Abstract
Brd4, a bromodomain protein capable of interacting with acetylated histones, is implicated in transmitting epigenetic memory through mitosis. It also functions as an associated factor and positive regulator of P-TEFb, a Cdk9-cyclin T1 heterodimer that stimulates transcriptional elongation by phosphorylating RNA polymerase II. In the present study, experiments were performed to determine whether these two functions of Brd4 are interrelated and, if so, how they may impact cell cycle progression. Our data demonstrate that while the P-TEFb level remains constant, the Brd4-P-TEFb interaction increases dramatically in cells progressing from late mitosis to early G1. Concurrently, P-TEFb is recruited to chromosomes, beginning around mid- to late anaphase and before nuclear envelope/lamina formation and nuclear import of other general transcription factors. Importantly, the recruitment of P-TEFb depends on Brd4. Abrogation of this process through Brd4 knockdown reduces the binding of P-TEFb to and expression of key G1 and growth-associated genes, leading to G1 cell cycle arrest and apoptosis. Because P-TEFb is synonymous with productive elongation, its recruitment by Brd4 to chromosomes at late mitosis may indicate those genes whose active transcription status must be preserved across cell division.
Eukaryotic transcription elongation is a highly regulated process important for not only the production of full-length RNA transcripts but also the coupling of transcription with other major gene expression events. The positive transcriptional elongation factor b (P-TEFb) plays a central role in this process. Consisting predominantly of a Cdk9-cyclin T1(CycT1) heterodimer, P-TEFb stimulates elongation by phosphorylating the carboxy-terminal domain of RNA polymerase II (Pol II) as well as negative elongation factors. These events allow Pol II to escape from promoter-proximal pausing and engage in productive elongation (16, 26).
Nuclear P-TEFb is maintained in a functional equilibrium through alternately interacting with its positive and negative regulators (26). Diverse signals, including those that impact cell growth and differentiation, can affect this equilibrium by altering the ratio of the negative and positive factors associated with P-TEFb (8, 26). These observations raise an intriguing possibility that the functional P-TEFb equilibrium is tightly linked to the cellular transcriptional demand and the global control of cell growth and differentiation (8, 26).
Whereas the associations with the HEXIM1 protein and 7SK snRNA sequester P-TEFb in an inactive complex, it is the interaction with the bromodomain protein Brd4 that forms the transcriptionally active P-TEFb (10, 24). Brd4 recruits P-TEFb to a promoter by contacting acetylated chromatin and the Mediator complex, and this process is essential for elongation (10, 24). While the Brd4-P-TEFb complex stimulates transcription in general, it is worth noting that Brd4 can also be found in a separate complex assembled by the human papillomaviruses E2 to silence the expression of human papillomavirus-encoded E6 and E7 oncoproteins (23). Another important feature of Brd4 is its ability to remain bound to chromosomes through mitosis. This function of Brd4, which is observed in a number of cell lines (2, 3, 13, 15, 25), has been proposed to play a key role in transmitting epigenetic memory across cell division (3, 15).
Cdk9 and CycT1 are constitutively expressed through the cell cycle (4). Moreover, the kinase activity of the isolated Cdk9-CycT1 heterodimer also remains constant in this process. Based on these observations, it is generally presumed that, unlike many other Cdk-cyclin pairs, P-TEFb plays no direct role in cell cycle progression. However, the recent identification of Brd4 as a major P-TEFb-associated factor and positive regulator makes it necessary to reconsider this presumption.
Here, we show that while the P-TEFb level displayed no major change through the cell cycle, the interaction of P-TEFb with Brd4 increased dramatically in cells progressing from late mitosis to early G1. Concurrent to this increase, P-TEFb became intimately associated with mitotic chromosomes, starting around mid- to late anaphase and before nuclear envelope/lamina formation. This early recruitment sets P-TEFb apart from virtually all other general transcription factors and Pol II, which are imported into the nucleus only after nuclear envelope formation. Importantly, the association of P-TEFb with mitotic chromosomes correlated closely with P-TEFb's interaction with Brd4. We show that the Brd4-dependent recruitment of P-TEFb to mitotic chromosomes before the M/G1 transition was crucial for the binding of P-TEFb to and expression of key G1 genes and progression of the cell cycle through G1. Since the P-TEFb-mediated productive elongation is a hallmark of a committed, robust transcription state, we propose that the Brd4-dependent recruitment of P-TEFb before the onset of fresh rounds of transcription in early G1 serves to mark those genes whose active transcription status needs to be preserved across cell division.
MATERIALS AND METHODS
Cell synchronization and mitotic shake-off.
To obtain populations of mitotic cells, HeLa cells were plated at 1.7 × 104 cells/cm2 and incubated for 24 h. Cells were then placed in fresh medium containing 2 mM thymidine. Upon incubation for 15 h, cells were separated from the medium, rinsed twice, and incubated in new medium without thymidine for another 12 h. Thymidine was then reintroduced into the medium, and cells were incubated for 12 h prior to the removal of thymidine by rinsing cells twice with normal, drug-free medium. Synchronized cells were incubated for 10 h to reach mitosis in normal medium. Next, the rounded-up mitotic cells were shaken off the culture dish and replated in fresh medium. At different time points following the start of replating, cells were harvested and lysed with lysis buffer (50 mM HEPES [pH 7.9], 150 mM NaCl, 0.5% NP-40, 5 mM EDTA, 3 mM dithiothreitol, and 0.25 mM phenylmethylsulfonyl fluoride).
Generation of a stable Brd4 knockdown cell line.
