteins used for photobleaching experiments were not fragmented.
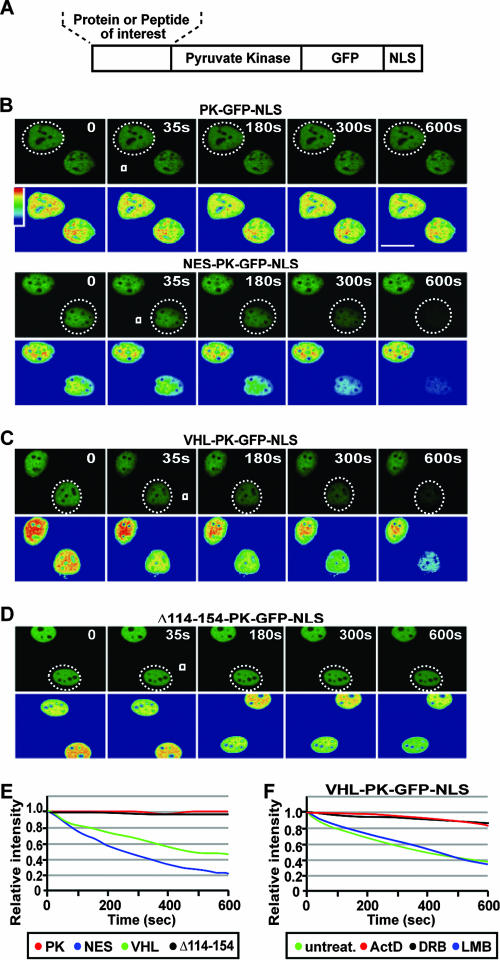
Larger GFP-tagged truncation mutants of VHL failed to provide insight on transcription-dependent nuclear export of VHL, since they exhibited export rates similar to GFP-GFP, which diffuses across the nuclear envelope (Fig. 1F). To further examine the nuclear export properties of VHL, we established a quantitative live cell nuclear export assay utilizing FLIP technology. A fusion protein consisting of the large and amorphous PK, GFP, and an NLS (29) derived from the simian virus 40 large T antigen (PK-GFP-NLS) was used to ensure that any movement across the nuclear envelope was not due to diffusion (Fig. 2A) (see Materials and Methods). Also, the presence of such a strong NLS would override an intrinsic nuclear import signal, ensuring that all fusion proteins being tested exhibited similar rates of nuclear import. PK-GFP-NLS strictly localized in the nucleus at steady state, owing to the strong NLS activity (Fig. 2B). PK-GFP-NLS did not undergo a significant loss of nuclear GFP fluorescence after cytoplasmic FLIP, as expected, since this protein fails to export from the nucleus (Fig. 2B and E). Addition of a strong, classical, and LMB-sensitive NES of the human immunodeficiency virus Rev (9) to the reporter protein (NES-PK-GFP-NLS) resulted in a rapid loss of nuclear GFP fluorescence during cytoplasmic FLIP, demonstrating that this fusion protein engages in nuclear export (Fig. 2B and E). The inability of PK-GFP-NLS to export from the nucleus compared to its NES-containing counterpart is not a consequence of unforeseen nuclear retention or a significant difference in nuclear dynamics between the two molecules as revealed by nuclear FLIP (see Fig. S2 in the supplemental material). Nuclear fluorescence was also similar for both fusion proteins, indicating that the difference in nuclear export capacity is not due to a large difference in expression levels (see Fig. S2 in the supplemental material). Full-length VHL was able to confer nuclear export activity to the PK-GFP-NLS reporter, confirming that this protein encodes nuclear export activity (Fig. 2C and E). The kinetics for the loss of nuclear signal of VHL-PK-GFP-NLS was very different compared to molecules that are able to passively diffuse, suggesting that the loss of nuclear signal is not a consequence of fragmentation of the fusion protein (see Fig. S2 in the supplemental material). ActD or DRB, but not LMB, inhibited nuclear export of VHL-PK-GFP-NLS (Fig. 2F; see also Fig. S2 in the supplemental material) without affecting intranuclear dynamics (see Fig. S2 in the supplemental material), similar to what was observed with VHL-GFP (Fig. 1) and confirming that inhibitors of RNA Pol II-mediated transcription significantly alter nuclear export of VHL. Furthermore, these results verify that the transcription-dependent nuclear export property of VHL was maintained in the fusion protein. Removal of the exon-2-encoded β-domain (Δ114-154-PK-GFP-NLS) abrogated the ability of VHL to export the reporter protein from the nucleus (Fig. 2D and E). This observation is consistent with previous reports demonstrating that nuclear accumulation of VHL upon treatment with RNA Pol II inhibitors requires the exon-2-encoded β-domain of VHL (4, 34).
FIG. 2.
A transcription-dependent nuclear export sequence is encoded within the exon-2-encoded β-domain of VHL. (A) Schematic diagram of the PK-GFP-NLS fusion protein used for the live cell FLIP nuclear export assay, describing the region where protein or peptide sequences were fused. (B to E) MCF-7 cells transiently expressing the indicated constructs were submitted to cytoplasmic FLIP analysis, in which a small cytoplasmic region (white squares) within specific cells (a dashed circle outlines the cell nucleus) was repeatedly bleached. Kinetics for the loss of nuclear fluorescence from images obtained in panels B, C, and D were calculated and plotted on a graph (E). PK refers to the PK-GFP-NLS reporter construct, and NES, VHL, and Δ114-254 indicate the sequences fused to PK-GFP-NLS. Bar, 10 μm. (F) Cells were transiently transfected with VHL-PK-GFP-NLS and were either treated with ActD (2 μM), DRB (25 μg/ml), or LMB (10 μM) for 1 h or left untreated. Cytoplasmic FLIP was performed as described above to verify nuclear export activity.
