Abstract
Replication-dependent histone mRNAs are the only eukaryotic cellular mRNAs that are not polyadenylated, ending instead in a conserved stem-loop. The 3′ end of histone mRNA is required for histone mRNA translation, as is the stem-loop binding protein (SLBP), which binds the 3′ end of histone mRNA. We have identified five conserved residues in a 15-amino-acid region in the amino-terminal portion of SLBP, each of which is required for translation. Using a yeast two-hybrid screen, we identified a novel protein, SLBP-interacting protein 1 (SLIP1), that specifically interacts with this region. Mutations in any of the residues required for translation reduces SLIP1 binding to SLBP. The expression of SLIP1 in Xenopus oocytes together with human SLBP stimulates translation of a reporter mRNA ending in the stem-loop but not a reporter with a poly(A) tail. The expression of SLIP1 in HeLa cells also stimulates the expression of a green fluorescent protein reporter mRNA ending in a stem-loop. RNA interference-mediated downregulation of endogenous SLIP1 reduces the rate of translation of endogenous histone mRNA and also reduces cell viability. SLIP1 may function by bridging the 3′ end of the histone mRNA with the 5′ end of the mRNA, similar to the mechanism of translation of polyadenylated mRNAs.
Metazoan replication-dependent histone mRNAs differ from other eukaryotic mRNAs in that they do not end in poly(A) tails. Instead, these mRNAs end in a conserved 26-nucleotide sequence that contains a stem-loop (3). The stem-loop binding protein (SLBP) binds to the stem-loop and is required for both histone pre-mRNA processing in the nucleus (4, 9, 26) and histone mRNA translation in the cytoplasm (7, 24). The efficient translation of polyadenylated mRNAs requires both the cap and the poly(A) tail (5). The role of the poly(A) tail in translation in both mammals and yeast (Saccharomyces cerevisiae) is well understood. The 5′ mRNA cap is bound by eukaryotic initiation factor 4E (eIF4E), which interacts with the large scaffold protein eIF4G. In mammals, the poly(A) tail is bound by PABP in the cytoplasm, and PABP also interacts directly with eIF4G (8). Direct biochemical experiments using purified yeast PABP, eIF4E, and eIF4G demonstrate that these three factors can effectively circularize the mRNA (34), and this structure is thought to both stabilize the mRNA and enhance the recycling of terminating ribosomes, thus increasing translation efficiency.
PABP can also interact with other factors, including PAIP1 (2) and PAIP2 (13), which act to stimulate and inhibit translation, respectively. PAIP1 and PAIP2 compete for the same site in the C-terminal region of PABP (13, 23), a site distinct from the binding site on PABP for eIF4G (12). The binding of the PAIP proteins to PABP acts to increase or decrease its affinity for eIF4G. When bound with PAIP2, PABP does not interact as strongly with eIF4G (10, 13), while PAIP1 bound to PABP stimulates the formation of the translation initiation complex.
Histone mRNAs, although they have a different 3′ end, may be translated by a similar mechanism as the “standard” polyadenylated mRNAs. In support of this possibility, the translation of histone mRNA in mammalian cells also requires both the cap and the stem-loop (6). Histone mRNA translation is uniquely regulated during oogenesis in Xenopus laevis. Interestingly, histone mRNAs with an oligo(A) tail added to the stem-loop are stored in a translationally inactive particle until oocyte maturation (1, 36). There are two SLBPs in Xenopus oocytes: xSLBP1, the orthologue of the mammalian SLBP, and xSLBP2, which is oocyte specific (32). There are no identified orthologues of xSLBP2 in mammals. Previous studies demonstrated that xSLBP1 is required for the efficient translation of histone mRNA and that xSLBP2 inhibits histone mRNA translation (24). xSLBP2 is bound to the stored translationally inactive histone mRNA in stage VI oocytes and is degraded during oocyte maturation, allowing the activation of translation by xSLBP1 (25, 32).
SLBP stimulates the translation of histone mRNA in Xenopus oocytes (7, 24) and in reticulocyte lysates (24). We identified a 15-amino-acid (aa) region in the amino-terminal domain of xSLBP1 (aa 68 to 83) which is essential for the activation of histone mRNA translation (24). Deletion or mutation of this sequence completely abolished the activity of xSLBP1 in translation. A region of xSLBP1 containing this sequence fused to xSLBP2 converted xSLBP2 to an activator, and an MS2-xSLBP1 or MS2-human SLBP (hSLBP) fusion protein activated the translation of a reporter mRNA ending in an MS2 site, indicating that the RNA binding domain (RBD) of xSLBP1 was not required for translation (7, 24).
Here we further characterize the specific sequences in SLBP required for translation and report the isolation of a protein, SLIP1 (SLBP-interacting protein 1), which specifically binds to the translation activation region of SLBP. We demonstrate that SLIP1, together with xSLBP1 or hSLBP, activates the translation of mRNAs ending in the histone stem-loop both in Xenopus oocytes and in mammalian cells. Further supporting a role for SLIP1 in translation, we find that SLIP1 interacts with eIF4GI and eIF4GII. We propose that SLIP1 stimulates histone mRNA translation by bridging between SLBP and the 5′ end of histone mRNA.
MATERIALS AND METHODS
Plasmid constructions.
Wild-type xSLBP1, xSLBP2, and hSLBP were cloned into the modified p64T vector pXFRM as previously described (32). Single or double amino acid substitutions to alanine were generated by site-directed mutagenesis. The xSLBP1 internal deletions xSLBP1Δ89-126 and xSLBP1Δ112-126 were generated by PCR. The chimeric mutant xSLBP1-(x2 89-126) was generated by deletion of 37 aa (aa 89 to 126) from xSLBP1 and insertion of 35 aa of xSLBP2 (aa 90 to 125) into the matching location. xSLBP 1-89-xSLBP2 has the first 89 aa of xSLBP1 fused to xSLBP2 starting at aa 90.
For directed yeast two-hybrid assays, xSLBP1 or indicated mutants were cloned into the pGBT8 vector by use of PCR, and hSLIP1 was cloned into pGAD10. Luc-poly(A), Luc-SL, Luc-TL, Luc-MS2, and CAT-A50 constructs have been previously described (6, 24).
hSLIP1 was cloned into the pGEX vector to generate GST-hSLIP1. Hemagglutinin (HA)-hSLIP1 was generated by cloning hSLIP1 into pcDNA3. To generate the siRNA1-resistant form of HA-hSLIP1 in pcDNA3.HA, we used primers to introduce conserved changes in the third bases of the five codons targeted by the small interfering RNA (siRNA).
In vitro transcription.
Luc-SL, Luc-TL, Luc-poly(A), Luc-MS2, and CAT-A50 were transcribed with T7 RNA polymerase in the presence (for experiments in oocytes) or the absence (for experiments in reticulocyte lysate) of a cap analogue as previously described (24). The clones encoding SLBPs and SLIP1 proteins from pXFRM (32) were linearized with EcoRI and transcribed with SP6 polymerase in the presence of a cap analogue.
In vitro translation.
SLBP, hSLIP1, eIF4GI, and eIF4GII were expressed in a reticulocyte lysate as described previously (24). One-third of the in vitro translation reactions were resolved by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE), and protein synthesis detected using a Storm 840 PhosphorImager. The remainder was used for the glutathione S-transferase (GST) pulldown experiments. hSLBP, hSLIP1, and MS2 viral coat proteins were expressed in reticulocyte lysate, and 1/20 of the reaction mixture was used for Western blot analysis against SLIP1 protein.
In vivo translation assay.
In vivo injection experiments and measurements of firefly luciferase activities in Xenopus oocytes were performed as described previously (24). The results were normalized to the expression of Luc-TL, which was set at 1. When Luc-poly(A) and Luc-SL mRNAs activities in Xenopus oocytes were compared, their activities were normalized to the amount of luciferase expressed in oocytes injected with buffer. RNA was isolated from oocytes by use of Trizol reagent (Invitrogen) as per the manufacturer's instructions. One oocyte equivalent of RNA was electrophoresed on a denaturing formaldehyde agarose gel and probed with a purified NcoI/XbaI-digested fragment of the Luc-SL plasmid containing the full-length firefly luciferase open reading frame.
Western blot analysis.
Proteins were extracted from oocytes as previously described (24). Proteins from HeLa cell lysates extracted in NP-40 lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 1× protease inhibitor cocktail [P8340; Sigma]) were used for Western blot analysis. Transfected or untransfected CG-1945 yeast cells were grown in yeast-peptone-dextrose medium to an optical density at 595 nm of 1.0 at 27°C, and protein extracts were analyzed on 12% SDS-PAGE gels and by Western blotting. A polyclonal antibody was generated in rabbits against full-length hSLIP1 as a GST fusion protein, and a second polyclonal antibody was generated in rabbit against a peptide containing the C terminus of hSLIP1 (CTPAAHKYYYSEVSD) (Pacific Immunology). Dilutions of antibodies used for Western blotting were as follows: rabbit polyclonal anti-xSLBP1 (32), rabbit polyclonal anti-hSLBP (33), rabbit polyclonal anti-hSLIP1 (full length), 1:500; rabbit polyclonal anti-hSLIP1 (C terminal), 1:1,000; mouse monoclonal anti-caspase 9 (MBL International), 1:1,000; rabbit polyclonal anti-eIF4GI (gift from Robert Rhoads, Louisiana State University Health Sciences Center); rabbit polyclonal anti-PTB (polypyrimidine tract binding protein), 1:2,000 (gift from Mariano Garcia-Blanco, Duke University); mouse monoclonal anti-HA, 1:1,000 (Covance); and horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G, 1:10,000 (GE Healthcare).
