Abstract
The basic helix-loop-helix (bHLH) protein DIMMED (DIMM) supports the differentiation of secretory properties in numerous peptidergic cells of Drosophila melanogaster. DIMM is coexpressed with diverse amidated neuropeptides and with the amidating enzyme peptidylglycine α-hydroxylating monooxygenase (PHM) in approximately 300 cells of the late embryo. Here we confirm that DIMM has transcription factor activity in transfected HEK 293 cells and that the PHM gene is a direct target. The mammalian DIMM orthologue MIST1 also transactivated the PHM gene. DIMM activity was dependent on the basic region of the protein and on the sequences of three E-box sites within PHM's first intron; the sites make different contributions to the total activity. These data suggest a model whereby the three E boxes interact cooperatively and independently to produce high PHM transcriptional activation. This DIMM-controlled PHM regulatory region displayed similar properties in vivo. Spatially, its expression mirrored that of the DIMM protein, and its activity was largely dependent on dimm. Further, in vivo expression was highly dependent on the sequences of the same three E boxes. This study supports the hypothesis that DIMM is a master regulator of a peptidergic cell fate in Drosophila and provides a detailed transcriptional mechanism of DIMM action on a defined target gene.
Neurons release numerous biologically active transmitters, including neuropeptides, which are derived from larger precursor proteins. Certain neurons termed neurosecretory cells (NSCs) are specialized in neuropeptide production and have greatly amplified secretory capabilities that are akin to those of peripheral endocrine cells. NSCs play essential roles in animal physiology by releasing biologically active peptides that can act at long distances. They represent a distinctive neuronal class because their secretory properties are amplified: such cells are specialized to produce, package, and release large amounts of such signaling molecules. Additionally, NSC properties can be strongly modified in response to changes in environment or internal homeostasis (9). Examining the intracellular regulatory pathways that organize and modulate these specialized properties is therefore critical to understanding NSC physiology.
In vertebrates, several transcription factors have been identified that are required for the proper differentiation of neuroendocrine cells in the brain and periphery, including Mash1, Sim1, Otp, and others (1, 8, 24, 26, 31, 32, 36, 47, 52). In the case of Sim1, this requirement involves early aspects of development prior to terminal differentiation (e.g., cell migration [56]). In Drosophila melanogaster, only the basic helix-loop-helix (bHLH) protein DIMMED (DIMM) has so far been implicated in the development of peptidergic cells (2, 17, 20, 21). The Drosophila genome contains at least 30 neuropeptide precursor-encoding genes; remarkably, more than 90% of the known or predicted Drosophila neuropeptides are amidated (22). In the Drosophila central nervous system (CNS), neuropeptides are expressed by cells at all axial levels and individual neuropeptide genes are typically expressed by small subsets among the approximately 10,000 neurons of the larval CNS. DIMM displays a highly regulated pattern of expression in approximately 300 diverse larval CNS neurons. DIMM neurons (and the few DIMM-positive peripheral cells) are chemically heterogeneous but share certain properties: (i) individual DIMM cells contain large amounts of neuropeptides (as indicated by immunocytochemistry), and (ii) DIMM cells all express high amounts of the critical neuropeptide biosynthetic enzyme peptidylglycine α-hydroxylating monooxygenase (PHM), which is required for peptide α-amidation.
The results of previous studies of DIMM have paralleled those for regulatory factors for other transmitters. For example, PET-1 (an ETS domain transcription factor) coordinates the terminal differentiation of the serotonergic phenotype and is precisely expressed by the precursors to the vast majority of serotonergic neurons, but not elsewhere (18). Likewise, the Paired-like homeodomain proteins Phox2a and Phox2b have been implicated as essential determinants of the noradrenergic phenotype (35, 41). However, in contrast to the situations for serotonergic and noradrenergic cell fates, in which all cells sharing the same fate express the same transmitter, the NSC cell fate represents the expression of many different secretory peptides by diverse cells. We estimate that there are at least 31 different neuropeptide genes in the Drosophila genome (51). Thus, NSC cell differentiation must include both cell type-specific features (e.g., neuropeptide selection) and generic features that support the expression, packaging, and release of amidated neuropeptides. Reflecting this fact, we previously concluded that, within single neurons, DIMM operates in two distinct mechanisms: one is termed combinatorial and the other a form of master regulation (3).
In Drosophila, the selection of an appropriate neuropeptide gene is controlled by combinations of transcription factors. The compositions of codes are different for different cells, but many include DIMM as a constant element to help drive the selection of different neuropeptide precursors. This DIMM activity represents the first mechanism by which it regulates NSC development. For example, in identified Tv neuroendocrine neurons of the CNS, dFMRFa neuropeptide expression depends on DIMM but also on the LIM homeodomain protein Apterous (AP) (6) and the zinc-finger protein SQUEEZE (SQZ) (3, 40). No single member of this three-factor code produces ubiquitous dFMRFa expression when broadly misexpressed, and the triple comisexpression of dimm, ap, and sqz is more effective at producing ectopic dFMRFa. Thus, different sets of positively acting factors combine with DIMM to drive specific neuropeptide gene expression in different neurons (19, 33, 40).
Independent of combinatorial codes, DIMM exerts a second critical influence, its so-called master regulatory role, on developing NSCs (2). DIMM displays singular control of properties held in common by diverse NSCs and has the ability to impose them onto non-NSCs. For example, DIMM misexpression throughout the entire CNS confers PHM expression (a common NSC property) on all neurons. Other common NSC features controlled by a master regulator like DIMM could involve mechanisms of precursor processing and routing and of dense-core secretory granule generation, trafficking, and accumulation. Therefore, the current conception of NSC cell organization supposes two interlocked regulatory networks (combinatorial codes and single master regulator) operating within single cells.
Previous work has demonstrated that DIMM controls the expression of the neuropeptide biosynthetic enzyme PHM (15, 28). This monooxygenase is most closely related to dopamine-β-hydroxylase (DBH) and is selective and required for secretory peptide C-terminal amidation (25). Loss-of-function genetic studies indicate that DIMM normally controls PHM (21), while gain-of-function studies show that it can confer ectopic PHM expression efficiently on all neurons (2). We used in vitro and in vivo experiments to show that DIMM directly activates PHM via three specific E boxes located in the first intron of PHM. Furthermore, we extend these results by showing that the same cis mechanism operates robustly in vivo and can explain in large part the specific high-level expression of PHM in peptidergic neurons and endocrine cells. Finally, we demonstrate that the mammalian orthologue Mist1 is capable of transactivating Drosophila PHM both in cell lines and in transgenic flies. Thus, DIMM and Mist1 share functional as well as sequence attributes, and the regulatory features we describe for specialized secretory cells in Drosophila are likely to be broadly applicable across animal phyla.
MATERIALS AND METHODS
Fly stocks.
The following fly lines were used in this study: dimm mutant allele (Rev4 and Rev8); UAS (upstream activation sequence)-dimm-myc (21); UAS-2X EGFP, UAS-mist1, and ap-gal4 lines. PHM-gal4, PHM-GFP, PHM-E1M-gal4, PHM-E2M-gal4, PHM-E4M-gal4, PHM-E24M-gal4, and PHM-E124M-gal4 lines were established for this study and are described below. Two plasmids, pPT-gal4 and pStinger (4), were used for gal4 and green fluorescent protein (GFP) constructs, respectively. Some PHM-gal4 transgenic flies were generated by a microinjection into w1118 embryos, and the other transgenic flies were established commercially by Model Systems Genomics (http://www.biology.duke.edu/model-system/).