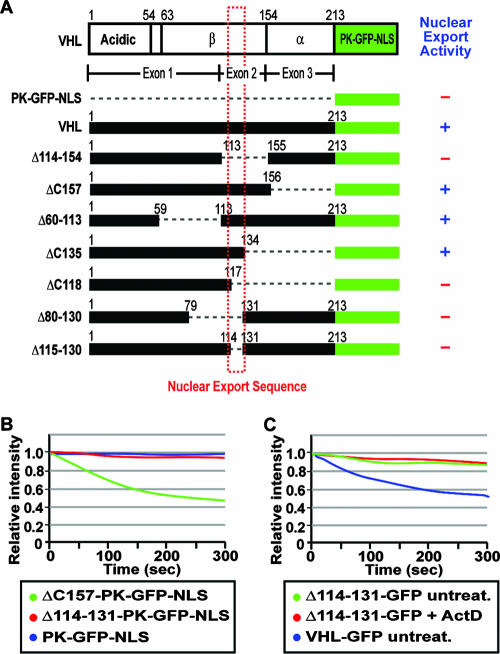
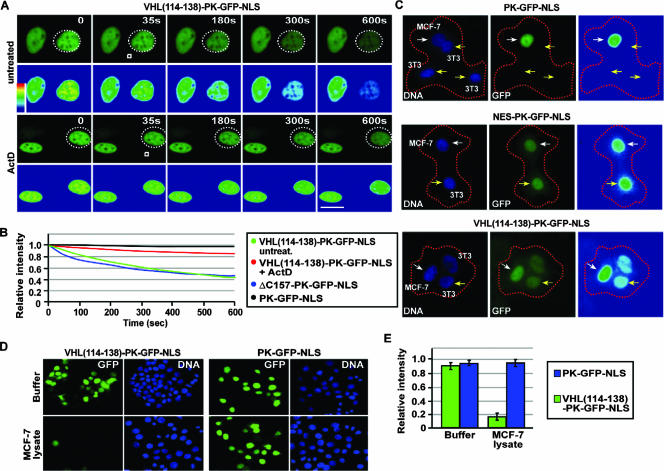
We used this in vivo FLIP nuclear export assay to map the region within the exon-2-encoded β-domain that mediates transcription-dependent nuclear export of VHL. Further mutagenesis analysis revealed that a discrete domain between residues 114 and 131 of VHL is required for efficient nuclear export of the PK-GFP-NLS reporter (Fig. 3A and B; see also Fig. S3 in the supplemental material). The deletion mutant Δ114-131-GFP (without PK and NLS) also displayed a markedly reduced rate of nuclear export compared to wild-type VHL-GFP (Fig. 3C; see also Fig. S3 in the supplemental material). The subcellular localization and the nuclear export dynamics of Δ114-131-GFP were unaffected by treatment with ActD or DRB (Fig. 3C and data not shown; see also Fig. S3 in the supplemental material) but rather exhibited a similar export rate to ΔC157-GFP after ActD treatment (Fig. 1E). More importantly, residues 114 to 138 alone were sufficient to confer efficient nuclear export properties to PK-GFP-NLS, which was abolished upon addition of ActD (Fig. 4A and B) or DRB (data not shown). The rate of export observed with residues 114 to 138 was similar to that of ΔC157, indicating that these residues alone confer nuclear export activity to the full-length protein (Fig. 4B). In a cellular fusion assay, this sequence was able to efficiently export the PK-GFP-NLS reporter protein from the donor nucleus to the acceptor nuclei compared to the PK-GFP-NLS control (Fig. 4C). Residues 114 to 138 also stimulated nuclear export of PK-GFP-NLS in an in vitro nuclear export assay (Fig. 4D and E). We have, therefore, identified a novel and discreet motif that mediates transcription-dependent nuclear export of VHL.
FIG. 3.
Residues 114 to 131 are required for nuclear export of VHL. (A) Map of the nuclear export domain of VHL. The schematic diagram indicates deletion mutants of VHL that were submitted to cytoplasmic FLIP to assess the nuclear export activity. + and - indicate the ability or inability of the fusion protein to engage in nuclear export. (B) MCF-7 cells transiently transfected with PK-GFP-NLS, ΔC157-PK-GFP-NLS, or Δ114-131-PK-GFP-NLS were submitted to cytoplasmic FLIP, in which a small cytoplasmic region of a cell was bleached repetitively. The loss of nuclear GFP fluorescence was monitored over time and plotted on a graph. (C) Cells transiently expressing Δ114-131-GFP were initially bleached in a large cytoplasmic region followed by repetitive bleaching in a small cytoplasmic region after being treated for 1 h with 2 μM ActD or left untreated. Kinetics for the loss of nuclear fluorescence were calculated and plotted on a graph.
FIG. 4.
Residues 114 to 138 mediate transcription-dependent nuclear export of VHL. (A and B) MCF-7 cells were transiently transfected to express VHL(114-138)-PK-GFP-NLS. Cells were treated with a final concentration of 8 μM ActD or left untreated for 1 h before being subjected to photobleaching. Cytoplasmic FLIP was performed by repetitively bleaching a small cytoplasmic region (white squares) of a cell (the dotted circle outlines the cell nucleus). Cells were imaged between pulses, and the corresponding kinetics for the loss of nuclear fluorescence were calculated and are graphed (B). The nuclear export kinetics of ΔC157-PK-GFP-NLS revealed by a cytoplasmic FLIP analysis were plotted (B). Bar, 10 μm. (C) VHL(114-138) can mediate export in a polykaryon fusion assay. MCF-7 cells (donors) were fused with NIH 3T3 cells (acceptors) using PEG, and the transfer of nuclear fluorescence from donor to acceptor cells was monitored. Donor and acceptor cells were differentiated by a cell-specific Hoechst staining pattern. White arrows indicate donor cells, and yellow arrows indicate acceptor cells. Bar, 10 μm. (D and E) VHL(114-138) can mediate export in an in vitro nuclear export assay. MCF-7 cells transiently expressing the indicated constructs were permeabilized with digitonin, after which they were incubated with transport buffer containing ATP, GTP, and an ATP-regenerating system in the presence of buffer or MCF-7 cell lysate. Relative loss in nuclear fluorescence was calculated and plotted on a graph (E).
VHL and PABP1 share a common transcription-dependent nuclear export motif.