The lentiviral vector pSico (a kind gift from Tyler Jacks, Massachusetts Institute of Technology) was used for the Cre-regulated RNA interference (20) against Brd4. Two DNA oligonucleotides (5′-TGAACCTCCCTGATTACTATAAGCTTCCTGTCACTTATAGTAATCAGGGAGGTTCTTTTTTC-3′ and 5′-TCGAGAAAAAAGAACCTCCCTGATTACTATAAGTGACAGGAAGCTTATAGTAATCAGGGAGGTTCA-3′) for expressing short hairpin RNA were annealed and then cloned into HpaI-XhoI-digested pSico vector for knocking down Brd4. For the generation of lentiviruses, 1 μg of recombinant pSico vector and 0.4 μg of each packaging vector were cotransfected into 293T cells in one well of a six-well plate. Supernatants were collected 48 h posttransfection, centrifuged at 20,000 × g for 5 min, and then used to infect HeLa cells supplemented with 8 μg/ml polybrene. Two rounds of infection 24 h apart were sufficient to infect most cells. The infected HeLa cells were serially diluted after 2 days of infection, and then the green fluorescent protein-positive clones were picked under a fluorescent microscope about 2 weeks later. The adenoviruses expressing Cre recombinase were purchased from the Gene Transfer Vector Core facility of the University of Iowa College of Medicine (Iowa City, IA). Two rounds of infection 24 h apart were performed by using 5 to 10 PFU of virus per cell. Brd4 knockdown was confirmed by Western blotting.
Immunoaffinity purification of Cdk9 and its associated factors.
The Cdk9-containing complexes were affinity purified from cell lysates by incubation at 4°C for 2 h with the anti-Cdk9 antibodies and protein A-Sepharose beads (Amersham Biosciences). After extensive washes with buffer D (20 mM HEPES-KOH [pH 7.9], 150 mM KCl, 15% glycerol, 0.2 mM EDTA, 0.1% NP-40, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride), the immunoprecipitated proteins were eluted off the beads with 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 4% 2-mercaptoethanol, 2% sodium dodecyl sulfate, 0.5× dye mix, and 10% glycerol) at 95°C.
Immunofluorescence staining.
Immunofluorescence staining was performed based on a protocol described previously (1) with minor modifications. Briefly, cells were grown on coverslips, fixed for 10 min in 2% formaldehyde, and permeabilized for 7 min in cold (−20°C) methanol. Cells were then rinsed with phosphate-buffered saline (PBS) and blocked for 30 min at room temperature in PBS containing 0.1% Triton X-100, 10% fetal bovine serum, and 3% bovine serum albumin. Cells were incubated at room temperature for 1 h with the indicated primary antibodies: anti-Brd4 (1:200), anti-cyclin T1 (1:200), anti-Cdk9 (1:400), anti-Cdk9 (monoclonal, 1:100), anti-TFIIB (1:200), anti-p34 (TFIIE subunit, 1:200), anti-RAP30 (TFIIF subunit, 1:100), anti-RNA Pol II (8WG16, 1:200), anti-lamin B (1:100), or anti-α-tubulin (1:200). After a washing step in PBS containing 0.1% Triton X-100 and 10% fetal bovine serum, secondary anti-species-specific antibodies were added for 1 h at room temperature. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Cells were mounted in FluorSave Reagent (Calbiochem) and examined using a Zeiss LSM510 Meta confocal microscope (Carl Zeiss Inc.) with a Plan Apochromat 100× lens (numerical aperture, 1.4) or Zeiss Axiophot epifluorescence microscope with a Plan NeoFLUAR 40× lens (numerical aperture, 0.75).
Flow cytometry analysis.
Cells (1 × 106) were washed twice with PBS and fixed in cold 70% ethanol (−20°C). For assessing DNA profiles, cells were then stained with propidium iodide (100 μg/ml) for 15 min at room temperature and analyzed by a Beckman Coulter Epics XL flow cytometer.
RT-PCR analysis.
Total RNA was prepared with a Qiagen RNAeasy kit following the manufacturer's instructions. Reverse transcription-PCRs (RT-PCRs) were performed with Superscriptase III (Invitrogen) using oligo(dT) primer (Ambion). The transcription mixture was then used in PCRs with primers specific for the indicated mRNA. Pilot experiments were performed first to ensure that PCRs were occurring in the linear range of amplification. The following primers were used for amplification: for c-Myc, 5′-CAGCTTGTACCTGCAGGATC-3′ and 5′-GTCGAGGAGAGCAGAGAATC-3′; for JunB, 5′-AACAGCAACGGCGTGATCAC-3′ and 5′-GCAGATCGTCCAGGGCTTTG −3′; for Cdk7, 5′-TTCAGTGCAGCAGGAGACGAC-3′ and 5′-CTGGACAGTTTGGTCTTGGC-3′; for cyclin D1, 5′-AAGATCGTCGCCACCTGGAT-3′ and 5′-CACTTGCATGTTCGTGGCCT-3′; for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-CGTCTTCACCACCATGGAGA-3′ and 5′-CGGCCATCACGCCACAGTTT-3′; for Ki67, 5′-TGACCAGAACAAGGGGAAGG-3′ and 5′-CCAGTAGTATACAGGTCTTTG-3′; for Cdk1, 5′-TCAGCTCGTTACTCAACTCC-3′ and 5′-TCCACTTCTGGCCACACTTC-3′; for Cdk2, 5′-TTGTCAAGCTGCTGGATGTC-3′ and 5′-AATGGCAGAAAGCTAGGCCC-3′; for cyclin B, 5′-AGAACCTGAGCCTGTTAAAG-3′ and 5′-GCATCCACATCATTTACTGC-3′; for cyclin E, 5′-TATTGCACCATCCAGAGGC-3′ and 5′-CACACCTCCATTAACCAATC-3′.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed based on a protocol described previously (21) with anti-Cdk9 antibody. After DNA purification, PCRs containing [α-32P]dCTP were carried out for 20 cycles, and the products were analyzed on a 6% polyacrylamide gel. Input and immunoprecipitated chromatin were analyzed first in pilot experiments to ensure that PCRs were occurring in the linear range of amplification. The distributions of Cdk9 on the c-myc promoter region between −93 and +87, exon 1 of the c-myc gene between +266 and +419, exon 2 between +4863 and +5003, and an interior region of GAPDH between +4233 and +4550 were detected.
RESULTS
P-TEFb becomes associated with chromosomes toward the end of mitosis.
