Abstract
Members of the PIAS (for protein inhibitor of activated STAT) family play critical roles in modulating the activity of a variety of transcriptional regulators. Zimp10, a novel PIAS-like protein, is a transcriptional coregulator and may be involved in the modification of chromatin through interactions with the SWI/SNF chromatin-remodeling complexes. Here, we investigate the biological role of Zimp10 in zimp10-deficient mice. Homozygosity for the Zimp10-targeted allele resulted in developmental arrest at approximately embryonic day 10.5. Analysis of knockout embryos revealed severe defects in the reorganization of the yolk sac vascular plexus. No significant abnormality in hematopoietic potential was observed in zimp10 null mice. Microarray and quantified reverse transcription-PCR analyses showed that the expression of the Fos family member Fra-1, which is involved in extraembryonic vascular development, was reduced in yolk sac tissues of zimp10 null embryos. Using fra-1 promoter/reporter constructs, we further demonstrate the regulatory role of Zimp10 on the transcription of Fra-1. This study provides evidence to demonstrate a crucial role for Zimp10 in vasculogenesis.
The PIAS proteins originally were identified as repressors of the STAT transcription factors and have been shown to function as transcriptional coregulators that modulate the activity of a diverse set of transcription factors, such as p53, the Smads, and steroid hormone receptors (11, 16, 25, 31). Members of the PIAS family share a high degree of sequence similarity and are characterized by the presence of a highly conserved zinc finger domain, called the Miz (Msx-interacting zinc finger) domain (37). This motif appears to be important for interactions with target proteins and is highly similar to the RING finger domain present in E3 ubiquitin ligases (31). Indeed, numerous studies have implicated a role for the PIAS proteins in the ubiquitin-like sumoylation pathway (12, 14, 15, 27).
Zimp10 is a novel PIAS-like protein that originally was identified as an androgen receptor (AR)-interacting protein (33). Zimp10 and its homolog, Zimp7, share the highly conserved Miz domain with other members of the PIAS family. These two Zimp proteins contain a strong intrinsic transactivation domain, and through it they augment the transcriptional activity of steroid hormone receptors and Smads (3, 19, 21, 33). It has been shown that Zimp10 and Zimp7 colocalize with SUMO-1 at subcellular replication foci, although the biological consequences of these interactions are unclear. An ortholog of Zimp10 and Zimp7, called tonalli, has been identified in Drosophila melanogaster, and it genetically interacts with the ATP-dependent SWI/SNF and Mediator complexes, suggesting a potential role for the Zimp proteins in chromatin remodeling (8). Additionally, recent data have shown that Zimp7 interacts with Brg-1 and BAF57, components of the mammalian SWI/SNF complexes (9).
The loss of PIAS1 in mice results in increased interferon signaling and enhanced protection against viral and microbial challenge, as well as hypersensitivity to lipopolysaccharide endotoxic shock (22). These animals also display partial perinatal lethality and appear 20 to 40% smaller than wild-type littermates. Disruption of the PIASx gene reduced the testis weight in mice, although the knockouts appear viable and fertile (30). However, PIASy knockout mice display no obvious phenotypic defects (36). Interestingly, no abnormality in SUMO conjugation was observed in the knockout mice described above. To investigate the biological role of Zimp10 in vivo, we took a loss-of-function approach by disrupting the zimp10 allele in mice. Mice homozygous for the zimp10-targeted allele display embryonic lethality at approximately embryonic day 10.5 (E10.5). Severe defects in yolk sac vascular development were observed in zimp10 null mice as early as E9.5, resulting in obvious anemia and growth retardation in these animals. No reduction in hematopoietic potential was identified in yolk sacs deficient for zimp10. Intriguingly, microarray analysis of zimp10 null yolk sacs showed that the expression levels of several genes involved in vascular development were decreased compared to those of wild-type yolk sacs. These data suggest a novel role for Zimp10 in the regulation of vascular development.
MATERIALS AND METHODS
Gene targeting and genotyping.
The targeting vector for the Zimp10 gene was constructed by ligating a PCR-amplified 1.3-kb fragment and a 9-kb lambda phage fragment (clone no. 23; Ingenious Targeting Labs, Stonybrook, NY), both homologous to the zimp10 genomic sequence, into a targeting vector containing a PGKneo cassette. This construct was designed to replace 2.7 kb of the zimp10 sequence containing exons 8 to 10 (amino acids 149 to 314) and to generate a frameshift mutation for the remaining Zimp10 transcript. The targeting vector was linearized and electroporated into 129 SvEv iTL1 embryonic stem cells, and recombinant clones were selected in medium containing G418. Targeted embryonic stem cells were confirmed by PCR, and two independent recombinant clones were microinjected into C57BL/6J blastocysts. Chimeric mice then were crossed with C57BL/6J mice to achieve germ line transmission of the neomycin-disrupted Zimp10 allele. For genotyping, mouse tail tips or embryo yolk sacs were incubated in lysis buffer (100 mM NaCl, 10 mM EDTA, 20 mM Tris-HCl, pH 7.5, 0.5% sodium dodecyl sulfate, 400 μg/ml proteinase K) overnight at 60°C, phenol extracted, and ethanol precipitated. Genomic DNA was dissolved in 0.5× Tris-EDTA and PCR amplified using three primers to distinguish the wild-type allele from the mutant allele: wild type, 5′-GGCAGCCTTGCAAGAGACACAGAACA-3′; neo cassette forward, 5′-TGCTGTCCATCTGCACGAGACTA-3′; common reverse, 5′-GCCTGGGTAAAATCCACAGA-3′. PCR fragments were amplified at 95°C for 5 min and then for 40 cycles at 95°C for 45 s, 63°C for 50 s, and 72°C for 45 s, followed by a final step at 72°C for 5 min. For the genotyping of histological sections, a needle was used to scrape tissue from individual sections, the tissue was deparaffinized by incubation with xylenes and centrifuged, and the genotyping procedure was performed as described above.
Western blot analysis.
E9.5 embryos from heterozygous intercrosses or lentivirus-infected HEK293 cells were collected, and protein was extracted in lysis buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 1% NP-40, 5 mM EDTA, 300 mM NaCl, 150 mM KCl, 1 mM dithiothreitol, 10 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 mM phenylmethylsulfonyl fluoride, and 0.5 mM sodium orthovanadate) on ice for 10 min. Samples then were sonicated for 5 s on ice using a sonic dismembrator (model 100; Fisher Scientific) at 30% power output. Lysates were cleared by centrifugation at 4°C, and supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Proteins were transferred to nitrocellulose membranes, blocked with 5% milk, and immunoblotted using appropriate primary and species-specific horseradish-peroxidase-conjugated secondary antibodies. Two homemade Zimp10 antibodies (αZimp10 KC and αZimp10 R1) were used at a 1:500 dilution in independent immunoblots of embryo lysates. The anti-α-tubulin antibody (clone DM1A; Neomarkers, Freemont, CA) was used at a 1:1,000 dilution. For Zimp10 small hairpin RNA (shRNA)-infected cells, the αZimp10 R1 antibody was used.