Directed yeast two-hybrid experiments.
The CG-1945 yeast strain was transformed with the pGBT8 bait plasmids encoding SLBPs under TRP1 selection and then with target plasmid pGAD10 bearing hSLIP1 (Leu). The cells were grown on medium lacking leucine and tryptophan. Four individual colonies for each interaction were plated on SD minimal medium lacking tryptophan, leucine, and histidine and containing 20 mM of 3-aminotriazole (33). The same colonies tested for the interaction were also analyzed for protein expression by Western blotting.
Immunoprecipitation.
HeLa cells stably expressing HA-hSLIP1 were grown to 70 to 80% confluence on 150-mm plates containing Dulbecco's modified Eagle medium (DMEM) including 10% fetal bovine serum and 1% Geneticin. Six plates were used for each experiment. In some experiments, cells were treated with 5 mM hydroxyurea (HU) 1 h prior to lysis. Cells were lysed on ice for 15 min in NP-40 lysis buffer and clarified by centrifugation at 15,000 × g for 10 min. Lysates were then incubated with either anti-HA (Covance) or anti-myc (Invitrogen) for 2 h at 4°C. Protein G beads were equilibrated with phosphate-buffered saline (PBS) including 0.07% NP-40 and resuspended in an equal volume of PBS. Fifty microliters of protein G beads was added to lysates and incubated for 1 h at 4°C. The beads were washed with lysis buffer and resuspended in 2× SDS sample buffer, and the proteins were detected by Western blotting. RNase treatment was carried out either by adding 10 μg/ml RNase A to the lysates prior to immunoprecipitation (11) or by treating the protein G beads with RNase after immunoprecipitation. Identical results were obtained with each approach.
Transfection with siRNAs and measurement of cell growth.
Two siRNAs, siRNA1 (UUAUACUCCUCUCUACUGGdTdT) and siRNA2 (AUGAUGGCGUAGCACAUGCdTdT) for hSLIP1 (lowercase letters indicate deoxynucleotides), were purchased from Dharmacon, Inc. The control siRNA C2 and hSLBP siRNA were previously described (29). Cells were harvested 48 h after transfection. Levels of the proteins were determined by Western blot analysis. For growth analysis, 2.5 × 105 HeLa cells/well in a six-well plate were cotransfected with the combinations of 180 nM control siRNA or HA-hSLIP1 siRNA and 500 ng of siRNA-resistant forms of HA-hSLIP1 or green fluorescent protein (GFP) expression constructs. Transfections were performed with Lipofectamine Regular (Life Tech) according to the manufacturer's instructions. At 24, 48, and 72 h posttransfection, cells were collected and viabilities determined by trypan blue exclusion. Cell cycle analysis of siRNA-treated cells was determined by propidium iodide staining followed by flow cytometry as previously described (29).
Cell culture and transient transfection.
Culturing 293T cells and transfection of cDNAs were carried out as described previously (28). Cell lines expressing HA-hSLIP1 were isolated by first transfecting cells with HA-hSLIP1 cloned in pcDNA3, and a stable population of cells resistant to G418 was selected. The cells were plated at low density and clones were isolated. These clones were expanded and the cells expressing HA-hSLIP1 were identified by Western blotting and immunofluorescence, and cultures with 100% of the cells expressing SLIP1 were utilized in the experiments. Apoptosis was induced in HeLa cells through a 6-h treatment of cells with 1 μM staurosporine, while cells treated with an equivalent volume of dimethyl sulfoxide served as a negative control.
Protein expression and purification.
Expression of hSLBP-His, GST-hSLBP-His, GST-hSLBPΔ1-68-His, GST-hSLBPΔ1-81-His, and GST-hSLBPΔ1-126-His in Sf9 insect cells was previously described (9), and these were purified as described previously (4).
GST-fused hSLIP1 proteins were expressed from pGEX vector in BL21(λDE3) cells. The proteins were purified from a clarified cell lysate incubated for 2 h at 4°C with 1 ml of a slurry of glutathione-Sepharose beads equilibrated with lysis buffer per liter of culture. Proteins were eluted with 5 mM glutathione (G4251; Sigma) and were quantified by SDS-PAGE using Coomassie staining or a Bio-Rad assay with bovine serum albumin as a standard.
GST pulldown assay.
Five to 10 micrograms of Escherichia coli-expressed GST-hSLIP1 was incubated with 5 to 8 μl [35S]methionine-labeled SLBP proteins on ice for 30 min. Protein mixtures were then added with 50 μl (50% [vol/vol]) glutathione 4B Sepharose beads (Amersham) equilibrated in PBS and incubated for an additional 30 min on ice. Beads were washed three times with PBS supplemented with 200 mM NaCl. Proteins bound to beads were recovered in 40 μl 2× SDS sample buffer. Reciprocal experiments were performed with the same strategy as described above, in which baculovirus-expressed GST-hSLBP and its mutants were utilized to pull down hSLIP1 labeled with [35S]methionine. The GST proteins were isolated on glutathione beads, and the proteins were solubilized by boiling in sample buffer and then resolved by SDS-PAGE. The gel was dried and the proteins were detected by autoradiography or with a PhosphorImager. The direct pulldown of baculovirus-expressed His-hSLBP by GST-hSLIP1 was analyzed by Western blotting.
Pulldowns of [35S]methionine-labeled eIF4GI, eIF4GII, and their fragments were performed with GST-hSLIP1 as described above. The GST proteins were isolated on glutathione beads, and the proteins solubilized by boiling in sample buffer and then resolved by SDS-PAGE. The gel was dried and the proteins were detected by autoradiography or with a PhosphorImager.
Expression of the GFP-SL reporter.
293T cells were transfected with plasmids encoding either LacZ (control) or myc-tagged ZFP100 (28) by use of Lipofectamine 2000 (Invitrogen, CA) according to the manufacturer's protocols. The following day, 2 μg of GFP-SL reporter (31) was cotransfected with increasing amounts of plasmid encoding hSLIP1 and decreasing amounts of plasmid encoding LacZ by use of Lipofectamine reagent (Invitrogen, CA). Note that there was a total of 4 μg of plasmid present in all transfections, thus allowing for equivalent transfection efficiencies. Two days after the second transfection, cells were harvested and measured by fluorescence-activated cell sorter (FACS) analysis and analyzed using Summit software (Fort Collins, CO). Total cell RNA was analyzed for reporter mRNA with an S1 nuclease protection assay (31) or using a Northern blot analysis of cellular RNA (2 μg) that was separated by electrophoresis using a 6% denaturing acrylamide gel.
Pulse-labeling of cellular proteins.
HeLa cells were transfected with siRNAs as described above. Following the siRNA treatment for 72 h, cells were starved for methionine for 30 min at 37°C in DMEM starvation medium (DMEM lacking methionine and supplemented with 1 mM sodium pyruvate, 0.25 mM HEPES [pH 7.3], and 2 mM l-glutamine). Cells were then exchanged into starvation medium supplemented with [35S]methionine at 10 μl/ml (0.1 mCi/ml) and labeled for 10 min at 37°C. Cells were washed three times with cold 1× PBS, scraped into 1× PBS, and pelleted at 2,000 rpm for 5 min at 4°C. The cell pellet was lysed in a nuclear isolation buffer (250 mM sucrose, 1 mM CaCl2, 2 mM MgCl2, 1% Triton X-100, 10 mM Tris-HCl [pH 8], and 2 mM phenylmethylsulfonyl fluoride) for 60 min on ice. Nuclei were pelleted at 5,000 rpm for 5 min at 4°C. The nuclear pellet was resuspended in 0.4 N HCl and incubated on ice for 30 min. Nuclear samples were centrifuged at 15,000 rpm for 15 min at 4°C and acid-soluble proteins precipitated in 20% trichloroacetic acid at −20°C overnight. Samples were then centrifuged at 15,000 rpm for 30 min at 4°C. Protein pellets were washed three times with cold acetone, dried, and resuspended in sample buffer for SDS-PAGE analysis and autoradiography.
RESULTS
The region between aa 85 and the RBD is not required for translation.
Our initial experiments had defined a 15-aa region of xSLBP1 (aa 68 to 83) required for translation and demonstrated that the amino-terminal 68 aa and C-terminal 57 aa were not essential for translation (24). Using an MS2-SLBP fusion protein, Gorgoni and coworkers demonstrated that aa 1 to 127 fused to MS2 had the same stimulatory activity on histone mRNA translation as the full-length protein (7). We had not tested whether there might also be a role for aa 83 to 126, located between the translation activation region and the start of the RBD. We created mutants in SLBP to address whether this region also plays a role in translation using Xenopus oocytes.