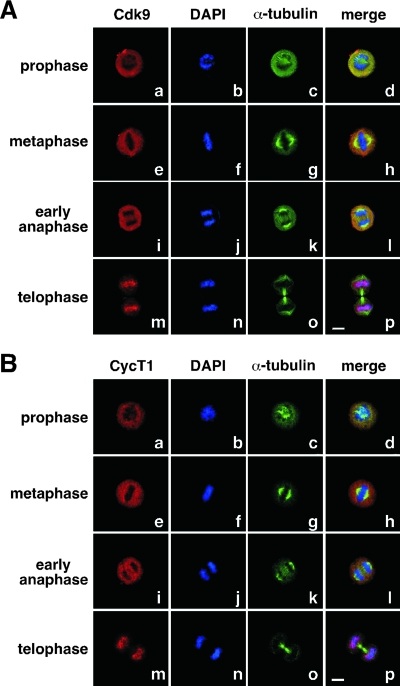
Brd4 has been shown to remain bound to chromosomes throughout mitosis in a number of human and murine cell lines (2, 3, 13, 15, 25). Given that P-TEFb has been demonstrated as a major associated factor of Brd4, we examined the localizations of Cdk9 and CycT1 at different stages of mitosis in HeLa cells by immunofluorescence staining followed by confocal microscopy. Different from what has been described for Brd4, both Cdk9 and CycT1 were displaced from chromosomes (as revealed by DAPI staining) and dispersed into the cytoplasm during prophase, metaphase, and what appeared to be early anaphase of mitosis (Fig. 1). This observation is nevertheless consistent with the global shut-down of transcription during mitosis, when most other transcription and RNA processing factors are known to dissociate from chromosomes (6, 14, 18). Interestingly, as mitosis progressed further toward telophase and two daughter cells were about to pinch off, as indicated by the presence of the cleavage furrow (Fig. 1, anti-α-tubulin staining in frames o), Cdk9 and CycT1 became abruptly reassociated with chromosomes and costained with DAPI (frames m to p). Importantly, this behavior of Cdk9 and CycT1 was observed not only in HeLa cells but also in the mouse mammary tumor cell line C127 (see below), implicating it as a rather general phenomenon.
FIG. 1.
Reloading of P-TEFb onto mitotic chromosomes toward the end of mitosis. HeLa cells were cultured on coverslips, fixed, and immunostained to reveal the subcellular localizations of Cdk9 (A) and CycT1 (B) at various stages of mitosis by confocal microscopy (red). The localizations of chromosomal DNA (DAPI staining; blue) and α-tubulin (green) in the same cells are also shown. Bar, 5 μm.
P-TEFb associates with chromosomes before nuclear envelope/lamina reassembly.
To determine at what stage of mitosis P-TEFb becomes associated with chromosomes, we examined the temporal relationship between P-TEFb localization on mitotic chromosomes and the process of nuclear envelope/lamina reassembly. To this end, we double-stained HeLa cells with antibodies against CycT1 and lamin B1. As a component of the nuclear lamina, lamin B1 is involved in nuclear envelope/lamina assembly. The nuclear envelope/lamina of higher eukaryotes breaks down during prophase and is reconstituted around chromosomes during late anaphase to telophase, reestablishing the boundary of the interphase nucleus (5, 7).
In agreement with this description, the nuclear lamina was disassembled, and lamin B1 was dispersed into the cytoplasm during metaphase (Fig. 2A, frames a to d) and early or mid-anaphase (frames e to h). Meanwhile, CycT1 was also excluded from chromosomes and showed a similarly diffused pattern. However, as mitosis proceeded further, lamin B1 and CycT1 began to have drastically different localizations. While CycT1 was already localized on chromosomes, lamin B1 was still mostly dispersed throughout the cytoplasm (frames i to l). Based on the fact that the reconstitution of the nuclear envelope/lamina had not occurred at this moment and would not do so until late anaphase to telophase, P-TEFb was judged to begin its association with chromosomes some time around mid- to late anaphase. Consistent with this analysis, after the nuclear envelope/lamina was completely reconstituted at telophase, lamin B1 finally displayed the characteristic ring-like structure that tightly surrounded chromosomes, which were yet to undergo complete decondensation (Fig. 2A, frames m to p).
FIG. 2.
Unlike most other general transcription factors and Pol II, P-TEFb is recruited to mitotic chromosomes before nuclear envelope/lamina reassembly. (A) Coimmunofluorescence followed by confocal microscopic analyses of the localizations of CycT1 (red) and lamin B1 (green) in HeLa cells at various stages of mitosis (left) was performed. Staining of chromosomal DNA by DAPI (blue) and a merge of all three stainings in the same cells are also shown. Bar, 5 μm. (B) The localizations of lamin B1, TFIIB, TFIIF (Rap30), Pol II (the cells under discussion are indicated by an arrow), and Cdk9 in HeLa cells at mid- to late anaphase were examined by coimmunofluorescence as in panel A.
P-TEFb differs from other transcription factors in terms of the timing of entry into daughter nuclei.
It is important to emphasize that the interaction of P-TEFb with chromosomes before nuclear envelope/lamina reassembly sets P-TEFb apart from virtually all other general transcription and RNA processing factors. These factors enter daughter nuclei only after nuclear envelope/lamina formation and presumably depend on the nuclear import machinery for this effect (17). For example, when analyzed under the same conditions as used for Cdk9 and CycT1, TFIIB, TFIIE (anti-p34) (data not shown), TFIIF (anti-RAP30), and RNA Pol II (monoclonal antibody 8WG16) were found still dispersed in the cytoplasm when lamina and the nuclear envelope were beginning to form around chromosomes (Fig. 2B). In fact, for these general transcription factors and Pol II, we did not find a single cell showing a situation similar to that depicted in frames a to d of Fig. 2B, where Cdk9 was already localized on condensed chromosomes even before nuclear envelope/lamina formation. Thus, unlike these factors, P-TEFb must rely on a specialized recruitment mechanism rather than the default nuclear import process to become associated with chromosomes.
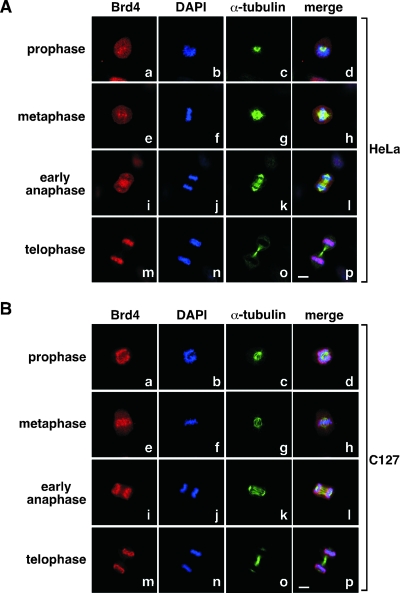
Cell-type-specific Brd4 localization patterns during mitosis.
