Abstract
Promyelocytic leukemia protein (PML) is an important regulator due to its role in numerous cellular processes including apoptosis, viral infection, senescence, DNA damage repair, and cell cycle regulation. Despite the role of PML in many cellular functions, little is known about the regulation of PML itself. We show that PML stability is regulated through interaction with the peptidyl-prolyl cis-trans isomerase Pin1. This interaction is mediated through four serine-proline motifs in the C terminus of PML. Binding to Pin1 results in degradation of PML in a phosphorylation-dependent manner. Furthermore, our data indicate that sumoylation of PML blocks the interaction, thus preventing degradation of PML by this pathway. Functionally, we show that in the MDA-MB-231 breast cancer cell line modulating levels of Pin1 affects steady-state levels of PML. Furthermore, degradation of PML due to Pin1 acts both to protect these cells from hydrogen peroxide-induced death and to increase the rate of proliferation. Taken together, our work defines a novel mechanism by which sumoylation of PML prevents Pin1-dependent degradation. This interaction likely occurs in numerous cell lines and may be a pathway for oncogenic transformation.
Recent work has suggested a role for PML (promyelocytic leukemia protein) in many cellular processes including apoptosis, viral infection, transcription regulation, cell cycle regulation, and DNA damage repair (2, 5, 29). The role of PML in different cellular processes largely depends on its ability to form PML nuclear bodies (PML NBs). PML NBs are discrete nuclear structures which are thought to be organizing centers that serve to bring together proteins in a manner that allows more efficient regulation of cellular outcomes. Proteins localized to PML NBs include p53, CBP/p300, Daxx, BLM, Myc, and pRB (3, 56). Formation of PML NBs is thought to require PML based on their absence in PML−/− primary cells (3, 18, 23, 56) and their disruption in blast cells derived from acute promyelocytic leukemia patients that express PML-retinoic acid receptor α (RARα) (8, 10, 21).
Since its discovery, PML has been implicated in playing a role in carcinogenesis. PML was identified due to its involvement in acute promyelocytic leukemia as part of a chromosomal translocation with the RARα gene (30). The transformation potential of PML-RARα is dependent on the RBCC/TRIM motif found in the N terminus of PML, which is comprised of a RING finger, two B boxes, and a predicted coiled-coil (RBCC) domain (19). Subsequent studies show that PML−/− mice are prone to develop tumors in chemical and physical models of carcinogenesis (45). This may be due to defects in apoptotic responses to insults such as irradiation and oxidative stress (22, 28). Furthermore, PML protein expression is reduced or greatly diminished in tumor cell lines derived from prostate adenocarcinomas, colon adenocarcinomas, breast carcinomas, lung carcinomas, lymphomas, central nervous system tumors, and germ cell tumors; however, there is no change to PML transcript levels in these tissues compared to their normal counterparts (14).
Regulation of PML levels is critical to maintain proper cellular functions. Expression of PML can be regulated at both the transcriptional and posttranslational levels. PML mRNA can be increased by interferon treatment, which signals through Jak/Stat and interferon-stimulated response elements at the PML promoter (4, 24, 40). Ras transformation of mouse embryonic fibroblasts also induces PML in a p53-dependent manner (7, 12). On the other hand, there are various mechanisms that reduce cellular PML protein levels. PML degradation in Chinese hamster ovary cells can be induced by As2O3, which leads to PML sumoylation and proteosome-dependent proteolysis (23, 31). A recent report proposes that As2O3 induces PML phosphorylation in an extracellular signal-regulated kinase 2 (ERK2)-dependent manner, consequently leading to PML sumoylation (16), although the proteins responsible for PML sumoylation in this response are not well defined. The increased PML sumoylation, which results in additional PML NB formation, is thought to increase the expression of apoptotic genes. PML sumoylation is thus an intriguing regulatory target in carcinogenesis (22, 28). Due to PML's integral role in regulating PML NB formation as well as its other roles in the cell, it is important to understand how PML is regulated.
To uncover cellular factors that may directly regulate PML protein levels, we focused on proteins that are overexpressed in other cancers and that can directly affect the stability of their target proteins. One intriguing target is the peptidyl-prolyl isomerase Pin1, which is overexpressed in many human cancers and can function to facilitate the timing of cell proliferation (49).
Pin1 is a member of the pavrulin family of peptidyl-prolyl cis-trans isomerases (PPIases) (46). It is composed of an N-terminal WW domain, which is a protein-protein interaction domain, and a C-terminal PPIase domain. The WW domain of Pin1 preferentially binds to peptides containing a phospho-Ser/Thr-Pro (pS/T-P), whereas the PPIase domain catalyzes cis-trans isomerization of the peptide bond on the amino-terminal side of the proline residue (32, 38, 53). Through associations with its targets, Pin1 has been found to affect phosphorylation status, protein-protein interactions, subcellular localization, and protein stability (13, 15, 20, 25, 35, 36, 43, 51). Furthermore, Pin1 overexpression has been correlated with oncogenesis (51), leading us to hypothesize that Pin1 overexpression may decrease PML protein levels, thereby decreasing apoptotic potential and increasing cellular proliferation.
