Abstract
By combining a tissue-specific microarray screen with mouse uniparental duplications, we have identified a novel imprinted gene, Dopa decarboxylase (Ddc), on chromosome 11. Ddc_exon1a is a 2-kb transcript variant that initiates from an alternative first exon in intron 1 of the canonical Ddc transcript and is paternally expressed in trabecular cardiomyocytes of the embryonic and neonatal heart. Ddc displays tight conserved linkage with the maternally expressed and methylated Grb10 gene, suggesting that these reciprocally imprinted genes may be coordinately regulated. In Dnmt3L mutant embryos that lack maternal germ line methylation imprints, we show that Ddc is overexpressed and Grb10 is silenced. Their imprinting is therefore dependent on maternal germ line methylation, but the mechanism at Ddc does not appear to involve differential methylation of the Ddc_exon1a promoter region and may instead be provided by the oocyte mark at Grb10. Our analysis of Ddc redefines the imprinted Grb10 domain on mouse proximal chromosome 11 and identifies Ddc_exon1a as the first example of a heart-specific imprinted gene.
Genomic imprinting refers to the differential epigenetic programming of male and female gametes that confers parent-of-origin-dependent allele-specific expression upon a small subset of genes in somatic cells (11, 65). Imprinted genes regulate a variety of physiological functions, though a significant number play pivotal roles in fetal growth and development (12, 18, 19, 21, 45, 52, 68). The imprinting mechanism is not yet fully understood, but it is clear that differential epigenetic marking of the parental alleles primarily involves DNA methylation (11, 38) and may also involve the modification of histone proteins as well as the recruitment of chromatin-associated factors, such as Polycomb group (PcG) proteins (40), insulator proteins (7), and the transcription of antisense RNA (61, 62). The study of tissue-specific imprinted genes can be expected to yield new information upon imprint establishment and maintenance, as well as revealing important insights into the misregulation of imprinting in human disorders. The identification of novel tissue-specific imprinted genes is therefore a priority for a comprehensive understanding of imprinting at the molecular level.
Dopa decarboxylase (DDC) is a multifunctional enzyme that plays an essential role in the biosynthesis of catecholamine neurotransmitters and serotonin (15). Perturbations in DDC expression have been reported in a range of neurodegenerative and psychiatric disorders, including Parkinson's disease (30), bipolar affective disorder (9, 10, 32), and attention deficit hyperactivity disorder (24, 33), suggesting a critical role in correct neuronal functioning in adults. However, DDC is expressed not only in dopaminergic and serotonergic neurons of the central and peripheral nervous systems but also in several nonneuronal tissues, with high levels present in liver, pancreas, kidney, and intestine, thus setting it apart from other catecholamine pathway enzymes. Promoter switching and alternative splicing have been shown to direct tissue-specific DDC expression in neuronal and nonneuronal lineages (2, 3, 13, 20, 29, 31, 37).
In mice, Ddc maps to proximal chromosome 11 at a distance of approximately 25 kb from the imprinted Grb10 gene. Major Grb10 isoforms are expressed preferentially or exclusively from the maternal allele in the majority of peripheral tissues, while an alternative promoter generates a paternally expressed transcript in brain with the reciprocal imprinting of these transcripts arising from the differential reading of a maternal germ line methylation mark (5, 25) and the establishment of repressive histone modifications (70). In humans, DDC and GRB10 are located in a region of conserved linkage on chromosome 7p12.2 that is associated with the growth disorder Silver-Russell syndrome (SRS) (28). In this study, by means of a tissue-specific microarray screen of mouse uniparental duplications (UpDps), we identify Ddc as a novel imprinted gene. Imprinting at this locus involves a transcriptional variant, Ddc_exon1a, which is expressed exclusively from the paternal allele in the developing heart. Finally, by analysis of DNA methyltransferase 3-like gene (Dnmt3L)-deficient embryos, we reveal a link between Ddc imprinting and maternal germ line methylation suggesting the existence of coordinate imprinting control with neighboring maternally methylated Grb10.
MATERIALS AND METHODS
Tissue sources.
For the UpDp microarray expression analysis, mice carrying maternal (MatDp11) and paternal (PatDp11) duplications of the proximal region of chromosome 11 were generated using the reciprocal translocation T(7:11)65H strain as described elsewhere (6). Brain, liver, heart, and carcass (residual body cavity with the head and all internal organs including heart removed) tissues were dissected from day 1 animals. Whole embryos and placentae were collected at embryonic day 13.5 (e13.5). For the Dnmt3L microarray analysis, wild-type and Dnmt3Lmat/+ mutant embryos (11) were collected at e8.5.
Microarray hybridization and data analysis.
Affymetrix mouse U74v2 (UpDp analysis) and 430v2 (UpDp analysis and Dnmt3L analysis) GeneChips were used as described previously (60). Total RNA extracts for microarray hybridizations were prepared by cesium chloride density gradient centrifugation as described elsewhere (47) and quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies). Biotinylated cRNA probes were generated and hybridized to the Affymetrix GeneChip arrays. Hybridization signals were measured with an Affymetrix Scanner 3000 and analyzed using the GeneChip operating software (GCOS). Additional bioinformatic analyses were performed as described previously (60).
Allele-specific RT-PCR assays.
Allele-specific expression was determined by semiquantitative reverse transcription and PCR (RT-PCR) combined with direct sequencing using reagents and conditions essentially as described elsewhere (60). For mouse Ddc, parental allele identity was inferred via a G→A single-nucleotide polymorphism (SNP) between the Mus mus musculus C57BL6 (B6) and Mus mus castaneus CAST/Ei (CAST) strains in exon 6 at nucleotide 645 (GenBank accession NM_016672). Products encompassing this SNP were recovered by RT-PCR from reciprocal B6 × CAST intersubspecies hybrid tissues and sequenced to determine the allelic expression. Ddc_exon1a and Ddc_exon1 transcripts were distinguished by combining either of the exon-specific forward primers EXON1A-F (5′-TCACCAAGGAGAGAGAGAGAGC-3′) and EXON1-F (5′-AGAGTGGACCTGTGAAGAATCC-3′), respectively, with a common reverse primer, EXON6-R (5′-GACCACAAAGAATGGAATCAGG-3′) with cycling parameters of 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, followed by a final extension step of 72°C for 5 min.
Northern blotting.
Northern blotting and hybridization were performed according to standard protocols (57). For the Ddc probe, a 1.2-kb cDNA fragment spanning exons 6 to 15 was generated by PCR with the primers DDC-F8 (5′-CTGGGTTAATTGGTGGAATAAAGC-3′) and DDC-R8 (5′-TCTGAAGGTAAGACCAAAGACTGC-3′) and cloned into the pGEM-T vector (Promega). Probe template DNA was then isolated from pGEM-T by digestion with NotI, purified with the QiaQuick gel extraction kit (Qiagen), and used to generate [α-32P]dCTP-labeled probe with the Hi-prime random prime labeling kit (Roche) according to the manufacturer's protocol. Hybridization intensity signals were quantified with a FLA-3000 PhosphorImager system (Fujifilm).
qPCR.
cDNA was prepared from 2 μg of total RNA, using the Superscript first-strand system (Invitrogen). A 1/40 portion of each 20-μl reverse transcription reaction mixture was used for the subsequent PCR step. Quantitative PCR (qPCR) was performed using a Taqman 7900HT Fast real-time PCR system, TaqMan gene expression master mix (Applied Biosystems), and cycling conditions as directed by the manufacturer. Primers were designed to span the 3′-terminal intron of murine Ddc. Primer and probe sequences were as follows: forward primer, 5′-AGGGCAGAGAAAGAATGAAAGCA-3′; reverse primer, 5′-GGAGTGGTAGTTATTTTTCTCTTTCCAGTTT-3′; probe, 5′-6-carboxyfluorescein-CTGCTTCAGAGATCAAAAG-nonfluorescent quencher-3′.
