Abstract
Transcriptional activation of histone subtypes is coordinately regulated and tightly coupled with the onset of DNA replication during S-phase entry. The underlying molecular mechanisms for such coordination and coupling are not well understood. The cyclin E-Cdk2 substrate NPAT has been shown to play an essential role in the transcriptional activation of histone genes at the G1/S-phase transition. Here, we show that NPAT interacts with components of the Tip60 histone acetyltransferase complex through a novel amino acid motif, which is functionally conserved in E2F and adenovirus E1A proteins. In addition, we demonstrate that transformation/transactivation domain-associated protein (TRRAP) and Tip60, two components of the Tip60 complex, associate with histone gene promoters at the G1/S-phase boundary in an NPAT-dependent manner. In correlation with the association of the TRRAP-Tip60 complex, histone H4 acetylation at histone gene promoters increases at the G1/S-phase transition, and this increase involves NPAT function. Suppression of TRRAP or Tip60 expression by RNA interference inhibits histone gene activation. Thus, our data support a model in which NPAT recruits the TRRAP-Tip60 complex to histone gene promoters to coordinate the transcriptional activation of multiple histone genes during the G1/S-phase transition.
Histone proteins are integral components of eukaryotic chromatin and play crucial roles in virtually all cellular processes that involve chromosomal DNA, such as DNA replication, transcription, DNA repair, recombination, and chromosome segregation (27, 28). The bulk of the histones are assembled with genomic DNA into chromosomes, and the biosynthesis of multiple histone subtypes (replication-dependent histones) is tightly coordinated and coupled with DNA synthesis during the S phase of the cell cycle (20, 36, 45, 51). Perturbation of such coordination and coupling can lead to the loss of chromosomes, DNA damage, and developmental arrest (39, 55, 62, 64), underscoring the importance of the proper regulation of these events.
The rate of histone synthesis in S phase is regulated at both the transcriptional and the posttranscriptional levels (20, 36, 45, 51). Histone gene transcription increases from 3- to 10-fold as cells enter S phase (20). The transcription of each histone subtype (H1, H2A, H2B, H3, and H4) in S phase is likely regulated by proteins or protein complexes that interact directly with the subtype-specific regulatory elements (SSREs) in the promoters of replication-dependent histone genes (20, 45). Indeed, it has been shown that Oct1 and its coactivator complex OCA-S interact with the H2B SSRE to activate H2B transcription, while HiNF-P interacts with the H4 SSRE to stimulate H4 expression (14, 40, 67). Components of OCA-S include nuclear p38/glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lactate dehydrogenase, and the activity of OCA-S is regulated by NAD+ and NADH, suggesting a link between the histone gene transcription and the cellular metabolic state/redox status (67). The molecular mechanism underlying the regulation of H2B transcription by Oct1/OCA-S and cellular metabolic/redox states, however, is not yet clear. The mechanism of HiNF-P action is also not fully understood. Additional protein factors, such as YY1, HIRA, FLASH, and BZAP45, have also been implicated in the regulation of histone gene transcription with unknown molecular mechanisms (3, 12, 19, 31, 41, 61).
We and others have demonstrated previously that the cyclin E-Cdk2 substrate NPAT associates with histone gene promoters in S phase (66, 67). The overexpression of NPAT activates promoters of multiple histone genes through the SSREs within the promoters (66). The suppression of NPAT expression through RNA interference or conditional knockout impedes expression of all histone subtypes (16, 63). The promoter DNA sequences of different histone subtypes are quite divergent, and direct DNA binding by NPAT has not been detected. Therefore, it was proposed that coordination of the transcription of multiple histone subtypes by NPAT probably occurs through the interaction of NPAT with factors that regulate transcription of the individual subtypes (66). Indeed, both the physical and the functional interactions of NPAT with Oct-1/OCA-S and HiNF-P have been demonstrated (40, 67). Additionally, it has been shown that both the association of NPAT with histone promoters and the NPAT-mediated histone transcriptional activation are regulated by cyclin E-Cdk2 phosphorylation (34, 53, 66). Hence, NPAT functions as a key global regulator of coordinated transcriptional activation of multiple histone subtypes during the G1/S-phase transition and links the cell cycle machinery to the regulation of histone gene expression. In addition to histone gene expression, NPAT has been shown to play a critical role in S-phase entry (16, 63, 65). Thus, NPAT may also play a role in coupling histone expression with the onset of DNA synthesis.
Despite recent advances in our understanding of the regulation of histone gene transcription, the molecular mechanisms underlying NPAT function, as well as the coordinated transcriptional activation of histone subtypes and the coupling of histone gene expression with DNA replication, have remained largely unknown. Given the importance of NPAT in histone gene transcription, an understanding of NPAT function would likely advance the elucidation of these mechanisms.
The transformation/transactivation domain-associated protein (TRRAP) was initially identified as a factor that interacts with the N terminus of c-Myc, as well as with the transactivation domain of E2F (37). TRRAP is an essential cofactor for oncogenic transformation by c-Myc and E1A through its direct interaction with these proteins (7, 37, 46). TRRAP also functions as an essential cofactor for E2F-mediated transcriptional activation (30). TRRAP, as well as its Saccharomyces cerevisiae homolog Tra1, has been shown to be a key component of several multiprotein histone acetyltransferase (HAT) complexes (4, 6, 8, 24, 35, 52, 58) and is likely involved in the recruitment of one or more of these HAT complexes to target gene promoters through its interaction with transcriptional activators, such as c-Myc, E2Fs, and p53 (2, 5, 15, 38, 56). In addition to transcription-related functions, the TRRAP-containing HAT complexes are also directly involved in double-stranded DNA break repair (6, 24, 42, 47, 50). Thus, TRRAP plays crucial roles in both the proliferation control and the maintenance of genomic integrity.
To elucidate the molecular mechanism by which NPAT regulates transcriptional activation of histone genes, we carried out a systematic deletion analysis to identify the domain(s) critical for NPAT function. Here, we describe the identification of a novel amino acid sequence motif termed the DLFD motif, which is conserved in the NPAT, E2F, and E1A proteins and is involved in the functions of these proteins. Our data, together with previous observations, indicate that the DLFD motif is required for the interaction of NPAT, and likely that of E2F and E1A, with the cofactor TRRAP. Moreover, our results show that TRRAP and Tip60 are recruited to histone gene promoters at the G1/S-phase boundary by NPAT and are required for the transcriptional activation of histone genes. Consistent with the NPAT-dependent recruitment of TRRAP-Tip60, histone H4 acetylation at histone promoters also increases during the G1/S-phase transition in an apparently NPAT-dependent manner. These results suggest a mechanism by which the transcriptional activation of histone genes is coordinately regulated during S-phase entry.
MATERIALS AND METHODS
Cell culture, transfection, immunoprecipitation, luciferase reporter assays, Northern blotting, and antibodies.
Human osteosarcoma U2OS cells and human embryonic kidney 293T (HEK293T) cells were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). Human colon carcinoma HCT116 cells carrying one conditional allele of NPAT and one wild-type allele (referred to as NPATflox/− [63]) were generously provided by Wade Harper. Transient transfections were carried out either by calcium precipitation or by using FuGENE6 (Roche) as described previously (65, 66). Luciferase reporter assays, chromatin immunoprecipitation (ChIP) assays, and Northern blotting were performed essentially as described previously (16, 53, 65, 66).
Antibodies specific for the NPAT protein were described previously (65, 66). Antibodies against TRRAP (catalog no. T-17), Tip60 (catalog no. N-17 and K-17), and the GAL4 DNA binding domain (catalog no. RK5C1) were from Santa Cruz Biotechnology. Antibodies for γ-tubulin (catalog no. GTU88) and Flag tag (catalog no. M2) were from Sigma. Antibody specific for histone H4 acetylated at lysines 5, 8, 12, and 16 (catalog no. 06-866) was from Upstate Biotechnology.
Expression plasmids and construction of fusion proteins.