Our work shows that Pin1 is capable of binding to PML, resulting in decreased levels of PML. This interaction is mediated by four key serine residues in the C-terminal half of PML that are phosphorylated in mammalian cells. Furthermore, knockdown of Pin1 levels in MDA-MB-231 breast cancer cells, where Pin1 is normally overexpressed, results in an increase in PML levels. Our work indicates that the interaction between Pin1 and PML in these cells is at least partially responsible for increasing their ability to resist hydrogen peroxide-induced death as well as the rate of proliferation.
MATERIALS AND METHODS
Plasmid construction.
The construct CMX-HA-PML4 (where CMX is a cytomegalovirus-based promoter and HA is hemagglutinin) was cloned by conventional methods from a previously characterized PML construct (9) and follows the PML isoform labeling as outlined previously (19). The single PML mutants used (S403A, S518A, S527A, and S530A) have been previously described (52). The PML fragments used (residues 1 to 325 and 331 to 633) have been previously described (48). The mutants CMX-HA-PML4 (4X mutant, S403/505/518/527A; 3KR mutant, K65/160/490R) were created by multiple rounds of PCR site-directed mutagenesis using Pfu Turbo on the template CMX-HA-PML4. pGEX4T-1-Pin1 was cloned in our laboratory from a HeLa cDNA library. CMX-FLAG-SUMO1 was derived from a construct generously provided by Hsiu-Ming Shih. CMX-HA-PML-RARα has been described previously (33).
Cell culture.
CV-1 cells were maintained at 37°C in 7% CO2 in Dulbecco's modified Eagle medium supplemented with 10% charcoal-stripped fetal bovine serum, 50 units/ml of penicillin G, and 50 μg/ml of streptomycin sulfate. MDA-MB-231 cells were grown under similar conditions in 1× Dulbecco's modified Eagle medium with 4.5g/liter glucose, l-glutamine, and sodium pyruvate (Cellgro) supplemented with 10% charcoal-stripped fetal bovine serum, 50 units/ml of penicillin G, and 50 μg/ml of streptomycin sulfate. MDA-MB-231 and HeLa control short hairpin RNA (shRNA) and pSuper-shPin1 cell lines were created as described in Yi et al. (55). These were grown in the same medium as regular MDA-MB-231 cells with the addition of 0.5 μg/ml puromycin to maintain selection for cells with stably integrated DNA.
GST pull-down assays.
For glutathione S-transferase (GST) pull-down assays, cells were transfected with ≤10 μg of the indicated plasmid DNA. In cases where multiple pull-down assays were compared, all lanes received equal amounts of DNA. The transfections were carried out according to the manufacturer's protocol using Lipofectamine 2000 (Invitrogen). Whole-cell lysates were prepared 48 h after transfection using radioimmunoprecipitation assay (RIPA) buffer (1× phosphate-buffered saline [PBS], 1% NP-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) plus protease inhibitors. Whole-cell lysates were incubated for 60 min on a Nutator at 4°C with GST-Pin1-conjugated glutathione-Sepharose beads in NETN buffer (20 mM Tri-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.1% NP-40) with a mixture of protease inhibitors (Sigma). After incubation, the beads were washed three times with NETN and collected by centrifugation; the proteins were eluted and denatured by placing the samples at 100°C for 5 min and then run on 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels. The products were visualized by immunoblot analysis with anti-HA-horseradish peroxidase (HRP) (Roche) followed by detection using an ECL detection kit (Pierce). For the experiment shown in Fig. 2B, prior to pull-down, the lysates were incubated with calf intestinal phosphatase (CIP) for 30 min at 30°C for concentrations from 0 to 25% of the total volume of CIP (10 U/ml).
FIG. 2.