Brd4 has been identified as a major P-TEFb-associated factor (10, 24). With the demonstration of a controlled reloading of P-TEFb onto chromosomes toward the end of mitosis, we next examined the localization of Brd4 in mitotic cells. To our surprise, in HeLa cells, Brd4 was not always localized on chromosomes throughout mitosis (Fig. 3A), which contrasted sharply with the observations made in a number of other human and mouse cell lines (2, 3, 13, 15, 25). First, in HeLa cells that were at either pro- or metaphase, Brd4 was not preferentially associated with mitotic chromosomes but rather showed relatively dispersed distributions throughout the entire cell (Fig. 3A, frames a to h). At early anaphase, Brd4 was even largely excluded from chromosomes (frames i to l). However, at telophase when two daughter cells were pinching off through cytokinesis, Brd4 was found to colocalize with chromosomes (frames m to p), a behavior reminiscent of that of P-TEFb as described above.
FIG. 3.
Brd4 displays cell-type-specific localization patterns during mitosis. Distributions of Brd4 in HeLa (A) and C127 cells (B) at various stages of mitosis were examined by coimmunofluorescence with antibodies against either Brd4 (red) or α-tubulin (green). Chromosomes in the same cells were stained with DAPI (blue). Cell images were obtained with a confocal microscope. Bar, 5 μm.
To be sure that our immunofluorescence procedure did not prevent the detection of Brd4 on chromosomes until late mitosis in HeLa cells, we performed the same procedure on mouse C127 cells, where Brd4 had previously been reported to localize on chromosomes at metaphase (13). Indeed, a major portion of Brd4 remained associated with chromosomes through the entire mitotic process (Fig. 3B), although there appeared to be further enrichment of the protein on chromosomes at telophase (Fig. 3B, compare frame m with frames a, e, and i). Thus, the association of Brd4 with chromosomes during most of mitosis is controlled differently in HeLa and C127 cells. However, at late mitosis, Brd4 displayed the same intimate association with chromosomes in both cell lines.
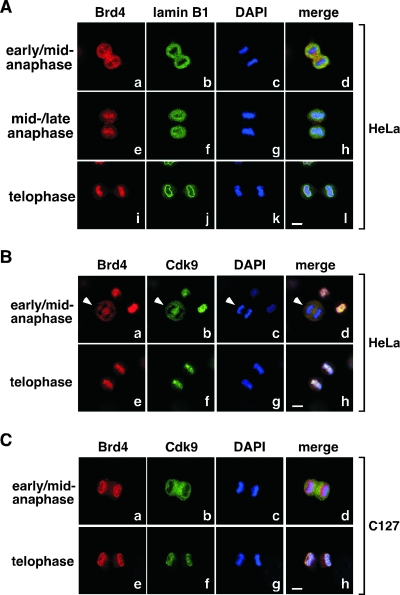
Colocalization of Brd4 with P-TEFb on mitotic chromosomes before nuclear envelope/lamina reassembly.
Just like P-TEFb, Brd4 also began its reloading onto mitotic chromosomes around mid- to late anaphase right before nuclear envelope/lamina reassembly in HeLa cells (Fig. 4A, frames e to h). In fact, both proteins were in the cytoplasm around early to mid-anaphase (Fig. 4B) but then colocalized on chromosomes immediately afterward (Fig. 4B, telophase). In mouse C127 cells, however, the situation was somewhat different. Despite the fact that Brd4 and Cdk9 were localized differently from each other at early to mid-anaphase (Fig. 4C), Cdk9 eventually joined Brd4 to become associated with chromosomes toward the end of mitosis (Fig. 4C, telophase). These results indicate that although Brd4 displayed distinct localization patterns during most of mitosis in HeLa and C127 cells, it became intimately associated with mitotic chromosomes in both cell lines right before nuclear envelope/lamina formation, which could be responsible for the loading of P-TEFb onto chromosomes at precisely the same moment.
FIG. 4.
Colocalization of Brd4 and P-TEFb on mitotic chromosomes before nuclear envelope/lamina reassembly. (A) The localizations of Brd4 (red) and lamin B1 (green) in HeLa cells at various stages of mitosis (indicated on the left) were examined by coimmunofluorescence and confocal microscopy. Chromosomal DNA in the same cells was stained by DAPI (blue). Bar, 5 μm. (B and C) HeLa and C127 cells at the indicated stages of mitosis were immunostained with antibodies against Brd4 (red) and Cdk9 (green) and examined with a confocal microscope. DAPI staining of chromosomes (blue) in the same cells is also shown. Bar, 5 μm.
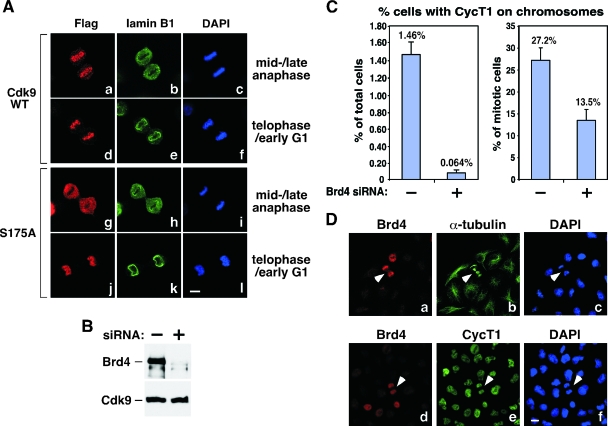
P-TEFb's association with mitotic chromosomes correlates with its ability to bind Brd4.
To test whether Brd4 played a key role in the recruitment of P-TEFb to mitotic chromosomes, a stable HeLa-based cell line expressing the Flag-tagged Cdk9 mutant S175A-f, which fails to bind to Brd4 (24), was established. Except for Brd4, this mutant is able to bind to all other known Cdk9-associated factors including CycT1, HEXIM1, and 7SK (24). Coimmunofluorescence staining with anti-Flag and anti-lamin B1 antibodies indicated that wild-type Cdk9-f, like its endogenous counterpart, was properly localized to chromosomes around mid- to late anaphase and before nuclear envelope/lamina reassembly (Fig. 5A, frames a to c). In contrast, S175A-f failed to display chromosome-association at this stage (frames g to i). It was imported into the nucleus only after nuclear envelope/lamina was completely reconstituted in late telophase/early G1 (frames j to l), most likely through the conventional nuclear import machinery. These data suggest that the ability to bind Brd4 was crucial for P-TEFb to become associated with chromosomes before nuclear envelope/lamina formation.