Luciferase assay.
PHM fragments were subcloned into the pGL3 and sv40-pGL3 luciferase (luc) vectors (Promega, Madison, WI). To avoid nonsense-mediated decay, the translation start site of PHM was mutated by site-directed mutagenesis. Drosophila bHLH proteins (DIMM, DIMM-MB, Daughterless [DA], Atonal [ATO], HLH4C, and NAUTILUS [NAU]), the LIM homeodomain protein APTEROUS (AP), and the mammalian bHLH Mist1 were subcloned into the hemagglutinin (HA)-tagged pCDNA3 vector. dimm-MB encodes a DIMM protein with a mutated basic region that is projected to display minimal DNA binding. Three amino acids of the basic region of DIMM were changed to glycines (R164G, E165G, and R165G); the primers used for the site-directed mutagenesis are listed in Table S1 in the supplemental material. For luciferase assays, HEK 293 cells were transiently transfected with 2.1 μg of DNA using Lipofectamine 2000 (Invitrogen, San Diego, CA) and the activity of cell lysates was measured with a Victor-Wallac2 plate reader (Perkin-Elmer, CA). For each experiment, a vector containing the thymidine kinase-Renilla luciferase gene (TK-Rluc) was cotransfected for normalization, and each transfection was performed at least three times independently.
Site-directed mutagenesis.
We performed site-directed mutagenesis following the manufacturer's protocols (Stratagene) using the set of primers listed in Table S1 in the supplemental material. Mutagenesis was performed in the pGEM-TA vector, and sequence variants were then subcloned into the pGL3 vector (both from Promega, Madison, WI). All constructs were confirmed by sequencing.
DNA-protein interaction assays.
Electrophoretic mobility shift assays (EMSAs) were performed as previously described (29). A double-stranded DNA probe corresponding to the E1 E box (see below) was radiolabeled with T4 polynucleotide kinase and [γ-32P]ATP and then column purified. In vitro-translated proteins were prepared following the manufacturer's protocols using a TNT kit (Promega, Madison, WI). The binding reaction mixtures contained 5 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol, 2% glycerol, 0.5 μg of poly(dI-dC), 3 μl of in vitro-translated protein, and radiolabeled DNA probe (25-μl final volume) and were incubated at room temperature for 30 min. For competition assays, the reaction mixtures included 20 pmol of cold competitor. For supershift assays, the reaction mixtures included 1 μl of antibody and were preincubated for 30 min on ice. After incubation, all reaction mixtures were analyzed by 6% nondenaturing gel electrophoresis, which was run at 200 V at 4°C for 4 h. Signals were detected by autoradiography. The primers are listed in Table S1 in the supplemental material.
Antibodies, immunocytochemistry, and fluorescence imaging.
Affinity-purified guinea pig anti-Dimm (1:200) (2), mouse monoclonal anti-GFP 3E6 (1:800; Molecular Probes, Carlsbad CA), rabbit anti-Mist1 (1:250) (29), rabbit anti-FMRFamide (1:1,000) (50), and rabbit anti-GFP (1:500, rabbit polyclonal, catalogue no. AB3080; Chemicon, Temecula, CA) as primary antibodies and Cy3-conjugated or Alex-488-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) were used for immunocytochemistry. Immunostaining methods were as previously described (21). Images were acquired on an Olympus FV500 confocal laser scanning microscope and manipulated by Adobe Photoshop software to adjust contrast and/or levels.
RESULTS
DIMM activates PHM in vitro.
Our previous data indicated that dimm activity is required for the expression of PHM. To test the possibility that such regulation is direct, we tested four genomic regions around the PHM locus for enhancer activity by creating luciferase fusion constructs and transfecting them into mammalian HEK 293 cells (Fig. 1A). These regions included the complete intergenic domain (249 bp) between PHM and its nearest annotated neighbor and the first intron, which includes an unexpectedly large number of E boxes (see below). The two larger constructs (containing ∼0.8 and 1.0 kb of PHM, respectively) displayed fivefold or greater increases in luciferase activity when dimm was cotransfected. In contrast, the two smaller constructs (∼0.2 and 0.4 kb of PHM) did not respond to cotransfected dimm. We then asked whether cotransfection of dimm and the proneural E protein bHLH daughterless gene (da) would increase PHM transactivation since da encodes a putative heteromeric binding partner (10, 11, 13, 14). DA did not interact with DIMM in our previous in vitro biochemical measures (2). While the cotransfection of dimm with da increased PHM transactivation compared to the levels in controls, it was always to a level lower than that of the response to transfection of dimm alone (Fig. 1B). Next, we evaluated the specificity of DIMM's transcriptional activity by comparing the response to this PHM fragment to that for cotransfection with other transcription factors. All four additional bHLH proteins tested (DA, NAU, ATO, and HLH4C) failed to activate the 0.8-kb PHM fragment, as did the LIM-HD protein AP (Fig. 1C). The mammalian protein Mist1 includes a bHLH domain with >90% identity to that of DIMM (29). Mist1 increased PHM-luc activity significantly, although not to the same extent as DIMM (Fig. 1C). Finally, to ask whether DIMM's DNA binding properties were required for its specific transactivating properties, we mutated the basic region within the binding domain of DIMM; we observed little residual activity (Fig. 1D).
FIG. 1.
Transactivation of PHM fragments by DIMM in mammalian HEK 293 cells. (A) Schematic diagram of the PHM gene and PHM-luciferase constructs used for these experiments. When present, the PHM ATG site (shown by X) was mutated. (B) The activities of PHM promoter constructs with and without cotransfected effectors (DIMM or DIMM and DA). (C) Evaluating the specificity of PHM activation by DIMM. NAU, NAUTILUS; ATO, ATONAL; AP, APTEROUS; Mist1, murine Mist1. (D) PHM transactivation depends on the presence of a wild-type basic (DNA binding) domain in DIMM. Histograms represent means and standard errors of the means of the results. *, P < 0.05; **, P < 0.01. P values in comparison to the control values were obtained by using Student's t test.
In summary, we found that a small PHM fragment is transactivated by DIMM following transient transfection in a heterologous cell line. Furthermore, these activities are specific, they are not augmented by cotransfection of a class I (E class) bHLH, and they appear to require DNA binding by DIMM.
Three E boxes within the first intron of PHM are necessary for activation by DIMM.
Next, we searched for critical cis-regulatory elements within the PHM genomic fragment by functional assay and by DIMM binding. We found that the ∼400-bp first intron of the PHM gene, when placed upstream of a heterologous promoter, displayed significant, orientation-independent enhancer activity (Fig. 2B). While both orientations were active, the antisense construct was more so: in response to DIMM transactivation, the PHM-E71 construct (antisense) produced an increase similar to that from the wild-type PHM promoter; however, the PHM-E17 construct (sense) produced approximately half that level of activity. Generally, most bHLH proteins are known to bind a canonical 6-bp sequence, CANNTG, called the E box (7), although noncanonical sites have also been observed (54). Therefore, we searched for and found seven E boxes (putative DIMM binding sites) within this PHM intronic fragment; by random estimate, less than two E boxes are expected within this ∼400-bp domain. We named them E1 to E7, from 5′ to 3′ within the intron (Fig. 2A). E boxes E1 to E7 display at least some sequence conservation among related Drosophila species (see Fig. S1 in the supplemental material).