Plasmids carrying the wild-type NPAT cDNA sequence were described previously (65, 66). For the construction of mammalian expression plasmids encoding the yeast GAL4 DNA binding domain fused with various NPAT sequences, DNA fragments encoding the indicated NPAT sequences were generated either by restriction enzyme digestion or by PCR and cloned in frame into the vector pFA-CMV (Stratagene). Bacterial glutathione S-transferase (GST) fusion constructs were generated by cloning the indicated NPAT fragments, prepared by PCR, into the vector pGEX (Amersham). The point mutation constructs were generated by PCR and cloned into appropriate expression plasmids. The PCR products were sequenced to confirm that the correct DNA sequences were obtained.
Preparation of GST-NPAT fusion proteins.
GST fusion proteins containing either the wild-type or the mutant transactivation domain of NPAT were expressed in Escherichia coli BL21 cells. The bacterial cells were lysed by sonication. The lysates were incubated with glutathione-agarose beads (Sigma). After the GST fusion proteins were washed with a buffer of 50 mM Tris-HCl (pH 8.0), 1 M NaCl, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), they were eluted from the beads with reduced glutathione (Sigma) in phosphate-buffered saline. The eluted proteins were then loaded onto to an SP-Sepharose column (Amersham) equilibrated with SP buffer (5 mM phosphate [pH 7.0], 1 mM EDTA, 5% glycerol, 0.5 mM PMSF). The flowthrough was loaded onto a DEAE-Sepharose column (Amersham) equilibrated with buffer D (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5 mM PMSF). The proteins were eluted from the column by a 100 mM to 1 M NaCl gradient in D buffer. Fractions containing the fusion proteins were combined and dialyzed against phosphate-buffered saline.
Purification of nuclear proteins that interact with the transactivation domain of NPAT.
HeLa S3 cells (from the National Cell Culture Center) were lysed in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 10 mM EDTA, 0.4% NP-40; Panomics) and centrifuged to collect the nuclei. The nuclei were treated with buffer B (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA, 10% glycerol; Panomics) and centrifuged to collect nuclear proteins in the supernatant. To remove proteins that interact with GST or with the NPAT transactivation domain independently of the DLFD motif, the nuclear extract was precleared by incubation with a mutant NPAT transactivation domain fusion protein (amino acids [aa] 262 to 329 or aa 262 to 350 [AAA]) and glutathione beads. The precleared nuclear extract was then incubated with GST-NPAT aa 262 to 338 or GST-NPAT aa 262 to 350 and glutathione beads. The beads were washed with buffer B with 0.1% NP-40. The proteins associated with the GST-NPAT fusion were eluted from the beads with 0.2% Sarkosyl in buffer B (43). The eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with colloidal Coomassie blue (Invitrogen). Protein bands specific for samples purified from the GST-NPAT aa 262 to 338 or from GST-NPAT aa 262 to 350 beads were excised for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis at Taplin Biological Mass Spectrometry Facility of Harvard Medical School.
RNA interference by shRNA and quantitative real-time PCR analyses.
For generating small hairpin RNAs (shRNAs) targeting human TRRAP mRNA, two sequences (shTRRAP-1, GGCTCGAAAGGAATGGATCC; and shTRRAP-2, GGCGACTACCTGGAGAACAT) were cloned into the pBS/U6 vector (54). The pBS/U6 plasmid expressing an shRNA specific for the firefly luciferase mRNA sequence (ACAAGACAATTGCACTGA) (57) was used as a control with Northern blotting experiments. The pBS/U6/Ski plasmid expressing an shRNA specific for the mouse Ski mRNA sequence (GCTGGAGGCAGAGTTGGAG) was also used as a control. For the Tip60 knockdown experiment, we employed two pLKO.1 constructs expressing Tip60-specific shRNAs, the RNA interference Consortium clones TRCN0000020314 (shTip60-1) and TRCN0000020315 (shTip60-2), targeting the sequences CGTCCATTACATTGACTTCAA and CCTCAATCTCATCAACTACTA, respectively. The pLKO.1 vector expressing a scrambled sequence (GTTCTCCGAACGTGTCACG) was used as a control.
After the knockdown, Tip60 mRNA levels were measured by using real-time quantitative PCR (qPCR). RNA was isolated using TRIzol (Invitrogen) reagent and then used as a template for cDNA synthesis by Moloney murine leukemia virus reverse transcriptase (Invitrogen) as described by the manufacturer. qPCRs using iQ SYBR green Supermix (Bio-Rad) were performed in triplicate with either the Tip60-specific primers TCCAGGCAATGAGATTTACCG and TCTTATGGTCAAGGAAACACTTGG or the GAPDH-specific primers CATGGGTGTGAACCATGAGA and CAGTGATGGCATGGACTGTG. Gene expression was normalized to GAPDH.
ChIP assays.
The ChIP assays were carried out essentially as described previously (66). Briefly, HCT116 NPATflox/− cells were cross-linked with 1% formaldehyde for 10 min at room temperature. After undergoing cross-linking, the cells were lysed, and the chromatin was sonicated into 400- to 1,000-bp fragments, using a Branson 450 sonicator. The lysates were cleared by centrifugation at 11,000 rpm for 5 min at room temperature. A fraction of the lysates was saved as an input control, and the remaining lysates were diluted 10-fold with the dilution buffer. The diluted lysates were precleared with protein G beads (protein G-Sepharose; Amersham) and then incubated with antibodies specific for either NPAT, TRRAP, TIP60, acetylated H4, or the Flag tag and 15 μl of protein G-Sepharose at 4°C. Immunoprecipitated samples were washed three times with the dilution buffer and twice with a wash solution (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM EDTA, 0.5% Triton X-100) and then eluted with 1% SDS and 10 mM Tris-EDTA buffer at 65°C. The samples were treated with proteinase K at 50°C for 1 h. The input control samples were treated with 10 μg RNase at 37°C for 3 h and then 100 μg proteinase K at 50°C for 1 h. Reversion of cross-linking was carried out at 65°C overnight. DNA was phenol-chloroform extracted and ethanol precipitated. The DNA samples were subsequently quantitated by real-time PCR with SYBR green. Data analysis was performed using the method described previously (33). For the amplification of H4 sequences, the primers (5′-CTATTTCGGTTTGGCCCTTT-3′ and 5′-CTGAGGCAGCGCCTTTATAC-3′) were used to cover about a 120-bp promoter region of the H4/e gene. For the amplification of the H2B sequence, the previously described primer set (66), which covers a 180-bp promoter sequence upstream of the initiation codon ATG, was used.
RESULTS
NPAT contains a transactivation domain with a motif conserved in the E2F and E1A proteins.
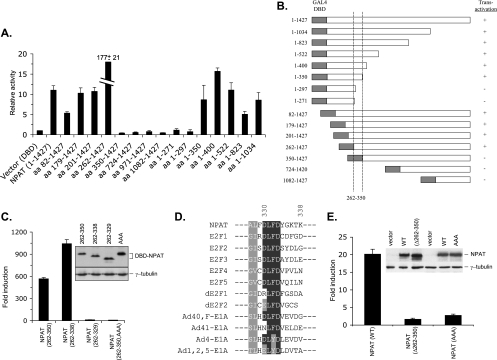
In order to elucidate the mechanism by which NPAT coordinates histone gene transcription during the G1/S-phase transition, we set out to identify the domain(s) in NPAT that is required for transcriptional activation. Short deletions from either the N terminus or the C terminus of NPAT abolished its ability to activate histone promoters (data not shown) (60), hampering the analysis of the internal domain(s) required for histone gene expression by the use of histone promoter-reporter assays. To circumvent this difficulty and to facilitate the identification of the domain involved in the transactivation, we fused full-length NPAT or various NPAT terminal deletions with the DNA binding domain (DBD) of the yeast transcriptional activator GAL4 protein and tested the fusions' abilities to activate a GAL4-responsive reporter. Fusion proteins containing either full-length NPAT or regions encompassing aa 262 to 350 of NPAT exhibited significant transcriptional activity (Fig. 1A and B), suggesting that a transcriptional activation domain resides in this region. The fusion of aa 262 to 350 of NPAT with the DBD activated the GAL4 reporter dramatically (Fig. 1C), confirming that this region carries a transcriptional activation function. To further define the transcriptional activation domain, we generated additional deletion constructs and tested their abilities to activate transcription. The domain that consists of aa 262 to 338 functions as a potent transcriptional activator (for convenience, it was referred to as the transactivation domain of NPAT). In contrast, the fusion containing aa 262 to 329, though expressed at a level similar to that of the fusion containing aa 262 to 338, has a greatly decreased ability to activate transcription (Fig. 1C). These data indicate that aa 329 to 338 are crucial for transcriptional activation.
