Abstract
Focal adhesion kinase (FAK) is an essential kinase that regulates developmental processes and functions in the pathology of human disease. An intramolecular autoinhibitory interaction between the FERM and catalytic domains is a major mechanism of regulation. Based upon structural studies, a fluorescence resonance energy transfer (FRET)-based FAK biosensor that discriminates between autoinhibited and active conformations of the kinase was developed. This biosensor was used to probe FAK conformational change in live cells and the mechanism of regulation. The biosensor demonstrates directly that FAK undergoes conformational change in vivo in response to activating stimuli. A conserved FERM domain basic patch is required for this conformational change and for interaction with a novel ligand for FAK, acidic phospholipids. Binding to phosphatidylinositol 4,5-bisphosphate (PIP2)-containing phospholipid vesicles activated and induced conformational change in FAK in vitro, and alteration of PIP2 levels in vivo changed the level of activation of the conformational biosensor. These findings provide direct evidence of conformational regulation of FAK in living cells and novel insight into the mechanism regulating FAK conformation.
Focal adhesion kinase (FAK) is an essential non-receptor tyrosine kinase, since FAK-null mice exhibit embryonic lethality (21). In endothelial cells, FAK is required for the proper development of the vasculature (5, 51), and in neurons FAK regulates netrin-mediated axon outgrowth and dendrite formation (41, 45). In addition to these roles in development, FAK is also implicated in the pathology of disease. FAK expression is required in cardiomyocytes to promote hypertrophy and fibrosis in response to cardiac stress and potentially plays a role in the development of heart disease (18, 42). Overexpression of FAK is observed in many types of cancer (22), and experiments using animal models have implicated FAK in tumor formation and metastasis in a number of neoplasms, including cancer of the brain, breast, and skin (38, 39, 58).
Despite the importance of FAK in controlling multiple developmental and pathological events, the molecular mechanism of regulation remains incompletely elucidated. Like many kinases posttranslation modification, particularly phosphorylation, is a major regulatory mechanism. Tyrosine 397 is the major site of autophosphorylation, and mutation of this site abrogates the biological activity of FAK (47). This site primarily serves a scaffolding function, providing a docking site for a number of proteins containing SH2 domains, including Src and phosphatidylinositol 3-kinase (47). The interaction with FAK occupies both the SH2 and SH3 domains of Src preventing intramolecular inhibitory interactions resulting in stabilization of Src in its active conformation (54). Complex formation also serves to direct Src to its substrates, which include FAK itself. Activated Src phosphorylates FAK on multiple sites, including two tyrosine residues in the activation loop, Y576 and Y577, which function in regulating catalytic activity (6).
A number of protein-protein interactions also regulate FAK activity. FIP200 is a negative regulator of kinase activity (57). The interaction of FIP200 with FAK is complex, with multiple FIP200 binding sites in FAK (1). This complex dissociates upon cell adhesion, which is a stimulus leading to FAK activation (1). The interaction of FAK with growth factor receptors positively regulates FAK signaling. Stimulation of fibroblasts promotes FAK binding to the epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) receptors, which is required for EGF- and PDGF-induced chemotaxis (52). Stimulation of cells with hepatocyte growth factor (HGF) promotes the direct association of FAK with the Met receptor, and this interaction is required for HGF induced activation of FAK (14). A potential ligand-binding site within the N-terminal domain of FAK, consisting of a series of basic residues, is required for maximal activation of FAK and subsequent downstream signaling following cell adhesion to fibronectin (20). The same site forms the binding site for tyrosine-phosphorylated Met receptor (14), and thus this sequence is required for activation of FAK in response to diverse stimuli.
Multiple studies support a model of regulation whereby the FAK FERM domain interacts with the FAK catalytic domain to impair catalytic activity in the inactive state. Truncation of the FERM domain increases FAK tyrosine phosphorylation and/or kinase activity (11, 27, 48, 49, 56). Further, the FERM domain can interact in trans with the catalytic domain in vivo and inhibit its activity (16). Mutation of K38 within the FERM domain disrupts the FERM/kinase domain interaction, and when introduced into the full-length molecule activates FAK (15). The molecular details of the FERM-catalytic domain interaction were revealed by X-ray crystallography (32). In this complex, the FERM domain blocks the active site of the kinase domain inhibiting access to the ATP and substrate binding sites. Further, the activation loop lies within the cleft between the FERM and kinase domains. These studies support a model of FAK activation entailing a switch from autoinhibited to active conformation, however, the challenging task of testing this model under physiological conditions in living cells has yet to be undertaken.
In this paper, we report the development of two biosensors that can measure two important facets of FAK activation in live cells: (i) phosphorylation of Y397, which reflects a FAK scaffolding function; and (ii) a conformational change associated with FAK activation. The novel conformational biosensor directly demonstrates that a conformation change in FAK occurs in living cells upon FAK activation. The biosensor was further used to probe the spatial regulation of FAK and mechanisms involved in controlling conformation changes. Acidic phospholipids are identified as novel ligands for FAK. Phosphatidylinositol 4,5-bisphosphate (PIP2)-containing vesicles alter FAK conformation in vitro and modulating PIP2 levels alters FAK conformation in vivo. These findings support the model that FAK is regulated via interaction with lipid ligands and provide mechanistic insight into the observation that acidic phospholipids are required for FAK activation (7, 28, 34, 36).
MATERIALS AND METHODS
Cell culture.
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) F-12 medium containing 10% fetal bovine serum (FBS) and HeLa cells in DMEM containing 10% FBS. Cells were transfected using Lipofectamine Plus (Invitrogen) according to manufacturer's instructions. HeLa cells were starved in DMEM overnight prior to stimulation with ligands. To measure adhesion-dependent activation cells were maintained adhered to substrate or incubated in suspension at 37°C for 1 h prior to lysis.
Molecular biology.
To generate CFAK, the full-length FAK cDNA was inserted into the enhanced cyan fluorescent protein (CFP) construct ECFP-C1, in frame with CFP. Citrine was generated from the enhanced yellow fluorescent protein (YFP) construct EYFP-C1 vector by mutation of glutamine 69 to methionine (25). To generate citrine-dSH2, two Src SH2 domains were sequentially introduced into the citrine construct. This was achieved by PCR amplification of the c-Src SH2 domain (positions 553 to 894) and insertion of two copies of the SH2 coding sequence between the XhoI and PstI sites within citrine vector. To generate FAK conformation probes, six-nucleotide insertions encoding a NotI site were introduced into different sites of CFAK. Citrine was amplified with primers containing NotI sites, and the PCR product was introduced into the NotI site within CFAK. All mutations were generated by the QuickChange mutagenesis strategy. The full-length constructs were analyzed by sequencing to verify the intended mutations and ensure that no unintended mutations were present. The expression construct for PIPKIα was the generous gift of Richard Anderson (University of Wisconson) and a YFP-SopB construct was a generous gift of Jorge Galán (Yale University). For this study, the SopB encoding sequences were amplified by PCR and subcloned into pcDNA3.1.
Fluorometric measurements of FRET.
For live-cell fluorometric measurements of fluorescence resonance energy transfer (FRET), 293 cells expressing different constructs were typsinized and suspended in phosphate-buffered saline (PBS). The cells were analyzed using a Fluorolog SPEX 168 fluorometer or a spectrofluorophotometer RF-1501 (Shimadzu). The cell suspension was excited at 425 nm, and an emission scan was acquired from 450 to 550 nm. The spectra of different samples in a single experiment were normalized to CFP emission of a reference spectrum.
Measuring FRET by microscopy.
HeLa cells expressing biosensors were plated on fibronectin (5 μg/ml [Sigma])-coated coverslips (Lucas Highland). After incubation at 37°C for 1 h, the cells were transferred to the microscope's heated chamber (Warner Instrument Corporation TC-344B) in Ham's F-12 K medium without phenol red (Biosource), 25 mM HEPES (pH 7.4), and 5% FBS. Images were collected using a Zeiss 100× 1.3-numerical aperture lens, a Zeiss Axiovert 100TV microscope, a Cool Snap ES digital camera, and Metamorph software (Universal Imaging). The filters and dichroic mirror setting for FRET images were as follows: (i) CFP, D436/20 and D470/40; (ii) FRET, D436/20 and HQ535/30; and (iii) YFP, HQ500/20 and HQ535/30 (44). The CFP, FRET, and YFP images were recorded using 2 × 2 binning. For ligand stimulation, transfected HeLa cells were serum starved overnight, transferred to the microscope's heated chamber, and imaged, before and after ligand stimulation, in Hank's balanced salt solution buffer with 20 mM HEPES (pH 7.4) and 2 g/liter glucose. At each time point, images were collected using a Zeiss 40× 1.3-numerical aperture Plan NeoFluor lens and 4 × 4 binning.