FIG. 2.
DIMM transactivates the PHM via the subsets of E boxes in its first intron and binds the E1 box directly. (A) A schematic of the first PHM intron indicating the positions of seven E boxes, E1 to E7 (see Table 1 for sequences of wild-type and mutated E boxes). Arrows indicate the original orientation of the DNA fragments. (B) Activity of the two constructs and the control vector diagrammed in panel A. In response to DIMM cotransfection, the first intron of PHM displays strong enhancer activity when fused to a heterologous simian virus 40 (SV40) promoter. (C to F) Effects on PHM transactivation by DIMM following site-directed mutagenesis of different E-box sequences within PHM's first intron. A gray box indicates a particular E-box mutation site. Ratios were calculated by dividing values resulting from dimm cotransfection by those resulting from no cotransfection. The histogram represents the means and standard errors of the means of the results. *, P < 0.05; **, P < 0.01. P values in comparison to the wild type were obtained by Student's t test.
To examine the functional contributions of these individual E-box elements to transactivation by DIMM, we performed selective site-directed mutagenesis and tested the resulting levels of enhancer activity in HEK 293 cells. Different E boxes made different contributions to DIMM activity. Sequence alteration of the single E box E1 strongly reduced responsiveness to DIMM. In contrast, sequence alteration to either E6 or E7 had no such effect (Fig. 2C). Likewise, the alteration of either E3 or E5 did not lessen DIMM responsiveness (Fig. 2D), and in fact, the result with the E5 variant was significantly elevated over that with the wild-type sequence control (Fig. 2D). Simultaneously altering four E boxes (E2, E3, E4, and E5) strongly reduced responsiveness to DIMM, to a level equal to that from altering E1 alone (Fig. 2C). Sequence alteration of E2 or of E4 individually resulted in a decrease of approximately 50% (Fig. 2D). Thus, the three E boxes E1, E2, and E4 appeared to make the biggest contributions; we therefore compared their activities directly to the activity of the vector only (Fig. 2E). The single E2 or E4 mutation displayed about half the activity of the wild-type sequence, while the single E1 mutation and the double E2/E4 mutation had even greater effects and displayed only about 20% of that level. All the activities were significantly different from that of the empty vector control. Finally, to determine whether (i) the E-box position or (ii) its specific sequence was the critical determinant of functionality, we tested additional E-box sequence variants (Fig. 2F). Exchanging the sequence of E1 (plus its four adjacent bp [TCCATATGGA]) for that of the nonfunctional E7 (CCCATTTGAA) (Table 1) resulted in the loss of all DIMM responsiveness. Likewise, the exchange of both E2 and E3 with E7 or both E4 and E5 with E7 produced a PHM-enhancer fragment with half of its normal DIMM responsiveness (Fig. 2F), mimicking the effect of the loss of the paired sites.
TABLE 1.
Wild-type and mutant sequence probes representing the seven E box sites present in the first PHM introna
| E box(es) and probe | Sequence |
|---|---|
| E1 | |
| E1 box | 5′-cgc tcCATATGga tga-3′ |
| E1M box | 5′-cgc tcTATACTga tga-3′ |
| E1 to 2 box | 5′-cgc ccCAGCTGca tga-3′ |
| E1 to 7 box | 5′-cgc ccCATTTGaa tga-3′ |
| E2 and E3 | |
| E23 box | 5′-tcg ccCAGCTG CAGCTGtg cgc-3′ |
| E23M box | 5′-tcg ccTAGCCT TAGCCTtg cgc-3′ |
| E2M box | 5′-tcg ccTAGCCT CAGCTGtg cgc-3′ |
| E3M box | 5′-tcg ccCAGCTG TAGCCTtg cgc-3′ |
| E23 to 7 box | 5′-tcg ccCATTTG CATTTGaa cgc-3′ |
| E4 and E5 | |
| E45 box | 5′-ttt tcCATATGt tCAGTTGta ccc-3′ |
| E45M box | 5′-ttt tcTATACTt tTAGTCTta ccc-3′ |
| E4M box | 5′-ttt tcTATACTt tCAGTTGta ccc-3′ |
| E5M box | 5′-ttt tcCATATGt tTAGTCTta ccc-3′ |
| E45 to 7 box | 5′-tcg ccCATTTGt tCATTTGaa cgc-3′ |
| E6 | |
| E6 box | 5′-cca ccCACATGta agg-3′ |
| E6M box | 5′-cca ccTACACTta agg-3′ |
| E7 | |
| E7 box | 5′-act ccCATTTGaa taa-3′ |
| E7M box | 5′-act ccTATTCTaa taa-3′ |
The 6-bp E-box sequences are in uppercase and in order as named in column 1. Sequence alterations are shown in bold. Underlines indicate the final extent of sequence matching to the target E-box site (e.g., E1 to 2 box is the 10 bp surrounding E2).
In summary, the first intron of the PHM gene has seven E boxes. Three of these, E1, E2, and E4, proved especially important for the transactivation by DIMM protein in HEK 293 cells, but our experiments indicated that they make different contributions (E1 > E2 = E4). Additional data implicated the specific sequences within each of these E boxes, and not simply their positions, as critical determinants of their specific contributions. Together, these data indicate that the three cis regions within the first intron represent critical elements of a putative DIMM response element.
DIMM binds the E1 box of the PHM gene in vitro.
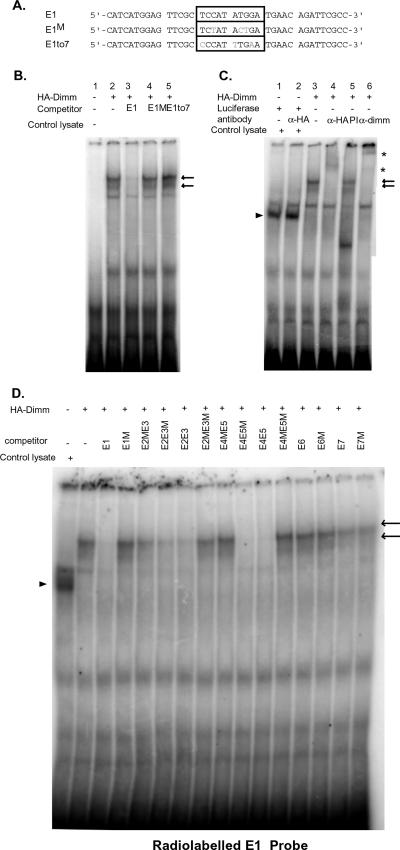
Next, we examined DIMM's ability to bind the relevant E boxes within the PHM intron (Fig. 3A to C). We performed EMSAs to address this question, using an E1-box probe. In vitro-translated DIMM bound the E1 probe specifically: the wild type, but not the sequence-mutated E1 probe, was able to compete such binding efficiently (Fig. 3B, lanes 2 to 4). Likewise, we observed poor competition by a cold E1 box probe in which the E-box sequence had been switched to that of E7 (Fig. 3B, lane 5). Supershift assays using either anti-HA or anti-DIMM antibodies confirmed the presence of DIMM protein (Fig. 3C).