Immunohistochemistry.
Dissected embryos and tissues were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS) and then embedded in paraffin wax, and sections were cut at a 6-μm thickness. Sections were dewaxed in xylene and rehydrated by passage through a graded ethanol-water series. Antigen retrieval was performed by incubating in 10 mM citric acid (pH 6) at 100°C for 30 min, after which endogenous tissue peroxidases were inactivated by incubation in 0.3% hydrogen peroxide at room temperature for 30 min. Sections were blocked in 10% normal goat serum (Dako) in PBS for 1 h at room temperature and then incubated with a rabbit polyclonal anti-bovine DDC antibody (ab 3905; Abcam) diluted 1:500 in 10% normal goat serum-PBS for 1 h at room temperature. Sections were washed three times for 10 min each in PBS, 0.1% Tween and then incubated with a biotinylated goat anti-rabbit immunoglobulin G secondary antibody (Vector Laboratories) diluted 1:500 in 10% normal goat serum-PBS for 1 h at room temperature. Sections were washed three times for 10 min each in PBS, 0.1% Tween and then incubated with a streptavidin-horseradish peroxidase conjugate (Vector Laboratories) for 30 min at room temperature. Bound streptavidin-horseradish peroxidase complexes were detected by incubation in 0.05% diaminobenzidine (Sigma), after which the sections were counterstained in Harris' hematoxylin and mounted.
Bisulfite mutagenesis and sequencing.
Bisulfite conversions were performed essentially as described elsewhere (14). Genomic DNA was prepared using the DNeasy tissue kit (Qiagen), digested overnight with HindIII (New England Biolabs), and purified by phenol-chloroform extraction followed by ethanol precipitation. DNA strands were denatured by incubation in 0.3 M NaOH at 37°C for 15 min, and the reaction mixtures were combined with a final concentration of 3.25 M sodium metabisulfite and 0.93 M hydroxyquinone (Sigma), overlaid with mineral oil, and incubated at 55°C for 6 h in the dark. Bisulfite-treated DNA fragments were purified on QIAEX II beads (Qiagen), desulfonated by incubation in 0.3 M NaOH at 37°C for 15 min, recovered by ethanol precipitation, and resuspended in nuclease-free water (Ambion). Primer sequences for bisulfite PCR amplification were designed using MethPrimer (http://www.urogene.org/methprimer/index1.html). For mouse Ddc methylation analysis, the 419-bp region immediately upstream of the Ddc_exon1a transcription start site was amplified by PCR in bisulfite-converted DNA with the primers DDC_METHP1A-F (5′-AGTGAGATTTAGGTTTGGTATTTTTAGTTT-3′) and DDC_METHP1A-R (5′-CTCTTACTCTCTCTCTCTCCTTAATAACAA-3′) with cycling conditions of 94°C for 3 min, followed by 40 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, and a final extension step of 72°C for 5 min.
PCR products were purified on QiaQuick min-elute PCR purification columns (Qiagen) and then cloned into the pGEM-T vector (Promega). Recombinant clones were sequenced from pUC/M13 −20 and reverse primer sites using ABI Big Dye v3.1 reagents and protocols (Applied Biosystems). The parental origin of the DNA strands was inferred using a C→A polymorphism (nucleotide 56912; GenBank accession AL645803.19) in reciprocal B6 × CAST hybrid tissues. Random nonconverted cytosine residues were used to determine that each clone originated from a unique DNA strand.
Bioinformatics.
Sequence alignments between the mouse and human Ddc/DDC were performed and analyzed using VISTA (http://pipeline.lbl.gov). Regions of the mouse and human genomes aligned are as follows: mouse chr11, 11,779,879 to 11,798,950 (UCSC Mouse, February 2006); human chr7, 50,578,440 to 50,601,478 (UCSC Human, March 2006).
Microarray data accession number.
Microarray data were deposited in GEO and assigned accession number GSE4870.
RESULTS
Ddc_exon1a is a novel heart-specific imprinted gene.
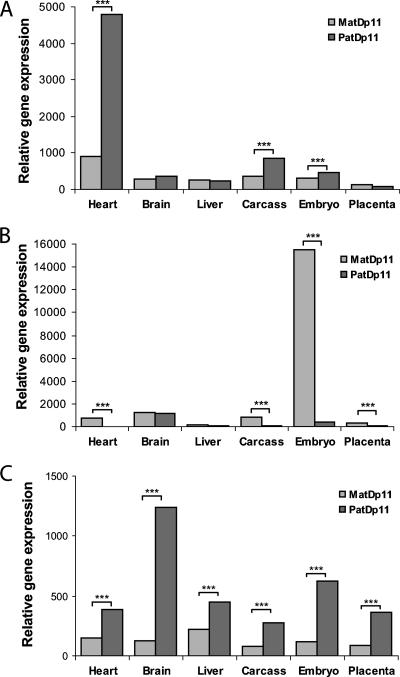
We have identified a novel imprinted gene from a microarray screen of mouse tissues carrying UpDps within the proximal regions of chromosomes 11 and 7 (Fig. 1A). Microarray probe sets corresponding to the Ddc gene detected an increase in paternal expression between PatDp11 versus MatDp11 newborn heart (GCOS/MAS5 signal log ratio [SLR], 2.1; GCOS/MAS5 change, P < 0.0001), consistent with preferential expression from the paternal allele. To a smaller degree, this was noted in PatDp11 versus MatDp11 e13.5 whole embryo (SLR, 0.9; P < 0.0001) and in newborn carcass (SLR, 1.4; P < 0.0001). The observed differential expression in the embryo is likely attributable to a strong paternal Ddc signal in heart. The same cannot be argued for newborn carcass, since its constituent heart was removed prior to the array analysis and this result may reflect imprinting in a tissue still contained within the carcass. Ddc was not differentially expressed in PatDp11 versus MatDp11 newborn brain (SLR, 0.2; P = 0.5), liver (SLR, −0.1; P = 0.08909), or e13.5 placenta (SLR, 0.5; P = 0.5). In addition to Ddc, the arrays detected as differentially expressed two known imprinted genes, Grb10 (Fig. 1B) and U2af1-rs1 (Fig. 1C), from within the same UpDp region on proximal mouse chromosome 11 in multiple UpDp tissues, demonstrating the validity of our microarray-based approach.
FIG. 1.
UpDp proximal chromosome 11 microarray tissue expression profiles. (A) Ddc expression levels in T65H proximal MatDp11 versus PatDp11 mouse tissues as quantified using an Affymetrix U74v2 array probe set 160074_at (newborn heart, liver, carcass, and e13.5 placenta) and 430v2 array probe set 1426215_at (newborn brain, e13.5 embryo). Both probe sets are complementary to the same target sequence in exon 15. UpDp tissue expression profiles recovered for the two known imprinted genes Grb10 (B) and U2af1-rs1 (C) contained within the same UpDp region are shown for comparison. Where present, asterisks indicate statistically significant differential expression (GCOS change P ≤ 0.003) between MatDp11 and PatDp11 tissues.
