Abstract
Hbo1 is a histone acetyltransferase (HAT) that is required for global histone H4 acetylation, steroid-dependent transcription, and chromatin loading of MCM2-7 during DNA replication licensing. It is the catalytic subunit of protein complexes that include ING and JADE proteins, growth regulatory factors and candidate tumor suppressors. These complexes are thought to act via tumor suppressor p53, but the molecular mechanisms and links between stress signaling and chromatin, are currently unknown. Here, we show that p53 physically interacts with Hbo1 and negatively regulates its HAT activity in vitro and in cells. Two physiological stresses that stabilize p53, hyperosmotic shock and DNA replication fork arrest, also inhibit Hbo1 HAT activity in a p53-dependent manner. Hyperosmotic stress during G1 phase specifically inhibits the loading of the MCM2-7 complex, providing an example of the chromatin output of this pathway. These results reveal a direct regulatory connection between p53-responsive stress signaling and Hbo1-dependent chromatin pathways.
The dynamic regulation of chromatin structure and function is essential for normal cell proliferation and differentiation. This regulation is mediated by several overlapping pathways, including the posttranslational enzymatic modification of histones, the alteration of nucleosome structure by DNA-dependent ATPase complexes, and changes in the histone variant composition of chromatin (4, 34, 58). Alterations in histone modification enzymes, particularly histone acetyltransferase (HAT) enzymes, have been linked to human cancer (23, 73). Viral oncoproteins, such as adenovirus E1A or simian virus 40 large T antigen, target a number of these enzymes, including p300, CBP, and PCAF. Furthermore, in addition to modifying histones, these HATs can also directly acetylate and activate tumor suppressors and key growth control transcription factors such as p53, Rb, and E2F (24). The MYST family of histone acetyltransferases, named for the four founding proteins in the family (67), can also contribute to carcinogenesis and tumor progression. The MYST proteins are part of large multisubunit HAT complexes conserved from yeast to humans, and they have diverse roles in gene expression, DNA replication, and DNA repair (72). The human MOZ gene, encoding one of the human MYST enzymes, was first identified as a translocation fusion with CBP in acute myeloid leukemias (9). Subsequently, a number of other translocation fusions involving the MYST HATs MOZ and MORF and partners including CBP, p300, MLL, and TIF2 have been identified. It is thought that the mislocalization or misregulation of the HAT activities of these fusions contributes to tumor formation or progression (72).
Hbo1 is a member of the MYST family of HAT enzymes and is conserved from flies to humans. It has essential roles in DNA replication and transcription (1, 12, 22, 32, 55, 59) and is the catalytic subunit of at least two protein complexes comprised of JADE1/JADE2/JADE3 paralogs, hEaf6, and either ING4 or ING5, two members of the “inhibition-of-growth” (ING) tumor suppressor protein family (17). Hbo1 was originally identified through its physical interactions with the human DNA replication proteins ORC1 and MCM2 (12, 32). A critical step in DNA replication is the formation of a prereplicative complex (pre-RC) involving the sequential assembly of the origin recognition complex, Cdc6/Cdcl8, Cdtl, and the minichromosome maintenance (MCM2-7) complex. The assembly of the pre-RC on replication origins confers a license for subsequent replication initiation. Disassembly of the pre-RC following initiation ensures that replication occurs only once per cell cycle (41). There is increasing evidence that chromatin modulation plays important roles in DNA replication (for a review, see reference 63). Recently, we discovered that Hbo1 is required for the chromatin loading of the MCM2-7 complex, the final step in pre-RC assembly and DNA replication licensing (31). Depletion of Hbo1 in human cells and in Xenopus egg extracts specifically blocked MCM2-7 assembly into the pre-RC and inhibited DNA replication. Furthermore, this defect could be corrected in egg extracts by the addition of excess Cdt1, a key positive regulator of pre-RC assembly. Thus, Hbo1 function regulates the pathway, ensuring that DNA replication occurs once, and only once, per cell division cycle.
Members of the JADE and ING protein families are thought to function, at least in part, by interacting with other human tumor suppressors. For example, ING4 and ING5 have been shown to physically interact with the human tumor suppressor p53 and potentiate its activity (19, 56, 57). Similarly, Jade1 has been found to stabilize pVHL, the product of the von Hippel-Lindau tumor suppressor gene, which in turn can stabilize and activate p53 (51, 76). The tumor suppressor p53 is a stress-induced regulatory protein which, upon a variety of cellular stresses, can trigger cell cycle arrest or apoptosis (69). These functions are significantly compromised by somatic mutations in cancers, and approximately half of all human tumors carry mutated alleles of p53. The best-understood role of p53 is as a sequence-specific DNA-binding transcription factor, which regulates a number of key cell cycle and apoptotic genes (28, 69). However, p53 may also function through transcription-independent pathways to maintain genome integrity (33). The best studied of these is its cytoplasmic role in regulating apoptosis (45), but transcription-independent functions in homologous recombination, replication, and DNA damage checkpoint responses have also been suggested (5, 15, 25, 49, 54, 75).
There are strong connections between chromatin regulation and p53 function. The stability and activity of p53 are known to be regulated by several histone modification factors, including the p300/CBP and PCAF HAT complexes and the Set9 and Smyd2 methylase complexes (2, 13, 24, 26, 30, 39, 53). Recently, two different members of the MYST family HATs, Tip60 and Mof, were shown to acetylate p53 and change its transcriptional targets from genes that favor cell cycle arrest to those that promote apoptosis (62, 64). Considerably less is known about downstream p53-dependent pathways that might, in turn, regulate the activity of histone modification complexes. Thus, at present, it is unclear how such potent cellular factors as Hbo1, ING4/5, Jade1/Jade2/Jade3, and p53 interact to regulate cell proliferation, and there are currently no known links between Hbo1 and p53. While we were investigating the roles of Hbo1 in DNA replication, we discovered that p53 copurifies with protein complexes immunoprecipitated with Hbo1 antisera. Here, we report studies designed to characterize the regulation of Hbo1, its physical and functional interactions with p53, and the stress signals that feed into the pathway. The results of these experiments show that p53 physically interacts with Hbo1 and down-regulates its HAT activity both in vitro and in cells. Furthermore, we show that physiological stresses that activate p53 are coupled to decreased Hbo1 HAT activity in a p53-dependent manner. In particular, these results define a previously unrecognized pathway linking the cell stress response to replication control at the level of pre-RC assembly via p53 and Hbo1.
MATERIALS AND METHODS
Mouse Hbo1 cDNA.
A mouse dbEST cDNA (dbEST accession number 8230062; GenBank accession number BG519441), which showed extensive homology with the human Hbo1 protein by blastX search, was purchased (Research Genetics).
Plasmids.
His-tagged Hbo1 truncations were cloned into plasmid pET19Spe (a gift from Masumi Hidaka). His-Hbo1(G485A) was created by PCR using mutated primers. Glutathione S-transferase (GST)-tagged p53 deletion mutants were PCR amplified from a wild-type human p53 plasmid (a gift from Takashi Kohno) or mouse p53 and cloned into plasmid pET19GST (a gift from Masumi Hidaka). The Hbo1 and p53 truncations were verified by DNA sequencing.
Antibodies and peptides.
Rabbit antibodies to human Orc2 and Mcm2 were provided by B. Stillman, and a rabbit antibody to Cdtl was obtained from H. Nishitani. The following reagents were obtained commercially: anti-histidine epitope tag (H-3), polyclonal antigeminin (FL-209), monoclonal anti-Cdc6 (D-l), and anti-Mek2 (N-20) (Santa Cruz Biotechnology); monoclonal anti-p53 (BP53-12), polyclonal anti-H2B, and monoclonal anti-ING1 (CAb3) (Upstate Biotechnology); anti-PMS2, anti-Mcm6, monoclonal anti-p53 (polyclonal antibody [PAb] 1801), and monoclonal anti-C-terminal p53 (PAb 122) (BD Pharmingen); mouse monoclonal antibody against α-tubulin (DM1A) and γ-tubulin (GTU-88) (Sigma); polyclonal anti-ING4 (Abcam); polyclonal anti-ING5 (Rockland Immunochemicals); monoclonal anti-p21 (EA10) (Calbiochem); monoclonal anti-p53 (PAb 122) (BD Biosciences); and monoclonal anti-Mdm2 (D-7) (Santa Cruz). Peptide L74-1 (residues 158 to 172 of human Hbo1) (32) and peptide MBP4-14 (human myelin basic protein, residues 4 to 14) were synthesized by Sawady Technology (Tokyo, Japan). Affinity-purified or crude rabbit polyclonal antiserum (CS445) against Hbo1 peptide L74-1 (32) was used for Western blots. Fusion protein GST-p53(1-393) was purchased from Santa Cruz.
Protein purification.
His-tagged Hbo1(1-611) and His-Hbo1(311-611) were expressed in bacteria and purified on heparin-Sepharose (Amersham Pharmacia) and Ni-nitrilotriacetic acid-agarose (Qiagen). GST-tagged p53 truncations were expressed in bacteria and purified on glutathione-Sepharose (Amersham Pharmacia).