FIG. 3.
Direct binding of DIMM to the E1 E box. (A) E1, the sequence of the probe representing the wild-type E1-box region; E1M and E1to7, the sequences of two E1-box variants that were used as competitors for EMSA. The box demarcates the E box and 2 bp on either side. (B) HA-tagged DIMM binds to the radiolabeled E1-box probe directly and specifically. (C) The E1-DIMM complex was supershifted by its specific antibody, and not by control antibodies. Arrows indicate the shifted bands; asterisks indicate supershifted bands. α, anti. (D) The E1 box competes differently with the other E boxes of the first intron of the PHM gene. Arrowheads in panels C and D indicate nonspecific bands observed when lysate (control or experimental) was added. These assays were performed twice, with similar results. +, present; −, absent.
We also examined the degree to which the DIMM-E1 interaction could be competed by the six other PHM intronic E-box sequences, E2 to E7. DIMM-E1 interaction was dose dependent (see Fig. S2 in the supplemental material). DIMM binding with E1 was competed efficiently by probes containing both E2 and E3 and also ones containing both E4 and E5 (Fig. 3D). Within the E2/E3 pair, the competition derived primarily but not exclusively from E2; within the E4/E5 pair, all the competing activity derived from E4 (Fig. 3D). We note that E2 and E3 share the same core 6-bp sequence, while the E4 and E5 sequences are different from each other (Table 1). The E6 and E7 oligonucleotides competed poorly with E1 binding and at levels that did not differ when they were tested following mutation of their specific E-box sequences (Fig. 3D). Next, we investigated the sequence requirements within the 10-bp E1 sequence (NNCANNTGNN) in greater detail by using site-directed mutagenesis. We found that specific sequences outside the canonical 6-bp E box (CANNTG) were required for activation by DIMM. Specifically, the nucleotides at the ±1, but not at the ±2 region, could not be altered without loss of DIMM activity. The results also indicated that, within the 6-bp sequence, DIMM prefers TA (and to a lesser extent, GC) to AT or CG for the internal NN sequences (see Fig. S3 in the supplemental material).
In summary, we found that DIMM directly binds E box E1 located within the first intron of PHM; this binding was efficiently competed by oligonucleotides corresponding to E2 and E4, but not by those corresponding to E3, E5, E6, or E7. These results further demonstrate differential E-box contributions to transactivation by DIMM, ranking them in an order consistent with the results from the luciferase reporter assays.
A 1-kb enhancer fragment from the first intron of PHM recapitulates the expression of the endogenous gene in vivo and is dependent on DIMM.
We next asked whether the PHM first intronic fragment contains sufficient cis-regulatory information to drive a normal PHM-like expression in vivo, particularly within NSCs that are strongly DIMM expressing. We established transgenic flies that contain the ∼1.0-kb PHM fragment as a GAL4 fusion (PHM-WT-GAL4) and crossed them into a UAS-GFP reporter background. Gene expression was studied by immunocytochemistry in the third instar larval brain. We found that PHM-WT-GAL4 was expressed normally throughout the brain (Fig. 4). GFP-expressing cells were predominantly DIMM positive (Fig. 4B). Forty-one percent of GFP+ cells in the brain and 91% of GFP+ cells in the ventral nerve cord (VNC), respectively, were colocalized with DIMM (Fig. 4D). Notably, most DIMM-positive cells were strongly GFP positive (in one representative specimen, we counted 189 GFP-positive cells among the 192 DIMM-positive cells [98.4%] throughout the entire CNS). The DIMM-negative, GFP-positive brain cells were primarily Kenyon cells of the larval mushroom bodies (MBs). PHM is found in moderate levels in both larval and adult MBs (49).
FIG. 4.
High coincidence between the expression pattern of the ∼1.0-kb PHM transgenic reporter and that of DIMM in the third instar larval CNS. The diagram at the top describes the PHM-gal4 construct. Arrows indicate the orientation of gene transcription, and the X indicates mutation of the PHM AUG sequence. (A to C) Double-staining with anti-DIMM and anti-GFP in the third instar larval brain of PHM-WT line. (D) Quantification of the coincidence of patterns in PHM-WT flies. About 41% of GFP+ [GFP(+)] cells in the brain were also DIMM+ (n = 112 total cells); more than 90% of those that were DIMM+ in the VNC are GFP+ (n = 156 total cells). The number of GFP−/DIMM+ cells was typically less than 1% and, for clarity, was not reported in the graphs.
Next, we wished to determine whether DIMM protein regulates the expression of the PHM transgene as it does the endogenous PHM locus, and so performed dimm loss-of-function analyses. The strongest dimm loss-of-function alleles display lethality in the early larval stages (21): in the second instar larval CNS, we observed a strong correspondence between DIMM protein expression and PHM-GAL4 transgene expression (Fig. 5A to C). This activity was greatly reduced in a strong dimm mutant background (Fig. 5D to F). Some residual GFP activity was detectable in a few neurons in mutant tissues; several of these cells were DIMM positive, likely reflecting that, even in the severe dimm hypomorphic background, a small number of DIMM-positive neurons remain. PHM transgene activity in Kenyon cells was not apparent at that age and so could not be scored in the dimm loss-of-function state. Together these data indicate that the DIMM-responsive ∼1.0-kb PHM fragment also drives patterned gene expression in vivo. The reporter activities are largely coincident with the expression of DIMM and are largely dependent upon normal DIMM expression.
FIG. 5.
The activity of the ∼1.0-kb transgenic PHM reporter largely depends on DIMM. PHM-GAL4 expression in the second instar larval CNS is much stronger in dimm heterozygotes (Rev8/+) (A to C) than in dimm transheterozygotes (Rev8/Rev4) (D to F). At least 10 samples were tested per genotype. Note that the PHM-GAL4 activity is not as prevalent in these second instar tissues as in the older tissues displayed in Fig. 4.
The PHM transgene also responds strongly to DIMM overexpression in vivo.
To analyze the effects of DIMM gain-of-function in vivo, we constructed PHM-GFP transgenic reporter lines using the same ∼1-kb PHM fragment. In the larval CNS, we again found high-level GFP expression largely coincident with and dependent on DIMM expression (see Fig. S4 in the supplemental material). Second, we found that within the CNS, misexpressed DIMM protein produced strong, ectopic PHM transgene expression (Fig. 6). In the VNC, AP is expressed in three cell groups: (i) the four-cell Tv cluster (Tv, Tva, Tvb, and Tvc), (ii) the ventral AP pair (vAP), and (iii) the dorsal single Aplet cell (dAP) (2, 40). Within the Tv cluster, DIMM is normally found in two cells, Tv and Tvb, but not in Tva or Tvc (the latter are here called Tv3 and Tv4 due to lack of identifying markers [Fig. 6]). Likewise, DIMM is normally found in the dAP, but not in vAP, cells. Using an ap-GAL4 driver, we found that PHM-GFP expression was responsive to DIMM overexpression in vivo throughout all four neurons of the Tv cluster (Fig. 6D) and in both vAP neurons (data not shown). This is consistent with the results of previous studies demonstrating that DIMM misexpression throughout all these AP neurons directs ectopic PHM protein in them all (2, 16). Notably, the mammalian homologue, MIST1, also showed an ability to activate the PHM-GFP reporter in vivo throughout all four cells of the Tv cluster (Fig. 6E and F) and in the vAP cells (data not shown).