FIG. 5.
Brd4-dependent recruitment of P-TEFb to mitotic chromosomes. (A) Wild-type (WT) but not mutant Cdk9 S175A becomes associated with mitotic chromosomes before nuclear envelope/lamina reassembly. Stable HeLa-based cell lines expressing Flag-tagged wild-type Cdk9 or Cdk9 mutant S175A were immunostained at the indicated cell cycle stages (right) with antibodies against Flag (red) and lamin B1 (green). Chromosomes in the same cells were stained with DAPI (blue). Bar, 5 μm. (B) siRNA-mediated depletion of Brd4 in HeLa cells. The levels of Brd4 and Cdk9 in cells containing either an empty vector (−) or the Brd4-specific siRNA (+) were analyzed by Western blotting. (C) Reduced association of CycT1 with mitotic chromosomes in Brd4 knockdown cells. HeLa cells with (+) or without (−) the expressed Brd4 siRNA were immunostained with antibodies against CycT1 and α-tubulin to examine the behaviors of CycT1 in mitotic cells. Quantification of the percentages of cells with CycT1 on chromosomes is shown against either total cell population (left) or only mitotic cells (right). Error bar represents the mean ± standard deviation. (D) Association of CycT1 with mitotic chromosomes in cells with incomplete Brd4 depletion. Coimmunofluorescence staining was performed with anti-Brd4 (red) and anti-α-tubulin (green) or anti-CycT1 (green) antibodies in Brd4 knockdown cells. Chromosomes were stained with DAPI (blue). Arrowheads indicate cells with detectable levels of Brd4 that were in mitosis. Bar, 10 μm.
Brd4 knockdown prevents the recruitment of P-TEFb to mitotic chromosomes.
To further investigate the role of Brd4 in recruiting P-TEFb to mitotic chromosomes, we knocked down Brd4 levels in HeLa cells by using a lentivirus-based system that uses the Cre-loxP control switch for inducible small interfering RNA (siRNA) expression (20). Upon the introduction of the Cre recombinase, cellular Brd4 levels were reduced by about 90% compared to control cells containing an empty vector (Fig. 5B).
We then performed coimmunostaining with antibodies against CycT1 and α-tubulin to examine the behaviors of CycT1 in control and Brd4 knockdown cells that were in mitosis. Inspections of several thousand cells in each group indicated that when the total cell population was used as the base, the Brd4 siRNA reduced the association of CycT1 with chromosomes by about 23-fold (from 1.46% to 0.064%) (Fig. 5C, left graph). However, when only the mitotic cells were selected (more than 150 cells from each group), the knockdown reduced CycT1 on chromosomes by twofold (from 27.2% to 13.5%) (Fig. 5C, right graph). This discrepancy is caused by the fact that the percentage of cells able to reach mitosis due to siRNA-induced cell cycle arrest and apoptosis (see below) was much smaller within the knockdown population than in control cells.
Further examination of the situations in knockdown cells revealed that the twofold decrease in CycT1's association with chromosomes was most likely a gross underestimation of the true extent of the siRNA-mediated reduction. This is because the siRNA did not uniformly reduce Brd4 levels in all cells. Compared to either the α-tubulin or DAPI staining that was ubiquitous in all cells, a small percentage of cells from the Brd4 knockdown population still contained Brd4 at levels similar to those of control cells (Fig. 5D, frames a and d; also data not shown). Moreover, almost all the cells that were able to reach late telophase, as indicated by the formation of the cleavage furrow, also contained detectable levels of Brd4 (Fig. 5D, cells indicated by arrows in frames a to c). Thus, it is likely that the association of CycT1 with mitotic chromosomes observed in these cells was actually due to the presence of Brd4 (or lack of siRNA effect) in them (Fig. 5D, arrows in frames d to f). In light of these considerations, we believe that the true effect of Brd4 siRNA in reducing the association of P-TEFb with chromosomes should be much greater than the above-mentioned twofold difference. This analysis, together with the data shown in Fig. 5A employing the Cdk9 mutant S175A, is consistent with the notion that Brd4 is required for reloading P-TEFb onto chromosomes at late mitosis.
Significantly enhanced Brd4-P-TEFb interaction in cells progressing from late mitosis to early G1.
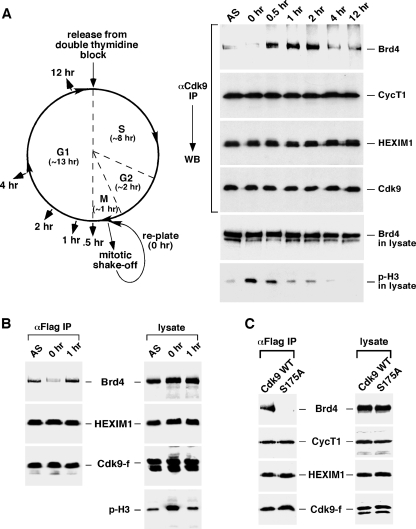
Next, we investigated whether the Brd4-dependent recruitment of P-TEFb to mitotic chromosomes would involve any increase in the interaction between the two proteins and, if so, how long this increase would last. To address these two questions, we first arrested HeLa cells at the G1/S boundary by the method of a double-thymidine block (Fig. 6A, left panel). Synchronized cells were then released from the arrest and allowed to proceed for about 10 h to reach mitosis. To collect mitotic cells, the rounded-up, detached cells were shaken off the culture dish and replated in fresh medium at time zero (Fig. 6A, left). Cells were subsequently harvested at the indicated time points as they were progressing from mitosis into G1.
FIG. 6.
