Abstract
Histone H3 methylation at Lys27 (H3K27 methylation) is a hallmark of silent chromatin, while H3K4 methylation is associated with active chromatin regions. Here we report that a Drosophila JmjC family member, dUTX, specifically demethylates di- and trimethylated but not monomethylated H3K27. dUTX localization on chromatin correlates with the elongating form of RNA polymerase II (Pol II), and dUTX can associate with Pol II. Furthermore, heat shock induction results in the recruitment of dUTX to the hsp70 gene, like that of several other Pol II elongation factors. Our data indicate that dUTX is intimately associated with actively transcribed genes and may provide a paradigm for how H3K27 demethylation is required for the activation of preinitiated Pol II on transcriptionally poised genes.
Histone H3 methylation at lysine 27 (H3K27 methylation) is a chromatin modification that is enriched at silent genes and is mediated by Polycomb (PcG) group proteins (24, 28). PcG proteins are required for the stable maintenance of HOX gene silencing during animal development. In contrast, the trithorax (trxG) group of proteins mediates Lys4 methylation of H3 (H3K4 methylation), which is associated with the maintenance of open chromatin at HOX genes to ensure proper expression of these genes during development (28). The roles of these proteins in cellular memory, together with the stability of histone methylation relative to other histone modifications, have led to the assumption that H3K4 and H3K27 methylation marks are also stable. However, the recent discovery of histone lysine demethylases has led to a reappraisal of the stability of many histone methylation marks (18, 25, 27).
Previously, we and others demonstrated that the trithorax group gene little imaginal discs (lid) encodes a JmjC domain-containing histone demethylase that demethylates trimethylated H3K4 (H3K4me3) (9, 22, 26). While lid is genetically classified as a trxG member in Drosophila melanogaster, a human homolog of Lid, Jarid1D, has been found to associate with PcG proteins (20), suggesting that the cellular machinery responsible for the maintenance of Lys27 methylation and gene silencing works, in part, by demethylation of the activating H3K4 trimethyl mark. In a continuing effort to understand the cellular and molecular functions of the JmjC demethylases, we have been studying the biochemical and genetic properties of these factors. In this article, we describe the initial characterization of the Drosophila UTX homolog, a JmjC domain-containing histone demethylase that demethylates H3K27me3, a mark associated with Polycomb repression. We find that dUTX is closely associated with RNA polymerase II (Pol II) on actively transcribed genes, including heat shock loci, suggesting a possible role for H3K27 demethylation on preinitiated Pol II on transcriptionally poised genes.
MATERIALS AND METHODS
Antibodies.
Commercially available antibodies used in this study are as follows. Anti-mono-, di-, and trimethyl H3K27 (07448, 07452, and 07449), anti-trimethyl H3K4 (07473), and anti-trimethyl H3K9 (07442) were purchased from Upstate/Millipore. The anti-trimethyl H3K36 (ab9050), anti-trimethyl H3K79 (ab2621), and anti-trimethyl H4K20 (ab9053) antibodies were purchased from Abcam. The anti-FLAG antibody (F3165) was from Sigma. H5 and H14 (recognizing Ser2- and Ser5-phosphorylated Pol II) were purchased from Covance (MMS-129R and MMS-134R). Rabbit anti-dUTX custom antibodies were prepared against synthetic peptides at Pocono Rabbit Farm and Lab. The dUTX immunogen was the peptide Gly-Val-Glu-Ile-Arg-Phe-Asn-Gly-Arg-Gly-Lys-Asn-Glu-Ala-Ser-His-Tyr-Cys. The anti-dUTX antibody was affinity purified with Sulfolink resin (Pierce) bearing the immunizing peptide by using the manufacturer's protocol. Anti-HP1 monoclonal antibody C1A9 was a gift from Sarah Elgin.
Immunofluorescence.
Salivary gland polytene chromosome squashing and staining were done essentially as previously described (8).
Immunoprecipitation.
Immunoprecipitations were done as previously described (30), using 0.35 M NaCl in the buffer for nuclear extraction and washing of immunoprecipitates.
Heat shock chromatin immunoprecipitation.