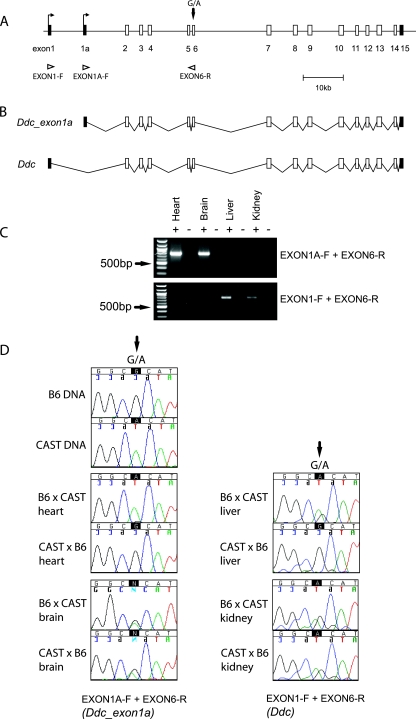
Imprinted genes generally occur in clusters, and the location of Ddc, at approximately 25 kb proximal to the imprinted Grb10 gene on mouse chromosome 11, was consistent with this expectation and prompted further study. Mouse Ddc contains at least 16 exons spanning approximately 84 kb of genomic DNA sequence (Fig. 2A). A single 2-kb transcript (hereafter referred to as Ddc) that initiates from a 5′ noncoding exon is currently annotated in the Ensembl mouse genome browser (ENSMUST00000066237; NCBI 36 genome assembly). Extensive mouse expressed sequence tag evidence predicted a second transcript of similar size but initiating from an alternative 5′ noncoding exon (hereafter referred to as Ddc_exon1a), embedded within intron 1 of the related Ddc transcript. RT-PCR and sequencing confirmed the expression and splice junctions of the predicted Ddc_exon1a transcript (data not shown). The structures of the novel Ddc_exon1a and the related Ddc transcript are shown (Fig. 2B). Molecular characterization revealed that Ddc_exon1a is highly expressed in heart and brain yet is not expressed at levels detectable by RT-PCR in liver or kidney. By contrast, the related Ddc transcript is expressed in liver and kidney, which we note are tissues with a significant endoderm component, but is not detectable in heart (mesoderm) or brain (ectoderm) (Fig. 2C). The two transcripts therefore show reciprocal tissue expression profiles, with the novel Ddc_exon1a transcript being predominant in heart.
FIG. 2.
Tissue- and transcript-specific imprinting of mouse Ddc. (A) Map of the ∼84-kb genomic region containing the Ddc locus. Coding exons are shown as open boxes, noncoding exons are shown as filled boxes, and open triangles indicate the positions of primers used in allele-specific RT-PCR assays. The location of a G→A SNP in exon 6 between the Mus mus musculus (B6) and Mus mus castaneus (CAST) subspecies is shown by a vertical arrow. (B) Deduced structures of the Ddc_exon1a transcript variant and the related Ddc transcript in their respective genomic contexts. (C) Exon-specific RT-PCR in newborn mouse tissues. The primers EXON1A-F and EXON6-R amplify the Ddc_ exon1a transcript (786-bp product); EXON1-F and EXON6-R amplify the related Ddc transcript (762-bp product). The 500-bp marker is indicated for a reference (arrows). (D) Allele-specific RT-PCR assays in newborn mouse tissues. cDNA fragments were recovered from reciprocal B6 × CAST intersubspecies hybrid tissues by RT-PCR and directly sequenced using the exon-specific primer combinations described above. The G→A SNP (arrows) in exon 6 was used to infer the parental origin of the expressed allele.
We surmised that Ddc_exon1a is the most likely candidate for imprinting. To confirm the UpDp microarray predictions, we analyzed the imprinting status of both major Ddc transcripts in allele-specific RT-PCR assays. An SNP was used to distinguish the parental alleles in mouse intersubspecies hybrids and demonstrated that Ddc_exon1a is expressed exclusively from the paternal allele in newborn heart (Fig. 2D). By contrast, Ddc_exon1a was biallelically expressed in newborn brain, and the related endoderm-specific Ddc transcript was also biallelically expressed in newborn liver, kidney (Fig. 2D), and small intestine (data not shown). We can therefore confirm Ddc_exon1a as a novel and heart-specific imprinted gene that is reciprocally expressed relative to mouse Grb10.
Human DDC.
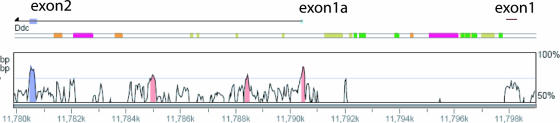
Human DDC is located on chromosome 7p12.2 in a region of conserved linkage with GRB10 and consists of 15 exons spanning 107 kb (64). Two DDC transcripts, a neuronal isoform and a nonneuronal (endoderm-specific) isoform, have been characterized in humans, and like mouse Ddc these differ only in having alternative 5′ noncoding exons that initiate from distinct promoters (29, 31, 63). VISTA plot analysis between mouse and human Ddc/DDC genomic regions (Fig. 3) showed that promoter regions of imprinted mouse exon1a and the 5′ noncoding exon of the neuronal DDC isoform are orthologous and share 77.9% sequence homology over 131 bp. We hypothesized that the neuronal DDC isoform might show a similar heart-specific imprinting profile in humans. An earlier study established that DDC/Ddc is biallelically expressed in several human and mouse fetal tissues, but those authors did not analyze the heart in either species (26). Seven fetal genomic DNAs were sequenced for expressed polymorphisms in DDC (data not shown), but only one heterozygous fetus of 10 weeks gestational age was identified with two T→C SNPs in exon 15. RT-PCR combined with sequencing across these polymorphic sites indicated monoallelic DDC expression in fetal heart (data not shown), but the n of 1 was not considered significant enough to draw conclusions from human samples and parental origin could not be assigned because maternal DNA samples were not available and all samples available to us have been tested.
FIG. 3.
Conserved genomic organization of mouse and human Ddc/DDC. The mouse Ddc genomic region was used as the base sequence for construction of a VISTA plot on the human genome. For simplicity, only the exon1-exon2 interval is shown. A 75% sequence similarity threshold was applied and is shown on the plot by a thin bar. Conserved coding exons are purple, conserved noncoding exons are light blue, and conserved noncoding nonexonic sequences are pink.
Ddc_exon1a is progressively silenced during postnatal development.
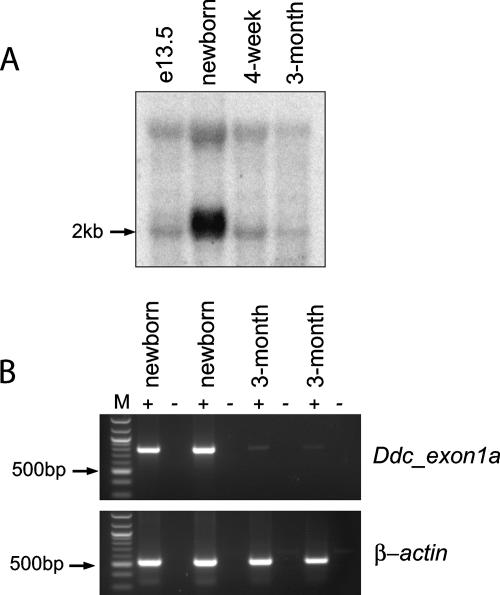
From a developmental perspective, we were keen to establish whether imprinted expression of mouse Ddc_exon1a persists throughout postnatal development and into the adult. Northern blotting showed that while Ddc is expressed at high levels in newborn heart tissue, its expression diminishes considerably in 4-week-old heart and is weak or absent in 3-month-old heart (Fig. 4A). RT-PCR analysis of these same tissues confirmed a sharp decline in the abundance of Ddc_exon1a transcripts to virtually undetectable levels in 3-month-old heart (Fig. 4B). This indicates that Ddc_exon1a expression is progressively downregulated in the heart during postnatal development and occurs only at low basal levels in the adult heart.
FIG. 4.
Progressive silencing of Ddc_exon1a during mouse postnatal development. (A) Northern blot analysis of mouse Ddc expression in total RNA extracted from e13.5 embryo, newborn heart, 4-week heart, and 3-month heart, showing a 2-kb transcript. (B) RT-PCR analysis of Ddc_exon1a expression in newborn heart and 3-month heart cDNAs using the primers EXON1A-F and EXON6-R (see Materials and Methods), which amplify a 786-bp product. The 500-bp marker is indicated for a reference (arrows). RNA integrity was verified by amplification of β-actin sequences from each sample, and samples treated with (+) and without (-) reverse transcriptase are indicated.