The Brd4-P-TEFb interaction significantly increases in cells progressing from late mitosis to early G1. (A) Schematic representation (left) the experimental setup. HeLa cell growth was synchronized by a double-thymidine block. Ten hours after the release from the block, mitotic cells were collected by shake-off and replated in fresh medium. Cell lysates were prepared at indicated time points and subjected to anti-Cdk9 immunoprecipitation. The lysates and the immunoprecipitates were analyzed by Western blotting (right) for the various components, as indicated to the right. (B) Mitotic cells were also collected by shake-off at 0 h from the asychronized (AS) HeLa-based cell line F1C2 expressing Flag-tagged Cdk9-f and replated in fresh medium for 1 h. The levels of the indicated components in either the cell lysate or the anti-Cdk9-f immunoprecipitates were analyzed by Western blotting. (C) The enhanced Brd4-P-TEFb interaction at late M/early G1 is disrupted by the S175A mutation in Cdk9. Stable cell lines expressing Flag-tagged wild-type Cdk9 or S175A were collected by shake-off and replated in fresh medium for 1 h. The cell lysates and anti-Flag immunoprecipitates were analyzed by Western blotting as in panel B. IP, immunoprecipitation; WT, wild type; WB, Western blotting.
Western analysis of the levels of phosphorylated histone 3, which served as an M phase indicator (15), in the lysates of the harvested cells indicated that more than 50% of cells exited the M phase and entered early G1 after about 0.5 h in fresh medium (Fig. 6A, right panel). By 2 h, very few cells were still in mitosis, which was consistent with DNA content analysis by flow cytometry (data not shown). Interestingly, although Western analyses indicate that the levels of Cdk9, Brd4, HEXIM1, and CycT1 in cell lysates remained unchanged during the progression from mitosis to G1 (Fig. 6A, right panel; also data not shown), the interaction of the immunoprecipitated Cdk9 with Brd4 but not CycT1 or HEXIM1 underwent significant changes (Fig. 6A, right panel). For example, while the Cdk9-Brd4 interaction was barely detectable during most of the M phase, it was dramatically enhanced when cells were reaching the end of mitosis, exiting mitosis, and then entering G1. The elevated Cdk9-Brd4 interaction lasted for about 1.5 h (0.5 to 2 h from the start of replating) (Fig. 6A, right panel). However, as cells were progressing into late G1, this interaction decreased again and remained at an intermediate level for the rest of G1.
The interaction of P-TEFb with Brd4 can be affected by a number of stress-inducing agents (24). To rule out the possibility that the observed fluctuation in this interaction was caused by the thymidine treatment of cells, we collected natural, untreated mitotic cells and replated them in fresh medium. Consistent with the results described above, the association of P-TEFb with Brd4 was markedly reduced when cells were in mitosis (Fig. 6B, left panel, 0 h) and increased significantly 1 h later when they were entering G1. Again, the levels of Brd4 and Cdk9 remained unchanged throughout the entire process (Fig. 6B, right panel). Finally, the enhanced Brd4-P-TEFb interaction observed at late mitosis and early G1 was completely disrupted by the S175A mutation in Cdk9 (Fig. 6C). This result mirrors the situation described previously in unsynchronized cells (24).
Brd4 depletion causes G1 cell cycle arrest and apoptosis.
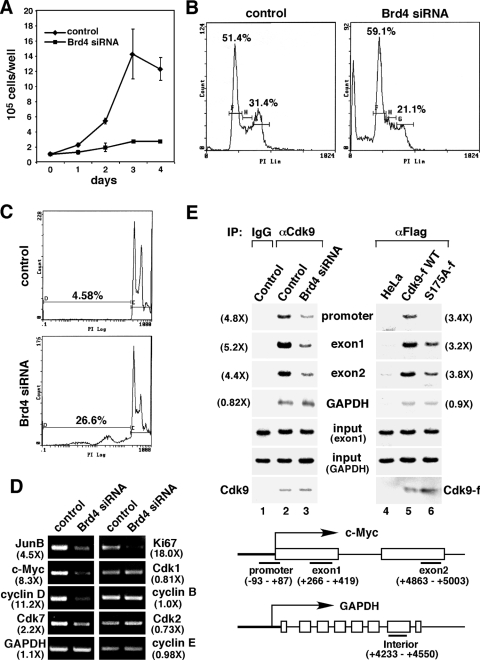
What could be the biological significance of the enhanced Brd4-P-TEFb interaction as well as the Brd4-dependent recruitment of P-TEFb to chromosomes during the cell cycle progression from late mitosis to early G1? To address this question, we first examined the effect of the Brd4 siRNA on HeLa cell growth. Compared to control cells containing an empty vector, induction of siRNA expression significantly retarded cell growth (Fig. 7A), consistent with the previous finding that Brd4-null cells do not grow in culture (15). Next, DNA profiles of control and Brd4 knockdown cells were analyzed by flow cytometry. Compared to control cells, the expression of the Brd4 siRNA resulted in an increase in the percentage of cells in G1 (from 51.4% to 59.1%) (Fig. 7B) and a concurrent reduction in the fraction of cells in G2/M (from 31.4% to 21.1%). Further analyses by BrdU incorporation to determine cells in S phase and phospho-H3 immunoblotting to identify mitotic cells also revealed a 14.4% and 33.6% reduction, respectively, in the number of cells in these two phases as a result of Brd4 knockdown (data not shown). These data are consistent with the notion that Brd4 depletion and the resulting lack of recruitment of P-TEFb to chromosomes at the M/G1 boundary caused more cells to be arrested at G1. Along with—and also probably a direct result of—this arrest, a significant increase in the sub-G1 population containing fragmented DNA was also observed (from 4.58% to 26.6%) (Fig. 6C). This latter result is consistent with the previous observation that Brd4 depletion leads to apoptosis (15).
FIG. 7.