For photobleaching assays, HeLa cells expressing the FAK biosensor were illuminated to excite CFP and imaged in both the CFP and FRET channels at 11-s intervals. The YFP acceptor was photobleached by pulse illumination for 6 s at each 11-s interval. The mean intensity from the whole cell was measured at each time point and normalized to the zero time point at the initiation of photobleaching. For laser-scanning confocal microscopy, 293 cells expressing the donor (CFAK), acceptor (citrine-dSH2), or both were analyzed. Images were recorded using a LSM 510 laser-scanning confocal microscope (Zeiss) in lambda stack mode. For photobleaching, the cells were photobleached at 488 nm. The emission spectra following stimulation at 458 nm were recorded before and after photobleaching.
Image processing and analysis.
Images were processed as previously described (10, 13, 29, 44). CFP, FRET, and citrine images were background subtracted and registered. For the FAK conformation biosensor, the background subtracted CFP image was divided by the background subtracted FRET image to get a pixel-to-pixel CFP/FRET ratio image. The CFP images were thresholded to generate a binary mask with a value of 0 outside the cell and a value of 1 inside the cell. The CFP/FRET ratio images were multiplied with the mask and displayed in pseudocolors scaled from the lowest to the highest signal within the cell, eliminating pixels outside the 5 to 95% range on the intensity histogram, to provide a more reasonable estimate of the biosensor's useful dynamic range. For the FAK autophosphorylation biosensor, the images were processed with the equation FRETC = FRET − (a·CFP) − (b·citrine) (59), where FRETC is corrected FRET, a is the percentage of CFP bleed through (determined from cells expressing only CFP-FAK), and b is the percentage of citrine bleed through (determined from cells expressing only expressing citrine-dSH2).
To quantify the focal adhesion and cytoplasm FRET values, two segmented images were generated using a threshold function. Two segmented images were used to generate binary masks for focal adhesions and the cytoplasm. The FRET ratio images were multiplied with the two masks to generate the ratio images for focal adhesions and cytoplasm. To determine the relationship between focal adhesion FRET value and distance to the cell margin, the focal adhesion binary mask was used to generate an individual segmented region around each focal adhesion. The segmented regions for focal adhesions were loaded to the focal adhesion CFP/FRET ratio image. The individual focal adhesion parameters, including average intensity and x and y values of each focal adhesion, were exported to Excel to generate a focal adhesion database. The cell margin was plotted based on the registered CFP image and transformed to a set of x and y values using Image J. The set of margin x and y values were imported into Excel. The minimal margin distance was measured by determining the minimal value of , where x1 and y1 are the coordinates of each focal adhesion and x2 and y2 are the coordinates of each point at the margin of the cell.
, where x1 and y1 are the coordinates of each focal adhesion and x2 and y2 are the coordinates of each point at the margin of the cell.
Lipid binding assays.
Lipid vesicles were prepared by mixing chloroform-dissolved phospholipids (Avanti Polar Lipids) in the appropriate ratios keeping the PE/PC mass ratio as 3:1, while other lipids were supplemented to the final concentration indicated. The mixture was dried using a speed vacuum for 15 min. The dried lipid mixture was suspended and sonicated in lipid binding buffer (40 mM HEPES [pH 7.5], 2 mM dithiothreitol,150 mM NaCl) to a final concentration of 2.5 μg/μl. For cosedimentation, 4 μg of glutathione S-transferase (GST) fusion protein was incubated with 250 μg of lipid vesicles on ice for 1 h. The mixtures were subsequently centrifuged at 110,000 × g for 1 h at 4°C. The supernatants were collected. The pellets were suspended in 100 μl sodium dodecyl sulfate (SDS) buffer and boiled for 2 min to dissolve. Both the supernatants and pellets were mixed with Laemmli sample buffer and boiled for 5 min. The samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining. For fluorescence polarization, the purified FERM domain (9) was incubated with BODIPY (dipyrromethene boron difluoride)-labeled phosphoinosotides with C6-acyl chains (8). Increasing concentrations of purified protein was added to 12.5 nM fluorescent phosphoinositide in buffer containing 20 mM HEPES [pH 7.0], 150 mM NaCl, and 5 mM β-mercaptoethanol. Anisotropy measurements were taken at 21°C using a Beacon 2000 fluorescence polarization instrument. Binding curves and dissociation constants were determined using the program Prism (GraphPad Software Inc.).
Protein purification and kinase assay.
The GST-fusion proteins were induced and purified as previously described (20, 37). For immune complex kinase assays, FAK and its variants were immunoprecipitated using the BC4 polyclonal antiserum or green fluorescent protein (GFP) antibody and the immune complexes incubated with 2 μg of GST-paxillin-N-C3 using previously described kinase reaction conditions (37). The reaction was terminated by the addition of sample buffer and phosphorylation of paxillin examined by Western blotting.
Expression of the recombinant fragment of FAK containing the FERM and catalytic domains has been described previously (32). For in vitro kinase assays using recombinant proteins, 200 ng of the recombinant FAK fragment and 1 μg Src (SH3 plus SH2 plus kinase) were incubated with liposomes in kinase reaction buffer (100 mM Tris [pH 7.5], 100 mM MgCl2, 50 μM ATP) for 30 min. The reaction was terminated by the addition of sample buffer, and phosphorylation of recombinant FAK was examined by Western blotting.
Protein analysis.
Cells were lysed in ice-cold modified radioimmunoprecipitation assay buffer. Lysates were clarified, and protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, IL). Immunoprecipitations were performed using a polyclonal FAK antibody (BC4) or a GFP monoclonal antibody (Roche). For Western blotting, the FAK antibody (447) and phosphotyrosine antibody (4G10) were from Millipore, the PY576 antibody was from Biosource, and the paxillin polyclonal antibody was previously described (53).
RESULTS
Development of a FAK autophosphorylation biosensor.
Tyrosine 397 is the major FAK autophosphorylation site, which is critical for FAK function and serves as a scaffolding site to recruit other proteins into complex. To visualize FAK autophosphorylation status in living cells, a genetically encoded FRET-based biosensor was developed. As a similar biosensor has been described (4), it will only be briefly introduced. CFP was fused to the FAK N terminus to serve as the FRET donor and citrine, a variant of YFP, was fused to two Src SH2 domains to serve as the FRET acceptor (Fig. 1A). Since phosphorylation of tyrosine 397 regulates Src SH2 domain binding to this site, CFAK should only interact with citrine-dSH2 and produce a FRET signal when the former is phosphorylated at tyrosine 397. The interaction between CFAK and citrine-dSH2 was confirmed by coimmunoprecipitation (X. Cai and M. D. Schaller, unpublished data). Citrine-dSH2, but not citrine alone, coimmunoprecipitated with CFAK, and this interaction was abolished when tyrosine 397 in CFAK was substituted for with phenylalanine. Upon coexpression in 293 cells, CFAK and citrine-dSH2 colocalized peripherally near the membrane (Cai and Schaller, unpublished). In contrast, CFAK-Y397F and citrine dSH2 were not colocalized. These data demonstrate that the interaction between CFAK and citrine-dSH2 depends upon phosphorylation of tyrosine 397. FRET was measured in several ways. Initially, cells were scraped from the tissue culture dish and FRET was measured in cells in suspension using a fluorometer. These measurements were made rapidly upon removing the cells from the dish (within 10 min), before exogenously expressed FAK could be inactivated and dephosphorylated. No FRET signal was detected in 293 cells expressing CFAK or citrine-dSH2 alone, whereas a strong FRET signal was detected in cells coexpressing both proteins (Fig. 1B). Notably, no FRET signal was detected in cells coexpressing CFAK-Y397F and citrine-dSH2. Note that similar results were obtained when adherent cells were lysed and the lysates, rather than suspended cells, were analyzed (Cai and Schaller, unpublished). Expression levels of the wild type and mutant biosensor were comparable (Cai and Schaller, unpublished data). FRET was also measured microscopically. As a further control, acceptor photobleaching was used to validate the FRET signal in vivo. HEK293 cells coexpressing CFAK and citrine-dSH2 were photobleached using an argon laser (488 nm). A series of fluorescent images were collected by laser-scanning confocal microscopy, and the average pixel intensity of the photobleached area at the CFP and FRET emission wavelengths was determined. As shown in Fig. 1C, photobleaching the FRET acceptor resulted in a decrease in the acceptor signal and an increase in the CFP donor signal, which was caused by the interruption of energy transfer from CFAK to citrine-dSH2.