Reverse transcription-PCR (RT-PCR) and microarray analysis.
Total RNA was extracted from E9.5 yolk sacs using RNAbee RNA isolation reagent (Tel-Test inc., Friendswood, TX) according to the manufacturer's recommendations. One to 2 μg of RNA was reverse transcribed using AMV reverse transcriptase (Promega, Madison, WI) with oligo(dT) and random primers in a total volume of 20 μl. Primers for amplifying Zimp10 were 5′-GCACAGTGATGGGTCATTTC-3′ (forward) and 5′-CAGGGTAACTGGCCCCAAAG-3′ (reverse); those for Fra-1 were 5′-TCTGGCCTATCCCCAGTACA-3′ (forward) and 5′-CCTTCTGCTTCTGCAGCTCT-3′ (reverse). The Zimp10 and Fra-1 cDNA was amplified at 95°C for 5 min, followed by three cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and then 30 (Zimp10) or 33 (Fra-1) cycles of 95°C for 1 min, 58°C for 1 min, and 72°C for 1 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified with the following primers: 5′-CCATGGAGAAGGCTGGGG-3′ (forward) and 5′-CAAAGTTGTCATGGATGACC-3′ (reverse) as described previously (21).
For microarray analysis, wild-type and zimp10 null E10 yolk sacs were isolated from a heterozygous intercross, and RNA samples were isolated as described above and purified using the RNeasy kit (Qiagen, Valencia, CA). Three RNA samples from each genotype were pooled, amplified, and labeled using the Affymetrix one-cycle target-labeling protocol (Santa Clara, CA). Samples then were hybridized to Affymetrix mouse 430 2.0 oligonucleotide arrays.
Whole-mount in situ hybridization, immunohistochemistry, and histology.
Wild-type E9.5, E10.5, E11.5, and E12.5 embryos were isolated and fixed in 10% neutral-buffered formalin at 4°C overnight. The in situ hybridization procedure was performed as described previously (26a). Two human Zimp10 fragments for either the N-terminal region (between amino acids 152 and 238) or the C-terminal untranslated region (between nucleotides 3383 and 3863) were generated by PCR with specific primers and were cloned into T Easy vector (Promega). Either sense or antisense RNA probes were generated using the Roche DIG RNA labeling kit (SP6/T7; Palo Alto, CA).
For immunohistochemistry, embryos and yolk sacs were isolated and fixed in 10% neutral-buffered formalin overnight at 4°C and processed through a gradient of 25, 50, 75, and 100% methanol. Endogenous peroxidase activity was blocked by incubating samples in 5% hydrogen peroxide-methanol for 5 h. Samples were washed twice in methanol, processed through a methanol gradient to phosphate-buffered saline (PBS), blocked twice for 1 h each in PBSMT (3% nonfat milk, 0.1% Triton X-100 in PBS), and incubated with a rat anti-PECAM antibody at a 1:50 dilution (Roche) or a chicken anti-Zimp10 antibody at a 1:1,500 dilution overnight at 4°C. Samples then were washed five times for 1 h each in PBSMT, incubated with either a goat anti-rat antibody-horseradish peroxidase (1:200 dilution; Roche) or a rabbit anti-chicken antibody-horseradish peroxidase (1:2,000 dilution; Promega), and exposed to DAB substrate (Vector Laboratories, Burlingame, CA) for visualization. For histological analysis, 7-μm serial sections were processed from xylene to water through a decreasing ethanol gradient, stained with hematoxylin and eosin, and processed back to xylene through an increasing alcohol gradient. Coverslips were mounted using Permount mounting medium (Fisher Scientific). Images for all experiments in this study were acquired on a Leica dissecting microscope (model MZ95) using a Zeiss Axiocam and Axiovision software.
Methocellulose colony formation assay.
For the methocellulose colony formation assay, yolk sacs were isolated, washed in Iscove's medium containing 2% fetal bovine serum (FBS), and disaggregated in 0.25% collagenase at 37°C for 10 min. Cells (5 × 104) were plated in methocult semisolid medium (Stem Cell Technologies). Erythroid colonies were counted on day 4, and myeloid colonies were counted on day 11 (18).
Ink injection.
E9.5 embryos were dissected in PBS and immediately injected with 20% India ink diluted in PBS-Tween 20 (0.1%) using a 5-μm microcapillary device and a mouth pipette (34). To allow for the visualization of the yolk sac vasculature, yolk sacs were dissected away from the embryos but remained attached. Images were acquired immediately after ink injection using the Leica imaging system described above.
Proliferation assay.
Mouse embryo fibroblast (MEF) isolation and the Zimp10-specific shRNA have been described previously (21). The Zimp10 shRNA lentiviruses were generated using the pLentiSuper vector and were produced in HEK293T cells. MEFs were plated in 96-well dishes in full medium, and cell viability was determined over a 4-day time course using the MTS proliferation assay kit (Promega). HEK293 cells were transduced in 6-well plates with Zimp10 shRNA or green fluorescent protein (GFP) control lentiviruses for 3 days and then were replated in 96-well dishes. Cell viability was determined over an 8-day time course. Prior to MTS reagent addition, bright-field and fluorescence images were captured for each time point. The above-described cells also were plated in full medium in 12-well dishes for colony formation assays as described previously (35).
Luciferase reporter assays.
The fra-1 promoter constructs were a kind gift from Sekhar Reddy, Johns Hopkins University (1). HEK293 cells were maintained in 10% FBS-Dulbecco's modified Eagle's medium and seeded in 48-well plates at a density of approximately 1 × 105 cells/well. Transient transfection and luciferase assays were performed as described previously (33).
RESULTS
Expression of Zimp10 during development.
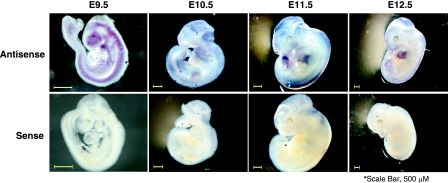
Using in situ hybridization approaches, we assessed the expression of Zimp10 during mouse development using wild-type embryos. As shown in Fig. 1, Zimp10 expression was specifically detected throughout the embryo at E9.5 by an antisense probe derived from the N terminus of Zimp10. The areas of intense Zimp10 staining included the forebrain, the midbrain, and the mandibular brachial arch. In addition, Zimp10 displayed widespread dorsal expression, which persisted throughout development and appeared to become more localized to the periphery of the embryo at later stages. Craniofacial expression of Zimp10 was apparent at all stages examined and also became more localized to the periphery of the embryo as development progressed. Interestingly, the embryo limb buds displayed significant Zimp10 staining, which was most pronounced at E12.5. Similar staining patterns also were observed with a C-terminal Zimp10 antisense probe on E9.5 to E12.5 embryos (data not shown). However, no staining was observed with the sense probes used as negative controls (Fig. 1, bottom row).
FIG. 1.