Dmel-2 cells, a derivative of Schneider line 2 from Invitrogen, were grown to near-confluence in a 100-mm-diameter dish in SFX insect medium (HyClone). For heat shock, the medium was removed from a dish of cells growing at room temperature, and 10 ml of fresh medium prewarmed to 37°C was added. Cells were placed in a 37°C incubator for 5 min. The medium was aspirated from the dish, and 10 ml of room temperature medium was added. Two hundred seventy microliters of 37% formaldehyde was added to the dish, and cells were fixed for 10 min at room temperature to ensure that heat-shocked and non-heat-shocked cells cross-linked with the same efficiency. Quenching with glycine and the rest of the chromatin immunoprecipitation procedure were done as previously described (29). Hsp70 primers at 1 kb into the hsp70 transcribed region were CATCGACGAGGGATCTCTGTTC and GGCGCGAGGGTTGGA (4). Rp49 primers were AGAGTTCTTGTAACGTGGTCGGAATA and CAATGGTGCTGCTATCCCAATC. DNA obtained from the chromatin immunoprecipitation was measured by SYBR green real-time PCR using a Bio-Rad iCycler.
Histone demethylation assay.
Bulk histones (4 μg; Sigma) were incubated with 1 to 2 μg of recombinant proteins in a histone demethylase assay buffer [20 mM Tris-HCl (pH 7.3), 150 mM NaCl, 100 μM (NH4)2(SO4)2, 1 mM α-ketoglutarate, 2 mM ascorbate, 5% glycerol, and 0.2 mM phenylmethylsulfonyl fluoride] in a final volume of 10 μl for 5 to 7 h at 37°C. The reaction mixture was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting.
Fly stocks and crosses.
The UTX RNA interference (RNAi) fly stock (5640-R-2) was obtained from the National Institute of Genetics (Japan) and corresponds to an upstream activation sequence-Gal4-driven 500-nucleotide hairpin targeting all known isoforms of CG5640 (dUTX). RNAi transgene males were crossed to yw; Act5c-Gal4/CyO, y+ females. For polytene staining from dUTX knockdown larvae, we chose male larvae with y− mouth hooks. Adult flies were used for knockdown and control Western blot analysis. Flies were separated on the basis of straight or curly wings.
RESULTS AND DISCUSSION
Recently, the human UTX protein has been shown to function as an H3K27 demethylase (21). In order to learn about the cellular and molecular properties of this enzyme, we have identified its Drosophila homolog, dUTX (Fig. 1A). UTX was originally discovered as an X-linked homolog of a ubiquitously transcribed gene on the mammalian Y chromosome, UTY (14). In addition to a C-terminal JmjC domain, both UTX and UTY contain an N-terminal domain with several tetratricopeptide repeats, versatile protein binding modules found in a variety of proteins with diverse cellular functions (6). Remarkably, UTX was recently identified as a component of trithorax-related MLL3 and MLL4 complexes that mediate the methylation of H3 on lysine 4 (5, 16). Together, these findings reveal a new level of complexity in the regulation of gene expression by Polycomb and trithorax proteins through histone methylation and demethylation.
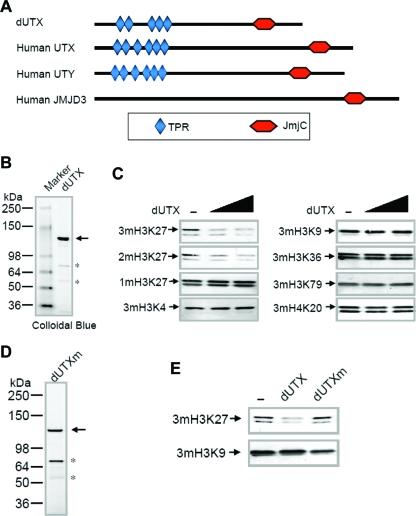
FIG. 1.
