Abstract
Our current concept postulates that histone acetylation is required for the recruitment of bromodomain-containing transcription complexes, such as the chromatin-remodeling machine SWI/SNF and the basal transcription factor TFIID. We generated simple NF-κB-dependent enhancers of increasing transcriptional strengths and found that the histone acetylation requirements for activation of transcription depended on the strengths of these enhancers. All enhancers function by recruiting SWI/SNF and TFIID to induce nucleosome sliding, a prerequisite for transcriptional activation. However, histone acetylation, although it occurs, is dispensable for TFIID and SWI/SNF recruitment by the strong enhancers, indicating that strong activators can overcome the chromatin barrier by directly recruiting the necessary transcriptional complexes. Weak enhancers depend on histone acetylation for recruitment, and this requirement is independent of a histone acetylation code. Thus, the need for nucleosome modifications is imposed on genes and translated according to the quality and strengths of the activators.
Regulation of gene transcription is the main mechanism by which cells control gene expression during development and adult life and in response to environmental signals and stresses (29). Activation or repression of transcription relies on the coordinated actions of many proteins, ranging from transcription factors to basal transcription factors and chromatin modifiers. The process of regulation of transcription begins with the binding of transcription factors to enhancers and promoters, forming higher-order nucleoprotein complexes termed enhanceosomes (25, 36). According to our current understanding, enhanceosomes work by recruiting to nearby promoters the various proteins required for the mechanics of RNA synthesis, along with coactivators of transcription and chromatin modifiers (21, 30). These recruiting reactions depend on simple adhesion interactions between complementary surfaces on activators and the recruited proteins (26). The question arises as to how so many different protein complexes unite to activate genes. Some of these recruited complexes, by modifying chromatin, increase the affinity for other complexes and thereby make the further recruiting task of enhanceosomes easier (13, 14, 35). However, the order of recruitment varies from gene to gene, since in some cases histone acetylation precedes and prepares the chromatin for remodeling (1, 8, 12), whereas in other cases remodeling precedes histone acetylation (5, 19, 20). Furthermore, there are cases in which components of the basal transcriptional machinery associate with nucleosomal DNA long before chromatin modification and remodeling (24, 32).
In the case of the human beta interferon (IFN-β) gene, the enhanceosome assembles on the nucleosome-free enhancer DNA and functions by initially recruiting the GCN5 histone acetyltransferase (HAT) complex. GCN5 acetylates a nucleosome masking the core promoter at specific lysine residues on histones H3 and H4, and this acetylation mark prints a code decrypted by transcriptional-regulatory proteins bearing bromodomains as follows. The nucleosome bearing acetylated histone H4 at K8, together with the enhanceosome-bound CBP, builds a three-dimensional adhesive surface used to recruit the SWI/SNF chromatin-remodeling machine, which modifies the histone-DNA contacts, thus allowing the subsequent recruitment of TFIID (1, 2). Recruitment of TFIID is also mediated by bivalent interactions with the acetylated histone H3 at K9 and K14 and the enhanceosome. The radical DNA bend induced by TFIID induces sliding of the SWI/SNF-modified nucleosome 36 nucleotides downstream to expose the core promoter, a prerequisite for initiation of transcription (22).
It was believed that the elaborate recruitment programs of chromatin modifiers and other transcriptional complexes contribute to temporal and cell-type-specific transcription (6, 10). However, this view was challenged by observations demonstrating that the transcriptional identity of the human IFN-β gene was lost when the local promoter chromatin structure was switched from inhibitory to permissive (23). In the permissive chromatin environment, the enhanceosome's adhesive surface instructs a recruitment program indistinguishable from the one operating in the wild-type (WT) gene, but in this case, the requirements for transcription were shifted. Thus, the inherent accessibility of the IFN-β core promoter allowed the assembly of the basal transcriptional machinery and initiation of transcription before the completion of the recruitment program of chromatin modifiers. Thus, the recruitment programs operating at the enhancers and promoters are hardwired in the enhanceosome's adhesive surface, but they do not suffice to determine the specificity of the transcriptional response (23). In addition, these experiments raised the question of the general role of nucleosome modifications in the regulation of gene transcription. If, indeed, covalent nucleosome modifications, such as histone acetylation, work by providing additional surfaces for the subsequent recruitment of other proteins, then one would expect that an increase in the recruiting power of activators (a stronger enhancer) would alleviate the requirement for histone acetylation (reviewed in reference 11). Previous studies of Saccharomyces cerevisiae have suggested that GAL4 working from strong sites does not require GCN5 (33) and that the requirement for SWI/SNF depends on the local chromatin structure (4). However, in these early studies, the actual role of histone acetylation and its direct relationship to chromatin remodeling were not investigated (4, 7, 33). Therefore, the question remained as to whether there are constraints imposed on genes regarding the general requirements and the interplay between nucleosome modifiers and chromatin remodeling in gene transcription. Here, we have addressed this problem by generating a series of synthetic enhancers of increasing transcriptional strengths composed of one, two, or four copies of the NF-κB site taken from the human IFN-β gene and placed upstream of the IFN-β core promoter bearing the inhibitory nucleosome. We found that upon activation this inhibitory nucleosome slides, as in the case of the natural IFN-β gene, to allow gene transcription. However, although the same set of complexes is recruited to the natural and synthetic genes, their individual roles differ. Strong synthetic enhancers can induce nucleosome sliding in the absence of histone acetylation, whereas weaker enhancers require histone acetylation. Furthermore, we found that chromatin remodeling is the rate-limiting step in the initial stages of transcriptional activation, whereas at the later stages, reinitiation of transcription determines the magnitude of the response, at least in part.
MATERIALS AND METHODS
Plasmid construction.
The PRDII1, PRDII2, and PRDII4 reporters were generated by cloning a double-stranded DNA oligonucleotide containing the PRDII element (5′-GGGAAATTCC-3′) upstream of the IFN-β core promoter (22). Constructs containing either the engineered NcoI site at +147 or the IFN-βslid core promoter, bearing a nucleosome artificially relocated to the +22 position, were made by replacing the IFN-β enhancer from the original template (22, 23) with each of the synthetic enhancers. The mammalian and bacterial expression vectors for WT p65, CBP, GCN5, and their derivatives have been described previously (26, 27, 38).