FIG. 1.
Identification of a functional DLFD motif. (A) Deletion analysis of the GAL4-NPAT fusion proteins. pFA-CMV vector, which expresses the GAL4 DBD, or a pFA-CMV plasmid that expresses the indicated DBD-NPAT fusion protein was transfected together with the GAL4-responsive luciferase reporter pFR-Luc (Stratagene) into U2OS cells. For the normalization of transfection efficiency, pCMV-LacZ, which expresses β-galactosidase, was also cotransfected. Twenty-four hours posttransfection, the cells were lysed and the activities of luciferase and β-galactosidase were assayed. The relative activity was calculated as previously described (66). The mean results and standard deviations from three independent experiments are depicted. (B) Schematic representation and summary of results shown in panel A. The positions of the first and last amino acids of the fused NPAT sequences are indicated on the left. Full-length NPAT contains 1,427 amino acids. The dotted lines indicate the region of NPAT required for transactivation. (C) The DLFD sequence is required for transactivation. The indicated GAL4 DBD-NPAT fusion proteins were tested for their ability to activate the pFR-Luc reporter. The expression levels of the fusion proteins were analyzed by Western blotting using an antibody specific for the GAL4-DBD. The Western blot of γ-tubulin was used as an equal loading control. (D) Conservation of the DLFD motif in NPAT, E2F and E1A proteins. The sequences surrounding the amino acids DLFD from human NPAT, human and Drosophila E2F and several human adenovirus E1A proteins are aligned. Identical amino acid residues are shaded black, and similar residues are shaded gray. The numbers indicate the locations of the amino acids in NPAT. (E) Essential role for the DLFD motif in NPAT-mediated histone promoter activation. Full-length wild-type NPAT, a mutant NPAT with the deletion of amino acid residues 262 to 350, and a mutant NPAT with the LFD sequence in the transactivation domain replaced by AAA sequence were tested for their ability to activate a histone H4 promoter (pGLH4(65)) as previously described (66). The expression of the wild-type and mutant NPAT was analyzed by Western blotting with an NPAT-specific antibody. The Western blot of γ-tubulin was used as a loading control.
The sequence aa 329 to 338 of NPAT contains a sequence motif that is also present in the transcriptional activation domain of E2Fs (9, 26, 49), as well as in the conserved region 1 (CR1) of the adenovirus E1A protein that was shown to be essential for E1A-mediated transformation (10, 11) (Fig. 1D). The most conserved residues in this motif are the amino acids DLFD, and thus we refer to this element as the DLFD motif. Previous studies suggest that the region containing the DLFD motif is involved in E2F and E1A function (7, 13). To assess the significance of the DLFD motif in NPAT-mediated transcriptional activation, we replaced the amino acids LFD with alanine residues in the NPAT (262-350)-GAL4-DBD fusion protein. This replacement had no effect on the expression of the fusion protein but severely compromised its transcriptional activation ability (Fig. 1C), suggesting that the DLFD motif is critical for the function of the NPAT transactivation domain. To determine whether the region consisting of aa 262 to 350 is also required for NPAT to activate histone promoters, we constructed a mutant NPAT protein with aa 262 to 350 deleted. This mutant NPAT was unable to activate a histone H4 promoter reporter, indicating that these residues are essential for histone gene activation by NPAT (Fig. 1E). To further demonstrate the role of the DLFD motif in NPAT-mediated histone gene activation, we changed the amino acids LFD to AAA in the full-length NPAT and tested histone promoter activation by the mutant NPAT. Similar to the GAL4 DBD fusion proteins, mutation of the LFD sequences in NPAT, which had no effect on NPAT expression, resulted in a marked decrease in histone promoter activation (Fig. 1E). These results indicate that the DLFD motif is critical for NPAT to regulate histone gene activation. Thus, the DLFD motif may represent an important functional domain conserved in NPAT, E2F, and E1A.
TRRAP interacts with the transactivation domain of NPAT via the DLFD motif.
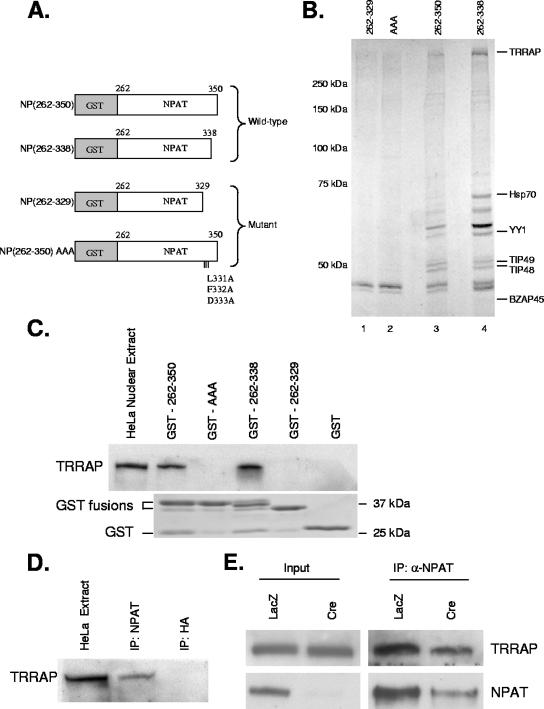
Having identified a transactivation domain in NPAT that contains a conserved functional DLFD motif, we sought next to identify proteins that interact with this transactivation domain through the DLFD motif. In order to facilitate this investigation, two wild-type and two mutant forms of the transactivation domain were constructed and fused to GST (Fig. 2A). These GST fusion proteins were purified from bacteria and subsequently incubated with HeLa nuclear extract. Proteins that interacted with these NPAT transactivation domain fusion proteins were affinity purified, separated by SDS-PAGE, and visualized by Coomassie staining. The staining revealed that the wild-type transactivation domain binds to a number of proteins that showed weak or no interaction with the mutant transactivation domain (Fig. 2B, compare lanes 3 and 4 with lanes 1 and 2). The identities of the purified interacting proteins were determined by mass spectrometry.
FIG. 2.
Interaction of TRRAP with NPAT. (A) Schematic representation of GST-NPAT fusion proteins used for affinity purification. (B). Identification of nuclear proteins interacting with functional transactivation domain of NPAT. HeLa nuclear extract was incubated with the indicated GST fusion proteins bound to the glutathione-agarose beads. Proteins associated with the fusion proteins were eluted and separated on an 8% SDS-PAGE gel. The proteins were visualized by colloidal Coomassie blue staining. The proteins were then identified by LC-MS/MS. (C) Interaction of TRRAP with NPAT transactivation domain requires the DLFD motif. The indicated GST fusion proteins were incubated with HeLa nuclear extract. Interaction of TRRAP with the fusion proteins was analyzed by Western blotting using a TRRAP-specific antibody (top panel). Ten percent of the input was loaded for analysis of the TRRAP level in HeLa nuclear extract. (Lower panel) a Coomassie blue-stained SDS-PAGE gel indicating equal amounts of GST fusion proteins were used for the pull-down experiments. (D) In vivo interaction of TRRAP with NPAT. HeLa nuclear extracts were immunoprecipitated with a monoclonal antibody specific for NPAT (DH3) or HA tag (12CA5). The immunoprecipitates were analyzed by Western blotting with a TRRAP-specific antibody. Five percent of the input was loaded for analysis of TRRAP in HeLa nuclear extract. One hundred percent of the immunoprecipitates was loaded for the analysis. (E) Confirmation of in vivo interaction of TRRAP with NPAT. HCT116 NPATflox/− cells were infected with either Ad-LacZ or Ad-Cre (Baylor Vector Development Laboratory) as indicated. Three days after infection, the whole cell lysates were immunoprecipitated with an NPAT-specific antibody. The presence of TRRAP in the immunoprecipitates was analyzed by Western blotting with a TRRAP-specific antibody. The input lanes were loaded with 2% (for TRRAP) and 5% (for NPAT) of the starting material used for the immunoprecipitation. One hundred percent of the immunoprecipitated material was analyzed for IP samples. Under the experimental conditions, the NPAT knockout efficiency is about 80% as previously reported (63).