Ddc_exon1a expression is specific to trabecular cardiomyocytes.
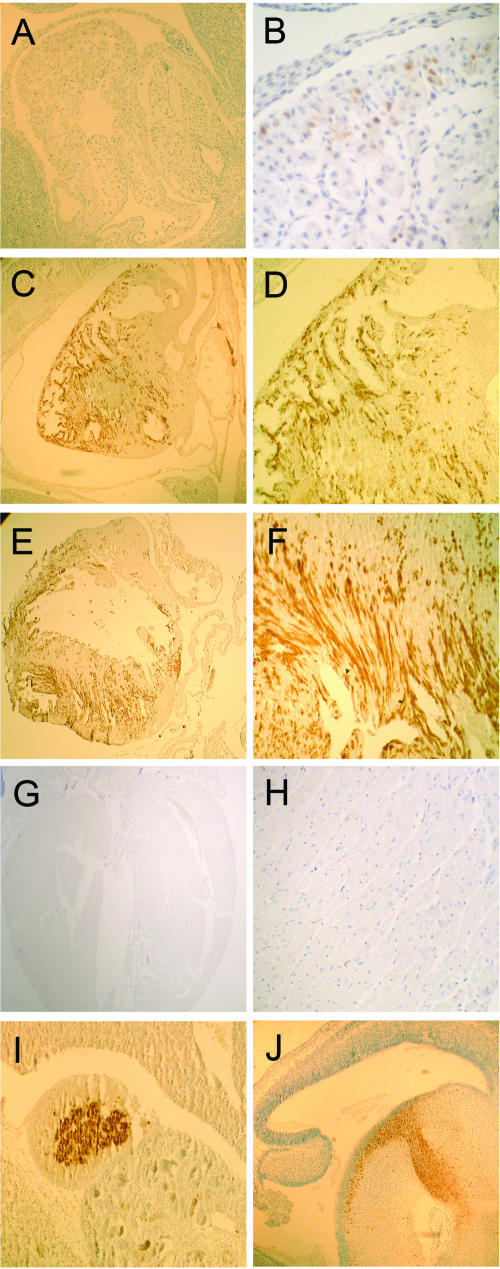
The temporal and spatial expression characteristics of Ddc_exon1a are striking and suggest the existence of a link between imprinting and cardiac development. This is intriguing, because other catecholamine/monoamine pathway enzymes have established roles in cardiogenesis (34, 55) and serotonin signaling pathways, which are dependent upon Ddc activity and play pivotal roles in myocardial proliferation and differentiation, as well as in cardiac functioning in adults (49, 50). To further explore the relationship between Ddc_exon1a and cardiogenesis, we performed immunohistochemistry to discern cell types that express Ddc enzyme in the developing mouse heart. At e10.5, isolated clusters of Ddc-immunoreactive cells were clearly seen in the differentiating myocardium of the early heart (Fig. 5A and B), and aside from marked staining in midbrain neurons and ganglia (not shown) expression was generally absent at other sites in the embryo. At e14.5, Ddc expression had intensified dramatically in the heart, with strong staining apparent in a scattered cell population throughout the myocardium that we identified as trabecular cardiomyocytes (Fig. 5C and D). In the newborn heart, Ddc expression continued to mark trabecular cardiomyocytes but had declined in some areas of the myocardium compared with the preceding embryonic stages (Fig. 5E and F). At 3 months, and consistent with the RNA expression data, Ddc expression had essentially disappeared from the heart altogether (Fig. 5G and H). Staining at other known sites of Ddc expression confirmed the specificity of the primary antibody, notably, in secretory cells of the adrenal medulla (Fig. 5I) and midbrain neurons (Fig. 5J).
FIG. 5.
Immunohistochemical localization of Ddc expression during mouse heart development. Sites of positive Ddc immunoreactivity are marked by the brown signals. The hematoxylin counterstain appears as blue nuclear staining. (A) Parasagittal view of the e10.5 heart with positive Ddc staining in a small number of cardiomyocytes. Magnification, ×200. (B) Higher-magnification view of the e10.5 heart. Magnification, ×400. (C) Parasagittal view of the e14.5 heart, showing widespread Ddc staining in scattered trabecular cardiomyocytes. Magnification, ×200. (D) Higher-magnification view of the e14.5 heart. Magnification, ×400. (E) Parasagittal view of the newborn heart, showing persistent Ddc staining in trabecular cardiomyocytes. Magnification, ×10. (F) Higher-magnification view of the newborn heart. Magnification, ×400. (G) Low-magnification view of the 3-month heart showing an absence of Ddc staining. Magnification, ×10. (H) Higher-magnification view of the 3-month heart. Magnification, ×400. (I and J) Other sites of Ddc expression in e14.5 embryos: adrenal gland with Ddc staining in the secretory adrenomedullary cells (I) and Ddc expression in differentiating pancreatic acinar cells (J). Magnification (both panels), ×200. Control sections incubated with anti-rabbit immunoglobulin G secondary antibody alone did not produce signals (not shown).
No evidence for imprinting at adjacent Fignl1 or Cobl.
Since imprinted genes generally occur in clusters, we considered the possibility of additional imprinted genes in the regions flanking Ddc/Grb10. In both humans and mice, Ddc and Grb10 are flanked by the Fidgetin-like 1 (Fignl1) and Cordon-bleu (Cobl) genes in their proximal and distal aspects, respectively. Examination of UpDp microarray data revealed that Cobl was not differentially expressed in any of the mouse UpDp tissues we analyzed (data not shown), a finding consistent with an earlier study that reported biallelic COBL/Cobl expression in a range of human and mouse fetal tissues (26). Fignl1 probe sets detected a minor paternal expression bias in MatDp11 versus PatDp11 newborn heart, but this was not corroborated by independent allele-specific RT-PCR assays in intersubspecies hybrids that showed biallelic expression in heart (data not shown). While we cannot exclude the possibility of imprinting in highly discrete tissues or at developmental stages not represented in our UpDp microarray experiments, our data indicate that Fignl1 and Cobl are unlikely to be imprinted in the mouse.
Ddc is regulated by Dnmt3L-dependent maternal DNA methylation.
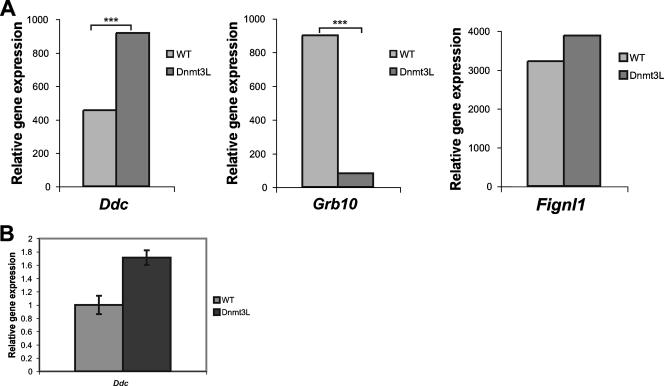
In many imprinted gene clusters, the imprinting of the entire cluster is dependent upon a single cis-acting imprinting element that coincides with a germ line differentially methylated region (21, 39, 66, 71). Likewise, we postulated that Ddc and Grb10 might be regulated by a shared imprinting element. A bioinformatic scan of mouse Ddc revealed an absence of CpG-dense sequence features, which typically attract differential methylation at other imprinted loci (data not shown). The adjacent Grb10 gene harbors a germ line differentially methylated CpG island, CGI2, which acquires methylation in oocytes and is differentially methylated in somatic lineages (5, 25) and could potentially act to imprint both genes. The acquisition of methylation imprints in the maternal germ line, including this oocyte mark at Grb10, is dependent upon the Dnmt3L gene. Dnmt3L-deficient mice display a maternal effect phenotype in which methylation imprints fail to be established in oocytes (4, 11). This has two major consequences in Dnmt3Lmat−/+ heterozygous progeny: (i) maternally repressed imprinted genes are biallelically expressed and (ii) imprinted genes depending on maternal methylation for expression are silenced. Crucially, imprinted genes controlled by paternal germ line methylation and nonimprinted genes are not expected to change in expression (11). To address a role for maternal methylation, potentially involving the CGI2 region, we examined Ddc and Grb10 expression profiles in e8.5 Dnmt3Lmat−/+ embryos versus wild-type embryos by microarray analysis (Fig. 6A). As expected, Grb10, which depends on maternal methylation for expression, was significantly downregulated in Dnmt3Lmat−/+ embryos compared to wild-type controls (SLR, −3.6; P = 1.0000). By contrast, Ddc was twofold overexpressed in Dnmt3Lmat−/+ embryos compared to controls (SLR, 1.0; P < 0.0001), consistent with reactivation of a maternally repressed imprinted allele. A qPCR assay designed at the same location as the Affymetrix probe (which assays both Ddc transcripts) confirmed the array data by detecting a 1.7-fold increase in expression of Ddc in the absence of Dnmt3L (Fig. 6B). The adjacent Fignl1 gene, which is not imprinted, was not differentially expressed on the microarray (SLR, 0.0; P = 0.0196) (Fig. 6A). Ddc and Grb10 are therefore reciprocally regulated by Dnmt3L-dependent maternal methylation, suggesting the existence of a shared imprinting mechanism.