Zimp10 expression during development. Wild-type embryos were isolated at E9.5, E10.5, E11.5, and E12.5 and subjected to in situ hybridization using an RNA probe specific for the Zimp10 sequence (amino acids 152 to 238) as described in Materials and Methods. The magnification settings were adjusted according to the stage so that the embryo filled the field of view. Top panel, antisense probe; bottom panel, sense probe. Scale bar, 500 μm.
Disruption of the Zimp10 gene.
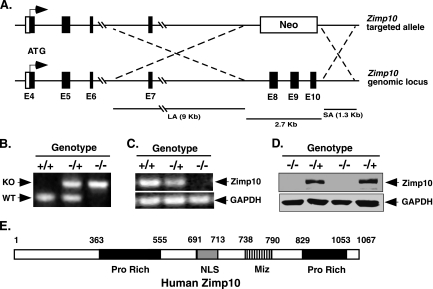
To investigate the biological role of Zimp10 in vivo, we took a loss-of-function approach by generating a zimp10-deficient mouse strain. The mouse zimp10 gene (also named zmiz1) is localized to chromosome 14 and is organized into 24 exons spanning an approximately 205-kb region of genomic DNA. A 2.7-kb fragment encompassing the region between exons 8 and 10 (amino acids 149 to 314) was replaced with a PGKneo cassette through homologous recombination, which also resulted in a missense mutation in the remaining Zimp10 transcript (Fig. 2A). Two independent embryonic stem cell clones heterozygous for the disrupted allele were injected into C57BL/6 blastocysts, and germ line transmission was achieved by crossing chimeric mice with wild-type C57BL/6J mice.
FIG. 2.
Targeted disruption of the Zimp10 gene. (A) Schematic representation of the Zimp10 targeting strategy. The Zimp10 locus was disrupted with a targeting vector that replaced approximately 2.7 kb of the Zimp10 genomic sequence containing exons 8 to 10 with a neomycin (Neo) resistance gene. The top line represents the neomycin targeting vector, and the bottom line represents the Zimp10 genomic sequence. The ATG translational start site is indicated with arrows. LA, long homologous recombination arm; SA, short homologous recombination arm. (B) PCR-based genotyping of E9.5 embryos using primers specific for the wild-type or neomycin-targeted allele. Knockout (KO) and wild-type (WT) bands are indicated. (C) RT-PCR analysis of Zimp10 transcripts in E9.5 embryos using primers specific for the N-terminal region of Zimp10. GAPDH cDNA was amplified as a control. (D) Western blot using affinity-purified anti-Zimp10 antibody to examine the expression of Zimp10 proteins in E9.5 embryos. (E) A schematic representation of the human Zimp10 protein. The numbers correspond to the amino acid sequence of the protein. The putative functional domains are marked as the proline-rich domain (Pro Rich), the nuclear localization signal (NLS), and the Miz domain (Miz).
Mice heterozygous for the disrupted Zimp10 allele (zimp10+/−) were viable and fertile and displayed no phenotypic abnormalities compared to the phenotype of wild-type littermates (zimp10+/+). The presence of the Neo-disrupted allele was confirmed by PCR analysis of genomic DNA isolated from the yolk sacs of E9.5 embryos (Fig. 2B). RT-PCR analysis revealed that the transcription of Zimp10 was disrupted, resulting in the absence of the portion of the Zimp10 transcript starting at exon 8 (Fig. 2C). Using a C-terminal anti-Zimp10 antibody, we failed to detect full-length Zimp10 protein in embryos homozygous for the zimp10-disrupted allele (zimp10−/−) by Western blot analysis (Fig. 2D), which was further confirmed by reblotting the same embryo lysates with another anti-Zimp10 antibody (data not shown). Taken together, the above data demonstrate that the expression of Zimp10 protein was disrupted in zimp10 null mice.
Loss of functional Zimp10 protein results in embryonic lethality around E10.5.
Both heterozygous males and females were fertile and phenotypically indistinguishable from wild-type littermates. However, no homozygous mutants were produced from heterozygous intercrosses, suggesting that homozygosity for the disrupted Zimp10 allele results in embryonic lethality (Table 1). To assess the dynamics of the embryonic lethality, embryos from heterozygous intercrosses were analyzed at different stages of gestation. zimp10−/− embryos were obtained with the expected Mendelian frequency at E9.5 or earlier. By E10.5, the number of viable homozygous embryos was significantly reduced. The knockout embryos appeared to be severely underdeveloped and in the process of dying (Fig. 3A). No viable homozygous embryos were observed at E11.5 or later (Table 1). The above results suggest that fetuses lacking functional Zimp10 proteins died in utero at around E10.5 of gestation. In addition, heterozygous mutant mice were underrepresented among weaned pups (Table 1) but appeared otherwise indistinguishable from wild-type littermates. No neonatal lethality was seen among zimp10+/− mice, suggesting that a fraction of the heterozygous embryos died in utero.
TABLE 1.
Zimp10 homozygotes die at E10.5 of gestation
| Time point | No. (%) of animals
|
Total | ||
|---|---|---|---|---|
| Zimp10+/+ | Zimp10+/− | Zimp10−/− | ||
| E9.5 | 122 (25) | 248 (50) | 122 (25) | 492 |
| E10.5 | 18 (27) | 33 (50) | 15a (23) | 66 |
| E11.5-E15.5 | 8 (28) | 21 (72) | 0 (0) | 29 |
| Postnatal | 224 (40) | 340 (60) | 0 (0) | 564 |
Dying embryos.
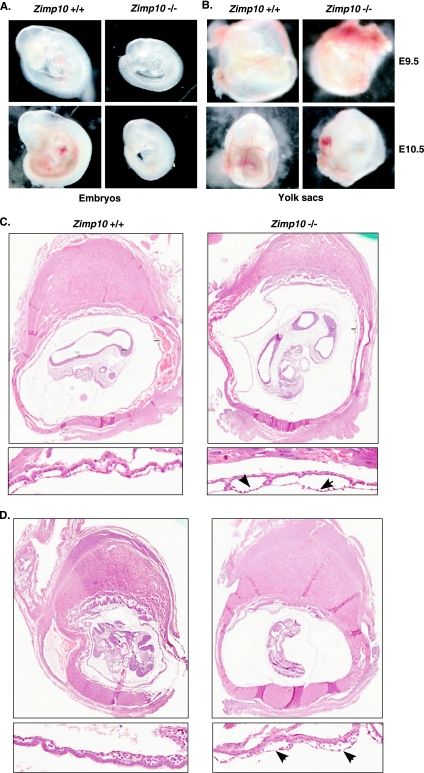
FIG. 3.