VHL and PABP1 are implicated in distinct molecular networks but share the characteristic of engaging in a nuclear export pathway that requires ongoing RNA Pol II activity (1, 34). Similar to VHL, the subcellular trafficking dynamics of the highly cytoplasmic PABP1 are affected by treatment with ActD (Fig. 5A and B), but not LMB (Fig. 5A) (1). Sequence alignment revealed a striking similarity between residues 114 to 138 of VHL and 296 to 317 of PABP1 that consisted of DXGX2DX2L (Fig. 5C). Consistent with a possible role in nuclear export, removal of residues 296 to 317 from full-length PABP1 caused a drastic shift in the steady-state localization from exclusively cytoplasmic to highly nuclear (Fig. 5D). This deletion mutant of PABP1 was also insensitive to ActD treatment compared to wild type (Fig. 5D). Residues 296 to 317 of PABP1 were sufficient to mediate nuclear export of the PK-GFP-NLS reporter as efficiently as residues 114 to 138 of VHL in our FLIP nuclear export assay (Fig. 5E; see also Fig. S4 in the supplemental material). In addition, ActD or DRB, but not LMB, abolished the ability of residues 296 to 317 of PABP1 to mediate nuclear export of the PK-GFP-NLS reporter (Fig. 5E; see also Fig. S4 in the supplemental material). Residues 296 to 317 of PABP1 were able to efficiently export the reporter protein from the donor nucleus to the acceptor nuclei in a cellular fusion assay (Fig. 5F), similar to results obtained with residues 114 to 138 of VHL (Fig. 4C). This led us to postulate that the DXGX2DX2L consensus sequence might act as a common motif for transcription-dependent nuclear export of proteins. A search of sequence data banks identified several proteins that contain a DXGX2DX2L motif. We randomly selected the cell cycle regulator, cyclin C, whose nuclear-cytoplasmic shuttling profile had not been previously investigated, to test the activity of its potential nuclear export sequence. Full-length cyclin C was fused to PK-GFP-NLS to prevent any passive diffusion through nuclear pores. Cyclin C was able to confer nuclear export activity to the PK-GFP-NLS reporter in a process that was inhibited by addition of ActD (Fig. 5G). Furthermore, residues 158 to 179 of cyclin C, which encode the DXGX2DX2L motif, were able to export the reporter PK-GFP-NLS from the nucleus in an ActD-sensitive manner (Fig. 5H). Further characterization of cyclin C, and other proteins, will reveal whether the DXGX2DX2L motif is involved in their subcellular trafficking profiles. Nonetheless, we have identified a novel motif that mediates efficient transcription-dependent nuclear export of VHL, PABP1, and perhaps cyclin C. For the sake of simplicity, the DXGX2DX2L motif is referred to as a TD-NEM.
FIG. 5.
VHL and PABP1 share a common transcription-dependent nuclear export motif. (A) PABP1 exports by a transcription-dependent mechanism. MCF-7 cells transiently expressing PABP1-GFP, NES-GFP, or GFP alone were either untreated or treated with 8 μM ActD or 10 μM LMB for 3 h. Insets are the corresponding Hoechst staining of the cells. Bars, 10 μm. (B) Endogenous PABP1 is also sensitive to ActD treatment. MCF-7 cells were either left untreated or were treated with 8 μM ActD. Endogenous PABP1 was detected by immunofluorescence using an anti-PABP1 antibody. Insets show nuclei stained with Hoechst stain. A primary antibody exclusion (no 1ary Ab) control is also shown. Bar, 10 μm. (C) Schematic diagram depicting a region of alignment between the nuclear export sequence of VHL and PABP1. Conserved residues are indicated in red. (D) MCF-7 cells transiently expressing PABP1(Δ296-317)-GFP or PABP1-GFP were treated as described for panel A. Insets are the corresponding Hoechst staining of the cells. Bar, 10 μm. (E) Residues 296 to 317 of PABP1 encode a transcription-dependent nuclear export sequence. Transfected MCF-7 cells were treated with 2 μM ActD or 10 μM LMB for 1 h before being submitted to cytoplasmic FLIP. The corresponding loss of nuclear fluorescence was monitored, measured, and plotted on a graph. (F) PABP1(296-317) can mediate export in a polykaryon fusion assay. MCF-7 cells (donors) were fused with NIH 3T3 cells (acceptors) using PEG, and the transfer of nuclear fluorescence from donor to acceptor cells was monitored. Donor and acceptor cells were differentiated by a cell-specific Hoechst staining pattern. White arrows indicate donor cells, and yellow arrows indicate acceptor cells. Bar, 10 μm. (G) Full-length cyclin C can export the PK-GFP-NLS reporter in a transcription-dependent manner. Cells were treated with 2 μM ActD for 1 h or untreated, and the loss of nuclear fluorescence after cytoplasmic FLIP was plotted on a graph. (H) Residues 158 to 179 of cyclin C encode a transcription-dependent nuclear export motif. The indicated constructs were transiently expressed in MCF-7 cells and treated the same as for panel G. Cells were subjected to cytoplasmic FLIP, and the corresponding kinetics for loss of nuclear fluorescence were plotted on a graph.
Conserved residues of TD-NEM are essential for mediating efficient nuclear export of proteins.
Analysis of the TD-NEM sequences from VHL, PABP1, and cyclin C failed to identify additional conserved residues surrounding the basal DXGX2DX2L motif (Fig. 6A), suggesting that these four amino acids may encode transcription-dependent nuclear export activity. Replacement of the four key residues with alanines abolished nuclear export activity of residues 114 to 138 from VHL (Fig. 6A and B; see also Fig. S5 in the supplemental material). Replacement of the aspartic acid, leucine, or glycine residues with alanine reduced nuclear export activity by approximately 50 to 70% compared to the basal sequence (Fig. 6A and C; see also Fig. S5 in the supplemental material). In addition, replacing glycine 123 with alanine in full-length VHL, tagged to GFP and not PK-GFP-NLS, shifted the steady-state localization towards a more predominantly nuclear localization (Fig. 6D) and reduced its ability to export from the nucleus (Fig. 6E), further demonstrating that DxGx2Dx2L is a nuclear export motif. These data demonstrate that all four conserved residues in TD-NEM play a central role in mediating nuclear export.
FIG. 6.
