Abstract
Intramembrane proteolysis by presenilin-dependent γ-secretase produces the Notch intracellular cytoplasmic domain (NCID) and Alzheimer disease-associated amyloid-β. Here, we show that upon Notch signaling the intracellular domain of Notch-1 is cleaved into two distinct types of NICD species due to diversity in the site of S3 cleavage. Consistent with the N-end rule, the S3-V cleavage produces stable NICD with Val at the N terminus, whereas the S3-S/S3-L cleavage generates unstable NICD with Ser/Leu at the N terminus. Moreover, intracellular Notch signal transmission with unstable NICDs is much weaker than that with stable NICD. Importantly, the extent of endocytosis in target cells affects the relative production ratio of the two types of NICD, which changes in parallel with Notch signaling. Surprisingly, substantial amounts of unstable NICD species are generated from the Val→Gly and the Lys→Arg mutants, which have been reported to decrease S3 cleavage efficiency in cultured cells. Thus, we suggest that the existence of two distinct types of NICD points to a novel aspect of the intracellular signaling and that changes in the precision of S3 cleavage play an important role in the process of conversion from extracellular to intracellular Notch signaling.
Presenilin (PS)-dependent γ-secretase (PS/γ-secretase) mediates the degradation of transmembrane domains (TMs) in many type 1 receptors, including Notch and β-amyloid protein precursor (βAPP) (18, 52). Degradation of these receptors is characterized by sequential endoproteolysis: following shedding by cleavage in the extracellular juxtamembrane region, the receptors undergo PS-dependent intramembrane proteolysis, releasing amyloid-β (Aβ)-like peptides and intracellular cytoplasmic domains (ICDs) (5, 14). At least in the cases of βAPP, Notch, and CD44, cleavages of the C termini of Aβ-like peptides and of the N termini of ICDs in the TM are distinct. This process of cleavage at two sites is known as “dual cleavage” (33). The process of Aβ generation has been intensively studied, and it is suggested that Aβ is released by a series of sequential cleavages followed by the ICD generation (25, 36, 53). An unusual characteristic of this intramembrane proteolysis is that some of the cleavage sites can vary (42). The precision of cleavage can therefore be defined as the ratio of the cleavage at each site. For example, PS-dependent cleavage of βAPP at the γ site, which is associated with Alzheimer disease (AD), occurs mainly at residue 40 (γ40), producing Aβ40, and at residue 42 (γ42), producing Aβ42. A small increase in the proportion of γ42 to γ40 cleavage is consistently observed in many familial AD (FAD)-associated PS or βAPP mutants (42), but it is unclear whether such changes in the precision of PS-dependent intramembrane proteolysis have any biological effects.
Notch signaling, which is essential for development, is a type of local-cell signaling that participates in neurodegeneration and tumorigenesis (1). The canonical Notch pathway is mediated by the regulated intramembrane proteolysis pathway, in which Notch receptors undergo ligand-dependent sequential endoproteolysis via a series of enzymes, including PS/γ-secretase (8). The Notch-1 ICD (NICD), which is produced by PS/γ-secretase-mediated cleavage at site 3 (S3), translocates to the nucleus and participates in transactivation of target genes (40). Elimination of PS function results in the Notch phenotype, which includes disruption of segmentation during the development of many kinds of animals, demonstrating the importance of NICD generation (41).
The intensity of Notch signaling is crucial for cell fate decisions. For example, Notch haplo-insufficiency causes the “notched-wing” phenotype in Drosophila (9). Reduced Notch activity favors the γδ T-cell fate over the αβ T-cell fate, whereas a constitutively activated form of Notch produces a reciprocal phenotype (48). The endocytosis of Notch and its ligands plays a key role in the regulation of the signaling intensity (40), but the biochemical aspects regulating this process have not been well studied. N-terminal amino acid sequencing revealed that S3 in mouse Notch-1 lies between Gly1743 and Val1744 (murine Notch-1 numbering) (39). Whether the site of S3 cleavage can vary has not been examined previously.
In this study, we found that there is diversity in the site of S3 cleavage, resulting in the production of two types of NICD with apparently distinct stability and ability to transmit Notch signaling in cultured cells. Our results suggest that the precision of PS/γ-secretase-mediated cleavage is important for determining the intensity of Notch signaling.
MATERIALS AND METHODS
Antibodies.
To generate affinity-purified polyclonal N-terminal capping antibodies to NICD-S (anti-NT-S), rabbits were immunized with a synthetic peptide (SRKRR) corresponding to the N terminus of NICD-S(+3). We also prepared two kinds of affinity columns in which the N-terminal peptide of NICD-V (VLLSRKRR) or NICD-S (SRKRR) was conjugated to Sepharose 4B (Amersham). We isolated the fraction of the anti-NT-S antiserum that bound to the NICD-S(+3) column but not the NICD-V column (32). Anti-NT-L antiserum was raised against a synthetic peptide (LLSRKRR) corresponding to the N terminus of NICD-L(+1) and then purified by affinity chromatography on Sepharose 4B conjugated to the N-terminal peptide of NICD-L (LLSRKRR), followed by a second step of affinity chromatography on Sepharose 4B conjugated to the NICD-S peptide (SRKRR). Other antibodies were purchased from commercial sources as follows: anti-NT-V (V1744 antibody) from Cell Signaling; antinicastrin and antibody mN1A against Notch-1 from Sigma-Aldrich; antibody H114 against Jagged-1 and antitubulin from Santa Cruz Biotechnology; anti-early endosome antigen 1 and anti-GM130 from BD Transduction Laboratories; antibody 12CA5 against the HA epitope from Roche Diagnostics, Inc.; antibody 9E10 against the myc epitope from Zymed; and anti-Na-K ATPase from Upstate Biotechnology.
cDNA constructs.
The cDNA encoding the mouse Notch-1 variant NEXT was previously described (33). NEXTΔC was generated by PCR-based mutagenesis using the QuikChange-II kit (Stratagene) with NEXT cDNA as a template. The mutant versions of NEXT and NEXTΔC were generated using the same kit. To generate expression constructs for polypeptides NICD-V, NICD-L (+1), and NICD-S (+3), cDNAs encoding Val, Leu(+1), and Ser(+3) as the N termini were subcloned into the pASK-IBA6 vector (IBA). HES-1-luc (a kind gift from Alain Israel) (16) and pGa981-6 (a kind gift from Georg W. Bornkamm) (21) were used as described. For more sensitive detection of HES-1 promoter transactivation, we newly generated a hairy and enhancer of split-Y (HES-Y) construct containing four sequential RBP-Jκ binding sites in the HES-1 promoter region.
Cell culture.
We generated HEK293 cells expressing PS1 R278I (a kind gift from M. Nishimura) (27) or PS1 G384A (a kind gift from H. Steiner) (44). HEK293 cells expressing either wild-type (wt) or mutants of PS1 were previously described (31). These cells were transfected with wt, mutant NEXT, or mutant NEXTΔC. HeLa cells expressing Dyn-1 K44A (a kind gift from S. Schmid) were used as described and stably transfected with NEXT or NEXTΔC. CHO(r) cells (a gift from S. Shirahata) (29) were stably transfected with mouse Notch-1 or Jagged-1.
Cell-free Notch-1 cleavage assay.
To obtain crude membrane fractions (CMFs), cells were homogenized in buffer (0.25 M sucrose and 10 mM HEPES, pH 7.4) containing a protease inhibitor cocktail (Roche), followed by centrifugation at 1,000 × g for 5 min. The postnuclear supernatant was further centrifuged at 100,000 × g for 30 min, and the resulting pellet was collected. This CMF was resuspended in 150 mM sodium citrate buffer (pH 6.4) containing a 4× concentration of protease inhibitor cocktail (Sigma-Aldrich) and 5 mM 1,10-phenanthroline (Sigma-Aldrich), incubated for 20 min at 37°C, and then centrifuged at 100,000 × g for 30 min (11).
Pulse-chase experiments.
Pulse-chase experiments were performed as described previously (11, 30, 31, 33, 34).
Immunoprecipitation/MALDI-TOF MS.
Immunoprecipitation and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis was carried out as described previously (11, 31, 33). The heights of the MS peaks and molecular weights were calibrated using angiotensin and bovine insulin β-chain as standards (Sigma-Aldrich).
Immunoprecipitation-immunoblotting and immunoprecipitation-autoradiography.