This analysis identified a number of proteins that appear to interact with the NPAT transactivation domain through the DLFD motif. Among the proteins identified were YY1 and BZAP45 (Fig. 2B), which are proteins that had previously been implicated in the regulation of histone gene transcription (12, 31, 41, 61). Mass spectrometric analysis also revealed the presence of heat shock protein 70 (Hsp70), which was previously shown to be a component of the OCA-S coactivator complex (67). These findings suggest that this approach identifies authentic proteins involved in the transcriptional regulation of histone genes. In addition to these previously described regulators, we identified several components of the Tip60/hNuA4 histone acetyltransferase complex, TRRAP, Tip48, and Tip49 (6, 8) (Fig. 2B). Interestingly, TRRAP has previously been characterized as a cofactor that is essential for the functions of both E2F and E1A (7, 29, 30, 37, 46). Moreover, the DLFD motifs in E2F and E1A are apparently involved in their interactions with TRRAP (7, 30). Hence, TRRAP may also be an important cofactor for NPAT-mediated transactivation, and its interaction with NPAT is likely mediated through the DLFD motif.
To confirm the interaction between NPAT and TRRAP, observed with the initial mass spectrometry analysis, the GST fusion proteins were once again incubated with HeLa nuclear extract, and the interaction of TRRAP with the GST-fusion proteins was determined by Western blotting analysis with a TRRAP-specific antibody. As shown in Fig. 2C, the wild-type NPAT transactivation domain, but not the mutant domain, interacts with TRRAP, validating the observation that TRRAP interacts with the transactivation domain of NPAT via the DLFD motif. To determine whether endogenous NPAT and TRRAP interact, nuclear extracts prepared from HeLa cells were subjected to immunoprecipitation with an NPAT-specific antibody. TRRAP protein can be readily detected in the immunoprecipitates of the NPAT antibody but not in the immunoprecipitates of a control antibody (Fig. 2D), indicating that NPAT associates with TRRAP in vivo. Endogenous NPAT in anti-TRRAP immunoprecipitates was not detected, likely due to the low affinity of the available TRRAP antibody (data not shown). To verify that the coimmunoprecipitation of TRRAP results from the in vivo interaction between TRRAP and NPAT, rather than examine nonspecific immunoprecipitation by the anti-NPAT antibody, we compared coimmunoprecipitations of TRRAP by the NPAT-specific antibodies in both the wild-type NPAT and the HCT116 conditionally null NPAT cells. These latter cells (referred to as NPATflox/− cells) carry one mutant NPAT allele and one wild-type allele with exon 2 flanked by loxP sites. Infection of the NPATflox/− cells with adenovirus that expresses Cre recombinase (Ad-Cre) induces the excision of exon 2 of the wild-type allele, generating NPAT-deficient cells (63). As shown in Fig. 2E, the amount of TRRAP that coimmunoprecipitated is reduced in the Ad-Cre-infected NPATflox/− cells, where NPAT levels are decreased but not completely depleted under the experimental conditions. Coimmunoprecipitation of TRRAP with NPAT is due to the specific interaction of these two proteins, rather than a nonspecific association of these proteins with an antibody, as the antibody specific for the hemagglutinin (HA) tag immunoprecipitates neither NPAT nor TRRAP (data not shown). Together, these data show that NPAT associates with TRRAP in vivo, likely through the DLFD motif.
NPAT recruits TRRAP to histone promoters during the G1/S-phase transition.
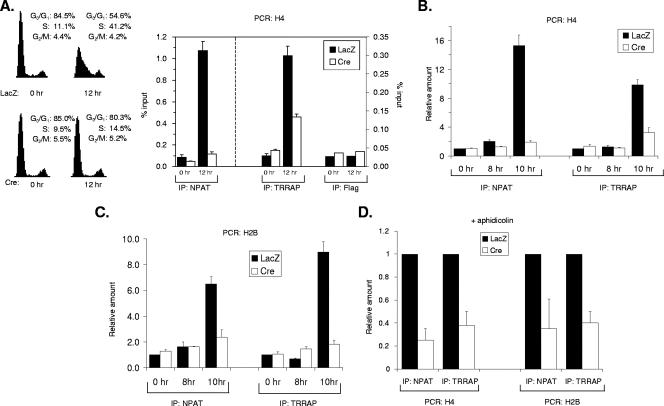
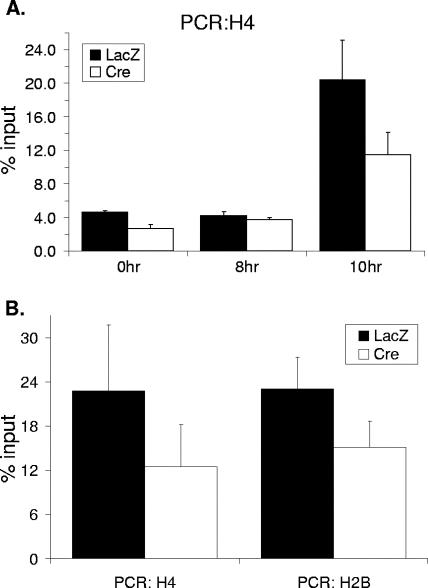
We and others have previously shown that NPAT physically associates with histone promoters in vivo and that this association increases upon S-phase entry (66, 67). As NPAT interacts with TRRAP, which has been shown to be recruited by Myc and E2F to their respective target promoters (15, 38, 46, 56), it is possible that NPAT may recruit TRRAP to histone promoters to activate histone gene transcription during the G1/S-phase transition. To test this possibility, we investigated the association of TRRAP with histone promoters in the HCT116 NPATflox/− cells under different conditions by ChIP assays. Similar to the cell cycle-dependent association of NPAT with the histone promoters (Fig. 3A) (67), the association of TRRAP with a histone H4 promoter increases about 10-fold in early S-phase cells compared to that with quiescent cells (Fig. 3A). In contrast, the association of TRRAP with the H4 promoter in vivo is compromised in the NPAT-deficient (Ad-Cre-infected) cells. Thus, TRRAP associates with histone promoters in S phase, and this association depends on NPAT. Under the experimental conditions used, the NPAT knockout efficiency is approximately 80 to 85% (63) Fig. 2 (and data not shown). The small increase in the association of TRRAP with the H4 promoter in the Ad-Cre-infected cells upon S-phase entry is likely due to the effect of the remaining NPAT protein. To determine more precisely the cell cycle stage at which TRRAP becomes associated with histone promoters, we analyzed the association of TRRAP with histone promoters at additional time points during the G1-to-S-phase transition. As shown in Fig. 3B and Table 1, the association of TRRAP with the H4 promoter in vivo occurs at the G1/S-phase boundary, as does the association of NPAT with the H4 promoter. The association of TRRAP and that of NPAT with a histone H2B promoter follow virtually identical kinetics (Fig. 3C). Thus, the associations of both NPAT and TRRAP with histone promoters occur during the G1/S-phase transition, a point when S-phase-specific histone gene transcription is initiated. Again, the increased association of TRRAP with the histone promoters relies on NPAT (Fig. 3B and C). Thus, TRRAP associates with histone promoters in vivo at the G1/S-phase boundary and in an NPAT-dependent manner.
FIG. 3.
NPAT recruits TRRAP to histone promoters. (A) NPAT-dependent association of TRRAP with a histone H4 promoter at early S phase. (Left panel) HCT116 NPATflox/− cells were infected with either Ad-LacZ or Ad-Cre as indicated. Sixty hours after infection, the cells were starved for 36 h in medium containing 0.1% FBS. The cells were then stimulated to enter the cell cycle with 10% FBS. The cell cycle distribution of the cells at the indicated times was analyzed as previously described (53, 66). (Right panel) ChIP assays on serum-starved (0 h) or early S-phase cells (12 h). The cross-linked chromatin was immunoprecipitated with the indicated antibodies, and the presence of the H4 promoter DNA sequence in the immunoprecipitates was analyzed as described in Materials and Methods. The antibody specific for the Flag tag was used as a negative control. (B) NPAT-dependent association of TRRAP to an H4 promoter at the G1/S-phase boundary. The ChIP assays were carried out as described for panel A, except that the cells were harvested at 0, 8 and 10 h after serum stimulation. The amounts of DNA immunoprecipitated at different time points were compared, relative to the amounts at 0 h set as 1. (C) NPAT-dependent association of TRRAP to an H2B promoter at the G1/S-phase boundary. The ChIP assays were performed as described for panel B, except that primers specific for an H2B promoter were used. (D) NPAT-dependent, but S-phase-independent association of TRRAP with the H4 and H2B promoters. The NPATflox/− cells were treated as described for panel A, except that 5 μg/ml aphidicolin was added into the medium at the same time when the cells were stimulated with 10% FBS to block S-phase entry (53). The cells were harvested 18 h after serum and aphidicolin addition, and the ChIP assays were carried out as described for panels B and C. Under these experimental conditions, the levels of association of NPAT and TRRAP with histone promoters are 50 to 55% of those observed at 12 h after serum starvation.