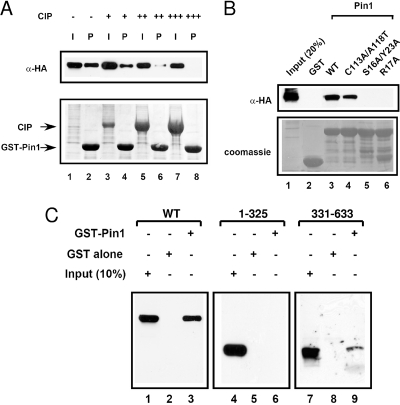
Mapping the motifs required for the interaction between PML and Pin1. (A) The interaction between PML and Pin1 is phosphorylation dependent. CV-1 cells were transfected with HA-PML4, and the resulting cell lysates were subjected to digestion with increasing amounts of CIP followed by GST pull-down with GST-Pin1 beads. The inputs (I) are 20%. The top panel shows an immunoblot of the pull-down products (P) probed with anti-HA antibody, and the lower panel shows Coomassie staining of the beads and CIP used in the experiments. (B) Pin1 must have an intact WW domain to interact with PML. CV-1 cells were transfected with HA-PML4, and whole-cell lysates were used in GST pull-down experiments with either wild-type or the indicated mutants of GST-Pin1 or GST alone. Immunoblot analyses were performed with anti-HA antibody and are shown in the upper panel. The lower panel shows Coomassie staining of the membrane to indicate equal loading of the GST beads. (C) The C-terminal region of PML is required for interaction with Pin1. Whole-cell lysates from CV-1 cells transfected with the PML fragments shown were subjected to GST pull-down with GST alone or GST-Pin1 and analyzed by Western blotting with anti-PML antibodies. WT, wild type; α, anti.
MS.
CV-1 cells were transfected with 10 μg of FLAG-PML4 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Whole-cell lysates were made using NETN buffer with protease inhibitors (Sigma) and lysed by sonication. The resulting lysates were immunoprecipitated in NETN using red anti-FLAG (M2) affinity gel (Sigma) for 4 h at 4°C. The products were then washed and separated on a 10% SDS-PAGE gel. The gel was stained using Coomassie stain, and the band corresponding to PML4 was excised and sent for mass spectrometry (MS) analysis by Mike Kinter at the Cleveland Clinic Foundation. Briefly, the gel pieces were washed with water, dehydrated with acetonitrile, reduced with dithiothreitol, and alkylated with iodoacetamide. In-gel proteolytic digestion was performed using trypsin overnight at room temperature. The resulting products were eluted using 50% acetonitrile and 5% formic acid and then processed down to 1% acetic acid for use in liquid chromatography-MS.
Immunoprecipitation.
To detect endogenous interactions, lysates of MDA-MB-231 cells were prepared using RIPA buffer with a mixture of protease inhibitors (Sigma). The resulting lysates were immunoprecipitated in NETN buffer for 4 h with antibody (anti-PML [5261; Santa Cruz] or anti-Pin1 [Upstate Biotechnology]). The immunoprecipitated complexes were resolved by SDS-PAGE and immunoblotted with the same antibodies. For overexpression immunoprecipitation analysis, CV-1 cells were transfected with 10 μg of DNA composed of CMX-FLAG-Pin1 and CMX-HA-PML4 or CMX alone using Lipofectamine 2000 following the published protocol from Invitrogen. Forty-eight hours after transfection, whole-cell lysates were made using RIPA buffer plus protease inhibitors and then immunoprecipitated by RIPA using red anti-FLAG (M2) affinity gel (Sigma) for 4 h at 4°C. The immunoprecipitates were analyzed by immunoblotting using anti-FLAG (Sigma) and anti-HA-HRP (Roche). Corresponding secondary antibodies were used, and visualization of the products was done using an ECL detection kit (Pierce).
Confocal microscopy.
Immunostaining was performed on endogenous proteins using MDA-MB-231 wild-type, control shRNA, or Pin1 shRNA cell lines. The cells were fixed in 3.7% paraformaldehyde in 1× PBS for 30 min at room temperature and permeabilized in 1× PBS with the addition of 0.1% Triton X-100 and 10% goat serum for 10 min. The cells were washed three times with 1× PBS and incubated in a PBS-10% goat serum-0.1% Tween-20 solution (ABB) for 60 min. Incubation with primary antibodies was carried out for 120 min in ABB. The cells were washed three times in 1× PBS, and the secondary antibodies were added for 30 to 60 min in the dark at room temperature in ABB. Coverslips were mounted to slides using Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) (H-1200; Vector Laboratories, Inc.). The primary antibodies used were anti-Pin1 (purified in our own laboratory) and anti-PML (mouse) (Santa Cruz). All confocal images were acquired using a Zeiss LSM 510 inverted laser scanning confocal microscope. A 63× (numerical aperture, 1.4) oil immersion Plan Apochromat objective was used for all experiments. To investigate the localization of the endogenous Pin1, images of Alexa Fluor 488 were collected using a 488-nm excitation light from an argon laser, a 488-nm dichroic mirror, and 500- to 550-nm band-pass barrier filter. For endogenous PML, images of Alexa Fluor 594 were collected using a 633-nm excitation light from an He/Ne2 laser, a 633-nm dichroic mirror, and 650-nm long-pass filter. All DAPI-stained nuclear images were collected using a Coherent Mira-F-V5-XW-220 (Verdi 5W) Ti-Sapphire laser tuned at 750 nm, a 700-nm dichroic mirror, and a 390- to 465-nm band-pass barrier filter.