FIG. 6.
Misexpressed DIMM drives ectopic expression of the PHM-GFP reporter in vivo. (A) Top diagram indicates the positions of two of the three normal ap cell groups present in three thoracic segments of the larval VNC: the four-cell Tv cluster (red circles) and the pair of vAP neurons (blue circles). Two of the Tv cluster neurons in the larval stage (called Tv and Tvb) are peptidergic, and both express DIMM and PHM (3, 44). The bottom diagram schematizes the expression of DIMM and PHM in the Tv cluster in the wild type (left) and following DIMM misexpression driven by ap-GAL4 (right). (B) ap-gal4 reporter (UAS-2X EGFP line) is expressed in all four cells of the Tv cluster. (C) Without ap-GAL4 activity, PHM-GFP displays normal restriction in the DIMM+ Tv and Tvb cells and not in the Tv3 or Tv4 cells and is not found in vAPs (not shown). (D) When DIMM is misexpressed in all ap neurons, PHM-GFP is seen in all four Tv cluster neurons and in the two vAP neurons (data not shown). Anti-MYC recognizes the epitope-tagged DIMM (UAS-dimm-myc). (E and F) Misexpressed MIST1, the mammalian homologue of DIMM, also drives PHM-GFP throughout the four-cell Tv cluster. (E) An example is shown without the UAS-Mist1 gene. (F) When the UAS-Mist1 gene was included, GFP was found to be elevated in all four Tv cells. The results shown are representative for at least five specimens studied per experiment. α, anti. In designations of genotypes, + indicates the wild type.
In summary, we found that in vivo, dimm regulated the activity of an ∼1-kb PHM regulatory fragment in a manner consistent with its regulation of the endogenous PHM protein. This further validates this ∼1-kb fragment as a primary response element mediating the transcriptional control of PHM by DIMM. Additionally, the mammalian orthologue Mist1 also activated this same PHM fragment in vivo, supporting the hypothesis that these related factors share functional properties.
Specific E-box sequences within the first PHM intron are essential for the activity of the PHM transgene.
We further asked which cis-regulatory sequences within this ∼1-kb PHM fragment contribute to its activity as a transgene in vivo. Based on the functional analysis in HEK 293 cells, three E boxes (E1, E2, and E4) are required for activation by DIMM. We therefore asked whether these specific cis-regulatory sequences also contribute to the activity of this PHM fragment in vivo. In the third instar larval brain, all activity was lost in flies bearing a GAL4 fusion of the 1-kb PHM fragment in which E1, E2, and E4 E boxes were mutated (n = 10; Fig. 7A to C). We note that activity was lost equally from the DIMM-positive and DIMM-negative (primarily MB) cells.
FIG. 7.
The three E boxes E1, E2, and E4 are essential for PHM-GFP activity in vivo. The in vivo activities of three transgenic PHM-gal4 lines were evaluated with UAS-2X EGFP in the CNS of third instar larvae. Xs in diagrams on the left indicate positions of site-directed alterations in the ∼1.0-kb PHM genomic fragment; all constructs altered the PHM AUG to prevent initiation at that site. PHM-WT was otherwise wild type in sequence. PHM-E1M, PHM-E2M, PHM-E4M, PHM-E24M, and PHM-E124M harbor alterations of specific E-box sequences. Each altered PHM transgene displayed significant reductions of promoter activity, as indicated by the results shown in the histogram on the right for the average number of GFP+ neurons (n = 5) observed in a representative line for each construct. Error bars show the standard errors of the means. **, P < 0.01. For each construct, there was good consensus in the results for independent transgenic lines: PHM-WT (5 lines), PHM-E1M (8 lines) PHM-E2M (3 lines), PHM-E4M (3 lines), PHM-E24M (3 lines), and PHM-E124M (10 lines).
To investigate contributions by individual E boxes or subsets of them, we also established four different PHM-gal4 lines harboring either single (E1M, E2M, and E4M) or double (E24M) E-box mutations. Analysis of the single E-box mutations revealed that reporter activity was very low in each, with, typically, a few weakly expressing cells at most (Fig. 7). The reporter activity in the double-mutant line was likewise very low. These results suggest that all three E boxes (E1, E2, and E4) are required for the normal activities of this PHM fragment in vivo and that the contribution of these E boxes is crucial.
DISCUSSION
The experiments reported here address the mechanisms underlying DIMM's regulatory functions within peptidergic NSCs in Drosophila. We have shown that the tissue-restricted bHLH factor DIMM works as a transcriptional activator and that PHM is a direct DIMM target both in heterologous mammalian cells and in vivo. We have also resolved the enhancer activity of PHM to a region that contains its first intron and further shown that three tandem E boxes within that intron are essential for full DIMM activation.
DIMM is a transcription factor that regulates PHM directly.
Class I bHLH factors (e.g., E12, E47, and HEB) are widely expressed, while class II bHLH proteins (e.g., MyoD, myogenin, Mash1, and NeuroD) exhibit tissue-restricted expression profiles. Class I and II bHLH proteins function as heterodimer complexes to regulate the expression of target genes by binding to E-box (CANNTG) DNA elements (16, 30). Some bHLH proteins also form homodimers; for example, HAIRY is thought to form homodimers exclusively (39, 46). Likewise, the Drosophila mesoderm regulator TWIST forms heterodimers with DA but also forms homodimers: these different TWIST molecular pairs produce different transcriptional readouts in vivo (12).
At present, we favor the hypothesis that the master regulatory functions of DIMM (such as its control of PHM) represent actions as a homomeric dimer or oligomer. Four lines of evidence support this conclusion. (i) Cotransfection of PHM-luc with dimm was sufficient to transactivate PHM in heterologous (mammalian) cells, (ii) whereas cotransfection with dimm and da produced a less robust transactivation (Fig. 2). (iii) In vitro, DIMM did not bind DA, but did bind itself efficiently when tested as a GST fusion protein or via coimmunoprecipitation (2). (iv) In the absence of a class I bHLH, DIMM was nevertheless able to bind to the PHM E1 probe in vitro, presumably as a homodimer (Fig. 2). It may be significant that the three E-box sequences indicated by functional analysis display palindromic core sequences, which is consistent with binding by a homomeric bHLH dimer. We also note that the DIMM mammalian sequence orthologue (called Mist1) forms heterodimers with class I bHLH factors under certain conditions but also forms homodimers to directly regulate the fate of developing acinar cells of the pancreas (29, 45, 56).
The significance of PHM as a direct regulatory target of the prosecretory factor DIMM.
In the context of establishing transmitter phenotypes, there is special significance to having established direct transcriptional regulation of PHM by DIMM. DIMM and PHM, among those neurons that display high levels of neuropeptide expression, display highly congruent patterns of expression within the nervous system (2, 21). PHM is a monooxygenase that is required for neuropeptide amidation and whose closest homologue is the hydroxylase DBH. Phox2a and Phox2b are critical regulators of the noradrenergic phenotype and have been shown to directly regulate DBH transcription (27, 48, 53, 55). Likewise, PET-1, which is a critical regulator of the serotonergic phenotype, directly regulates the transcription of tryptophan hydroxylase, which encodes the rate-limiting enzyme for serotonin biosynthesis (18). In each case cited, genes encoding biosynthetic hydroxylases appear to be critical points of regulation. Thus, direct transcriptional control of such enzymes by highly dedicated developmental regulators represents a mechanistic parallel between aminergic and peptidergic transmitter cell fates.