TABLE 1.
Cell cycle distribution before and after serum stimulationa
| Cell cycle | Cell distribution (%) at the indicated time (h) of stimulation
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0
|
8
|
10
|
12
|
|||||
| LacZ | Cre | LacZ | Cre | LacZ | Cre | LacZ | Cre | |
| G0/G1 | 77.4 | 78.4 | 75.5 | 76.6 | 71.5 | 73.2 | 50.2 | 69.3 |
| S | 13.0 | 9.6 | 12.3 | 9.7 | 16.0 | 10.9 | 41.6 | 18.0 |
| G2/M | 9.6 | 12.0 | 12.2 | 13.7 | 12.5 | 15.8 | 8.1 | 12.7 |
Since NPAT is required for S-phase entry, the decreased association of TRRAP with the histone promoters in NPAT-deficient cells could result from a cell cycle block at the G1/S-phase transition, rather than from the direct requirement for NPAT to recruit TRRAP to these promoters. To distinguish these two possibilities, the cells were treated with aphidicolin, a DNA polymerase-specific inhibitor, to block S-phase entry, and the association of TRRAP with the H2B and H4 promoters was then analyzed. In the presence of aphidicolin, both the control (Ad-LacZ-infected) and the NPAT-deficient (Ad-Cre-infected) cells were blocked at the G1/S-phase boundary (Table 2) (53). Similar to the results with untreated cells (Fig. 2A and B), the association of TRRAP with the H2B and H4 promoters was markedly decreased in Ad-Cre-infected (NPAT-deficient) cells compared with that in the Ad-LacZ-infected cells (Fig. 3D), indicating that the association of TRRAP with histone promoters requires NPAT directly. These results, together with the above observation that NPAT-dependent association of TRRAP with histone promoters is greatly increased in late G1 phase (Fig. 3A and B), demonstrate that NPAT recruits TRRAP to histone gene promoters during the G1-to-S-phase transition.
TABLE 2.
Cell cycle distribution in the presence of aphidicolina
| Stimulus | Cell distribution (%) at the indicated cycle
|
||
|---|---|---|---|
| G0/G1 | S | G2/M | |
| LacZ | 88.6 | 10.4 | 1.0 |
| Cre | 84.3 | 12.7 | 3.0 |
NPAT-dependent association of Tip60 with histone promoters.
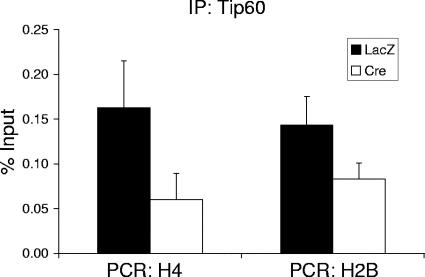
TRRAP is a component of several multiprotein histone acetyltransferase complexes and is involved in the recruitment of these HAT complexes to chromatin (5, 6, 8, 38, 46). Thus, the association of TRRAP with histone promoters suggests that one or more HAT complexes also associate with the histone promoters. Since the transactivation domain of NPAT interacts with other components of the Tip60 complex, Tip48 and Tip49 (Fig. 2B), we hypothesized that NPAT may interact with the Tip60 complex and recruit Tip60 to histone promoters during the G1/S-phase transition. To test this hypothesis, we investigated the association of Tip60 with histone promoters by using ChIP assays. The results shown in Fig. 4 indicate that, similar to TRRAP, Tip60 associates with histone promoters in an NPAT-dependent fashion. Furthermore, the NPAT-dependent association of Tip60 with histone promoters likely results from a direct requirement that NPAT recruit Tip60 to the histone promoters, rather than from an indirect cell cycle effect, as the experiments were carried out in the presence of aphidicolin as described above. Thus, our data suggest that NPAT recruits Tip60, and likely the Tip60 HAT complex as well, to the histone gene promoters at the G1/S-phase boundary.
FIG. 4.
NPAT-dependent association of Tip60 with histone promoters. The treatment of NPATflox/− cells and ChIP assays with a Tip60-specific antibody were carried out as described for Fig. 3D.
Effect of NPAT on histone acetylation at the histone gene promoters.
Since it appears that NPAT recruits Tip60 to histone promoters, NPAT may regulate histone acetylation at histone promoters. To test this possibility, we investigated histone acetylation of chromatin surrounding the sites where NPAT associates with an H4 promoter. It had been reported previously that Tip60, like its yeast homolog Esa1, preferentially acetylates histone H4 (1, 24), and thus, we focused our study on H4 acetylation. Similar to the associations of NPAT, TRRAP, and Tip60 to the H4 promoter, the acetylation of histone H4 is greatly increased at the G1/S-phase boundary (Fig. 5A and Table 1). This increase in H4 acetylation is partially inhibited in the NPAT-deficient cells (Fig. 5A and see Discussion), suggesting that NPAT plays a role in the H4 acetylation at the histone promoters. Again, the NPAT-dependent H4 acetylation is independent of S-phase entry (Fig. 5B), reminiscent of the NPAT-dependent association of TRRAP and Tip60 to histone promoters. Thus, histone H4 acetylation relies on NPAT and correlates with the association of TRRAP and Tip60 at histone promoters during the G1-to-S-phase transition.
FIG. 5.
NPAT-dependent increase in histone H4 acetylation at histone promoters at the G1/S-phase boundary. (A) Increase in histone H4 acetylation at the G1/S-phase boundary. The treatment of the cells and ChIP analysis using an antibody specific for acetylated histone H4 were performed as described for Fig. 3B. (B) S-phase-independent increase in histone H4 acetylation. The treatment of cells and the ChIP assays with an antibody specific for acetylated histone H4 were carried out as described for Fig. 3D.
TRRAP and Tip60 are required for histone promoter activation.
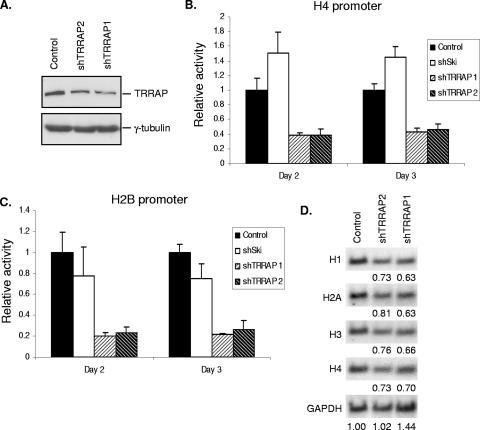
Our observations that TRRAP interacts with NPAT and associates with histone promoters in vivo in an NPAT-dependent manner suggest that TRRAP also functions as a cofactor for histone gene transcription. To test this idea directly, we examined the requirement for TRRAP in histone gene promoter activation. For this purpose, we designed two shRNA constructs targeting distinct sequences in human TRRAP mRNA. Using these constructs, we were able to achieve a moderate knockdown of TRRAP expression (50 to 60%) (Fig. 6A). Interestingly, TRRAP knockdown resulted in a significant reduction of promoter activity from both the H4 and the H2B promoters (Fig. 6B and C), while it had no effect on either the CMV promoter or mutant histone promoters (see Fig. S1 in the supplemental material). These data suggest that TRRAP is required for the activation of multiple histone gene promoters. The observation that a moderate depletion of TRRAP protein by TRRAP-specific shRNA results in a significant inhibition of histone promoter activation raises the possibility that TRRAP is a limiting factor for histone gene activation.
