Abstract
The proto-oncogenes c-, L-, and N-myc can all be translated by the alternative method of internal ribosome entry whereby the ribosome is recruited to a complex structural element (an internal ribosome entry segment [IRES]). Ribosome recruitment is dependent upon the presence of IRES-trans-acting factors (ITAFs) that act as RNA chaperones and allow the mRNA to attain the correct conformation for the interaction of the 40S subunit. One of the major challenges for researchers in this area is to determine whether there are groups of ITAFs that regulate the IRES-mediated translation of subsets of mRNAs. We have identified four proteins, termed GRSF-1 (G-rich RNA sequence binding factor 1), YB-1 (Y-box binding protein 1), PSF (polypyrimidine tract binding protein-associated splicing factor), and its binding partner, p54nrb, that bind to the myc family of IRESs. We show that these proteins positively regulate the translation of the Myc family of oncoproteins (c-, L-, and N-Myc) in vivo and in vitro. Interestingly, synthesis from the unrelated IRESs, BAG-1 and Apaf-1, was not affected by YB-1, GRSF-1, or PSF levels in vivo, suggesting that these three ITAFs are specific to the myc IRESs. Myc proteins play a role in cell proliferation; therefore, these results have important implications regarding the control of tumorigenesis.
The proteins encoded by the myc gene family function as sequence-specific transcription factors that regulate target genes integral to the processes of cell proliferation, differentiation, and cell death (2, 9). Although many of the functions of the three major myc genes are overlapping, unique properties have been ascribed to individual Myc proteins. For example, each of the Myc proteins can restore the growth and proliferative defects of c-myc null fibroblasts and can promote apoptosis following growth factor deprivation (23, 30). In addition, all three proteins can regulate known myc target genes (23). Unique roles for these proteins are supported by the following observations: during embryogenesis and in adult tissues, the expression patterns of c-, L-, and N-Myc are distinct (52); homozygous null c- or N-myc mice die early in development, whereas targeted disruption of both L-myc alleles is not lethal; and in some instances, L-myc displays distinct cell transformation and transcriptional properties (1, 38, 39).
It is not surprising, given the role of the Myc family of proteins in proliferation and apoptosis, that the expression of these proteins is highly regulated at the levels of both transcription and translation. Indeed, deregulated Myc expression, through either of these mechanisms, has been associated with tumorigenesis (12, 35, 36, 49, 50).
Previous studies have shown that the 5′ untranslated regions (UTRs) of c-, L-, and N-myc encoded by exon 1 each contain a complex RNA structural element known as an internal ribosome entry segment (IRES). Consequently, synthesis of the Myc family proteins can occur by the process of internal ribosome entry (18, 19, 28, 45). In this mechanism of translation initiation, the IRES, in conjunction with IRES trans-acting factors (ITAFs), recruits the 40S ribosomal subunit. Cellular IRESs promote the selective synthesis of certain proteins during situations when cap-dependent translation is compromised. For example, the c-myc IRES functions during apoptosis (6, 44), during development (10), and during genotoxic stress (47). ITAFs regulate the activity of cellular IRESs (43), and moreover, it has been proposed that there are both specific ITAFs that control the activity of related groups of IRESs and general ITAFs (e.g., polypyrimidine tract binding protein [PTB]) (26) that are required by the majority of cellular IRESs (6, 26, 43). In contrast, c-myc and BAG-1 IRESs have been shown to require members of the poly(rC) binding protein family for activity (12, 37), while the fibroblast growth factor 2 IRES requires hnRNPA1 (4) and the XIAP IRES requires La and hnRNPC for function (16). However, to date, no comprehensive study has been carried out on a group of related IRESs to determine whether there are defined sets of ITAFs that control their activity. To address this question, we have used an affinity chromatography approach to identify ITAFs that interact with the myc family of IRESs. We identified four proteins—PSF (PTB-associated splicing factor); its binding partner, p54nrb (3, 40); GRSF-1 (G-rich RNA sequence binding factor 1) (32); and YB-1 (Y-box binding protein 1) (22)—that interacted and stimulated myc family IRES activity but not that of other unrelated IRESs. In addition, a reduction in the levels of some of these ITAFs in vivo was found to reduce the expression of endogenous c-Myc protein without altering the levels of the corresponding mRNA, strongly suggesting that these factors play a role in controlling c-Myc synthesis. Regulation of c-Myc expression via these proteins has important implications for the understanding of neoplasia.
MATERIALS AND METHODS
Materials.
Media and sera were purchased from Gibco BRL; luciferase assay kits, Stop & Glo substrate, and rabbit reticulocyte lysates were purchased from Promega. The GalactoLight Plus assay system was purchased from Tropix. HeLa and NIH 3T3 cells were all obtained originally from the American Type Culture Collection. FuGene 6 was purchased from Roche Molecular Biochemicals. All other chemicals were purchased from Sigma.
Plasmid constructs.
The plasmids pRF, pRMF, pRLF, and pRNF, which harbor the myc family of IRESs, have previously been described (18, 19, 45). The plasmid pSKL is based upon the vector pBluescript SK+ (Stratagene); the c-, L- and N-myc 5′ UTRs were cloned into this vector upstream of the firefly luciferase gene. For the expression of the proteins used, the cDNAs for YB-1, GRSF-1, and p54nrb were cloned into pET21a vectors, enabling the expression of protein in Escherichia coli and the subsequent purification of the protein. Recombinant PSF protein was expressed in E. coli from the vector pET15-PSF (kind gift of James G. Patton, Vanderbilt University) by the method described previously (33).
To knock down endogenous levels of PSF and GRSF-1, inverted repeat hairpin sequences targeted the mouse GRSF-1 and PSF coding sequences (5′-AAAGCACAGGGAAGAAATTGGTA-3′ and 5′-GATATGGTAGAGGGAGAGAAG-3′, respectively) were cloned into the plasmid mU6pro (a gift from D. L. Turner, University of Michigan) in order to express small interfering RNAs (siRNAs) against these proteins. Levels of p54nrb/NONO were reduced using the vector pSuper, which harbors a hairpin sequence against mouse p54nrb/NONO (kind gift of Steven Brown, Geneva, Switzerland) (5). The vector pSuperDuper, which harbors a tail-to-tail inverted repeat sequence targeted against the human YB-1 coding sequence (5′-GGTCATCGCAACGAAGGTTTT-3′) (kind gift of Peter R. Mertens, Aachen, Germany), was used to express an siRNA against YB-1.