Cell culture and transfection.
HeLa cells were transfected by the calcium phosphate method, as previously described (36). In the transfection experiments, followed by chromatin immunoprecipitation and nucleosome mapping, the transfection cocktail for 150-mm plates contained 10 μg of reporter plasmid and 20 μg of expression vectors. To achieve a constant amount of transfected DNA, vector DNA was added in every case, as necessary.
In vivo nucleosome mapping.
HeLa cell monolayers (2.5 × 107) that had been transfected with the reporter plasmids and infected with Sendai virus or cotransfected with expression vectors were fixed with 1.1% formaldehyde for 45 min at 37°C. The cells were harvested and disrupted in a Dounce homogenizer in 0.3 M sucrose, 2 mM magnesium acetate, 3 mM calcium chloride, 1% Triton X-100, and 10 mM HEPES (pH 7.9). The lysate was spun twice through a pad of 25% glycerol, 5 mM magnesium acetate, 0.1 mM EDTA, pH 8.0, and 10 mM HEPES, pH 7.4, at 3,000 rpm for 5 min (4°C). The optical density at 260 nm, corresponding to DNA absorbance, was calculated for each sample in a small aliquot after reversing of the cross-link, proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. Next, nuclei corresponding to an optical density at 260 nm of 100 were resuspended in a buffer containing 25 mM KCl, 4 mM MgCl2, 1 mM CaCl2, 12.5% glycerol, 50 mM Tris, pH 7.4, as well as micrococcal nuclease at a 200 U/ml final concentration. Samples were incubated at 37°C for 10 min, and the reactions were stopped by adding an equal volume of 0.2 M NaCl, 10 mM EDTA, pH 8.0, 10 mM EGTA, pH 8.0, 50 mM Tris, pH 8.0, 2% sodium dodecyl sulfate, and 100 μg/ml proteinase K and further incubation at 37°C for 2 h. The cross-link was reversed by heating the mixture at 65°C overnight. The DNA was extracted, and the samples were incubated with RNase A (100 μg/ml), followed by phenol-chloroform extraction and ethanol precipitation. The DNA was separated on a 1.7% agarose gel, and fragments of an average size of 150 bp were purified, denatured, and hybridized with 2 ng of end-labeled primer. Primer extension was performed at 37°C using 1.25 units Sequenase in 1× Sequenase buffer supplemented with 0.13 mM deoxynucleoside triphosphates and 6.7 mM dithiothreitol. The sequence of the primer used was 5′-CAACGGTGGTATATCCAGTG-3′. The products were analyzed in 6% polyacrylamide sequencing gels and visualized with a phosphorimager.
Chromatin immunoprecipitation.
Chromatin preparation and immunoprecipitation experiments were carried out as previously described (1, 2). The following antibodies were purchased from Santa Cruz Biotechnology, Inc.: p65 (sc-109), BRG1 (sc-10768), GCN5 (sc-6303), CBP (sc-369), PolII (sc-9001), and TBP (sc-273). The following histone antibodies were obtained from Upstate Biotechnology: H4ac (06-866), H3ac (06-599), H4K5ac (06-759), H4K8 (06-760), H4K12ac (06-761), H4K16ac (06-762), H3K9ac (06-942), and H3K14ac (06-911). Precipitated promoters were detected by PCR analysis using the T7 and chloramphenicol acetyltransferase primers. The sequence of the chloramphenicol acetyltransferase primer was 5′-CCG TAA TAT CCA GCT GAA CGG TCT GGT-3′.
In vitro transcription, restriction site accessibility assay, and in vitro recruitment assay.
In vitro transcription was carried out exactly as previously described (38), except that templates bound by recombinant NF-κB were first incubated with the HeLa nuclear extracts (HNEs) for 30 min at 30°C to allow preinitiation complex (PIC) assembly, and then nucleoside triphosphates (NTPs) were added and the reaction mixtures were incubated at 30°C for another 30 min. Preparation of templates immobilized on paramagnetic beads was done by standard PCRs using ∼10 pmol from each primer and 10 ng plasmid template. The 5′ primer was biotinylated at the 5′ end. Products were run on 1.5% agarose gels, and bands corresponding to the correct size were extracted using the Qiagen gel extraction kit. Roughly 500 ng of purified linear template was used for coupling onto M280 streptavidin Dynabeads according to the manufacturer's instructions by using 50 to 100 μg of beads per reaction. Next, the templates were reconstituted to nucleosomes as described previously (1, 2, 22) and washed extensively before the addition of recombinant NF-κB. The assay was completed essentially as described by Yie et al. (37, 38).
In the case of restriction site accessibility assays, a biotinylated, 3′-radiolabeled fragment was prepared by using [γ-32P]ATP (10 mCi/ml; 6,000 Ci/mmol; NEN). After reconstitution to nucleosomes (1, 2, 22), the templates were incubated with HNEs for 30 min at 30°C and washed twice before digestion with NcoI. Digestions were carried out by incubation with 50 U/ml NcoI for 10 min at 37°C. Reactions were stopped by adding an equal volume of 0.4 M sodium acetate, 10 mM EDTA, 0.2% sodium dodecyl sulfate, 5 μg tRNA, and 20 μg proteinase K and further incubation at 37°C. Finally, the samples were phenol-chloroform extracted and ethanol precipitated before being loaded on a denaturing (5 M urea) 8% polyacrylamide gel.
RESULTS
Uncoupling chromatin remodeling from transcriptional synergy.
To investigate whether there is a direct correlation between the degree of chromatin remodeling and the magnitude of transcriptional response, we generated synthetic reporter constructs bearing one, two, or four copies of the IFN-β NF-κB site (PRDII) cloned immediately upstream of the IFN-β core promoter. These plasmids were introduced into HeLa cells, and their transcriptional activities were determined after virus infection, a treatment that induces NF-κB. Figure 1A shows that a single copy of PRDII responded marginally to virus infection, whereas two or four PRDII copies strongly induced transcriptional activity in response to virus infection. Importantly, the amount of transcription induced by four PRDII copies was approximately an order of magnitude higher than the amount induced by two PRDII copies, indicating transcriptional synergy.