FIG. 6.
TRRAP is required for transcriptional activation of histone genes. (A) Knockdown of TRRAP expression by TRRAP-specific shRNAs. HEK293T cells were transfected with pBS/U6 vector (control) or pBS/U6 plasmids expressing shRNAs specific for human TRRAP mRNA (shTRRRAP1 and shTRRAP2, respectively). pBabe-puro that carries a puromycin resistance gene was also cotransfected for selection with puromycin. Twenty-four hours posttransfection, puromycin was added into the culture medium to select for transfected cells. Three days after selection, the cells were harvested and expression of TRRAP was analyzed by Western blotting using a TRRAP-specific antibody. (B) Effect of TRRAP knockdown on H4 promoter activation. 293T cells were transfected with an H4 promoter-luciferase reporter (66), together with the pBS/U6 vector, pBS/U6 expressing a mouse Ski-specific shRNA or pBS/U6 expressing a TRRAP-specific shRNA. To normalize for transfection efficiency, the cells were also cotransfected with pCMV-LacZ. At the indicated time after transfection, the cells were harvested and the activities of luciferase and β-galactosidase were assayed as previously described (66). (C) Effect of TRRAP knockdown on H2B promoter activation. Activation of an H2B promoter (pGLH2B) (66) in cells transfected with the vector or plasmids expressing TRRAP-specific shRNAs or mouse Ski-specific shRNA was assayed as described for panel B. (D) Effect of TRRAP knockdown on the expression of endogenous histones. HEK293T cells were transfected with pBS/U6 expressing shRNAs specific for the luciferase gene (control) or TRRAP (shTRRAP-1 and shTRRAP-2). pBabe-puro was also cotransfected for selection. Twenty-four hours after transfection, puromycin was added into the medium to select for transfected cells. Two days after puromycin selection, the cells were harvested and mRNA levels of multiple histone genes were analyzed by Northern blotting, and the hybridization signals were quantitated using a phosphorimager as previously described (16). The levels of GAPDH were used as loading controls. The relative levels of the indicated histone mRNA in shTRRAP-expressing cells compared with those in control cells, after normalization with GAPDH signal, are shown below each Northern blot.
As it was previously shown that the knockout of TRRAP leads to proliferation arrest (23), one possible scenario would be that the decrease observed for the histone promoter activity when TRRAP expression is perturbed might result from a cell cycle effect rather than from a direct requirement for TRRAP in the activation of histone gene transcription. To assess this possibility, we examined the cell cycle distribution in cells that express the TRRAP-specific shRNAs in parallel with the analysis of the histone promoter activation. Flow cytometry analysis revealed that the S-phase populations of cells transfected with the TRRAP shRNA constructs were similar to that of the vector-transfected cells (Table 3). This observation indicates that the decreased levels of histone promoter activity in cells with perturbed TRRAP expression are not a result of cell cycle arrest but rather are due to the requirement for TRRAP in histone gene promoter activation.
TABLE 3.
Cell cycle distribution of cells transfected with indicated shRNA constructsa
| Cell cycle | Cell distribution (%) ± SD at the indicated time
|
|||||
|---|---|---|---|---|---|---|
| Day 2
|
Day 3
|
|||||
| Control | shTRRAP-1 | shTRRAP-2 | Control | shTRRAP-1 | shTRRAP-2 | |
| G0/G1 | 37.1 ± 3.8 | 35.4 ± 6.4 | 27.4 ± 3.9 | 34.2 ± 3.0 | 37.4 ± 2.1 | 30.7 ± 7.1 |
| S | 40.5 ± 2.1 | 39.5 ± 2.2 | 45.7 ± 4.8 | 44.7 ± 1.2 | 37.4 ± 5.1 | 37.5 ± 6.1 |
| G2/M | 22.4 ± 3.0 | 25.0 ± 4.6 | 26.9 ± 4.2 | 21.2 ± 2.1 | 25.3 ± 3.7 | 31.8 ± 11.0 |
To determine whether TRRAP is required for endogenous histone gene expression, we examined the levels of histone mRNAs in TRRAP-knocked-down cells by Northern blotting analysis. Suppression of TRRAP expression leads to about a 20 to 35% reduction in the levels of endogenous histone mRNAs (Fig. 6D). This effect is less than that seen with H4 and H2B promoter-reporter assays (Fig. 6B and C). However, this is not unexpected, as it is known that posttranscriptional mechanisms also play a major role in the regulation of histone mRNA levels in mammalian cells (20, 36, 45). Together, these data indicate that TRRAP is a critical cofactor for histone gene transcription.
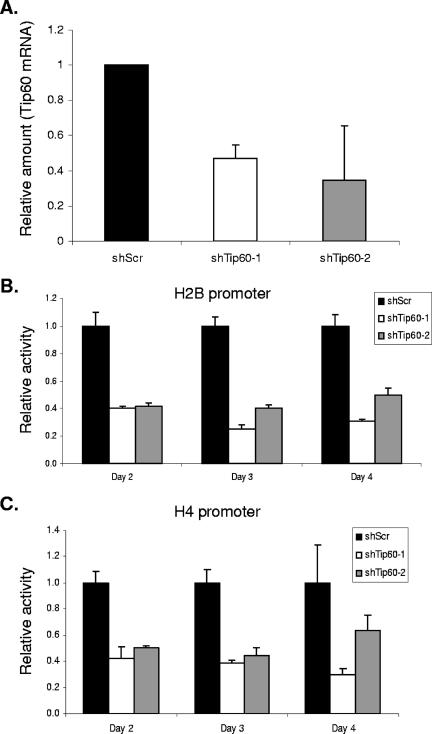
Similar to TRRAP, Tip60 also was observed to associate with histone promoters. Therefore, we investigated whether Tip60 is involved in histone promoter activation. We employed two shRNAs targeting two distinct sequences in Tip60 mRNA to knock down the expression of Tip60 (Fig. 7A). As shown in Fig. 7B and C, the suppression of Tip60 expression inhibits both the H2B and the H4 promoter activities, suggesting that Tip60 is also required for histone gene transcriptional activation.
FIG. 7.
Tip60 is required for histone promoter activation. (A) Tip60 expression is reduced by two Tip60-specific shRNA constructs. U2OS cells were transfected with pLKO.1 constructs carrying Tip60-specific shRNA sequences or a control nonspecific scrambled sequence. pLKO.1 carries a puromycin resistance gene allowing for selection of transfected cells. Twenty-four hours after transfection, puromycin was added to culture media and transfected cells were selected. The cells were harvested three days after selection. Relative Tip60 mRNA levels were determined by real-time qPCR. The qPCR data were normalized to expression of GAPDH. (B) Effect of Tip60 knockdown on H2B promoter activation. U2OS cells were transfected with an H2B promoter luciferase reporter (66), together with pLKO.1/Scramble (shScr) or pLKO.1 expressing shRNAs specific for human Tip60 mRNA (shTip60-1 and shTip60-2, respectively). To normalize for transfection efficiency, the cells were cotransfected with pCMV-LacZ. At the indicated time after transfection, the cells were harvested and the activities of luciferase and β-galactosidase were assayed as described previously (66). (C) Effect of Tip60 knockdown on H4 promoter activation. Activation of an H4 promoter luciferase reporter (66) in cells transfected with pLKO.1/Scramble (shScr), pLKO.1/shTip60-1 or pLKO.1/shTip60-2 was assayed as described for panel B.