FIG. 1.
(A) Design of the FAK autophosphorylation biosensor. When tyrosine 397 is unphosphorylated, CFAK does not interact with citrine-dSH2 and FRET does not occur. When tyrosine 397 is phosphorylated, CFAK interacts with citrine-dSH2 and FRET occurs. (C, CFP; Y, citrine). (B) The FAK wild-type autophosphorylation biosensor and the CFAK-Y397F control biosensor were expressed in HEK293 cells and analyzed by fluorometry. CFP was selectively excited at 425 nm, and the resulting emission spectra were normalized to the CFP emission peak. (C) FRET in the FAK biosensor was verified by acceptor photobleaching in living cells. HEK293 cells were photobleached using a 488-nm laser. The emission spectra produced by CFP excitation were recorded before and after photobleaching. Shown are the average results from three experiments ± standard error.
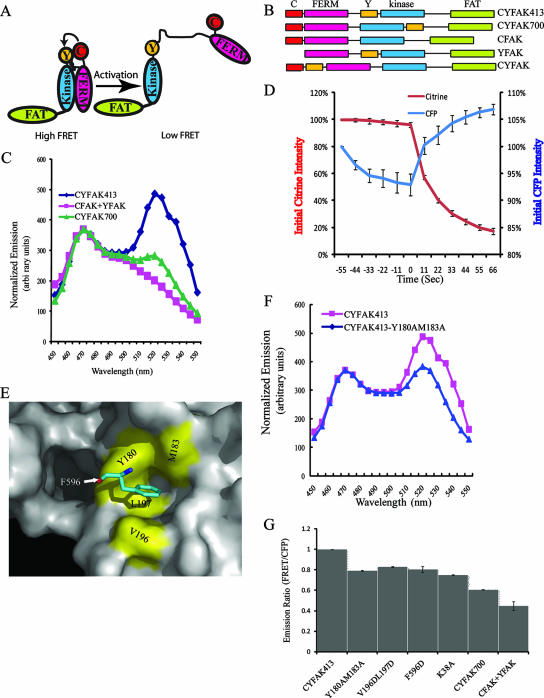
Development of a FAK conformational biosensor.
The structure of the FERM/catalytic domain complex confirms the role of the FERM domain in autoinhibition of catalytic activity and suggests that a transition from the autoinhibitory conformation to an open, active conformation is a required step for FAK activation. A FRET-based FAK probe was developed to monitor such a conformational change in living cells. As shown in Fig. 2A and B, CFP was fused in the N terminus of FAK and citrine was inserted into the linker between the FERM and catalytic domains to generate the CYFAK413 biosensor. As a control, citrine was inserted at the C terminus of the catalytic domain to generate the CYFAK700 construct. The emission spectrum of these constructs following stimulation with the CFP excitation wavelength was determined using a fluorometer. Again, measurements were made rapidly after taking the cells into suspension and similar results were found using lysates from adherent cells. A strong FRET peak was detected in cells expressing CYFAK413 (Fig. 2C). In contrast a very weak FRET signal was detected in cells expressing CFAK700. Coexpression of CFAK and YFAK, in which the CFP and citrine are presented in trans, did not yield a FRET signal in this assay. In addition to the fluorometer assay, CYFAK413 was characterized by microscopy and FRET was validated by acceptor photobleaching. Upon photobleaching, the citrine intensity decreased and the CFP signal increased, confirming that FRET was occurring (Fig. 2D). The 12% increase in CFP signal upon acceptor photobleaching is similar to that reported for the vinculin conformation biosensor (13).
FIG. 2.
(A) Design of the FAK conformation biosensor (CYFAK413). In the inactive conformation, the CFP and citrine are in proximity and FRET occurs. In the open conformation, the CFP and citrine are further apart and can rotate more freely, which will result in a reduced FRET signal. (B) Schematic structure of the FAK biosensor and control biosensors. CFP is fused to the N terminus. The numbers in the construct name refer to the citrine insertion site. (C) Normalized spectra of the FAK wild-type biosensor and control biosensors were measured by fluorometry. (D) FRET in the FAK conformation biosensor was verified by acceptor photobleaching in living cells. HeLa cells expressing the biosensor were imaged in both CFP and citrine channels following excitation of CFP at 11-s intervals. The acceptor (citrine) was photobleached by pulse illumination for 6 s at each 11-s interval, after the zero time point. The mean intensity from whole cells was measured at each time point and normalized to the zero time point. Shown are the average results from three experiments ± standard error. (E) Representation of the FERM/kinase domain interface was created using Pymol and illustrates the key interaction between F596 in the kinase domain and a hydrophobic pocket in the F2 subdomain of the FERM domain (17). (F) Normalized spectra of the FAK wild-type biosensor and a FAK biosensor with mutations designed to disrupt the FERM/kinase domain interaction are shown. (G) Normalized FRET/CFP emission ratios of FAK biosensors containing different mutations are shown. The constructs were analyzed as in panel C, and the FRET/CFP ratios of each were normalized to the FRET/CFP ratio of the wild-type biosensor. Shown are the average results from at least three experiments ± standard error. The results were analyzed by one-way analysis of variance (P < 0.0001) and Tukey's multiple comparison post test (CYFAK413 versus each construct; P < 0.001).
To test whether the FRET signal of CYFAK413 was regulated by conformational change, mutational studies were performed. The major contacts between the FERM and catalytic domains are centered around F596 of the catalytic domain, which binds in a hydrophobic pocket on the FERM domain (Fig. 2E). Mutation of Y180 and M183, which form part of the hydrophobic pocket of the FERM domain, results in activation of FAK in vitro and in vivo (32). These activating mutations were engineered into CY413FAK, and as shown in Fig. 2F, this mutant showed reduced FRET compared to the wild-type probe. In addition to this mutant, other activating mutations that disrupt the FERM/catalytic domain interaction were also introduced into the FAK conformational probe (Fig. 2G). Each of these mutants also exhibited a low FRET/CFP ratio compared with the wild-type probe. The expression of these mutant biosensors was comparable to that of the wild-type biosensor (Cai and Schaller, unpublished). These findings demonstrate that the FRET signal of this biosensor depends upon the interaction between the FERM and catalytic domains and that changes in FRET reflect changes in conformation.
Lysine 38 is a FERM domain residue that is required for the interaction between the FERM and kinase domains (15). Thus, it was surprising when the crystal structure of the autoinhibited FERM/catalytic domain complex revealed that K38 did not directly contact the catalytic domain. Instead, K38 appears to interact with acidic residues within the linker extending between the FERM and catalytic domains (32). To further explore the role of K38 in FAK regulation, the K38A mutation was introduced into the CYFAK413 biosensor. Interestingly, this mutant also showed a low FRET/CFP ratio similar to that seen in CYFAK413 variants with mutations that directly disrupt the FERM/catalytic domain interaction (Fig. 2G). This finding supports a role for K38 in stabilizing the inactive conformation of FAK, perhaps through interactions with the linker.
The biochemical characteristics of CYFAK413 were examined to determine if incorporation of the fluorescent probes altered catalytic activity and if CYFAK413 was regulated in a manner similar to wild-type FAK. The catalytic activity of CYFAK413 was tested using an in vitro, immune complex kinase assay. As shown in Fig. 3A, the kinase activity of CYFAK413 was modestly higher than that of wild-type FAK, presumably due to the insertion of citrine in the linker, since CYFAK413 kinase activity was also higher than CFAK, which has CFP fused to the N terminus of FAK. Notably, the activity of CYFAK413 was much lower than the activated variant CYFAK413-Y180A/M183A. To examine cell adhesion-dependent regulation, cells expressing CYFAK413 or CFAK were lysed and tyrosine phosphorylation was examined following immunoprecipitation and Western blotting. Both CFAK and CYFAK413 were phosphorylated on tyrosine in adherent cells and exhibited reduced phosphotyrosine when cells were held in suspension (Fig. 3B). Thus, tyrosine phosphorylation of CYFAK413 was regulated by cell adhesion, similar to wild-type FAK. In addition, the biochemical response of the biosensor to lysophosphatidic acid (LPA) stimulation was measured to further validate that this probe is regulated similar to wild-type FAK. Following LPA stimulation, the biosensor exhibited increased tyrosine phosphorylation (Fig. 3C). Despite a modest increase in its catalytic activity, the fact that the biosensor was regulated similarly to wild-type FAK in response to multiple stimuli suggests that CYFAK413 is a suitable biosensor for monitoring conformational changes in FAK in living cells.