FIG. 1.
Synergistic activation is independent of nucleosome sliding. (A) HeLa cells were transfected with luciferase reporter plasmids bearing one, two, or four copies of PRDII placed upstream of the IFN-β core promoter. Twenty-four hours later, the cells were mock or virus infected for 10 h before being harvested. The error bars indicate standard deviations. (B) HeLa cells were transfected as in panel A, except that after virus infection they were cross-linked with formaldehyde and treated with micrococcal nuclease to isolate mononucleosomes. Mononucleosomal DNA was purified and annealed with radiolabeled primer A, followed by extension. A sequencing gel containing the extended products run side by side with a DNA-sequencing reaction serving as size markers is shown. The diagram at the bottom of the figure depicts the chromatin architecture of the synthetic templates before and after virus infection. The efficiency of nucleosome sliding was calculated by measuring the intensities of the bands using a phosphorimager.
To determine the degree of chromatin remodeling in these synthetic templates, we carried out nucleosome-mapping experiments. We have previously shown that the nucleosomal organization of transfected IFN-β promoters is identical to that of the endogenous gene (22, 23). HeLa cells were transfected with the PRDII1, PRDII2, and PRDII4 reporters, followed by cross-linking of the histone DNA contacts and treatment with micrococcal nuclease of the isolated nuclei. The isolated mononucleosomal DNA was hybridized with primer A, followed by primer extension. Figure 1B shows that primer A produced a fragment of 118 bp when the mononucleosomes used were prepared from uninfected HeLa cells transfected with either PRDII1, PRDII2, or PRDII4 (lanes 1, 3, and 5), indicating that the nucleosome's 5′ border is at −15, a position identical with that of the nucleosome in the natural IFN-β promoter (22). However, virus infection produced four new major fragments, indicating nucleosome sliding, only at the templates bearing two or four PRDII copies (lanes 4 and 6). No detectable nucleosome sliding was observed at the template bearing a single PRDII element (lane 2). In contrast to the intact IFN-β promoter, on which the nucleosome slides to a fixed position at +20, we found that in the synthetic templates the nucleosome slid at four preferred positions (+19, +21, +24, and +25). The relative nucleosome-sliding efficiencies of PRDII1, PRDII2, and PRDII4 were 0.1%, 31%, and 52%, respectively. Thus, chromatin remodeling/nucleosome sliding are not the only mechanisms explaining the synergistic activation of transcription of PRDII4 compared to PRDII2.
To investigate the mechanisms of transcriptional synergy, we carried out in vitro transcription experiments using the synthetic enhancers in the presence or in the absence of chromatin. The DNA templates were incubated with saturating amounts of recombinant NF-κB, followed by incubation with HeLa nuclear extracts for 30 min to allow the assembly of preinitiation complexes. Next, NTPs were added, followed by the addition of Sarkosyl, which prevents the reassembly of initiation complexes, or buffer, and the samples were incubated for 30 min. Correctly initiated transcripts were detected by primer extension. Figure 2 shows that on naked DNA templates, a linear increase in the number of NF-κB binding sites (from one to four) led to an exponential increase in the magnitude of transcription. This was due both to an increase in the number of PICs assembled on the different promoters and to an increased number of rounds of transcription reinitiation. Interestingly, the presence of chromatin did not alter the above-mentioned ratios, but it decreased the overall amounts of transcription. Thus, the transcriptional synergy is an inherent property of the mechanisms of transcriptional activation, and it is not directly related to pathways involved in chromatin remodeling.
FIG. 2.
Transcriptional synergy is independent of chromatin remodeling. Shown is an in vitro transcription experiment using the synthetic PRDII reporters, naked or assembled into chromatin. Following the addition of recombinant NF-κB, the reaction mixtures were incubated with HNEs and Sarkosyl was added as indicated before the addition of NTPs. Correctly initiated transcripts were detected by autoradiography and quantitated using a phosphorimager. rel., relative.
Bypassing the requirement for histone acetylation in chromatin remodeling by increasing the strength of an enhancer.
The experiments described above suggest that NF-κB-dependent chromatin remodeling does not correlate with transcriptional synergy. NF-κB activates transcription by recruiting the CBP and GCN5 coactivators (26, 31, 39). Since both complexes possess HAT activity, we carried out experiments to determine the requirement for their HAT activities in transcriptional activation. HeLa cells were transfected with the PRDII1, PRDII2, and PRDII4 reporters, along with a small fixed amount of the p65 expression vector in the presence or in the absence of expression vectors for CBP and GCN5. Figure 3 (lane 2) shows that NF-κB activated transcription synergistically as the number of NF-κB molecules bound on the templates was increased, a result consistent with the experiments described above (Fig. 1). Coexpression of CBP, but not GCN5, led to a dramatic increase in p65-dependent transcription (Fig. 3, compare lane 1 with 3 and 5). However, replacement of the WT CBP expression vector with a vector expressing a CBP mutant lacking HAT activity (27) in the transfection experiments led to the abolishment of CBP's coactivation ability on the template bearing a single NF-κB site, but it had no effect on the template bearing four NF-κB sites (lane 4). Interestingly, CBP's HAT activity is only partially required for the synthetic enhancer bearing two NF-κB sites (lane 4). Taken together, these experiments suggest that strong NF-κB-dependent enhancers utilize CBP as a classical coactivator of transcription, whereas weak enhancers require CBP's HAT activity, presumably to modify chromatin.
FIG. 3.
Differential use of CBP's HAT activity by weak and strong enhancers. HeLa cells were transfected with the PRDII1, PRDII2, and PRDII4 reporters, along with expression vectors for p65, in the presence of WT CBP or GCN5 or derivatives bearing catalytic inactivating mutations in their HAT domains. Shown is the average of six independent experiments; the variability from experiment to experiment was less than 30%. The error bars indicate standard deviations.