DISCUSSION
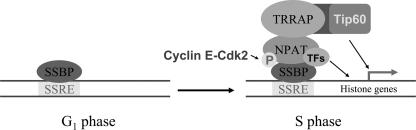
The work presented here provides mechanistic insights into the coordinated transcriptional activation of multiple histone subtypes by NPAT at the G1/S-phase transition. Our data indicate that the TRRAP-Tip60 complex interacts with NPAT and becomes associated with histone promoters at the G1/S-phase boundary in an NPAT-dependent manner. Moreover, consistent with the presence of the TRRAP-Tip60 HAT complex at histone promoters, histone H4 acetylation, which is associated with transcriptional activation (25, 59), increases at histone promoters during the G1/S-phase transition. This increase in histone H4 acetylation also depends on NPAT. Suppression of TRRAP or Tip60 expression through RNA interference leads to the inhibition of histone gene transcriptional activation. These results, together with the previous observations that the association of NPAT with histone promoters as well as NPAT-mediated histone promoter activation is regulated by cyclin E-Cdk2 phosphorylation (34, 53, 66), suggest there is a mechanism underlying the coordinated transcription of histone subtypes during the G1/S-phase transition (Fig. 8): NPAT becomes phosphorylated by cyclin E-Cdk2 at the G1/S-phase boundary, and this phosphorylation promotes the association of NPAT with histone promoters, likely through its interactions with the subtype-specific factors, such as Oct1/OCA-S and HiNF-P, which bind to specific DNA sequences (e.g., SSREs) in the promoters of the different histone gene subtypes. As a result, a TRRAP-containing HAT complex(s), for example, the Tip60 HAT complex, is recruited to the promoters of multiple histone genes. This HAT complex recruitment in turn leads to histone acetylation and, subsequently, transcriptional activation of multiple histone promoters.
FIG. 8.
A model for coordinated transcriptional activation of histone subtypes by cyclin E-Cdk2 substrate NPAT. SSRE, subtype-specific regulator element; SSBP, proteins, such as Oct-1 and HiNF-P that directly bind SSRE elements within the promoters of a histone subtype; TFs, transcription factors, P, phosphorylation. See text for details.
It was previously shown that the phosphorylation of NPAT by cyclin E-Cdk2 regulates its activity in histone gene activation (34, 66). It appears that cyclin E-Cdk2 modulates NPAT function by regulating the localization of NPAT at histone gene clusters (53). It is not clear whether cyclin E-Cdk2 regulates NPAT through additional mechanisms. Our preliminary results indicate that NPAT interacts with TRRAP throughout the cell cycle, suggesting that the interaction of NPAT with TRRAP may be independent of cyclin E-Cdk2 activity.
The results shown in Fig. 3 and 4 suggest that the majority of TRRAP/Tip60 recruitment to histone gene promoters occurs immediately prior to S-phase entry. It is possible that TRRAP/Tip60 recruitment may involve a two-step mechanism in which additional recruitment of these factors takes place in S phase, as intermediate levels of association of NPAT and TRRAP with histone gene promoters were observed in the presence of aphidicolin. In addition to NPAT, the recruitment of the TRRAP/Tip60 complex to histone gene promoters may also involve other histone gene transcription factors.
According to the model proposed in Fig. 8, one might expect the NPAT (the LFD-to-AAA) mutant to be dominant negative. Our results, however, indicate that this mutant apparently has no inhibitory activity on histone promoter activation (Fig. 1E). The exact reason why the NPAT (AAA) mutant protein fails to function as a dominant-negative mutant is not clear. One possible explanation is that one or more of the proteins that interact with the NPAT transactivation domain may be involved in stabilizing the interaction of NPAT with histone promoters. Without this stabilizing interaction, the presence of the mutant at the promoter may be merely transient, thus resulting in a failure of the NPAT (AAA) mutant to be dominant negative.
In this study, we have identified a domain in NPAT that possesses intrinsic transactivation potential. Interestingly, this domain, referred to as the transactivation domain of NPAT, contains a DLFD motif that is required for NPAT-mediated transcriptional activation and is functionally conserved in E2F and adenovirus E1A proteins. Our results clearly demonstrate that the DLFD motif is crucial for the interaction of NPAT with TRRAP, as well as for NPAT-mediated transcriptional activation (Fig. 1 and 2). The DLFD motif also appears to be crucial for the interaction of several E2F proteins with TRRAP and for their transcriptional activation function. It was previously observed that deletion of the DLFD sequence in a transactivation domain of E2F1 (residues 389 to 422) fused to the GAL4 DNA-binding domain resulted in an almost complete loss of its transcriptional activation capability (13). Consistent with this observation, the LFD-to-AAA mutation in the transcriptional activation domain of E2F3 (residues 391 to 465) results in the loss of transactivation when the mutant domain is fused to the GAL4 DBD (our unpublished observation). Moreover, the replacement of the LFD sequence with AAA in the transactivation domain abolishes the interaction of E2F3 with TRRAP (our unpublished observation). It was reported that the last seven amino acids of E2F4, which include the second aspartic acid residue in the DLFD motif, are critical for its interaction with TRRAP and E2F4-mediated reporter activation (30). E1A may also utilize the DLFD motif to interact with TRRAP. It was shown that the deletion of E1A from the CR1 region, which includes the DLFD motif, abolishes both TRRAP binding and transformation (7). Thus, the DLFD motif functions as a TRRAP-interacting module that is conserved in NPAT, E2F, and E1A proteins. Several other TRRAP-interacting proteins, such as c-Myc, p53, and BRCA1 (2, 37, 44), apparently lack the DLFD motif and therefore likely interact with TRRAP through a different sequence motif(s). The existence of multiple TRRAP-interacting motifs may allow the recruitment of TRRAP-containing complexes by distinct factors to be differentially regulated. It is interesting to note that the DLFD motif is also part of the sequences in E2F proteins shown to interact with the retinoblastoma protein pRB (13, 21, 22, 32, 48). Hence, pRB may inhibit E2F function by preventing the association of E2F with TRRAP-containing HAT complexes.
TRRAP has been shown to be a component of a number of HAT complexes, including the GCN5/PCAF and Tip60 complexes (6, 8). We focused on the TRRAP-Tip60 HAT complex in this study because we observed an interaction between the NPAT transactivation domain and two other components of the Tip60 HAT complex, Tip48 and Tip49, in our initial mass spectrometric analysis (Fig. 2B). It is possible that, similar to E2F and c-Myc, which recruit Tip60 as well as GCN5 complexes to their target promoters, NPAT may interact with and recruit additional TRRAP-containing HAT complexes to histone promoters in vivo. Compared with the NPATflox/− cells infected with Ad-LacZ, the Ad-Cre-infected NPATflox/− cells showed only a moderate (30 to 55%) reduction in histone H4 acetylation at histone promoters at the G1/S-phase boundary. This might be due to the fact that some residual NPAT protein remains in these cells and can still recruit the TRRAP-Tip60 complex to the histone promoters (Fig. 2E, 3, and 4). Alternatively, other protein factors might also recruit a HAT complex (or complexes) to the histone gene promoters to induce histone acetylation in concert with, but independent of, NPAT. Since proteins other than histones can also be the substrates of HATs (17, 18), the NPAT-recruited HAT(s) may also play a role in histone gene transcription by acetylating nonhistone proteins at histone promoters.
In addition to components of the Tip60 complex (Fig. 2B), NPAT appears to interact with YY1, BZAP45, and Hsp70, which have been implicated in histone gene transcription (12, 31, 41, 61, 67). Although an in vivo interaction of NPAT with these proteins remains to be determined, the observation raises the possibility that these proteins may participate in regulation of histone gene transcription through their cooperation with NPAT. The transactivation domain of NPAT apparently interacts with a number of additional proteins (Fig. 2B), which have not been shown to be involved in histone gene transcription. Further studies are needed to determine their interactions with NPAT in vivo, as well as their roles in transcriptional activation of histone genes. Such studies may shed new light on the coordinated regulation of histone gene transcription.
Supplementary Material
Acknowledgments
We thank Hartmut Land and Dirk Bohmann for helpful discussions and critical reading of the manuscript. We also thank Wade Harper for providing HCT116 NPATflox/− cells, Yang Shi for providing the pBS/U6 plasmid, Dennis McCance for providing the pBS/U6/Luc plasmid, Yin Sun for providing the pBS/U6/Ski plasmid, and Luojing Chen for providing the pLKO.1 nonspecific control. We are grateful to Peter Keng for help with FACS analysis, Anders Naar for suggestions on protein purification, and Taplin Biological Spectrometry Facility for mass spectrometric analysis.