FIG. 3.
Biochemical characterization of the FAK conformational biosensor. (A) HEK293 cells expressing empty vector (Mock) or the indicated FAK constructs were lysed and immunoprecipitated using a FAK antibody. The immune complexes were incubated in an in vitro kinase assay utilizing recombinant GST-paxillin-N-C3 as an exogenous substrate. Phosphorylation of paxillin was detected using the 4G10 phosphotyrosine (P-Tyr) antibody. Equal amounts of substrate were verified by blotting for paxillin using a polyclonal antiserum. FAK in the immune complexes was verified by blotting for FAK. (B) The wild-type and mutant biosensors were transiently expressed in HeLa cells, and adherent cells (Ad) or cells incubated in suspension (Sus) at 37°C for 1 h were lysed. The biosensors were immunoprecipitated (IP) using a GFP antibody and the immune complexes were analyzed by Western blotting for phosphotyrosine (P-Tyr) using 4G10. Equal amounts of FAK in the immune complexes were verified by blotting for FAK. (C) HeLa cells expressing the FAK biosensor were serum starved and stimulated with LPA (200 ng/ml) for 5 min. The biosensor was immunoprecipitated from cell lysates and blotted with phosphotyrosine or a FAK antibody.
Monitoring spatial regulation of FAK activity in living cells.
To monitor phosphorylation of tyrosine 397 in live cells, HeLa cells transiently coexpressing CFAK and citrine-dSH2 were plated onto fibronectin-coated coverslips and imaged by fluorescence microscopy. Cells were illuminated with a CFP excitation wavelength and the CFP and citrine (i.e., FRET) emissions were captured. The FRETc/CFP ratio image is shown in Fig. 4A. CFAK localized prominently to focal adhesions, although there was also cytoplasmic localization. The distribution of FRET indicated that phosphorylation of FAK at tyrosine 397 occurs at focal adhesions. Importantly, CFAK-397F exhibited a similar localization to CFAK, but there was no FRET signal produced in cells coexpressing CFAK-397F and citrine-dSH2. Expression levels of the wild-type and mutant biosensors were comparable (Cai and Schaller, unpublished data). The localization of FAK phosphorylated at tyrosine 397 to these sites was anticipated from studies using phospho-specific antibodies for immunofluorescence and results using a similar biosensor from Geiger's laboratory (4, 46). Strikingly, there appeared to be a heterogeneous distribution of phosphorylated FAK among the CFAK-positive focal adhesions (Fig. 4A). A similar heterogeneous pattern of autophosphorylated FAK was observed by staining with a PY397 phospho-specific antibody (Cai and Schaller, unpublished).
FIG. 4.
(A) HeLa cells expressing the FAK autophosphorylation biosensor or the 397F control were trypsinized and plated on fibronectin-coated coverslips. CFP, FRET, and citrine images were sequentially captured by fluorescence microscopy. CFP and FRETC/CFP ratio images of the cells are shown. The ratio images are pseudocolored so that hotter colors reflect an increase in FAK autophosphorylation. Shown are representative images (n > 5 cells). The boxed area in the “CFAK+Citrine-dSH2 FRETc/CFP” image is shown at higher magnification to the right. (B) HeLa cells expressing the FAK biosensor or the constitutively active mutant were trypsinized and plated on fibronectin-coated coverslips. CFP, FRET, and citrine images were sequentially captured by fluorescence microscopy. CFP and CFP/FRET ratio images of the cells are shown. Note that the ratio images are pseudocolored so that hotter colors reflect an increase in the open, active conformation. Representative images are shown (n > 15 cells).
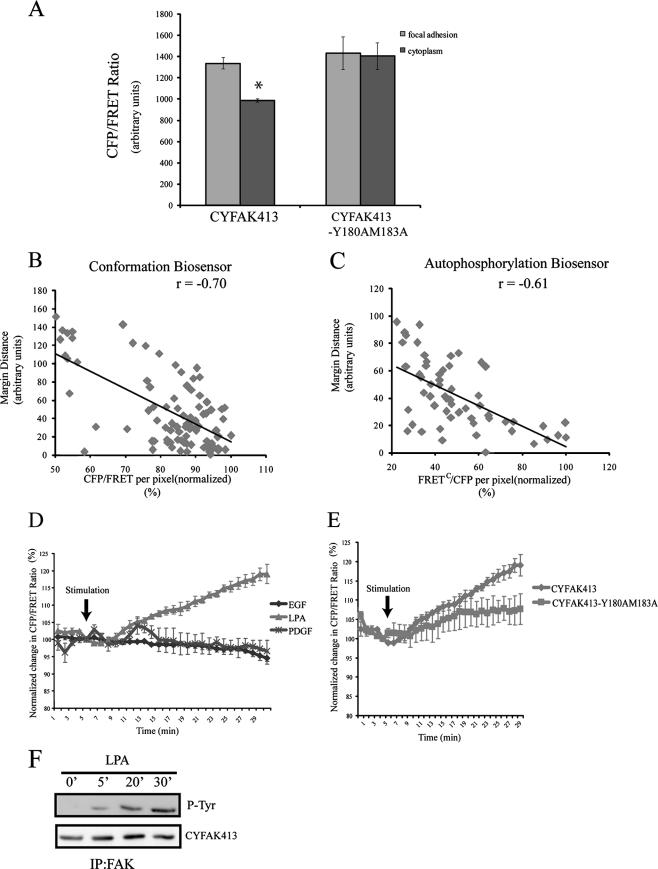
The current model of FAK regulation entails an autoinhibitory interaction between the FERM and catalytic domains, and thus activation requires a conformational change relieving this inhibition. The development of the FAK conformational biosensor allows a direct test of this model in living cells. HeLa cells transiently expressing CYFAK413 were plated onto fibronectin-coated coverslips and analyzed by fluorescence microscopy. Stimulation with the CFP excitation wavelength produced emission of both a CFP and a citrine signal. A representative CFP image and CFP/FRET ratio image are shown in Fig. 4B. (Note that the ratio image shows the open conformation as “hot” and the closed conformation as “cold.”) The CFP/FRET ratio was elevated in focal adhesions, compared with the ratio seen in the cytoplasm. As a control, the activated variant CYFAK413-Y180A/M183A was analyzed in parallel. The CFP/FRET ratio was high both in focal adhesions and in the cytoplasm in cells expressing this control. Expression levels of the wild-type and mutant biosensors were comparable (Cai and Schaller, unpublished data). This result demonstrates that the differences in CFP/FRET ratio in the CYFAK413-expressing cells were due to differences in conformation. The CFP/FRET ratio in focal adhesions and in the adjacent cytoplasm was quantified. In CYFAK413-Y180A/M183A-expressing cells, this ratio was the same in focal adhesions and in the cytoplasm (Fig. 5A). In CYFAK413-expressing cells, the CFP/FRET ratio in focal adhesions was comparable to that seen in CYFAK413-Y180A/M183A-expressing cells, but the cytoplasmic CFP/FRET ratio was significantly lower. These data demonstrate that FAK exists in different conformations in different regions of the cell. While it is not surprising that FAK is active in focal adhesions, the CYFAK413 biosensor allows visualization of conformation changes in live cells and thus provides strong support for the current model of FAK regulation.
FIG. 5.
(A) Average CFP/FRET ratio of the cytoplasm or focal adhesions in HeLa cells expressing the wild-type biosensor or the constitutively active variant (average of n = 4 cells). The focal adhesion and cytoplasmic values were analyzed using an unpaired t test (*, P < 0.0005). (B and C) The relationship between biosensor activity and focal adhesion location is shown. The distance of each focal adhesion from the cell margin is plotted versus the average CFP/FRET or FRETC/CFP ratio, where the highest ratio has been normalized to 100%. Representative data from single cells are shown (n = 4). r, correlation coefficient. (D and E) HeLa cells expressing the FAK biosensor were serum starved and simulated with LPA (200 ng/ml), EGF (50 ng/ml), or PDGF (50 ng/ml). CFP and FRET images were sequentially captured at 1-min intervals. The mean CFP/FRET ratio of cells was calculated and normalized to the ratio at the time of stimulation. The change in the emission ratio of the FAK biosensor following EGF, PDGF, or LPA stimulation is shown in panel A. The change in the emission ratio of the wild-type FAK biosensor is compared with that of the constitutively active FAK biosensor mutant following LPA stimulation in panel B. Shown is the average response ± standard error (n = 3 cells). (F) HeLa cells expressing the biosensor were stimulated with LPA and lysed at the indicated times. The biosensor was immunoprecipitated (IP) and blotted with a phosphotyrosine (P-Tyr) antibody (top) or FAK antibody as a loading control (bottom).