In vitro transcription reactions.
Vector DNA was linearized by restriction digestion using a site downstream of the region of interest (HpaI for dicistronic, NcoI for monocistronic); transcripts were synthesized in a reaction mixture containing 1× transcription buffer (40 mM HEPES-KOH [pH 7.9], 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol [DTT], 10 mM NaCl), 40 U RNAguard or RNasin, 1 mM ATP, 1 mM UTP, 1 mM CTP, 0.5 mM GTP, 1 μM m7G(5′)ppp(5′)G, 1 μg DNA template, and 20 U T7 or T3 RNA polymerase to a final volume of 50 μl. For radiolabeled RNAs, CTP was replaced with 50 μCi [α-32P]CTP. For biotinylated RNA, the transcription reaction mixtures were supplemented with 0.1 mM biotin UTP. The reaction mixture was incubated at 37°C for 1.5 h, and the RNA purified.
Affinity purification.
Cytoplasmic HeLa extracts were obtained from 4C Biotech (Belgium). We incubated them with biotinylated RNA transcripts (generated as described above) in a buffer containing 1.2 mM MgCl2, 1.65 mM ATP, 30 mM KCl, 0.02% Tween 20, 10 mM Tris (pH 7.5), 10 μg/ml heparin, 10 μg/ml tRNA, and 10 units RNasin for 30 min at 4°C. Then these were incubated with paramagnetic streptavidin beads for a further 10 min, and the RNA and bound proteins were isolated using a magnet. The complexes were washed with binding buffer and then again with the same buffer but containing 100 mM NaCl and 200 mM NaCl and finally eluted with 300 mM NaCl. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by staining with silver (41), and bands were excised from the gel. To achieve digestion, proteins within the gel pieces were first reduced, carboxyamidomethylated, and then digested to peptides using trypsin on a MassPREP station (Waters, Manchester, United Kingdom). The resulting peptides were subjected to liquid chromatography-tandem mass spectrometry (LC-MS-MS). For LC-MS-MS, the reverse-phase liquid chromatographic separation of peptides was achieved with a PepMap C18 reverse-phase, 75-μm (inside diameter), 15-cm column (LC Packings, Amsterdam, The Netherlands) on a capillary LC system (Eksigent) attached to an LTQ Orbitrap (Thermo Fisher) MS. The MS-MS fragmentation data achieved was used to search the National Center for Biotechnology Information database, using the MASCOT search engine (Matrix Science). Probability-based MASCOT scores were used to evaluate identifications. Only matches with P values of <0.05 for random occurrence were considered significant (for further explanation of MASCOT scores, contact Matrix Science).
Protein expression.
Proteins were overexpressed in E. coli from the pET21a vector by the addition of isopropyl-β-d-thiogalactopyranoside to the growth medium. The proteins that contained a His tag were purified using a nickel affinity column (Qiagen). Cells were harvested and lysed in phosphate-buffered saline containing 0.1% Triton X-100, and the tagged protein was purified on a nickel affinity column.
Cell culture and transient transfections.
Cells were typically grown in Dulbecco's modified Eagle's medium (Gibco-BRL) containing 10% fetal calf serum, under a humidified atmosphere containing 5% CO2. Cells were transfected using FuGene 6 (Roche) as specified by the manufacturer. Lysates were prepared from transfected cells using 1× passive lysis buffer. Firefly and Renilla luciferase activities were measured using the Stop & Glo dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions, with the exception that only 25 μl of each reagent was used. Light emission was measured over 10 seconds using an Optocomp I luminometer. The activity of the β-galactosidase transfection control was measured using a GalactoLight Plus assay system (Tropix). Relative IRES activity is calculated as firefly luciferase activity relative to either the transfection control β-galactosidase (since this protein has a half-life of around 30 h) or total protein levels. All transfections were carried out in triplicate on at least three independent occasions.
Thymidine incorporation.
Cells were seeded at 0.3 × 106 cells per well of a six-well plate and the next day transfected with 5 μg of either control plasmids or plasmids harboring sequences targeted against YB-1, p54nrb, or GRSF-1. Transfected cells were grown for 48 h; after this period, 5 μCi/ml of [3H]thymidine was added to each well with fresh media, and then cells were incubated for a further 24 h. The cells were then washed twice with phosphate-buffered saline before the addition of 10% trichloroacetic acid. Cells were collected and precipitated on ice for 30 min before being filtered through nitrocellulose with extensive washing. Filters were dried, and scintillation readings taken. Cell proliferation is expressed as [3H]thymidine incorporation relative to the number of cells.
Immunoblotting.
For analysis of c-Myc, Apaf-1, BAG-1, PSF, p54nrb, and YB-1 expression, cell pellets were solubilized in electrophoresis buffer (50 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 1 mM EDTA, 10% glycerol, and 0.01% bromophenol blue, supplemented with 1% aprotinin, 1 μg/ml leupeptin, and 1 μg/ml N-α-p-tosyl-l-lysine chloromethyl ketone [TLCK] immediately before use) by sonication. Cell extracts (equal numbers of cells per lane) were then analyzed by SDS-PAGE and electroblotted. Blots were probed with antibodies purchased from Abcam and used at dilutions of 1:1,000 (PSF), 1:100 (GRSF-1), 1:500 (YB-1), and 1:1,600 (p54nrb). Those from Cell Signaling were used at dilutions of 1:2,000 (BAG-1), 1:1,000 (c-myc, 9E10), and 1:1,000 (Apaf-1). Antiactin monoclonal antibody was purchased from Sigma and used at a dilution of 1:2,000. The blots were then incubated with peroxidase-conjugated secondary antibodies raised against mouse or rabbit immunoglobulin and developed using the chemiluminescence reagent ECL (Amersham).
In vitro translation reactions.
The Promega Flexi rabbit reticulocyte lysate in vitro translation system was primed with 5 ng/μl RNA and used according to the manufacturer's instructions. The reaction was performed with a final volume of 12.5 μl, and 0.5 to 2 μg of each protein was added where appropriate (see Fig. S1 in the supplemental material). Luciferase activities were assayed as described above, and the firefly and Renilla luciferase values expressed relative to the control plasmid pRF, which was assigned a value of 1. All experiments were performed in triplicate on at least three independent occasions.
UV cross-linking analysis.