Cycloheximide.
For cycloheximide assays, MDA-MB-231 cells were transfected with either wild-type or mutant HA-PML4 as indicated (see Fig. 5) using Lipofectamine 2000 following the manufacturer's protocol (Invitrogen). Twenty-four hours after transfection, the transfected cells were collected and split evenly into five 35-mm plates. The next day (48 h after transfection), the cells were treated with cycloheximide (20 μg/ml; Biomol) for the indicated times (see Fig. 5). Following treatment, whole-cell lysates were prepared using RIPA buffer with protease inhibitors. Equal amounts of protein for each time point were then run on an 8% SDS-PAGE gel, and immunoblotting was performed for visualization. The primary antibodies used were anti-HA-HRP (Roche) and anti-α-tubulin (Sigma). Quantification was performed using a VersaDoc imager (Bio-Rad). Graphs are representative of at least three replicate experiments.
FIG. 5.
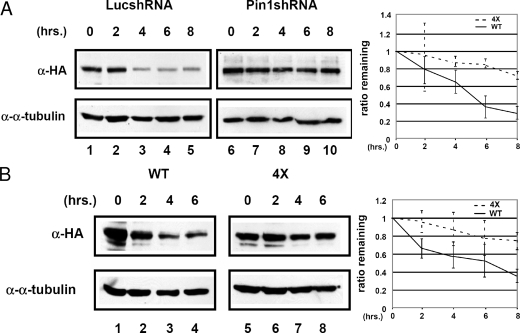
Interaction with Pin1 decreases PML stability. (A) PML has a longer half-life when Pin1 levels are decreased. MDA-MB-231 control or Pin1 shRNA cell lines were transfected with an HA-PML4 construct and treated with cycloheximide. Whole-cell lysates were prepared at the times indicated, and immunoblot analysis was performed with anti-HA and anti-α-tubulin antibodies. The panel on the right indicates the amount of PML remaining relative to α-tubulin. Results are representative of three replications. The estimated half-life in the MDA-MB-231 Luc shRNA lines is about 5.5 h, and in the MDA-MB-231 cell lines it is about 16 h. (B) A PML mutant that cannot interact with Pin1 is more stable than wild-type Pin1. MDA-MB-231 cells were transfected with either HA-PML4 wild-type or 4X mutant DNA, and cycloheximide experiments were performed and analyzed as described in panel A. The panel on the right indicates the amount of PML remaining relative to α-tubulin. Results are representative of three replications. The estimated half-life for wild-type PML4 is about 5.5 h, and for the PML4 4X mutant it is about 19 h. α, anti; WT, wild type.
Cell proliferation.
MDA-MB-231 control and Pin1 shRNA cell lines were grown to 30 to 40% confluence in six-well cell culture plates in the medium noted above without the antibiotics. Cells were treated with either a control random small interfering RNA (siRNA) oligonucleotide or siRNA directed against PML obtained from Dharmacon, according to the manufacturer's protocol using the Dharmafect 1 reagent. Each well received 10 μg of siRNA. Forty-eight hours after introduction of the siRNA, the cells were trypsinized and plated into 96-well plates. At times 0, 24, 48, 72, and 96 h after plating, cell proliferation was measured using an Aqueous One CellTiter reagent according to the manufacturer's protocol (Promega). Briefly, 20 μl of the CellTiter reagent was placed in each well and placed at 37°C for 1.5 h, after which the total volume of liquid was brought to 200 μl using deionized water; the absorbance was then read using a SpectraMax M2 plate reader at A490. Each time point was determined in triplicate, and the results are representative of at least three independent experiments. In parallel, a second set of six-well plates was similarly treated with siRNA, but after trypsinization, the cells were collected, and the RNA was isolated using an RNeasy RNA isolation kit from Qiagen. siRNA knockdown was assessed by comparing the amounts of RNA in the control and PML siRNA-treated cells by performing reverse transcription-PCR (RT-PCR) to analyze RNA levels of PML, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as a control. RT-PCR was performed using a One-step RT-PCR kit from Invitrogen. Primer sequences are available upon request.
Cell death.
MDA-MB-231 cell lines were treated with control oligonucleotide or siRNA directed against PML as described above. siRNA effectiveness was assayed by RT-PCR as described above. After 48 h of siRNA treatment, the cells were trypsinized and replated in equal numbers in six-well plates. Twenty-four hours later, the cells were treated with hydrogen peroxide (0, 50, 100, 150, or 200 μM). Twenty-four hours after treatment, cells and medium were collected and analyzed using trypan blue staining. Briefly, the cells were treated with trypan blue (MP Biochemicals) for 5 min at room temperature, and then the percentage of blue cells was counted using a hemocytometer. Each well was counted in triplicate, and data are representative of at least three independent experiments.