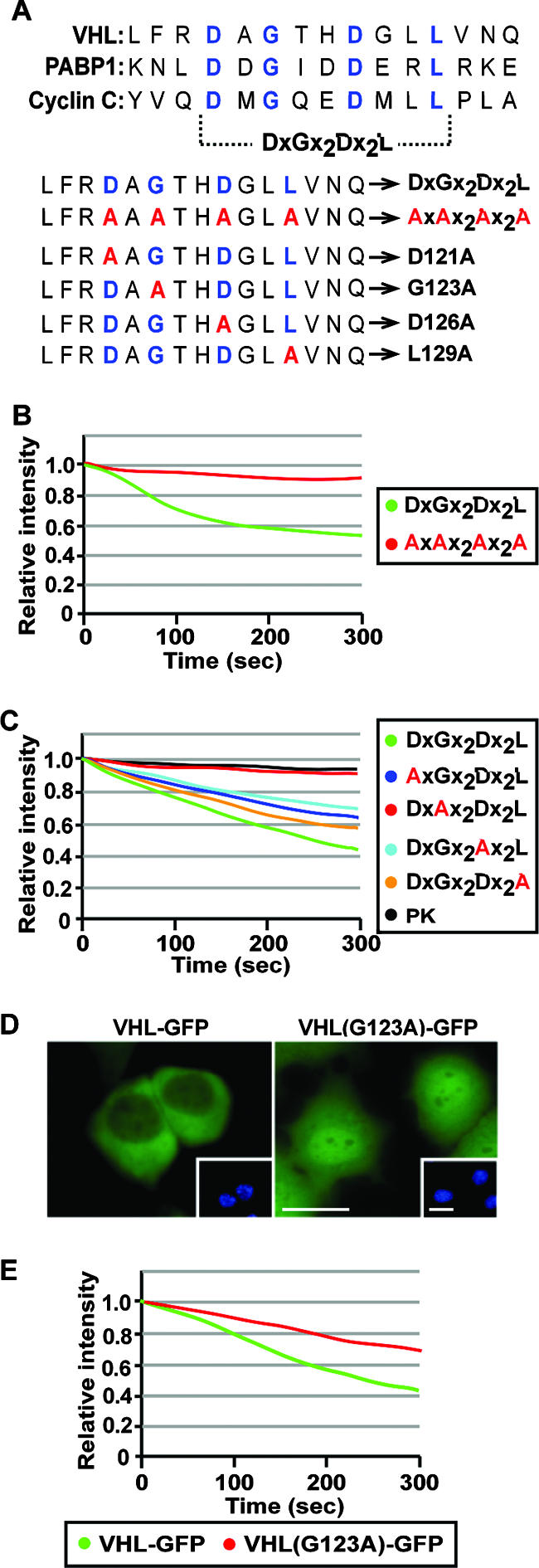
Identification of key residues that mediate transcription-dependent nuclear export. (A) Sequence alignment depicting conserved residues (blue) in the transcription-dependent nuclear export motifs of VHL, PABP1, and cyclin C. Residues in red indicate alanine substitutions at key residues in the TD-NEM sequence of VHL. (B) Conserved residues in the DXGX2DX2L consensus sequence are essential for nuclear export. MCF-7 cells were transiently transfected with VHL(115-130)-PK-GFP-NLS (DxGx2Dx2L) or VHL(115-130AAAA)-PK-GFP-NLS (AxAx2Ax2A), where the four key residues were replaced with alanines. Cytoplasmic FLIP was performed, and the loss of nuclear fluorescence was plotted on a graph. (C) Single alanine substitutions of key residues within the DXGX2DX2L consensus sequence have a differential effect on nuclear export activity. MCF-7 cells transiently expressing the indicated PK-GFP-NLS-tagged constructs were subjected to cytoplasmic FLIP. The graph represents the loss of nuclear fluorescence. (D and E) A single amino acid substitution of G123A in full-length VHL affects steady-state localization and nuclear export activity. Steady-state localization of VHL(G123A)-GFP expressed in MCF-7 cells compared to wild-type VHL-GFP is shown (D). Insets are the corresponding Hoechst staining of the cells. Cells expressing VHL-GFP or VHL(G123A)-GFP were subjected to cytoplasmic FLIP, and the loss of nuclear fluorescence was monitored and graphed (E).
Cancer-causing mutations within TD-NEM abrogate nuclear export of VHL and oxygen-dependent degradation of HIFα.
To study the functional consequence of TD-NEM mutations in the context of a full-length VHL protein, we searched for naturally occurring point mutations within the key TD-NEM residues that are associated with VHL disease. Replacement of the first aspartic acid residue of the consensus, DXGX2DX2L, with glycine (D121G) is a germ line mutation associated with type 2B VHL disease, characterized by a high risk of RCC (Fig. 7A) (15, 52, 61). This mutant is of particular interest, since previous studies have reported that it maintains its ability to form an E3 ubiquitin ligase complex and to bind and ubiquitylate HIFα in vitro (19). In addition, the second aspartic acid residue of the VHL TD-NEM has been reported to be replaced with tyrosine (D126Y) in individuals afflicted with polycythemia (Fig. 7A) (48, 49). Stable cell lines of D121G-GFP, D126Y-GFP, and G123A-GFP (not PK-GFP-NLS) were generated in VHL-defective 786-0 RCC to further study the effect of these point mutants on the ability of VHL to be exported from the nucleus and mediate oxygen-dependent degradation of HIFα. As previously reported, immunoprecipitation analysis revealed that the VHL mutant D121G is able to bind as efficiently to HIF2α as wild-type VHL (Fig. 7B). Likewise, VHL D126Y was also able to efficiently bind HIF2α (Fig. 7B). Cytoplasmic FLIP experiments revealed a markedly decreased rate of nuclear export of D121G and D126Y, of approximately 40% and 20%, respectively, compared to wild-type VHL (Fig. 7C). The function of VHL in oxygen-dependent degradation of HIFα has been linked to its ability to export from the nucleus (17). Thus, we decided to examine the effect of nuclear export-defective VHL mutants on oxygen-dependent degradation of HIFα. Cells were exposed to hypoxia to promote HIFα accumulation, followed by reoxygenation (Fig. 7D). VHL-defective RCC 786-0 cells did not display a decrease in HIF2α levels (786-0 cells express HIF2α but not HIF1α) following reoxygenation of hypoxic cells, as expected, since these cells do not express wild-type VHL (Fig. 7D). Reintroduction of wild-type VHL caused rapid degradation of HIF2α upon reoxygenation of hypoxic cells (Fig. 7D). In contrast, both the D121G and D126Y mutants were not as efficient in degrading HIF2α in reoxygenated cells (Fig. 7D). Mutants D121G and D126Y were eventually capable of degrading HIF2α after long periods of reoxygenation, consistent with their ability to assemble with HIFα and mediate ubiquitylation (Fig. 7E). These data support the hypothesis that VHL mutants that are defective in nuclear export display a reduced ability to degrade HIFα even though they are able to bind to and ubiquitylate HIFα.