Radiolabeled transcript was generated from pSKML, pSKNL, pSKLsL, or PSKGAPL linearized with NcoI. Approximately 2.5 pmol per reaction was incubated with 0.25 μg of protein in 1× UV cross-linking buffer (10 mM HEPES [pH 7.4], 3 mM MgCl2, 100 mM KCl, 5 mM creatine phosphate, 1 mM DTT, 1 mM ATP, 6% glycerol, 0.1 μg/μl tRNA, 5 μg heparin) for 15 min at room temperature. For competition assays, unlabeled competitor RNAs were added with labeled RNA. The reaction mixtures were UV irradiated using a 305-nm UV light source for 30 min on ice. RNase A, RNase T1, and RNase V1 (0.2 mg/ml) were added to the mixture to degrade any unprotected RNA by incubation at 37°C for 30 min. Sample buffer was added, and the samples were separated on a 10% polyacrylamide gel by SDS-PAGE. Gels were dried at 80°C under a vacuum for 2 h and analyzed on a Molecular Dynamics PhosphorImager.
Filter binding assays.
Approximately 23,000 cpm of labeled transcript was added to 10 μl of buffer mix containing 2 μl of 5× transcription buffer (200 mM Tris-HCl [pH 8.0], 40 mM MgCl2, 10 mM spermidine, 250 mM NaCl), 0.75 μl of 1 M DTT, 2 μl of tRNA (10 mg/ml), 1 μl of 10 mM rATP, and 40 U of RNasin. Recombinant PSF, p54nrb, YB-1, PTB, and GRSF-1 between the range of 0.05 μg and 2 μg were then incubated with the mixture at room temperature for 10 min. Reaction mixtures were then filtered through a nitrocellulose membrane (0.45-μm pore size), presoaked in 1× binding/wash buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 14.4 mM 2-mercaptoethanol). Membranes were washed with 0.5 ml of 1× binding/wash buffer. Membrane filters were air dried and then counted in a scintillation counter. A known volume of RNA was spotted onto a separate filter without washing to provide total input radioactivity. For the calculation of dissociation constants, the fraction of bound RNA was determined from the scintillation reading.
RESULTS
Identification of myc IRES family binding proteins.
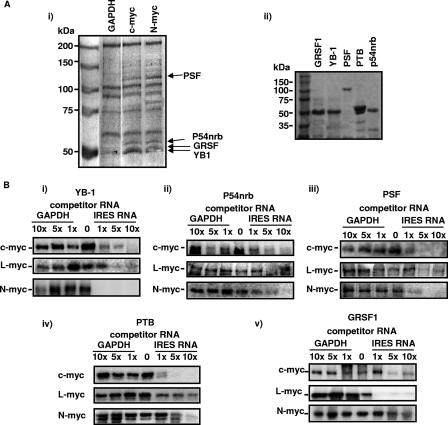
Biotinylated c-myc and N-myc IRES RNAs and control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA were transcribed from pSKML (34), pSKNL, or pSKGAPL (34) and incubated with HeLa cell cytoplasmic extracts, and RNA-protein complexes were purified using streptavidin paramagnetic beads. After extensive washing, bound proteins were eluted with buffer containing 300 mM NaCl. The products were separated by SDS-PAGE, and bands which were common to the N- and c-myc IRESs but not to the GAPDH mRNAs (Fig. 1Ai) were excised and identified by LC-MS-MS. The proteins that bound to both c- and N-myc IRESs—PSF (33); its binding partner, p54nrb (40); GRSF-1 (20, 32); and YB-1 (22)—are multifunctional RNA binding proteins that have roles in transcription, splicing, and translation. hnRNPC was identified as a protein that bound the c-myc IRES but not the other IRESs (data not shown), in agreement with previous data (21). cDNAs that encoded the corresponding proteins were obtained by reverse transcription-PCR and then subcloned into expression vectors, and the resultant proteins purified for use in subsequent experiments (Fig. 1Aii).
FIG. 1.
PSF, p54nrb, GRSF-1, and YB-1 bind specifically to the myc IRESs. (Ai) Biotinylated RNAs were incubated with HeLa cell extracts, and protein-RNA complexes were isolated on streptavidin magnetic beads. Proteins were separated by PAGE and identified by LC-MS-MS. (Aii) Proteins were subcloned and overexpressed in E. coli. A polyacrylamide gel of the purified proteins used in subsequent experiments is shown. (B) UV cross-linking assays were performed using radiolabeled c-, L- or N-myc IRES RNAs, which were incubated with 0.2 μg of YB-1 (i), p54nrb (ii), PSF (iii), PTB (iv), or GRSF-1 (v) in the presence/absence of unlabeled GAPDH RNA or IRES RNA. Products were separated on a 10% polyacrylamide gel. In all cases, unlabeled IRES RNA competed for binding, while no competition was observed with GAPDH RNA, even at a 10-fold (10×) molar excess.
PSF, p54nrb, GRSF-1, YB-1, and PTB bind to myc family IRESs in vitro.
To confirm the interactions of the four proteins identified in the screen and PTB, which we have shown previously to be a general ITAF (26), with all three myc IRESs, UV cross-linking analysis was performed. Thus, radiolabeled IRES RNAs were generated and incubated with one of the five purified proteins (Fig. 1Aii), samples were exposed to UV light, any RNA not bound to protein was digested with RNase, and the products were separated by PAGE (Fig. 1Bi through Bv). All five proteins bound to the three IRESs, and in each case, there was a reduction in the binding of protein to the radiolabeled transcripts with a 1 M higher concentration of unlabeled IRES RNA but not to those with a 5- to 10-fold higher concentration of GAPDH RNA (Fig. 1Bi through Bv). To test the strength of the interactions, filter binding assays were performed (Table 1). These data show that the proteins bind to these RNA elements with different affinities. For example, YB-1 binds ∼10-fold more tightly to the c- and N-myc IRESs than to the L-myc IRES, while in contrast, the L-myc IRES interacts more strongly with PSF (Table 1). It has been suggested that PSF and p54nrb act as a dimer; therefore, assays were performed in the presence of both proteins to determine whether this increased the strength of the interactions. However, there was only a small increase in the binding constant in each case, suggesting that these proteins are not acting in concert in this situation (data not shown).
TABLE 1.