The Drosophila homolog of UTX is an H3 lysine 27 demethylase. (A) Schematic representation of the domain organization of Drosophila UTX (CG5640) and its human homologs. Members of the UTX family have a conserved JmjC domain at the C terminus and 5 to 6 tetratricopeptide repeats (TPR) at the N terminus. Human UTX and UTY are X and Y chromosome-linked homologs, respectively, from a pseudoautosomal portion of the sex chromosomes and are therefore highly similar. Since the Y-linked copy of such X-Y pairs generally mutates at a higher rate (13), the X-linked copy, UTX, can be considered more orthologous to the Drosophila homolog. dUTX shares higher overall sequence identity with UTX than with UTY or JMJD3. (B) Expression of recombinant dUTX in a baculovirus system. N-terminally Flag-tagged dUTX was purified from SF21 cells. Asterisks indicate nonspecific polypeptides. (C) Different amounts of recombinant dUTX (1 and 2 μg) were used in histone demethylation assays using bulk histones as substrates, and reaction mixtures were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting with antibodies to various sites of histone methylation. 1m, monomethylated; 2m, dimethylated; 3m, trimethylated. (D) Expression of a mutant version of dUTX (dUTXm) that bears double mutations (H883A and E885A) in the predicted iron-binding sites of the JmjC domain. Recombinant dUTXm was purified as described for panel B. (E) Comparison of H3K27 demethylase activities with 2 μg each of dUTX or dUTXm. Only the wild-type version of dUTX can demethylate H3K27me3, indicating that the observed enzymatic activity is intrinsic to the dUTX polypeptide.
To determine whether dUTX is a histone demethylase in vitro, recombinant dUTX was expressed in insect cells by using a baculovirus expression system (Fig. 1B). dUTX specifically demethylated di- and trimethylated but not monomethylated H3K27 when presented with a mixture of total histones (Fig. 1C). Furthermore, dUTX is not capable of removing other repressive chromatin marks, such as H3K9me3 or H4K20me3, or the sites of methylation marks correlated with active transcription, such as H3K4me3 and H3K36me3, in vitro (Fig. 1C). While H3K27 di- and trimethylated forms are found primarily at repressive chromatin sites, the monomethylated form of H3K27 is found in coding regions of transcribed genes (3). Polycomb has a strong preference for binding the trimethylated form of H3K27, suggesting that demethylation down to the di- or monomethylated form could disrupt Polycomb binding (10). To ensure that the observed demethylase activity was intrinsic to dUTX and not a copurifying protein, a dUTX mutant bearing double mutations in the JmjC domain was engineered and expressed in the baculovirus system (Fig. 1D). The purified mutant enzyme lacked catalytic activity, demonstrating that dUTX is an H3K27 demethylase (Fig. 1E).
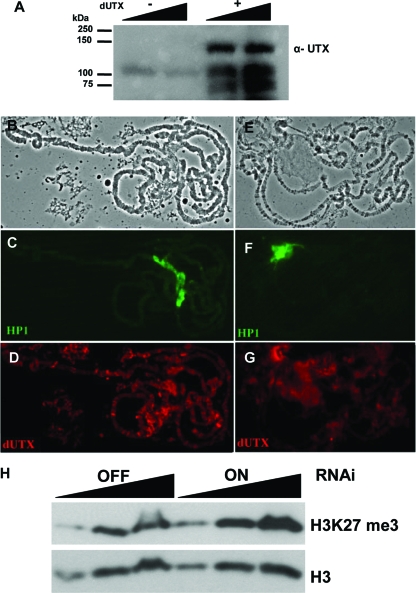
H3K27 methylation has been classically associated with the stable maintenance of transcriptional silencing. Therefore, it was of interest to determine the genomic distribution of dUTX. Anti-dUTX antibodies were made against a C-terminal peptide and tested for reactivity against full-length dUTX. The antibody recognized dUTX in SF21 cells infected with the Drosophila UTX baculovirus (Fig. 2A). To help validate the specificity of the antibodies, we used a dUTX-targeted RNAi transgenic line obtained from the National Institute of Genetics (Japan). Significant reductions in levels of immunostaining with an anti-dUTX antiserum were observed for knockdown compared to control polytene chromosomes from salivary glands (Fig. 2B to G). This RNAi line was also used to assess H3K27 methylation levels in extracts from adult flies. Modest levels of enrichment of H3K27 methylation were observed in total histones from adult flies after normalization to total-H3 levels (Fig. 2H). However, we were unable to detect significant changes in H3K27 methylation levels by immunofluorescence on polytene chromosomes of 3rd-instar larvae (data not shown). This may indicate that other factors in addition to dUTX are involved in H3K27 demethylation in vivo or that dUTX may function at specific loci throughout development.