While conformationally active FAK was found in focal adhesions, there was heterogeneity in the CFP/FRET ratio seen in focal adhesions. This did not correlate with focal adhesion size or amount of FAK in individual focal adhesions but rather with location. FAK activity was higher in focal adhesions at the cell margin compared with internal focal adhesions, and a similar pattern was observed using the autophosphorylation biosensor. This relationship was examined quantitatively by plotting the minimal distance of each focal adhesion from the cell margin versus the CFP/FRET and FRETc/CFP ratio of the focal adhesion (Fig. 5B and C). This analysis supports the hypothesis that there is a tendency of focal adhesions near the periphery of the cell to have elevated levels of active/autophosphorylated FAK compared with focal adhesions further removed from the cell margin. Interestingly, an asymmetric FAK conformation spatial pattern was identified in many cells: i.e., FAK activation was elevated along one margin of the cell relative to its activation at the opposite margin. These findings suggest that FAK activity is regulated differentially in different focal adhesions, which may play a role in the determination of cell polarity and directional migration.
Temporal regulation of FAK conformation in living cells.
To study FAK conformation changes in response to soluble ligands, HeLa cells expressing the conformational biosensor were serum starved and then stimulated with LPA. The average CFP/FRET ratio in the cell was determined over time and was found to increase following stimulation (Fig. 5D). The kinetics of conformation change in FAK paralleled the kinetics of tyrosine phosphorylation of the biosensor (Fig. 5F). Stimulation with PDGF produced a small, transient change in the CFP/FRET ratio, while EGF stimulation did not produce a change in FRET under these conditions. The CYFAK413-Y180A/M183A constitutively active biosensor exhibited a constant CFP/FRET ratio before and after stimulation with LPA (Fig. 5E). These findings demonstrate that conformational changes occur in FAK in response to soluble ligands.
A FERM domain basic patch is required for conformational change.
A basic patch in the F2 subdomain of the FERM domain of FAK was identified as a key motif for FAK activation in vivo in response to cell adhesion and stimulation of the Met receptor (14, 20). This motif is on the surface of the FERM/catalytic domain complex and is close to the catalytic domain interaction site in the crystal structure. Thus, this motif is a potential site for controlling FAK conformational regulation. To determine if this basic region was required for conformational regulation of FAK, alanine substitutions for basic residues in this region were engineered into the conformational biosensor to create the K216A/K218A/R221A/K222A mutant (KAKTLRK). The wild-type and mutant biosensors were transiently expressed, and FRET was examined by fluorometric analysis. The mutant exhibited a higher FRET/CFP ratio than the wild-type biosensor, suggesting that the integrity of the basic patch was required for optimal induction of the active conformation of FAK (Fig. 6A).
FIG. 6.
Acidic phospholipids bind the basic patch within the FAK FERM domain. (A) Normalized FRET/CFP emission ratios of the wild-type FAK biosensor and the basic patch mutant are shown. The mutant FRET/CFP ratio was normalized to FRET/CFP ratio of the wild-type biosensor. Shown is a representative experiment (n = 3). (B) The purified recombinant FERM domain was incubated with BODIPY-labeled phospholipids, and binding was measured by fluorescence polarization. Anisotropy (mP, millipolarization units) is plotted against FERM domain concentration. The average of three experiments ± standard deviation is shown. (C) PE/PC vesicles containing increasing amounts of PIP2 were incubated with a GST-FERM fusion protein. The vesicles were sedimented by centrifugation, and the amount of fusion protein in the vesicle containing pellet (P) and the supernatant (S) was determined by SDS-PAGE and Coomassie blue staining. (D) PE/PC vesicles containing 10% (mass ratio) of the indicated lipids were incubated with the GST-FERM fusion protein and analyzed as in panel A. Buf, buffer. (E) PE/PC vesicles containing PIP2 or phosphatidylinositol (PI) were incubated the wild-type GST-FERM domain or a basic patch mutant (KAKTLRK) and analyzed as in panel A. (F and G) HeLa cells expressing a YFP-FERM domain fusion protein or the basic patch mutant (KAKTLRK) were fixed, permeabilized, and then stained with rhodamine-phalloidin. Fixed cells were observed by laser-scanning confocal microscopy. (F) The percentages of cells containing the YFP-FERM constructs in ruffles or at the membrane at the edge of the cell were scored. wt, wild type. Results represent the average from three experiments ± standard deviation with >100 cells counted per experiment. **, P < 0.005; *, P < 0.05. (G) Representative images of cells expressing the YFP-FERM domain (two left panels) or the YFP-FERM KAKTLRK mutant (right panel) are shown. Arrows indicate YFP-FERM localization in ruffles (left panel) and at the membrane at the periphery of the cell (middle panel).
Acidic phospholipids bind the basic patch within the FAK FERM domain.
Given the basic nature of this region, potential binding partners are likely to be acidic. Other FERM domains exhibit high-affinity binding to PIP2 via positively charged motifs (23). The structure of the FAK FERM domain precludes PIP2 binding via the precise mechanism utilized by other FERM domains (9); however, an interaction via the basic patch of the F2 subdomain remains a possibility. To examine binding to PIP2, a fluorescence polarization assay was used. Purified FERM domain was titrated into a solution of BODIPY-labeled PIP2 containing C6-acyl chains, and anisotropy was measured. Anisotropy was plotted against protein concentration, and the dissociation constant (Kd) was calculated (Fig. 6B). The Kd for PIP2 was 31.52 ± 1.7 μM, and that for phosphatidylinositol was >200 μM. The interaction of the FERM domain with lipids was further validated using a vesicle cosedimentation assay. PE/PC vesicles containing increasing amounts of PIP2 were incubated with a GST-FERM fusion protein. The vesicles were sedimented, and the amounts of fusion protein in the vesicle-containing pellet and the supernatant were determined by SDS-PAGE and Coomassie blue staining. While the FERM domain bound poorly to PE/PC vesicles, approximately 50% of the FERM domain bound to vesicles containing 1% PIP2, and virtually all of the fusion protein associated with vesicles containing 5% and 10% PIP2 (Fig. 6C). To further characterize lipid binding, the interaction of the GST-FERM domain fusion protein with vesicles containing other lipids was examined. Under conditions where the protein was completely found in the pellet with PIP2-containing vesicles, GST-FERM associated weakly with phosphatidylinositol-containing vesicles (Fig. 6D). In contrast, the GST-FERM fusion protein bound quite well to phosphatidylserine-containing vesicles. Thus, the data suggest that the interaction of GST-FERM with lipids is dictated by charge and that the structure of the head group is less important for binding in vitro.
The most likely lipid binding site on the FERM domain is the F2 subdomain basic patch as it is the most basic feature of the domain. To test its role in lipid binding, the KAKTLRK mutations were engineered into the FERM domain. While the wild-type GST-FERM domain associated strongly with PIP2-containing vesicles, this mutant was defective for binding (Fig. 6E). If the FERM domain was capable of binding lipids in vivo the domain might be recruited to the membrane. The FERM domain of FAK was transiently expressed in HeLa cells as a YFP fusion protein, and its localization was examined by confocal microscopy (Fig. 6G). The FERM domain partially colocalized with F-actin in ruffles and was observed at the membrane at the periphery of the cell. This pattern of localization suggests that the FERM domain is capable of associating with the membrane of the cell. Notably, KAKTLRK mutant exhibited a cytoplasmic localization and was not frequently observed in ruffles or at the membrane at the periphery of the cell (Fig. 6F). Expression of the wild-type and mutant FERM domains was comparable (Cai and Schaller, unpublished). These data suggest that the FAK FERM domain can interact with acidic lipids in vitro and associate with the membrane in vivo, and the basic patch on the F2 subdomain is required for both of these activities.
PIP2-containing vesicles can activate FAK via conformational change.