Filter binding assays were performed to obtain dissociation constants (Kd) of the c-, L-, and N-myc IRESs with p54nrb, PSF, PTB, GRSF-1, and YB-1a
| Protein |
Kd (M) of the indicated IRESb
|
||
|---|---|---|---|
| c-myc | L-myc | N-myc | |
| p54nrb | 1.85 (±0.96) × 10−7 | 9.8 (±0.52) × 10−6 | 1.78 (±0.55) × 10−5 |
| PSF | 2.38 (±0.18) × 10−6 | 5 (±0.043) × 10−7 | 6.6 (±1.3) × 10−6 |
| PTB | 3.16 (±0.9) × 10−6 | 1.76 (±0.15) × 10−5 | 8.3 (±0.33) × 10−6 |
| GRSF-1 | 2.9 (±0.23) × 10−6 | 3.72 (±0.523) × 10−5 | 1.87 (±0.73) × 10−5 |
| YB-1 | 2.94 (±0.1) × 10−7 | 5.33 (±0.106) × 10−6 | 5.20 (±0.17) × 10−7 |
To test the strength of the interactions of the c-, L-, and N-myc IRESs with p54nrb, GRSF-1, PSF, PTB, and YB-1, filter binding assays were performed. Radiolabeled transcripts were incubated with a range of concentrations of recombinant proteins. Protein-RNA complexes were isolated on nitrocellulose filters, and the fractions of bound RNA were determined from the scintillation readings.
Standard deviations are given in parentheses.
PSF, p54nrb, GRSF-1, and YB-1 are required for the myc family of IRESs to function in vitro and in vivo.
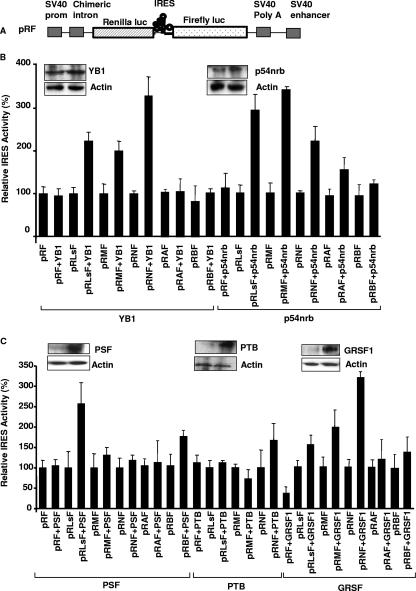
In general, cellular IRESs work very inefficiently (if at all) in vitro, but we have shown that it is possible to stimulate certain cellular IRESs by the addition of known trans-acting factors (12, 25, 27). Therefore, to determine whether the ITAFs identified could stimulate IRES activity in vitro, dicistronic RNAs were generated (from pRF and pRIRESF) (Fig. 2A) and used to prime reticulocyte lysates that were supplemented with the putative trans-acting factors (Fig. 2B; also see Fig. S1 in the supplemental material). The myc family of IRESs function very inefficiently in vitro, and no appreciable luciferase activity was detected beyond that produced from RNA derived from the vector pRF, which does not contain an IRES (Fig. 2B and C). However, increased production of firefly luciferase was observed in the presence of one of the four proteins identified, and the degrees of activation of the three IRESs were, in general, related to the binding affinities that these factors had for the individual RNAs. Thus, only the L-myc IRES was stimulated significantly by the addition of PSF to the reticulocyte lysates (Fig. 2C). The c-myc IRES and L-myc IRES were both stimulated more by the addition of p54nrb to the lysates than the N-myc IRES. Finally, YB-1 increased N-myc IRES-mediated translation 3.2-fold, while the L- and c-myc IRESs were stimulated 2.3- and 1.8-fold, respectively (Fig. 2B). We have shown previously that PTB is required by many cellular IRESs; however, additional factors are often required to enable this protein to bind to cellular IRESs (27, 37). In agreement with these data, only a small stimulation of the N-myc IRES was observed with this protein (Fig. 2C). To test whether the putative ITAFs identified in the screen were specific to the myc family of IRESs, in vitro translation reactions were also primed with dicistronic RNAs that contained the Apaf-1 and BAG-1 IRESs (7, 8). Neither of these IRESs was stimulated by YB-1, and only small increases in IRES activity were observed with the Apaf-1 IRES upon the addition of p54nrb and with the BAG-1 IRES upon the addition of PSF (Fig. 2B and C). Finally, combination studies were performed to assess whether there was a synergistic effect when these proteins were added together; however, no additional stimulation of IRES activity was observed (data not shown).
FIG. 2.
PSF, p54nrb, GRSF-1, and YB-1 stimulate myc IRESs in vitro. (A) Schematic representation of the dicistronic reporter constructs pRF and pRIRESF, where pRIRESF contains the c-myc (pRMF), L-myc (pRLsF), N-myc (pRNF), BAG-1 (pRBF), or Apaf-1 (pRAF) 5′ UTRs inserted into the vector pRF and fused in frame with the firefly luciferase gene. SV40 prom, simian virus 40 promoter; luc, luciferase. (B and C) In vitro translation reactions in reticulocyte lysates primed with 100 ng of capped pRIRESF RNA and 50 to 200 ng of PSF, p54nrb, GRSF-1, YB-1, or PTB (see Fig. S1 in the supplemental material) show that these proteins can stimulate IRES activity from these dicistronic plasmids, albeit to different extents. The greatest effects were observed with the addition of YB-1 and p54nrb. IRES activity is expressed as a ratio of the downstream cistron to the upstream cistron (firefly/Renilla luciferase). All experiments were performed in triplicate on three independent occasions.