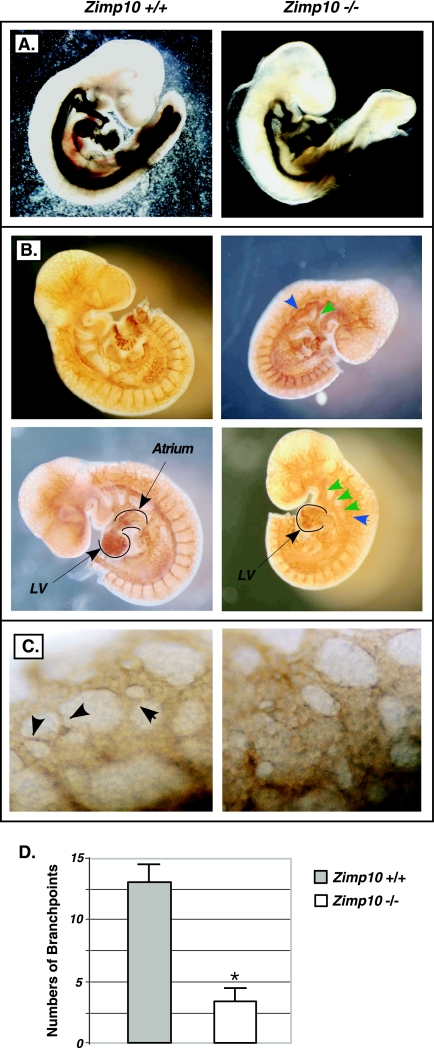
Morphology of Zimp10 knockout embryos. (A) Images of freshly dissected wild-type and zimp10−/− embryos at E9.5 and E10.5. Growth retardation and anemia of the zimp10−/− embryos is observed at E9.5 (top images) and is more pronounced at E10.5 (bottom images). (B) Images of yolk sacs from the embryos presented in panel A. Prominent defects in vascular development were noted in zimp10−/− yolk sacs at E10.5 (bottom images), which also are evident at earlier stages (top images). (C) Low- and high-magnification images of hematoxylin- and eosin-stained zimp10+/+ and zimp10−/− littermate embryo sections at E9.5. Arrows show dilated vessels in zimp10−/− yolk sacs. (D) A second set of embryo littermate sections was analyzed as described for panel C.
Embryos lacking zimp10 are retarded in growth and development.
In general, zimp10−/− embryos were indistinguishable from wild-type littermates at E8.5, but they were developmentally retarded to various degrees at the E9.5 stage. As shown in Fig. 3A, zimp10−/− embryos generally were pale and small compared to wild-type and zimp10+/− embryos, but no gross developmental abnormalities were observed under bright-field microscopy. By E10.5, knockout embryos were approximately half the size of wild-type littermates and appeared severely anemic (Fig. 3A). Beating hearts were observed in zimp10−/− embryos at E9.5 and E10.5. However, no branching vascular network was observed in zimp10−/− yolk sacs (Fig. 3B). Histological analysis of E9.5 embryos showed the presence of dilated blood vessels in zimp10−/− yolk sacs (arrows in figures), while placental development appeared normal (Fig. 3C and D). The above results provide the first line of evidence suggesting that zimp10 knockouts display abnormalities in vascular development.
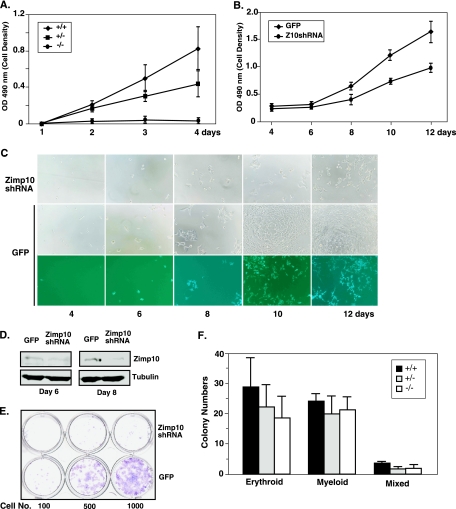
The retarded growth phenotype of zimp10−/− embryos suggests that cell proliferation is reduced during their development. To examine this possibility, we used several approaches to assess the potential effects of Zimp10 on cell growth and survival. First, we isolated MEFs of different genotypes from heterozygous intercrosses and tested their viability using the MTS assay. As shown in Fig. 4A, a significant reduction in cell growth was observed in zimp10−/− MEFs compared to that of wild-type or heterozygous MEFs (P < 0.05). To confirm the role of Zimp10 in cell growth and survival, we generated specific Zimp10 shRNA lentiviruses and infected them into HEK293 cells. Western blots showed that the level of endogenous Zimp10 protein was reduced in cells infected with the Zimp10 shRNA lentivirus compared to that of the ones infected with the GFP lentivirus, which were used as negative controls (Fig. 4D). Accordingly, the cell numbers were 35 and 45% lower at days 4 and 5, respectively, after Zimp10 shRNA infection compared to that of controls according to the MTS assay (P < 0.05) (Fig. 4B). The reduction in cell viability also was observed directly under bright-field microscopy (Fig. 4C). The effect of Zimp10 on cell growth and survival then was assayed using a colony formation assay. A series of dilutions of cells that either were infected by the Zimp10 shRNA or GFP lentiviruses were seeded in 6-well plates. After 14 days, cells were fixed and stained with crystal violet. A significant reduction in the number and size of colonies in the samples infected with the Zimp10 shRNA lentiviruses was observed compared to that of samples infected with the control viruses (Fig. 4E). Taken together, the above results suggest an important role for Zimp10 in cell proliferation and survival.
FIG. 4.
Effects of Zimp10 loss on cell viability and hematopoietic potential. (A) MTS cell proliferation assay using E9.5 MEFs isolated from zimp10 heterozygous intercrosses (+/+, n = 4; +/−, n = 4; −/−, n = 3). (B to E) HEK293 cells were infected with Zimp10 shRNA (Z10shRNA) lentivirus or GFP control virus and replated for the assessment of cell viability using the MTS cell proliferation assay (B), bright-field and fluorescence microscopy (C), or clonogenic assay (E). (D) Western blot analysis of the knockdown effect in cells after 6 or 8 days of infection with Zimp10 shRNA (top panel). The same blots were probed with an antitubulin antibody as a control. (F) E9.5 yolk sacs were isolated from heterozygous intercrosses, disaggregated, and plated for hematopoietic colony-forming ability on methocellulose semisolid medium plus hematopoietic growth factors (5 × 104 cells/well). Erythroid colonies were counted on day 4, and myeloid colonies were counted on day 11. Erythroid-myeloid mixed colonies were counted accordingly. Bars represent averages and standard deviations of results from multiple yolk sacs isolated from the same litter (Zimp10+/+, n = 6; Zimp10+/−, n = 7; Zimp10−/−, n = 6). OD 490 nm, optical density at 490 nm.
Normal myeloerythroid hematopoiesis in zimp10−/− yolk sacs.
The anemic appearance of zimp10 knockout embryos prompted us to investigate whether there was a reduction in their hematopoietic potential compared to that of wild-type littermates. Hematopoietic colony-forming assays were performed on E9.5 yolk sacs by plating cells onto methylcellulose-containing medium supplemented with the appropriate cytokines (18). As shown in Fig. 4F, no significant differences in the total number and content of the erythroid, myeloid, and mixed colonies were observed in homozygous, heterozygous, or wild-type yolk sacs. Therefore, it appears that Zimp10 does not play a primary role in the early differentiation and maturation of erythroid and myeloid lineages.
zimp10−/− embryos have abnormal vascular development.