This work was supported by NIH grant R01 GM65814 to J.Z.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 185108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ard, P. G., C. Chatterjee, S. Kunjibettu, L. R. Adside, L. E. Gralinski, and S. B. McMahon. 2002. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol. Cell. Biol. 225650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcaroli, D., L. Bongiorno-Borbone, A. Terrinoni, T. G. Hofmann, M. Rossi, R. A. Knight, A. G. Matera, G. Melino, and V. De Laurenzi. 2006. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA 10314808-14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 27418285-18289. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 2922333-2337. [DOI] [PubMed] [Google Scholar]
- 6.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19321-329. [DOI] [PubMed] [Google Scholar]
- 7.Deleu, L., S. Shellard, K. Alevizopoulos, B. Amati, and H. Land. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 208270-8275. [DOI] [PubMed] [Google Scholar]
- 8.Doyon, Y., and J. Cote. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14147-154. [DOI] [PubMed] [Google Scholar]
- 9.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 122245-2262. [DOI] [PubMed] [Google Scholar]
- 10.Dyson, N., P. Guida, C. McCall, and E. Harlow. 1992. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J. Virol. 664606-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson, N., and E. Harlow. 1992. Adenovirus E1A targets key regulators of cell proliferation. Cancer Surv. 12161-195. [PubMed] [Google Scholar]
- 12.Eliassen, K. A., A. Baldwin, E. M. Sikorski, and M. M. Hurt. 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 187106-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemington, E. K., S. H. Speck, and W. G. Kaelin, Jr. 1993. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA 906914-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher, C., N. Heintz, and R. G. Roeder. 1987. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51773-781. [DOI] [PubMed] [Google Scholar]
- 15.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., A. P. Bracken, K. Burkard, D. Pasini, M. Classon, C. Attwooll, M. Sagara, T. Imai, K. Helin, and J. Zhao. 2003. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell. Biol. 232821-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 36315-23. [DOI] [PubMed] [Google Scholar]
- 18.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90595-606. [DOI] [PubMed] [Google Scholar]
- 19.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 211854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heintz, N. 1991. The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta 1088327-339. [DOI] [PubMed] [Google Scholar]
- 21.Helin, K., E. Harlow, and A. Fattaey. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 136501-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helin, K., J. A. Lees, M. Vidal, N. Dyson, E. Harlow, and A. Fattaey. 1992. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell 70337-350. [DOI] [PubMed] [Google Scholar]
- 23.Herceg, Z., W. Hulla, D. Gell, C. Cuenin, M. Lleonart, S. Jackson, and Z. Q. Wang. 2001. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 29206-211. [DOI] [PubMed] [Google Scholar]
- 24.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102463-473. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 26.Kaelin, W. G., Jr., W. Krek, W. R. Sellers, J. A. DeCaprio, F. Ajchenbaum, C. S. Fuchs, T. Chittenden, Y. Li, P. J. Farnham, M. A. Blanar, et al. 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70351-364. [DOI] [PubMed] [Google Scholar]
- 27.Khorasanizadeh, S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116259-272. [DOI] [PubMed] [Google Scholar]
- 28.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98285-294. [DOI] [PubMed] [Google Scholar]
- 29.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 222836-2841. [DOI] [PubMed] [Google Scholar]
- 30.Lang, S. E., S. B. McMahon, M. D. Cole, and P. Hearing. 2001. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 27632627-32634. [DOI] [PubMed] [Google Scholar]
- 31.Last, T. J., A. J. van Wijnen, M. J. Birnbaum, G. S. Stein, and J. L. Stein. 1999. Multiple interactions of the transcription factor YY1 with human histone H4 gene regulatory elements. J. Cell. Biochem. 72507-516. [PubMed] [Google Scholar]
- 32.Lee, C., J. H. Chang, H. S. Lee, and Y. Cho. 2002. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 163199-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 34.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 142298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 216782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marzluff, W. F., and R. J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14692-699. [DOI] [PubMed] [Google Scholar]
- 37.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94363-374. [DOI] [PubMed] [Google Scholar]
- 38.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meeks-Wagner, D., and L. H. Hartwell. 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 4443-52. [DOI] [PubMed] [Google Scholar]
- 40.Miele, A., C. D. Braastad, W. F. Holmes, P. Mitra, R. Medina, R. Xie, S. K. Zaidi, X. Ye, Y. Wei, J. W. Harper, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2005. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 256140-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitra, P., P. S. Vaughan, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 2001. Purification and functional analysis of a novel leucine-zipper/nucleotide-fold protein, BZAP45, stimulating cell cycle regulated histone H4 gene transcription. Biochemistry 4010693-10699. [DOI] [PubMed] [Google Scholar]
- 42.Murr, R., J. I. Loizou, Y. G. Yang, C. Cuenin, H. Li, Z. Q. Wang, and Z. Herceg. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 891-99. [DOI] [PubMed] [Google Scholar]
- 43.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398828-832. [DOI] [PubMed] [Google Scholar]
- 44.Oishi, H., H. Kitagawa, O. Wada, S. Takezawa, L. Tora, M. Kouzu-Fujita, I. Takada, T. Yano, J. Yanagisawa, and S. Kato. 2006. An hGCN5/TRRAP histone acetyltransferase complex co-activates BRCA1 transactivation function through histone modification. J. Biol. Chem. 28120-26. [DOI] [PubMed] [Google Scholar]
- 45.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60827-861. [DOI] [PubMed] [Google Scholar]
- 46.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 151619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robert, F., S. Hardy, Z. Nagy, C. Baldeyron, R. Murr, U. Dery, J. Y. Masson, D. Papadopoulo, Z. Herceg, and L. Tora. 2006. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell. Biol. 26402-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan, B., T. Durfee, and W. H. Lee. 1996. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc. Natl. Acad. Sci. USA 93679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shan, B., X. Zhu, P. L. Chen, T. Durfee, Y. Yang, D. Sharp, and W. H. Lee. 1992. Molecular cloning of cellular genes encoding retinoblastoma-associated proteins: identification of a gene with properties of the transcription factor E2F. Mol. Cell. Biol. 125620-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squatrito, M., C. Gorrini, and B. Amati. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16433-442. [DOI] [PubMed] [Google Scholar]
- 51.Stein, G. S., J. L. Stein, A. J. Van Wijnen, and J. B. Lian. 1996. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol. Int. 2041-49. [DOI] [PubMed] [Google Scholar]
- 52.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su, C., G. Gao, S. Schneider, C. Helt, C. Weiss, M. A. O'Reilly, D. Bohmann, and J. Zhao. 2004. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 231133-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sui, G., C. Soohoo, B. el Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 995515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taubert, S., C. Gorrini, S. R. Frank, T. Parisi, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2004. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 244546-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thrash, B. R., C. W. Menges, R. H. Pierce, and D. J. McCance. 2006. AKT1 provides an essential survival signal required for differentiation and stratification of primary human keratinocytes. J. Biol. Chem. 28112155-12162. [DOI] [PubMed] [Google Scholar]
- 58.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell. 2869-875. [DOI] [PubMed] [Google Scholar]
- 59.Wang, Y., W. Fischle, W. Cheung, S. Jacobs, S. Khorasanizadeh, and C. D. Allis. 2004. Beyond the double helix: writing and reading the histone code. Novartis Found. Symp. 2593-17. (Discussion, 17-21 and 163-169.) [PubMed] [Google Scholar]
- 60.Wei, Y., J. Jin, and J. W. Harper. 2003. The cyclin E/Cdk2 substrate and Cajal body component p220NPAT activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 233669-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, F., and A. S. Lee. 2001. YY1 as a regulator of replication-dependent hamster histone H3.2 promoter and an interactive partner of AP-2. J. Biol. Chem. 27628-34. [DOI] [PubMed] [Google Scholar]
- 62.Ye, X., A. A. Franco, H. Santos, D. M. Nelson, P. D. Kaufman, and P. D. Adams. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 11341-351. [DOI] [PubMed] [Google Scholar]
- 63.Ye, X., Y. Wei, G. Nalepa, and J. W. Harper. 2003. The cyclin E/Cdk2 substrate p220NPAT is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 238586-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao, J. 2004. Coordination of DNA synthesis and histone gene expression during normal cell cycle progression and after DNA damage. Cell Cycle 3695-697. [PubMed] [Google Scholar]
- 65.Zhao, J., B. Dynlacht, T. Imai, T. Hori, and E. Harlow. 1998. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 12456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 142283-2297. [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng, L., R. G. Roeder, and Y. Luo. 2003. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114255-266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.