Metabolically labeled or unlabeled lysates were lysed in radioimmunoprecipitation assay buffer (1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) containing a protease inhibitor mix (Sigma-Aldrich). The cell lysates were centrifuged at 10,000 × g for 15 min, and the supernatant fractions were immunoprecipitated as indicated. Following 8% Tris-glycine (Tefco) or 10 to 20% Tris-Tricine (Invitrogen) SDS-polyacrylamide gel electrophoresis (SDS-PAGE), the gels were either transferred to a polyvinylidene difluoride membrane and probed with the indicated antibodies or dried and analyzed by autoradiography. To quantitatively measure the levels of NICD species in cultured cells, several doses of each NICD polypeptide were separated together with samples by SDS-PAGE and analyzed by immunoblotting with the corresponding N-terminal capping antibody. The chemiluminescence intensities were measured using an LAS3000 scanner, followed by analysis with Multi Gauge Ver3.0 software (Fuji Film). Biotinylated transferrin was semiquantitatively measured by chemiluminescence using the scanner, followed by analysis with the software.
Cell-cell association assay.
For the detection of de novo NICD species, CHO(r) cells stably expressing Notch-1 were grown to confluence in 150-mm dishes (2 × 107 cells per dish) in triplicate. Next, 3 × 107 CHO(r) cells stably expressing Jagged-1 were spread over the Notch-1-expressing cells. After 8 h of coculture, the cells were collected. For the reporter assay, the procedure was the same, although it was carried out in a 12-well plate and the number of cells was reduced accordingly.
cDNA transfection and reporter assay.
To examine the intensity of Notch signaling, we used a dual luciferase reporter assay system (Promega) as described by the manufacturer (31). Briefly, cells expressing Notch-1 or its derivatives in a 12-well plate were transiently transfected with 125 ng of HES-Y or pGa981-6 and 1.25 ng of the control Renilla luciferase reporter plasmid pRL-TK. The reporter assay was performed on the next day. Dyn-1 K44A expression was induced with various concentrations of tetracycline 24 h prior to transfection with HES-Y. Cell-cell association was performed 24 h after transfection with HES-Y.
Preparation of nuclear extract from mouse tissues.
A nuclear complex co-IP kit (Active Motif) was used to obtain nuclear extracts from C57BL/6 (Japan SLC) mouse tissues. Homogenized adult mouse brain or fetal mouse tissues without internal organs (embryonic day 12) were treated according to the manufacturer's instructions. Subsequently, the nuclear extracts were diluted using the immunoprecipitation buffer included in the kit, precleared three times with protein G- or protein A-Sepharose, and immunoprecipitated according to the manufacturer's instructions.
Purification of polypeptides.
NICD-V, NICD-L(+1), and NICD-S(+3) polypeptides fused with strept-tag-II followed by an N-terminal factor Xa cleavage site were obtained by transforming Escherichia coli (BL21) with pASK-IBA6 (IBA) encoding each polypeptide. Briefly, after the expression was induced, the cells were collected by centrifugation at 4,500 × g for 12 min, resuspended in ice-cold TSE buffer (10 mM Tris-HCl [pH 7.4], 20% sucrose, and 2.5 mM Na-EDTA), and then incubated on ice for 10 min. The cells were again collected by centrifugation, resuspended in ice-cold water, incubated for 10 min, briefly sonicated, and sedimented by centrifugation at 14,000 × g for 15 min (osmotic shock fractionation). The supernatant was passed through a Strep-Tactin Sepharose column (IBA). Bound polypeptides were eluted with phosphate-buffered saline containing 2.5 mM desthiobiotin. Eluted polypeptides were treated with factor Xa (Sigma-Aldrich), and the solution was passed through the column again to remove uncleaved polypeptide (34). The purity of the polypeptides was confirmed by 6% Tris-glycine SDS-PAGE, followed by staining with Coomassie brilliant blue.
Loading of polypeptides.
The polypeptides obtained as described above were loaded into cells using Chariot protein transfection reagent (Active Motif) according to the manufacturer's instructions. Briefly, cells were grown and transfected with reporter genes in a 12-well plate. The cells were then loaded with 5 μg of each polypeptide or bovine serum albumin (BSA) along with 0.5 μg of β-galactosidase, using 15 μl of Chariot reagent per well. Finally, the cells were stained for β-galactosidase or used for the reporter assay.
In vitro degradation assay.
NICD polypeptides (0.2 μg) were mixed with 60 μl of fresh rabbit reticulocyte lysate (Promega) and incubated at 37°C. Clasto-lactacystin (10 μM), MG262 (100 nM), and 4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone (NLVS) (10 μM) were added to inhibit the action of the proteasome.
Transferrin uptake assay.
To estimate the rate of endocytosis, the levels of internalized and surface-bound biotinylated-transferrin were measured as described previously (11).
Subcellular fractionation.
Linear gradients of 2.5% to 25% iodixanol (Optiprep; Axis-Shield) were prepared, and fractionation was performed as previously described (11).
Statistical analysis.
Experiments were performed at least three times unless otherwise indicated. Representative results are shown for cell-free immunoprecipitation/MALDI-TOF MS, immunoblotting, immunoprecipitation-autoradiography, and immunocytochemistry. The statistical significance of differences was determined by Student's t test.
RESULTS
A cell-free Notch-1 cleavage assay indicates diversity in the site of S3 cleavage.
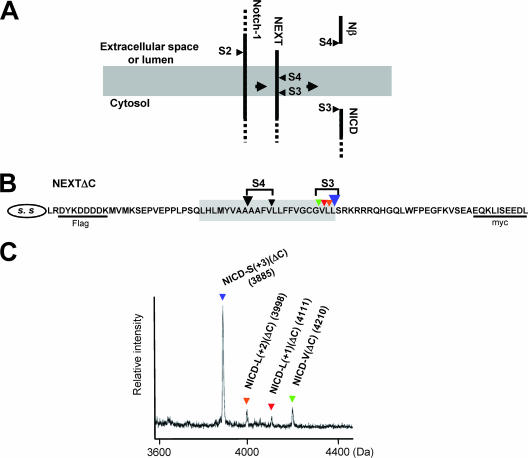
The cleavage of the Notch-1 TM occurs at least at two sites, one at S3, which determines the N terminus of intracellularly liberated NICD (39), and the other at S4, which determines the C terminus of extracellularly secreted Nβ (Fig. 1A) (31, 33). We first examined the diversity of the S3 cleavage site. We constructed NEXTΔC, a mouse Notch-1 derivative that lacks the majority of its extracellular and intracellular domains (Fig. 1B), and we established a cell-free Notch-1 cleavage assay using the CMF from cells stably expressing this construct (11). The de novo-generated NICD(ΔC) was immunoprecipitated with anti-myc antibodies and then analyzed by MALDI-TOF MS (Fig. 1C). Strikingly, proteolysis at S3 did not occur at a unique site but rather occurred at multiple sites, as indicated by the presence of multiple sizes of NICD(ΔC) (see Table S1 in the supplemental material). Specifically, proteolysis at S3 occurred at the following sites: S3-L(+1), between Val1744 and Leu1745; S3-L(+2), between Leu1745 and Leu1746; S3-S(+3), between Leu1746 and Ser1747; and the previously reported S3-V, between Gly1743 and Val1744 (39) (Fig. 1B; see Fig. S1A in the supplemental material). Unexpectedly, the highest peak was for NICD-S(+3)(ΔC) rather than NICD-V(ΔC) (Fig. 1C), suggesting that S3-S(+3) is the major site of S3 cleavage under these assay conditions. Addition of the PS/γ-secretase inhibitors eliminated the cleavage at both S3-V and S3-S(+3) (see Fig. S1B in the supplemental material). Moreover, we did not observe generation of these shorter NICD(ΔC) species from the longer NICD(ΔC) (see Fig. S1C in the supplemental material). Therefore, the results are consistent with the possibility that all the fragments are produced by PS-dependent S3 cleavage in the Notch-1 TM.
FIG. 1.
MALDI-TOF MS analysis of NICD(ΔC) produced by the cell-free Notch-1 cleavage assay. (A) Schematic representation of sequential endoproteolysis of Notch-1. S2 to S4 and the gray area represent the proteolytic sites and the putative TM, respectively. (B) Schematic representation of the NEXTΔC construct used in the cell-free Notch-1 cleavage assay. Colored inverted triangles show the S3 and S4 proteolytic sites. SS, signal sequence. (C) MS spectrum of de novo NICD(ΔC) generated in the cell-free assay. CMF was derived from K293 cells stably expressing NEXTΔC. The molecular mass of each species is indicated. To inhibit degradation by proteases other than aspartyl proteases, BSA and a mixture of metallo-, serine, and cysteine protease inhibitors were added to the cell-free assay buffer. Colored inverted triangles indicate the NICD(ΔC) species produced by cleavage at the sites shown in panel B.
Diversity in the site of S3 cleavage in living cells.