To confirm the dual role of CBP in transcriptional-activation mechanisms, we determined the chromatin architectures of the PRDII1, PRDII2, and PRDII4 templates transfected with p65 in the presence or in the absence of WT CBP or CBP HAT−. Figure 4A shows that the low levels of transcription obtained from a single NF-κB binding site by transfected NF-κB are due to the absence of nucleosome sliding (compare lanes 1 and 2). However, overexpression of WT CBP, but not CBP HAT−, induced nucleosome sliding (compare lanes 3 and 4), a result consistent with the high and low levels of transcriptional activation, respectively (Fig. 3). Interestingly, NF-κB bound to the PRDII2 template induced nucleosome sliding, and this was maximized by expressing WT CBP, but not CBP HAT− (Fig. 4A, lanes 5 to 8). Finally, maximum nucleosome sliding was observed on the PRDII4 template even in the absence of transfected WT CBP or CBP HAT− (lanes 9 to 12). In summary, these experiments strongly suggest that the HAT activity of CBP is required for nucleosome sliding on weak enhancers only but that it is dispensable for nucleosome sliding and the transcriptional activity of strong enhancers.
FIG. 4.
Strong enhancers can induce chromatin remodeling and nucleosome sliding in the absence of histone acetylation. (A) HeLa cells were transfected with the PRDII1 (lanes 1 to 4), PRDII2 (lanes 5 to 8), and PRDII4 (lanes 9 to 12) reporters, along with expression vectors for p65, in the presence of WT CBP or CBP HAT−. Thirty-six hours after transfection, the cells were treated with formaldehyde, and the isolated nuclei were incubated with micrococcal nuclease. The nucleosome position was determined as in Fig. 1B. The two bands labeled with asterisks denote nonspecific products appearing in some experiments. (B) HeLa cells were transfected as described for panel A, except that following formaldehyde cross-linking, the isolated nuclei were sonicated and chromatin was purified and immunoprecipitated with the indicated antibodies. The precipitated DNA was subjected to PCR analysis using [α-32P]dCTP and plasmid-specific primers as indicated.
To investigate how CBP is used differentially by weak and strong enhancers, we carried out chromatin immunoprecipitation experiments using chromatin prepared from HeLa cells transfected with the synthetic templates in the absence or in the presence of WT CBP or CBP HAT−. Figure 4B (compare lanes 1 and 2) shows that transfected NF-κB bound to the PRDII1 template but was unable to recruit CBP, GCN5, BRG1, and basal transcription factors, an observation consistent with the absence of nucleosome sliding (Fig. 4A) and the very low levels of activated transcription (Fig. 3). However, overexpression of CBP led to its recruitment to the PRDII1 enhancer by NF-κB, together with GCN5, BRG1, and basal transcription factors (lane 3). As expected, the recruitment of CBP and GCN5 was correlated with histone H3 and H4 acetylation (lane 3). Replacement of WT CBP with CBP HAT− in the transfection cocktail did not induce histone acetylation, despite the fact that GCN5 was efficiently recruited to the promoter. The absence of histone acetylation was correlated with the lack of recruitment of BRG1 and basal factors (lane 4). These experiments further underscore the unique requirement for CBP's HAT activity in transcriptional activation by the weak PRDII1 NF-κB-dependent enhancer.
In contrast to PRDII1, NF-κB recruited the endogenous CBP and the associated GCN5 to the PRDII2 template, and this recruitment led to H3 and H4 acetylation and to the recruitment of BRG1 and basal transcription factors (Fig. 4B, lanes 5 to 8). Overexpression of WT CBP enhanced its recruitment to the promoter, and this was also correlated with increased histone acetylation and association of BRG1 and basal factors with the promoter, a result consistent with the increased levels of transcriptional activation. Unexpectedly, overexpression of CBP HAT− led to the recruitment of BRG1 and basal transcription factors in the absence of histone acetylation in both PRDII2 and PRDII4 templates (lanes 8 and 12). Most likely, the high levels of transfected CBP HAT− compete with the endogenous WT CBP for recruitment to the promoter. These results are in contrast with previous observations on the natural IFN-β enhancer showing an absolute requirement for histone acetylation for the recruitment of the bromodomain complexes BRG1 and TFIID and sliding of the nucleosome obstructing the core promoter (2).
Additional support for these findings was provided by the transfection experiment shown in Fig. 5. We generated reporter constructs bearing one, two, or four NF-κB copies placed upstream of the IFN-βslid core promoter, in which the nucleosome has been artificially relocated to the +22 position and therefore does not obstruct access of the basal transcriptional machinery (23). As seen in the figure, p65 activates transcription at higher levels from these templates than from those containing the natural IFN-β core promoter (compare Fig. 5 with Fig. 3). CBP coactivates transcription from all reporters, and this activation does not require its HAT activity, even from the PRDII1 template (Fig. 5, compare lanes 2, 3, and 4). Thus, even a weak enhancer, when placed in the appropriate chromatin environment, does not require nucleosome modifications for activation of transcription.
FIG. 5.
HeLa cells were transfected with reporter constructs bearing PRDII1, PRDII2, or PRDII4 cloned immediately upstream of the IFN-βslid core promoter, along with expression vectors for p65, in the presence of WT CBP or GCN5 or derivatives bearing catalytic inactivating mutations in their HAT domains. The error bars indicate standard deviations.
Nucleosome sliding in the absence of a histone acetylation code.
Further insights into the distinct molecular mechanisms leading to activation of the synthetic templates as opposed to the natural IFN-β enhancer was provided by the time course experiment shown in Fig. 6A. HeLa cells were transfected with the PRDII4 template and infected with Sendai virus for different amounts of time, followed by chromatin immunoprecipitation using the antibodies indicated in the figure. As seen in the figure, when the levels of bound NF-κB reached the maximum at 3 h postinfection (Fig. 6A, lane 4), CBP, BRG1, acetylated histones, and basal factors were detected on the promoter simultaneously (lane 4). Surprisingly, GCN5 was recruited at a later time point (lane 5), suggesting that its recruitment depends on another protein(s), presumably CBP. In the natural IFN-β enhancer case, GCN5 was recruited before CBP and acetylated histones, followed by the ordered recruitment of CBP, PolII, SWI/SNF, and TFIID (1). Thus, although similar complements of factors and cofactors mediate transcriptional activation by both the IFN-β enhanceosome and the synthetic NF-κB enhancer complex, their orders of recruitment and functions appear to be different, despite the fact that the chromatin architectures on the two promoters are identical.