The assay used to measure the activity of the stem-loop reporters (24) is diagrammed in Fig. 1A. Control oocytes were injected with buffer or with mRNA encoding xSLBP2, which is inactive in translation. Briefly, oocytes were injected with synthetic capped mRNAs encoding the proteins of interest, incubated for 16 h to allow protein expression, and then injected with the reporter mRNAs, luciferase mRNAs ending in either a stem-loop (Luc-SL) or a tetraloop which does not bind SLBP (Luc-TL), or a poly(A) tail [Luc-poly(A)]. Sixteen hours later, the oocytes were harvested and the luciferase activity was measured. The activity of the Luc-TL was used as a control, and its activity was set at 1. The levels of the expressed proteins were determined by Western blotting. Note that there is some xSLBP1 in the oocyte, and thus oocytes injected with buffer show a small activation of translation of the Luc-SL reporter (24) (Fig. 1C).
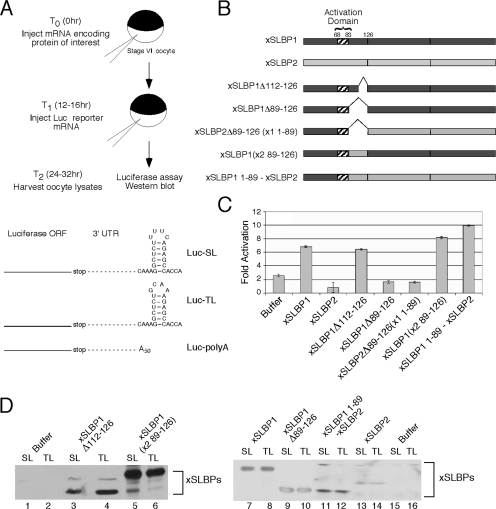
FIG. 1.
Analysis of activity of deletion mutants of xSLBP1 in Xenopus oocytes. (A) Schematic of Xenopus oocyte system used as an in vivo translation assay (24). The structures of the luciferase reporters Luc-SL, Luc-TL, and Luc-poly(A) are shown. Abbreviations: ORF, open reading frame; UTR, untranslated region. (B) The structure of the mutant proteins. The central domain of the xSLBPs contains the RBD, beginning with amino acid 126, and the translation activation domain of xSLBP1 is indicated (amino acids 68 to 83). (C) Synthetic mRNAs encoding each of these proteins were injected into Xenopus oocytes, and 16 h later the Luc-SL or Luc-TL mRNAs were injected. Sixteen hours later, the oocytes were harvested and luciferase activity was measured. Results are normalized to the Luc-TL activity. (D) The levels of the expressed proteins in the experiment shown in panel C were analyzed by Western blotting using anti-xSLBP1 antibody (which cross-reacts weakly with xSLBP2 (lanes 13 and 14) to demonstrate the expression of the mutant proteins.
To determine whether the region between the translation activation domain and the RBD of SLBP is required for translation, we created the SLBP mutants shown in Fig. 1B. We deleted the region between aa 112 and 126 (xSLBP1Δ112-126) and also that between 89 and 126 (xSLBP1Δ89-126). We replaced aa 1 to 88 of xSLBP2 with the same region of xSLBP1 (xSLBP1 1-89-xSLBP2), which resulted in a fusion protein that contained just the first 89 aa from xSLBP1. We then deleted the region between aa 89 and the RBD in this clone to give xSLBP2Δ89-126 (x1 1-89). To determine whether a specific sequence was required at positions 89 to 126, we replaced aa 89 to 126 of xSLBP1 with those from xSLBP2 [xSLBP1-(x2 89-126)].
Deletion of the 37 aa from 89 to 126 (xSLBP1Δ89-126) abolished translation activation activity in Xenopus oocytes, while deletion of only 14 aa (the serine-rich linker region) from 112 to 126 (xSLBP1Δ112-126) had no effect on translation (Fig. 1C). However, when the 37 aa from xSLBP2 were substituted for aa 89 to 126 of xSLBP1 [xSLBP1-(x2 89-126)], the resulting protein had complete activity, even though there is no similarity between xSLBP1 and xSLBP2 in this region. This result demonstrates that the effect of the deletion is likely to alter the spacing between the translation activation region and the RBD and that specific sequences in this region are not required for translation. Similarly, a protein that contains only the first 89 aa of xSLBP1 followed by the remainder of the xSLBP2 protein (xSLBP1 1-89-xSLBP2) had full activity in translation (Fig. 1C), while a protein that has aa 89 to 126 deleted from this protein [xSLBP2Δ89-126 (x1 1-89)] was inactive. All of these proteins were overexpressed in the oocytes as assayed by Western blotting using xSLBP1 antibody (Fig. 1D).
These results demonstrate that specific sequences in xSLBP1 between aa 83 and 126 were not required for translation, but an appropriate spacing between the RBD and the translation activation region was important for translation activation. Since we have previously shown that the removal of the first 67 aa of xSLBP1 and the entire C-terminal domain has no effect on translation activity (24), we conclude that the only essential region for translation lies between aa 68 and 83.
Identification of critical residues in the translation activation region of SLBP.
Comparison of the SLBP sequences from vertebrates reveals that there is extensive conservation of much of the SLBP sequence throughout the protein (33). However, when we included sea urchin (22) and Ciona intestinalis (sea squirt) in the comparison, then the similarity among the SLBPs is only in the RBD and the region we identified as important for translation. The sequences in this region for human, Xenopus, sea urchin, and Ciona are compared in Fig. 2A. Extending the comparison to more evolutionarily distant organisms (Drosophila melanogaster and Caenorhabditis elegans) results in a loss of obvious similarity among the SLBPs other than in the RBD, and the Drosophila SLBP is not active in translation in Xenopus oocytes (Sanchez and Marzluff, unpublished). The sequence comparison suggested that a consensus core sequence required for translation might be DWX3VEE, with the invariant amino acids underlined.
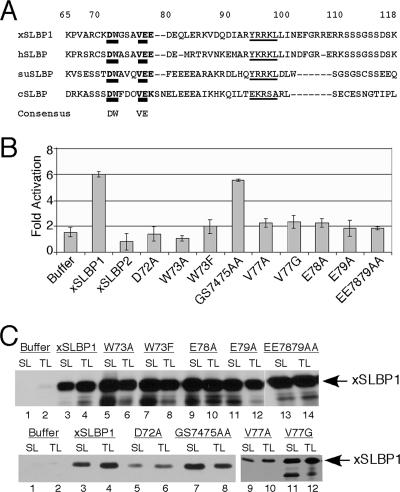
FIG. 2.
Identification of amino acids in the translation activation region of SLBP required for activity. (A) The sequences of xSLBP1, hSLBP, sea urchin SLBP (suSLBP), and Ciona SLBP (cSLBP) are compared in the region just before the RBD. The numbering is for xSLBP1. The thickly underlined region(s) within 72 to 79 is the consensus core translation activation region, and the thinly underlined region from 95 to 99 is the cyclin binding site required for SLBP degradation (38). (B) Wild-type xSLBP1, xSLBP2, or the indicated mutants of xSLBP1 were expressed in Xenopus oocytes by injection of synthetic capped mRNA as described in Fig. 1A. Results are expressed relative to the expression of the Luc-TL mRNA, which was set at 1. Oocytes injected with buffer served as a control. They have activity higher than 1 because of the endogenous xSLBP1 in the oocyte. For each point, 12 oocytes were pooled. The results are an average of three independent experiments using oocytes from different frogs. (C) Protein from the equivalent of 0.5 oocyte that had been injected with either Luc-SL or Luc-TL reporter in Fig. 2B was resolved on 12% SDS-PAGE. The amounts of wild-type xSLBP1 and xSLBP1 mutants were determined by Western blot analysis.
Based on this analysis, we made a number of point mutants in the consensus sequence in xSLBP1 and tested them for activity in translation. We assayed the ability of the mutant xSLBP1s to enhance translation of a luciferase reporter mRNA ending with a histone stem-loop in Xenopus oocytes (24) by use of the experimental system shown in Fig. 1A. Synthetic mRNAs encoding xSLBP1 mutants were injected into stage VI oocytes to express the SLBP. Control oocytes were injected with buffer or with mRNA encoding xSLBP2, which binds the stem-loop but is inactive in translation. Oocytes were then injected with the Luc-SL or Luc-TL reporter mRNA and 16 h later assayed for luciferase activity (Fig. 2B). The expression of all the proteins was monitored by Western blotting (Fig. 2C).
Point mutations in the conserved residues reduced activity of xSLBP1 in this assay (Fig. 2B). Mutation of aspartic acid to alanine (D72A), tryptophan to phenylalanine or alanine (W73F and W73A), valine to glycine or alanine (V77G and V77A), or glutamic acid to alanines (E78A, E79A, and EE7879AA) abolished the translation stimulation activity of SLBP. In contrast, mutation of the nonconserved amino acids glycine and serine to alanines (GS7475AA) did not affect the ability of SLBP to enhance translation of the stem-loop reporter mRNA. These results demonstrate that each of the conserved amino acids is essential in this system. All of the mutant proteins were expressed in excess of endogenous xSLBP1 in Xenopus oocytes (Fig. 2C), although the D72A and V77A mutants were expressed at lower levels than the other proteins.
Identification of a factor that binds the region of SLBP required for translation activation.