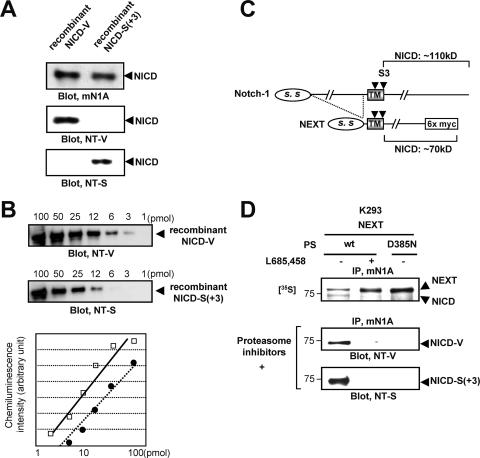
To identify the N terminus of NICD molecules in vivo, we prepared two N-terminal capping antibodies, anti-NT-V (anti-V1744) and anti-NT-S (32), and corresponding recombinant NICD species with distinct N termini (Fig. 2A; see Fig. S2A in the supplemental material). We found that (i) the anti-NT-V antibody specifically recognizes recombinant NICD-V but not NICD-S(+3) (34) (Fig. 2A, middle panel; see Fig. S2B in the supplemental material), whereas the anti-NT-S antibody has the opposite specificity (Fig. 2A; lower panel, see also Fig. S2B in the supplemental material), and (ii) these antibodies can be used to determine the relative amounts of NICD-V and NICD-S(+3) generated, that is, the extents of S3-V and S3-S(+3) cleavage, respectively (Fig. 2B).
FIG. 2.
Characterization of capping antibodies to the N terminus of NICD and detection of distinct NICD species in cultured cells. (A) Specificities of the anti-NT-V and the anti-NT-S antibodies. Immunoblotting with antibody mN1A confirmed that equal amounts of the polypeptides were loaded in each lane (upper panel). (B) Affinities of the anti-NT-V and anti-NT-S antibodies. The indicated amounts of NICD-V or NICD-S(+3) were separated by SDS-PAGE and analyzed by immunoblotting with the anti-NT-V or anti-NT-S antibody, respectively. The graph shows the chemiluminescence intensity versus the concentration of NICD-V (squares) or NICD-S(+3) (circles). Each antibody detected the respective polypeptide in a dose-dependent manner. (C) Schematic representation of Notch-1 and NEXT constructs. The C-terminal portion of the NEXT construct is replaced by myc6, while full-length Notch-1 has no modification. Note that the molecular mass of NICD generated from NEXT-myc6 is ∼70 kDa, while that of NICD generated from unmodified Notch-1 is ∼110 kDa. (D) Generation of NICD-V and NICD-S(+3) in cultured cells. Proteasome inhibitors lactacystin (10 μM), MG262 (100 nM), and NLVS (10 μM) were added to the medium 12 h prior to cell collection.
Using these capping antibodies, we examined the diversity in the site of S3 cleavage in living cells. The Notch extracellular truncation (NEXT) (Fig. 2C), which lacks the majority of the extracellular domain of Notch-1, undergoes constitutive ligand-independent intramembrane proteolysis by PS/γ-secretase (24). We prepared cells stably expressing NEXT and wt or a dominant-negative form of PS1 (PS1 D385N) (51). A 30-min pulse with [35S]methionine followed by a 2-h chase revealed production of an ∼70-kDa NICD band that completely disappeared upon elimination of PS/γ-secretase function (Fig. 2D, upper panel). Because degradation of NICD is mediated by the ubiquitin-proteasome pathway (8, 40, 41), we added a potent proteasome inhibitor mixture consisting of lactacystin, MG262, and NLVS (Fig. 2D, middle and lower panels). The resulting cell lysates were immunoprecipitated with antibody mN1A, separated by SDS-PAGE, and analyzed by immunoblotting with anti-NT-V or anti-NT-S. The anti-NT-V and anti-NT-S antibodies specifically detected the PS-dependent production of NICD-V and NICD-S(+3), respectively (Fig. 2D, middle and lower panels; see Fig. S2C in the supplemental material). We also confirmed that both NICD-V and NICD-S(+3) are produced in cells in the absence of the proteasome inhibitor mixture (see Fig. S2D in the supplemental material).
S3 cleavage during Notch signaling produces distinct molecular species of NICD.
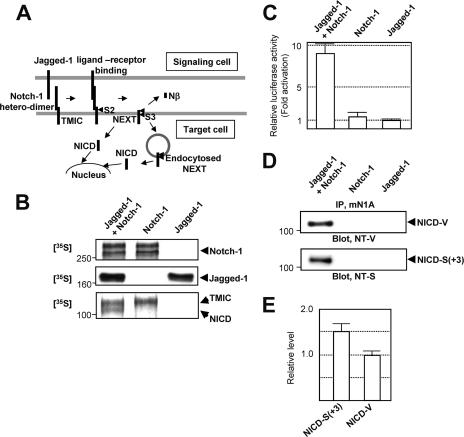
Because ligand-induced degradation of Notch receptors is initiated at the plasma membrane (PM), intramembrane proteolysis of Notch-1 is thought to occur at restricted subcellular locations, such as the PM and endocytosed vesicles (13, 17, 20) (Fig. 3A). We prepared cells stably expressing either Jagged-1 (a Notch ligand) or full-length Notch-1 and then cocultured them. We determined the extent of Notch signaling using a luciferase reporter assay in cells expressing a newly improved construct, HES-Y (see Materials and Methods for details). Pulse-chase experiments revealed that upon coculture, sequential endoproteolysis of Notch-1 occurs, producing the NICD band (Fig. 3B, bottom panel). Moreover, when the cells were cocultured, we observed concomitant activation of the HES-1 promoter (Fig. 3C). The results therefore demonstrate Notch-1 signaling in cell culture. Using this assay system, we investigated whether NICD-S(+3) and NICD-V are indeed generated during Notch signaling. Strikingly, upon coculture, both NICD-V and NICD-S(+3) were detected (Fig. 3D, upper and lower panels). The intensities of the bands indicated that 1.5-fold more NICD-S(+3) than NICD-V was produced (Fig. 3E). Therefore, the results indicated that there are multiple forms of NICD produced during Notch signaling.
FIG. 3.
Detection of different NICD species during Notch signaling. (A) Schematic representation of Notch signaling. (B) Notch signaling in cell culture. CHO(r) cells stably expressing Jagged-1 or Notch-1 were used. Expression of Notch-1 (top panel) and Jagged-1 (middle panel) was determined by a 1-h pulse experiment, followed by immunoprecipitation using antibodies mN1A and H114, respectively. The precipitated proteins were analyzed by SDS-PAGE, followed by autoradiography. A 1-h pulse/2-h chase experiment detected an ∼110-kDa NICD band only when the cells were cocultured (bottom panel). (C) Notch signaling was measured using a dual luciferase assay. The relative luciferase activity of Jagged-expressing cells was defined as 1.0. Values represent means ± standard deviations (n = 3). (D) In the presence of the proteasome inhibitors, both NICD-V (upper panel) and NICD-S(+3) (lower panel) were detected during Notch signaling. The bands were detected as described for Fig. 2D. (E) Relative levels of NICD-S(+3) and NICD-V generated during Notch signaling. The relative levels were calculated based on the standard curve shown in Fig. 2B.
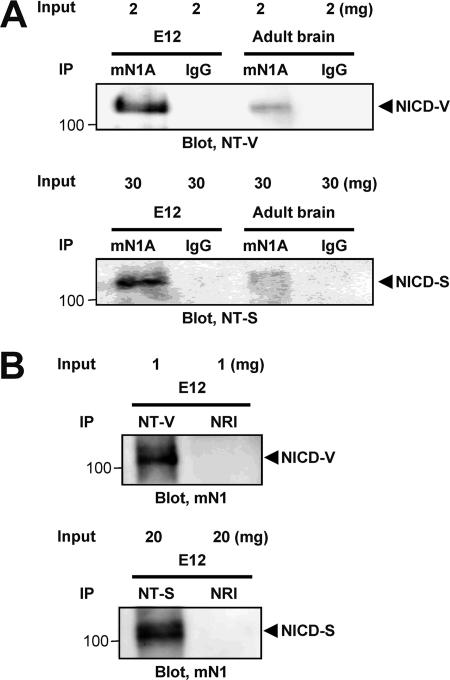
We next tried to detect the NICD-V and NICD-S(+3) in fetal (embryonic day 12, whole embryo without internal organs) and adult (brain) mouse tissues (Fig. 4). Nuclear extracts from these tissues were immunoprecipitated with mN1A, which specifically recognizes intracellular domain of Notch-1 but not Notch-2, -3, or -4. The precipitated proteins were then analyzed by SDS-PAGE, followed by immunoblotting with anti-NT-V or anti-NT-S antibodies. As shown in Fig. 4A, we clearly found both NICD-V (upper panel) and NICD-S(+3) species (lower panel) in the fetal mouse tissues, but these species were barely detectable in adult brains. This is consistent with the finding of a high level of Notch signaling in fetal mouse tissue (22). Moreover, we successfully detected the same NICD-V and NICD-S bands in the mouse embryo even when the combination of antibodies used for immunoprecipitation and immunoblotting was swapped (Fig. 4B). Therefore, the results indicated that multiple forms of NICD are produced in vivo.