GST pull-down assay.
Two microliters of settled and washed glutathione beads (Amersham Pharmacia) was incubated with 2 μg of GST-tagged protein in 0.5 ml buffer (20 mM Tris-Cl [pH 7.5], 0.2 M NaCl, 10% glycerol, 0.05% NP-40, 1 mM dithiothreitol) for 2 h at 4°C. After extensive washing with buffer, the beads were resuspended in 0.5 ml buffer and incubated with 0.5 μg of input protein for 4 h at 4°C. After extensive washing, bound proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and blotted with antibody to the His epitope tag.
Cell lines, transfection, immunoprecipitation, and HAT assays.
HeLa, MCF7, Saos-2, and NIH 3T3 cell lines were purchased from the ATCC. For immunoprecipitation and HAT assays or Western blot assays, 1 μg of DNA was transfected per 10-cm plate using Lipofectamine Plus according to the manufacturer's instructions (Life Technologies). Forty-eight hours posttransfection, cells were lysed with buffer (20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40). Immunoprecipitation and HAT assays were performed as reported previously (32). Human Hat1 holoenzyme, purified from 293 cells (68), was a gift from Alain Verreault. Ethidium bromide (400 μg/ml) was added to immunoprecipitation reaction mixtures and washes to prevent protein-nucleic acid interactions (38). For peptide competition in immunoprecipitation, 2 mg/ml of peptides was included in the immunoprecipitation and the first washing step. For assays of HAT inhibition, GST-p53 (Santa Cruz Biotechnology) was added to reaction mixtures prior to the addition of HAT enzymes. For p53 induction, MCF7 cells were treated with either 1.5 mM hydroxyurea (HU) (Sigma) or 0.26 M NaCl, and mouse embryo fibroblast (MEF) cells were treated with either 10 mM HU or 0.24 M NaCl. Acetylation reactions and Western blot assays were quantified by digitizing developed films using an Epson Perfection 4180 photo scanner and integration of the pixel intensities of the bands.
Adenoviruses and siRNA.
Information on the construction of the adenoviruses is available upon request. Replication-deficient recombinant viruses were created as described previously (27). Adenoviral stocks were maintained as described previously (47) and purified by cesium chloride density gradient centrifugation. Viruses were used at a multiplicity of infection of 100. SignalSilence p53 small interfering RNA (siRNA) (human specific) (catalog number 6231) was purchased from Cell Signaling and used for the inhibition of p53 expression. The control oligonucleotide for the siRNA experiments was siCONTROL Non-Targeting siRNA no. 1 (catalog number D-001210-01-05), which was purchased from Dharmacon. MCF7 cells were transfected with siRNA oligonucleotides using RNAiMAX (Invitrogen) and incubated for 48 to 72 h to achieve the inhibition of p53 expression.
Growth of MEFs.
Wild-type p53+/+ and homozygous p53−/− MEFs were prepared from C57BL/6 wild-type and p53 knockout mice (Taconic) as described previously (42). Cells were collected for HAT assays and protein analysis at passage 3. For HAT assays, a total of 5 × 106 cells were lysed with M-PER (Pierce) supplemented with 400 mM NaCl and a 1:100 dilution of protease inhibitor cocktail for mammalian tissues (Sigma). A total of 2 × 105 cells were resuspended in 2× Laemmli buffer with β-mercaptoethanol for whole-cell protein and Western blot analyses.
Nucleotide sequence accession number.
The sequences determined here were deposited in the GenBank database under accession number DQ076247.
RESULTS
Hbo1 interacts with p53 in human cells.
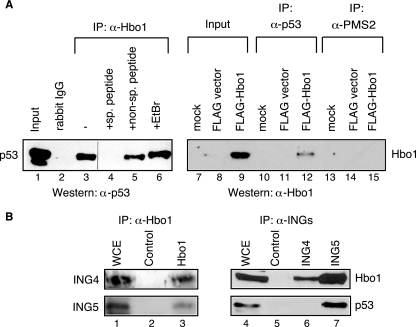
We reasoned that tumor suppressor p53 was a strong candidate for interactions with Hbo1, given the role of Hbo1 in cell proliferation and its association with ING4 and ING5 (17, 31). To test this hypothesis, anti-Hbo1 immunoprecipitates were prepared from whole-cell extracts. The proteins were resolved by SDS-PAGE and assayed by Western blotting using a monoclonal antibody against p53 (BP53-12). As shown in Fig. 1A (lanes 1 to 6), Hbo1 antibody specifically coimmunoprecipitated p53 from extracts of MCF7 cells, a cell line with wild-type p53 expression. This coimmunoprecipitation was blocked by a peptide for the Hbo1 antigenic epitope but not by an unrelated peptide or the inclusion of ethidium bromide to disrupt protein-DNA interactions (38).
FIG. 1.
Hbo1, ING4, ING5, and p53 associate in shared protein complexes in cells. (A) Hbo1 and p53 specifically coprecipitate from MCF7 whole-cell extracts. (Left) Results of Western blotting using a monoclonal anti-p53 antibody (α-p53) to detect the presence of p53 in the labeled samples. Lane 1, 13% of the input MCF7 whole-cell extract used for the other immunoprecipitations; lane 2, control immunoprecipitation (IP) prepared using purified rabbit IgG; lane 3, precipitates prepared with anti-Hbo1 antibody alone; lane 4, Hbo1 antibody plus the specific antigenic peptide L74-1; lane 5, Hbo1 antibody plus a nonspecific peptide, MBP4-14; lane 6, Hbo1 antibody plus ethidium bromide (EtBr). (Right) Results of Western blotting using anti-Hbo1 antibody to detect the presence of Hbo1 in the labeled samples. Lanes 7, 10, and 13, MCF7 cells that were mock transfected; lanes 8, 11, and 14, cells transfected with FLAG vector without insert; lanes 9, 12, and 15, cells transfected with vector expressing FLAG-tagged Hbo1; lanes 7 to 9, samples of 4% of the input extracts used for the other immunoprecipitations; lanes 10 to 12, immunoprecipitates prepared with anti-p53 antibody (PAb 1801) (α-p53); lanes 13 to 15, immunoprecipitates prepared with anti-PMS2 antibody of the same IgG1 subtype as PAb 1801 (α-PMS2). (B) Hbo1 and ING4 coprecipitate, and Hbo1, ING5, and p53 coprecipitate. (Left) Results of Western blotting of anti-Hbo1 immunoprecipitations using anti-ING4 and anti-ING5 antibodies (lanes 1 to 3). Lane 1, whole-cell extract (WCE) which was first immunoprecipitated with anti-ING4 antibody (top row) or anti-ING5 antibody (bottom row) and then run together with the control (lane 2) and anti-Hbo1 immunoprecipitates (lane 3) for Western blotting; lane 2, immunoprecipitates prepared with control preimmune serum; lane 3, immunoprecipitates prepared with anti-Hbo1 antibody. (Right) Results of Western blotting using anti-Hbo1 and anti-p53 antibodies (lanes 4 to 7). Lane 4, 1% of the whole-cell extract; lane 5, immunoprecipitates prepared with control goat IgG; lane 6, immunoprecipitates prepared with anti-ING4 antibody; lane 7, immunoprecipitates prepared with anti-ING5 antibody.
In reciprocal experiments, we were unable to detect endogenous Hbo1 directly in immunoprecipitates prepared with anti-p53 antibody, presumably due to the large number of p53 binding partners, the nature of the interactions, or limitations in the sensitivities of available antibodies. Similar limitations were reported previously for other p53 binding factors (18, 51, 60). However, when we ectopically expressed FLAG-Hbo1, coimmunoprecipitation with p53 was readily detected (Fig. 1A, lanes 7 to 15). Direct analysis of cell lysates by Western blotting with rabbit anti-Hbo1 confirmed that FLAG-Hbo1 was detected only in cells transfected with the FLAG-Hbo1 construct and not after mock transfection or transfection with the empty vector. Furthermore, a mouse monoclonal antibody to the unrelated protein PMS2, but of the same idiotype (immunoglobulin G1 [IgG1]) as that of the antibody to p53, did not immunoprecipitate Hbo1. Together, these results demonstrate that Hbo1 and p53 are subunits of one or more of the same protein complexes in MCF7 cells.
Endogenous Hbo1, p53, and ING4/ING5 coprecipitate.
Our finding that p53 physically associates with Hbo1 in cells suggested that Hbo1, p53, and ING4 or ING5 might all be components of the same protein complexes in cells (17, 57). Therefore, we sought to test this coassociation and ensure that the interactions could be detected among the endogenous proteins and not simply ectopically expressed tagged proteins. As shown in Fig. 1B, immunoprecipitation of MCF7 whole-cell extracts with anti-Hbo1 antibody specifically coprecipitated both ING4 and ING5 (lanes 1 to 3). In contrast, we found that anti-Hbo1 antibody failed to coprecipitate ING1 (data not shown), which is part of a protein complex known to lack Hbo1 (17, 36). In reciprocal experiments, immunoprecipitation with either anti-ING4 or anti-ING5 antibodies also coprecipitated Hbo1 (Fig. 1B, lanes 4 to 7). In addition, p53 was clearly present in anti-ING5 immunoprecipitates but not in anti-ING4 immunoprecipitates. However, we cannot rule out association of p53 with Hbo1-ING4 complexes since the antigenic epitope used to raise the anti-ING4 antibody may overlap with the p53 binding domain of ING4 (74). In any case, these results demonstrate the pairwise coassociations of normal endogenous levels of Hbo1, ING4, and ING5 and show that p53 also physically associates with complexes containing Hbo1 and ING5.