FIG. 7.
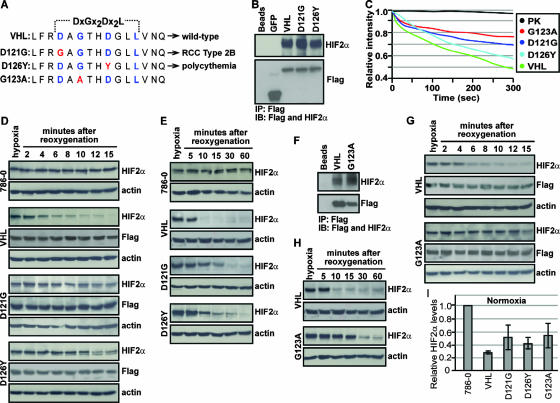
Cancer-causing mutations within TD-NEM of VHL abrogate nuclear export and oxygen-dependent degradation of HIF2α. (A) Sequence alignment depicting cancer-causing mutations (red) in the key TD-NEM residues of VHL that lead to RCC type 2B (D121G) and polycythemia (D126Y) in humans. G123A, which exhibits a defect in nuclear export as shown in Fig. 6, was also used to test its ability to mediate HIF degradation. (B) D121G and D126Y retain the ability to bind to HIF. Cells stably expressing Flag-tagged VHL-GFP, D121G-GFP, D126Y-GFP, and GFP were placed under hypoxic conditions and treated with 10 μM MG132 for 2 h before being harvested. Cell lysates were immunoprecipitated with anti-Flag beads and immunoblotted with anti-Flag and anti-HIF2α antibodies. (C) Cancer-causing mutants D121G and D126Y in TD-NEM decrease the nuclear export activity of VHL. Cells stably expressing VHL-GFP, D121G-GFP, D126Y-GFP, or G123A-GFP or transiently expressing PK-GFP-NLS were submitted to cytoplasmic FLIP as described previously. The loss of nuclear fluorescence was monitored and plotted on a graph. (D and E) Cells expressing D121G or D126Y exhibit a deficiency in HIF degradation. Stable cell lines of VHL-GFP, D121G-GFP, D126Y-GFP, and the VHL-defective cell line 786-0 were incubated for 20 h under hypoxic conditions before being reoxygenated by placing them in a normoxic environment, for the indicated time. Cells were lysed with 4% SDS and submitted to Western blot analysis using an anti-HIF2α antibody. Levels of VHL or its mutant counterparts were monitored using anti-Flag antibody, and actin was used to ensure equal loading of lysates. (F) G123A retains the ability to bind to HIF. Cells stably expressing the VHL point mutant, G123A, were treated the same as described for panel B. (G and H) Cells expressing G123A exhibit a deficiency in HIF degradation. G123A stably expressing cells were treated in the same manner as described for panels D and E. (I) D121G, D126Y, and G123A stable cells express higher normoxic HIF levels compared to wild-type VHL. Stable cell lines incubated under normoxic conditions were lysed with 4% SDS and submitted to Western blot analysis using anti-HIF2α and antiactin antibodies. HIF2α levels were normalized to actin and values, calculated relative to HIF2α levels in 786-0 cells, were plotted on a graph.
In light of these results, we were interested in testing if the nuclear export-defective G123A, although not a known disease-causing mutation, would also exhibit a decreased efficiency in degrading HIFα (Fig. 7A). Stably expressed G123A maintained its ability to interact with HIF2α (Fig. 7F) but displayed a reduced nuclear export activity (Fig. 7C). The G123A mutant displayed a striking defect in degradation of HIF2α upon reoxygenation of hypoxic cells (Fig. 7G and H). We noticed that 786-0 cells expressing the nuclear export-defective mutants D121G, D126Y, and G123A generally displayed higher levels of HIF2α than wild-type VHL, but levels were still lower than with the VHL-defective 786-0 cells (Fig. 7I). This may be explained by the fact that the nuclear export-defective mutants are still able to bind and ubiquitylate HIFα and partially able to be exported from the nucleus. Nonetheless, the data shown in Fig. 7 suggest that nuclear export of VHL is required for efficient oxygen-dependent degradation of HIFα.
TD-NEM mediates efficient transcription-dependent nuclear export of proteins.
Efficient nuclear export of proteins is required for cellular homeostasis and survival. The classical NES/CRM1 pathway was the first general and discreet motif identified which mediates nuclear export of a wide array of different proteins (9, 60). The herein-described TD-NEM is a discreet motif that is sensitive to drugs that inhibit RNA Pol II activity but operates independently of the classical NES pathway, since it is insensitive to LMB, as demonstrated in our live cell nuclear export assay (Fig. 8A and B). This is in contrast to what is observed with the classical NES, whose activity is inhibited by LMB but not by RNA Pol II inhibitors (Fig. 8C and D). VHL and PABP1 TD-NEM display nuclear export activities nearly as efficient as the Rev NES, which is thought to be the strongest NES yet characterized (Fig. 8E) (21). Based on these results, we suggest that TD-NEM is an efficient nuclear export motif.
FIG. 8.