RT-PCR.
Total RNA was isolated from MDA-MB-231 luciferase (Luc) shRNA and MDA-MB-231 Pin1 shRNA cell lines using an RNeasy Mini Kit (Qiagen). RT-PCR was performed using a One-Step RT-PCR kit from Invitrogen according to the manufacturer's protocol. Sequences for PML and GAPDH primers are available upon request. A total of 200 ng of RNA was used in each reaction mixture.
RESULTS
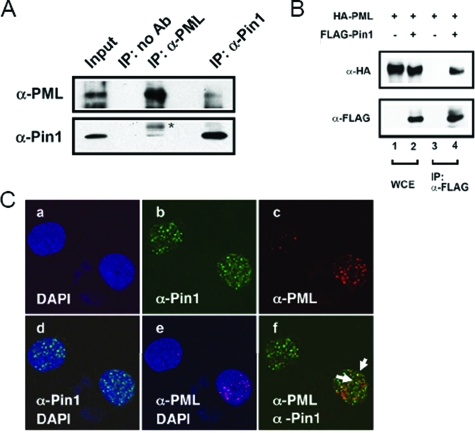
We hypothesized that for Pin1 to promote cell growth, it may need to down-regulate factors that promote cell death, such as PML. To evaluate if Pin1 may control PML activity, we first tested whether Pin1 and PML can interact in mammalian cells. We performed endogenous coimmunoprecipitation experiments using anti-Pin1 or anti-PML antibodies (Fig. 1A). We found that immunoprecipitation with either of the antibodies was able to pull down the other protein, indicating that they interact in mammalian cells. The association was observed in all cell lines tested including Hep2 and HeLa cells (data not shown). In order to further investigate this interaction, we cotransfected cells with HA-PML4 and FLAG-Pin1 and performed immunoprecipitations on the whole-cell lysates to determine if PML and Pin1 interact when overexpressed (Fig. 1B). The results confirm that Pin1 interacts with PML when these proteins are cooverexpressed. Furthermore, using confocal immunofluorescence microscopy, we observed that Pin1 is a mostly nuclear protein and that the majority of PML is localized in discrete nuclear speckles typical of PML NBs (Fig. 1C). When the images for Pin1 and PML were overlaid, we observed partial colocalization. Taken together, these results indicate that PML and Pin1 interact and can colocalize in mammalian cells.
FIG. 1.
PML interacts with Pin1. (A) Pin1 can interact with PML in cells. Whole-cell lysates from MDA-MB-231 cells were immunoprecipitated with anti-Pin1 antibodies. The immunopellets were separated by SDS-PAGE, and immunoblotting was performed with anti-PML and anti-Pin1 antibodies. Ten percent input is shown. The asterisk indicates a nonspecific band. (B) Overexpressed Pin1 and PML interact. CV-1 cells were transfected with HA-PML4 and FLAG-Pin1 constructs, and whole-cell lysates were immunoprecipitated with anti-FLAG antibodies. The products were analyzed by immunoblotting with anti-HA and anti-FLAG antibodies. Ten percent input is shown. (C) PML partially colocalizes with Pin1. MDA-MB-231 cells were immunostained with anti-PML and anti-Pin1 antibodies, and images were taken by confocal microscopy. DAPI shows staining for DNA. Arrows indicate a few points of colocalization. α, anti; IP, immunoprecipitation; Ab, antibody; WCE, whole-cell extract.
Since Pin1 interacts with proteins in a phosphorylation-dependent manner and catalyzes cis-trans isomerization of selected prolyl bonds, we determined whether the interaction between PML and Pin1 was dependent on PML phosphorylation. Whole-cell lysates from cells transfected with HA-PML4 were treated with increasing amounts of phosphatase and analyzed for their ability to bind to GST-Pin1 in a pull-down experiment (Fig. 2A). As expected, increasing the concentration of phosphatase resulted in decreased interaction between PML and Pin1 (lane 7 versus lane 2), suggesting that the interaction is phosphorylation dependent. Next, we performed a GST pull-down using three previously characterized Pin1 mutants to determine which ones interacted with PML. The S16A/Y23A and R17A mutations are located in the WW domain and have been shown to interrupt the ability of Pin1 to bind to target proteins (27, 39). The C113A/A118T mutations are located in the PPIase domain of Pin1 and result in decreased catalytic activity but can still interact with target proteins (27, 39). We used lysates from cells transfected with HA-PML4 and performed a pull-down using either wild-type or mutant GST-Pin1s (Fig. 2B). We found that both wild-type Pin1 and the PPIase mutant were capable of interacting with PML (lanes 3 and 4), while the two WW domain mutants could not interact with PML (lanes 5 and 6). Thus, the nature of the interaction between PML and Pin1 appears to be similar to that established between Pin1 and other targets. Next, we evaluated which portions of PML are required for the interaction with Pin1 by performing GST pull-down assays with fragments of PML (Fig. 2C). Deletion of the C-terminal region (starting at amino acid 325) almost completely abrogated the interaction with Pin1 (lanes 4 to 6), indicating that the C terminus is required for the interaction. When the N-terminal region of PML (residues 331 to 633) was deleted, there was also decreased interaction with Pin1, although binding was not completely abrogated (lanes 7 to 9). This suggests that the N terminus may play a role in stabilizing the protein-protein interaction. Taken together, our results indicate that the interaction between PML and Pin1 is phosphorylation dependent and requires the WW domain of Pin1 and the C-terminal region of PML.