FIG. 6.
The histone acetylation codes and the orders of recruitment differ between synthetic and natural IFN-β enhancers. (A) HeLa cells were transfected with the PRDII4 reporter construct, followed by virus infection for different amounts of time. Cross-linked chromatin was immunoprecipitated with the indicated antibodies, and the precipitated promoter sequences were detected by PCR using [α-32P]dCTP in the reaction mixture, along with plasmid-specific primers. (B) HeLa cells were transfected with PRDII1 (lanes 1 to 3) or PRDII4 (lanes 4 to 6), followed by chromatin immunoprecipitation using the indicated histone acetylation site-specific antibodies. Since the PRDII1 template cannot be efficiently activated by virus infection (Fig. 1) or p65 (Fig. 3) alone, we included in the transfection cocktail the CBP-expressing vector (lane 3) to promote high levels of transcription and to correlate this transcription with the acetylation of specific lysines in histones H3 and H4. Histone acetylation on the PRDII4 template was determined after its activation by either virus infection (lane 5) or p65 expression (lane 6).
So far, our results could be summarized as follows. (i) The PRDII1 template, which is bound by a single NF-κB molecule, is a weak transcriptional enhancer because it cannot efficiently recruit HAT proteins (CBP and GCN5), chromatin remodelers (BRG1), and basal transcription factors. (ii) An increase in the amount of CBP by transfection leads to its recruitment to PRDII1 by NF-κB and subsequently to transcriptional activation. (iii) Although CBP and GCN5 are both recruited to the synthetic enhancers, it is the CBP's HAT activity that acetylates the histones. However, in the context of the natural IFN-β enhancer/promoter, it is the HAT activity of GCN5 that prints the histone acetylation code (2). (iv) Powerful NF-κB-dependent enhancers can recruit the bromodomain-containing factors BRG1 and TFIID to induce nucleosome sliding in vivo and high levels of transcription in the absence of histone acetylation.
Since the biochemical cascade of activation by the synthetic enhancer appears to be different from that of the natural IFN-β enhanceosome, we reasoned that the histone acetylation code deciphered in the IFN-β case (2) would not be useful for activating the synthetic enhancers, although the local chromatin structures are identical. To address this point, we determined whether a specific histone acetylation pattern could be observed during activation of the PRDII1 and PRDII4 templates by virus infection or by expressing p65. Figure 6B shows that every lysine residue tested on histones H3 and H4 was acetylated by the recruited CBP protein.
To investigate the role of histone acetylation in chromatin remodeling, we carried out in vitro nucleosome-sliding experiments using assays of restriction site accessibility to immobilized templates to monitor the state of the remodeled nucleosomes (22). Linear DNA templates bearing one, two, or four copies of the PRDII element were biotinylated at one end and radiolabeled at the other end (Fig. 7, bottom). Next, the templates were attached to paramagnetic beads, reconstituted to nucleosomes using donor chromatin, and incubated with HeLa nuclear extracts in the presence or in the absence of recombinant NF-κB, followed by digestion with NcoI (Fig. 7). The released radioactive fragments were analyzed by polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography. These templates bear a natural NcoI site at −10 and a second engineered NcoI site at +147 (22). We had previously shown that the −10 and +147 sites are inaccessible and accessible, respectively, in the absence of transcription, but their accessibility is reversed due to nucleosome sliding and transcriptional activation (22). Figure 7 (lanes 1 and 2) shows that NF-κB bound to the PRDII1 template cannot induce nucleosome sliding after incubation with the HNE, a result consistent with the inability of a single NF-κB molecule to recruit CBP and promote histone acetylation in vivo. Interestingly, incubation of the chromatin templates with recombinant GCN5 or CBP and acetyl-coenzyme A (AcCoA) prior to incubation with the HNEs strongly induced nucleosome sliding (lanes 3 and 4), since the accessibility of the −10 site was increased and accessibility at the +147 site was strongly reduced. Furthermore, NF-κB bound to the PRDII2 template induced nucleosome sliding to some degree, and this was further enhanced by histone acetylation (lanes 5 to 8). Finally, NF-κB bound to the PRDII4 template fully induced nucleosome sliding, even in the absence of histone preacetylation (lanes 9 to 12). Taken together, these observations strongly suggest that the inability of weak enhancers to induce nucleosome sliding and transcriptional activation is due to their inability to promote histone acetylation. On the other hand, strong enhancers, like PRDII4, can induce nucleosome sliding and transcription in the absence of histone acetylation. To exclude the possibility that the basal level of acetylation of the donor chromatin prepared from HeLa cells might facilitate nucleosome sliding by the strong PRDII4 enhancer, we repeated the sliding experiment using recombinant nucleosomes assembled from bacterially expressed histones (2). Figure 7 (lanes 13 to 16) shows that the strong PRDII4 enhancer induces nucleosome sliding on chromatin templates containing recombinant histones, thus verifying that histone acetylation is dispensable for this process.
FIG. 7.
Nucleosome sliding in the absence of “epigenetic” modifications. DNA templates bearing one, two, or four copies of PRDII were reconstituted to nucleosomes using HeLa donor chromatin (lanes 1 to 12), acetylated by recombinant GCN5, and incubated with HeLa nuclear extracts, followed by extensive washes and NcoI digestion according to the scheme shown at the top. In lanes 13 to 16, the HeLa donor chromatin was replaced by recombinant core particles in the reconstitution reaction using the PRDII4 template. After NcoI digestion, the beads were concentrated, and the radioactive supernatant was analyzed by PAGE and detected by autoradiography. The diagram at the bottom depicts the restriction map, nucleosome positions, and locations of the detected radioactive fragments.
Recruitment of TFIID is the rate-limiting step in nucleosome sliding.