Hbo1 directly interacts with p53 in vitro.
We next sought to determine if p53 could interact directly with Hbo1 or whether its association might be entirely explained by its binding to ING5. A series of full-length and truncated forms of p53, representing known functional domains, were expressed as GST fusions in bacteria and purified, and equivalent amounts of protein were tested for binding to His-Hbo1 using GST pull-down assays (Fig. 2A). As shown in Fig. 2B, full-length GST-p53, but not GST alone, was able to pull down full-length His-Hbo1 (lanes 1 to 3). Further mapping demonstrated that this interaction is complex. Deleting the extreme C-terminal 30 amino acids of p53 in derivative GST-p53(1-363) improved the relative recovery of His-Hbo1 (Fig. 2A, lanes 4 to 6), while the most efficient copurification was observed with the 108-residue C-terminal GST-p53(286-393) fragment (lanes 7 and 8). A substantial portion of the latter interaction could be mapped to the 33-residue fragment expressed in GST-p53(361-393) (Fig. 2A, lane 16). Thus, this C-terminal region appears to partially restrict binding to Hbo1 in the context of full-length p53 but favors interactions with Hbo1 by itself and in the context of other C-terminal-region fragments.
FIG. 2.
p53 binds to Hbo1 directly in vitro. (A) Schematic showing the locations of known functional domains of human p53 (shaded boxes). Shown below are the extents of the GST-p53 fusion fragments tested for their interactions with Hbo1, labeled “A1” to “A8.” (B) Mapping of the Hbo1 interaction domains of human p53 by GST pull-down. Full-length His6-Hbo1 protein was loaded at 5% (lane 1) and 10% (lanes 4, 7, and 10) of the amounts used in the accompanying binding reaction mixtures. Glutathione beads carrying GST only (lanes 2, 5, 8, and 11) or various GST-p53 deletion mutants (lanes 3, 6, 9, and 12 to 16) were incubated with purified His-Hbo1(1-611) protein. After extensive washing, the bound proteins were resolved by SDS-PAGE (10% gel) and assayed by Western blotting using rabbit antiserum to the His6 epitope tag. The individual GST-p53 fusions are labeled “A1” to “A8” and correspond to the constructs depicted above (A). (C) The Hbo1 constructs illustrated were expressed as His6-tagged proteins in bacterial cells and affinity purified. Hbo1(G485A) carries a point mutation predicted to be catalytically dead based on the structure of the MYST domain (71). The constructs are labeled “C1” to “C5.” ZF, zinc finger. (D) Results of HAT assays using the subclones shown above (C). The affinity-purified proteins (12 pmol each) were assayed using chicken core histones as a substrate in a filter binding assay. The plotted values represent means and standard errors of the incorpora tion of [3H]acetyl-CoA into histone (n = 3). A control without Hbo1 enzyme is labeled “N,” and the different Hbo1 enzyme fragments used in the reaction mixtures are labeled “C1” to “C5,” corresponding to the fragments shown above (C). (E) The C-terminal domain of human p53 binds the MYST domain of Hbo1. Glutathione beads carrying GST only (lanes 2 and 5), full-length GST-p53(1-393) protein A1 (lane 3), or C-terminal-domain GST-p53(328-393) protein A3 (lane 6) were incubated with the MYST domain His-Hbo1(311-611) and processed as described above. Five percent of the input His6-Hbo1(311-611) is shown in lane 1, and 10% of the input is shown in lane 4.
We carried out similar mapping studies on Hbo1. Full-length and truncated versions of His-HBO1 were expressed in bacteria, purified, and assayed using equivalent amounts of protein in each reaction mixture (Fig. 2C and D). As expected, constructs containing the entire MYST domain retain HAT activity, whereas those that lack the MYST domain do not. Interestingly, Hbo1(311-611) is more active than Hbo1(1-611) in vitro, suggesting that the N-terminal domain of HBO1 may negatively regulate the HAT activity of the C-terminal MYST domain. Surprisingly, GST pull-down experiments revealed that p53 interacts with the catalytic MYST domain of Hbo1. As shown in Fig. 2E, full-length GST-p53 was able to interact with the Hbo1(311-611) MYST domain fragment (lanes 1 to 3). Indeed, just the C-terminal domain represented by GST-p53(328-393) also specifically bound to the MYST domain of Hbo1 (Fig. 2E, lanes 4 to 6). Previous work showed that p53 also interacts with CBP, another histone acetyltransferase, but in that case, p53 binds to the noncatalytic region of CBP (26, 53).
p53 inhibits Hbo1 HAT activity in vitro.
The finding that p53 binds the catalytic domain of Hbo1 suggested that it might regulate the HAT activity of Hbo1. To test this prediction, we first compared the HAT activities of purified His-Hbo1 in the presence and absence of GST-p53 in vitro. As shown in Fig. 3A, GST-p53 specifically inhibits the HAT activity of Hbo1 (lanes 2 to 6). To determine whether this inhibition correlated with Hbo1 binding, we assayed two fragment derivatives of p53: GST-p53(328-393), which binds strongly to Hbo1 in pull-down assays, and GST-p53(286-330), which binds poorly to Hbo1 (Fig. 2B, lanes 10, 11 and 14). Assays with GST-p53(328-393) demonstrated that this derivative is sufficient to inhibit the HAT activity of Hbo1 (Fig. 3A, lanes 9 to 12). In contrast, an identical concentration of GST-p53(286-330) is poor at inhibiting Hbo1 HAT activity (Fig. 3B, lanes 1 to 6). These results suggest that the inhibition of Hbo1 catalytic activity by p53 may be a direct consequence of its binding to the enzyme.
FIG. 3.
p53 inhibits Hbo1 HAT activity in vitro and in vivo. (A) Addition of p53 inhibits Hbo1 HAT activity in vitro. The results of HAT assays using [3H]acetyl-CoA and chicken-free core histones are shown. Control incubations with histones only (lanes 1 and 9) or with histones plus GST-p53 only (lane 7, 12.5 pmol GST-p53; lane 8, 50 pmol GST-p53) did not give histone acetylation. However, the addition of His-Hbo1 (14 pmol) to the reaction mixtures gave strong histone acetylation (lanes 2 and 10). This activity was inhibited by the addition of GST-p53 (lane 3, 12.5 pmol; lane 4, 50 pmol) but not GST alone (lane 5, 12.5 pmol; lane 6, 50 pmol). Inhibition activity was retained in only the C-terminal regulatory domain represented on GST-p53(328-393) (lane 11, 12.5 pmol; lane 12, 50 pmol). (B) The inhibition of Hbo1 by p53 in vitro is specific. Hbo1 HAT activity is not inhibited by GST-p53(286-330), a fragment that does not bind well to the enzyme (Fig. 2B). HAT assays with and without the enzyme served as positive and negative controls (lanes 1 and 2). His-Hbo1 (14 pmol) was incubated with the GST-p53(286-330) protein (lanes 3 and 4) or GST alone (lanes 5 and 6) at 12.5 pmol (lanes 3 and 5) and 50 pmol (lanes 4 and 6). A different histone H4 acetyltransferase, Hat1, is not inhibited by p53 in vitro (lanes 7 to 12). Purified human Hat1 (0.7 pmol) was incubated with free chicken core histones and [3H]acetyl-CoA in the absence (lane 8) or presence (lanes 9 and 10) of GST-p53 or GST alone (lanes 11 and 12) at 12.5 pmol (lanes 9 and 11) or 50 pmol (lanes 10 and 11). Reactions were separated by SDS-PAGE and assayed by measuring the incorporation of [3H]acetate into histones (“Fluorogram”). The amounts of histone substrate were measured by Coomassie staining of the gels (“Stain”). Histone acetylation activity in these reaction mixtures required the addition of the Hat1 enzyme as expected (lane 7). (C) Expression of p53 in cells inhibits Hbo1 acetylation activity. Saos-2 cells (p53−/−) were transiently transfected with either empty pcDNA3.1Zeo(+) plasmid (lane 1) or a pcDNA3.1Zeo(+) clone expressing p53 (lane 2). Whole-cell extracts were separated by PAGE and assayed by Western blotting with antibodies against p53, Hbo1, Mdm2, p21, and α-tubulin. Immunoprecipitates pulled down from the extracts with anti-Hbo1 antibody were assayed for HAT activity using [3H]acetyl-CoA incorporation (“Fluorogram”) with chicken core histones as a substrate (“Stain”). (D) Expression of the C-terminal domain of p53 is sufficient to inhibit the HAT activity of Hbo1 in cells. Saos-2 cells were transfected with either adenovirus expressing GFP (lane 1) or adenovirus expressing only the C-terminal regulatory domain of p53, residues 328 to 393 (lane 2). Whole-cell extracts were probed by Western blotting as described above (B), with the exception that p53C was detected by slot blot Western analysis of the chromatin-enriched fractions using an antibody (PAb 122) specific for the C-terminal region of p53. Immunoprecipitation (IP) and HAT assays were carried out as described above (B).