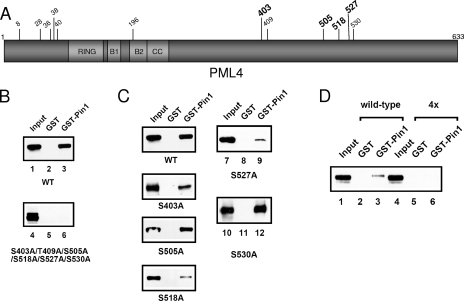
Pin1 is known to target S/T-P motifs, when the serine or threonine preceding the proline is phosphorylated (32, 53). There are 12 S/T-P motifs in PML4 (Fig. 3A). Since the C-terminal region of PML is required for the interaction with Pin1, we mutated all of the S/T residues that followed a P in this region of PML to A and investigated the ability of this nonphosphorylated mutant to bind to Pin1 in GST pull-down assays (Fig. 3B). We found that mutation of the six S/T sites in the C-terminal half of PML led to a significant loss of binding between PML and Pin1, indicating that at least some of these residues are involved in the interaction with Pin1. In order to further define which of these residues are important, we questioned which residues of PML were phosphorylated in vivo at levels high enough to detect. To answer this question, we overexpressed FLAG-PML4, immunoprecipitated the protein with anti-FLAG antibodies, isolated the band corresponding to PML, and analyzed the protein by MS. Our results show that four S/T residues of S/T-P motifs are phosphorylated in vivo, as indicated in bold in the schematic shown in Fig. 3A. Three of these sites are located in the region of PML found to be required for the interaction with Pin1, S403, S505, and S527, and one more residue, S518, may be phosphorylated at low levels, but the MS peak was not strong enough for an unambiguous identification. We used GST pull-down assays to test the effect of single mutations of each of these residues on their ability to bind to Pin1 (Fig. 3C). Our data show that mutation of S403 and S518 led to a partial decrease in the ability of PML to bind to Pin1, while S505A did not appear to significantly affect binding when mutated alone. Due to the close proximity of S527 to another S/T-P site (S530), we first generated and evaluated a double mutant and found a significant decrease in binding (data not shown). However, when single mutants containing S530A or S527A were made, S527A showed a significant loss of binding whereas S530A did not, indicating that only S527 appears to be involved in binding to Pin1. Finally, we generated an HA-PML4 4X mutant and tested its ability to interact with Pin1. In this assay, we performed the pull-down on lysates from MDA-MB-231 cells; we had previously observed an interaction between endogenous PML and Pin1 (Fig. 1C) in these cells, which overexpress Pin1. The MDA-MB-231 cells were transfected with either wild-type or mutant HA-PML4 (Fig. 3D). As shown in lane 6, the HA-PML4 4X mutant displayed reduced binding to Pin1 (lane 6 versus lane 3). We conclude that phosphorylation of one or more of these four residues is likely required for Pin1 binding. It is important to note that overall binding of transfected wild-type PML to Pin1 is lower in MDA-MB-231 cells than in CV-1 cells based on previous data. This decrease may reflect the amount of steady-state phosphorylation of PML in the different cell lines or other cell-type-specific differences. Taken together, we have identified residues important for interaction of Pin1 with PML.
FIG. 3.