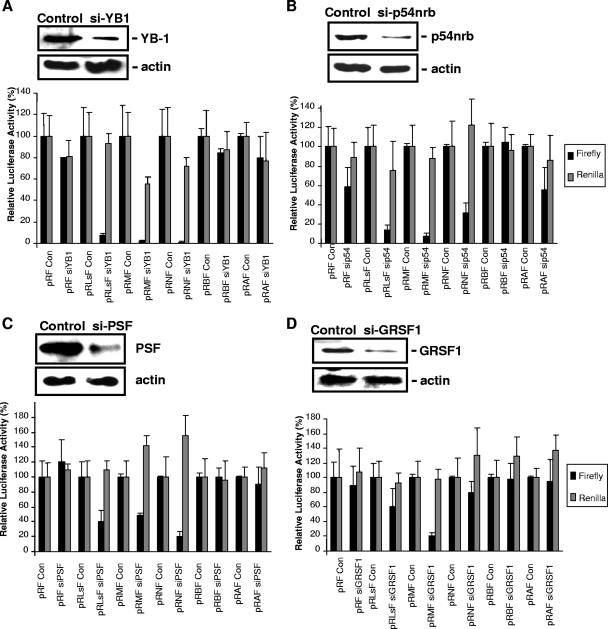
The reticulocyte lysate system is known to lack many other factors required for IRES-mediated translation, since the activity of cellular IRESs in vitro, even in the presence of these ITAFs, is much lower than cellular IRES activity in vivo. Therefore, it was important to test whether modulating the levels of these factors also alters IRES activity in vivo. Cells were transfected with plasmids that express siRNA duplexes that would reduce the expression of p54nrb, PSF, GRSF-1, and YB-1 or control plasmids (Fig. 3) and then transfected with the IRES-containing constructs, as described previously (7, 8, 18, 19, 45) (Fig. 2A, and see Fig. S2 in the supplemental material). Cell extracts were harvested, and Western analysis showed that in each case, the level of the targeted proteins produced was reduced to less than 20% of that expressed in the control cells (Fig. 3A through D). Luciferase assays were then performed, and there was a reduction in firefly luciferase produced in each case, albeit to different extents. For example, in cells that have reduced expression of p54nrb, the level of firefly luciferase produced via c-myc IRES-mediated initiation was decreased to 5%, while N-myc IRES-mediated expression of luciferase was still 30% (Fig. 3B). In cells with a reduction in the levels of GRSF-1, the activity of the c-myc IRES was decreased to 10%, with only a small inhibitory effect on the L- and N-myc IRESs (Fig. 3D). Interestingly, all three IRESs were strongly affected by the reduction in YB-1 levels, and the firefly luciferase activity produced was reduced to less than 5% of that found in control cells in all cases (Fig. 3A). Again, to test whether the factors identified interacted with other cellular IRESs, the Apaf-1 and BAG-1 IRESs were similarly tested (Fig. 3). These data agree with those obtained in vitro, in that YB-1 and GRSF-1 appear to be specific to the myc family of IRESs (Fig. 3A and D), while the Apaf-1 IRES activity was decreased only in cells that have a reduction in p54nrb (Fig. 3B). There are some differences between the in vitro and in vivo data, and this is most likely due to the differences in the levels of these proteins in vitro compared to the levels in vivo. For example, the level of GRSF in vitro is very low, and the N-myc IRES is stimulated by the addition of this protein (Fig. 2B). However, there is a much smaller effect on IRES activity following a reduction in the level of this protein in vivo.
FIG. 3.
Reductions in the expression of p54nrb, PSF, YB-1, and GRSF-1 correlate with decreases in IRES activity in cultured cells. Cells were transfected with plasmids that reduce the expression of p54nrb, PSF, GRSF-1, or YB-1 and then transfected with the plasmids harboring the c-, L-, or N-myc (pRMF, pRLsF, pRNF, respectively); Apaf-1 (pRAF); or BAG-1 (pRBF) IRES. Cells were harvested and lysed, and a proportion of the resulting material was immunoblotted using antibodies against small interfering YB-1 (si-YB1) (A), p54nrb (B), PSF (C), or GRSF-1 (D); actin was used as a loading control (Con). In each case, the level of the protein was reduced to less than 20% of the value in the control cells. The samples were then assayed for firefly and Renilla luciferase activities, and the relative IRES activity was calculated as firefly activity relative to the transfection control β-galactosidase. All experiments were carried out in triplicate and performed on at least three independent occasions. All three myc IRESs were affected by a reduction in the levels of these four proteins, although the greatest decrease was observed when the level of YB-1 was reduced, causing the activity of all three IRESs to be reduced by 95%. In contrast, only a small reduction in Apaf-1 IRES activity was observed in cells with a decreased expression of p54nrb, and no effect was observed in BAG-1 IRES activity.
Following a reduction in the levels of YB-1 and, to a lesser extent, p54nrb, there was a decrease in the level of Renilla luciferase produced by cap-dependent translation; however, the decrease observed was much less than the decrease in IRES-mediated translation. In this regard, it has previously been shown that cap-dependent translation is regulated in a dose-dependent manner by YB-1 (14).
Translational regulation of the c-Myc protein by p54nrb, PSF, GRSF-1, and YB-1.
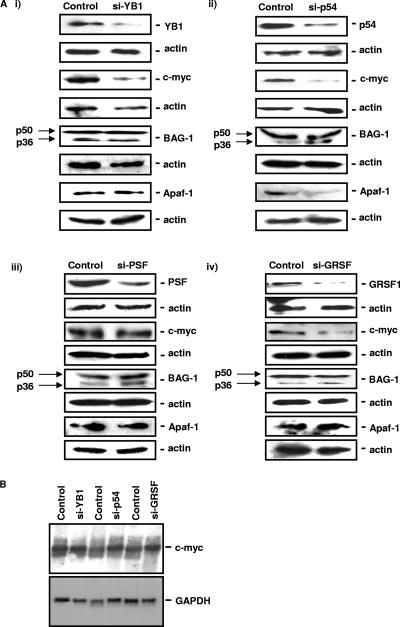
Having identified a series of proteins that are capable of modulating the activity of the myc family of IRESs, it was then important to determine how these ITAFs affect the levels of endogenous proteins. Therefore, NIH 3T3 cells were transfected with plasmids that express siRNA duplexes directed against p54nrb, PSF, GRSF-1, and YB-1 RNAs and harvested; cell extracts were separated by SDS-PAGE; and Western blotting was performed to determine the levels of the c-Myc, Apaf-1, and BAG-1 proteins (Fig. 4Ai through Aiv). Following a reduction in the levels of YB-1 and p54nrb, there was a large decrease in the expression of the c-Myc protein (Fig. 4Ai and Aii) and a smaller but reproducible decrease in c-Myc protein levels in cells with reduced expression of GRSF-1 (Fig. 4Aiii), suggesting that these three ITAFs are indeed important for regulating c-Myc synthesis (Fig. 4Ai through Aiii). However, there was no reduction in endogenous c-Myc expression in cells with reduced levels of PSF (Fig. 4Aiv). In addition, there was no effect on the synthesis of the p36 isoform of BAG-1 (which can be IRES-mediated [7]) by a reduction in the levels of any of these proteins, and Apaf-1 protein levels were reduced only in cells that expressed lower levels of p54nrb (in agreement with the data shown in Fig. 3A).
FIG. 4.