FIG. 4.
Detection of NICD-V and NICD-S(+3) in vivo. The indicated amounts of nuclear extracts were loaded for immunoprecipitation. IgG and NRI, isotype-matched immunoglobulin and normal rabbit immunoglobulin, respectively.
Transactivation of the HES-1 promoter by NICD-S(+3) is much weaker than transactivation by NICD-V in living cells.
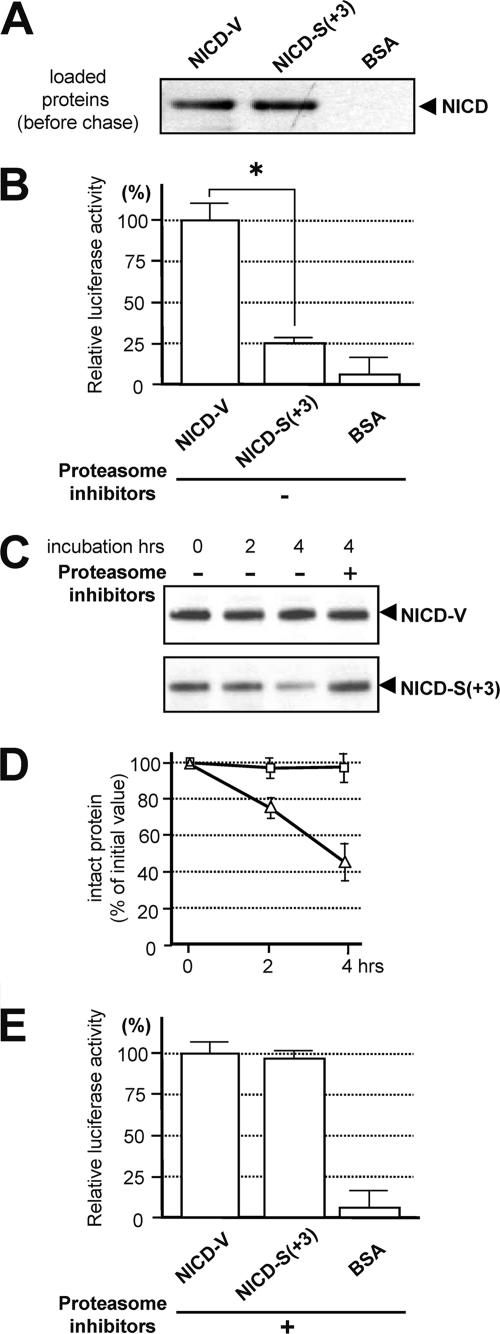
The stability of polypeptides degraded by the ubiquitin-proteasome pathway depends on the N-end rule, where an N-terminal valine is a stabilizing residue and an N-terminal serine is destabilizing (2, 12). Therefore, we examined whether the intensity of Notch signaling differs for NICD-V and NICD-S(+3) in living cells. We loaded HES-Y-transfected cells with equal amounts of purified NICD-V or NICD-S(+3) (see Fig. S3 in the supplemental material). After a 1-h loading period, the cells contained similar levels of each NICD species (Fig. 5A). Induction of Notch signaling by chasing the loaded cells for 4 h (signaling period) resulted in much less luciferase activity in the NICD-S-loaded cells than in NICD-V-loaded cells, demonstrating that NICD-S(+3) is much weaker than NICD-V at activating the promoter in living cells (Fig. 5B, left panel).
FIG. 5.
Characterization of the NICD species. (A) Loading of cells with NICD. HeLa cells (2.5 × 105) were loaded with 5 μg of purified NICD-V, NICD-S(+3), or BSA (control). The cells were collected 1 h after the addition of the Chariot macromolecule complex (defined as the loading period). (B) Assay of Notch downstream signaling induced by the NICD species. The NICD-loaded cells from panel A were chased for 4 h and collected, and Notch downstream signaling was assayed. The values were corrected for background luciferase activity (0.5 μg of β-galactosidase-loaded cells), and the luciferase activity in the NICD-V-loaded cells was defined as 100%. Values represent means ± standard deviations (n = 3). The asterisk indicates that the relative luciferase activity in NICD-V-loaded cells is statistically different than that in NICD-S(+3)-loaded cells (P < 0.001). Similar results were obtained using cells expressing pGa981-6 (data not shown). (C) Stability of recombinant NICD species in rabbit reticulocyte lysate. Note that the degradation of NICD-S(+3) was inhibited in the presence of proteasome inhibitors. (D) Levels of intact NICDs in the lysates during in vitro degradation. The amount of intact polypeptide was determined using a standard curve of the chemiluminescence intensities of the bands versus their concentrations (data not shown). Squares and triangles indicate the means for NICD-V and NICD-S(+3), respectively. Values represent means ± standard deviations (n = 3). (E) Assay of Notch downstream signaling induced by the NICD species in the presence of the proteasome inhibitor mixture. Experiments were performed as described for panel B in the presence of the proteasome inhibitor mixture. The values were corrected for background luciferase activity (0.5 μg of β-galactosidase-loaded cells), and the luciferase activity in the NICD-V-loaded cells was defined as 100%. Values represent means ± standard deviations (n = 3).
We next investigated whether the rates of degradation by the proteasome pathway differ for the various species of NICD in an in vitro assay. We incubated recombinant NICD-V or NICD-S(+3) (34) with rabbit reticulocyte lysate (Promega) (12) and examined the levels of the two types of NICD by immunoblotting (Fig. 5C). Our results indicated that NICD-S(+3) is much less stable than NICD-V (Fig. 5D). Moreover, following the proteasome inhibitor treatment, the relative luciferase activities in NICD-S- and NICD-V-loaded cells turned out to be almost the same (Fig. 5E). Collectively the results suggest that unstable NICD-S(+3) may have a much weaker ability than stable NICD-V to mediate intracellular signaling.
The precision of S3 cleavage is distinct in the subcellular locations where it occurs.
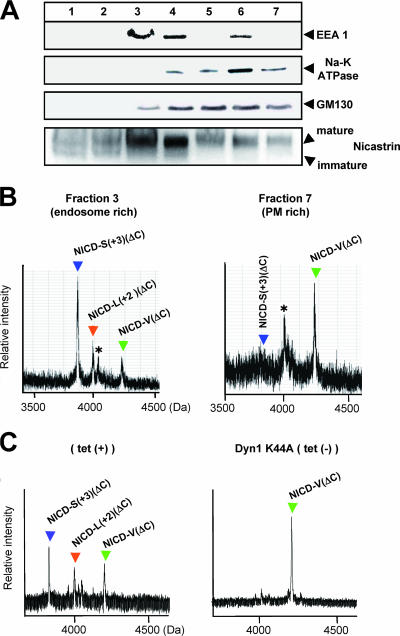
A previous study showed that the precision of ɛ cleavage of βAPP, which topologically corresponds to S3 cleavage of Notch-1, differs before and after endocytosis (11). To determine whether S3 cleavage precision differs on PM and endosomes, we performed the cell-free assay using organelles separated by iodixanol gradient fractionation from NEXTΔC-expressing HeLa cells (11). Fractions from a 2.5% to 25% linear iodixanol gradient were examined by immunoblotting with antibodies to early endosome antigen 1 (endosome marker), Na-K ATPase (PM marker), GM130 (Golgi marker), or nicastrin (a component of the PS complex) (Fig. 6A). NICD(ΔC) was generated in the cell-free assay using membranes collected by centrifugation from the endosome-rich (fraction 3) and the PM-rich (fraction 7) fractions and analyzed by immunoprecipitation/MALDI-TOF MS. Remarkably, the relative ratio of NICD-V(ΔC) to NICD-S(+3)(ΔC) was much higher in the PM-rich fraction than in the endosome-rich fraction (Fig. 6B). This indicates that cleavage at S3-V, which generates the longer NICD-V(ΔC), and at S3-S(+3), which generates the shorter NICD-S(+3)(ΔC), occurs predominantly on the PM and endosomes, respectively. This is very similar to the case of ɛ cleavage of βAPP (11). Subsequently, we investigated the effect of endocytosis on the precision of S3 cleavage. To down-regulate endocytosis, we used cells that express a dominant-negative mutant of dynamin-1 (Dyn-1 K44A) upon tetracycline withdrawal (Fig. 6C) or treatment with bafilomycin A1 (11) (see Fig. S4 in the supplemental material). When endocytosis was strongly inhibited, the precision of S3 cleavage changed drastically, so that the S3-V site instead of the S3-S(+3) site became the major site of cleavage in the cell-free assay (Fig. 6C; see Fig. S4A in the supplemental material). These results from the cell-free assay suggest that generation of stable NICD-V and unstable NICD-S occur predominantly on the PM and endosomes, respectively.