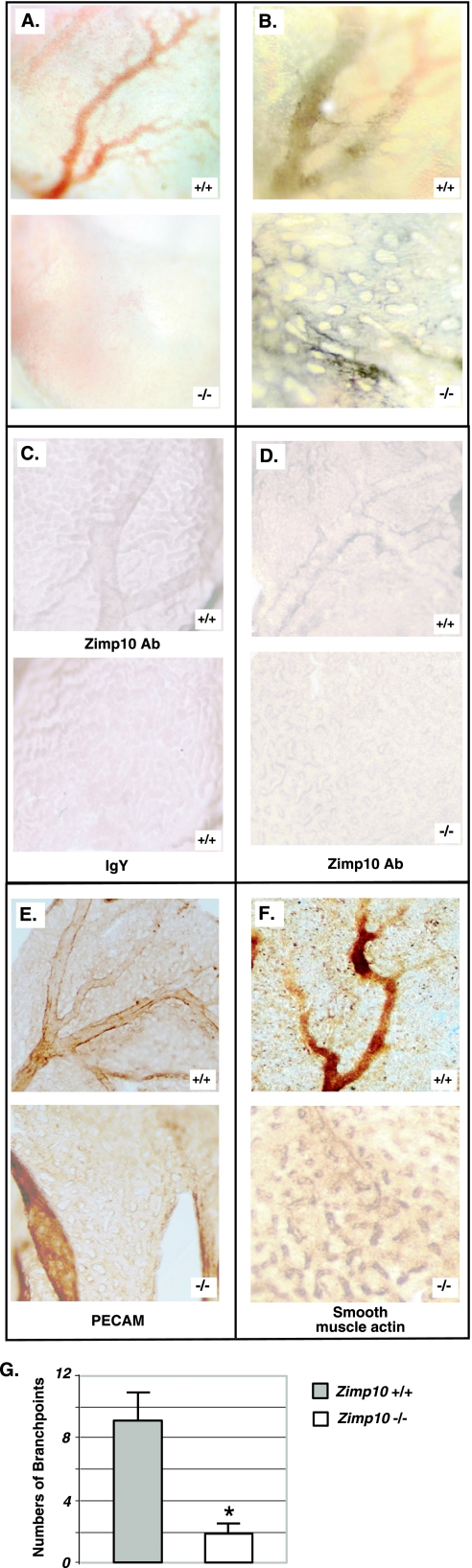
To continue searching for the possible mechanisms of the lethality of zimp10−/− mice, we investigated potential defects in vascular development in zimp10−/− embryos. To assess the integrity of the vasculature, we first injected India ink into the cardiac outflow tract of normal and knockout embryos at E9.5 to visualize the vascular network (34). As shown in Fig. 5A, the main vascular network in zimp10−/− embryos appeared relatively normal; however, the vessels within the branchial arches and dorsal aorta were smaller and less well defined. To further characterize the vascular network in detail, we stained E9.5 whole-mount embryos with an antibody specific for the endothelium-specific marker PECAM (4). We observed dorsal aortas in zimp10 null embryos that were dilated compared to those of the wild type (Fig. 5B). Although pharyngeal arch arteries are present in the knockouts, they were smaller than and not as well defined as those of the wild-type embryos (Fig. 5B, arrows). The hearts in the knockouts appeared abnormal. In particular, the left heart valve and atrium were smaller and underdeveloped in the knockouts (Fig. 5B, bottom images, marked areas). In addition, under high-magnification, zimp10−/− embryos appeared to have fewer small capillary vessels within the head region than wild-type embryos (Fig. 5C, arrows). To confirm this observation, the numbers of capillary branch points in the head region were quantified using PECAM-stained E9.5 embryos as described previously (29). The data from two independent experiments showed a significant difference between wild-type and knockout embryos (Fig. 5D). Taken together, the above results suggest the presence of vascular defects in zimp10−/− embryos.
FIG. 5.
Analysis of embryo vasculature. (A) India ink was injected into the cardiac outflow tract of E9.5 zimp10+/+ and zimp10−/− embryos to visualize the embryo vasculature. (B) Whole-mount immunostaining of E9.5 zimp10+/+ and zimp10−/− embryos using an antibody against the endothelium-specific marker PECAM. (C) High magnification of the head region of PECAM-stained embryos. Small capillaries are observed in wild-type embryos (arrows) but not in zimp10−/− embryos. (D) Branching capillaries within the perineural vascular plexus of PECAM-stained E9.5 embryos were quantified. Bars represent the averages and standard deviations from duplicate experiments. *, P < 0.05.
Embryos lacking zimp10 display severe defects in yolk sac vascular development.
The above defects in zimp10−/− embryos, although obvious and interesting, did not appear to be the primary cause of embryonic lethality. Since the development of an intact extraembryonic vasculature is essential for embryo viability during midgestation, we analyzed zimp10−/− extraembryonic tissues in further detail. Upon dissection of E9.5 and E10.5 embryos, the surface of the yolk sacs in zimp10 knockouts appeared pitted and pale (Fig. 3B and 6A). Intriguingly, we discovered the complete absence of a branching vascular network in zimp10−/− yolk sacs, which was most pronounced at E10.5. To identify the precise abnormalities of zimp10−/− yolk sacs, we systematically characterized yolk sac vascular development using E9.5 embryos isolated from heterozygous intercrosses. First, Zimp10 expression in the yolk sac vasculature of wild-type embryos was confirmed using whole-mount immunohistochemistry with a chicken antibody against the N terminus of Zimp10. As shown in Fig. 6C, staining of the yolk sac vascular network was observed with an anti-Zimp10 antibody (top image) but not with immunoglobulin Y, which was used as a control (bottom image). As expected, this vascular-network-specific staining was not observed in zimp10−/− yolk sacs using the Zimp10 antibodies (Fig. 6D, bottom image). Staining for the endothelial cell marker PECAM then was carried out. The data showed that endothelial cells were present in zimp10−/− yolk sacs but failed to organize into a vascular network (Fig. 6E). Following primitive endothelial tube formation, mesenchymal cells are recruited and differentiate into vascular smooth-muscle cells to establish mature blood vessels (28). Smooth-muscle actin staining showed that smooth-muscle cells in the zimp10−/− yolk sac are mislocalized and accumulate in a spotted, punctate pattern (Fig. 6F). In contrast, a branching network of vessels was clearly observed in normal yolk sacs by bright-field microscopy or immunostaining with antibodies against PECAM or smooth-muscle actin (Fig. 6A, E, and F). Analysis of the capillary branch point numbers in yolk sacs showed a significant reduction in the knockouts (Fig. 6G). To determine the integrity of intact vessel networks in zimp10−/− yolk sacs, the yolk sacs were deflected from the dissected E9.5 embryos that were injected with India ink through the cardiac outflow tract of the heart. Under bright-field microscopy, we observed that the ink traveled through the embryo vasculature and into the yolk sac vasculature. However, the vasculature was severely disorganized and lacked the typical branching pattern of vessels that was observed in zimp10+/+ yolk sacs (Fig. 6B). These results suggest a potential role for Zimp10 in the remodeling of the primitive yolk sac vascular plexus during embryogenesis, as a loss of Zimp10 function results in the observed defects in yolk sac vasculature during midgestation.