Four serine residues in the C terminus of PML are important for the interaction with Pin1. (A) A schematic representation of PML4 depicting all of the S/T-P motifs. Asterisks indicate residues that were found to be phosphorylated in vivo. Bold type indicates the four mutants found to be important for interaction with Pin1. The RBCC motif is included, noting the RING, B boxes 1 and 2 (B1 and B2), and coiled-coil domains (CC). (B) Mutation of the C-terminal S/T-P motifs in PML results in loss of binding to Pin1. CV-1 cells were transfected with wild-type PML or with a construct in which the six most C-terminal S/T-P motifs were mutated to alanines. The resulting lysates were used in a GST pull-down using either GST alone or GST-Pin1 beads. Immunoblot analysis was performed using anti-PML antibodies. (C) Single mutations at four of the C-terminal S/T-P motifs reduced the ability of PML to bind to Pin1. Experiments were performed as described for panel B using constructs harboring single serine-to-alanine mutations at the specified residues. (D) Mutation of four of the S/T-P motifs in combination abrogates PML binding to Pin1. MDA-MB-231 cells were transfected with constructs for wild-type or the HA-PML4 4X mutant, and GST pull-downs were performed with GST-Pin1 followed by immunoblotting with anti-HA antibodies. Note that the decrease in the amount of binding to Pin1 by wild-type PML in panel D compared to panel B is a result of the different cell lines used. WT, wild type.
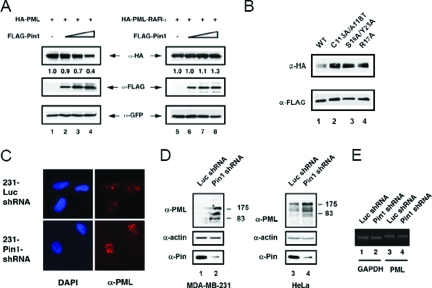
We next investigated whether increasing the amount of Pin1 affected PML protein levels in a cotransfection assay and found that as the amount of Pin1 increased, the amount of PML decreased (Fig. 4A, left panels). Pin1 had no effect on the levels of the oncogenic protein PML-RARα, which contains only the first 363 amino acids of PML (Fig. 4A, right panels). Furthermore, both the ability of Pin1 to bind to PML and the Pin1 PPIase activity are required for this effect because cotransfection of equal of amounts of mutant Pin1 (C115A/A118T has no PPIase activity; S16A/Y23A and R17A cannot bind to phospho-S/T-P motifs) with PML results in a higher level of PML expression than cotransfection with wild-type Pin1 (Fig. 4B, lanes 2 to 4 versus lane 1). In order to investigate whether Pin1 regulates the steady-state levels of PML, we utilized MDA-MB 231 cells that are stably transfected with either a control shRNA (Luc shRNA) or shRNA directed against Pin1 (Pin1 shRNA) and contain markedly reduced levels of Pin1. First, we performed immunofluorescence microscopy for PML in these cell lines to determine how knockdown of Pin1 affects PML localization and signal intensity. Figure 4C shows that when there is less Pin1, the intensity of PML is stronger and there are larger PML NBs, suggesting the presence of more PML in these cells. We did not observe any significant change in the localization of PML due to loss of Pin1. To confirm these observations, we performed immunoblot analysis with anti-PML and anti-Pin1 antibodies on whole-cell lysates from these cells. We also performed immunoblot analysis of HeLa cell lines that were stably transfected with the same control and Pin1 shRNA constructs (Fig. 4D). The multiple bands detected with the anti-PML antibody reflect multiple isoforms of PML (19). As a control to confirm that the changes we observed were not due to increased PML transcription in the absence of Pin1, we performed RT-PCR of total RNA isolated from the MDA-MB-231 Luc control and MDA-MB-231 Pin1 shRNA cell lines. There was no difference in PML mRNA levels, indicating that the Pin1-mediated regulation of PML expression occurs at the protein level (Fig. 4E). The results confirm the microscopy experiments and indicate that knocking down the amount of Pin1 in these cells results in an increase in PML levels. As a whole, these experiments suggest that Pin1 participates in the regulation of PML protein levels.
FIG. 4.
Pin1 decreases the steady-state levels of PML. (A) Increasing amounts of Pin1 result in decreasing amounts of PML but have no effect on PML-RARα. CV-1 cells were cotransfected with HA-PML or HA-PML-RARα constructs and increasing amounts of a FLAG-Pin1 expression construct. Vector DNA was used to maintain equal amounts of DNA transfected into each lane. The whole-cell lysates were analyzed by immunoblotting with anti-HA and anti-FLAG antibodies. A green fluorescent protein expression construct was also transfected and used as a control to show equal transfection efficiency and loading. The values under the top panel indicate the percentage of PML remaining compared to the green fluorescent protein control. (B) Pin1 must have intact WW and PPIase domains to affect PML levels. CV-1 cells were cotransfected with HA-PML and wild-type or mutant FLAG-Pin1 constructs. Whole-cell lysates were analyzed by immunoblotting with anti-HA and anti-FLAG antibodies. (C) Decreased Pin1 levels result in increased steady-state PML NB intensity. MDA-MB-231 cells stably transfected with either control Luc shRNA or shRNA directed against Pin1 were immunostained with anti-PML antibodies, and confocal microscopy images were taken. DAPI staining was used to detect DNA. (D) Decreased Pin1 levels result in increased steady-state PML protein levels. Whole-cell lysates of the control or Pin1 shRNA cell lines from both MDA-MB-231 cells and HeLa cells were analyzed by immunoblotting with anti-Pin1 and anti-PML antibodies. Immunoblotting with anti-actin antibody was used as a loading control. (E) PML levels are unaffected by knocking down Pin1 expression. Total RNA was isolated from the MDA-MB-231 Luc control shRNA and MDA-MB-231 Pin1 shRNA cells, and RT-PCR was performed to analyze PML and GAPDH protein levels. WT, wild type; α, anti.