Ordinarily, on the natural IFN-β promoter, nucleosome sliding is a two-step process (22). First, the enhancer complex recruits SWI/SNF, which alters the histone-DNA contacts, and this recruitment requires histone acetylation. Second, the enhancer complex recruits TFIID, whose interaction with the SWI/SNF-remodeled nucleosome also requires histone acetylation. TFIID DNA binding is a prerequisite for nucleosome sliding via a DNA-bending-dependent mechanism. However, the strong synthetic PRDII4 enhancer can induce nucleosome sliding and high levels of transcription in the absence of histone acetylation, suggesting that recruitment of SWI/SNF and TFIID can occur via alternative pathways that are independent of histone acetylation. To determine the roles of histone acetylation and TFIID DNA binding in nucleosome sliding, we generated p65 derivatives compromised in CBP or TFIID recruitment. We had previously shown that deletion of the region of p65 encompassing amino acids 322 to 458 prevented interaction with CBP (26). A biotinylated DNA fragment containing the PRDII4 enhancer/promoter extending to +184 and bearing the nucleosome with or without recombinant NF-κB was incubated with nuclear extract and washed extensively, and the bound proteins were detected by Western blotting. Figure 8A shows that WT NF-κB (p50/p65) recruited CBP, GCN5, and TFIID (compare lanes 2 and 3). However, deletion of the CBP-interacting domain of p65 nearly eliminated both CBP and GCN5 recruitment (lane 5). Furthermore, deletion of the last 43 amino acids of p65 reduced TFIID recruitment, but it had no affect in CBP recruitment (lane 4). Thus, distinct domains in p65 are used for the recruitment of HATs and TFIID. The in vitro transcription experiment of Fig. 8B demonstrates that deletion of either the CBP- or TFIID-interacting domain of p65 reduced its ability to activate transcription in vitro from naked DNA templates in a similar manner (lanes 1 to 4). Deletion of the CBP-interacting domain only weakly affected transcription from chromatin templates, but deletion of the TFIID-interacting domain eliminated transcription (lanes 5 to 8). These experiments suggest that direct recruitment of TFIID by p65 is an absolute requirement for transcription on chromatin templates (9, 17) and that TFIID can be recruited by other means on naked DNA.
FIG. 8.
Recruitment of TFIID is the rate-limiting step in nucleosome sliding. (A) A biotinylated promoter fragment (−182 to +184) bearing four copies of the PRDII element was reconstituted to nucleosomes using HeLa donor chromatin, followed by incubation with the indicated recombinant NF-κB proteins. After being washed to remove unbound proteins, the templates were incubated with HeLa nuclear extracts and the bound proteins were detected by Western blotting using the antibodies shown on the left. (B) An in vitro transcription experiment using the templates shown on the right, either naked (lanes 1 to 4) or plasmids assembled into chromatin (lanes 5 to 8). The activators used are shown at the top. Correctly initiated transcripts were detected by reverse transcription using the appropriate primer. (C) An in vitro transcription experiment using the PRDII4 template reconstituted to nucleosomes, followed by incubation with the indicated recombinant NF-κB proteins. Next, the templates were reacted with recombinant GCN5 HAT protein in the presence of AcCoA (lanes 4, 8, 12, and 16) or GCN5 HAT alone (lanes 3, 7, 11, and 15) or AcCoA alone (lanes 2, 6,10, and 14), followed by incubation with HeLa nuclear extracts and NTPs. Correctly initiated transcripts were detected by reverse transcription using the appropriate primer. (D) A DNA template bearing four copies of PRDII (−182 to +184) was reconstituted to nucleosomes using HeLa donor chromatin, followed by acetylation using recombinant GCN5 protein. After being washed, the templates were incubated with the indicated recombinant NF-κB proteins and HeLa nuclear extracts supplemented with ATP. Thirty minutes later, the templates were washed and incubated with NcoI, and the radioactive supernatant was analyzed by PAGE and detected by autoradiography.
Since recruitment of TFIID is stabilized by acetylated histones (2, 15), we tested whether the requirement for direct recruitment of TFIID by activators could be bypassed by histone acetylation. The in vitro transcription experiment of Fig. 8C shows that prior histone acetylation by GCN5 or the CBP HAT domain (not shown) rescues the ability of the TFIID recruitment-deficient p65 derivative to activate transcription (compare lanes 11 and 12). The in vitro nucleosome-sliding experiment of Fig. 8D verifies that the ability of p65 derivatives to activate transcription depends on their property of promoting nucleosome sliding. More specifically, in agreement with the in vitro transcription experiment shown in Fig. 8C, the TFIID recruitment-deficient p65 derivative induces nucleosome sliding only after prior acetylation of the histones (Fig. 8D, lanes 5 to 8).
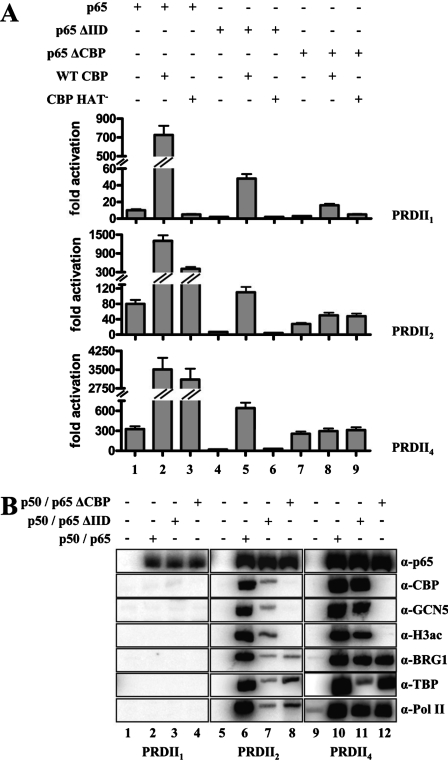
To determine whether a similar mechanism operates in vivo, we transfected cells with the PRDII1, PRDII2, and PRDII4 reporters in the presence or in the absence of the p65 derivatives and/or CBP. Figure 9A shows that deletion of the TFIID interaction domain in p65 dramatically reduced the levels of transcription (compare lanes 1 and 4), and these levels were increased by transfecting WT CBP, but not CBP HAT− (compare lanes 5 and 6). By contrast, the p65 derivative lacking the CBP interaction domain activated transcription from the PRDII4 template at nearly the same levels with WT p65, and these levels were not essentially affected by overexpressing CBP (lanes 7 to 9). However, there was a significant reduction of transcription from the PRDII2 template (compare lane 1 with 7), a result consistent with the requirement for CBP's HAT activity for full transcriptional activity (Fig. 3). The chromatin immunoprecipitation experiment of Fig. 9B verifies that the high levels of transcription obtained with p65 ΔCBP occurred in the absence of histone acetylation (lanes 8 and 12). Significantly, the low levels of transcription activated by p65 ΔIID correlated with the reduced recruitment of TFIID.