We performed a yeast two-hybrid screen with hSLBP lacking the last 27 aa as the bait (the last 27 aa contains an intrinsic trans-activation activity), against a HeLa cell cDNA library. One of the clones isolated, hSLIP1, encoded a 222-aa protein that has not previously been functionally characterized (Fig. 3A). Comparing this clone with the expressed sequence tag (EST) database as well as the mouse genome and EST databases suggested that it encoded a full-length polypeptide plus an extensive portion of the 5′ untranslated region which remained in frame with the initiator AUG. An antibody prepared to the C-terminal peptide of hSLIP1 interacts with a single polypeptide in HeLa cells, which is identical in mobility to the polypeptide produced from the cloned SLIP1 in reticulocyte lysates (Fig. 3B). Although there are potential alternative spliced forms of SLIP1 in the EST databases, we have no evidence for any alternatively spliced forms of SLIP1 expressed in significant amounts (Fig. 3B). The sequence of SLIP1 (Fig. 3A) is most similar to a number of translation factors. The structure of the zebrafish orthologue of SLIP1 has recently been determined in a structure genomics project (PDB 2I2OA and 2I2OB), and the protein contains a number of HEAT repeats which are similar to those found in the middle domain of eIF4G and CBP80 (17, 18).
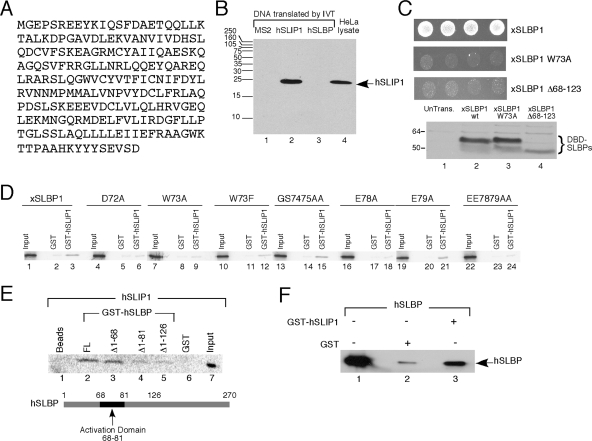
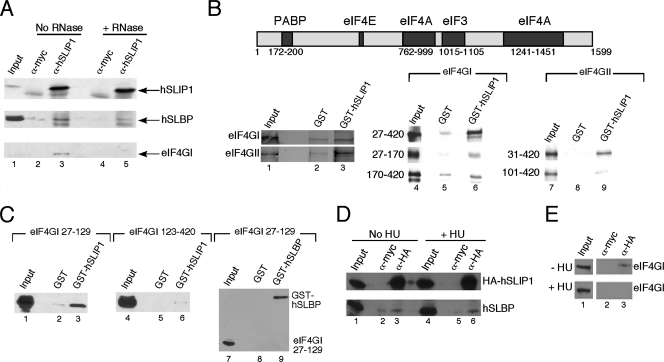
FIG. 3.
SLIP1 interacts with the translation activation domain of SLBP. (A) The predicted protein sequence of the SLIP1 protein (accession number EU287989). (B) MS2 viral coat protein (lane 1), hSLIP1 (lane 2), or hSLBP (lane 3) was synthesized by in vitro translation (IVT) in the reticulocyte lysate. An aliquot of the lysate was resolved by gel electrophoresis together with a lysate from HeLa cells (lane 4). hSLIP1 protein was detected by Western blotting using the antibody against the C-terminal peptide of hSLIP1. (C) Directed yeast two-hybrid assays between xSLBP1 and hSLIP1. Four independent yeast colonies containing the two-hybrid vectors were spotted on plates lacking L, W, and H in the presence of 20 mM 3-aminotriazole, and growth was observed. The wild-type (wt) xSLBP1, the W73A mutant, and a mutant with a deletion of aa 68 to 123 of xSLBP1 were analyzed. Expression levels of wild-type and mutant xSLBP1s were assayed by Western blotting. (D) Full-length xSLBP1 and the indicated mutant proteins were labeled with [35S]methionine by in vitro translation, GST (lanes 2, 5, 8, 11, 14, 17, 20, and 23) or GST-hSLIP1 (lanes 3, 6, 9, 12, 15, 18, 21, and 24) was added, and then the GST proteins were isolated on glutathione agarose. The bound proteins were resolved by gel electrophoresis and detected using a PhosphorImager. Lanes 1, 4, 7, 10, 13, 16, 19, and 22 are 10% of the input used in the pulldown assays. (E) hSLIP1 was labeled with [35S]methionine by in vitro translation and GST or the indicated GST-hSLBP fusion proteins were added and the bound proteins were detected as described for panel D. Lane 7 is 10% of the input for each reaction. A schematic of the hSLBP showing the translation activation domain is below the figure. (F) Recombinant His-hSLBP (lane 1) was incubated with GST (lane 2) or recombinant GST-hSLIP1 (lane 3). The proteins bound to glutathione agarose were detected by Western blotting using anti-hSLBP. The same amount of hSLBP (lane 1) was used in all reactions.
We confirmed the interaction of SLBP and SLIP1 and mapped the region of SLIP1 and SLBP that interact both by directed yeast two-hybrid experiments and by in vitro binding experiments using GST pulldown assays. xSLBP1 interacts with hSLIP1 in a yeast two-hybrid assay (Fig. 3C). A mutant of xSLBP1 which has aa 68 to 123 deleted (xSLBP1Δ68-123), including the region from 68 to 83 required for translation, did not interact with hSLIP1. Similarly, xSLBP1W73A, which contains a point mutation in the conserved tryptophan 73 critical for xSLBP1 function in translation, also did not interact with hSLIP1, although the deletion and mutant proteins were expressed at levels similar to those seen for the wild-type protein (Fig. 3C, lanes 2 to 4).
We tested the ability of GST-hSLIP1 expressed in E. coli to pull down 35S-labeled xSLBP1 labeled by in vitro translation. The xSLBP1 bound to GST-hSLIP1 (Fig. 3D, lane 3). Binding of the D72A, W73A, E78A, and EE7879AA mutants of xSLBP1 to hSLIP1 was much lower than seen for the wild type and similar to binding to the GST control (Fig. 3D, lanes 6, 9, 18, and 24), and the W73F mutant bound less strongly than wild-type xSLBP1 (Fig. 3D, lane 12). The GS7475AA and E79A mutants bound with efficiencies similar to that of wild-type xSLBP1 in this assay (Fig. 3D, lanes 15 and 21). These results are in agreement with the directed yeast two-hybrid data.
GST fusion proteins containing various N-terminal deletions of hSLBP were used in a reciprocal GST pulldown assay with 35S-labeled hSLIP1 (Fig. 3E). Both the full-length hSLBP and the hSLBPΔ1-68 protein bound hSLIP1 with equal efficiency (Fig. 3E, lanes 2 and 3), while hSLBPΔ1-81 and hSLBPΔ1-126, which have the entire translation activation domain deleted, bound very weakly to hSLIP1 (Fig. 3E, lanes 4 and 5). Purified recombinant His-tagged hSLBP also interacted with purified recombinant GST-hSLIP1 in vitro, demonstrating that there was no dependence on other proteins or RNAs for this interaction (Fig. 3F).
hSLIP1 cooperates with SLBP to stimulate translation of a reporter mRNA ending in the histone stem-loop in Xenopus oocytes.
To test whether hSLIP1 plays a role in the translation of histone mRNAs, we injected synthetic mRNAs encoding xSLBP1, hSLBP, or hSLIP1 and we compared the effect of expressing SLBP with the effect of coexpressing SLBP and hSLIP1 on the activation of the Luc-SL reporter. xSLBP1 stimulated expression of the Luc-SL more than hSLBP did, as previously reported (7, 24). The expression of hSLIP1 alone had little effect on the expression of the Luc-SL reporter (Fig. 4A and B), suggesting that in the presence of limiting amounts of xSLBP1 in the oocyte, xSLIP1 is not limiting. However, the expression of hSLIP1 together with xSLBP1 resulted in an additional 1.5- to 2-fold increase in total activity (Fig. 4A and B). Moreover, the expression of hSLIP1 together with hSLBP also resulted in an increase in activity, up to the same level as seen for xSLBP1, with hSLIP1 (Fig. 4A).
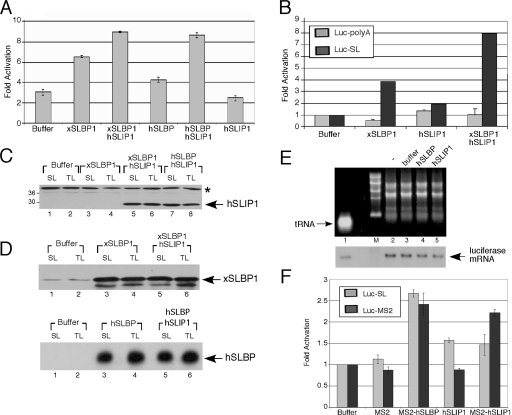
FIG. 4.