FIG. 6.
Dnmt3L-dependent regulation of Ddc expression. (A) Relative gene expression levels were compared in Dnmt3Lmat/+ mutant e8.5 embryos and wild-type controls using the Affymetrix mouse 430v2 GeneChip system. Hybridization intensity signals are shown for Ddc, Grb10, and the nonimprinted control gene Fignl1. Where present, asterisks indicate statistically significant differential expression (GCOS change P ≤ 0.0003 or ≥ 0.997) between Dnmt3Lmat−/+ embryos and wild-type controls. (B) qPCR shows overexpression of Ddc in e8.5 maternal imprint-free embryos (Dnmt3L) relative to wild-type (WT) controls. For each genotype, expression of Ddc is shown as a ratio of the mean Ct value in relation to the mean Ct value for β-actin. Ct values were transformed as described elsewhere (53), and the relative Ddc/β-actin expression ratio in WT embryos was set to 1. n = 3 for each sample type. The 95% confidence intervals are shown.
Ddc_exon1a is not differentially methylated in the heart.
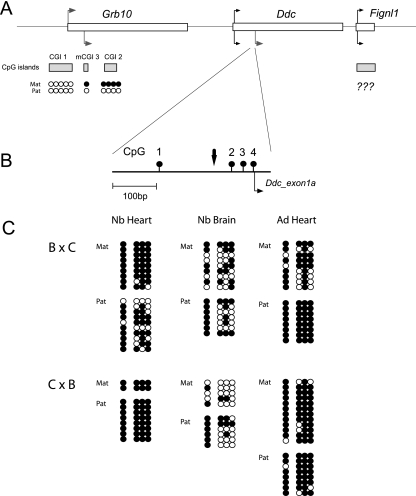
We considered the possibility that the Ddc locus is itself marked by maternal methylation. In particular, we asked whether allele-specific methylation exists over the Ddc_exon1a promoter region that could (i) distinguish the parental alleles in the heart, (ii) account for the difference in Ddc_exon1a imprinting status between newborn heart and brain, and (iii) orchestrate the progressive silencing of Ddc_exon1a that occurs in the postnatal heart. We performed bisulfite mutagenesis and sequencing to determine the allelic methylation status of the four available CpG dinucleotides in the promoter region immediately upstream of the Ddc_exon1a transcription start site (represented in Fig. 7A and B) in intersubspecies hybrids. Both parental alleles were highly methylated in newborn heart, though in brain, where Ddc_exon1a is biallelically expressed, both alleles were relatively hypomethylated (Fig. 7C). While differences in overall bulk methylation levels clearly exist between heart and brain, the allelic methylation profiles do not correlate with the imprinting status in these tissues, suggesting that the Ddc_exon1a promoter region does not carry imprinting information. Moreover, we observed no differences in the methylation status of the Ddc_exon1a promoter between newborn and adult heart tissue (Fig. 7C), suggesting that local methylation does not play a major role in silencing Ddc_exon1a during postnatal development. A small intronic CpG island that is located between exons 12 and 13 of human DDC but is not conserved in mouse and bisulfite analysis demonstrated biallelic hypomethylation of this region in cord blood DNA (data not shown).
FIG. 7.
DNA methylation analysis of the Ddc_exon1a promoter region. (A) Schematic of the Ddc/Grb10 genomic interval, showing relevant sequence features. Genes are shown as open boxes, CpG islands are shown as shaded boxes, and the CpG island allelic methylation status (where known) is indicated by solid (methylated) or open (unmethylated) circles. The position of the maternally methylated Grb10 CGI2 region (gDMR) is shown (5). (B) Enlarged view of the ∼400-bp Ddc_exon1a promoter region. Relevant CpG dinucleotides (numbered 1 to 5) are shown as filled lollipops. A C→A SNP between B6 and CAST strains was used to infer parental origin of the strands (vertical arrow). (C) Bisulfite DNA sequencing profiles of the Ddc_exon1a promoter region in newborn heart, newborn brain, and adult heart tissues derived from reciprocal B6 (B) × CAST (C) intersubspecies hybrids with the maternal (Mat) and paternal (Pat) origin of the strands indicated. Methylated and unmethylated CpGs are represented as solid and open circles, respectively.
DISCUSSION
Dopa decarboxylase is an important enzyme that plays a fundamental role in the biosynthesis of catecholamine neurotransmitters and serotonin. In this study we report genomic imprinting of the Ddc gene on mouse chromosome 11. Imprinting affects an alternative transcriptional variant, Ddc_exon1a, which is expressed exclusively from the paternal allele in trabecular cardiomyocytes of the developing heart. Ddc_exon1a expression is progressively silenced during postnatal development and is essentially absent in the adult heart, suggesting a link with cardiac development. Imprinting at this locus was unexpected, particularly since an earlier study reported biallelic Ddc/DDC expression in multiple human and mouse fetal tissues not including heart (26). This study therefore provides a sound demonstration of microarray-based methods for the detection of tissue-specific imprinted genes. The phenomenon of tissue- and transcript-specific imprinting is not novel and has been described for other organs (5, 14, 25, 46, 54); however, to our knowledge Ddc_exon1a is the first example of heart-specific imprinting in mammals.
Tissue-specific imprinting.
The tissue-restricted imprinting profile of Ddc_exon1a is striking and raises the question of how it might be controlled at the molecular level. We note that the alternative endoderm-specific Ddc transcript was biallelically expressed in all tissues analyzed (Fig. 2D). Promoter switching may, in part, provide a mechanism for Ddc_exon1a imprinting in heart, but biallelic transcription from the Ddc_exon1a promoter in brain argues that tissue-specific epigenetic modifications must also play a role. This is unlikely to involve local DNA methylation, since imprinting status was unrelated to allelic methylation of the Ddc_exon1a promoter, though in the heart analysis we cannot exclude the possibility of predominant methylation patterns in bulk cardiomyocyte populations that may have masked lineage-specific marks in trabecular cardiomyocytes. It is nonetheless conceivable that distinct epigenetic marks, such as histone modifications, are more important for defining allelic promoter architecture at Ddc_exon1a. Tandem (GA)n dinucleotide repeats overlap the EXON1A transcription start site in human DDC (9). These repeats are conserved and more extensive in mouse exon1a but do not occur at the (nonimprinted) endoderm-specific promoter in either species (data not shown). In Drosophila melanogaster, such (GA)n repeats have been shown to bind the GAGA factor (51, 69), a chromatin-associated protein which regulates transcription by modulating the repressive effects of histones and other chromatin remodelling proteins (1). This implicates a mechanism whereby the (GA)n repeats might regulate imprinting by targeting repressive (or active) chromatin modifications to the Ddc_exon1a promoter.