FIG. 9.
CBP recruitment is dispensable for transcriptional activation by strong enhancers. (A) HeLa cells were transfected with the PRDII1, PRDII2, and PRDII4 reporters, along with expression vectors for WT p65 or a p65 derivative lacking its CBP interaction domain (p65 ΔCBP) or a p65 derivative unable to interact with TFIID (p65 ΔIID) in the presence of WT CBP or CBP HAT−. Shown is the average of four independent experiments; the variability from experiment to experiment was less than 35%. The error bars indicate standard deviations. (B) HeLa cells were transfected with the PRDII1, PRDII2, and PRDII4 templates and the indicated NF-κB expression plasmids, followed by formaldehyde cross-linking. The isolated nuclei were sonicated, and the chromatin was purified and immunoprecipitated with the indicated antibodies. The precipitated DNA was subjected to PCR analysis using plasmid-specific primers and [α-32P]dCTP.
DISCUSSION
In this study, we used simple synthetic enhancers to answer many important questions related to the roles of epigenetic modifications in the mechanisms of transcriptional activation. Although a plethora of previous studies addressed the roles of epigenetic modifications in gene regulation, a significant number of important questions remained elusive. First, we asked whether the order of recruitment of factors, cofactors, and chromatin modifiers on promoters of genes in higher eukaryotes is determined by the local chromatin structure. In other words, is the order of recruitment predetermined by the local chromatin structure, or is it determined by the identity of the recruiter (enhanceosomes) and/or a combination thereof? Then, we addressed whether a well-characterized gene-specific inhibitory chromatin structure is always bypassed using the same histone modification “code” or whether there are multiple chromatin modification (epigenetic) “codes” that are printed and translated by the transcriptional machinery under the specific instructions of enhancer complexes. Thus, depending on the answers to these questions, it would be possible to draw rules regarding the roles of specific chromatin structures in regulating transcription and to test whether few, many, or a nearly infinite number of biochemical pathways operate during gene activation.
To address these points, we compared the pathways of transcriptional activation between synthetic genes, in which the natural IFN-β enhancer was replaced by simple enhancers of increasing transcriptional strengths (one, two, and four NF-κB sites), to that of the natural IFN-β gene. As is the case with the intact IFN-β gene, activation of transcription from these synthetic enhancers required nucleosome sliding to unmask the core promoter, thus allowing the assembly of a functional preinitiation complex. Thus, this specific chromatin barrier is bypassed by nucleosome sliding independently of the recruiter. However, the exact biochemical pathway and the requirements for activation are distinct from those of the natural enhancer, despite the fact that all bear the same inhibitory nucleosome. We found that the requirement for histone acetylation depends on the strength of each of the synthetic enhancers. That is, weak enhancers require histone acetylation for transcription, whereas strong enhancers can work independently of this modification, although both types of enhancers require nucleosome sliding for their functions. Furthermore, the synergistic activation of transcription obtained by increasing the strength of the enhancer (from one to two and four copies) is not directly related to the extents of chromatin remodeling and nucleosome sliding. Instead, we showed that chromatin remodeling and nucleosome sliding are correlated with the probability of a template promoting transcription, but they are not correlated with the magnitude of activated transcription. Part of the mechanism leading to high levels of transcription from the strong PRDII4 enhancer is related to its property of supporting more rounds of transcription than the moderate PRDII2 and the weak PRDII1 enhancers.
The inability of the weak enhancer bearing a single NF-κB copy to activate transcription is due to its inability to recruit chromatin modifiers and thus to promote chromatin remodeling and nucleosome sliding, prerequisites for activation of transcription. This recruitment inability is bypassed by increasing the concentration of cellular CBP by transfection, thus leading to its efficient recruitment, histone acetylation, and nucleosome sliding and therefore to high levels of transcription. The moderate enhancer PRDII2 depends on histone acetylation only partially, whereas the strong PRDII4 enhancer can work independently of histone acetylation. All types of enhancers induce nucleosome sliding and histone acetylation of the inhibitory nucleosome. We interpret these results as follows. A single molecule of an activator (NF-κB), albeit strong, cannot recruit in an ordered manner, or at once, all the necessary components of the chromatin modification and basal transcriptional machinery because the activating surface exposed for this recruitment is limited and presumably incapable of providing the minimal required surfaces to nucleate the assembly of a full PIC in the context of an inhibitory nucleosome. The order of recruitment in this case is critical, because the nucleosome needs to be modified and to slide before the assembly of a functional PIC. However, the same activating surface in the context of a permissive chromatin environment can induce transcription by recruiting the basal transcriptional machinery without the need for prior chromatin remodeling (Fig. 5). In the case of PRDII2, the higher concentration of activation domains located on the enhancer increases the probability of the simultaneous recruitment of both chromatin modifiers and general transcription factors (GTFs). Again, some of these recruited complexes might not lead to the assembly of a functional PIC because of the random appearance of recruited proteins. In this case, the HAT activity of CBP is critical for histone acetylation and for the subsequent recruitment of SWI/SNF and TFIID to set the stage for transcription. The case becomes radically different with the strong PRDII4 enhancer. This superactivator displays extended activation domain surfaces that can simultaneously recruit many protein complexes, including chromatin modifiers and GTFs. The chromatin modifiers alter the structure of the chromatin, allowing the already recruited GTFs to activate transcription.