FIG. 6.
Subcellular locations where S3-V and S3-S(+3) cleavages occur. (A) Fractions from a 2.5% to 25% linear iodixanol gradient examined by immunoblotting with the indicated antibodies. (B) MS spectra of NICD(ΔC) generated in the cell-free assay using membranes collected by centrifugation from the endosome-rich (fraction 3) and the PM-rich (fraction 7) fractions. Asterisks indicate nonspecific peaks. (C) MS spectra of NICD(ΔC) generated in the cell-free assay. CMFs from HeLa cells stably expressing NEXT(ΔC) and conditionally expressing Dyn-1 K44A were used. The precision of PS-dependent cleavage at the TM-cytoplasmic border in HeLa (left panel) and K293 (Fig. 1C) cells was different, in agreement with a previous report (11).
The precision of S3 cleavage changes in parallel with the rate of endocytosis in target cells.
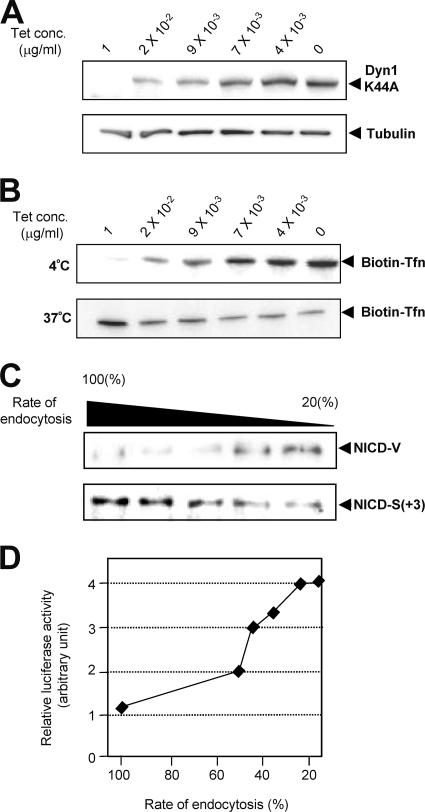
Next, we investigated whether these phenomena also occur in living cells. We examined the influence of the rate of endocytosis of NEXT by altering the expression of Dyn-1 K44A in living cells (Fig. 7A). Measurement of biotinylated transferrin uptake revealed various rates of endocytosis in Dyn-1 K44A-expressing cells (Fig. 7B). Strikingly, we found that the ratio of S3-V to S3-S(+3) is low in cells with a high rate of endocytosis and, conversely, that the ratio is high in cells with a low rate of endocytosis (Fig. 7C), which is consistent with the result from the cell-free assay (Fig. 6C). We observed a similar change in the S3 cleavage when endocytosis was blocked with bafilomycin A1 (see Fig. S4B in the supplemental material). Therefore, it appears that the precision of S3 cleavage changes in parallel with the rate of endocytosis, and relative generation of stable NICD-V increases as the rate of endocytosis decreases.
FIG. 7.
Parallel change in the rate of endocytosis and the precision of S3 cleavage. (A) Expression of Dyn-1 K44A at various concentrations of tetracycline. Dyn-1 K44A/NEXT-coexpressing HeLa cells were cultured in medium with the indicated concentrations of tetracycline, and cell lysates were examined by immunoblotting with antibody 12CA5 (upper panel) or antitubulin (lower panel). The levels of Dyn-1 K44A increase as the concentration of tetracycline is decreased. (B) Various rates of endocytosis in Dyn-1 K44A expressing cells. Transferrin (Tfn) uptake assays were performed to measure the rate of endocytosis. The ratio of internalized Tfn (37°C; lower panel) to surface-bound Tfn (4°C; upper panel) in cells cultured in medium containing 1 μg/ml of tetracycline was defined as 100%. The rate of endocytosis decreased to ∼15% when tetracycline was completely withdrawn. (C) Effect of the rate of endocytosis on the NICD species. The calculated rates of endocytosis were 100%, 41%, 37%, 21%, and 15% in lanes 1 to 5, respectively. (D) A plot of the relative Notch downstream luciferase activity versus the rate of endocytosis at various tetracycline concentrations.
Because Notch signaling is associated with endocytosis of Notch receptors and/or Notch ligands during development (3, 28, 35, 40, 43), we next examined the effect of the change in S3 precision on the intensity of Notch signaling. Surprisingly, we observed a higher intensity of Notch signaling in cells in which the rate of endocytosis of NEXT was lower (Fig. 7D; see Fig. S4C in the supplemental material). These results suggest that the rate of endocytosis in target cells could affect the intensity of intracellular Notch signaling by changing the precision of S3 cleavage and thus its stability.
Mutations around S3 can induce changes in the precision of the cleavage.
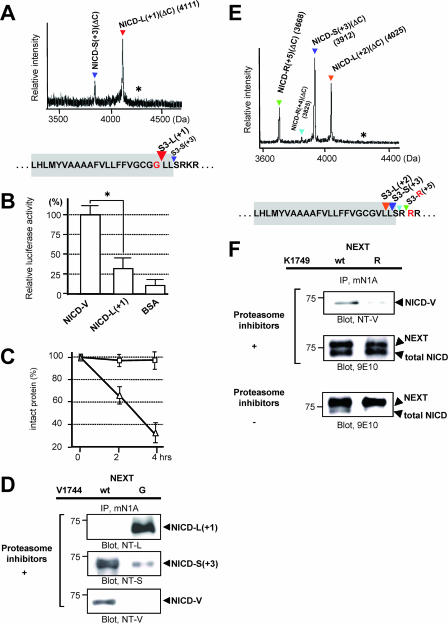
S3 mutations, such as the Val→Gly mutation (V1744G mutant) and the Lys→Arg mutation (K1749R mutant), cause a decrease in the NICD level (13, 15). To date, this reduction has been considered to be due to decreased NICD generation. However, since we revealed that multiple NICD species with different stabilities are generated, we investigated whether these mutants also change the S3 cleavage precision, which would accelerate degradation of NICD. First, we determined the precision of the S3 cleavage of the Val→Gly mutant version of NEXTΔC in the cell-free assay (Fig. 1C). Strikingly, this mutant was degraded mainly into NICD-L(+1)(ΔC), which has Leu1745 (murine Notch-1 numbering) at its N terminus (Fig. 8A). Therefore, it appears that the Val→Gly mutation causes not only a complete loss of stable NICD-V, but also a dramatic shift in the major product from NICD-S(+3) to NICD-L(+1). We next examined whether NICD-L(+1) behaved like NICD-S(+3) in cellular signal transduction. Like NICD-S(+3), NICD-L(+1) was much weaker than NICD-V at inducing promoter activation in living cells (Fig. 8B). Moreover, our in vitro degradation assay revealed that NICD-L(+1) was much less stable than NICD-V (Fig. 8C). Thus, our in vitro experiments suggested that NICD-L(+1) and NICD-S(+3) are both unstable and have similar effects on Notch signaling but that their effects are distinct from those of NICD-V. We further investigated whether the precision change in the mutant is also observed in living cells. To specifically detect NICD-L(+1), we generated anti-NT-L, an N-terminal capping antibody (see Fig. S5 in the supplemental material). As is clearly shown in Fig. 8D, although almost no NICD-V was detected, a substantial amount of NICD-L(+1) was detected in the Val→Gly version of NEXT-expressing cells. These results indicate that the relative production of unstable NICDs with respect to stable NICD increases in the mutant NEXT cells due to change of the S3 cleavage precision.
FIG. 8.
Characteristics of the NICD species generated from the Val→Gly and the Lys→Arg mutants in cultured cells. (A) MS spectrum of de novo NICD(ΔC) generated from the Val→Gly mutant of NEXTΔC in K293 cells. The asterisk indicates the position of molecular mass corresponding to NICD-G(ΔC) species. Colored letters and inverted triangles show the mutation and proteolytic sites, respectively. (B) Assay of Notch downstream signaling induced by NICD-L(+1). Experiments were performed as described for Fig. 5B. The luciferase activity in the NICD-V-loaded cells was defined as 100%. Values represent means ± standard deviations. (n = 3). The asterisk indicates that the relative luciferase activity in NICD-V-loaded cells is statistically different than that in NICD-L(+1)-loaded cells (P < 0.001). (C) Degradation of NICD-L(+1) species in vitro. Experiments were performed as described for Fig. 5C but using NICD-L(+1) (triangles) and NICD-V (squares). Values represent means ± standard deviations (n = 3). (D) Generation of NICD-L(+1) and NICD-S(+3) in the Val→Gly mutant NEXT cells. K293 cells expressing either wt or the Val→Gly mutant NEXT were analyzed as described for Fig. 2D. (E) MS spectrum of de novo NICD(ΔC) generated from the Lys→Arg mutant of NEXTΔC in K293 cells. The asterisk indicates the position of molecular mass corresponding to NICD-V(ΔC) species. Colored letters and inverted triangles show the mutation and proteolytic sites, respectively. (F) Generation of unstable NICD species in the Lys→Arg mutant NEXT cells. K293 cells expressing either wt or the Lys→Arg mutant NEXT were analyzed as described for Fig. 2D (top and middle panels).