Given that the basic patch mutants are defective for signaling in vivo and that this region can mediate binding to PIP2-containing vesicles, it is possible that this interaction might play a role in FAK activation. This was tested using a recombinant fragment of FAK containing the FERM and catalytic domains. The fragment adopts the inactive conformation in which the Src phosphorylation sites within the activation loop of FAK are protected from phosphorylation (32). The FAK fragment was incubated with purified, active Src in lipid vesicle binding buffer. Under these conditions, Src could weakly induce FAK phosphorylation (Fig. 7A). Incubation of the FAK fragment with PE/PC vesicles containing 10% PIP2 prior to phosphorylation by Src led to a dramatic increase in phosphorylation of FAK, whereas incubation with PE/PC vesicles did not (Fig. 7A). As a control, phosphorylation of GST fusion protein containing the activation loop peptide from FAK by Src was measured and did not change in the presence or absence of PIP2-containing vesicles. These results are consistent with the hypothesis that lipid vesicle binding leads to a conformational change in FAK exposing the activation loop for phosphorylation by Src.
FIG. 7.
(A) A recombinant fragment of FAK containing the FERM and catalytic domains was incubated with Src (SH3 plus SH2 plus kinase) in the presence of the indicated liposomes in kinase reaction buffer or in buffer alone (buf) for 30 min. As a control substrate, a GST fusion protein containing a peptide mimicking the activation loop of FAK was used. The reaction was terminated by the addition of sample buffer, and phosphorylation of the substrates was examined by Western blotting with a phosphotyrosine (P-Tyr) antibody. (B) The wild-type (WT) biosensor and the F2 basic patch mutant biosensor (KAKTLRK) were coexpressed with PIP5KIα or a catalytically defective mutant of PIP5KIα (KD). The CFP/FRET ratio in each case was determined by fluorometry. The average of three experiments ± standard error is shown. The emission ratios observed in the presence of wild-type and catalytically inactive PIP5KIα were analyzed using an unpaired t test (*, P < 0.05). (C) The average CFP/FRET ratio of the cytoplasm or focal adhesions in HeLa cells coexpressing the wild-type biosensor and wild-type SopB or the catalytically inactive mutant SopBcs is shown (average of n = 6 cells ± standard error). The focal adhesion values in SopB- and SopBcs-expressing cells were analyzed using an unpaired t test (*, P < 0.0025).
Modulation of PIP2 regulates FAK conformation in vivo.
To determine if perturbation of PIP2 levels in vivo could alter FAK activity, the conformational biosensor was used. To alter PIP2 levels, type I phosphatidylinositol kinase α (PIPKIα) was coexpressed with the biosensor and the active state of the biosensor compared with cells expressing the catalytically inactive mutant of PIPKIα. Coexpression with PIPKIα resulted in a higher CFP/FRET ratio (indicating an increase in activated FAK) than coexpression with the catalytically inactive variant of the enzyme (Fig. 7B). These results demonstrate that elevation of PIP2 levels in the cell promote the conversion of FAK into its active conformation. Further, the basic patch in the F2 subdomain of the FAK FERM domain is important for this effect since the KAKTLRK mutant lacking the basic patch was not responsive to altering the levels of PIP2 in the cell (Fig. 7B). To determine if PIP2 levels played a significant role in regulating FAK conformation under physiological conditions, SopB, an inositol polyphosphate 4-phosphatase, was coexpressed with the conformation biosensor and the effect upon the biosensor in adherent cells was examined. The conformation of the biosensor in focal adhesions was compared with its conformation in the cytoplasm by calculating the CFP/FRET ratio, where a higher ratio reflects the active conformation and a lower ratio reflects the inactive conformation (Fig. 7C). Expression of SopBcs, the catalytically inactive negative control, had little effect upon FAK conformation. These cells exhibited more activated FAK in focal adhesions than the cytoplasm, similar to the results shown in Fig. 5A (Fig. 7C). In contrast, SopB expression resulted in a decrease in activated FAK. In these cells, the level of FAK activation in focal adhesions was only slightly higher than the level of activation in the cytoplasm. These findings demonstrate that PIP2 depletion reduces FAK activation in focal adhesions, suggesting that PIP2 plays a role in regulating the conformation of FAK in cells adherent to fibronectin.
DISCUSSION
The current model of FAK regulation evokes an intramolecular autoinhibitory interaction between the N-terminal FERM domain and central catalytic domain. Mutational and biochemical studies support this hypothesis and the crystal structure of the FERM/catalytic domain complex provides insight into the mechanism of inhibition (15, 32). However, using these technologies it is not possible to probe protein conformation in vivo. FRET technology is a powerful tool to study protein-protein interactions in living cells. As FRET efficiency is dependent upon the distance between the two fluorophores and their relative orientation, changes in FRET correspond to changes in distance and orientation between the fluorescent probes, and in the case of a single protein fused to two fluorophores can reflect changes in conformation (35, 40, 43). Here, we have developed genetically encoded FRET biosensors to visualize changes in FAK conformation in vivo. These biosensors have provided two important pieces of evidence in support of the autoinhibitory model of FAK regulation. First, the analysis of the biosensors provides support for the crystal structure of the FERM/catalytic domain complex. CFP is encoded at the N terminus of FAK in these biosensors and citrine was inserted at two different sites. Based upon the crystal structure, the probe insertion sites in the CYFAK413 construct are approximately 20 Å apart, and in the CYFAK700 construct the insertion sites are predicted to be 70 Å apart. As predicted from the structure, the former exhibited a high FRET signal and the latter produced a weak FRET signal. Second, using the conformational biosensor, this study provides for the first time direct evidence that a FAK conformational switch occurs in vivo and is associated with FAK activation. In focal adhesions, the probe exhibited an open conformation, whereas probes located in the cytoplasm exhibited a closed conformation. This is consistent with the body of literature implicating FAK as a cell adhesion-regulated kinase and suggests that integrin signaling contributes to the FAK conformational change. Moreover, another stimulus of FAK activation, LPA, induced a change in the wild-type FAK biosensor conformation but had no effect upon a mutant biosensor that is constitutively in the open conformation. These findings demonstrate that multiple stimuli regulate the conformation of FAK and that conformational change is a general mechanism leading to FAK activation.
Spatial regulation of FAK.
The FAK biosensors provide unique tools to investigate the spatial regulation of FAK. Currently, immunostaining using FAK phosphorylation-specific antibodies is used for this purpose (46), but this approach has some disadvantages. Antibody specificity is a potential problem that is often not well controlled. Further, tyrosine phosphorylation of FAK does not strictly correlate with catalytic activity (50). It is impossible to probe FAK activity in live cells using these approaches, and it is currently impossible to monitor the conformation change of FAK by immunostaining.
The two biosensors report similar patterns of autophosphorylation/conformational activation. Both indicate elevated autophosphorylation/activation in peripheral focal adhesions compared with internal focal adhesions. Interestingly, this pattern is similar to the pattern of cell traction forces in spreading cells (i.e., greatest traction forces at peripheral focal adhesions) (3), which is consistent with a role for FAK in sensing mechanical stimuli. In addition, results using both biosensors indicate an asymmetry in autophosphorylation/activation of FAK in polarized cells with the highest levels in areas of cell protrusion. As FAK has been reported to regulate cell polarity (55), this asymmetric activation pattern may define the direction of polarization.
FAK and lipid binding.
The FERM domain of FAK was shown to bind acidic phospholipids. PIP2 binding is a mechanism of regulation of ezrin, radixin and moesin, but the molecular details of the interaction differ from FAK. In radixin, the PIP2 headgroup associates with a basic pocket between the F1 and F3 subdomains of the FERM domain, whereas the corresponding site in FAK lacks the basic pocket (9, 23). Although the F3 subdomain of FERM domains have a protein fold similar to pleckstrin homology (PH) domains, the F3 subdomain of FAK cannot bind phospholipids since it lacks the basic pocket that mediates the interaction of PH domains with phospholipids. Instead, FAK appears to bind acidic phospholipids through surface-exposed basic residues at the tip of the F2 subdomain. Not surprisingly, the FERM domain of FAK binds PIP2 with lower affinity than PH domains. In contrast with the 30 μM Kd of the FERM domain, the PH domains of phospholipase C-δ (PLC-δ) and SOS-1 are reported to bind PIP2 with Kds of ∼2 μM (PLC-δ and SOS-1) (26, 30, 31). The affinity of the PH domain of spectrin ranges from 15 to 50 μM, depending upon buffer conditions (24). Thus, the affinity of the FAK FERM domain for PIP2 is in the range of lower-affinity PH domain interactions. It is also notable that there is precedent for PIP2 binding to surface basic residues of proteins. For example, PIP2 associates with the tail domain of vinculin through surface basic residues rather than a discrete basic pocket (2, 12).