DIMM binding sites on PHM.
We propose that the high level of PHM expression found in Drosophila NSCs results from direct control by DIMM acting via specific PHM cis-regulatory elements (Fig. 8A). The full activity of Class 2 bHLH proteins often requires interactions with multiple tandem E boxes (37), although single E boxes are sometimes sufficient (54). In the current study, we identified a cluster of seven E boxes in the enhancer region of PHM, of which at least three (E1, E2, and E4) are critical. bHLH proteins exhibit specificity as to which E-box sequences they utilize (44). The loss of these three motifs in PHM resulted in a substantial reduction in luciferase reporter assays and transgenic flies, indicating that there is an essential requirement of multiple E boxes for PHM activation by DIMM. We observed small differences in the activities of mutated PHM regulatory regions when tested in vitro versus in vivo. Focusing on the in vivo results, we propose a simple model of synergistic or cooperative interactions between DIMM homodimers bound to E1, E2, and E4 (Fig. 8A).
FIG. 8.
A proposed mechanism and model for DIMM activity. (A) DIMM homodimers activate PHM directly via three specific E boxes (E1, E2, and E4). Presumed coactivators present in HEK 293 cells and in vivo are not shown. In vitro analysis showed that the three E boxes contribute differentially to the total amount of transactivation: the E1 box is absolutely required for in vitro activity, while E2 and E4 have only partial activity. To display full activity, the DIMM-bound E1 box must interact with E2-bound DIMM and E4-bound DIMM independently and cooperatively. However, in vivo, all three E boxes contribute equally to the activity. (B) DIMM plays two distinct roles in the regulation of neuroendocrine cell fate. (i) The left panel shows a “core program” of neuroendocrine differentiation that defines that set of genes that is utilized by all cells displaying an amidated peptide secretory cell fate. It is directly and completely controlled by DIMM. The core program consists of PHM and other as-yet-unidentified gene products. (ii) The right panel shows a “shell program” of neuroendocrine differentiation that defines that set of genes which varies among neuroendocrine cell types and which are controlled in combinatorial fashion. DIMM contributes partial control in concert with diverse other transcription factors which are symbolized as different shapes. Genes in the shell program include neuropeptide-encoding genes, as well as other as-yet-unidentified ones.
Mutation of the E3 and E5 E boxes produced elevated PHM-luc levels upon dimm cotransfections. The simplest explanation is that these E boxes bind one or more bHLH proteins that can repress PHM expression. The spacing of these potentially “inhibitory” E boxes, approximately one-half turn away from the “activating” E2 and E4 boxes, respectively, may be significant in effecting such negative regulation. Interestingly, Mist1 functions as a transcriptional repressor when bound as an E47 heterodimer to troponin E boxes (29). DA cotransfection reduced the DIMM transactivation of PHM: one possible explanation for this effect is that DA may bind to one or more of the “inhibitory” PHM E boxes. Whether and how other factors modulate the level of DIMM's activation by binding PHM E3 and/or E5 in vivo will require additional studies.
PHM regulatory elements tested in vivo.
We extended the scope of our analysis by considering the behavior of the ∼1-kb PHM fragment in vivo. Importantly, DIMM regulation of PHM in Drosophila cells paralleled its activity in HEK 293 cells. Although PHM regulation in vivo is complex and subject to multiple influences, these data suggest that much of PHM expression in Drosophila NSCs can be ascribed to DIMM direct regulation via the ∼1-kb PHM fragment that includes its first intron. Significantly, most DIMM-positive cells were PHM-GAL4-positive cells, suggesting a strong correlation between DIMM and the same 1-kb PHM fragment that displayed transactivation in vitro. In addition, the promoter activity that could be assayed in second instar larvae (before dimm mutant animals die) was essentially all DIMM dependent: the few resilient GFP-positive cells that remained in dimm mutants were those that also retained DIMM protein—the strongest dimm alleles are not protein nulls. As found in HEK 293 cells, specific E boxes appeared to be especially important in vivo as well. The same three PHM E boxes that were implicated from in vitro experiments (E1, E2, and E4) were again implicated as critical for supporting PHM regulatory activity in vivo. This work therefore defines the PHM enhancer region and the constituent cis-regulatory sequences therein that confer sensitivity to DIMM. The data also indicate additional complexity of PHM regulation. The reliable incidence of GFP-positive/DIMM-negative neurons (especially some or all MB cells in the brain lobes) indicated that other factors besides DIMM may regulate PHM expression in other domains of the CNS. Interestingly, the loss of PHM transgene expression by MB neurons in flies bearing mutated PHM E boxes suggests that the regulatory factor(s) driving PHM in MBs may also be a bHLH protein(s).
What is the precise role of DIMM in the differentiation of NSC?
In mammals, several transcription factors have been implicated in directing the differentiation of neuroendocrine lineages (1, 8, 24, 26, 31, 32, 36, 47, 52). Based on information so far available, DIMM does not resemble any known mammalian regulator of neuroendocrine development. For example, while DIMM is activated postmitotically in NSCs, Mash1 appears required for the generation of pulmonary neuroendocrine cells (8). Like dimm, Sim1 and Otp are thought to act in parallel as determinants of cell fate in hypothalamic neuroendocrine cells and are needed for cell generation or survival (1, 32, 52). Recently, Sim1 mutant cells were shown to reach their final cell division with normal display of their molecular phenotype but subsequently fail to differentiate as a function of altered cell migration (56). These factors, therefore, appear to act early and prior to the terminal differentiation of the peptidergic cells.
Based on several observations of normal expression and from genetic analyses, we consider DIMM to be the major regulator of terminal cell differentiation in Drosophila NSCs. Our previous work proposed two, nonexclusive mechanisms to explain dimm actions. One is a model of transcription factor combinatorial regulation to drive specific neuropeptide expression (e.g., dFMRFa and NPLP1) (2, 5) in different neurons. In this scenario, DIMM acts combinatorially with several locally expressed transcription factors to drive neuropeptide gene expression. In no case, however, has it been possible to detail the precise identities of any particular combination regulating one specific neuropeptide gene. DIMM also acts independently to regulate the transcription of genes that control common NSC traits. dimm is a “master regulator,” as its overexpression confers high-level PHM expression in all neurons of the CNS (2). In this dual manner, we speculate that dimm contributes both to specific cellular properties of diverse NSCs (e.g., neuropeptide gene selection—a “shell program” of neuroendocrine differentiation) and to more general ones involving generic NSC properties (e.g., PHM expression—the “core program” of neuroendocrine differentiation) (Fig. 8B). The present results in vitro and in vivo strongly support the second model of dimm actions: we have shown that the control of PHM protein expression derives from the very strong, direct regulation of PHM transcription. Given its potent transcriptional activity, we suspect that DIMM promotes a general NSC phenotype by also activating target genes besides PHM, but this supposition will require further analysis.
The relationship of DIMM and the mammalian bHLH Mist1.