FIG. 6.
Severe vascular defects in zimp10−/− yolk sacs. E9.5 yolk sacs were dissected away from the embryos as described in Materials and Methods and were used for the following analyses. (A) Bright-field microscopy; (B) India ink injection; (C) wild-type yolk sacs stained with the anti-Zimp10 antibody (Ab) or immunoglobulin Y (IgY) as a control; (D) normal or zimp10−/− yolk sacs stained with anti-Zimp10 antibody; (E) PECAM immunostaining; (F) smooth-muscle actin immunostaining. (G) Vascular branch points were quantified using PECAM-stained yolk sacs from wild-type or zimp10−/− animals. Bars represent averages and standard deviations from duplicate experiments. *, P < 0.05.
Zimp10 regulates expression of the Fra-1 transcription factor.
Previous studies have shown that Zimp10 is a transcriptional coregulator and modulates the transcription mediated by multiple transcriptional factors (21, 33). To search for potential molecular mechanisms by which Zimp10 regulates vascular development, we evaluated possible downstream targets of Zimp10 using microarray analysis. RNA samples from wild-type and knockout E10 yolk sacs were isolated, and expression was analyzed using Affymetrix mouse arrays (http://smd.stanford.edu). Interestingly, a variety of transcripts encoding proteins involved in vascular development were shown to be downregulated in zimp10−/− yolk sacs, including proliferator protein-activated receptor gamma (PPAR-γ), activator protein 2 gamma (AP-2γ), GATA-2, GATA-3, and Fra-1 (Table 2). Using RT-PCR analysis, we confirmed that the amount of Fra-1 transcript was reduced in zimp10−/− yolk sacs (Fig. 7A). To determine if residual Fra-1 expression in the zimp10−/− samples was due to Zimp7 expression, we infected zimp10−/− MEFs with a specific Zimp7 shRNA lentivirus or control virus but did not observe a further decrease in Fra-1 levels (data not shown). Fra-1 is a member of the fos family of transcriptional regulators and heterodimerizes with Jun family members to form the AP-1 transcription factor (38). fra-1 knockout embryos display embryonic lethality around E10, with defects in placental and yolk sac vasculature. Given the striking similarity between the phenotypes of zimp10 and fra-1 knockouts, we further investigated the potential role of Zimp10 in Fra-1 transcription. Using luciferase reporter constructs driven by a functional Fra-1 promoter (1), we assessed the regulatory role of Zimp10 on Fra-1 transcription. As shown in Fig. 7B, exogenous Zimp10 expression increased the activities of three fra-1 promoter/reporter constructs approximately three- to fivefold in HEK293 cells. Zimp10 in particular showed a high level of induction of the fra-1-570-luc reporter construct. Interestingly, Zimp7, a homolog of Zimp10, appeared to show an even greater enhancement on the fra-1 promoter/reporter plasmids than the Zimp10 expression construct. To confirm the role of Zimp10 in Fra-1 transcription, we knocked down the expression of endogenous Zimp10 using specific shRNA constructs in HEK293 cells as described previously (20, 21). As shown in Fig. 7C, a significant decrease of endogenous Fra-1 transcript was observed in samples infected with the Zimp10 shRNA viruses compared to that of the controls. Specific knockdown of the Zimp10 protein in this experiment was confirmed by Western blotting (Fig. 7D). These results provide evidence to demonstrate a potential role for Zimp10 in Fra-1 transcription, which may contribute to the vascular defects observed in zimp10 null mice.
TABLE 2.
Microarray analysis of zimp10−/− yolk sacs
| Gene | Role in vascular development | Fold change from level in wild type |
|---|---|---|
| PPAR-g | Endothelial tube formation | −6.3 |
| AP-2g | Transcription factor involved in trophoblast cell development | −5.9 |
| BMP8b | Ligand for TGF-b receptor | −5.7 |
| fra-1 | Early endothelial cell development | −4.7 |
| GATA-3 | Proliferation/differentiation of endothelial cells | −4 |
| GATA-2 | Proliferation/differentiation of endothelial cells | −3.9 |
FIG. 7.
Zimp10 regulates expression of the Fra-1 transcription factor. (A) RT-PCR analysis for Fra-1 expression in E9.5 yolk sacs isolated from zimp10+/+, zimp10+/−, or zimp10−/− yolk sacs. Specific primers were used to amplify Zimp10 (top), Fra-1 (middle), or GAPDH (bottom) transcripts. (B) HEK293 cells were transfected with luciferase (Luc) reporter constructs containing 861 (−861-Luc), 570 (−570-Luc), or 105 (−105-Luc) bp of the fra-1 promoter in the presence of pcDNA3-hZimp7 or pcDNA3-hZimp10 expression vector. A constitutive β-galactosidase (β-gal) vector also was included to control for transfection efficiency. After 48 h, cells were isolated and luciferase and β-gal values were determined. Bars represent luciferase/β-gal averages and standard errors from triplicate determinations. (C) The levels of Fra-1 transcripts were assessed in two independent experiments (Exp) by quantitative RT-PCR assays in HEK293 cells infected with Zimp10 shRNA or control lentiviruses. (D) Western blot analyses were performed using the whole-cell lysates isolated from the cells depicted in panel C.
DISCUSSION
Zimp10 is emerging as a transcriptional coregulator in a variety of important cellular pathways, including the AR, transforming growth factor β (TGF-β), and p53 signaling cascades (20, 21, 33). However, its biological role in vivo still remains unclear. To address this question, we have generated mice with a targeted disruption of the zimp10 gene. In this study, we show that embryos deficient for the Zimp10 allele die around E10.5 due to severe defects in yolk sac vessel organization, indicating that Zimp10 plays an essential role in the extraembryonic vascular development process. Subtle differences also were observed in aortic arch architecture, heart ventricle development, and the number of branching capillaries in the head region in zimp10 knockout embryos. Additionally, cell proliferation was inhibited in zimp10−/− embryo fibroblasts and in cells with reduced Zimp10 levels. Despite the anemic appearance of the zimp10−/− embryos, the hematopoietic potential appeared normal in zimp10−/− yolk sacs. Intriguingly, we provide evidence in this report demonstrating that the most severe defect in zimp10 knockouts is the failure of the yolk sac vascular plexus to form a mature vascular network, which is critical for proper development during embryogenesis.