Brd4 knockdown decreases the binding of P-TEFb to and expression of key early G1 genes, which leads to G1 cell cycle arrest and apoptosis. (A) HeLa cells expressing the Brd4 siRNA or containing an empty vector were placed at a low concentration (1 × 105 cells/well each) in culture medium on day 0. The numbers of cells were counted at the indicated days, with the error bars representing the mean ± standard deviation. (B) Control or Brd4 knockdown cells were stained with propidium iodide and analyzed by flow cytometry. Lin, linear. (C) Control and Brd4 knockdown cells were analyzed by flow cytometry as in panel B, with the areas corresponding to sub-G1 populations enlarged. (D) mRNAs were harvested from control or Brd4 knockdown cells and subjected to RT-PCR analyses with the primer sets specific for the indicated genes. Numbers in parentheses indicate the relative reduction in mRNA levels as a result of Brd4 knockdown. (E) Brd4 knockdown or the S175A mutation in Cdk9 reduces the binding of Cdk9 to the c-Myc gene in early G1. Control or Brd4 knockdown cells (lanes 1 to 3), HeLa (lane 4), or HeLa-based cell lines expressing Cdk9-f (lane 5) or S175A-f (lane 6) in early G1 were harvested and subjected to ChIP analysis. Three regions corresponding to the promoter, exon 1, and exon 2 of the c-Myc gene (bottom), as well as an interior region of GAPDH were amplified by PCR from the input chromatin. Cdk9 or Cdk9-f associated with the immunoprecipitated chromatin was detected by Western blotting. Numbers in parentheses indicate the relative reduction caused by Brd4 knockdown or the S175A mutation. IgG, immunoglobulin G; α, anti; IP, immunoprecipitation; WT, wild type; PI, propidium iodide.
Reduced expression of key G1 and growth-associated genes in Brd4 knockdown cells.
To determine whether the Brd4-mediated recruitment of P-TEFb to mitotic chromosomes is required for gene expression in early G1, we compared mRNA levels of a selected panel of genes in control and Brd4 knockdown cells. Mitotic cells were collected by shake-off. Upon incubation in fresh medium for 1 h, most cells were in early G1 (Fig. 6). Analysis by RT-PCR at this point indicates that the expression of two immediate-early transcription factors, JunB and c-Myc, as well as the G1-specific cyclin, cyclin D1—all of which are known to be induced in early G1 and important for cell cycle progression (9, 11)-was significantly reduced in Brd4 knockdown cells (Fig. 7D). Moreover, a diminished expression was also observed for Cdk7 (Fig. 7D), the catalytic subunit of the Cdk-activating kinase, whose expression is known to be stimulated at the M/G1 boundary (22). Finally, in agreement with the Brd4 siRNA-induced growth arrest, the level of Ki67, a proliferation marker expressed throughout the cell cycle but not in quiescent (G0) cells, was also significantly reduced in knockdown cells (Fig. 7D).
In contrast to these key early G1 and growth-associated genes, the mRNA level of a well-characterized housekeeping gene, GAPDH, was mostly unaffected by Brd4 knockdown (Fig. 7D). Moreover, several proteins that function at later time points in the cell cycle, such as the G2/M-specific Cdk1 and cyclin B as well as the G1/S-specific Cdk2 and cyclin E, also displayed no major change in their expression. Although a generic effect of Brd4 knockdown cannot be completely ruled out, these data are consistent with the model that Brd4 is important for the expression of key early G1 and growth-associated genes through recruiting P-TEFb to mitotic chromosomes prior to the M/G1 transition. This model agrees well with recent microarray analysis, which revealed that the induction of numerous genes at early G1 was markedly inhibited in Brd4 knockdown cells (K. Ozato, personal communication).
Reduced recruitment of P-TEFb to the c-Myc locus at early G1 in Brd4 knockdown cells.
To determine how the Brd4-mediated recruitment of P-TEFb to mitotic chromosomes would affect the loading of P-TEFb to a specific gene locus at early G1, we performed a ChIP assay to examine the association of Cdk9 with the promoter, exon 1, and exon 2 regions of the c-Myc gene in control and Brd4 knockdown cells. Consistent with the Brd4-dependent c-Myc expression described above, Brd4 knockdown markedly decreased the association of Cdk9 with all three regions of the c-Myc gene (Fig. 7E, lanes 1 to 3). In contrast, the interaction of Cdk9 with the GAPDH gene was very weak (an exposure two times longer was needed to see the bands despite the abundant input signals amplified with the GAPDH-specific primers) and also largely unaffected by Brd4 depletion (lanes 1 to 3). Consistent with these observations, the Cdk9 mutant S175A-f, which was defective in Brd4-binding in early G1 cells (Fig. 6C), displayed a significantly decreased occupancy at the c-Myc locus compared to wild-type Cdk9-f (Fig. 7E, lanes 4 to 6). As for GAPDH, occupancy of wild-type Cdk9-f at this locus was only slightly above the background level (obtained through anti-Flag immunoprecipitation with HeLa chromatin as the input) and largely unaffected by the S175A mutation (Fig. 7E, lanes 4 to 6). Thus, unlike the c-Myc gene, which depends on the Brd4-recruited P-TEFb for efficient transcription in early G1, the GAPDH transcription involves only a very small amount of P-TEFb, which is recruited via a Brd4-independent mechanism. Taken together, through analyzing c-Myc and several other key G1 and growth-associated genes by RT-PCR and ChIP, our data strongly suggest that the decreased recruitment of P-TEFb to and expression from these genes are the primary causes for the observed G1 arrest and apoptosis of Brd4 knockdown cells.
DISCUSSION
Whether the activity of P-TEFb is regulated through the cell cycle and, if so, how it is regulated and whether this regulation may affect cell cycle progression are key questions that have not yet been addressed in the past. In this study, a critical role for the bromodomain protein Brd4 in linking P-TEFb to cell cycle control has been revealed. Although the levels of Cdk9 and CycT1 as well as the kinase activity of the isolated P-TEFb heterodimer do not fluctuate throughout the cell cycle (4), our current data indicate that the interaction of P-TEFb with its positive regulator Brd4 as well as the Brd4-dependent recruitment of P-TEFb to chromosomes can, in fact, become significantly enhanced at the M/G1 boundary. Importantly, these changes allow P-TEFb to stimulate transcription of the genes that are essential for the cell cycle progression through G1.
Our current data, in conjunction with previous findings, also indicate that P-TEFb is different from virtually all other transcription and pre-mRNA processing factors in terms of the timing and manner of its entry into newly formed daughter nuclei after each cell division. In contrast to most other factors that are imported into daughter nuclei only after nuclear envelope/lamina reconstitution (17) (Fig. 2B) and thus likely depend on the standard nuclear import machinery, P-TEFb is recruited to mitotic chromosomes by Brd4 before nuclear envelope/lamina formation.