Among mammalian bHLH proteins, Mist1 is the most similar to DIMM (34). At the sequence level, the bHLH regions of DIMM and Mist1 exhibit greater than 90% sequence identity. Both proteins are enriched in cells that are specialized for secretion, and genetic analyses suggest that both dimm and Mist1 are required for the proper expression of secretory properties by the cells that express them (21, 42, 43, 44a). Also, we showed that Mist1 can transactivate Drosophila PHM in HEK 293 cells with 50% of the activity displayed by DIMM and was able to modestly activate the PHM-GFP reporter in vivo. Notably, no other transcription factors that we tested (including other, closely related Drosophila bHLHs) displayed any effect on PHM transcription in vitro.
Even within Drosophila tissues, dimm is not the sole regulator of all “peptidergic” NSC fates. MB neurons express PHM (49) and specific neuropeptides (as studied in other insects [38]), yet they do not contain DIMM. In addition, other known peptidergic neurons, like the small LNv neurons expressing PDF (49) or the proctolin-containing neurons (D. Park and P. H. Taghert, unpublished data) do not contain DIMM. Thus, the fly nervous and endocrine systems appear divisible into different peptidergic cell domains, as defined by different developmental transcriptional regulators. In Drosophila, the dimm expression domain is large and diverse, containing many different kinds of NSCs, but there must be at least one more domain whose regulatory transcription factors as yet await identification.
Our studies of DIMM transcriptional activities suggest the hypothesis that there exists a core regulatory program underlying the differentiation and organization of NSCs. Many proteins are enriched in neuroendocrine tissues, and some are coordinately regulated under different physiological states (23). These differentiated gene batteries result from the activity of complex regulatory circuits whose identities and details are largely unknown. Which of these many proteins are primary targets of neuroendocrine cell-type transcriptional control (like DIMM) and which are derivative targets? We anticipate that further analysis of the core gene expression program driven by DIMM will help to explain the organization and evolution of NSCs and other neuroendocrine cell types. DIMM protein persists in neurons through adulthood (D. Park and P. H. Taghert, unpublished observations); studies of DIMM may therefore also help to address mechanisms of peptidergic cell physiology and plasticity.
Supplementary Material
Acknowledgments
We thank Lou Muglia, Ross Cagan, and Joe Corbo for critically reading the manuscript. We thank Anneliese M. Schaefer, Jason Mills, Minho Lee, Claire Cronmiller, Stephen F. Konieczny, Susan Abmayr, James W. Posakony, and Erik C. Johnson for reagents and comments and Weihua Li and Jungsook Yoon for technical assistance. We also thank the Bloomington Stock center for flies and the Drosophila Genome Center for providing information.
This work was supported by NIH Neuroscience Blueprint Core grant P30 NS057105 to Washington University. Orie T. Shafer was supported by NIH grant no. F32 NS53222. Stacie P. Shepherd was supported by NIH institutional training grant no. T32 DK063706. This work was supported by a grant (NS21749) from the NINDS of the National Institutes of Health to P.H.T.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acampora, D., M. P. Postiglione, V. Avantaggiato, M. Di Bonito, F. M. Vaccarino, J. Michaud, and A. Simeone. 1999. Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the Orthopedia gene. Genes Dev. 132787-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, D. W., D. Park, S. E. St. Pierre, P. H. Taghert, and S. Thor. 2005. Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron 45689-700. [DOI] [PubMed] [Google Scholar]
- 3.Allan, D. W., S. E. St. Pierre, I. Miguel-Aliaga, and S. Thor. 2003. Specification of neuropeptide cell identity by the integration of retrograde BMP signaling and a combinatorial transcription factor code. Cell 11373-86. [DOI] [PubMed] [Google Scholar]
- 4.Barolo, S., L. A. Carver, and J. W. Posakony. 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29726-732. [DOI] [PubMed] [Google Scholar]
- 5.Baumgardt, M., I. Miguel-Aliaga, D. Karlsson, H. Ekman, and S. Thor. 2007. Specification of neuronal identities by feedforward combinatorial coding. PLoS Biol. 5e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benveniste, R. J., S. Thor, J. B. Thomas, and P. H. Taghert. 1998. Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development 1254757-4765. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand, N., D. S. Castro, and F. Guillemot. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3517-530. [DOI] [PubMed] [Google Scholar]
- 8.Borges, M., R. I. Linnoila, H. J. van de Velde, H. Chen, B. D. Nelkin, M. Mabry, S. B. Baylin, and D. W. Ball. 1997. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386852-855. [DOI] [PubMed] [Google Scholar]
- 9.Burbach, G. J., K. H. Kim, A. S. Zivony, A. Kim, J. Aranda, S. Wright, S. M. Naik, S. W. Caughman, J. C. Ansel, and C. A. Armstrong. 2001. The neurosensory tachykinins substance P and neurokinin A directly induce keratinocyte nerve growth factor. J. Investig. Dermatol. 1171075-1082. [DOI] [PubMed] [Google Scholar]
- 10.Cabrera, C. V., and M. C. Alonso. 1991. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 102965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campuzano, S., L. Carramolino, C. V. Cabrera, M. Ruiz-Gomez, R. Villares, A. Boronat, and J. Modolell. 1985. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell 40327-338. [DOI] [PubMed] [Google Scholar]
- 12.Castanon, I., S. Von Stetina, J. Kass, and M. K. Baylies. 2001. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 1283145-3159. [DOI] [PubMed] [Google Scholar]
- 13.Caudy, M., H. Vassin, M. Brand, R. Tuma, L. Y. Jan, and Y. N. Jan. 1988. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell 551061-1067. [DOI] [PubMed] [Google Scholar]
- 14.Cronmiller, C., P. Schedl, and T. W. Cline. 1988. Molecular characterization of daughterless, a Drosophila sex determination gene with multiple roles in development. Genes Dev. 21666-1676. [DOI] [PubMed] [Google Scholar]
- 15.Eipper, B. A., B. T. Bloomquist, E. J. Husten, S. L. Milgram, and R. E. Mains. 1993. Peptidylglycine alpha-amidating monooxygenase and other processing enzymes in the neurointermediate pituitary. Ann. N. Y. Acad. Sci. 680147-160. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, A., and M. Caudy. 1998. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays 20298-306. [DOI] [PubMed] [Google Scholar]
- 17.Gauthier, S. A., and R. S. Hewes. 2006. Transcriptional regulation of neuropeptide and peptide hormone expression by the Drosophila dimmed and cryptocephal genes. J. Exp. Biol. 2091803-1815. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks, T., N. Francis, D. Fyodorov, and E. S. Deneris. 1999. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J. Neurosci. 1910348-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero, P., M. Magarinos, L. Torroja, and I. Canal. 2003. Neurosecretory identity conferred by the apterous gene: lateral horn leucokinin neurons in Drosophila. J. Comp. Neurol. 457123-132. [DOI] [PubMed] [Google Scholar]
- 20.Hewes, R. S., T. Gu, J. A. Brewster, C. Qu, and T. Zhao. 2006. Regulation of secretory protein expression in mature cells by DIMM, a basic helix-loop-helix neuroendocrine differentiation factor. J. Neurosci. 267860-7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewes, R. S., D. Park, S. A. Gauthier, A. M. Schaefer, and P. H. Taghert. 2003. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development 1301771-1781. [DOI] [PubMed] [Google Scholar]