Endogenous c-Myc protein levels are reduced in cells that have decreased expression of p54nrb, GRSF-1, or YB-1. (A) Cells were transfected with a control plasmid or a plasmid that reduces the expression of YB-1 (i), p54nrb (p54) (ii), PSF (iii), or GRSF-1 (iv). Cells were harvested, and the subsequent lysates separated by SDS-PAGE and immunoblotted for the proteins indicated. c-Myc protein levels are reduced in cells that have low levels of YB-1, p54nrb, or GRSF-1 but not PSF. In contrast, BAG-1 protein levels (including the p36 isoform which can be translated by internal ribosome entry) are unaffected, and Apaf-1 protein levels are decreased only in cells with a reduction of p54nrb. (B) RNA was generated from cells transfected with plasmids which reduce the expression of p54nrb (p54), YB-1, and GRSF-1. The RNA was separated by denaturing agarose gel electrophoresis and transferred to nitrocellulose. This was then probed with radiolabeled DNA generated from GAPDH or c-myc coding regions. These Northern analyses show that the level of c-myc RNA is not reduced.
To ensure that these proteins were not modulating the levels of c-myc mRNA, Northern analysis was performed, and in all cases, c-myc mRNA levels remained constant, strongly suggesting that YB-1, p54nrb, and GRSF-1 directly affect the synthesis of the c-Myc protein (Fig. 4B).
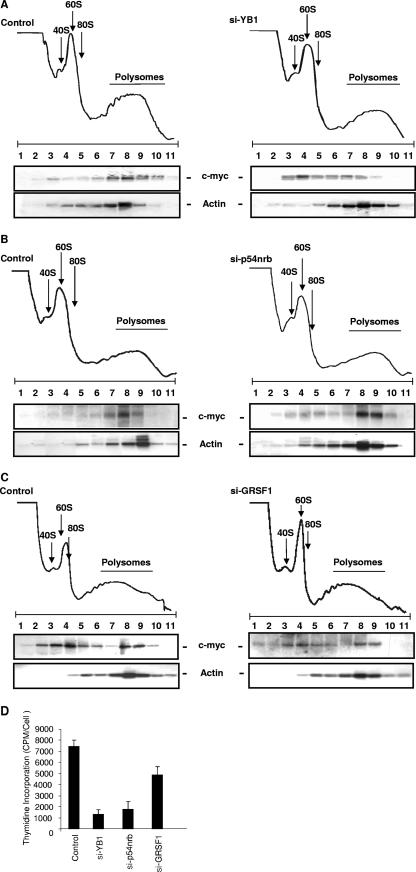
A decrease in YB-1 expression results in a relocalization of c-myc mRNA to the subpolysomes.
Polysome gradient analysis was then performed to investigate further the role of YB-1, p54nrb, and GRSF-1 in c-myc translation (Fig. 5). Thus, the levels of these proteins were reduced, cells lysed, and postnuclear extracts applied to 10 to 55% polysome gradients. These gradients were fractionated, the RNA was isolated, and Northern analysis was performed to identify the position in the gradients of c-myc or actin messages. The data show that in control cells, c-myc mRNA is, as expected, associated with the polysomes. In cells with a reduced level of YB-1, c-myc mRNA relocates to the inactive subpolysomal particles, strongly suggesting that YB-1 has a role in controlling the recruitment of c-myc mRNA to the polysomes (Fig. 5). In cells which have a reduced level of p54nrb, there was a relocalization of c-myc mRNA to the polysomes, albeit to a lesser extent. However, in cells with a reduction in GRSF-1 expression, there was no detectable difference in the polysomal location of c-myc mRNA (Fig. 5). These data are consistent with a smaller decrease in the endogenous c-Myc protein levels in cells with a reduction in the levels of GRSF-1 compared to cells with a reduction in the levels of YB-1 and p54nrb (Fig. 4). As c-Myc regulates target genes integral to the processes of cell proliferation (2, 9), the effect that a reduction in the levels of these proteins had on the cell proliferation was tested (Fig. 5D). Thus, the levels of YB-1, p54nrb, and GRSF-1 were decreased, as described above, and the degree of cell proliferation was measured (by determining the extent of thymidine incorporation). It can be seen that in each case, there is a general reduction in the ability of the cells to proliferate, albeit to different extents. The greatest effect was observed following a reduction in the level of YB-1, with a smaller decrease observed with GRSF-1 (Fig. 5D).
FIG. 5.
A reduction in YB-1 and p54nrb levels alters the association of c-myc mRNA with the polysomes. Cells were transfected with plasmids that reduce the expression of YB-1, p54nrb, or GRSF-1, and postnuclear extracts were separated on 10 to 55% sucrose density gradients. No overall changes in the polysome profiles were observed. RNA was generated from the individual fractions, and Northern analysis was performed using probes to c-myc and actin mRNA. When levels of YB-1 are reduced, there is a relocation of the c-myc mRNA from the polysomal region to the subpolysome particles (A). In cells with reduced expression of p54nrb, there is again a reduced polysomal location of c-myc mRNA (B), but no difference was observed in cells with a decreased expression of GRSF-1 (C). (D) To test the effect of the reduction in the levels of ITAFs on cell proliferation rates, the levels of these proteins were reduced as described above and cells were incubated with [3H]thymidine for 24 h and counted.
DISCUSSION
To obtain a better understanding of the function and mechanisms of action of cellular IRESs, it is essential that the trans-acting factors that mediate internal ribosome entry are elucidated. This is a complex task, since the data suggest that each cellular IRES has a distinct set of trans-acting-factor requirements (43). However, it is likely that there are also subsets of proteins that coordinately regulate the expression of defined groups of IRESs that have related functions. In the case of the myc family of IRESs, the data would suggest that specific ITAFs are required, since all three IRESs show marked differences in activities across a range of cell lines (18, 19, 46). Thus, the N-myc IRES is most active in neuronal cells (19), while the L-myc IRES is highly active in a range of cell types but especially in HeLa cells (18). Given the overlapping roles of the Myc family of proteins, it was very likely that there would be a subset of ITAFs that was required by the myc family IRESs. Our data strongly suggest that this is indeed the case, and we have identified four proteins that interact with all three IRESs (Fig. 1). These proteins are all multifunctional RNA binding proteins; however, our data show that they interact specifically with the three IRESs, albeit with different affinities (Table 1). All three IRESs are activated by p54nrb, GRSF-1, and YB-1, with only the L-myc IRES showing significant stimulation of activity in the presence of PSF (Fig. 2 and 3). Importantly, we have shown that the endogenous levels of c-Myc protein are regulated by these ITAFs (Fig. 4), and in particular, there was a large decrease in c-Myc expression observed in cells with low levels of YB-1 and p54nrb (Fig. 4). No large activation of the Apaf-1 or BAG-1 IRESs was observed in the presence of YB-1 or GRSF-1 in vitro (Fig. 3 and 4), and only the Apaf-1 IRES and the expression of the endogenous protein were affected by a reduction in p54nrb levels (Fig. 3 and 4).