FIG. 2.
Demethylase activity in vivo. (A) Western blot demonstrating that anti-dUTX antibodies (α-dUTX) recognize full-length dUTX protein. SF21 cells were infected with a dUTX baculovirus and probed with an affinity-purified rabbit polyclonal antibody recognizing dUTX. Two different amounts of total-cell extract from uninfected (−) and dUTX virus-infected (+) cells were loaded onto the gel. (B to G) Antibodies to dUTX were used to demonstrate reductions in dUTX protein levels after RNAi in Drosophila 3rd-instar larvae. Salivary glands from control (B to D) or RNAi (E to G) larvae were immunostained with anti-dUTX antibodies (D and G). HP1 levels for control and dUTX knockdown larvae were comparable (C and F). (B and E) Corresponding phase-contrast images. (H) H3K27 methylation levels are elevated in dUTX knockdown flies. Western blots of increasing amounts of total histone from lid RNAi knockdown (RNAi ON) or control (RNAi OFF) flies were probed with antibodies to H3K27 methylation and total histone H3.
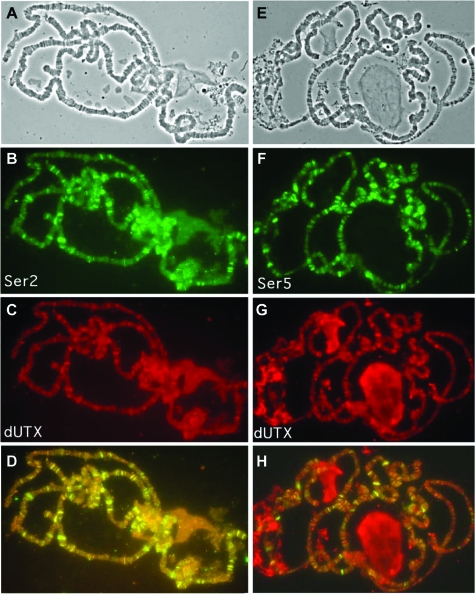
Since dUTX staining appeared to occur in interband regions, frequently sites of active transcription, we costained chromosomes with antibodies recognizing elongating and paused forms of Pol II (Fig. 3A to H). There was very extensive colocalization with the elongating form of RNA polymerase (Ser2-phosphorylated C-terminal domain [CTD]) and a lesser extent of colocalization with the engaged but paused form of RNA polymerase (Ser5-phosphorylated CTD). Although the Lys4 demethylase Lid also was present in interband regions, it did not colocalize with the elongating form of RNA polymerase (22), demonstrating distinct biological roles for these enzymes.
FIG. 3.
Colocalization of dUTX with the elongating form of RNA polymerase II (A through D), contrasted with the engaged but paused form (E through H). (B) Costaining for the Ser2-phosphorylated CTD of the elongating form of RNA polymerase. (C and G) Immunolocalization of dUTX on polytene chromosomes. (D) Merge of panels B and C, showing colocalization of Ser2-phosphorylated Pol II and dUTX. (F) Costaining for the engaged but paused Ser5-phosphorylated CTD of Pol II. (H) Merge of panels F and G, showing only partial colocalization. (A and E) Corresponding phase-contrast images.