The F2 basic patch is required for activation of FAK in response to cell adhesion and following HGF stimulation (14, 20). This sequence binds acidic phospholipids, lipid vesicles containing PIP2 alter the conformation of FAK in vitro, and elevating PIP2 levels in cells results in alteration of the conformation of FAK in vivo. Further, reduction of PIP2 levels by expression of SopB results in decreased activation of FAK in focal adhesions in cells adherent to fibronectin. These findings support the hypothesis that acidic phospholipids, including PIP2, are physiologically relevant FAK FERM domain ligands that regulate the release of autoinhibitory interactions allowing FAK to adopt its active conformation. Additional evidence from the literature demonstrates that acidic phospholipids are important for the activation of FAK. Activation of FAK by stimulation of some G protein-coupled receptors is apparently blocked by depletion of PIP2 levels (34). Interestingly, a splice variant of PIP5KIγ colocalizes with FAK at focal adhesions and by localized generation of PIP2 might regulation FAK function (19, 33). Other studies have suggested that the activity of phosphatidylinositol 3-kinase is required for activation of FAK in response to a number of different stimuli (7, 28, 36). We hypothesize that these acidic phospholipids might regulate FAK conformation through binding to the F2 basic patch. Following HGF stimulation, this same sequence mediates binding to tyrosine phosphorylated Met to facilitate FAK activation (14). These results suggest the intriguing hypothesis that a single site on the FERM domain of FAK can associate with either acidic phospholipids or phosphopeptides to trigger activation.
Acknowledgments
We thank Sue Craig and Hui Chen, who provided invaluable advice during the development and characterization of the biosensors. We also thank Sean Palmer for guidance in setting up the lipid vesicle cosedimentation assay.
This project was supported by NIH grant HL45100 (M.D.S.) and NIH Cell Migration Consortium grant GM0064346 (K.J. and K.M.H.).
Footnotes
Published ahead of print on 27 October 2007.
REFERENCES
- 1.Abbi, S., H. Ueda, C. Zheng, L. A. Cooper, J. Zhao, R. Christopher, and J. L. Guan. 2002. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol. Biol. Cell 133178-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakolitsa, C., J. M. de Pereda, C. R. Bagshaw, D. R. Critchley, and R. C. Liddington. 1999. Crystal structure of the vinculin tail suggests a pathway for activation. Cell 99603-613. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N. Q., U. S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi, and B. Geiger. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3466-472. [DOI] [PubMed] [Google Scholar]
- 4.Ballestrem, C., N. Erez, J. Kirchner, Z. Kam, A. Bershadsky, and B. Geiger. 2006. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J. Cell Sci. 119866-875. [DOI] [PubMed] [Google Scholar]
- 5.Braren, R., H. Hu, Y. H. Kim, H. E. Beggs, L. F. Reichardt, and R. Wang. 2006. Endothelial FAK is essential for vascular network stability, cell survival, and lamellipodial formation. J. Cell Biol. 172151-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calalb, M. B., T. R. Polte, and S. K. Hanks. 1995. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol. Cell. Biol. 15954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamassima, A., and E. Rozengurt. 1998. Insulin-like growth factor I stimulates tyrosine phosphorylation of p130(Cas), focal adhesion kinase, and paxillin. Role of phosphatidylinositol 3′-kinase and formation of a p130(Cas)·Crk complex. J. Biol. Chem. 27326149-26156. [DOI] [PubMed] [Google Scholar]
- 8.Ceccarelli, D. F., I. M. Blasutig, M. Goudreault, Z. Li, J. Ruston, T. Pawson, and F. Sicheri. 2007. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 28213864-13874. [DOI] [PubMed] [Google Scholar]
- 9.Ceccarelli, D. F., H. K. Song, F. Poy, M. D. Schaller, and M. J. Eck. 2006. Crystal structure of the FERM domain of focal adhesion kinase. J. Biol. Chem. 281252-259. [DOI] [PubMed] [Google Scholar]
- 10.Chamberlain, C. E., V. S. Kraynov, and K. M. Hahn. 2000. Imaging spatiotemporal dynamics of Rac activation in vivo with FLAIR. Methods Enzymol. 325389-400. [DOI] [PubMed] [Google Scholar]
- 11.Chan, P. Y., S. B. Kanner, G. Whitney, and A. Aruffo. 1994. A transmembrane-anchored chimeric focal adhesion kinase is constitutively activated and phosphorylated at tyrosine residues identical to pp125FAK. J. Biol. Chem. 26920567-20574. [PubMed] [Google Scholar]
- 12.Chandrasekar, I., T. E. Stradal, M. R. Holt, F. Entschladen, B. M. Jockusch, and W. H. Ziegler. 2005. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 1181461-1472. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H., D. M. Cohen, D. M. Choudhury, N. Kioka, and S. W. Craig. 2005. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 169459-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, S.-Y., and H.-C. Chen. 2006. Direct interaction of focal adhesion kinase (FAK) with Met is required for FAK to promote hepatocyte growth factor-induced cell invasion. Mol. Cell. Biol. 265155-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen, L. A., and J. L. Guan. 2005. Residues within the first subdomain of the FERM-like domain in focal adhesion kinase are important in its regulation. J. Biol. Chem. 2808197-8207. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, L. A., T.-L. Shen, and J.-L. Guan. 2003. Regulation of focal adhesion kinase by its amino-terminal domain through an autoinhibitory interaction. Mol. Cell. Biol. 238030-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLano, W. L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA.
- 18.DiMichele, L. A., J. T. Doherty, M. Rojas, H. E. Beggs, L. F. Reichardt, C. P. Mack, and J. M. Taylor. 2006. Myocyte-restricted focal adhesion kinase deletion attenuates pressure overload-induced hypertrophy. Circ. Res. 99636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Paolo, G., L. Pellegrini, K. Letinic, G. Cestra, R. Zoncu, S. Voronov, S. Chang, J. Guo, M. R. Wenk, and P. De Camilli. 2002. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 42085-89. [DOI] [PubMed] [Google Scholar]
- 20.Dunty, J. M., V. Gabarra-Niecko, M. L. King, D. F. J. Ceccarelli, M. J. Eck, and M. D. Schaller. 2004. FERM domain interaction promotes FAK signaling. Mol. Cell. Biol. 245353-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuta, Y., D. Ilic, S. Kanazawa, N. Takeda, T. Yamamoto, and S. Aizawa. 1995. Mesodermal defect in late phase of gastrulation by a targeted mutation of focal adhesion kinase, FAK. Oncogene 111989-1995. [PubMed] [Google Scholar]
- 22.Gabarra-Niecko, V., M. D. Schaller, and J. M. Dunty. 2003. FAK regulates biological processes important for the pathogenesis of cancer. Cancer Metastasis Rev. 22359-374. [DOI] [PubMed] [Google Scholar]
- 23.Hamada, K., T. Shimizu, T. Matsui, S. Tsukita, and T. Hakoshima. 2000. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 194449-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlan, J. E., H. S. Yoon, P. J. Hajduk, and S. W. Fesik. 1995. Structural characterization of the interaction between a pleckstrin homology domain and phosphatidylinositol 4,5-bisphosphate. Biochemistry 349859-9864. [DOI] [PubMed] [Google Scholar]
- 25.Heikal, A. A., S. T. Hess, G. S. Baird, R. Y. Tsien, and W. W. Webb. 2000. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine). Proc. Natl. Acad. Sci. USA 9711996-12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose, K., S. Kadowaki, M. Tanabe, H. Takeshima, and M. Iino. 1999. Spatiotemporal dynamics of inositol 1,4,5-trisphosphate that underlies complex Ca2+ mobilization patterns. Science 2841527-1530. [DOI] [PubMed] [Google Scholar]