Coordinate regulation of the Ddc/Grb10 domain.
A major conclusion of this study is that Grb10 should no longer be considered an isolated imprinted gene but is instead likely to form part of a novel cluster of jointly regulated imprinted genes. The reciprocal allelic expression (in heart) between Ddc_exon1a and Grb10 is indeed consistent with coordinate imprinting control, as shown for many other imprinted gene clusters (36, 42, 58, 62, 66). Based on current evidence, Grb10 imprinting is presumed to be primarily dependent upon the oocyte methylation mark at CGI2 (5, 25). The mechanism controlling Ddc_exon1a imprinting remains to be fully elucidated, but in Dnmt3Lmat−/+ embryos, where Ddc is overexpressed (Fig. 6A), maternal germ line methylation is likely to play a role. With no demonstrable local methylation mark at Ddc, it is reasonable to argue that Grb10 CGI2 carries the primary imprinting information for both Grb10 itself and Ddc_exon1a, since this element acquires its characteristic methylation in a Dnmt3L-dependent manner (4). At approximately 127 kb apart, it is not clear how a maternally methylated CGI2 allele might orchestrate transcriptional repression at Ddc_exon1a, although studies at other imprinted loci have revealed the existence of long-range silencing mechanisms that could act in this capacity. For example, imprinting of the Igf2 gene in the liver and gut endoderm is dependent upon methylation-sensitive binding of the insulator protein CTCF to the H19 differentially methylated domain located almost 90 kb away (7, 23, 59), with differential methylation at sites proximal to Igf2 playing no role in these tissues (17, 48). Mouse CGI2 contains at least two CTCF consensus sites, but these are not conserved in humans (25). Chromatin modifications also regulate imprinting in the Grb10 domain. On this line, Grb10 is one of a small number of paternally repressed imprinted genes known to be regulated by the PcG protein Embryonic ectoderm development (Eed). Grb10 was biallelically expressed in Eed−/− embryos and, we note, with no change in allelic methylation at CGI2 (40). More recent work has shown that silencing of the paternal Grb10 allele is correlated with allele-specific histone H3 lysine 27 methylation (H3K27) in the major promoter region (70).Therefore, imprinting at Grb10 is primarily dependent upon maternal germ line methylation, with secondary tissue-specific imprinting coordinated by the repressive effects of histones and PcG proteins. Future work should determine whether similar repressive histone modifications are important for tissue-specific imprinting of Ddc_exon1a.
Silver-Russell syndrome.
DDC/GRB10 display conserved linkage in humans and map to chromosome 7p12.2, a region associated with SRS, a heterogeneous growth disorder characterized by intrauterine and postnatal growth restriction and dysmorphia. Several modes of inheritance have been associated with SRS (28), including two recent reports of epigenetic anomalies within the 11p15.5 imprinted domain in at least some patients with classical SRS (8, 22). Maternal uniparental disomy of chromosome 7 (mUPD7) is consistently observed in about 10% of SRS patients (67), and segmental maternal duplications encompassing 7p11.2-p13 have been described in SRS families, providing strong evidence for the involvement of imprinting (43, 44). GRB10, a potent growth suppressor, has proven a logical though controversial candidate. Maternal deletion of Grb10 in mice results in significant embryonic overgrowth (12), thus demonstrating how perturbations in GRB10 dosage might account for the growth restriction seen in SRS. On the other hand, predominantly biallelic GRB10 expression in several human tissues is not easily reconciled with mUPD7 and, moreover, clinical studies have failed to identify GRB10 coding mutations or epigenetic aberrations in SRS patients, thus arguing against a major causative role in the disease (5, 27). The tissue and allele expression profile of DDC is broadly incompatible with the etiology of SRS, although it is noteworthy that a small number of SRS patients display cardiac defects as well as increased predisposition to congenital heart disease (16). Further studies to increase the number of human heart specimens tested for allele-specific expression would help decipher whether DDC expression in the developing heart, an organ that may indirectly influence fetal growth (35, 41, 56), could be involved in SRS at 7p12.2.
Functional significance of Ddc imprinting.
The physiological role of Ddc during embryonic development is not completely understood, since mouse knockout models have not yet been described. However, the imprinting and developmental regulation of Ddc_exon1a expression in the heart suggests fundamental roles in cardiogenesis and cardiac function. Accumulated evidence from gene targeting studies involving related catecholamine biosynthetic enzymes is strongly supportive of this assertion. For example, Tyrosine hydroxylase knockout mice, which are deficient in all catecholamine neurotransmitters, display cardiovascular disorganization and ventricular hypoplasia (34, 55). Serotonin signaling pathways also perform essential roles in heart development. Mice deficient in the serotonin Htr2b receptor exhibit a lack of cardiac trabeculae and profound ventricular hypoplasia due to defective cardiomyocyte proliferation and suffer midgestational to neonatal demise of variable penetrance (49). It has been proposed that some imprinted genes, by close physical proximity to imprinting control regions, might have passively acquired monoallelic expression as “innocent bystanders.” The well-characterized effects of catecholamine neurotransmitters upon cardiac development and function suggest a more active selection process for imprinting at Ddc.
Acknowledgments
This work was supported by grants from the Wellcome Trust (R.J.O.), The Guy's and St. Thomas' Charitable foundation (R.J.O.), The Biotechnology and Biological Sciences Research Council (R.J.O.), EMBO (R.S.), Wellbeing of Women (G.E.M.), and March of Dimes (G.E.M.).
We thank Timothy Bestor (Columbia State University) and Deborah Bourc'his (Inserm U741/Paris 7 University, France) for providing Dnmt3L mutant embryos. We are also grateful to Colin Beechey (MRC, Harwell, United Kingdom) for the T(7:11)65H translocation mouse strain. We thank Katarzyna Koltowska for technical assistance with the human bisulfite analysis.
Footnotes
Published ahead of print on 29 October 2007.