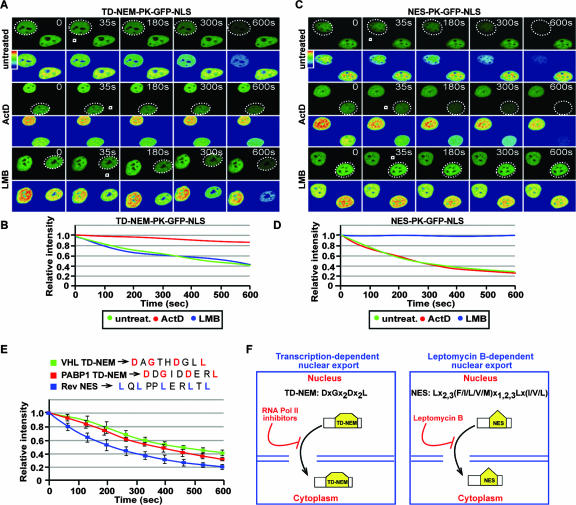
TD-NEM is a novel and efficient transcription-dependent nuclear export motif. (A to D) TD-NEM, contrary to the classical NES, mediates nuclear export in an ActD-sensitive but LMB-insensitive manner. Cells transiently expressing PK-GFP-NLS-tagged TD-NEM of VHL or NES of the Rev protein were either untreated or treated with ActD or LMB as previously described before being submitted to cytoplasmic FLIP. White squares indicate the bleached area, and the dotted circles outline the nucleus of the cell of interest. The loss of nuclear fluorescence was monitored, calculated, and plotted on a graph (B and D). (E) TD-NEM is an efficient nuclear export motif. Cells transiently expressing PK-GFP-NLS-tagged TD-NEM of VHL or PABP1, or the Rev NES, one of the strongest export signals, were submitted to cytoplasmic FLIP in order to compare the efficiency of nuclear export. (F) Model of nuclear export mediated by the TD-NEM consensus sequence compared to the classical NES (3). Nuclear export of TD-NEM is abrogated by RNA Pol II inhibitors, such as ActD and DRB, whereas NES activity is abrogated by LMB.
DISCUSSION
We report the identification of a novel and discreet motif, DXGX2DX2L, referred to as TD-NEM, which mediates efficient transcription-dependent nuclear export of VHL. This motif was also common among other proteins, including PABP1, where it mediated its respective transcription-dependent export from the nucleus. Nuclear export by TD-NEM requires ongoing RNA Pol II-mediated transcription and operates independently of the classical CRM1/NES-mediated export pathway (Fig. 8F). Key residues within TD-NEM of VHL are targeted by naturally occurring point mutations associated with renal carcinoma and polycythemia in humans. These disease-causing mutations, which alter the dynamic profile of VHL, restrain oxygen-dependent degradation of HIFα without affecting interaction between substrate and E3 ubiquitin ligase. Our results highlight the essential role of nuclear-cytoplasmic dynamics in protein function and provide evidence that mutations targeting subcellular trafficking can lead to disease.
It has become increasingly evident that degradation of nuclear proteins by the ubiquitylation pathway requires nuclear-cytoplasmic trafficking of the E3 ubiquitin-ligase as well as the substrate protein. Efficient degradation of nuclear proteins, such as p53, Smad3, and HIFα, is tightly linked to the ability of the E3 ligase to engage in nuclear export (13, 17, 34, 37). Thus, VHL, the ubiquitylation component of an E3 ubiquitin ligase complex that mediates oxygen-dependent degradation of HIFα, was an ideal subject for studying the functional role of nuclear-cytoplasmic trafficking of E3 ligases. We have previously shown that nuclear-cytoplasmic trafficking of VHL requires ongoing RNA Pol II-mediated transcription (17, 34). The initial goal of this study was to identify the sequence that mediates transcription-dependent nuclear export of VHL. We stumbled upon a sequence that may potentially mediate nuclear export of several proteins, including the mRNA nuclear export factor PABP1. VHL and PABP1 share the distinct ability to engage in constitutive and highly dynamic nuclear-cytoplasmic trafficking, utilizing a pathway that requires ongoing RNA Pol II activity. We have identified a new functional domain, TD-NEM, which is present in both VHL and PABP1 and mediates their transcription-dependent export from the nucleus. The rate of nuclear export of TD-NEM is approximately 70 to 80% of that observed for the Rev NES, perhaps the strongest LMB-sensitive NES so far identified, suggesting that this motif is highly efficient in mediating nuclear egression of molecules. Several lines of evidence support a role for TD-NEM as a nuclear export motif. First, removal of TD-NEM from full-length VHL and PABP1, or a single amino acid substitution of glycine to alanine, altered the steady-state distribution from mostly cytoplasmic to nuclear, accompanied by a reduction in sensitivity to ActD or DRB treatment and the rate of nuclear export. Also, TD-NEM alone from VHL and PABP1 was sufficient to confer transcription-dependent nuclear export properties to a large reporter protein (i.e., PK-GFP) in multiple living cells and in vitro assays. Based on these data, we suggest that TD-NEM represents a new, and possibly ubiquitous, nuclear export motif that operates independently of the known CRM1-NES pathway.
In this study we have shown that transcription-dependent nuclear export of VHL and PABP1 is mediated by a simple and linear sequence, DXGX2DX2L. Each of the four conserved residues of TD-NEM was found to be required for full activity of the export signal. It is not surprising that a mere four residues can act as a transport signal, considering that the classical NES, among others, relies on a consensus sequence of only a few conserved residues (9, 60). It would be of interest to examine the flexibility of this sequence by testing if naturally occurring conservative permutations of its key residues, such as aspartic acid to glutamic acid or leucine to isoleucine, retain activity. There were no other apparent conserved residues between the core TD-NEM of VHL, PABP1, and the putative TD-NEM of cyclin C; however, these three independent TD-NEMs displayed slightly different nuclear export activities, suggesting a functional role of the nonconserved residues in modulating the activity of TD-NEM. These observations are similar to the classical NES, which can display different export activities dependent on the surrounding amino acid context (32). Whether other substitutions or subtle differences of this TD-NEM, such as different spacing between the key residues, retain activity also remains to be tested. If functional combinations are found, this would simply provide additional evidence to support a ubiquitous nature of TD-NEM.
Discovery of LMB as an inhibitor of NES-mediated nuclear export has aided in both deciphering the components of this pathway and identifying many proteins that are exported from the nucleus (10, 14). Similarly, our use of transcriptional inhibitors, such as ActD and DRB, has led to the discovery of a nuclear export pathway that relies on the TD-NEM export signal. We envision that the discovery of TD-NEM through inhibitors of RNA Pol II-dependent transcription will help uncover another class of proteins that undergo nuclear export and the mechanism by which this occurs. It is still unclear as to how these drugs inhibit nuclear export of TD-NEM-containing proteins, but it is most likely through direct or indirect effects on either primary or secondary components of this pathway. It is well known that LMB directly interacts with and inhibits the action of CRM1, the exporter of NES-containing proteins (31). It will be interesting to see if ActD and DRB have a direct effect on a possible TD-NEM exporter or if they cause a secondary effect through transcriptional inhibition.