Activation of translation of reporter mRNAs ending in a histone stem-loop by SLIP1. (A) Xenopus oocytes were injected with buffer or with capped synthetic mRNA encoding the indicated protein(s), 16 h prior to the injection of Luc-SL or Luc-TL mRNA. Results are expressed relative to the expression of the Luc-TL mRNA. (B) An experiment similar to that described for panel A, except that the oocytes expressing the various proteins were injected with the either the Luc-SL or Luc-poly(A) reporter as well as Luc-TL. The relative expression of Luc-SL or Luc-poly(A) in the oocytes injected with buffer was set at 1. (C and D) Western blot analysis of the experiment whose results are shown in panel A. (C) Western blotting of the samples with the hSLIP1 antibody raised against the complete protein. The band indicated with an asterisk is a cross-reacting protein. (D) Shown are the same lysates analyzed with the xSLBP1 antibody (top) or the hSLBP antibody (bottom). (E) Oocytes were injected with buffer (lane 3) or the mRNA encoding the indicated proteins (lanes 4 and 5) and then injected with 3 ng of Luc-SL RNA, and RNA was prepared from the oocytes 16 h later. Lane 2 shows RNA from oocytes injected only with the reporter mRNA. One oocyte equivalent of RNA was analyzed by Northern blotting for luciferase mRNA. Three nanograms of in vitro-transcribed Luc-SL mRNA was mixed with an E. coli tRNA carrier as a control (lane 1). (F) We analyzed the effect of the indicated proteins on the translation of reporter histone mRNAs as previously described (24). The indicated proteins were synthesized in the reticulocyte lysate. We then added fresh lysate to each reaction and assayed the translation of uncapped Luc-SL, Luc-TL, or Luc-MS2 mRNA. The amount of luciferase activity was determined by luminometry. The Luc-SL, Luc-TL, and Luc-MS2 RNAs in the absence of in vitro-translated SLBP or SLIP1 produced similar amounts of luciferase, which was set at 1.
We further tested the effect of SLIP1 on the translation of a polyadenylated luciferase reporter mRNA [Luc-poly(A)]. While the expression of hSLIP1 together with xSLBP1 stimulated the translation of Luc-SL compared with what was seen for injection of xSLBP1 alone, hSLIP1 had no effect on the translation of the Luc-poly(A) mRNA (Fig. 4B). Thus, the effect of SLIP1 is specific for the Luc-SL mRNA.
Western blots confirmed that the hSLIP1 (Fig. 4C) and SLBP (Fig. 4D) proteins were expressed in the oocytes and that protein levels were not affected by the injection of reporter mRNAs or the injection of multiple mRNAs. We interpret the low activity level of hSLBP in oocytes compared to that seen for xSLBP1 to the fact that hSLBP may not interact as well with xSLIP1 as xSLBP1 does. We also confirmed that the expression of SLBP or SLIP1 had no effect on the stability of the reporter RNAs in the oocytes, as found previously for SLBP (24) (Fig. 4E). We conclude that SLIP1 together with SLBP stimulates the translation of mRNAs ending in the histone 3′ stem-loop.
The translation of a luciferase reporter ending in an MS2 stem-loop is stimulated by an MS2-SLBP fusion protein both in reticulocyte lysates (24) and in Xenopus oocytes (7). We tested the ability of an MS2-SLIP1 fusion protein to activate the translation of the luciferase reporter in reticulocyte lysates by use of the approach previously described (24). Expression of the hSLBP, hSLIP1, and MS2 fusion proteins and luciferase reporters were visualized by SDS-PAGE and luciferase quantitated by luminometry assay (Fig. 4F). In unsupplemented reticulocyte lysates, the Luc-SL and Luc-MS2 reporters were translated at the same level, which was unaffected by the addition of MS2 protein (Fig. 4F). The addition of MS2-hSLBP activated translation to the same extent on both reporters, as previously described (24). The addition of hSLIP1 had a small effect on the translation of the Luc-SL reporter and no effect on the Luc-MS2 reporter. In contrast, the addition of MS2-hSLIP1 had the same effect as SLIP1 on the Luc-SL reporter, and there was a much larger effect on the Luc-MS2 reporter, similar to that of MS2-SLBP. Thus, results suggest that the recruitment of SLIP1 to the 3′ end of the mRNA is sufficient to increase the translation of the Luc-MS2 reporter (Fig. 4F).
hSLIP1 activates the translation of histone mRNAs in mammalian cells.
To determine whether hSLIP1 also affected the translation of mRNAs ending in the histone stem-loop in mammalian cells, we utilized a GFP reporter gene ending in the histone stem-loop and quantified the expression of GFP by FACS analysis (31). We compared the expression of the GFP-SL gene to that of the GFP-poly(A) gene (Fig. 5A). Transfection of hSLBP specifically stimulated expression of the stem-loop reporter as previously reported (28). The transfection of hSLIP1 also stimulated the expression of the GFP-SL reporter but had a much smaller effect on the translation of the GFP-poly(A) reporter (Fig. 5A). The expression of both hSLIP1 and hSLBP together resulted in a similar increase in GFP expression as the expression of either protein alone.
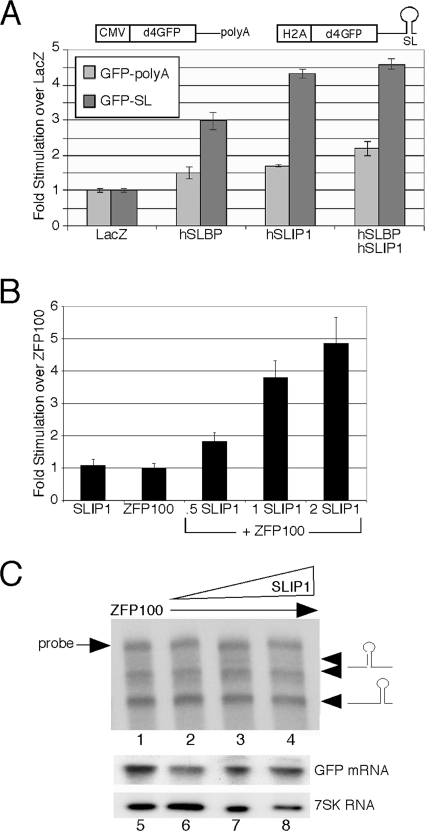
FIG. 5.
SLIP1 stimulates the translation of a histone stem-loop reporter in mammalian cells. (A) A reporter encoding GFP followed by the histone stem-loop and processing signals or a reporter encoding GFP followed by a polyadenylation signal (28, 31) was transfected into 293T cells together with plasmids expressing the indicated proteins. Forty-eight hours later, the cells were harvested and the amount of GFP expression was quantified using FACS analysis. Average GFP intensity for a population of cells transfected with either reporter was normalized to the average GFP intensity of the same reporter transfected into LacZ-expressing cells. (B) An experiment similar to that described for panel A, except that the ZFP100 plasmid was transfected into the cells together with the reporter construct to promote the expression of the processed GFP-SL mRNA (28). The hSLIP1 plasmid was then transfected 24 h later, the cells were harvested 48 h after the second transfection, and the expression of GFP was quantified. (C) (Top) Total cell RNA from the cells from panel B was analyzed using an S1 nuclease protection assay (28) that allows detection of the mRNAs processed at the 3′ end and the mRNAs resulting from readthrough past the 3′ end (arrows). The position of the probe is indicated. The RNAs were from cells transfected with ZFP100 (lane 1) and with ZFP100 plus increasing amounts of SLIP1 (lanes 2 to 4). (Bottom) Northern blotting was done on the same RNA samples to determine the amount of GFP mRNA and 7SK snRNA present in total cell RNA (lanes 5 to 8).
The production of substantial amounts of GFP-SL mRNA from this reporter requires the expression of ZFP100, a U7 snRNP protein that is limiting for the expression of histone mRNA (28). To further assess the role of SLIP1 in translation in mammalian cells, we transfected cells with both ZFP100 and the GFP-SL reporter. Twenty-four hours later, we transfected cells with increasing amounts of SLIP1 and then analyzed the levels of GFP-SL mRNA by Northern blotting and of GFP protein by use of FACS 48 h later. Increased levels of hSLIP1 increased the expression of the GFP protein up to fivefold in a concentration-dependent manner (Fig. 5B).
We determined whether the expression of hSLIP1 altered the amount of total GFP mRNA or processed GFP mRNA ending at the stem-loop. We measured the amount of GFP mRNA processed at the histone 3′ end in cells transfected with ZFP100 24 h before the transfection of hSLIP1 (28) by use of S1 nuclease mapping (Fig. 5C, lanes 1 to 4). There was no change in the amount of processed reporter mRNA or of readthrough reporter mRNA with transfection of increasing amounts of hSLIP1. We also performed Northern blot analysis for total GFP mRNA and 7SK RNA (as an internal control) on the same samples (Fig. 5C, lanes 5 to 8). The expression of hSLIP1 stimulated GFP expression but did not increase the levels of GFP-SL mRNA in either of these assays, indicating that the increased GFP levels are due to increased translation of the GFP-SL mRNA. There was no change in the cell cycle distribution after the expression of SLIP1 or SLBP in these cells (not shown), indicating that these results are not due to an increase in the number of S-phase cells.
SLIP1 interacts with both SLBP and eIF4G.
To determine whether SLBP and SLIP1 interact in cells, we used the anti-hSLIP1 antibody to the C-terminal peptide, which efficiently precipitates SLIP1 from total cell lysates from HeLa cells (Fig. 6A, top, lane 3). Western blotting of the immunoprecipitates for SLBP revealed that anti-hSLIP1 coprecipitates hSLBP (Fig. 6A, middle, lane 3). The addition of RNase to the lysate prior to the immunoprecipitation only slightly reduced the amount of SLBP that was coimmunoprecipitated (Fig. 6A, middle, lane 5). This demonstrates that endogenous SLBP and hSLIP1 interact in vivo and that the interaction is independent of RNA.
FIG. 6.