The Lys→Arg mutant NEXT (the K1749R mutant) is neither monoubiquitinated nor endocytosed (13). This mutant, like the Val→Gly mutant, causes decreased NICD levels in cell culture (13). Analysis to determine the precision of the S3 cleavage of the Lys→Arg mutant version of NEXTΔC was performed in the cell-free assay. Surprisingly, the Lys→Arg mutant was found to be degraded mainly into NICD-L(+2)(ΔC), NICD-S(+3)(ΔC), and NICD-R(+5)(ΔC) species, which have unstable Leu1746, Ser1747, and the mutated Arg1749 at the N terminus, respectively (Fig. 8E). These results suggest that due to a dramatic change in the S3 cleavage precision, the Lys→Arg mutation causes not only a decrease of NICD-V but also an increase of extra unstable NICD species besides NICD-S(+3). This finding is reminiscent of the S3 cleavage for the Val→Gly mutant NEXT. Moreover, we studied whether NICDs in living cells expressing the Lys→Arg mutant are composed mainly of unstable NICD species. Strikingly, in the cells stably expressing the mutant NEXT, the stable NICD-V was barely detectable (13), while substantial amounts of NICDs were generated in the presence of the proteasome inhibitor mixture (Fig. 8F, top and middle panels). Since almost no NICD is observed in the mutant cells without the inhibitor mixture, it is indicated that NICDs in the Lys→Arg mutant-expressing cells are composed of unstable species (Fig. 8F, bottom panel). Therefore, the changes in the S3 cleavage precision induced by these S3 mutations are at least partially responsible for the observed decrease in NICD level/Notch signaling in cultured cells.
Unlike several FAD-associated PS1 mutations, modifiers of PS/γ-secretase do not induce changes in the precision of S3 cleavage.
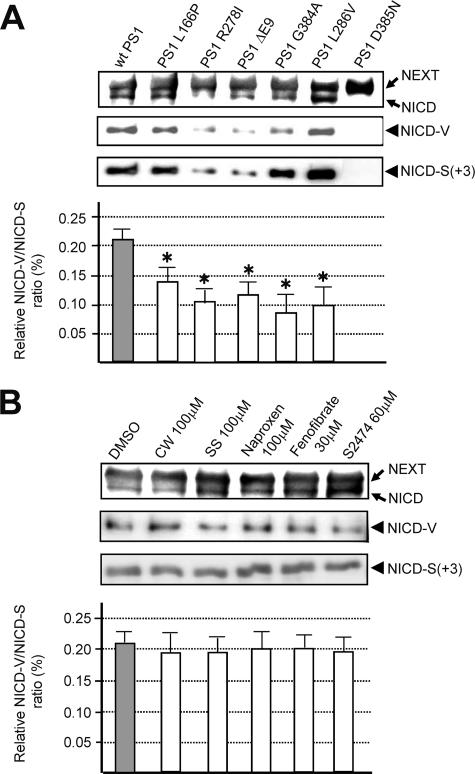
Many FAD PS mutations affect the precision of not only γ but also ɛ cleavages of βAPP and, therefore, generally increase the relative AICDɛ48/AICDɛ49 ratio as well as the Aβ42/Aβ40 ratio (38, 42). In addition, compounds that modify the activity of PS/γ-secretase, including a subset of nonsteroidal anti-inflammatory drugs, cause reciprocal changes in relative production of Aβ42 and Aβ38 (19, 31, 49). Therefore, we investigated whether FAD PS mutations or PS/γ-secretase modifiers affect the precision of S3 cleavage. To evaluate the precision change, we used the relative NICD-V/NICD-S ratio, mimicking the relative Aβ42/Aβ40 ratio in the case of γ cleavage. Immunoblotting using anti-NT-V or anti-NT-S revealed that cells coexpressing PS1 mutants and NEXT produce both of the NICD species (Fig. 9A, top three panels). We found that the relative ratio of S3-V to S3-S(+3) cleavage was significantly reduced in some mutants (Fig. 9A, bottom panel). Subsequently, we examined the effects of PS/γ-secretase modifiers and naproxen (Fig. 9B). Because the effective doses of the compounds for γ and S3 cleavage may differ, we performed dose-response experiments to select the highest working concentrations (data not shown). After confirming that the level of Aβ42 in the medium was altered following a 24-h incubation with each compound (data not shown), we analyzed the cell lysates for the presence of NICD-V and NICD-S(+3) (Fig. 9B). In contrast to the case for the FAD mutants, the relative ratio of S3-V to S3-S(+3) cleavage was unchanged by the modifiers. These results suggest that γ-secretase modifiers do not affect the precision of intramembrane proteolysis by PS/γ-secretase.
FIG. 9.
Changes in the precision of S3 cleavage induced by FAD mutations in PS1. (A) Effect of several FAD PS1 mutations on the precision of S3 cleavage. K293 cells expressing the indicated PS mutant were transiently transfected with NEXT and analyzed as for Fig. 2D. The production of total NICD (first panel), NICD-V (second panel), and NICD-S(+3) (third panel) was assessed. Note that amount of total NICD was greatly reduced in PS1 R278I- and Δexon 9-expressing cells. The ratio of NICD-V to NICD-S(+3) in cells expressing wt PS1 or FAD mutants is shown in the bottom panel. Values represent means ± standard deviations (n = 3). The asterisk indicates that the ratio of NICD-V to NICD-S(+3) in cells expressing PS1 FAD mutants is significantly different from that in wt PS1-expressing cells (P < 0.002). (B) Effect of PS/γ-secretase modifiers on the precision of S3 cleavage. K293 cells stably expressing NEXT were treated with several PS/γ-secretase modifiers at the indicated concentration for 24 h and analyzed as described for the first three panels in panel A. DMSO, dimethyl sulfoxide; CW, compound W (31); SS, sulindac sulfide (49). The ratio of NICD-V to NICD-S(+3) in the control and treated cells is shown in the bottom panel. Values represent means ± standard deviations (n = 3).
DISCUSSION
The current studies suggest a novel mode by which the intensity of Notch signaling is regulated. We found that intracellular Notch signaling molecules (NICDs) in target cells can be divided into a stable one that transmits a substantial signal and unstable ones that transmit a much weaker signal, depending on the specific site of S3 cleavage. Therefore, Notch signaling intensity transmitted by conversion of extracellular signaling into intracellular signaling could depend on the characteristics of the target cell. For example, although S3 cleavage occurs, cells might not receive a substantial Notch signal when predominantly unstable NICDs are generated. On the basis of our results, we propose that the precision of S3 cleavage by PS/γ-secretase in the target cell is an important factor in determining the signaling intensity.
PS-dependent proteolysis on the TM of Notch-1 and βAPP consists of dual cleavage at the S4/S3 and γ/ɛ sites, respectively (14, 31, 33). The finding of diversity in the site of S3 cleavage means that cleavage at all four sites in the TM of Notch-1 and βAPP can vary. Thus, we suggest that the existence of variability in both the site and precision of cleavage may be a common feature of PS-dependent intramembrane proteolysis. Furthermore, we found that the precision of cleavage at S3 changes according to the subcellular location. On the PM, cleavage is more likely at S3-V, whereas on endosomes, cleavage is more likely at S3-S(+3), which is more C terminal. Interestingly, we have obtained very similar results regarding the cleavage of βAPP at the ɛ site (11). The precision of cleavage at the ɛ site changes depending on the subcellular location (11). The ɛ49 cleavage, which topologically corresponds to S3-V, occurs mainly on the PM, whereas the more C-terminal ɛ51 cleavage, which topologically corresponds to S3-S, occurs mainly on endosomes. Therefore, we have demonstrated that such subcellular location-dependent changes in the precision of the cleavage by PS/γ-secretase are common to the substrates. These findings suggest that such changes reflect a functional alteration of PS/γ-secretase in each subcellular location.