Because p53 polypeptides were potential substrates for acetylation by Hbo1, we considered the possibility that p53 inhibited Hbo1 HAT activity by competing as substrate for limiting amounts of Hbo1 and [3H]acetyl coenzyme A (acetyl-CoA). To test this alternative, we assayed the ability of Hbo1 to acetylate these substrates individually. Hbo1 strongly prefers histone substrates, and p53 acetylation is detected only in reaction mixtures that do not contain histones, arguing against the competition model (data not shown). We also considered the possibility that p53 might appear to inhibit Hbo1 activity through indirect mechanisms such as an associated histone deacetylase activity or by blocking access to the N-terminal tails. In that case, p53 would appear to inhibit the acetylation activity of any HAT enzyme and not just Hbo1. However, we found that the HAT activity of Hat1, a different histone H4 acetyltransferase, was not altered in the presence of p53 (Fig. 3B, lanes 7 to 12), arguing against these alternatives. These results show that the inhibition of Hbo1 HAT activity by p53 in vitro is specific and is likely to be a direct consequence of the binding interactions.
p53 inhibits Hbo1 HAT activity in cells.
We next sought to determine whether p53 can inhibit Hbo1 HAT activity in human cells by expressing the exogenous p53 alleles in p53−/− Saos-2 osteosarcoma cells (16). Anti-Hbo1 immunoprecipitates were isolated from equivalent amounts of whole-cell extract, and both the p53 contents and HAT activities of these immunoprecipitates were compared. As shown in Fig. 3C, p53 protein was observed only in the cells transfected with the p53 expression construct. As expected, this also resulted in the increased expression of the Mdm2 and p21Waf1/Cip1 genes, two genes transcriptionally activated by p53. However, even though similar amounts of Hbo1 were present in the immunoprecipitates, the HAT activity of Hbo1 was inhibited more than 2.5-fold in cells expressing p53. We also transfected Saos-2 cells with an adenovirus expressing only the C-terminal 66 amino acids of p53 (p53C). This fragment lacks transcriptional activation potential, and, as expected, there were no changes in expression of the p53-activated Mdm2 or p21 gene in cells transfected with p53C. Nevertheless, the specific HAT activity of Hbo1 was almost entirely inhibited in cells expressing p53C (Fig. 3D). Together with our in vitro results, these experiments demonstrate that overexpressed p53 is capable of negatively regulating Hbo1 activity in cells through a transcription-independent pathway.
Hbo1 HAT activity in p53+/+ and p53−/− mouse cells.
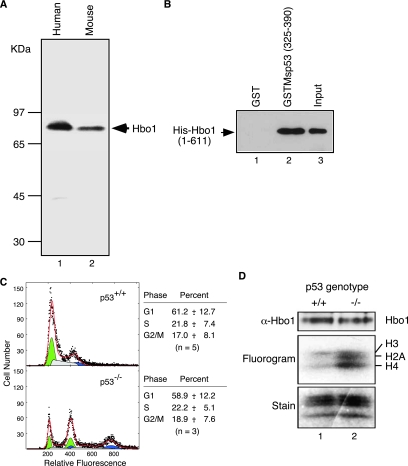
Since the HAT-specific activity of Hbo1 decreases when ectopic p53 or p53C is expressed in Saos-2 cells, we predicted that the opposite should also be true: Hbo1 HAT activity should increase when p53 is deleted from wild-type cells. MEFs isolated from p53+/+ and p53−/− mice provide a system to test this prediction. As a first step, we characterized key aspects of Hbo1 structure and function in the mouse. We subcloned and sequenced an expressed-sequence-tag cDNA (dbEST accession number 8230062) for Myst2, the mouse gene encoding Hbo1 (GenBank accession number DQ076247). The deduced amino acid sequence is identical to that of its human counterpart, except for the 63rd amino acid (Gln in human Hbo1 and Pro in mouse Myst2) and is identical to the coding sequences of the reference genomic Myst2 locus (GenBank accession number NT_039521). Western blots of NIH 3T3 cell extracts revealed an 83-kDa protein comigrating with human Hbo1 (Fig. 4A). Mouse Myst2 immunoprecipitates displayed HAT activity (data not shown), and a GST fusion with the C-terminal region of mouse p53 (amino acids 325 to 390) interacted with full-length His-Hbo1 (Fig. 4B).
FIG. 4.
Hbo1 is negatively regulated by p53 in MEFs. (A) Whole-cell extracts of human 293T cells (lane 1) and mouse NIH 3T3 cells (lane 2) were resolved by 12% SDS-PAGE, transferred onto a membrane, and immunoblotted with anti-Hbo1 antibody. (B) Mouse GST-tagged p53 or GST alone was incubated with His-Hbo1 and purified on glutathione beads. The bound proteins were separated by SDS-PAGE and immunoblotted for His-Hbo1. His-Hbo1 was used in these assays, rather than creating a His-Myst2 construct, since Hbo1 differs from Myst2 only at residue 63, which is outside the region of interaction with p53. (C) Flow cytometry of MEF cell cultures. Primary MEF cells cultured from wild-type mouse embryos (“p53+/+”) and p53 null embryos (“p53−/−”) were stained with propidium iodide and assayed for DNA content. The histograms of cell number versus relative DNA fluorescence are shown. The experimentally determined cell counts are plotted as solid dots. The histograms were fit to G1-phase (green), S-phase (gray), and G2/M-phase (blue) populations, which are plotted at half-scale below the experimental data. The overall fit is shown by the red line. Note that the p53−/− MEF cell cultures contained both diploid cells, cycling between 1C and 2C DNA content, and tetraploid cells, cycling between 2C and 4C DNA content (8, 21). Therefore, these cultures were treated as a mixture of cycling diploid and tetraploid cells with similar division cycle distributions. (D) Whole-cell extracts of the p53+/+ and p53−/− cells were probed with anti-Hbo1 antibody (top). Immunoprecipitates prepared with Hbo1 antibody were subjected to HAT assays with [3H]acetyl-CoA, and incorporation was monitored by fluorography (middle). The histone substrate levels were monitored by Coomassie blue staining (bottom).
To assess the role of p53 in regulating Hbo1 HAT activity in MEF cells, we examined early-passage primary embryonic fibroblast cells derived from p53+/+ and p53−/− animals. Analysis of these cultures by flow cytometry demonstrated that they had similar cell division cycle distributions (Fig. 4C), although the p53−/− cultures contained a subpopulation of tetraploid cells, as expected (8, 21). Whole-cell extracts were prepared from third-passage cell cultures, and Hbo1 complexes were immunoprecipitated. These Hbo1 immunocomplexes were then assayed for HAT activity (Fig. 4D). Western blot analysis showed that the total expression of Hbo1 was the same in both the p53+/+ and p53−/− cells. Nevertheless, the Hbo1 HAT activity of p53−/− cells was elevated by an average of 3.7-fold ± 1.2-fold (n = 3) compared to that of wild-type mouse cells. This increase is consistent with p53 negatively regulating Hbo1 and, moreover, shows that this role is conserved between mouse and human cells.
p53C inhibits pre-RC assembly.
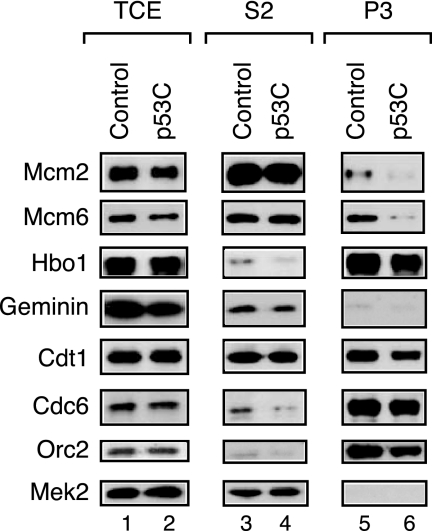
The direct inhibition of Hbo1 expression by antisense knockdown or immunodepletion specifically blocks MCM2-7 association with chromatin, a late step in pre-RC assembly (31). We reasoned that this step might also be inhibited by the down-regulation of Hbo1 by p53. To test this prediction, we transfected HeLa cells with either p53C adenovirus or green fluorescent protein (GFP) control adenovirus. After 48 h of infection, the cells were fractionated to give total cell extract and soluble cytoplasmic (S2), soluble nuclear (S3), and chromatin-enriched (P3) fractions as described previously (43). Good fractionation was achieved, as indicated by the predominant recovery of Mek2 in the cytoplasmic fraction and Hbo1 in the chromatin fraction (Fig. 5). These fractions were then subjected to SDS-PAGE, and the amounts of pre-RC components were determined for each fraction by Western blotting (Fig. 5). The overall levels of expression of pre-RC components remained unchanged, with or without p53C (Fig. 5, lanes 1 and 2). Nevertheless, the overexpression of p53C caused a sixfold decrease in the chromatin binding of Mcm2 and Mcm6 (lanes 5 and 6) without affecting the binding of Orc2, Cdc6, or Cdtl. This outcome matches the effect of depleting Hbo1 expression in human cells (31). Thus, these results suggest that p53 can be coupled to the control of pre-RC assembly in cells through its regulation of Hbo1 HAT activity.