SLIP1 interacts with SLBP and eIF4G in vitro and in vivo. (A) Lysates prepared from exponentially growing HeLa cells were immunoprecipitated with anti-myc (lanes 2 and 4) or anti-SLIP1 antibody against the C terminus of hSLIP1 (lanes 3 and 5). One-half of the lysate was treated with RNase prior to immunoprecipitation (lanes 4 and 5). Precipitates were probed by Western blotting with antibodies against the C terminus of hSLIP1, hSLBP, or eIF4GI. Lane 1 is 5% of the input. (B) A schematic of the structure of eIF4GI based on data from the study of Hinton and coworkers (7a) is shown at the top. Full-length eIF4GI (lanes 1 to 3, top) or eIF4GII (lanes 1 to 3, bottom) or the indicated fragments of eIF4GI (lanes 4 to 6) or eIF4GII (lanes 7 to 9) were labeled with [35S]methionine by in vitro translation. The lysates were incubated with either GST (lanes 2, 5, and 8) or GST-hSLIP1 (lanes 3, 6, and 9), and the bound proteins were resolved by electrophoresis and detected by autoradiography. Input protein (10%) was analyzed in lanes 1, 4, and 7. (C) The indicated fragments of His-tagged eIF4GI (lanes 1 to 3, 27 to 129; lanes 4 to 6, 123 to 420) were expressed in bacteria and incubated with GST (lanes 2 and 5) or GST-hSLIP1 (lanes 3 and 6). The proteins were recovered on GST-agarose and bound proteins detected by Western blotting for the histidine tag. The recombinant eIF4GI region from 27 to 129 was incubated with GST (lane 8) or GST-hSLBP (also His tagged; lane 9), and the proteins were recovered on GST-agarose. The bound proteins were detected by Western blotting for the His tag. (D and E) Lysates were prepared from exponentially growing HeLa cells stably expressing HA-tagged hSLIP1 or from cells treated with HU for 1 h. The lysates were immunoprecipitated with either anti-myc (lanes 2 and 5) or anti-HA (lanes 3 and 6) antibodies, and bound proteins were resolved by electrophoresis and detected by Western blotting. Samples shown in panel D were Western blotted against hSLIP1 and hSLBP. Lanes: 1 to 3, control; 4 to 6, HU treated. (E) Shown is an analysis of the same two samples Western blotted with eIF4GI antibody. Top, control; bottom, HU treated. α-, anti-.
Since PABP interacts with eIF4G to circularize mRNAs (8, 34), we also tested the SLIP1 immunoprecipitates for the presence of eIF4GI. eIF4GI was present in the immunoprecipitates (Fig. 6A, bottom, lane 3). However, treatment of the lysate with RNase prior to immunoprecipitation resulted in the loss of eIF4GI from the immunoprecipitates (Fig. 6A, bottom, lane 5).
Since the translation of polyadenylated mRNAs requires interaction between the 5′ end and the 3′ end of the mRNA mediated by PABP binding to eIF4G, we tested whether there was a direct interaction between SLIP1 and eIF4G. We synthesized eIF4GI and eIF4GII by in vitro translation and tested the ability of GST-hSLIP1 to bind to the 35S-labeled proteins (Fig. 6B). Both eIF4GI and eIF4GII bound to GST-hSLIP1 better than they bound to GST (Fig. 6B, lane 3). We tested various fragments of eIF4GI and eI4GII for their abilities to interact with GST-hSLIP1. Fragments containing approximately the first 420 aa of eIF4GI and eIF4GII each interacted with GST-hSLIP1. The binding activity was present in aa 27 to 170 of eIF4GI (Fig. 6B, middle, lane 6), while the fragment from 170 to 420 bound GST as well as it bound GST-hSLIP1 (Fig. 6B, bottom, lanes 5 and 6). Similarly, the binding of hSLIP1 to eIF4GII aa 101 to 420 was weaker than the binding to aa 31 to 420, suggesting that hSLIP1 binds within aa 31 to 101.
To determine whether the interaction could occur between recombinant proteins, we tested the ability of His-tagged eIF4GI to interact with GST-hSLIP1 (Fig. 6C). GST-hSLIP1 bound to recombinant eIF4GI(27-129) but not to eIF4GI(123-420) (Fig. 6C, lanes 3 and 6). Thus, hSLIP interacts with the N-terminal region of eIF4GI. The binding site for PABP is 172 to 200 in eIF4GI (8). Thus, the binding site for hSLIP1 on eIF4GI is close to, but does not overlap with, the binding site for PABP. Additionally, the 27 to 129 fragment did not bind to recombinant SLBP (Fig. 6C, lane 9), consistent with the inability of other investigators to detect a direct interaction between SLBP and eIF4G (7).
SLIP1 does not bind directly to RNA (unpublished results). Note that SLIP1 was active both in frog oocytes and in mammalian cells using reporter mRNAs that lacked any histone sequences other than the stem-loop. Thus, the likely role of SLIP1 is to help circularize the mRNA by interacting with both SLBP and eIF4G and not directly with the mRNA.
We also examined the effect of HU on the association of SLIP1 with SLBP and eIF4G. The treatment of cells with HU results in a rapid reduction in histone mRNA levels, with no effect on SLBP levels (35). We carried out immunoprecipitation experiments using cells stably expressing HA-hSLIP1. HA antibody was used to immunoprecipitate HA-tagged hSLIP1, and the presence of SLBP (Fig. 6D) or eIF4GI (Fig. 6E) was probed by Western blotting. Precipitation with an anti-myc antibody was used as a control. We detected SLBP associated with SLIP1, and this association was not affected by HU treatment (Fig. 6D, lanes 3 and 6). However, the treatment of cells with HU resulted in the loss of SLIP1 interaction with eIF4GI (Fig. 6E, lane 3), consistent with the loss of histone mRNA after HU treatment.
We interpret these experiments to imply that SLIP1 is bound to the translating histone mRNA and tethered to the 3′ end of the mRNA by SLBP and that it then interacts with eIF4GI at the 5′ end of the mRNA. The interaction between SLIP1 and SLBP is strong enough to be maintained in the absence of RNA. The interaction between SLIP1 and eIF4G is weaker, and the proteins do not remain associated during immunoprecipitation in the absence of RNA. These results are consistent with the results of Ling et al. that SLBP and eIF4G coimmunoprecipitate in an RNA-dependent manner (16). We note that similar results were obtained with PABP, which also interacts with eIF4G. Coimmunoprecipitation of eIF4G with PABP from cells is also RNase sensitive, although the proteins clearly interact in vitro (8).
SLIP1 is an essential protein in HeLa cells.
To establish whether SLIP1 is an essential gene, we used RNA interference (RNAi) to knock down hSLIP1 in HeLa cells (Fig. 7). As a control, we also used RNAi to knock down SLBP, which slows progression through S phase without affecting viability (29) (Fig. 7A). The treatment of HeLa cells with either siRNA1 or 2 against hSLIP1 resulted in a rapid depletion of hSLIP1 protein and cell death, with 50% of the cells dying within 24 h and over 75% dying by 72 h (Fig. 7A and B). HeLa cells treated with a control siRNA, C2 (29), were not affected. Thus, unlike SLBP, hSLIP1 is essential for the viability of HeLa cells and other cell lines that we tested (data not shown). Each siRNA effectively depleted both HA-tagged hSLIP1 expressed from a transfected gene (Fig. 7C, top, lanes 4 to 6) and the endogenous hSLIP1 (Fig. 7C, middle, lanes 4 to 6), using the antibody to the full-length SLIP1 protein. A Western blot against the PTB protein was used as a loading control (Fig. 7C, bottom).
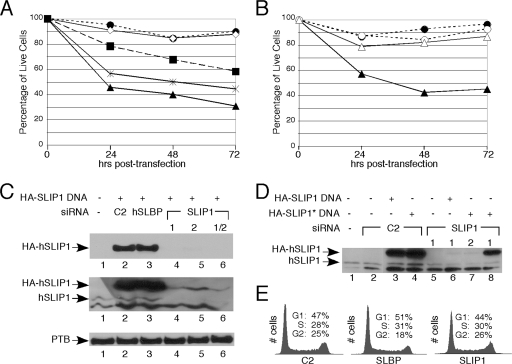
FIG. 7.
Knockdown of SLIP1 results in the death of HeLa cells. (A) Cells were transfected with two different siRNAs against SLIP1 (siRNA1, closed triangles; siRNA2, closed squares; siRNA 1 and 2 together, stars), a siRNA against SLBP (open diamonds), and a control siRNA (closed circles). Cell viability was assessed by trypan blue staining at the indicated times posttransfection. (B) Cells were transfected with siRNA1 against hSLIP1 or a control siRNA, together with either a plasmid expressing GFP (SLIP1 siRNA, closed triangles; control siRNA, closed circles) or a plasmid expressing an RNAi-resistant SLIP1 (SLIP1 siRNA, open triangles; control siRNA, open circles) constructed by mutating the third nucleotides of the codons in the region targeted by the siRNA. The viability of the cells was determined daily for 3 days. There were also fewer total cells in the siRNA-treated cultures without the RNAi-resistant SLIP1, suggesting that cell growth had also been affected and that dead cells that had lysed were no longer detectable. (C) Western blots were used to assess the abilities of the two different siRNAs to knock down endogenous SLIP1 protein and exogenously expressed SLIP1 (HA-tagged SLIP1) in HeLa cells (lanes 4 and 5) and a combination of the two SLIP1 siRNAs (lane 6). Cells were also transfected with a control siRNA C2 (lane 2) or a siRNA against SLBP (lane 3). Cell lysates were prepared, and equal amounts of protein resolved by gel electrophoresis were analyzed by Western blotting using an anti-HA antibody (top), the hSLIP1 antibody to the full-length protein (middle), or an antibody against PTB (30) as a loading control (bottom). (D) Cells were treated with either the control siRNA (lanes 2 to 4) or the indicated SLIP1 siRNA (lanes 5 to 8) together with either a plasmid expressing HA-tagged SLIP1 (lanes 3 and 6) or a plasmid expressing HA-SLIP1 resistant to siRNA1 but not siRNA2 (lanes 4, 7, and 8), and lysates were analyzed by Western blotting using anti-hSLIP1 antibody. (E) Cells treated with siRNAs against control (C2), SLBP, or SLIP1 as described above. Cells were harvested, stained with propidium iodide, and analyzed by FACS to determine cell cycle profiles.