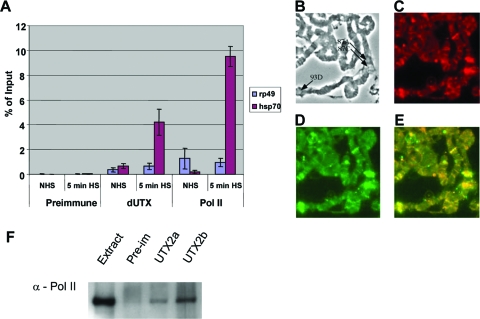
To test if dUTX could be recruited to a gene upon activation similarly to other elongation factors that we and others have tested (1, 4, 11, 12, 31), we used the hsp70 gene as a model inducible gene. Chromatin immunoprecipitation was performed before heat shock and after 5 min of heat shock of Schneider 2 cells. Antibodies directed to dUTX and RNA polymerase both immunoprecipitated increased levels of the hsp70 gene after heat shock (Fig. 4A). Consistent with the chromatin immunoprecipitation results from Schneider 2 cells, dUTX became highly enriched at major heat shock loci (puffs) on polytene chromosomes (Fig. 4B to E). Furthermore, we demonstrated that dUTX in cell extracts exists in complexes associated with Pol II (Fig. 4F).
FIG. 4.
dUTX is recruited to heat shock genes. (A) Chromatin immunoprecipitation from Schneider 2 (Dmal-2) cells before heat shock (no heat shock [NHS]) and after 5 min of heat shock (HS) with dUTX or RNA polymerase (monoclonal antibody 8WG16). Primers 1 kb into the heat shock gene were used to measure dUTX and Pol II recruitment to the heat shock gene. Rp49 levels are shown for comparison. The percentage of the input DNA recovered in the immunoprecipitate is shown along the y axis. Error bars represent ranges from two experiments. (B to E) Colocalization of dUTX on heat shock puffs of Drosophila salivary glands. (B) Phase-contrast image shows puffing at major heat shock loci, indicated by arrows. (C to E) dUTX (C) and the elongating, Ser2-phosphorylated form of Pol II (D) are colocalized (E) on the three heat shock loci indicated in panel B. (F) Two different antibodies to dUTX or a preimmune serum (Pre-Im) was used in an immunoprecipitation assay with a Drosophila nuclear extract. Interaction of Pol II with dUTX was detected with monoclonal antibody 8WG16.
The colocalization of an H3K27 demethylase with the elongating form of RNA polymerase was unexpected but is emblematic of how little we understand about the mechanisms of regulation of gene expression by histone modifications. The recent finding that human UTX associates with trithorax family members is consistent with a role for dUTX at transcribing genes (5, 16). Although no direct evidence exists for demethylation of Lys27 in coding regions, it is intriguing that a recent study found a large number of genes with paused polymerases (15). Lorincz and Schubeler (23) speculate that Polycomb group proteins could be responsible for the pausing of these polymerases. In this context, our finding of a Lys27 demethylase colocalizing with the elongating form of RNA polymerase suggests that demethylation could be a key step in the pathway by which these genes are activated.
ADDENDUM
While this article was under review, several groups reported demethylase activity for UTX and the related enzyme JMJD3 in humans (2, 7, 17, 19, 32).
Acknowledgments
We thank the National Institute for Genetics (Japan) for the UTX RNAi fly stock.
J.C.E. was supported by grants from the U.S. National Science Foundation (MCB 0131414 and MCB 0615831). R.S. was supported by a grant from the U.S. National Institutes of Health (GM61204). A.S. was supported by grants from the U.S. National Institutes of Health (CA089455 and GM069905).