We have previously shown that perturbing the nuclear-cytoplasmic trafficking profile of VHL is detrimental to its ability to mediate oxygen-dependent degradation of HIFα. Here we report that naturally occurring TD-NEM mutations D121G and D126Y, which lead to RCC and polycythemia, respectively, abrogate nuclear export of VHL. It has been puzzling as to how VHL cancer-causing mutations, such as D121G, retain the ability to bind and ubiquitylate HIFα in vitro yet are able to develop classical tumors associated with VHL disease (19). We showed that D121G and D126Y maintain the ability to bind to HIF2α, consistent with previously published data. Expression of these mutants leads to an extended HIF2α stability following reoxygenation of hypoxic cells, providing a correlation between the efficiency to mediate degradation of HIFα and nuclear export activity. We cannot exclude the possibility that other aspects of the ubiquitylation pathway may be affected in vivo; however, we did predict a defect in HIF2α degradation with G123A, a mutant that is restrained in its ability to export from the nucleus but retains its ability to bind to HIF2α. The G123A mutant further supports that the TD-NEM of the β-domain of VHL is not involved in HIFα binding or E3 ubiquitin ligase complex formation but plays an essential role in HIFα degradation by mediating nuclear export. We achieved low-expressing stable cell lines of the nuclear export-defective mutants, though expression was still higher than endogenous VHL. This raises the possibility that the nuclear export mutants may be partially rescued by overproduction and may have more pronounced defects in physiological settings. The differences in HIFα stability observed with the nuclear export-defective mutants may translate into different types of VHL disease, as observed with type 2A and type 2B mutants with respect to HIFα binding (36). Whether the prolonged presence and activity of HIFα provide an explanation for how patients with these mutations develop tumors remains to be tested. Nonetheless, our data illustrate that the dynamic profile of a protein is instrumental for functional integrity and provide evidence that mutations targeting subcellular trafficking can abrogate protein function.
In conclusion, we propose that TD-NEM is a novel nuclear export motif which utilizes a pathway that requires RNA Pol II-mediated transcription. There is emerging evidence that a large class of proteins is able to export by utilizing an NES-independent pathway. It would be interesting to test whether these proteins encode a TD-NEM and if they engage in transcription-dependent nuclear export. Future work aimed at elucidating the mechanism by which RNA Pol II inhibitors abolish the nuclear export activity of TD-NEM should yield interesting information as to the process involved in TD-NEM-mediated nuclear export and perhaps uncover another general nuclear export pathway.
Supplementary Material
Acknowledgments
This work is supported by a grant from the Canadian Institutes of Health Research to S.L. S.L. is the recipient of the National Cancer Institute of Canada Harold E. Johns Award. K.M. is a recipient of a Canadian Institutes of Health Research fellowship.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 27313015-13021. [DOI] [PubMed] [Google Scholar]
- 2.Blondel, M., J. M. Galan, Y. Chi, C. Lafourcade, C. Longaretti, R. J. Deshaies, and M. Peter. 2000. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 196085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 164207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonicalzi, M. E., I. Groulx, N. de Paulsen, and S. Lee. 2001. Role of exon 2-encoded beta-domain of the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 2761407-1416. [DOI] [PubMed] [Google Scholar]
- 5.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2941337-1340. [DOI] [PubMed] [Google Scholar]
- 6.Chen, F., T. Kishida, M. Yao, T. Hustad, D. Glavac, M. Dean, J. R. Gnarra, M. L. Orcutt, F. M. Duh, G. Glenn, et al. 1995. Germline mutations in the von Hippel-Lindau disease tumor suppressor gene: correlations with phenotype. Hum. Mutat. 566-75. [DOI] [PubMed] [Google Scholar]
- 7.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 27525733-25741. [DOI] [PubMed] [Google Scholar]
- 8.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 10743-54. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82475-483. [DOI] [PubMed] [Google Scholar]
- 10.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 11.Franovic, A., I. Robert, K. Smith, G. Kurban, A. Pause, L. Gunaratnam, and S. Lee. 2006. Multiple acquired renal carcinoma tumor capabilities abolished upon silencing of ADAM17. Cancer Res. 668083-8090. [DOI] [PubMed] [Google Scholar]
- 12.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 187288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuchi, M., T. Imamura, T. Chiba, T. Ebisawa, M. Kawabata, K. Tanaka, and K. Miyazono. 2001. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol. Biol. Cell 121431-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390308-311. [DOI] [PubMed] [Google Scholar]
- 15.Gallou, C., D. Chauveau, S. Richard, D. Joly, S. Giraud, S. Olschwang, N. Martin, C. Saquet, Y. Chretien, A. Mejean, J. M. Correas, G. Benoit, P. Colombeau, J. P. Grunfeld, C. Junien, and C. Beroud. 2004. Genotype-phenotype correlation in von Hippel-Lindau families with renal lesions. Hum. Mutat. 24215-224. [DOI] [PubMed] [Google Scholar]
- 16.Groulx, I., M. E. Bonicalzi, and S. Lee. 2000. Ran-mediated nuclear export of the von Hippel-Lindau tumor suppressor protein occurs independently of its assembly with cullin-2. J. Biol. Chem. 2758991-9000. [DOI] [PubMed] [Google Scholar]
- 17.Groulx, I., and S. Lee. 2002. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 225319-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunaratnam, L., M. Morley, A. Franovic, N. de Paulsen, K. Mekhail, D. A. Parolin, E. Nakamura, I. A. Lorimer, and S. Lee. 2003. Hypoxia inducible factor activates the transforming growth factor-alpha/epidermal growth factor receptor growth stimulatory pathway in VHL(-/-) renal cell carcinoma cells. J. Biol. Chem. 27844966-44974. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, W. J., M. Ohh, J. Moslehi, K. Kondo, W. G. Kaelin, and W. J. Welch. 2002. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol. Cell. Biol. 221947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, A. L. 2002. Hypoxia: a key regulatory factor in tumour growth. Nat. Rev. Cancer 238-47. [DOI] [PubMed] [Google Scholar]