By around E9.5, embryos no longer can survive by the direct exchange of gasses and nutrients, and thus the establishment of a functional extraembryonic vasculature becomes critical (10). The identification of a poorly organized yolk sac vasculature in zimp10 knockout embryos suggests that this defect is the primary cause of embryonic death around E10.5. Our results are consistent with those of previous studies showing similar embryonic lethal phenotypes between E9.5 and E10.5 that result from yolk sac defects (7, 17, 18, 34). Although we have observed that a primitive vascular network of large vessels is present in zimp10−/− yolk sacs, we demonstrated that the vessels fail to develop from a primitive vascular plexus into a mature vascular network. In addition, fewer branching capillary structures were observed in the head region of zimp10−/− embryos, and subtle differences in vessel and heart architecture were observed, suggesting that vascular abnormalities may exist in zimp10−/− embryos as well. The staining of zimp10−/− yolk sacs with a smooth-muscle actin antibody revealed that smooth-muscle cells are organized in a punctate pattern rather than the expected branching network seen in wild-type yolk sacs. This result provides evidence that a mature vessel network is unable to form in zimp10−/− yolk sacs, which may be one of the primary factors resulting in the embryonic lethality of zimp10−/− animals. Although the zimp10 knockout system is the first in vivo system to implicate a potential role for PIAS-like proteins in vascular development, previous studies have shown potential roles for the PIAS proteins in the regulation of angiogenesis in vivo and in smooth-muscle differentiation in cell culture systems through the STAT signaling pathways (2, 13). Thus, the roles of Zimp10 and other PIAS proteins in angiogenesis and vasculogenesis should be further explored.
Interestingly, disruptions of the Notch and TGF-β pathways in mice result in similar defects in vascular remodeling (6, 7, 17, 18, 34). In these models, abnormalities are observed around E9.5, and embryonic death occurs by E10.5. In particular, the TGF-β type I receptor knockout mice showed vascular defects very similar to those observed in zimp10−/− mice. Previous studies have shown a potential link between PIAS proteins and Smad proteins, downstream transducers of TGF-β pathways (23, 24). Interestingly, our recent works showed that Smad3/4 transcriptional activity is impaired in Zimp10 knockout embryos, implying a regulatory role for Zimp10 in Smad3- and Smad4-mediated transcription (21). However, we were unable to identify significant differences in the expression of notch components in the zimp10−/− yolk sacs by RT-PCR in this study (data not shown). Further study of the precise molecular mechanisms by which Zimp10 regulates the development of the vascular system and angiogenesis may help us to understand the interaction between Zimp10 and TGF-β pathways.
As we have reported previously, Zimp10 is a PIAS-like protein and shares structural and functional similarities with members of the PIAS family. The biological roles of PIAS proteins have been investigated in knockout mouse models. Disruption of the PIASx and PIASy genes did not show significant defects in mice (30, 36). In contrast, PIAS1 knockout mice display partial perinatal lethality before E17.5, and mice that survive are 20 to 40% smaller than wild-type littermates (22). In addition, these mice displayed enhanced interferon-mediated antiviral activity and increased protection against pathogenic infection. Given the fact that the PIAS proteins are very similar structurally, it is possible that other members of the PIAS family compensate for the function of PIAS1x and PIAS1y in their knockout mice. The creation of knockouts with multiple PIAS protein deletions may provide useful information in this regard. The phenotype of zimp10 null mice appears more severe than those observed for PIAS knockouts. These data suggest a unique and important role for Zimp10 in early development, which may be distinct from and cannot be compensated for by other PIAS proteins. While zimp10 knockout results in embryonic lethality by E10.5 with 100% penetrance, intriguingly, we observed that Zimp10 heterozygotes are born at less than the expected Mendelian ratio. It is currently unclear at what stage heterozygote lethality might occur, although several litters have been examined from E9.5 to E15.5 with no observable difference between wild-type and heterozygous embryos (data not shown). It will be very interesting to further identify the lethality of the heterozygotes, which may help us to further understand the biological role of Zimp10 in development.
Vascular development requires the coordinated activation of several transcription factors through a variety of different pathways, including the TGF-β, Notch, VEGF, Wnt, and BMP pathways (5, 26). The widespread implementation of knockout mouse models has led to the identification of a subset of transcription factors that are essential for vasculogenesis (28). To examine the precise role of Zimp10 in vascular development, we used a microarray approach to examine the transcription profiles of zimp10+/+ and zimp10−/− yolk sacs. A number of cDNA sequences encoding proteins that may contribute to vascular development were identified to be downregulated in zimp10 knockout yolk sacs, including Fra-1, PPARγ, AP2γ, GATA-2, and GATA-3 (Table 2). Using RT-PCR, we further confirmed the reduction of Fra-1 expression in Zimp10 knockouts. Previous studies have shown that fra-1 knockout mice display defects in the placental and yolk sac vasculature and die at around E10 (32). Using fra-1 promoter/reporter constructs, we further confirmed a regulatory role for Zimp10 in the fra-1 promoter in human HEK293 cells (Fig. 7B). In addition, quantitative RT-PCR experiments revealed that the fra-1 transcript is downregulated in HEK293 cells infected with a Zimp10-specific shRNA construct (Fig. 7C and D). Interestingly, Zimp7, a homolog of Zimp10, also affected the activity of the fra-1 promoter constructs in the in vitro reporter assays. However, as shown in this paper, Zimp7 appears to be unable to functionally compensate for Zimp10 in vivo. These observations suggest that Zimp7 is able to functionally compensate for Zimp10 in certain contexts, but that it may depend on the temporal and/or tissue-specific expression of the zimp7 gene. Indeed, our previous work has shown that Zimp7 and Zimp10 have different expression patterns in human tissues (9, 33). Alternatively, differential requirements for transcriptional cofactors that are essential for vascular development may explain this discrepancy. Studies are ongoing to clarify the functional interplay between Zimp10 and Zimp7 in the vasculature. Since Zimp10 has been identified to act as a transcriptional coregulator, our data implicate a possible mechanism by which Zimp10 regulates vascular development through the modulation of Fra-1 transcription.
In conclusion, this study demonstrates an important role for Zimp10 in development. Although we originally identified Zimp10 as an AR coactivator, our current data have elucidated a primary role for Zimp10 in vascular development in vivo. Specifically, homozygous disruption of the zimp10 allele results in defects in the remodeling of the yolk sac vascular plexus into a functional network of mature branching vessels and capillaries. This defect may be due, in part, to reduced expression of fra-1, which appears to be a novel Zimp10 target gene. Further study of the molecular mechanisms by which Zimp10 regulates vasculogenesis and angiogenesis should provide more insight into the role of Zimp10 in development and disease.
Acknowledgments
This work was supported by the National Institutes of Health Ruth L. Kirschstein national research service award 5T32 CA09302 (J.B.) and grants CA070297, CA087767, and DK061002 (Z.S.).
Footnotes
Published ahead of print on 29 October 2007.