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Adelman, K., W. Wei, M. B. Ardehali, J. Werner, B. Zhu, D. Reinberg, and J. T. Lis. 2006. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 26250-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agger, K., P. A. Cloos, J. Christensen, D. Pasini, S. Rose, J. Rappsilber, I. Issaeva, E. Canaani, A. E. Salcini, and K. Helin. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449731-734. [DOI] [PubMed] [Google Scholar]
- 3.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 4.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 237628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, Y. W., T. Hong, S. Hong, H. Guo, H. Yu, D. Kim, T. Guszczynski, G. R. Dressler, T. D. Copeland, M. Kalkum, and K. Ge. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 28220395-20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28655-662. [DOI] [PubMed] [Google Scholar]
- 7.De Santa, F., M. G. Totaro, E. Prosperini, S. Notarbartolo, G. Testa, and G. Natoli. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 1301083-1094. [DOI] [PubMed] [Google Scholar]
- 8.Eissenberg, J. C. 2006. Functional genomics of histone modification and non-histone chromosomal proteins using the polytene chromosomes of Drosophila. Methods 40360-364. [DOI] [PubMed] [Google Scholar]
- 9.Eissenberg, J. C., M. G. Lee, J. Schneider, A. Ilvarsonn, R. Shiekhattar, and A. Shilatifard. 2007. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 14344-346. [DOI] [PubMed] [Google Scholar]
- 10.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 171870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerber, M., J. Ma, K. Dean, J. C. Eissenberg, and A. Shilatifard. 2001. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J. 206104-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber, M., K. Tenney, J. W. Conaway, R. C. Conaway, J. C. Eissenberg, and A. Shilatifard. 2005. Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J. Biol. Chem. 2804017-4020. [DOI] [PubMed] [Google Scholar]
- 13.Goetting-Minesky, M. P., and K. D. Makova. 2006. Mammalian male mutation bias: impacts of generation time and regional variation in substitution rates. J. Mol. Evol. 63537-544. [DOI] [PubMed] [Google Scholar]
- 14.Greenfield, A., L. Carrel, D. Pennisi, C. Philippe, N. Quaderi, P. Siggers, K. Steiner, P. P. Tam, A. P. Monaco, H. F. Willard, and P. Koopman. 1998. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 7737-742. [DOI] [PubMed] [Google Scholar]
- 15.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Issaeva, I., Y. Zonis, T. Rozovskaia, K. Orlovsky, C. M. Croce, T. Nakamura, A. Mazo, L. Eisenbach, and E. Canaani. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 271889-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jepsen, K., D. Solum, T. Zhou, R. J. McEvilly, H. J. Kim, C. K. Glass, O. Hermanson, and M. G. Rosenfeld. 2007. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450415-419. [DOI] [PubMed] [Google Scholar]
- 18.Klose, R. J., and Y. Zhang. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8307-318. [DOI] [PubMed] [Google Scholar]
- 19.Lan, F., P. E. Bayliss, J. L. Rinn, J. R. Whetstine, J. K. Wang, S. Chen, S. Iwase, R. Alpatov, I. Issaeva, E. Canaani, T. M. Roberts, H. Y. Chang, and Y. Shi. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449689-694. [DOI] [PubMed] [Google Scholar]
- 20.Lee, M. G., J. Norman, A. Shilatifard, and R. Shiekhattar. 2007. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128877-887. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M. G., R. Villa, P. Trojer, J. Norman, K. P. Yan, D. Reinberg, L. Di Croce, and R. Shiekhattar. 2007. Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science 318447-450. [DOI] [PubMed] [Google Scholar]
- 22.Lee, N., J. Zhang, R. J. Klose, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2007. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat. Struct. Mol. Biol. 14341-343. [DOI] [PubMed] [Google Scholar]
- 23.Lorincz, M. C., and D. Schubeler. 2007. RNA polymerase II: just stopping by. Cell 13016-18. [DOI] [PubMed] [Google Scholar]
- 24.Ringrose, L., and R. Paro. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38413-443. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, J., and A. Shilatifard. 2006. Histone demethylation by hydroxylation: chemistry in action. ACS Chem. Biol. 175-81. [DOI] [PubMed] [Google Scholar]
- 26.Secombe, J., L. Li, L. Carlos, and R. N. Eisenman. 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21537-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, Y. 2007. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8829-833. [DOI] [PubMed] [Google Scholar]
- 28.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75243-269. [DOI] [PubMed] [Google Scholar]
- 29.Smith, E. R., C. D. Allis, and J. C. Lucchesi. 2001. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 27631483-31486. [DOI] [PubMed] [Google Scholar]
- 30.Smith, E. R., A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook, C. D. Allis, and J. C. Lucchesi. 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenney, K., M. Gerber, A. Ilvarsonn, J. Schneider, M. Gause, D. Dorsett, J. C. Eissenberg, and A. Shilatifard. 2006. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc. Natl. Acad. Sci. USA 10311970-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang, Y., Z. Zhu, G. Han, H. Lin, L. Xu, and C. D. Chen. 2007. JMJD3 is a histone H3K27 demethylase. Cell Res. 17850-857. [DOI] [PubMed] [Google Scholar]