S2 cleavage upon ligand binding should occur on the PM of the target cells, but whether the subsequent S3 cleavage occurs on the PM or after endocytosis remains controversial (13, 17, 20, 46). The results of the current study suggest that (i) S3 cleavage occurs in both subcellular fractions in cell culture and (ii) S3-V cleavage, which generates stable NICD-V, occurs predominantly on the PM, whereas S3-S(+3) cleavage, which generates unstable NICD-S(+3), occurs predominantly on endosomes. We also found that the intensity of Notch signaling changes along with the extent of endocytosis, perhaps due to a change in the precision of S3 cleavage. Therefore, our results suggest that NICD generation on the PM and endosomes increases and decreases the intensity of Notch signaling, respectively. Reports that Sanpodo positively regulates Notch signaling on the PM and that Numb, a negative regulator, promotes endocytosis of Notch receptors and/or Sanpodo are consistent with our findings (3, 28). A previous study showed that Drosophila with a mutation in shibire, which encodes a homolog of dynamin, has a phenotype indicating a loss of Notch function (43). Further study to solve the contradiction is necessary.
Elimination of either Notch or PS function causes a strong Notch loss-of-function phenotype in vivo (23, 41). Knock-in mice with the S3 (Val→Gly) mutant of Notch-1 display a hypomorphic Notch phenotype, possibly due to reduced Notch signaling caused by a decrease in the intracellular NICD level (6, 15, 39). This reduction in the level of NICD polypeptides could be due to decreased generation and/or increased degradation (4, 6). Previously, NICD was considered to be a single polypeptide mediating a single type of signaling, implying that the lower level of NICD is due to a decrease in its generation; however, we found that the N termini of NICDs may play critical roles in their stabilities and thus signaling intensities.
Among S3 mutants of Notch-1, the Lys→Arg mutant NEXT is neither monoubiquitinated at Arg1749 nor endocytosed (13). Concurrently, the NICD-V level in the cells expressing this mutant is low (13). In this paper, we demonstrated that the Lys→Arg mutation also causes a drastic change in the precision of the S3 cleavage, which results in a drastic decrease of NICD-V generation on the PM. This explanation, if valid, resolves the discrepancy between the two studies.
Both stabilizing (Val, Met, or Gly) and destabilizing amino acid residues seem to be conserved in Notch orthologs of various organisms (see Table S2 in the supplemental material). This suggests that both stable and unstable intracellular Notch signaling exists in many species. Previous studies have indicated that differences in the intensity, duration, and timing of Notch signaling in target cells affect cell fate decisions (7, 26). Furthermore, the signaling by a molecule is dependent on its lifetime in the cell, and the lifetime should be short enough for the target cell to be able to rapidly change the extent of signaling (22). Therefore, target cells in different contexts may convert extracellular signals to various relative amounts of short- and long-term Notch signals.
FAD PS mutations generally increase the generation of Aβ42 (42). This pathological gain of function of PS is due to a change in the precision of γ cleavage, resulting in an increase in cleavage at γ42 (42). PS/γ-secretase modifiers can up- or down-regulate the cleavage at γ42 (19, 31, 49). In both cases, the modifiers have reciprocal effects on the production of Aβ42 and Aβ38 (31, 49). In this study, we found that several FAD PS1 mutants change the precision of S3 cleavage but that the PS/γ-secretase modifiers do not. These findings are consistent with previous studies showing that the precision of ɛ cleavage in βAPP is altered by certain FAD PS mutations (38) and that Notch processing is unaffected by nonsteroidal anti-inflammatory drugs that can reduce Aβ42 generation (45, 49). PS/γ-secretase modifiers could thus affect the precision of the intramembrane proteolysis differently from PS FAD mutations.
Because up-regulation in Notch signaling is involved in a subset of malignancies (10, 37, 47, 50), γ-secretase inhibitors have been considered for the treatment of cancer; however, inhibitors would cause the accumulation of substrates (i.e., NEXT), inevitably producing a “rebound effect,” where the concentration of NICD would increase. Therefore, compounds that alter the precision of S3 cleavage and specifically inhibit the generation of stable NICD-V may be more effective therapeutic agents.
Supplementary Material
Acknowledgments
We thank J. Takeda, R. Kopan, M. Nishimura, H. Hasegawa, Y. Eguchi, Y. Tsujimoto, H. Steiner, and C. Haass for critically reading the manuscript and S. Shirahata, R. Kopan, J. S. Nye, A. Israel, S. L. Schmid, and G. W. Bornkamm for providing cDNAs, constructs, and cell lines.
M.O. conceived and designed the experiments. S.T. and others performed the experiments. M.O. wrote the paper.
We are grateful for funding from the Program for the Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (05-26) (to M.T., M.O., and S.T.), grants-in-aid for Scientific Research on Priority Areas-Advanced Brain Science Project (to M.O.) and KAKEN-HI from the Ministry of Education, Culture, Sports, Science, and Technology (to M.T., M.O., and S.T.), and grants-in-aid from the Japanese Ministry of Health, Labor and Welfare (to M.T. and M.O.).
We declare that no competing interests exist.
Footnotes
Published ahead of print on 29 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284770-776. [DOI] [PubMed] [Google Scholar]
- 2.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234179-186. [DOI] [PubMed] [Google Scholar]
- 3.Berdnik, D., T. Torok, M. Gonzalez-Gaitan, and J. A. Knoblich. 2002. The endocytic protein alpha-adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3221-231. [DOI] [PubMed] [Google Scholar]
- 4.Blat, Y., J. E. Meredith, Q. Wang, J. D. Bradley, L. A. Thompson, R. E. Olson, A. M. Stern, and D. Seiffert. 2002. Mutations at the P1′ position of Notch1 decrease intracellular domain stability rather than cleavage by gamma-secretase. Biochem. Biophys. Res. Commun. 299569-573. [DOI] [PubMed] [Google Scholar]
- 5.Cao, X., and T. C. Sudhof. 2001. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293115-120. [DOI] [PubMed] [Google Scholar]
- 6.Chandu, D., S. S. Huppert, and R. Kopan. 2006. Analysis of transmembrane domain mutants is consistent with sequential cleavage of Notch by gamma-secretase. J. Neurochem. 96228-235. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398518-522. [DOI] [PubMed] [Google Scholar]
- 9.Fehon, R. G., K. Johansen, I. Rebay, and S. Artavanis-Tsakonas. 1991. Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J. Cell Biol. 113657-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fre, S., M. Huyghe, P. Mourikis, S. Robine, D. Louvard, and S. Artavanis-Tsakonas. 2005. Notch signals control the fate of immature progenitor cells in the intestine. Nature 435964-968. [DOI] [PubMed] [Google Scholar]
- 11.Fukumori, A., M. Okochi, S. Tagami, J. Jiang, N. Itoh, T. Nakayama, K. Yanagida, Y. Ishizuka-Katsura, T. Morihara, K. Kamino, T. Tanaka, T. Kudo, H. Tanii, A. Ikuta, C. Haass, and M. Takeda. 2006. Presenilin-dependent gamma-secretase on plasma membrane and endosomes is functionally distinct. Biochemistry 454907-4914. [DOI] [PubMed] [Google Scholar]
- 12.Gonda, D. K., A. Bachmair, I. Wunning, J. W. Tobias, W. S. Lane, and A. Varshavsky. 1989. Universality and structure of the N-end rule. J. Biol. Chem. 26416700-16712. [PubMed] [Google Scholar]
- 13.Gupta-Rossi, N., E. Six, O. LeBail, F. Logeat, P. Chastagner, A. Olry, A. Israel, and C. Brou. 2004. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J. Cell Biol. 16673-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haass, C., and H. Steiner. 2002. Alzheimer disease gamma-secretase: a complex story of GxGD-type presenilin proteases. Trends Cell Biol. 12556-562. [DOI] [PubMed] [Google Scholar]
- 15.Huppert, S. S., A. Le, E. H. Schroeter, J. S. Mumm, M. T. Saxena, L. A. Milner, and R. Kopan. 2000. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 405966-970. [DOI] [PubMed] [Google Scholar]
- 16.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377355-358. [DOI] [PubMed] [Google Scholar]