- 27.Jacamo, R. O., and E. Rozengurt. 2005. A truncated FAK lacking the FERM domain displays high catalytic activity but retains responsiveness to adhesion-mediated signals. Biochem. Biophys. Res. Commun. 3341299-1304. [DOI] [PubMed] [Google Scholar]
- 28.King, W. G., M. D. Mattaliano, T. O. Chan, P. N. Tsichlis, and J. S. Brugge. 1997. Phosphatidylinositol 3-kinase is required for integrin-stimulated AKT and Raf-1/mitogen-activated protein kinase pathway activation. Mol. Cell. Biol. 174406-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraynov, V. S., C. Chamberlain, G. M. Bokoch, M. A. Schwartz, S. Slabaugh, and K. M. Hahn. 2000. Localized Rac activation dynamics visualized in living cells. Science 290333-337. [DOI] [PubMed] [Google Scholar]
- 30.Kubiseski, T. J., Y. M. Chook, W. E. Parris, M. Rozakis-Adcock, and T. Pawson. 1997. High affinity binding of the pleckstrin homology domain of mSos1 to phosphatidylinositol (4,5)-bisphosphate. J. Biol. Chem. 2721799-1804. [DOI] [PubMed] [Google Scholar]
- 31.Lemmon, M. A., K. M. Ferguson, R. O'Brien, P. B. Sigler, and J. Schlessinger. 1995. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA 9210472-10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lietha, D., X. Cai, D. F. Ceccarelli, Y. Li, M. D. Schaller, and M. J. Eck. 2007. Structural basis for the autoinhibition of focal adhesion kinase. Cell 1291177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling, K., R. L. Doughman, A. J. Firestone, M. W. Bunce, and R. A. Anderson. 2002. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 42089-93. [DOI] [PubMed] [Google Scholar]
- 34.Linseman, D. A., S. D. Sorensen, and S. K. Fisher. 1999. Attenuation of focal adhesion kinase signaling following depletion of agonist-sensitive pools of phosphatidylinositol 4,5-bisphosphate. J. Neurochem. 731933-1944. [PubMed] [Google Scholar]
- 35.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2444-456. [DOI] [PubMed] [Google Scholar]
- 36.Lymn, J. S., S. J. Rao, G. F. Clunn, K. L. Gallagher, C. O'Neil, N. T. Thompson, and A. D. Hughes. 1999. Phosphatidylinositol 3-kinase and focal adhesion kinase are early signals in the growth factor-like responses to thrombospondin-1 seen in human vascular smooth muscle. Arterioscler. Thromb. Vasc. Biol. 192133-2140. [DOI] [PubMed] [Google Scholar]
- 37.Lyons, P. D., J. M. Dunty, E. M. Schaefer, and M. D. Schaller. 2001. Inhibition of the catalytic activity of cell adhesion kinase beta by protein-tyrosine phosphatase-PEST-mediated dephosphorylation. J. Biol. Chem. 27624422-24431. [DOI] [PubMed] [Google Scholar]
- 38.McLean, G. W., N. H. Komiyama, B. Serrels, H. Asano, L. Reynolds, F. Conti, K. Hodivala-Dilke, D. Metzger, P. Chambon, S. G. Grant, and M. C. Frame. 2004. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev. 182998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra, S. K., S. T. Lim, A. Chi, and D. D. Schlaepfer. 2006. Intrinsic focal adhesion kinase activity controls orthotopic breast carcinoma metastasis via the regulation of urokinase plasminogen activator expression in a syngeneic tumor model. Oncogene 254429-4440. [DOI] [PubMed] [Google Scholar]
- 40.Miyawaki, A. 2003. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev. Cell 4295-305. [DOI] [PubMed] [Google Scholar]
- 41.Moeller, M. L., Y. Shi, L. F. Reichardt, and I. M. Ethell. 2006. EphB receptors regulate dendritic spine morphogenesis through the recruitment/phosphorylation of focal adhesion kinase and RhoA activation. J. Biol. Chem. 2811587-1598. [DOI] [PubMed] [Google Scholar]
- 42.Peng, X., M. S. Kraus, H. Wei, T. L. Shen, R. Pariaut, A. Alcaraz, G. Ji, L. Cheng, Q. Yang, M. I. Kotlikoff, J. Chen, K. Chien, H. Gu, and J. L. Guan. 2006. Inactivation of focal adhesion kinase in cardiomyocytes promotes eccentric cardiac hypertrophy and fibrosis in mice. J. Clin. Investig. 116217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertz, O., and K. M. Hahn. 2004. Designing biosensors for Rho family proteins—deciphering the dynamics of Rho family GTPase activation in living cells. J. Cell Sci. 1171313-1318. [DOI] [PubMed] [Google Scholar]
- 44.Pertz, O., L. Hodgson, R. L. Klemke, and K. M. Hahn. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 4401069-1072. [DOI] [PubMed] [Google Scholar]
- 45.Round, J., and E. Stein. 2007. Netrin signaling leading to directed growth cone steering. Curr. Opin. Neurobiol. 1715-21. [DOI] [PubMed] [Google Scholar]
- 46.Ruest, P. J., S. Roy, E. Shi, R. L. Mernaugh, and S. K. Hanks. 2000. Phosphospecific antibodies reveal focal adhesion kinase activation loop phosphorylation in nascent and mature focal adhesions and requirement for the autophosphorylation site. Cell Growth Differ. 1141-48. [PubMed] [Google Scholar]
- 47.Schaller, M. D. 2001. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim. Biophys. Acta 15401-21. [DOI] [PubMed] [Google Scholar]
- 48.Schlaepfer, D. D., M. A. Broome, and T. Hunter. 1997. Fibronectin-stimulated signaling from a focal adhesion kinase-c-Src complex: involvement of the Grb2, p130cas, and Nck adaptor proteins. Mol. Cell. Biol. 171702-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlaepfer, D. D., and T. Hunter. 1996. Evidence for in vivo phosphorylation of the Grb2 SH2-domain binding site on focal adhesion kinase by Src-family protein-tyrosine kinases. Mol. Cell. Biol. 165623-5633. (Erratum, 16:7182-7184.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlaepfer, D. D., K. C. Jones, and T. Hunter. 1998. Multiple Grb2-mediated integrin-stimulated signaling pathways to ERK2/mitogen-activated protein kinase: summation of both c-Src- and focal adhesion kinase-initiated tyrosine phosphorylation events. Mol. Cell. Biol. 182571-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, T. L., A. Y. Park, A. Alcaraz, X. Peng, I. Jang, P. Koni, R. A. Flavell, H. Gu, and J. L. Guan. 2005. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J. Cell Biol. 169941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sieg, D. J., C. R. Hauck, D. Ilic, C. K. Klingbeil, E. Schaefer, C. H. Damsky, and D. D. Schlaepfer. 2000. FAK integrates growth-factor and integrin signals to promote cell migration. Nat. Cell Biol. 2249-256. [DOI] [PubMed] [Google Scholar]
- 53.Thomas, J. W., M. A. Cooley, J. M. Broome, R. Salgia, J. D. Griffin, C. R. Lombardo, and M. D. Schaller. 1999. The role of focal adhesion kinase binding in the regulation of tyrosine phosphorylation of paxillin. J. Biol. Chem. 27436684-36692. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, J. W., B. Ellis, R. J. Boerner, W. B. Knight, G. C. White, and M. D. Schaller. 1998. SH2- and SH3-mediated interactions between focal adhesion kinase and Src. J. Biol. Chem. 273577-583. [DOI] [PubMed] [Google Scholar]
- 55.Tilghman, R. W., J. K. Slack-Davis, N. Sergina, K. H. Martin, M. Iwanicki, E. D. Hershey, H. E. Beggs, L. F. Reichardt, and J. T. Parsons. 2005. Focal adhesion kinase is required for the spatial organization of the leading edge in migrating cells. J. Cell Sci. 1182613-2623. [DOI] [PubMed] [Google Scholar]
- 56.Toutant, M., A. Costa, J.-M. Studler, G. Kadaré, M. Carnaud, and J.-A. Girault. 2002. Alternative splicing controls the mechanisms of FAK autophosphorylation. Mol. Cell. Biol. 227731-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueda, H., S. Abbi, C. Zheng, and J. L. Guan. 2000. Suppression of Pyk2 kinase and cellular activities by FIP200. J. Cell Biol. 149423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, D., J. R. Grammer, C. S. Cobbs, J. E. Stewart, Jr., Z. Liu, R. Rhoden, T. P. Hecker, Q. Ding, and C. L. Gladson. 2000. p125 focal adhesion kinase promotes malignant astrocytoma cell proliferation in vivo. J. Cell Sci. 1134221-4230. [DOI] [PubMed] [Google Scholar]
- 59.Xia, Z., and Y. Liu. 2001. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 812395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]