Clearly, the translational regulation of the myc family of oncogenes by these proteins has important implications for tumorigenesis, and YB-1 has been studied most widely in this regard. This protein has previously been shown to regulate translation in a dose-dependent manner, with low concentrations stimulating translation and high concentrations having an inhibitory effect on global translation rates (14). It is thought that the inhibition of translation occurs at the initiation stage of translation by preventing the association of eukaryotic initiation factor 4G (eIF4G) with the mRNAs (29) and displacing eIF4E from the cap structure (31). Recent data have shown that YB-1 binds and selectively activates the translation of a subset of silent mRNAs and that this is dependent on this protein's phosphorylation state, which is regulated by AKT (13). Our identification of YB-1 as an ITAF leads us to examine these data further, and interestingly, many of the mRNAs present in this screen contain known IRESs, e.g., cyclin T1 (6), cyclin D1 (42), vascular endothelial growth factor (17), and fibroblast growth factor (48), suggesting that YB-1 could be an ITAF that is associated with mRNAs that have a role in cell proliferation (13). In agreement with this observation, the overexpression of YB-1 in tumors is associated with a resistance to chemotherapy (15, 24) and poor prognosis (51). This suggests a model where under conditions of cell stress (including the exposure of cells to chemotherapeutic agents), there is a decrease in global translation rates (11). YB-1 could replace eIF4E and allow the translation of a subset of mRNAs by internal ribosome entry.
Supplementary Material
Acknowledgments
This work was supported by grants from the BBSRC (L.C.C., K.A.S., H.C.D., S.J.H., and M.B.).
We thank James G. Patton for all his advice and help with the expression and purification of PSF.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aalto, Y., W. El-Rifai, L. Vilpo, J. Ollila, B. Nagy, M. Vihinen, J. Vilpo, and S. Knuutila. 2001. Distinct gene expression profiling in chronic lymphocytic leukemia with 11q23 deletion. Leukemia 151721-1728. [DOI] [PubMed] [Google Scholar]
- 2.Atchley, W. R., and W. M. Fitch. 1995. Myc and Max: molecular evolution of a family of proto-oncogene products and their dimerization partner. Proc. Natl. Acad. Sci. USA 9210217-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bladen, C. L. 2005. Identification of the polypyrimidine tract binding protein-associated splicing factor·p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2805205-5210. [DOI] [PubMed] [Google Scholar]
- 4.Bonnal, S., C. Schaeffer, L. Creancier, S. Clamens, H. Moine, A. C. Prats, and S. Vagner. 2003. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 27839330-39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S. A., J. Ripperger, S. Kadener, F. Fleury-Olela, F. Vilbois, M. Rosbash, and U. Schibler. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308693-696. [DOI] [PubMed] [Google Scholar]
- 6.Bushell, M., M. Stoneley, Y.-W. Kong, T. Hamilton, K. A. Spriggs, H. C. Dobbyn, and A. E. Willis. 2006. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell 23401-412. [DOI] [PubMed] [Google Scholar]
- 7.Coldwell, M. J., M. L. deSchoolmeester, C. A. Fraser, B. M. Pickering, G. Packham, and A. E. Willis. 2001. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene 204095-4100. [DOI] [PubMed] [Google Scholar]
- 8.Coldwell, M. J., S. A. Mitchell, M. Stoneley, M. MacFarlane, and A. E. Willis. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19899-905. [DOI] [PubMed] [Google Scholar]
- 9.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 182916-2942. [DOI] [PubMed] [Google Scholar]
- 10.Créancier, L., P. Mercier, A.-C. Prats, and D. Morello. 2001. c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong ranslational repression in adult transgenic mice. Mol. Cell. Biol. 211833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobbyn, H. C., K. Hill, T. L. Hamilton, K. A. Spriggs, B. M. Pickering, M. J. Coldwell, C. H. de Moor, M. Bushell, and A. E. Willis. 13 August 2007. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. [Epub ahead of print.] doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed]
- 12.Evans, J. R., S. A. Mitchell, K. A. Spriggs, J. Ostrowski, K. Bomsztyk, D. Ostarek, and A. E. Willis. 2003. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene 228012-8020. [DOI] [PubMed] [Google Scholar]
- 13.Evdokimova, V., P. Ruzanov, M. S. Anglesio, A. V. Sorokin, L. P. Ovchinnikov, J. Buckley, T. J. Triche, N. Sonenberg, and P. H. B. Sorensen. 2006. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell. Biol. 26277-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evdokimova, V. M., E. A. Kovrigina, D. V. Nashchekin, E. K. Davydova, J. W. B. Hershey, and L. P. Ovchinnikov. 1998. The major core protein of messenger ribonucleoprotein particles (p50) promotes initiation of protein biosynthesis in vitro. J. Biol. Chem. 2733574-3581. [DOI] [PubMed] [Google Scholar]
- 15.Gluz, O., N. Harbeck, R. E. Kates, M. Schmitt, K. Mengele, H. D. Royer, N. Eckstein, M. Svjetlana, E. Ting, C. Poremba, U. Nitz, and R. Diallo-Danebrock. 2006. YB-1 protein correlates with high-risk tumor characteristics and response to high dose chemotherapy in breast cancer. Breast Cancer Res. Treat. 100S136. [Google Scholar]
- 16.Holčík, M., B. W. Gordon, and R. G. Korneluk. 2003. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 23280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huez, I., L. Créancier, S. Audigier, M.-C. Gensac, A.-C. Prats, and H. Prats. 1998. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 186178-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jopling, C. L., K. A. Spriggs, S. A. Mitchell, M. Stoneley, and A. E. Willis. 2004. L-Myc protein synthesis is initiated by internal ribosome entry. RNA 10287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jopling, C. L., and A. E. Willis. 2001. N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene 202664-2670. [DOI] [PubMed] [Google Scholar]
- 20.Kash, J. C., D. M. Cunningham, M. W. Smit, Y. Park, D. Fritz, J. Wilusz, and M. G. Katze. 2002. Selective translation of eukaryotic mRNAs: Functional molecular analysis of GRSF-1, a positive regulator of influenza virus protein synthesis. J. Virol. 7610417-10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. H., Y. Paek, K. Choi, T.-D. Kim, B. Hahm, K.-T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohno, K., H. Izumi, T. Uchiumi, M. Ashizuka, and M. Kuwano. 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25691-698. [DOI] [PubMed] [Google Scholar]