To demonstrate that cell death was a result of knocking down hSLIP1, we created a gene expressing an RNAi-resistant form of hSLIP1 mRNA (mutating the siRNA1 target site). Expressing the RNAi-resistant form of hSLIP1 restored both cell growth (Fig. 7B) and expression of the SLIP1 protein (Fig. 7D, lane 8) in cells treated with siRNA1 but not siRNA2, confirming that the cell death phenotype was due to SLIP1 depletion. There was no change in cell cycle distribution in the HeLa cells as a result of the knockdown of SLIP1 (Fig. 7E). Cell death was likely due to apoptosis, based on the loss of full-length caspase 9 (data not shown), and the accumulation of cells containing less than 2 N DNA content (data not shown). While the biological lesion in the cell that results in the activation of the apoptotic pathway is not known, knocking down SLBP, which has a larger effect on histone protein biosynthesis (Fig. 8), does not result in cell death, suggesting that there may be another pathway for which SLIP1 is essential.
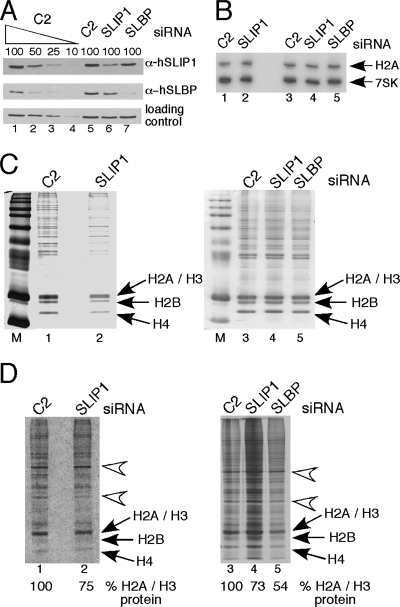
FIG. 8.
Effect of SLIP1 and SLBP knockdown on histone protein synthesis. (A) HeLa cells were treated with siRNA for SLIP1 or SLBP for 4 days. Western blots (top, anti-SLIP1; middle, anti-SLBP; bottom, loading control) were done to assess the knockdown of each protein. C2 is a control siRNA, and the loading control is a cross-reacting band with the hSLIP1 antibody. Lanes 1 to 4 are serial dilutions of the C2-treated lysate. Lanes 5 to 7 are extracts of cells treated with C2 siRNA, SLIP1 siRNA, and SLBP siRNA, respectively. (B) The levels of histone mRNA and 7SK RNA were determined by Northern blotting from the cells from panel A. (C) The total nuclear protein from the cells from panel A was resolved by electrophoresis on 15% SDS-polyacrylamide gels and stained with Coomassie blue. The histone proteins are indicated. (D) The gels from panel C were dried and autoradiographed. The H2a/H3 histones were quantified on a PhosphorImager, as were the two bands indicated by arrows. The ratio of the two nonhistone bands indicated by arrows to each other was constant (±5%), and the levels of histone protein synthesis are indicated. α-, anti-.
We also assessed the effect of SLIP1 depletion on endogenous histone mRNA levels and histone protein synthesis. For these experiments, we knocked down SLIP1 in HeLa cells for the minimal time that gave substantial knockdown (about 70%) (Fig. 8A, top, lane 6) but retained cell viability. There was only a small decrease in SLBP protein levels in the SLIP1 knockdown cells (Fig. 8A, middle, lane 6). We used the antibody against the C terminus of hSLIP1 for these analyses. We also knocked down SLBP in a parallel culture of cells and carried out the same analysis. The SLBP knockdown was about 75% effective (Fig. 8A, middle, lane 7) and there was no change in SLIP1 levels in the SLBP knockdown cells (Fig. 8A, top, lane 7). A cross-reacting band is shown as a loading control (Fig. 8A, bottom).
We analyzed the levels of histone mRNA in these cells relative to the level of 7SK RNA, and there was not a significant change in histone mRNA levels in either SLBP knockdown or SLIP1 knockdown cells (Fig. 8B), consistent with there being no change in cell cycle distribution. A reduction of histone mRNA levels due to SLBP knockdown requires longer treatment with siRNA and a more complete knockdown of SLBP (E. J. Wagner and W. F. Marzluff, unpublished data).
To assess the rate of histone protein synthesis, we starved the siRNA-treated cells for methionine for 30 min, labeled the cells for 10 min with [35S]methionine, and resolved total nuclear proteins by SDS-PAGE. We also analyzed the cytoplasm of these same cells, and there was no newly synthesized histone protein in this fraction (not shown). We consistently saw higher levels of total methionine incorporation in the SLIP1 knockdown cells for unknown reasons and thus performed analyses loading either equal amounts of radioactive protein (Fig. 8C and D, lanes 1 and 2) or equal amounts of total protein (Fig. 8C and D, lanes 3 to 5). We quantified the amount of radiolabeled H2a/H3 histone using a PhosphorImager and compared the levels of the histone proteins to two prominent labeled bands of nonhistone protein (Fig. 8D). There was no change in the relative intensity of the nonhistone protein bands, as judged either by Coomassie staining (Fig. 8C) or by autoradiography (Fig. 8D). In the SLBP knockdown, there was a decrease of 50% in histone protein synthesis, consistent with a role for SLBP in translation. There was reproducibly a 20 to 30% decrease in histone protein synthesis when SLIP1 was knocked down. The ratio of the two control bands varied by only 5% among the various samples.
These experiments, together with the stimulation of the translation of the histone reporter mRNA (Fig. 5), demonstrate a role for SLIP1 in the translation of mRNAs ending in the histone stem-loop in mammalian cells. The modest effect on translation in the SLBP and SLIP1 knockdown cells is likely due to the residual proteins present in the cells. Because SLIP1 depletion results in cell death while SLBP depletion does not, our data suggest that SLIP1 may function in other important cellular processes in addition to histone mRNA translation.
DISCUSSION
All known eukaryotic cellular mRNAs, with the exception of the metazoan replication-dependent histone mRNAs, are polyadenylated. Efficient translation of polyadenylated mRNAs requires both the 5′ cap and 3′ poly(A) tail of the mRNAs. The cap at the 5′ end is bound by eIF4E and the poly(A) tail at the 3′ end is bound by PABP. These two proteins bind to eIF4G, allowing eIF4G to bind and recruit eIF3 and the small ribosomal subunit, resulting in the initiation of translation.
While all eukaryotic mRNAs, including the metazoan replication-dependent histone mRNAs, require the 5′ cap and 3′ end for efficient translation, some viral RNAs utilize a specialized 3′ end for translation. The rotavirus genome mRNA does not end in a poly(A) tail and is translated in mammalian cells. Its translation is mediated by the viral protein NSP3, which binds both to the 3′ end of viral mRNA and to the same region on eIF4G as PABP binds. NSP3 may circularize the viral RNA (21, 27), although this role for NSP3 has recently been questioned (19). Many viral mRNAs which infect plants are also not polyadenylated, ending instead in a complex RNA pseudoknot (15). The 3′ ends of these mRNAs are also required for their translation, and some of these viruses (e.g., the alfalfa mosaic virus) encode proteins that bind to the 3′ end of their mRNA and eIF4G (14, 20).
SLIP1 binds SLBP and stimulates histone mRNA translation.
The 3′ end of histone mRNA is required for the efficient translation of histone mRNA both in mammalian cells (6) and in Xenopus oocytes (24). SLBP stimulates the translation of reporter mRNAs ending in the histone 3′ end both in Xenopus oocytes and in a rabbit reticulocyte lysate (24). We identified a 15-aa region in SLBP essential for translation that includes a highly conserved motif, DWX3VEE, and identified a novel protein, SLIP1, that interacts with this sequence. Point mutations in the conserved core translation region of SLBP prevent the activation of translation of a reporter ending in a histone stem-loop (Fig. 2) and also inhibit the binding of SLBP to SLIP1 (Fig. 3), strongly supporting a role for SLIP1 in histone mRNA translation.
SLIP1 is a 25.4-kDa protein that has putative orthologues in all metazoans but no obvious orthologue in plants or fungi. Plants and fungi express polyadenylated histone mRNAs and thus would not require SLIP1 for histone mRNA translation. The crystal structure of the Danio rerio orthologue indicates that SLIP1 consists of multiple HEAT domains and is similar to the middle domain of eIF4G in this respect (17). SLIP1 is not cell cycle regulated and is present in both the nucleus and the cytoplasm, as judged by immunofluorescence of HA-SLIP1 with anti-HA antibodies as well as measurement of endogenous SLIP1 by Western blotting after cell fractionation (R. S. Lerner and W. F. Marzluff, unpublished data).