FIG. 5.
A C-terminal fragment of p53 is sufficient to inhibit pre-RC assembly of the MCM2-7 complex. HeLa cells were transfected with adenovirus expressing either control GFP or p53C, the C-terminal regulatory domain of p53 (amino acids 328 to 393). Cells were harvested and fractionated as described previously (43). Fractions were separated by SDS-PAGE and assayed by Western blotting with antibodies against the proteins indicated. The results are shown for the total cell extracts (TCE), the soluble cytoplasmic fractions (S2), and the chromatin-enriched fractions (P3).
Activation of p53 by hyperosmotic stress inhibits Hbo1.
A strong prediction of this model is that physiological stress conditions that activate p53 should, in turn, inhibit Hbo1 and its downstream functions such as pre-RC formation. To test this prediction, we first examined the consequences of high salt, a stress known to stabilize and activate p53 (35), which can be imposed during G1 phase at the time of pre-RC assembly. MCF7 cells were arrested at metaphase with nocodazole for 16 h and then released into fresh medium. Cells were allowed to recover for 2 h, and the cultures were adjusted to 0.26 M NaCl to activate p53 and incubated for an additional 6.5 h. Flow cytometry confirmed that cells were arrested at metaphase by nocodazole and that both salt-treated cells and their untreated controls exited metaphase and were in G1 phase at the time of harvest (Fig. 6A). Analysis of whole-cell extracts demonstrated that salt treatment increased p53 levels approximately 1.7-fold, as expected (Fig. 6B). We also observed a reproducible decrease of roughly 2.5-fold in Mdm2 and p21 after 6.5 h of salt treatment. The decrease in Mdm2 is in agreement with data from previous experiments that demonstrated a transient decrease in Mdm2 in response to osmotic shock, with a minimum at approximately 6 h posttreatment (35). This decrease in Mdm2 contributes to the stabilization of p53. The relative expression of p21 following salt stress in synchronized MCF7 cells has not been examined previously, but our results presumably reflect the kinetics of activation of p53 by phosphorylation of Ser33 by p38MAPK (35). Hbo1 immunoprecipitates from these extracts were then assayed for p53 association and HAT activity. As seen in Fig. 6C, although similar amounts of Hbo1 were recovered in the precipitates from control and salt-treated cells, salt stress increased the amount of p53 recovered in Hbo1 complexes by approximately 3.3-fold. Furthermore, the HAT activity of Hbo1 in the treated cells was inhibited proportionally by 3.4-fold, consistent with the activation of p53.
FIG. 6.
Salt stress in G1 phase activates p53 expression, inhibits Hbo1 HAT activity, and blocks chromatin loading of the MCM2-7 complex. (A) MCF7 cells were arrested at metaphase with nocodazole for 16 h (“Noc Arrest”). The synchronized cell cultures were then split into fresh medium without nocodazole and allowed to release from arrest. After 2 h of recovery, one set of cells was left untreated (“Released Untreated”), while the other was treated with 0.26 M NaCl (“Released +NaCl”). Cells were harvested from the cultures after 6.5 h, stained for DNA, and assayed for their cell cycle distributions by flow cytometry. (B) Effect of salt stress on the amounts of key proteins. Whole-cell extracts were prepared from untreated (lane 1) (“−”) and NaCl-shocked (lane 2) (“+”) cultures described above (A). The extracts were separated by PAGE and assayed by Western blotting with antibodies against p53, Hbo1, Mdm2, p21, and γ-tubulin. (C) Immunoprecipitates were prepared using anti-Hbo1 antibody and whole-cell extracts of untreated cells (lane 1) and NaCl-shocked cells (lane 2) as described above (A). Portions of the Hbo1 immunoprecipitates were then assayed by Western blotting for the presence of Hbo1 and p53 (“p53/Hbo1 in the complex”) and for HAT activity by [3H]acetate incorporation (“Fluorography”) into chicken core histones (“Stain”). (D) MCF7 cells, either untreated or NaCl shocked as described above (A), were fractionated into total cell extract (TCE), a soluble cytoplasmic fraction (S2), a soluble nuclear fraction (S3), and a chromatin-enriched fraction (P3) as described previously (43). Fractions were separated by PAGE and assayed by Western blotting with antibodies against p53, Mcm2, Mcm6, Hbo1, Cdt1, Orc2, and Mek2.
We next assayed the state of pre-RC assembly in control and salt-treated cells by monitoring the chromatin loading of pre-RC component proteins. Total cell extracts were fractionated as described above and assayed for p53 levels and pre-RC subunits by Western blotting (Fig. 6D). We observed either no change or a slight increase in the overall relative level of expression of Hbo1 and pre-RC components in salt-treated cells (Fig. 6D, lanes 1 and 2). As expected, salt treatment increased p53 levels in the total cell extract and increased its recovery in the chromatin fraction by 2.4-fold (Fig. 6D, lanes 1 and 2 and 7 and 8). The chromatin-bound levels of Orc2 and Cdt1 were unaltered by salt treatment. However, the chromatin-bound amounts of Mcm2 and Mcm6 were strongly inhibited by at least 2.6-fold in extracts from the treated cells, consistent with down-regulation of Hbo1 by activated p53 (Fig. 6D, lanes 7 and 8). These results show that physiological stresses that stabilize p53 during the time of DNA replication licensing result in the inhibition of Hbo1 HAT activity and the association of MCM2-7 with chromatin.
Activation of p53 by HU arrest inhibits Hbo1.
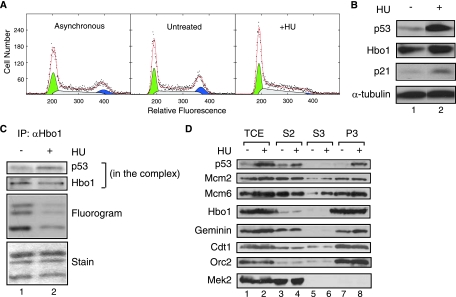
After pre-RC assembly, DNA replication is initiated by a series of regulated steps that result in the disassembly of the pre-RC and the release of MCM2-7 to travel with the DNA replication fork (3). We were curious whether p53 activation following replication initiation would also inhibit Hbo1 activity and whether it would affect the chromatin association of MCM2-7. HU, which inhibits ribonucleotide reductase, blocks DNA synthesis within S phase by limiting deoxynucleotide triphosphates (66). This stress, in turn, stabilizes p53, leading to increased protein levels (25, 65). To test the effect of this pathway on Hbo1 activity in S phase, we treated MCF7 cells with HU and examined p53 and Hbo1 functions at time intervals after the block. Flow cytometry of these cells showed that they were predominantly in G1 and S phases with underreplicated DNA (Fig. 7A). The expression of p53 was significantly increased after 6 h of HU treatment, and p21 expression was increased as well (Fig. 7B), consistent with previous observations (29). Hbo1 immunoprecipitates were then assayed for HAT activity and their Hbo1 and p53 protein levels (Fig. 7C). Equivalent amounts of Hbo1 were recovered in the immunoprecipitates from control and HU-treated cells; however, the precipitates from HU-arrested cells were enriched approximately twofold in the p53 protein. Along with this increased p53 association, we observed a 2.6-fold decrease in the relative HAT activity of immunocomplexes recovered from cells arrested with HU. Thus, as with hyperosmotic shock, the activation of p53 by HU stress correlated with an inhibition of Hbo1 HAT activity. Whole-cell extracts were then fractionated as described above to assess MCM2-7 loading under HU stress conditions (Fig. 7D). As expected, HU treatment increased p53 binding in the chromatin faction. However, in this case, the chromatin levels of Mcm2 and Mcm6 did not decrease, despite the down-regulation of Hbo1 activity (Fig. 7D, lanes 7 and 8). The only pre-RC factor apparently affected by HU stress was geminin, which displayed increased expression and association with chromatin (Fig. 7D, lanes 1, 2, 7, and 8). This is likely due to the normal increase in its expression during S phase and enrichment of those cells by HU treatment (48). The continued association of MCM2-7 with chromatin in the face of decreased Hbo1 activity is consistent with the disassembly of the pre-RC and relocation of MCM2-7 to the replication fork following replication initiation (7, 11, 37, 40).
FIG. 7.