- 17.Kaether, C., S. Schmitt, M. Willem, and C. Haass. 2006. Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic 7408-415. [DOI] [PubMed] [Google Scholar]
- 18.Koo, E. H., and R. Kopan. 2004. Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nat. Med. 10(Suppl.)S26-S33. [DOI] [PubMed] [Google Scholar]
- 19.Kukar, T., M. P. Murphy, J. L. Eriksen, S. A. Sagi, S. Weggen, T. E. Smith, T. Ladd, M. A. Khan, R. Kache, J. Beard, M. Dodson, S. Merit, V. V. Ozols, P. Z. Anastasiadis, P. Das, A. Fauq, E. H. Koo, and T. E. Golde. 2005. Diverse compounds mimic Alzheimer disease-causing mutations by augmenting Abeta42 production. Nat. Med. 11545-550. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Schier, H., and D. St. Johnston. 2002. Drosophila nicastrin is essential for the intramembranous cleavage of notch. Dev. Cell 279-89. [DOI] [PubMed] [Google Scholar]
- 21.Minoguchi, S., Y. Taniguchi, H. Kato, T. Okazaki, L. J. Strobl, U. Zimber-Strobl, G. W. Bornkamm, and T. Honjo. 1997. RBP-L, a transcription factor related to RBP-Jκ. Mol. Cell. Biol. 172679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto, M., Y. Takahashi, M. Endo, and Y. Saga. 2005. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435354-359. [DOI] [PubMed] [Google Scholar]
- 23.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228151-165. [DOI] [PubMed] [Google Scholar]
- 24.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5197-206. [DOI] [PubMed] [Google Scholar]
- 25.Munter, L. M., P. Voigt, A. Harmeier, D. Kaden, K. E. Gottschalk, C. Weise, R. Pipkorn, M. Schaefer, D. Langosch, and G. Multhaup. 2007. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 261702-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa, M., M. Ichikawa, K. Kumano, S. Goyama, M. Kawazu, T. Asai, S. Ogawa, M. Kurokawa, and S. Chiba. 2006. AML1/Runx1 rescues Notch1-Null mutation-induced deficiency of para-aortic splanchnopleural hematopoiesis. Blood 1083329-3334. [DOI] [PubMed] [Google Scholar]
- 27.Nakaya, Y., T. Yamane, H. Shiraishi, H. Q. Wang, E. Matsubara, T. Sato, G. Dolios, R. Wang, B. De Strooper, M. Shoji, H. Komano, K. Yanagisawa, Y. Ihara, P. Fraser, P. St George-Hyslop, and M. Nishimura. 2005. Random mutagenesis of presenilin-1 identifies novel mutants exclusively generating long amyloid beta-peptides. J. Biol. Chem. 28019070-19077. [DOI] [PubMed] [Google Scholar]
- 28.O'Connor-Giles, K. M., and J. B. Skeath. 2003. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell 5231-243. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi, H., and T. Sudo. 1994. Efficient expression of a transfected foreign gene by Cos1 cells in serum-free medium. Biosci. Biotechnol. Biochem. 58758-759. [DOI] [PubMed] [Google Scholar]
- 30.Okochi, M., S. Eimer, A. Bottcher, R. Baumeister, H. Romig, J. Walter, A. Capell, H. Steiner, and C. Haass. 2000. A loss of function mutant of the presenilin homologue SEL-12 undergoes aberrant endoproteolysis in Caenorhabditis elegans and increases abeta 42 generation in human cells. J. Biol. Chem. 27540925-40932. [DOI] [PubMed] [Google Scholar]
- 31.Okochi, M., A. Fukumori, J. Jiang, N. Itoh, R. Kimura, H. Steiner, C. Haass, S. Tagami, and M. Takeda. 2006. Secretion of the Notch-1 Abeta-like peptide during Notch signaling. J. Biol. Chem. 2817890-7898. [DOI] [PubMed] [Google Scholar]
- 32.Okochi, M., K. Ishii, M. Usami, N. Sahara, F. Kametani, K. Tanaka, P. E. Fraser, M. Ikeda, A. M. Saunders, L. Hendriks, S. I. Shoji, L. E. Nee, J. J. Martin, C. Van Broeckhoven, P. H. St George-Hyslop, A. D. Roses, and H. Mori. 1997. Proteolytic processing of presenilin-1 (PS-1) is not associated with Alzheimer's disease with or without PS-1 mutations. FEBS Lett. 418162-166. [DOI] [PubMed] [Google Scholar]
- 33.Okochi, M., H. Steiner, A. Fukumori, H. Tanii, T. Tomita, T. Tanaka, T. Iwatsubo, T. Kudo, M. Takeda, and C. Haass. 2002. Presenilins mediate a dual intramembranous gamma-secretase cleavage of Notch-1. EMBO J. 215408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okochi, M., J. Walter, A. Koyama, S. Nakajo, M. Baba, T. Iwatsubo, L. Meijer, P. J. Kahle, and C. Haass. 2000. Constitutive phosphorylation of the Parkinson's disease associated alpha-synuclein. J. Biol. Chem. 275390-397. [DOI] [PubMed] [Google Scholar]
- 35.Parks, A. L., K. M. Klueg, J. R. Stout, and M. A. Muskavitch. 2000. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 1271373-1385. [DOI] [PubMed] [Google Scholar]
- 36.Qi-Takahara, Y., M. Morishima-Kawashima, Y. Tanimura, G. Dolios, N. Hirotani, Y. Horikoshi, F. Kametani, M. Maeda, T. C. Saido, R. Wang, and Y. Ihara. 2005. Longer forms of amyloid beta protein: implications for the mechanism of intramembrane cleavage by gamma-secretase. J. Neurosci. 25436-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radtke, F., and K. Raj. 2003. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3756-767. [DOI] [PubMed] [Google Scholar]
- 38.Sato, T., N. Dohmae, Y. Qi, N. Kakuda, H. Misonou, R. Mitsumori, H. Maruyama, E. H. Koo, C. Haass, K. Takio, M. Morishima-Kawashima, S. Ishiura, and Y. Ihara. 2003. Potential link between amyloid beta-protein 42 and C-terminal fragment gamma 49-99 of beta-amyloid precursor protein. J. Biol. Chem. 27824294-24301. [DOI] [PubMed] [Google Scholar]
- 39.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393382-386. [DOI] [PubMed] [Google Scholar]
- 40.Schweisguth, F. 2004. Notch signaling activity. Curr. Biol. 14R129-R138. [PubMed] [Google Scholar]
- 41.Selkoe, D., and R. Kopan. 2003. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 26565-597. [DOI] [PubMed] [Google Scholar]
- 42.Selkoe, D. J. 2001. Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 81741-766. [DOI] [PubMed] [Google Scholar]
- 43.Seugnet, L., P. Simpson, and M. Haenlin. 1997. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192585-598. [DOI] [PubMed] [Google Scholar]
- 44.Steiner, H., M. Kostka, H. Romig, G. Basset, B. Pesold, J. Hardy, A. Capell, L. Meyn, M. L. Grim, R. Baumeister, K. Fechteler, and C. Haass. 2000. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nat. Cell Biol. 2848-851. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, Y., I. Hayashi, Y. Tominari, K. Rikimaru, Y. Morohashi, T. Kan, H. Natsugari, T. Fukuyama, T. Tomita, and T. Iwatsubo. 2003. Sulindac sulfide is a noncompetitive gamma-secretase inhibitor that preferentially reduces Abeta 42 generation. J. Biol. Chem. 27818664-18670. [DOI] [PubMed] [Google Scholar]
- 46.Tarassishin, L., Y. I. Yin, B. Bassit, and Y. M. Li. 2004. Processing of Notch and amyloid precursor protein by gamma-secretase is spatially distinct. Proc. Natl. Acad. Sci. USA 10117050-17055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Es, J. H., M. E. van Gijn, O. Riccio, M. van den Born, M. Vooijs, H. Begthel, M. Cozijnsen, S. Robine, D. J. Winton, F. Radtke, and H. Clevers. 2005. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435959-963. [DOI] [PubMed] [Google Scholar]
- 48.Washburn, T., E. Schweighoffer, T. Gridley, D. Chang, B. J. Fowlkes, D. Cado, and E. Robey. 1997. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell 88833-843. [DOI] [PubMed] [Google Scholar]
- 49.Weggen, S., J. L. Eriksen, P. Das, S. A. Sagi, R. Wang, C. U. Pietrzik, K. A. Findlay, T. E. Smith, M. P. Murphy, T. Bulter, D. E. Kang, N. Marquez-Sterling, T. E. Golde, and E. H. Koo. 2001. A subset of NSAIDs lower amyloidogenic Abeta42 independently of cyclooxygenase activity. Nature 414212-216. [DOI] [PubMed] [Google Scholar]
- 50.Weng, A. P., A. A. Ferrando, W. Lee, J. P. t. Morris, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306269-271. [DOI] [PubMed] [Google Scholar]
- 51.Wolfe, M. S., W. Xia, B. L. Ostaszewski, T. S. Diehl, W. T. Kimberly, and D. J. Selkoe. 1999. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 398513-517. [DOI] [PubMed] [Google Scholar]
- 52.Xia, W., and M. S. Wolfe. 2003. Intramembrane proteolysis by presenilin and presenilin-like proteases. J. Cell Sci. 1162839-2844. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, G., M. Z. Cui, G. Mao, Y. Dong, J. Tan, L. Sun, and X. Xu. 2005. Gamma-cleavage is dependent on zeta-cleavage during the proteolytic processing of amyloid precursor protein within its transmembrane domain. J. Biol. Chem. 28037689-37697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.