SLIP1 stimulates the translation of a reporter mRNA ending in a stem-loop when it is expressed together with SLBP in Xenopus oocytes, although the expression of hSLIP1 has little effect on the translation of the reporter RNA when only endogenous levels of xSLBP1 and xSLIP1 are present in Xenopus oocytes (Fig. 4A). SLIP1 also stimulates the translation of a reporter mRNA ending in a stem-loop in mammalian cells but has little effect on the translation of a polyadenylated reporter mRNA in either Xenopus oocytes or mammalian cells, demonstrating that it does not affect the translation of all mRNAs (Fig. 4 and 5). hSLBP has lower activity than xSLBP1 in oocytes, suggesting that it does not interact optimally with the Xenopus translation factors, such as xSLIP1 (24) (Fig. 4A). However, the expression of hSLIP1 together with hSLBP resulted in maximal activation of the reporter (Fig. 4A). Importantly, there was no change in the levels of the reporter mRNAs in either oocytes or mammalian cells after SLIP1 expression (Fig. 4E and 5C), while there was increased expression of reporter protein, consistent with a role of SLIP1 in translation.
SLIP1 interacts with eIF4G, bridging the 3′ and 5′ ends of the mRNA.
Previously, Ling and coworkers showed that eIF4G could be coimmunoprecipitated from cell lysates with SLBP, although this interaction was RNase sensitive (16). No direct interaction between SLBP and eIF4G has been detected despite extensive efforts (7). Our results strongly suggest that SLIP1 provides the bridge between SLBP and eIF4G, bridging the 5′ and 3′ ends of the mRNAs. SLIP1 interacts with the amino-terminal portion of eIF4GI and eIF4GII, in the same general region as PABP binds, although the binding sites are distinct, since SLIP1 interacts with a fragment of eIF4G that does not contain the PABP binding site (Fig. 6B and C). While SLIP1 clearly interacts with eIF4G, it is possible that it also contacts other components of the translation initiation machinery.
SLIP1 is an essential protein in cultured mammalian cells.
Knocking down hSLIP1 in HeLa cells (Fig. 7) by RNAi results in rapid cell death. This phenotype is a result of the hSLIP1 knockdown, since the expression of an RNAi-resistant hSLIP1 rescues cell viability. This phenotype was unexpected, since knocking down other proteins required for histone mRNA metabolism, such as SLBP (29, 37), ZFP100, Lsm10, or Lsm11 (28), does not affect cell viability but rather causes cell cycle arrest. hSLIP1 is not cell cycle regulated (Lerner and Marzluff, unpublished) and is also present in reticulocytes which do not contain nuclei. These results suggest that SLIP1 likely has other function(s) in addition to participating in histone mRNA translation.
Mechanism of histone mRNA translation.
The efficient translation of polyadenylated mRNAs requires that both PABP and eIF4E bind to the 3′ and 5′ ends of the mRNA, respectively, and each of these proteins then binds to eIF4G to circularize the mRNA. It is likely that the 3′ end of the histone mRNA interacts with the 5′ end of the mRNP, since there is a requirement for both the cap and the 3′ end for translation in vivo in mammalian cells (6). These results strongly suggest that SLIP1 is a critical factor in histone mRNA translation. SLBP does not interact directly with eIF4G, but the data presented here suggest that SLIP1 binds to both SLBP and eIF4G and allows circularization of histone mRNAs (7). It is likely that the mechanisms of translation of polyadenylated mRNAs and histone mRNAs are similar, and it is possible that the role of SLIP1 is to help bring the 5′ end and the 3′ end of histone mRNAs together to facilitate translation initiation.
Acknowledgments
This work was supported by NIH grant GM58921 to W.F.M. E.J.W. was supported by NIH grant F32 GM070101-02 and a Cottrell fellowship, and R.S.L was supported by NIH grant T32CA09156.
We thank Bob Duronio for helpful discussions and critical comments on the manuscript.
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Adamson, E. D., and H. R. Woodland. 1977. Changes in the rate of histone synthesis during oocyte maturation and very early development of Xenopus laevis. Dev. Biol. 57136-149. [DOI] [PubMed] [Google Scholar]
- 2.Craig, A. W., A. Haghighat, A. T. Yu, and N. Sonenberg. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392520-523. [DOI] [PubMed] [Google Scholar]
- 3.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 2391-14. [DOI] [PubMed] [Google Scholar]
- 4.Dominski, Z., L.-X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 193561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 52108-2116. [DOI] [PubMed] [Google Scholar]
- 6.Gallie, D. R., N. J. Lewis, and W. F. Marzluff. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 241954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorgoni, B., S. Andrews, A. Schaller, D. Schumperli, N. K. Gray, and B. Muller. 2005. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA 111030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Hinton, T. M., M. J. Coldwell, G. A. Carpenter, S. J. Morley, and V. M. Pain. 2007. Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 2821695-1708. [DOI] [PubMed] [Google Scholar]
- 8.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 177480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingledue, T. C., Z. Dominski, R. Sanchez, J. A. Erkmann, and W. F. Marzluff. 2000. Dual role for the RNA binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA 61635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karim, M. M., Y. V. Svitkin, A. Kahvejian, G. De Crescenzo, M. Costa-Mattioli, and N. Sonenberg. 2006. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl. Acad. Sci. USA 1039494-9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaygun, H., and W. F. Marzluff. 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12794-800. [DOI] [PubMed] [Google Scholar]
- 12.Kessler, S. H., and A. B. Sachs. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 1851-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khaleghpour, K., A. Kahvejian, G. De Crescenzo, G. Roy, Y. V. Svitkin, H. Imataka, M. O'Connor-McCourt, and N. Sonenberg. 2001. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 215200-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krab, I. M., C. Caldwell, D. R. Gallie, and J. F. Bol. 2005. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J. Gen. Virol. 861841-1849. [DOI] [PubMed] [Google Scholar]
- 15.Leathers, V., R. Tanguay, M. Kobayashi, and D. R. Gallie. 1993. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol. 135331-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling, J., S. J. Morley, V. M. Pain, W. F. Marzluff, and D. R. Gallie. 2002. The histone 3′ terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eucaryotic initiation factor 4G (eIF4G) and eIF3. Mol. Cell. Biol. 227853-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcotrigiano, J., I. B. Lomakin, N. Sonenberg, T. V. Pestova, C. U. Hellen, and S. K. Burley. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7193-203. [DOI] [PubMed] [Google Scholar]
- 18.Marintchev, A., and G. Wagner. 2005. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry 4412265-12272. [DOI] [PubMed] [Google Scholar]
- 19.Montero, H., C. F. Arias, and S. Lopez. 2006. Rotavirus nonstructural protein NSP3 is not required for viral protein synthesis. J. Virol. 809031-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neeleman, L., H. J. Linthorst, and J. F. Bol. 2004. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen. Virol. 85231-240. [DOI] [PubMed] [Google Scholar]
- 21.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 175811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson, A. J., J. T. Howard, Z. Dominski, B. J. Schnackenberg, J. L. Sumerel, J. J. McCarthy, J. A. Coffman, and W. F. Marzluff. 2004. The sea urchin stem-loop-binding protein: a maternally expressed protein that probably functions in expression of multiple classes of histone mRNA. Nucleic Acids Res. 32811-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy, G., G. De Crescenzo, K. Khaleghpour, A. Kahvejian, M. O'Connor-McCourt, and N. Sonenberg. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 223769-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez, R., and W. F. Marzluff. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 227093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez, R., and W. F. Marzluff. 2004. The oligo(A) tail on histone mRNA plays an active role in translational silencing of histone mRNA during Xenopus oogenesis. Mol. Cell. Biol. 242513-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vende, P., M. Piron, N. Castagné, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 747064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner, E. J., and W. F. Marzluff. 2006. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol. Cell. Biol. 266702-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner, E. J., A. Berkow, and W. F. Marzluff. 2005. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem. Soc. Trans. 33471-473. [DOI] [PubMed] [Google Scholar]
- 30.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 213281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, E. J., J. K. Ospina, Y. Hu, M. Dundr, A. G. Matera, and W. F. Marzluff. 2006. Conserved zinc fingers mediate multiple function of ZFP100, a U7 snRNP associated protein. RNA 121206-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Z.-F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, Z.-F., M. L. Whitfield, T. I. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 103028-3040. [DOI] [PubMed] [Google Scholar]
- 34.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2135-140. [DOI] [PubMed] [Google Scholar]
- 35.Whitfield, M. L., H. Kaygun, J. A. Erkmann, W. H. D. Townley-Tilson, Z. Dominski, and W. F. Marzluff. 2004. SLBP is associated with histone mRNA on polyribosomes as a component of histone mRNP. Nucleic Acids Res. 324833-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodland, H. R. 1982. The translational control phase of early development. Biosci. Rep. 2471-491. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, X., S. Killop-Smith, and B. Muller. 2004. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 1176043-6051. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, L.-X., Z. Dominski, X. Yang, P. Elms, C. S. Raska, C. H. Borchers, and W. F. Marzluff. 2003. Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 231590-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]