DNA replication arrest in S phase with HU activates p53 expression and inhibits Hbo1 HAT activity. (A) An asynchronously dividing culture was split and either left untreated or arrested with 1.5 mM HU treatment. Cells were harvested from the cultures after 6 h of HU treatment and assayed by flow cytometry. The DNA content histograms are plotted as shown. (B) MCF7 cells were either left untreated (lane 1) (“−”) or treated for 6 h with 1.5 mM HU (lane 2) (“+”) as described above (A). Whole-cell extracts were separated by PAGE and assayed by Western blotting with antibodies against p53, Hbo1, p21, and α-tubulin as a loading control. The pairs of Western blot bands presented in columns 1 and 2 are from nonadjacent lanes of the same gel images. (C) MCF7 whole-cell extracts as described above (A) were immunoprecipitated with anti-Hbo1 antibody. Portions of the Hbo1 immunoprecipitates were then assayed by Western blotting for the presence of Hbo1 and p53 (“p53/Hbo1 in the complex”) and for HAT activity by [3H]acetate incorporation (“Fluorography”) into chicken core histones (“Stain”). (D) MCF7 cells, either untreated or HU arrested as described above (A), were fractionated into total cell extract (TCE) (lanes 1 and 2), a soluble cytoplasmic fraction (S2) (lanes 3 and 4), a soluble nuclear fraction (S3) (lanes 5 and 6), and a chromatin-enriched fraction (P3) (lanes 7 and 8) (43). Fractions were separated by PAGE and assayed by Western blotting with antibodies against p53, Mcm2, Mcm6, Hbo1, geminin, Cdt1, Orc2, and Mek2.
The coupling of stress signals to Hbo1 is p53 dependent.
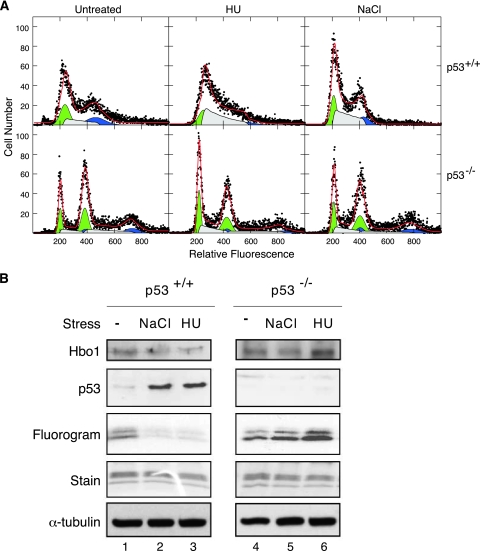
If p53 is mediating the Hbo1 response to salt and HU stress signals, then the deletion of p53 should abolish this response. To test this prediction, we first examined the effect of these stresses on p53+/+ and p53−/− MEF cells. Primary early-passage cell cultures were treated with either NaCl for 3 h or HU for 6 h. The DNA content histograms of treated and control cells are shown in Fig. 8A. Whole-cell extracts were prepared from these cultures and assayed for protein levels by Western blotting and HAT activity in Hbo1 immunoprecipitates. As shown in Fig. 8B, both NaCl and HU stresses induced the activation of p53 in wild-type p53+/+ cells greater than sixfold and caused an inhibition of Hbo1 HAT activity of over 2.5-fold. In contrast, the HAT activity of Hbo1 was not inhibited by either high salt or HU arrest in p53−/− cells. These results demonstrate that the inhibition of Hbo1 HAT activity caused by hyperosmotic stress and DNA replication arrest is dependent on p53 function in MEFs.
FIG. 8.
Stress regulation of Hbo1 is p53 dependent in MEFs. (A) Flow cytometry of MEF cells. MEF cell cultures from wild-type mouse embryos (top row) and p53 null embryos (bottom row) were split into three portions and either left untreated, treated with 10 mM HU for 6 h, or salt shocked with 0.24 M NaCl for 3 h. Cells were harvested, stained for DNA, and assayed by flow cytometry. The DNA content histograms are plotted as described for Fig. 7A. (B) Primary MEF cell cultures from p53+/+ and p53−/− animals were left untreated (lanes 1 and 4), stressed for 3 h with 0.24 M NaCl (lanes 2 and 5), or treated for 6 h with 10 mM HU (lanes 3 and 6). Whole-cell extracts were probed for levels of Hbo1, p53, and α-tubulin, as a loading control, by Western blot assays. Immunoprecipitates were prepared from the extracts with anti-Hbo1 antibody and assayed for HAT activity by [3H]acetate incorporation (“Fluorogram”) using chicken core histones as a substrate (“Stain”).
To test the p53 dependence of the pathway in human cells, we used siRNA to block p53 expression and assayed the inhibition of pre-RC assembly by osmotic shock. MCF7 cells were transfected with an siRNA oligonucleotide directed against p53 expression, or a nontargeted control siRNA oligonucleotide, and were incubated for 48 to 72 h. The cells were then arrested at metaphase by treatment with 50 ng/ml nocodazole for 16 h, and the rounded mitotic cells were collected by detaching them from the culture surface by shaking. Flow cytometry confirmed that the recovered cell population was synchronized in mitosis with a G2 DNA content (Fig. 9A). The cells were then incubated in medium without extra salt for 2.5 h to allow recovery from mitotic arrest. Cells were then treated with 0.26 M NaCl for 6.5 h and then collected for biochemical fractionation. As shown in Fig. 9A, both the control siRNA and p53 siRNA cell populations exited from mitosis and displayed similar flow cytometry profiles, with predominantly G1 DNA contents at harvest. In cells transfected with control siRNA oligonucleotides, osmotic shock increased the amount of p53 in the whole-cell extracts, and the levels of MDM2 and p21 decreased (Fig. 9B, lanes 1 and 2). The chromatin association of p53 in the P3 fraction also increased about twofold, and in two independent experiments, the chromatin association of Mcm2 and Mcm6 specifically decreased by approximately twofold (Fig. 9B, lanes 10 and 11). These results are in agreement with the above-described osmotic shock experiments (Fig. 6) demonstrating that pretreatment with siRNA did not affect the p53-Hbo1 regulatory pathway. The transfection of cells with p53 siRNA successfully blocked p53 expression in total cell extracts and also reduced the association of p53 in the P3 chromatin fractions (Fig. 9B, lanes 2, 3, 11, and 12). However, osmotic shock failed to inhibit the chromatin loading of Mcm2 and Mcm6 when p53 expression was knocked down by siRNA (Fig. 9A, lanes 10 to 12). These results show that p53 is required to couple the Hbo1-dependent regulation of pre-RC assembly to osmotic stress.
FIG. 9.
Osmotic stress regulation of MCM2-7 loading is p53 dependent. (A) Flow cytometry of synchronized cultures. MCF7 cells were transfected with either control siRNA or p53-specific siRNA (“p53 siRNA”) and cultured asynchronously for 72 h (“Asynchronous”). Cells were then arrested in mitosis with nocodazole for 16 h (“Noc Arrest”). The synchronized cell cultures were then split into fresh medium without nocodazole and allowed to release from arrest. After 2 h of recovery, cells were then treated with 0.26 M NaCl (“Released +NaCl”). Cells were harvested from the cultures after 6.5 h, stained for DNA, and assayed for their cell cycle distributions by flow cytometry. (B) MCM2-7 chromatin loading under osmotic stress is p53 dependent. MCF7 cells were fractionated into total cell extract (TCE) and soluble cytoplasmic, soluble nuclear, and chromatin-enriched fractions as described previously (43). Fractions were separated by PAGE and assayed by Western blotting with antibodies against p53, Mcm2, Mcm6, Hbo1, Cdt1, Orc2, and Mek2. Whole-cell extracts were also assayed for MDM2 and p21 by Western blotting. The standard cell population was transfected with control siRNA but not shocked with NaCl (lanes 1, 4, 7, and 10). The stress control cell population was transfected with a control siRNA and treated with 0.26 M NaCl (lanes 2, 5, 8, and 11). The stress- and p53-depleted cell population was transfected with p53 siRNA and treated with 0.26 M NaCl (lanes 3, 6, 9, and 12).
DISCUSSION
Our results identify a previously unrecognized pathway that links stress response signaling through p53 to the output of a histone acetyltransferase complex. In the case of salt stress during G1 phase, this pathway results in the inhibition of pre-RC assembly. These results are consistent with the previous observation that p53 can inhibit nuclear DNA replication in vitro in Xenopus egg extracts in a transcription-independent manner (15). In addition to p53, the tumor suppressors RB and p16 also regulate the initiation of DNA replication (10, 50). The Rb protein, which binds to Mcm7, inhibits DNA unwinding in vitro in Xenopus extracts. The tumor suppressor p16 inhibits the chromatin association of minichromosome maintenance proteins at G1 phase, presumably through the inhibition of G1 cyclin-dependent kinase activity. Thus, tumor suppressor proteins employ diverse molecular mechanisms to modulate the initiation of DNA replication.
Using antibodies against native proteins, we found that endogenous Hbo1 coprecipitates with endogenous ING4 and ING5, in agreement with previous work using epitope-tagged constructs (17). In addition, the Hbo1/ING5 complex also associates with p53 in MCF7 cells. We considered the possibility that p53 and ING4/ING5 compete for binding to Hbo1. However, this is unlikely since the interaction between ING5 and Hbo1 was not disrupted by the stabilization of p53 following HU treatment (data not shown). At present, the functions of ING4 and ING5 in the complexes are unclear. On one hand, their properties strongly suggest that they are tumor suppressor proteins, formally analogous to p53 in their actions (52, 57). On the other hand, the inhibition of ING4 and ING5 expression by RNA interference in HeLa cells causes phenotypes similar to those produced by the loss of Hbo1 (17, 31); that is, ING4 and ING5 apparently act as positive factors in their respective Hbo1/ING/Jade complexes and yet also act in parallel with p53, which negatively regulates the Hbo1 HAT activities of the complexes. Resolving this apparent paradox through further molecular and genetic experiments promises to provide novel insights into the control of cell growth and proliferation.