- 23.Landay, M., S. K. Oster, F. Khosravi, L. E. Grove, X. Yin, J. Sedivy, L. Z. Penn, and E. V. Prochownik. 2000. Promotion of growth and apoptosis in c-myc nullizygous fibroblasts by other members of the myc oncoprotein family. Cell Death Differ. 7697-705. [DOI] [PubMed] [Google Scholar]
- 24.Mantwill, K., N. Kohler-Vargas, A. Bernshausen, A. Bieler, H. Lage, A. Kaszubiak, P. Surowiak, T. Dravits, U. Treiber, R. Hartung, B. Gansbacher, and P. S. Holm. 2006. Inhibition of the multidrug-resistant phenotype by targeting YB-1 with a conditionally oncolytic adenovirus: implications for combinatorial treatment regimen with chemotherapeutic agents. Cancer Res. 667195-7202. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell, S. A., E. C. Brown, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2001. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 213364-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell, S. A., K. A. Spriggs, M. Bushell, J. R. Evans, M. Stoneley, J. P. C. le Quesne, R. V. Spriggs, and A. E. Willis. 2005. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 191556-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, S. A., K. A. Spriggs, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11757-771. [DOI] [PubMed] [Google Scholar]
- 28.Nanbru, C., I. Lafon, S. Audiger, M. C. Gensac, S.Vagner, G. Huez, and A.-C. Prats. 1997. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 27232061-32066. [DOI] [PubMed] [Google Scholar]
- 29.Nekrasov, M. P., M. P. Ivshina, K. G. Chernov, E. A. Kovrigina, V. M. Evdokimova, A. A. M. Thomas, J. W. B. Hershey, and L. P. Ovchinnikov. 2003. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 27813936-13943. [DOI] [PubMed] [Google Scholar]
- 30.Nikiforov, M. A., I. Kotenko, O. Petrenko, A. Beavis, L. Valenick, I. Lemischka, and M. D. Cole. 2000. Complementation of Myc-dependent cell proliferation by cDNA library screening. Oncogene 184828-4831. [DOI] [PubMed] [Google Scholar]
- 31.Ovchinnikov, L. P., M. A. Skabkin, P. V. Ruzanov, and V. M. Evdokimova. 2001. Major mRNP proteins in the structural organization and function of mRNA in eukaryotic cells. Mol. Biol. 35462-471. [PubMed] [Google Scholar]
- 32.Park, Y. W., J. Wilusz, and M. G. Katze. 1999. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. USA 966694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadalginard. 1993. Cloning and characterization of PSF, a novel premessenger RNA splicing factor. Genes Dev. 7393-406. [DOI] [PubMed] [Google Scholar]
- 34.Paulin, F. E. M., S. A. Chappell, and A. E. Willis. 1998. A single nucleotide change in the c-myc internal ribosome entry segment leads to enhanced binding of a group of protein factors. Nucleic Acids Res. 263097-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulin, F. E. M., M. J. West, N. F. Sullivan, R. L. Whitney, L. Lyne, and A. E. Willis. 1996. Aberrant translational control of the c-myc gene in multiple myeloma. Oncogene 13505-513. [PubMed] [Google Scholar]
- 36.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2764-776. [DOI] [PubMed] [Google Scholar]
- 37.Pickering, B. M., S. A. Mitchell, K. A. Spriggs, M. Stoneley, and A. E. Willis. 2004. BAG-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 245595-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prochownik, E. V., and J. Kukowska. 1986. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. Nature 322848-850. [DOI] [PubMed] [Google Scholar]
- 39.Prochownik, E. V., and M. E. VanAntwerp. 1993. Differential patterns of DNA binding by Myc and Max proteins. Proc. Natl. Acad. Sci. USA 90960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shav-Tal, Y., and D. Zipori. 2002. PSF and p54nrb/NonO—multi-functional nuclear proteins. FEBS Lett. 531109-114. [DOI] [PubMed] [Google Scholar]
- 41.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 42.Shi, Y. J., A. Sharma, H. Wu, A. Lichtenstein, and J. Gera. 2005. Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J. Biol. Chem. 28010964-10973. [DOI] [PubMed] [Google Scholar]
- 43.Spriggs, K. A., M. Bushell, S. A. Mitchell, and A. E. Willis. 2005. Internal ribosome entry segments-mediated translation during apoptosis; the role of IRES-trans-acting factors. Cell Death Diff. 12585-591. [DOI] [PubMed] [Google Scholar]
- 44.Stoneley, M., S. A. Chappell, C. L. Jopling, M. Dickens, M. MacFarlane, and A. E. Willis. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 201162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoneley, M., F. E. M. Paulin, J. P. C. Le Quesne, S. A. Chappell, and A. E. Willis. 1998. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16423-428. [DOI] [PubMed] [Google Scholar]
- 46.Stoneley, M., T. Subkhankulova, J. P. C. Le Quesne, M. J. Coldwell, C. L. Jopling, G. J. Belsham, and A. E. Willis. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subkhankulova, T., S. A. Mitchell, and A. E. Willis. 2001. Internal ribosome entry segment-mediated initiation of c-Myc protein synthesis following genotoxic stress. Biochem. J. 359183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vagner, S., M.-C. Gensac, A. Maret, F. Bayard, F. Amalric, H. Prats, and A.-C. Prats. 1995. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1535-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West, M. J., M. Stoneley, and A. E. Willis. 1998. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17769-780. [DOI] [PubMed] [Google Scholar]
- 50.West, M. J., N. F. Sullivan, and A. E. Willis. 1995. Translational upregulation of the c-myc oncogene in Bloom's syndrome cell lines. Oncogene 132515-2524. [PubMed] [Google Scholar]
- 51.Wu, J., C. Lee, D. Yokom, H. Jiang, M. C. U. Cheang, E. Yorida, D. Turbin, I. M. Berquin, P. R. Mertens, T. Iftner, C. B. Gilks, and S. E. Dunn. 2006. Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2. Cancer Res. 664872-4879. [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman, K. A., G. D. Yancopoulos, R. G. Collum, R. K. Smith, N. E. Kohl, K. A. Denis, M. M. Nau, O. N. Witte, D. Toran-Allerand, C. E. Gee, J. D. Minna, and F. W. Alt. 1986. Differential expression of myc family genes during murine development. Nature 319780-783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.