- 22.Hewes, R. S., and P. H. Taghert. 2001. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 111126-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hindmarch, C., S. Yao, G. Beighton, J. Paton, and D. Murphy. 2006. A comprehensive description of the transcriptome of the hypothalamoneurohypophyseal system in euhydrated and dehydrated rats. Proc. Natl. Acad. Sci. USA 1031609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosoya, T., Y. Oda, S. Takahashi, M. Morita, S. Kawauchi, M. Ema, M. Yamamoto, and Y. Fujii-Kuriyama. 2001. Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes Cells 6361-374. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, N., A. S. Kolhekar, P. S. Jacobs, R. E. Mains, B. A. Eipper, and P. H. Taghert. 2000. PHM is required for normal developmental transitions and for biosynthesis of secretory peptides in Drosophila. Dev. Biol. 226118-136. [DOI] [PubMed] [Google Scholar]
- 26.Keith, B., D. M. Adelman, and M. C. Simon. 2001. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc. Natl. Acad. Sci. USA 986692-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, H. S., H. Seo, C. Yang, J. F. Brunet, and K. S. Kim. 1998. Noradrenergic-specific transcription of the dopamine beta-hydroxylase gene requires synergy of multiple cis-acting elements including at least two Phox2a-binding sites. J. Neurosci. 188247-8260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolhekar, A. S., M. S. Roberts, N. Jiang, R. C. Johnson, R. E. Mains, B. A. Eipper, and P. H. Taghert. 1997. Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J. Neurosci. 171363-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemercier, C., R. Q. To, R. A. Carrasco, and S. F. Konieczny. 1998. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 171412-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michaud, J. L., C. DeRossi, N. R. May, B. C. Holdener, and C. M. Fan. 2000. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 90253-261. [DOI] [PubMed] [Google Scholar]
- 32.Michaud, J. L., T. Rosenquist, N. R. May, and C. M. Fan. 1998. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 123264-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miguel-Aliaga, I., D. W. Allan, and S. Thor. 2004. Independent roles of the dachshund and eyes absent genes in BMP signaling, axon pathfinding and neuronal specification. Development 1315837-5848. [DOI] [PubMed] [Google Scholar]
- 34.Moore, A. W., S. Barbel, L. Y. Jan, and Y. N. Jan. 2000. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc. Natl. Acad. Sci. USA 9710436-10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin, X., H. Cremer, M. R. Hirsch, R. P. Kapur, C. Goridis, and J. F. Brunet. 1997. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron 18411-423. [DOI] [PubMed] [Google Scholar]
- 36.Nakai, S., H. Kawano, T. Yudate, M. Nishi, J. Kuno, A. Nagata, K. Jishage, H. Hamada, H. Fujii, K. Kawamura, et al. 1995. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 93109-3121. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima, Y., M. Morimoto, Y. Takahashi, H. Koseki, and Y. Saga. 2006. Identification of Epha4 enhancer required for segmental expression and the regulation by Mesp2. Development 1332517-2525. [DOI] [PubMed] [Google Scholar]
- 38.Nassel, D. R., and U. Homberg. 2006. Neuropeptides in interneurons of the insect brain. Cell Tissue Res. 3261-24. [DOI] [PubMed] [Google Scholar]
- 39.Ohsako, S., J. Hyer, G. Panganiban, I. Oliver, and M. Caudy. 1994. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 82743-2755. [DOI] [PubMed] [Google Scholar]
- 40.Park, D., M. Han, Y. C. Kim, K. A. Han, and P. H. Taghert. 2004. Ap-let neurons: a peptidergic circuit potentially controlling ecdysial behavior in Drosophila. Dev. Biol. 26995-108. [DOI] [PubMed] [Google Scholar]
- 41.Pattyn, A., X. Morin, H. Cremer, C. Goridis, and J. F. Brunet. 1999. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature 399366-370. [DOI] [PubMed] [Google Scholar]
- 42.Pin, C. L., A. C. Bonvissuto, and S. F. Konieczny. 2000. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat. Rec. 259157-167. [DOI] [PubMed] [Google Scholar]
- 43.Pin, C. L., J. M. Rukstalis, C. Johnson, and S. F. Konieczny. 2001. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J. Cell Biol. 155519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell, L. M., P. I. Zur Lage, D. R. Prentice, B. Senthinathan, and A. P. Jarman. 2004. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell. Biol. 249517-9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Ramsey, V. G., J. M. Doherty, C. C. Chen, T. S. Stappenbeck, S. F. Konieczny, and J. C. Mills. 2007. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development 134211-222. [DOI] [PubMed] [Google Scholar]
- 45.Rukstalis, J. M., A. Kowalik, L. Zhu, D. Lidington, C. L. Pin, and S. F. Konieczny. 2003. Exocrine specific expression of Connexin32 is dependent on the basic helix-loop-helix transcription factor Mist1. J. Cell Sci. 1163315-3325. [DOI] [PubMed] [Google Scholar]
- 46.Rushlow, C. A., A. Hogan, S. M. Pinchin, K. M. Howe, M. Lardelli, and D. Ish-Horowicz. 1989. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 83095-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schonemann, M. D., A. K. Ryan, R. J. McEvilly, S. M. O'Connell, C. A. Arias, K. A. Kalla, P. Li, P. E. Sawchenko, and M. G. Rosenfeld. 1995. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 93122-3135. [DOI] [PubMed] [Google Scholar]
- 48.Swanson, D. J., E. Zellmer, and E. J. Lewis. 1997. The homeodomain protein Arix interacts synergistically with cyclic AMP to regulate expression of neurotransmitter biosynthetic genes. J. Biol. Chem. 27227382-27392. [DOI] [PubMed] [Google Scholar]
- 49.Taghert, P. H., R. S. Hewes, J. H. Park, M. A. O'Brien, M. Han, and M. E. Peck. 2001. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J. Neurosci. 216673-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taghert, P. H., and L. E. Schneider. 1990. Interspecific comparison of a Drosophila gene encoding FMRFamide-related neuropeptides. J. Neurosci. 101929-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taghert, P. H., and J. A. Veenstra. 2003. Drosophila neuropeptide signaling. Adv. Genet. 491-65. [DOI] [PubMed] [Google Scholar]
- 52.Wang, W., and T. Lufkin. 2000. The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev. Biol. 227432-449. [DOI] [PubMed] [Google Scholar]
- 53.Yang, C., H. S. Kim, H. Seo, C. H. Kim, J. F. Brunet, and K. S. Kim. 1998. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J. Neurochem. 711813-1826. [DOI] [PubMed] [Google Scholar]
- 54.Yoo, S. H., C. H. Ko, P. L. Lowrey, E. D. Buhr, E. J. Song, S. Chang, O. J. Yoo, S. Yamazaki, C. Lee, and J. S. Takahashi. 2005. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA 1022608-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zellmer, E., Z. Zhang, D. Greco, J. Rhodes, S. Cassel, and E. J. Lewis. 1995. A homeodomain protein selectively expressed in noradrenergic tissue regulates transcription of neurotransmitter biosynthetic genes. J. Neurosci. 158109-8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, L., T. Tran, J. M. Rukstalis, P. Sun, B. Damsz, and S. F. Konieczny. 2004. Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Mol. Cell. Biol. 242673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.