Based on our data, and those reported previously by others, we speculate that Hbo1 and p53 may serve to integrate and balance conflicting mitogenic and stress signals that impinge upon cell proliferation. Several observations support the idea that Hbo1 responds positively to mitogenic signaling pathways to promote replication licensing and proliferation. For example, the cyclin-dependent kinase CDK11p58 has recently been found to bind Hbo1 and simulate its HAT activity (77). This fits well with our observation that removing the N-terminal serine-rich domain increases Hbo1 HAT activity (Fig. 2D). Counterbalancing these proliferative responses, growth-inhibitory stress signals would result in p53 activation and thus the inhibition of Hbo1 activity. Pre-RC assembly provides a model example of this regulation. The activation of Hbo1 during G1 phase serves to facilitate MCM2-7 loading onto chromatin and ensure the efficient completion of pre-RC assembly (31). However, if cells experience hyperosmotic shock during G1 phase, this activates p53 expression, which in turn inhibits Hbo1 activity, blocking MCM2-7 loading and delaying pre-RC assembly. Another example of this regulatory pathway may be the cellular response to human cytomegalovirus infection, which both increases the expression of p53 and prevents MCM2-7 loading and replication licensing (6, 46, 70).
The extent to which this regulatory pathway influences cell proliferation remains to be determined, but it is likely to affect more than just replication licensing. Blocking DNA replication fork progression with HU causes a p53-dependent down-regulation of Hbo1 activity (Fig. 8). However, in this case, the MCM2-7 complex remains bound to chromatin. While this result makes sense given the importance of MCM2-7 for stalled replication forks and subsequent replication elongation following restart (7, 11, 37, 40), it leaves open the question of why Hbo1 is inhibited under these conditions. One possibility is that Hbo1 has additional unknown roles during S phase, and the results of experiments using RNA interference to inhibit ING4 and ING5 expression support this interpretation (17). In addition, it is also likely that other functions of Hbo1, such as its role as a transcriptional coactivator (22, 44, 55), must be kept in check by p53 during cellular stress responses.
The negative regulation of Hbo1 by p53 suggests that this HAT may be a candidate oncogene, and several observations are consistent with this idea. Hbo1 maps to 17q21, a locus that shows frequent allelic gain in a variety of human tumors. Moreover, the Hbo1 gene has been found to be amplified or overexpressed in several breast cancer cell lines (14). As p53 is deficient in many breast cancers, this would exacerbate the oncogenic potential of Hbo1 if amplified or overexpressed. Interestingly, retroviral tagging screens have identified two independent tumor lines in which the site of retroviral insertion mapped to locations consistent with a dominant activation of Hbo1 expression (61). Finally, the acetylation pattern of histone H4 in many cancers is consistent with the substrate specificity of Hbo1 in vivo (17, 20).
Acknowledgments
We thank Jacques Côté, Herbert Cohen, and Daniel Engel for discussions; Jacques Côté for communicating research results prior to publication; and Alain Verreault, Bruce Stillman, and Hideo Nishitani for reagents.
We thank Rong Li for financial support to M.I. M.I. was supported in part by UVA P30 CA44579 and the Kincaid Charitable Trust. This work was supported by a grant-in-aid for the 2nd Term Comprehensive 10-Year Strategy for Cancer Control and a research grant for human genome and gene therapy from the Ministry of Health and Welfare of Japan to T.S. and grants from the National Institutes of Health to C.D.A. (GM53512) and M.M.S. (GM60444).
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Aggarwal, B., and B. Calvi. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430372-376. [DOI] [PubMed] [Google Scholar]
- 2.Barlev, N., L. Liu, N. Chehab, K. Mansfield, K. Harris, T. Halazonetis, and S. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 81243-1254. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71333-374. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12142-148. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand, P., Y. Saintigny, and B. S. Lopez. 2004. p53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 20235-243. [DOI] [PubMed] [Google Scholar]
- 6.Biswas, N., V. Sanchez, and D. H. Spector. 2003. Human cytomegalovirus infection leads to accumulation of geminin and inhibition of the licensing of cellular DNA replication. J. Virol. 772369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borel, F., F. Lacroix, and R. Margolis. 2002. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J. Cell Sci. 1152829-2838. [DOI] [PubMed] [Google Scholar]
- 8.Borel, F., O. Lohez, F. Lacroix, and R. Margolis. 2002. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA 999819-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrow, J., V. Stanton, Jr., J. Andresen, R. Becher, F. Behm, R. Chaganti, C. Civin, C. Disteche, I. Dube, A. Frischauf, D. Horsman, F. Mitelman, S. Volinia, A. Watmore, and D. Housman. 1996. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 1433-41. [DOI] [PubMed] [Google Scholar]
- 10.Braden, W. A., J. M. Lenihan, Z. Lan, K. S. Luce, W. Zagorski, E. Bosco, M. F. Reed, J. G. Cook, and E. S. Knudsen. 2006. Distinct action of the retinoblastoma pathway on the DNA replication machinery defines specific roles for cyclin-dependent kinase complexes in prereplication complex assembly and S-phase progression. Mol. Cell. Biol. 267667-7681. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Branzei, D., and M. Foiani. 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17568-575. [DOI] [PubMed] [Google Scholar]
- 12.Burke, T., J. Cook, M. Asano, and J. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 27615397-15408. [DOI] [PubMed] [Google Scholar]
- 13.Chuikov, S., J. K. Kurash, J. R. Wilson, B. Xiao, N. Justin, G. S. Ivanov, K. McKinney, P. Tempst, C. Prives, S. J. Gamblin, N. A. Barlev, and D. Reinberg. 2004. Regulation of p53 activity through lysine methylation. Nature 432353-360. [DOI] [PubMed] [Google Scholar]
- 14.Clark, J., S. Edwards, M. John, P. Flohr, T. Gordon, K. Maillard, I. Giddings, C. Brown, A. Bagherzadeh, C. Campbell, J. Shipley, R. Wooster, and C. Cooper. 2002. Identification of amplified and expressed genes in breast cancer by comparative hybridization onto microarrays of randomly selected cDNA clones. Genes Chromosomes Cancer 34104-114. [DOI] [PubMed] [Google Scholar]
- 15.Cox, L., T. Hupp, C. Midgley, and D. Lane. 1995. A direct effect of activated human p53 on nuclear DNA replication. EMBO J. 142099-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diller, L., J. Kassel, C. Nelson, M. Gryka, G. Litwak, M. Gebhardt, B. Bressac, M. Ozturk, S. Baker, B. Vogelstein, and S. Friend. 1990. p53 functions as a cell cycle control protein in osteosarcomas. Mol. Cell. Biol. 105772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyon, Y., C. Cayrou, M. Ullah, A. Landry, V. Côté, W. Selleck, W. Lane, S. Tan, X. Yang, and J. Côté. 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2151-64. [DOI] [PubMed] [Google Scholar]
- 18.Enari, M., K. Ohmori, I. Kitabayashi, and Y. Taya. 2006. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 201087-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, X., Y. Hara, and K. Riabowol. 2002. Different HATS of the ING1 gene family. Trends Cell Biol. 12532-538. [DOI] [PubMed] [Google Scholar]
- 20.Fraga, M., E. Ballestar, A. Villar-Garea, M. Boix-Chornet, J. Espada, G. Schotta, T. Bonaldi, C. Haydon, S. Ropero, K. Petrie, N. Iyer, A. Perez-Rosado, E. Calvo, J. Lopez, A. Cano, M. Calasanz, D. Colomer, M. Piris, N. Ahn, A. Imhof, C. Caldas, T. Jenuwein, and M. Esteller. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37391-400. [DOI] [PubMed] [Google Scholar]
- 21.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 2711744-1747. [DOI] [PubMed] [Google Scholar]
- 22.Georgiakaki, M., N. Chabbert-Buffet, B. Dasen, G. Meduri, S. Wenk, L. Rajhi, L. Amazit, A. Chauchereau, C. Burger, L. Blok, E. Milgrom, M. Lombés, A. Guiochon-Mantel, and H. Loosfelt. 2006. Ligand-controlled interaction of HBO1 with the N-terminal transactivating domain of progesterone receptor induces SRC-1-dependent co-activation of transcription. Mol. Endocrinol. 202122-2140. [DOI] [PubMed] [Google Scholar]
- 23.Gibbons, R. 2005. Histone modifying and chromatin remodelling enzymes in cancer and dysplastic syndromes. Hum. Mol. Genet. 14(Spec. No. 1)R85-R92. [DOI] [PubMed] [Google Scholar]
- 24.Goodman, R., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 141553-1577. [PubMed] [Google Scholar]
- 25.Gottifredi, V., S. Shieh, Y. Taya, and C. Prives. 2001. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl. Acad. Sci. USA 981036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, W., X. Shi, and R. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387819-823. [DOI] [PubMed] [Google Scholar]