- 21.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell. Res. 256213-224. [DOI] [PubMed] [Google Scholar]
- 22.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 23.Ivan, M., and W. G. Kaelin, Jr. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 1127-34. [DOI] [PubMed] [Google Scholar]
- 24.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292464-468. [DOI] [PubMed] [Google Scholar]
- 25.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 9612436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292468-472. [DOI] [PubMed] [Google Scholar]
- 27.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2673-682. [DOI] [PubMed] [Google Scholar]
- 28.Kaelin, W. G., Jr., and E. R. Maher. 1998. The VHL tumour-suppressor gene paradigm. Trends Genet. 14423-426. [DOI] [PubMed] [Google Scholar]
- 29.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39499-509. [DOI] [PubMed] [Google Scholar]
- 30.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science 2691444-1446. [DOI] [PubMed] [Google Scholar]
- 31.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 969112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutay, U., and S. Guttinger. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell. Biol. 15121-124. [DOI] [PubMed] [Google Scholar]
- 33.Latif, F., K. Tory, J. Gnarra, M. Yao, F. M. Duh, M. L. Orcutt, T. Stackhouse, I. Kuzmin, W. Modi, L. Geil, et al. 1993. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 2601317-1320. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S., M. Neumann, R. Stearman, R. Stauber, A. Pause, G. N. Pavlakis, and R. D. Klausner. 1999. Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 191486-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leroux, M. R., and F. U. Hartl. 2000. Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr. Biol. 10R260-R264. [DOI] [PubMed] [Google Scholar]
- 36.Li, L., L. Zhang, X. Zhang, Q. Yan, Y. A. Minamishima, A. F. Olumi, M. Mao, S. Bartz, and W. G. Kaelin, Jr. 2007. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2a and type 2b Vhl mutations. Mol. Cell. Biol. 275381-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindstrom, M. S., A. Jin, C. Deisenroth, G. White Wolf, and Y. Zhang. 2007. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell. Biol. 271056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lisztwan, J., G. Imbert, C. Wirbelauer, M. Gstaiger, and W. Krek. 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 131822-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lonergan, K. M., O. Iliopoulos, M. Ohh, T. Kamura, R. C. Conaway, J. W. Conaway, and W. G. Kaelin, Jr. 1998. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 18732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maher, E. R., and W. G. Kaelin, Jr. 1997. von Hippel-Lindau disease. Medicine (Baltimore) 76381-391. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399271-275. [DOI] [PubMed] [Google Scholar]
- 42.Mekhail, K., L. Gunaratnam, M. E. Bonicalzi, and S. Lee. 2004. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 6642-647. [DOI] [PubMed] [Google Scholar]
- 43.Mekhail, K., M. Khacho, A. Carrigan, R. R. Hache, L. Gunaratnam, and S. Lee. 2005. Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 170733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mekhail, K., M. Khacho, L. Gunaratnam, and S. Lee. 2004. Oxygen sensing by H+: implications for HIF and hypoxic cell memory. Cell Cycle 31027-1029. [PubMed] [Google Scholar]
- 45.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 691237-1245. [DOI] [PubMed] [Google Scholar]
- 46.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T.-Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the [bgr]-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2423-427. [DOI] [PubMed] [Google Scholar]
- 47.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362857-860. [DOI] [PubMed] [Google Scholar]
- 48.Pastore, Y., K. Jedlickova, Y. Guan, E. Liu, J. Fahner, H. Hasle, J. F. Prchal, and J. T. Prchal. 2003. Mutations of von Hippel-Lindau tumor-suppressor gene and congenital polycythemia. Am. J. Hum. Genet. 73412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pastore, Y. D., J. Jelinek, S. Ang, Y. Guan, E. Liu, K. Jedlickova, L. Krishnamurti, and J. T. Prchal. 2003. Mutations in the VHL gene in sporadic apparently congenital polycythemia. Blood 1011591-1595. [DOI] [PubMed] [Google Scholar]
- 50.Pause, A., S. Lee, R. A. Worrell, D. Y. Chen, W. H. Burgess, W. M. Linehan, and R. D. Klausner. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 942156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404604-609. [DOI] [PubMed] [Google Scholar]
- 52.Rasmussen, A., S. Nava-Salazar, P. Yescas, E. Alonso, R. Revuelta, I. Ortiz, S. Canizales-Quinteros, M. T. Tusie-Luna, and M. Lopez-Lopez. 2006. Von Hippel-Lindau disease germline mutations in Mexican patients with cerebellar hemangioblastoma. J. Neurosurg. 104389-394. [DOI] [PubMed] [Google Scholar]
- 53.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheffner, M. 1999. Moving protein heads for breakdown. Nature 398103-104. [DOI] [PubMed] [Google Scholar]
- 55.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3721-732. [DOI] [PubMed] [Google Scholar]
- 56.Smith, K., L. Gunaratnam, M. Morley, A. Franovic, K. Mekhail, and S. Lee. 2005. Silencing of epidermal growth factor receptor suppresses hypoxia-inducible factor-2-driven VHL-/- renal cancer. Cancer Res. 655221-5230. [DOI] [PubMed] [Google Scholar]
- 57.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 901041-1050. [DOI] [PubMed] [Google Scholar]
- 58.Tomoda, K., Y. Kubota, and J. Kato. 1999. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398160-165. [DOI] [PubMed] [Google Scholar]
- 59.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2169-178. [DOI] [PubMed] [Google Scholar]
- 60.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82463-473. [DOI] [PubMed] [Google Scholar]
- 61.Whaley, J. M., J. Naglich, L. Gelbert, Y. E. Hsia, J. M. Lamiell, J. S. Green, D. Collins, H. P. Neumann, J. Laidlaw, F. P. Li, et al. 1994. Germ-line mutations in the von Hippel-Lindau tumor-suppressor gene are similar to somatic von Hippel-Lindau aberrations in sporadic renal cell carcinoma. Am. J. Hum. Genet. 551092-1102. [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 989630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.