REFERENCES
- 1.Adiseshaiah, P., S. R. Papaiahgari, H. Vuong, D. V. Kalvakolanu, and S. P. Reddy. 2003. Multiple cis-elements mediate the transcriptional activation of human fra-1 by 12-O-tetradecanoylphorbol-13-acetate in bronchial epithelial cells. J. Biol. Chem. 27847423-47433. [DOI] [PubMed] [Google Scholar]
- 2.Battle, T. E., R. A. Lynch, and D. A. Frank. 2006. Signal transducer and activator of transcription 1 activation in endothelial cells is a negative regulator of angiogenesis. Cancer Res. 663649-3657. [DOI] [PubMed] [Google Scholar]
- 3.Beliakoff, J., and Z. Sun. 2006. Zimp7 and Zimp10, two novel PIAS-like proteins, function as androgen receptor coregulators. Nucl. Recept. Signal. 4e017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante-Alcalde, D., L. Hernandez, H. Le Stunff, R. Maeda, H. S. Lee, C. Gang, Jr., V. A. Sciorra, I. Daar, S. Spiegel, A. J. Morris, and C. L. Stewart. 2003. The lipid phosphatase LPP3 regulates extra-embryonic vasculogenesis and axis patterning. Development 1304623-4637. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara, N., K. Carver-Moore, H. Chen, M. Dowd, L. Lu, K. S. O'Shea, L. Powell-Braxton, K. J. Hillan, and M. W. Moore. 1996. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380439-442. [DOI] [PubMed] [Google Scholar]
- 6.Fischer, A., N. Schumacher, M. Maier, M. Sendtner, and M. Gessler. 2004. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gale, N. W., M. G. Dominguez, I. Noguera, L. Pan, V. Hughes, D. M. Valenzuela, A. J. Murphy, N. C. Adams, H. C. Lin, J. Holash, G. Thurston, and G. D. Yancopoulos. 2004. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA 10115949-15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutiérrez, L., M. Zurita, J. A. Kennison, and M. Vazquez. 2003. The Drosophila trithorax group gene tonalli (tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development 130343-354. [DOI] [PubMed] [Google Scholar]
- 9.Huang, C. Y., J. Beliakoff, X. Li, J. Lee, X. Li, M. Sharma, B. Lim, and Z. Sun. 2005. hZimp7, a novel PIAS-like protein, enhances androgen receptor-mediated transcription and interacts with SWI/SNF-like BAF complexes. Mol. Endocrinol. 192915-2929. [DOI] [PubMed] [Google Scholar]
- 10.Iso, T., Y. Hamamori, and L. Kedes. 2003. Notch signaling in vascular development. Arterioscler. Thromb. Vasc. Biol. 23543-553. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, P. K. 2001. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 153053-3058. [DOI] [PubMed] [Google Scholar]
- 12.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8713-718. [DOI] [PubMed] [Google Scholar]
- 13.Kawai-Kowase, K., M. S. Kumar, M. H. Hoofnagle, T. Yoshida, and G. K. Owens. 2005. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol. Cell. Biol. 258009-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 27730283-30288. [DOI] [PubMed] [Google Scholar]
- 15.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 225222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotaja, N., M. Vihinen, J. J. Palvimo, and O. A. Janne. 2002. Androgen receptor-interacting protein 3 and other PIAS proteins cooperate with glucocorticoid receptor-interacting protein 1 in steroid receptor-dependent signaling. J. Biol. Chem. 27717781-17788. [DOI] [PubMed] [Google Scholar]
- 17.Krebs, L. T., J. R. Shutter, K. Tanigaki, T. Honjo, K. L. Stark, and T. Gridley. 2004. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 182469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsson, J., M. J. Goumans, L. J. Sjostrand, M. A. van Rooijen, D. Ward, P. Leveen, X. Xu, P. ten Dijke, C. L. Mummery, and S. Karlsson. 2001. Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J. 201663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J., J. Beliakoff, and Z. Sun. 2007. The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor. Nucleic Acids Res. 354523-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, T. H., H. Zhao, Y. Peng, J. Beliakoff, J. D. Brooks, and Z. Sun. 2007. A promoting role of androgen receptor in androgen-sensitive and -insensitive prostate cancer cells. Nucleic Acids Res. 352767-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X., G. Thyssen, J. Beliakoff, and Z. Sun. 2006. The novel PIAS-like protein hZimp10 enhances Smad transcriptional activity. J. Biol. Chem. 28123748-23756. [DOI] [PubMed] [Google Scholar]
- 22.Liu, B., S. Mink, K. A. Wong, N. Stein, C. Getman, P. W. Dempsey, H. Wu, and K. Shuai. 2004. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 5891-898. [DOI] [PubMed] [Google Scholar]
- 23.Long, J., I. Matsuura, D. He, G. Wang, K. Shuai, and F. Liu. 2003. Repression of Smad transcriptional activity by PIASy, an inhibitor of activated STAT. Proc. Natl. Acad. Sci. USA 1009791-9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long, J., G. Wang, I. Matsuura, D. He, and F. Liu. 2004. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3). Proc. Natl. Acad. Sci. USA 10199-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megidish, T., J. H. Xu, and C. W. Xu. 2002. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem. 2778255-8259. [DOI] [PubMed] [Google Scholar]
- 26.Miyazono, K., S. Maeda, and T. Imamura. 2005. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 16251-263. [DOI] [PubMed] [Google Scholar]
- 26a.Nagy, A., M. Gersenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Nishida, T., and H. Yasuda. 2002. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 27741311-41317. [DOI] [PubMed] [Google Scholar]
- 28.Oettgen, P. 2001. Transcriptional regulation of vascular development. Circ. Res. 89380-388. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer, A., T. Kessler, S. Silletti, D. A. Cheresh, and I. M. Verma. 2000. Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc. Natl. Acad. Sci. USA 9712227-12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santti, H., L. Mikkonen, A. Anand, S. Hirvonen-Santti, J. Toppari, M. Panhuysen, F. Vauti, M. Perera, G. Corte, W. Wurst, O. A. Janne, and J. J. Palvimo. 2005. Disruption of the murine PIASx gene results in reduced testis weight. J. Mol. Endocrinol. 34645-654. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, D., and S. Muller. 2003. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol. Life Sci. 602561-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber, M., Z. Q. Wang, W. Jochum, I. Fetka, C. Elliott, and E. F. Wagner. 2000. Placental vascularisation requires the AP-1 component fra1. Development 1274937-4948. [DOI] [PubMed] [Google Scholar]
- 33.Sharma, M., X. Li, Y. Wang, M. Zarnegar, C. Y. Huang, J. J. Palvimo, B. Lim, and Z. Sun. 2003. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 226101-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urness, L. D., L. K. Sorensen, and D. Y. Li. 2000. Arteriovenous malformations in mice lacking activin receptor-like kinase-1. Nat. Genet. 26328-331. [DOI] [PubMed] [Google Scholar]
- 35.Verras, M., J. Brown, X. Li, R. Nusse, and Z. Sun. 2004. Wnt3a growth factor induces androgen receptor-mediated transcription and enhances cell growth in human prostate cancer cells. Cancer Res. 648860-8866. [DOI] [PubMed] [Google Scholar]
- 36.Wong, K. A., R. Kim, H. Christofk, J. Gao, G. Lawson, and H. Wu. 2004. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol. Cell. Biol. 245577-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, L., H. Wu, L. Ma, F. Sangiorgi, N. Wu, J. R. Bell, G. E. Lyons, and R. Maxson. 1997. Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech. Dev. 653-17. [DOI] [PubMed] [Google Scholar]
- 38.Young, M. R., and N. H. Colburn. 2006. Fra-1 a target for cancer prevention or intervention. Gene 3791-11. [DOI] [PubMed] [Google Scholar]