REFERENCES
- 1.Adkins, N. L., T. A. Hagerman, and P. Georgel. 2006. GAGA protein: a multi-faceted transcription factor. Biochem. Cell Biol. 84559-567. [DOI] [PubMed] [Google Scholar]
- 2.Aguanno, A., R. Afar, and V. R. Albert. 1996. Tissue-specific expression of the nonneuronal promoter of the aromatic L-amino acid decarboxylase gene is regulated by hepatocyte nuclear factor 1. J. Biol. Chem. 2714528-4538. [DOI] [PubMed] [Google Scholar]
- 3.Aguanno, A., M. R. Lee, C. M. Marden, M. Rattray, A. Gault, and V. R. Albert. 1995. Analysis of the neuronal promoter of the rat aromatic L-amino acid decarboxylase gene. J. Neurochem. 651944-1954. [DOI] [PubMed] [Google Scholar]
- 4.Arnaud, P., K. Hata, M. Kaneda, E. Li, H. Sasaki, R. Feil, and G. Kelsey. 2006. Stochastic imprinting in the progeny of Dnmt3L−/− females. Hum. Mol. Genet. 15589-598. [DOI] [PubMed] [Google Scholar]
- 5.Arnaud, P., D. Monk, M. Hitchins, E. Gordon, W. Dean, C. V. Beechey, J. Peters, W. Craigen, M. Preece, P. Stanier, G. E. Moore, and G. Kelsey. 2003. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 121005-1019. [DOI] [PubMed] [Google Scholar]
- 6.Beechey, C. V., S. T. Ball, K. M. Townsend, and J. Jones. 1997. The mouse chromosome 7 distal imprinting domain maps to G-bands F4/F5. Mamm. Genome 8236-240. [DOI] [PubMed] [Google Scholar]
- 7.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 8.Bliek, J., P. Terhal, M. J. van den Bogaard, S. Maas, B. Hamel, G. Salieb-Beugelaar, M. Simon, T. Letteboer, J. van der Smagt, H. Kroes, and M. Mannens. 2006. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am. J. Hum. Genet. 78604-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borglum, A. D., T. G. Bruun, T. E. Kjeldsen, H. Ewald, O. Mors, G. Kirov, C. Russ, B. Freeman, D. A. Collier, and T. A. Kruse. 1999. Two novel variants in the DOPA decarboxylase gene: association with bipolar affective disorder. Mol. Psychiatry 4545-551. [DOI] [PubMed] [Google Scholar]
- 10.Borglum, A. D., G. Kirov, N. Craddock, O. Mors, W. Muir, V. Murray, I. McKee, D. A. Collier, H. Ewald, M. J. Owen, D. Blackwood, and T. A. Kruse. 2003. Possible parent-of-origin effect of Dopa decarboxylase in susceptibility to bipolar affective disorder. Am. J. Med. Genet. B 11718-22. [DOI] [PubMed] [Google Scholar]
- 11.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 2942536-2539. [DOI] [PubMed] [Google Scholar]
- 12.Charalambous, M., F. M. Smith, W. R. Bennett, T. E. Crew, F. Mackenzie, and A. Ward. 2003. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc. Natl. Acad. Sci. USA 1008292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatelin, S., R. Wehrle, P. Mercier, D. Morello, C. Sotelo, and M. J. Weber. 2001. Neuronal promoter of human aromatic L-amino acid decarboxylase gene directs transgene expression to the adult floor plate and aminergic nuclei induced by the isthmus. Brain Res. Mol. Brain Res. 97149-160. [DOI] [PubMed] [Google Scholar]
- 14.Choi, J. D., L. A. Underkoffler, A. J. Wood, J. N. Collins, P. T. Williams, J. A. Golden, E. F. Schuster, Jr., K. M. Loomes, and R. J. Oakey. 2005. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol. Cell. Biol. 255514-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christenson, J. G., W. Dairman, and S. Udenfriend. 1972. On the identity of DOPA decarboxylase and 5-hydroxytryptophan decarboxylase (immunological titration-aromatic L-amino acid decarboxylase-serotonin-dopamine-norepinephrine). Proc. Natl. Acad. Sci. USA 69343-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole, R. B., and S. E. Levin. 1973. Congenital heart disease associated with the Russell-Silver syndrome. S. Afr. Med. J. 47989-990. [PubMed] [Google Scholar]
- 17.Constancia, M., W. Dean, S. Lopes, T. Moore, G. Kelsey, and W. Reik. 2000. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 26203-206. [DOI] [PubMed] [Google Scholar]
- 18.Constancia, M., M. Hemberger, J. Hughes, W. Dean, A. Ferguson-Smith, R. Fundele, F. Stewart, G. Kelsey, A. Fowden, C. Sibley, and W. Reik. 2002. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417945-948. [DOI] [PubMed] [Google Scholar]
- 19.DeChiara, T. M., E. J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64849-859. [DOI] [PubMed] [Google Scholar]
- 20.Dugast-Darzacq, C., S. Egloff, and M. J. Weber. 2004. Cooperative dimerization of the POU domain protein Brn-2 on a new motif activates the neuronal promoter of the human aromatic L-amino acid decarboxylase gene. Brain Res. Mol. Brain Res. 120151-163. [DOI] [PubMed] [Google Scholar]
- 21.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32426-431. [DOI] [PubMed] [Google Scholar]
- 22.Gicquel, C., S. Rossignol, S. Cabrol, M. Houang, V. Steunou, V. Barbu, F. Danton, N. Thibaud, M. Le Merrer, L. Burglen, A. M. Bertrand, I. Netchine, and Y. Le Bouc. 2005. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat. Genet. 371003-1007. [DOI] [PubMed] [Google Scholar]
- 23.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 24.Hawi, Z., D. Foley, A. Kirley, M. McCarron, M. Fitzgerald, and M. Gill. 2001. Dopa decarboxylase gene polymorphisms and attention deficit hyperactivity disorder (ADHD): no evidence for association in the Irish population. Mol. Psychiatry 6420-424. [DOI] [PubMed] [Google Scholar]
- 25.Hikichi, T., T. Kohda, T. Kaneko-Ishino, and F. Ishino. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes: roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 311398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitchins, M. P., L. Bentley, D. Monk, C. Beechey, J. Peters, G. Kelsey, F. Ishino, M. A. Preece, P. Stanier, and G. E. Moore. 2002. DDC and COBL, flanking the imprinted GRB10 gene on 7p12, are biallelically expressed. Mamm. Genome 13686-691. [DOI] [PubMed] [Google Scholar]
- 27.Hitchins, M. P., D. Monk, G. M. Bell, Z. Ali, M. A. Preece, P. Stanier, and G. E. Moore. 2001. Maternal repression of the human GRB10 gene in the developing central nervous system: evaluation of the role for GRB10 in Silver-Russell syndrome. Eur. J. Hum. Genet. 982-90. [DOI] [PubMed] [Google Scholar]
- 28.Hitchins, M. P., P. Stanier, M. A. Preece, and G. E. Moore. 2001. Silver-Russell syndrome: a dissection of the genetic aetiology and candidate chromosomal regions. J. Med. Genet. 38810-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichinose, H., and T. Nagatsu. 1993. Molecular genetics of aromatic L-amino acid decarboxylase. Yakubutsu Seishin Kodo 13251-256. (In Japanese.) [PubMed] [Google Scholar]
- 30.Ichinose, H., T. Ohye, K. Fujita, F. Pantucek, K. Lange, P. Riederer, and T. Nagatsu. 1994. Quantification of mRNA of tyrosine hydroxylase and aromatic L-amino acid decarboxylase in the substantia nigra in Parkinson's disease and schizophrenia. J. Neural Transm. Park. Dis. Dement. Sect. 8149-158. [DOI] [PubMed] [Google Scholar]
- 31.Ichinose, H., C. Sumi-Ichinose, T. Ohye, Y. Hagino, K. Fujita, and T. Nagatsu. 1992. Tissue-specific alternative splicing of the first exon generates two types of mRNAs in human aromatic L-amino acid decarboxylase. Biochemistry 3111546-11550. [DOI] [PubMed] [Google Scholar]
- 32.Jahnes, E., D. J. Muller, T. G. Schulze, C. Windemuth, S. Cichon, S. Ohlraun, H. Fangerau, T. Held, W. Maier, P. Propping, M. M. Nothen, and M. Rietschel. 2002. Association study between two variants in the DOPA decarboxylase gene in bipolar and unipolar affective disorder. Am. J. Med. Genet. 114519-522. [DOI] [PubMed] [Google Scholar]
- 33.Kirley, A., Z. Hawi, G. Daly, M. McCarron, C. Mullins, N. Millar, I. Waldman, M. Fitzgerald, and M. Gill. 2002. Dopaminergic system genes in ADHD: toward a biological hypothesis. Neuropsychopharmacology 27607-619. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi, K., S. Morita, H. Sawada, T. Mizuguchi, K. Yamada, I. Nagatsu, T. Hata, Y. Watanabe, K. Fujita, and T. Nagatsu. 1995. Targeted disruption of the tyrosine hydroxylase locus results in severe catecholamine depletion and perinatal lethality in mice. J. Biol. Chem. 27027235-27243. [DOI] [PubMed] [Google Scholar]