This explanation is consistent with the observed lack of an order of recruitment of enzymatic activities on the strong synthetic PRDII4 template. The hypothesis was that the need to recruit a series of enzymatic activities in an orderly manner comes from the fact that regulation of transcription requires several steps, each one depending on a distinct type of chromatin-modifying activity used for the recruitment and/or stabilization of different sets of transcriptional-regulatory proteins (3, 6, 10, 18). For example, in the best-characterized case, the IFN-β enhancer/promoter, the initial recruitment of GCN5 is determined by the content of the early enhanceosomes, which display a higher affinity for GCN5 than for CBP and/or SWI/SNF. As the enhanceosome content changes dynamically over time (28) due to the arrival and departure of transcription factors, the “late” enhanceosomes recruit CBP and PolII, followed by the recruitment of SWI/SNF and TFIID. Thus, in this case, the order of recruitment is defined by the unique activation surface displayed each time by the enhanceosome in a way that ensures the sequential modification of chromatin, culminating with nucleosome sliding and activation of transcription. In sharp contrast, the homogeneous but extended activation surface exposed by the homopolymer PRDII4 enhancer leads to the simultaneous recruitment of all factors required for transcription. The adhesive surface of this enhancer ensures that many bromodomain-containing complexes (SWI/SNF, TFIID, etc.) that ordinarily require histone acetylation for their recruitment to the natural IFN-β enhancer (2) can be recruited directly to the synthetic enhancer without the need for prior histone acetylation. Thus, it is possible that the bromodomains present in SWI/SNF and TFIID may not be required for the recruitment and/or function of these complexes on the strong synthetic enhancers. Interestingly, weakening of the NF-κB activation domain by removing the CBP interaction domain did not significantly affect transcription in vivo or in vitro. This was because the remaining surfaces could efficiently recruit TFIID and SWI/SNF to promote nucleosome sliding. By contrast, deletion of the TFIID interaction domain abolished transcription, which could be recovered by prior histone acetylation. Thus, transcription would be activated by several other means as long as the bromodomain-containing complexes TFIID and SWI/SNF could be recruited to induce nucleosome sliding.
Finally, even in the case of CBP HAT-dependent activation of transcription from PRDII1, where histone acetylation is critical for nucleosome sliding, we found that every single lysine residue was acetylated, thus excluding the possibility of using the “code” as defined in the IFN-β case (2). These observations suggest that lysine-specific modifications occur only in the context of specific combinations of enhancer complexes and local chromatin, thus excluding the initial hypothesis that these modifications are always interpreted similarly by the transcriptional machinery (16, 34). Rather, many of these modifications will occur as long as the enzymes are recruited and have access to the chromatin template, but their roles would be dependent on the other corecruited proteins and the overall spatial structure of the transcriptional machinery.
Acknowledgments
We thank Mark Ptashne, Stavros Lomvardas, George Panayotou, and Ethan Ford for critical reading of the manuscript and helpful suggestions.
This work was supported by the Greek Secretariat for Research and Technology (PENED 01ED225), the Leukemia and Lymphoma Society of America, the March of Dimes, the Human Frontiers Science Program, Philip Morris USA Inc., and Philip Morris International.
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103667-678. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111381-392. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 4.Burns, L. G., and C. L. Peterson. 1997. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol. Cell. Biol. 174811-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97299-311. [DOI] [PubMed] [Google Scholar]
- 6.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10227-236. [DOI] [PubMed] [Google Scholar]
- 7.Dhasarathy, A., and M. P. Kladde. 2005. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 252698-2707. (Erratum, 26:388, 2006.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-driven chromatin remodeling activity and histone acetyl-transferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell 61049-1058. [DOI] [PubMed] [Google Scholar]
- 9.Dorris, D. R., and K. Struhl. 2000. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 124350-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Featherstone, M. 2002. Coactivators in transcription initiation: here are your orders. Curr. Opin. Genet. Dev. 2149-155. [DOI] [PubMed] [Google Scholar]
- 11.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodelling enzymes: who's on first? Curr. Biol. 11185-197. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Hörz. 1999. Chromatin remodeling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 186407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104817-827. [DOI] [PubMed] [Google Scholar]
- 14.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111369-379. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 2881422-1425. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Klein, C., and K. Struhl. 1994. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science 266280-282. [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 19.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 131412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102587-598. [DOI] [PubMed] [Google Scholar]
- 21.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 22.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106685-696. [DOI] [PubMed] [Google Scholar]
- 23.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110261-271. [DOI] [PubMed] [Google Scholar]
- 24.Martens, J. H., M. Verlaan, E. Kalkhoven, and A. Zantema. 2003. Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 231808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 2205-208. [DOI] [PubMed] [Google Scholar]
- 26.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 2277-287. [DOI] [PubMed] [Google Scholar]
- 27.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 4457-467. [DOI] [PubMed] [Google Scholar]
- 28.Munshi, N., T. Agalioti, S. Lomvardas, M. Merika, G. Chen, and D. Thanos. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 2931133-1136. [DOI] [PubMed] [Google Scholar]
- 29.Ptashne, M., and A. Gann. 2002. Genes and signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386569-577. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard, K. A., D. W. Rose, Z. K. Haque, R. Kurokawa, E. McInerney, S. Westin, D. Thanos, M. G. Rosenfeld, C. K. Glass, and T. Collins. 1999. Transcriptional activation by NF-κB requires multiple coactivators. Mol. Cell. Biol. 196367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 2951901-1904. [DOI] [PubMed] [Google Scholar]
- 33.Stafford, G. A., and R. H. Morse. 2001. GCN5 dependence of chromatin remodeling and transcriptional activation by the GAL4 and VP16 activation domains in budding yeast. Mol. Cell. Biol. 214568-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 35.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404414-417. [DOI] [PubMed] [Google Scholar]
- 36.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 831091-1100. [DOI] [PubMed] [Google Scholar]
- 37.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMGI(Y) in the assembly and function of the IFN-β enhanceosome. EMBO J. 183074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-β enhanceosome activates transcription. Proc. Natl. Acad. Sci. USA 9613108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 5661-671. [DOI] [PubMed] [Google Scholar]