What could be the benefit to have P-TEFb, a transcription elongation factor, reach chromatin templates before most transcription initiation factors and RNA Pol II, which are required to set up preinitiation complexes in early G1? We believe the answer may lie in P-TEFb's unique position in the transcription cycle as well as the demonstrated role of Brd4 in transmitting transcriptional memory through mitosis (3, 15). Compared to transcription initiation and early elongation, which are frequently aborted due to the presence of strong negative signals and/or the absence of positive stimuli (19), productive elongation is a much more reliable indicator of a highly committed and robust transcription state. Since P-TEFb is one of the most important activators of elongation, its early recruitment by Brd4 to chromatin templates may help mark those genes whose transcription cycle must be completed, and thus active expression status preserved, across cell division.
Brd4 has been shown to interact with the acetylated histones H3 and H4 (2). Interestingly, despite the demonstration of a general chromatin hypoacetylation in mitotic cells (12), several key lysine residues on H3 and H4 that are recognized by Brd4 remain acetylated on mitotic chromosomes (12). With the acetylated histones H3 and H4 serving as a permanent mark for transcriptional memory, it is perhaps not necessary for Brd4 to remain associated with chromosomes at all stages of mitosis. It is, however, more important for it to know precisely when and where to recruit P-TEFb to chromatin templates. Consistent with this analysis, Brd4 was found in HeLa and several other cell types (e.g., mouse cell lines NIH 3T3 and C2C12) to migrate to chromosomes together with P-TEFb only at late mitosis (Fig. 3A; also data not shown). This is different from the continuous association of Brd4 with mitotic chromosomes observed in a number of other cell lines (2, 3, 13, 15, 25) (Fig. 3B). The mechanism and biological significance of such cell-type-specific difference are yet to be elucidated.
It is important that the mere presence of Brd4 on chromosomes is insufficient to recruit P-TEFb. This is evident in C127 cells, where Cdk9 was recruited only toward the end of mitosis, despite the fact that Brd4 was continuously present on chromosomes throughout the entire process. Our preliminary data indicate that other factors, such as variations in posttranslational modifications on Brd4 and/or P-TEFb at different stages of mitosis, could be additionally involved in controlling the dissociation and then reassociation of P-TEFb with Brd4/chromatin as cells progress from mitosis to G1 (data not shown). Currently, a major effort is under way to test this hypothesis by determining the nature of the modifications and mapping the modification sites. Although the signals and mechanism that control the cell cycle-dependent interaction between Brd4 and P-TEFb are yet to be determined, the P-TEFb and Brd4 dynamics described in this work may already serve as a new paradigm to study the contributions of these two molecules to control of the cell cycle and to the transmission of transcriptional memory across cell division.
Acknowledgments
We thank Wanichaya Ramey for technical assistance.
This work was supported by grants from the National Institutes of Health (AI41757) and The Susan G. Komen Breast Cancer Foundation (BCTR71506) to Q.Z.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Bonifacino, J. S., J. B. Dasso, J. Harford, K. M. Lippincott-schwartz, Yamada, and K. S. Morgan (ed.). 1998. Current protocols in cell biology. John Wiley and Sons, New York, NY.
- 2.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 1008758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garriga, J., S. Bhattacharya, J. Calbo, R. M. Marshall, M. Truongcao, D. S. Haines, and X. Grana. 2003. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell. Biol. 235165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldman, R. D., Y. Gruenbaum, R. D. Moir, D. K. Shumaker, and T. P. Spann. 2002. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16533-547. [DOI] [PubMed] [Google Scholar]
- 6.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22197-202. [DOI] [PubMed] [Google Scholar]
- 7.Haraguchi, T., T. Koujin, T. Hayakawa, T. Kaneda, C. Tsutsumi, N. Imamoto, C. Akazawa, J. Sukegawa, Y. Yoneda, and Y. Hiraoka. 2000. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113779-794. [DOI] [PubMed] [Google Scholar]
- 8.He, N., A. C. Pezda, and Q. Zhou. 2006. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol. Cell. Biol. 267068-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer, V. R., M. B. Eisen, D. T. Ross, G. Schuler, T. Moore, J. C. Lee, J. M. Trent, L. M. Staudt, J. Hudson, Jr., M. S. Boguski, D. Lashkari, D. Shalon, D. Botstein, and P. O. Brown. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 28383-87. [DOI] [PubMed] [Google Scholar]
- 10.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19523-534. [DOI] [PubMed] [Google Scholar]
- 11.King, K. L., and J. A. Cidlowski. 1998. Cell cycle regulation and apoptosis. Annu. Rev. Physiol. 60601-617. [DOI] [PubMed] [Google Scholar]
- 12.Kruhlak, M. J., M. J. Hendzel, W. Fischle, N. R. Bertos, S. Hameed, X. J. Yang, E. Verdin, and D. P. Bazett-Jones. 2001. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J. Biol. Chem. 27638307-38319. [DOI] [PubMed] [Google Scholar]
- 13.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 798920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muchardt, C., J. C. Reyes, B. Bourachot, E. Leguoy, and M. Yaniv. 1996. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 153394-3402. [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiyama, A., A. Dey, J. Miyazaki, and K. Ozato. 2006. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol. Biol. Cell 17814-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 17.Prasanth, K. V., P. A. Sacco-Bubulya, S. G. Prasanth, and D. L. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell 141043-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segil, N., M. Guermah, A. Hoffmann, R. G. Roeder, and N. Heintz. 1996. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 102389-2400. [DOI] [PubMed] [Google Scholar]
- 19.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 182437-2468. [DOI] [PubMed] [Google Scholar]
- 20.Ventura, A., A. Meissner, C. P. Dillon, M. McManus, P. A. Sharp, L. Van Parijs, R. Jaenisch, and T. Jacks. 2004. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA 10110380-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, G., M. A. Balamotis, J. L. Stevens, Y. Yamaguchi, H. Handa, and A. J. Berk. 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17683-694. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield, M. L., G. Sherlock, A. J. Saldanha, J. I. Murray, C. A. Ball, K. E. Alexander, J. C. Matese, C. M. Perou, M. M. Hurt, P. O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 131977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19535-545. [DOI] [PubMed] [Google Scholar]
- 25.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, Q., and J. H. Yik. 2006. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70646-659. [DOI] [PMC free article] [PubMed] [Google Scholar]