- 35.Kramer, H. H., H. J. Trampisch, S. Rammos, and A. Giese. 1990. Birth weight of children with congenital heart disease. Eur. J. Pediatr. 149752-757. [DOI] [PubMed] [Google Scholar]
- 36.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 92079-2089. [DOI] [PubMed] [Google Scholar]
- 37.Le Van Thai, A., E. Coste, J. M. Allen, R. D. Palmiter, and M. J. Weber. 1993. Identification of a neuron-specific promoter of human aromatic L-amino acid decarboxylase gene. Brain Res. Mol. Brain Res. 17227-238. [DOI] [PubMed] [Google Scholar]
- 38.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366302-303. [DOI] [PubMed] [Google Scholar]
- 39.Lin, S. P., N. Youngson, S. Takada, H. Seitz, W. Reik, M. Paulsen, J. Cavaille, and A. C. Ferguson-Smith. 2003. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 3597-102. [DOI] [PubMed] [Google Scholar]
- 40.Mager, J., N. D. Montgomery, F. P. de Villena, and T. Magnuson. 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33502-507. [DOI] [PubMed] [Google Scholar]
- 41.Malik, S., M. A. Cleves, W. Zhao, A. Correa, and C. A. Hobbs. 2007. Association between congenital heart defects and small for gestational age. Pediatrics 119e976-e982. [DOI] [PubMed] [Google Scholar]
- 42.Mancini-Dinardo, D., S. J. Steele, J. M. Levorse, R. S. Ingram, and S. M. Tilghman. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 201268-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monk, D., L. Bentley, M. Hitchins, R. A. Myler, J. Clayton-Smith, S. Ismail, S. M. Price, M. A. Preece, P. Stanier, and G. E. Moore. 2002. Chromosome 7p disruptions in Silver Russell syndrome: delineating an imprinted candidate gene region. Hum. Genet. 111376-387. [DOI] [PubMed] [Google Scholar]
- 44.Monk, D., E. L. Wakeling, V. Proud, M. Hitchins, S. N. Abu-Amero, P. Stanier, M. A. Preece, and G. E. Moore. 2000. Duplication of 7p11.2-p13, including GRB10, in Silver-Russell syndrome. Am. J. Hum. Genet. 6636-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon, Y. S., C. M. Smas, K. Lee, J. A. Villena, K. H. Kim, E. J. Yun, and H. S. Sul. 2002. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol. 225585-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, T., M. Constancia, M. Zubair, B. Bailleul, R. Feil, H. Sasaki, and W. Reik. 1997. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl. Acad. Sci. USA 9412509-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mostoslavsky, R., N. Singh, T. Tenzen, M. Goldmit, C. Gabay, S. Elizur, P. Qi, B. E. Reubinoff, A. Chess, H. Cedar, and Y. Bergman. 2001. Asynchronous replication and allelic exclusion in the immune system. Nature 414221-225. [DOI] [PubMed] [Google Scholar]
- 48.Murrell, A., S. Heeson, L. Bowden, M. Constancia, W. Dean, G. Kelsey, and W. Reik. 2001. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 21101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nebigil, C. G., D. S. Choi, A. Dierich, P. Hickel, M. Le Meur, N. Messaddeq, J. M. Launay, and L. Maroteaux. 2000. Serotonin 2B receptor is required for heart development. Proc. Natl. Acad. Sci. USA 979508-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nebigil, C. G., and L. Maroteaux. 2001. A novel role for serotonin in heart. Trends Cardiovasc. Med. 11329-335. [DOI] [PubMed] [Google Scholar]
- 51.Omichinski, J. G., P. V. Pedone, G. Felsenfeld, A. M. Gronenborn, and G. M. Clore. 1997. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat. Struct. Biol. 4122-132. [DOI] [PubMed] [Google Scholar]
- 52.Ono, R., K. Nakamura, K. Inoue, M. Naruse, T. Usami, N. Wakisaka-Saito, T. Hino, R. Suzuki-Migishima, N. Ogonuki, H. Miki, T. Kohda, A. Ogura, M. Yokoyama, T. Kaneko-Ishino, and F. Ishino. 2006. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38101-106. [DOI] [PubMed] [Google Scholar]
- 53.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plagge, A., and G. Kelsey. 2006. Imprinting the Gnas locus. Cytogenet. Genome Res. 113178-187. [DOI] [PubMed] [Google Scholar]
- 55.Portbury, A. L., R. Chandra, M. Groelle, M. K. McMillian, A. Elias, J. R. Herlong, M. Rios, S. Roffler-Tarlov, and D. M. Chikaraishi. 2003. Catecholamines act via a beta-adrenergic receptor to maintain fetal heart rate and survival. Am. J. Physiol. Heart Circ. Physiol. 284H2069-H2077. [DOI] [PubMed] [Google Scholar]
- 56.Rosenthal, G. L., P. D. Wilson, T. Permutt, J. A. Boughman, and C. Ferencz. 1991. Birth weight and cardiovascular malformations: a population-based study. The Baltimore-Washington Infant Study. Am. J. Epidemiol. 1331273-1281. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Schmidt, J. V., P. G. Matteson, B. K. Jones, X. J. Guan, and S. M. Tilghman. 2000. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 141997-2002. [PMC free article] [PubMed] [Google Scholar]
- 59.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 3366-69. [DOI] [PubMed] [Google Scholar]
- 60.Schulz, R., T. R. Menheniott, K. Woodfine, A. J. Wood, J. D. Choi, and R. J. Oakey. 2006. Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res. 34e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sleutels, F., G. Tjon, T. Ludwig, and D. P. Barlow. 2003. Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J. 223696-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sleutels, F., R. Zwart, and D. P. Barlow. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415810-813. [DOI] [PubMed] [Google Scholar]
- 63.Sumi-Ichinose, C., S. Hasegawa, H. Ichinose, H. Sawada, K. Kobayashi, M. Sakai, T. Fujii, H. Nomura, T. Nomura, I. Nagatsu, et al. 1995. Analysis of the alternative promoters that regulate tissue-specific expression of human aromatic L-amino acid decarboxylase. J. Neurochem. 64514-524. [DOI] [PubMed] [Google Scholar]
- 64.Sumi-Ichinose, C., H. Ichinose, E. Takahashi, T. Hori, and T. Nagatsu. 1992. Molecular cloning of genomic DNA and chromosomal assignment of the gene for human aromatic L-amino acid decarboxylase, the enzyme for catecholamine and serotonin biosynthesis. Biochemistry 312229-2238. [DOI] [PubMed] [Google Scholar]
- 65.Surani, M. A., S. C. Barton, and M. L. Norris. 1986. Nuclear transplantation in the mouse: heritable differences between parental genomes after activation of the embryonic genome. Cell 45127-136. [DOI] [PubMed] [Google Scholar]
- 66.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 123693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wakeling, E. L., S. Abu-Amero, S. M. Price, P. Stanier, R. C. Trembath, G. E. Moore, and M. A. Preece. 1998. Genetics of Silver-Russell syndrome. Horm. Res. 49(Suppl. 2)32-36. [PubMed] [Google Scholar]
- 68.Wang, Z. Q., M. R. Fung, D. P. Barlow, and E. F. Wagner. 1994. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 372464-467. [DOI] [PubMed] [Google Scholar]
- 69.Wilkins, R. C., and J. T. Lis. 1998. GAGA factor binding to DNA via a single trinucleotide sequence element. Nucleic Acids Res. 262672-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamasaki-Ishizaki, Y., T. Kayashima, C. K. Mapendano, H. Soejima, T. Ohta, H. Masuzaki, A. Kinoshita, T. Urano, K. Yoshiura, N. Matsumoto, T. Ishimaru, T. Mukai, N. Niikawa, and T. Kishino. 2007. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 27732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, T., T. E. Adamson, J. L. Resnick, S. Leff, R. Wevrick, U. Francke, N. A. Jenkins, N. G. Copeland, and C. I. Brannan. 1998. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat. Genet. 1925-31. [DOI] [PubMed] [Google Scholar]