- 27.Hardy, S., M. Kitamura, T. Harris-Stansil, Y. Dai, and M. Phipps. 1997. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 711842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, S. L., and A. J. Levine. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 242899-2908. [DOI] [PubMed] [Google Scholar]
- 29.Ho, C. C., W. Y. Siu, A. Lau, W. M. Chan, T. Arooz, and R. Y. Poon. 2006. Stalled replication induces p53 accumulation through distinct mechanisms from DNA damage checkpoint pathways. Cancer Res. 662233-2241. [DOI] [PubMed] [Google Scholar]
- 30.Huang, J., L. Perez-Burgos, B. J. Placek, R. Sengupta, M. Richter, J. A. Dorsey, S. Kubicek, S. Opravil, T. Jenuwein, and S. L. Berger. 2006. Repression of p53 activity by Smyd2-mediated methylation. Nature 444629-632. [DOI] [PubMed] [Google Scholar]
- 31.Iizuka, M., T. Matsui, H. Takisawa, and M. Smith. 2006. Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol. 261098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iizuka, M., and B. Stillman. 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 27423027-23034. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, T. M., E. M. Hammond, A. Giaccia, and L. D. Attardi. 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat. Genet. 37145-152. [DOI] [PubMed] [Google Scholar]
- 34.Kamakaka, R., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19295-310. [DOI] [PubMed] [Google Scholar]
- 35.Kishi, H., K. Nakagawa, M. Matsumoto, M. Suga, M. Ando, Y. Taya, and M. Yamaizumi. 2001. Osmotic shock induces G1 arrest through p53 phosphorylation at Ser33 by activated p38MAPK without phosphorylation at Ser15 and Ser20. J. Biol. Chem. 27639115-39122. [DOI] [PubMed] [Google Scholar]
- 36.Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33ING1. Mol. Cell. Biol. 22835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labib, K., J. A. Tercero, and J. F. Diffley. 2000. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 2881643-16437. [DOI] [PubMed] [Google Scholar]
- 38.Lai, J., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 896958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 191202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupardus, P., T. Byun, M. Yee, M. Hekmat-Nejad, and K. Cimprich. 2002. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 162327-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machida, Y., J. Hamlin, and A. Dutta. 2005. Right place, right time, and only once: replication initiation in metazoans. Cell 12313-24. [DOI] [PubMed] [Google Scholar]
- 42.Maier, B., W. Gluba, B. Bernier, T. Turner, K. Mohammad, T. Guise, A. Sutherland, M. Thorner, and H. Scrable. 2004. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 18306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Méndez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 208602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miotto, B., and K. Struhl. 2006. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol. Cell. Biol. 265969-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moll, U. M., S. Wolff, D. Speidel, and W. Deppert. 2005. Transcription-independent pro-apoptotic functions of p53. Curr. Opin. Cell Biol. 17631-636. [DOI] [PubMed] [Google Scholar]
- 46.Muganda, P., O. Mendoza, J. Hernandez, and Q. Qian. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 688028-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevins, J., J. DeGregori, L. Jakoi, and G. Leone. 1997. Functional analysis of E2F transcription factor. Methods Enzymol. 283205-219. [DOI] [PubMed] [Google Scholar]
- 48.Nishitani, H., S. Taraviras, Z. Lygerou, and T. Nishimoto. 2001. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 27644905-44911. [DOI] [PubMed] [Google Scholar]
- 49.Notterman, D., S. Young, B. Wainger, and A. Levine. 1998. Prevention of mammalian DNA reduplication, following the release from the mitotic spindle checkpoint, requires p53 protein, but not p53-mediated transcriptional activity. Oncogene 172743-2751. [DOI] [PubMed] [Google Scholar]
- 50.Pacek, M., and J. C. Walter. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 233667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roe, J. S., H. Kim, S. M. Lee, S. T. Kim, E. J. Cho, and H. D. Youn. 2006. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol. Cell 22395-405. [DOI] [PubMed] [Google Scholar]
- 52.Russell, M., P. Berardi, W. Gong, and K. Riabowol. 2006. Grow-ING, age-ING and die-ING: ING proteins link cancer, senescence and apoptosis. Exp. Cell Res. 312951-961. [DOI] [PubMed] [Google Scholar]
- 53.Scolnick, D., N. Chehab, E. Stavridi, M. Lien, L. Caruso, E. Moran, S. Berger, and T. Halazonetis. 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 573693-3696. [PubMed] [Google Scholar]
- 54.Sengupta, S., and C. Harris. 2005. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 644-55. [DOI] [PubMed] [Google Scholar]
- 55.Sharma, M., M. Zarnegar, X. Li, B. Lim, and Z. Sun. 2000. Androgen receptor interacts with a novel MYST protein, HBO1. J. Biol. Chem. 27535200-35208. [DOI] [PubMed] [Google Scholar]
- 56.Shi, X., and O. Gozani. 2005. The fellowships of the INGs. J. Cell. Biochem. 961127-1136. [DOI] [PubMed] [Google Scholar]
- 57.Shiseki, M., M. Nagashima, R. Pedeux, M. Kitahama-Shiseki, K. Miura, S. Okamura, H. Onogi, Y. Higashimoto, E. Appella, J. Yokota, and C. Harris. 2003. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 632373-2378. [PubMed] [Google Scholar]
- 58.Smith, C., and C. Peterson. 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65115-148. [DOI] [PubMed] [Google Scholar]
- 59.Stedman, W., Z. Deng, F. Lu, and P. Lieberman. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 7812566-12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sui, G., E. B. Affar, Y. Shi, C. Brignone, N. R. Wall, P. Yin, M. Donohoe, M. P. Luke, D. Calvo, S. R. Grossman, and Y. Shi. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117859-872. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki, T., H. Shen, K. Akagi, H. Morse, J. Malley, D. Naiman, N. Jenkins, and N. Copeland. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32166-174. [DOI] [PubMed] [Google Scholar]
- 62.Sykes, S. M., H. S. Mellert, M. A. Holbert, K. Li, R. Marmorstein, W. S. Lane, and S. B. McMahon. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 24841-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabancay, A. P., and S. L. Forsburg, Jr. 2006. Eukaryotic DNA replication in a chromatin context. Curr. Top. Dev. Biol. 76129-814. [DOI] [PubMed] [Google Scholar]
- 64.Tang, Y., J. Luo, W. Zhang, and W. Gu. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24827-839. [DOI] [PubMed] [Google Scholar]
- 65.Taylor, W., M. Agarwal, A. Agarwal, D. Stacey, and G. Stark. 1999. p53 inhibits entry into mitosis when DNA synthesis is blocked. Oncogene 18283-295. [DOI] [PubMed] [Google Scholar]
- 66.Timson, J. 1975. Hydroxyurea. Mutat. Res. 32115-132. [DOI] [PubMed] [Google Scholar]
- 67.Utley, R., and J. Côté. 2003. The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 274203-236. [DOI] [PubMed] [Google Scholar]
- 68.Verreault, A., P. Kaufman, R. Kobayashi, and B. Stillman. 1998. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 896-108. [DOI] [PubMed] [Google Scholar]
- 69.Vogelstein, B., D. Lane, and A. Levine. 2000. Surfing the p53 network. Nature 408307-310. [DOI] [PubMed] [Google Scholar]
- 70.Wiebusch, L., R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus prevents replication licensing by inhibiting MCM loading onto chromatin. EMBO Rep. 442-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan, Y., N. Barlev, R. Haley, S. Berger, and R. Marmorstein. 2000. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 61195-1205. [DOI] [PubMed] [Google Scholar]
- 72.Yang, X. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, K., and S. Dent. 2005. Histone modifying enzymes and cancer: going beyond histones. J. Cell. Biochem. 961137-1148. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, X., K. S. Wang, Z. Q. Wang, L. S. Xu, Q. W. Wang, F. Chen, D. Z. Wei, and Z. G. Han. 2005. Nuclear localization signal of ING4 plays a key role in its binding to p53. Biochem. Biophys. Res. Commun. 3311032-1038. [DOI] [PubMed] [Google Scholar]
- 75.Zhou, J., and C. Prives. 2003. Replication of damaged DNA in vitro is blocked by p53. Nucleic Acids Res. 313881-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, M. I., R. L. Foy, V. C. Chitalia, J. Zhao, M. V. Panchenko, H. Wang, and H. T. Cohen. 2005. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc. Natl. Acad. Sci. USA 10211035-11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zong, H., Z. Li, L. Liu, Y. Hong, X. Yun, J. Jiang, Y. Chi, H. Wang, X. Shen, Y. Hu, Z. Niu, and J. Gu. 2005. Cyclin-dependent kinase 11(p58) interacts with HBO1 and enhances its histone acetyltransferase activity. FEBS Lett. 5793579-3588. [DOI] [PubMed] [Google Scholar]