To evaluate if Pin1 mediates changes in PML stability, we performed a translation shut-off experiment in MDA-MB-231 Luc shRNA control and MDA-MB-231 Pin1 shRNA cells. Cells were treated with cycloheximide, and then PML was detected by immunoblotting of whole-cell lysates to investigate the half-life of PML protein in the presence or absence of Pin1. Our results indicate that Pin1 can decrease the half-life of PML in the cell as when Pin1 expression is knocked down, the half-life of PML is increased about threefold (Fig. 5A). In order to test this hypothesis and ensure that the change in stability between the two cell lines is a result of the loss of Pin1 and not a nonspecific effect of the shRNA, we performed a similar cycloheximide study on MDA-MB-231 cells transfected with wild-type HA-PML4 and HA-PML4 4X (Fig. 5B). We found that the HA-PML4 4X mutant, which is unable to bind to Pin1, has a 3.5- to 4-fold longer half-life than wild-type PML, further suggesting that the effects we observe on PML stability are due to Pin1 binding to PML.
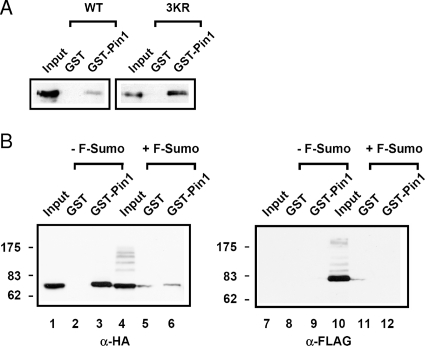
Because the binding of Pin1 to PML is dependent on the phosphorylation of PML and because studies indicate a link between phosphorylation and sumoylation of PML, we investigated how PML sumoylation may affect the ability of Pin1 to bind to PML. To do this, we transfected cells with either HA-PML4 or HA-PML4 3KR; the three previously characterized lysines on PML that are SUMOylated (K65/160/490) were mutated to arginine to prevent sumoylation. Whole-cell lysates from these cells were then used in a GST pull-down assays to compare the ability of wild-type and mutant PML to bind to Pin1 (Fig. 6A). We found that there was more HA-PML4 3KR bound to Pin1 than wild-type HA-PML4. These results are consistent with the hypothesis that sumoylation of PML inhibits binding to Pin1. To further test this hypothesis, we transfected cells with HA-PML4 with or without FLAG-SUMO1 and investigated the ability of SUMO1 to affect binding of PML to Pin1 by GST pull-down assays (Fig. 6B). CV-1 cells were chosen for the experiment because we observed a higher percentage of binding between PML and Pin1 in these cells, allowing changes in the amount of binding to be more easily observed. As shown in Fig. 6B, the presence of overexpressed SUMO1 and sumoylated PML species, indicated by the higher molecular weight bands (lane 4), results in a large decrease in the amount of PML that interacts with GST-Pin1 (lane 6 versus 3). The presence of sumoylated PML species was confirmed by stripping the membrane and reprobing with a FLAG antibody (lanes 7 to 12). Furthermore, the lack of FLAG signal in any of the other lanes indicates that any PML which was bound to Pin1 in this assay was not SUMOylated. Taken together, these results indicate that sumoylation of PML precludes Pin1 from binding to PML and suggest that sumoylated PML blocks Pin1-dependent degradation.
FIG. 6.
Sumoylation of PML inhibits the interaction with Pin1. (A) A PML mutant which is unable to be SUMOylated interacts with Pin1 more strongly than wild type. MDA-MB-231 cells were transfected with either wild-type or 3KR mutant HA-PML4 constructs, and cell lysates were used in GST pull-down experiments with either GST alone or GST-Pin1. Immunoblot analyses were performed with anti-HA antibodies. (B) Increased sumoylation of PML decreases the amount of PML that interacts with Pin1. CV-1 cells were cotransfected with DNA for HA-PML and either FLAG vector alone (−F-Sumo) or FLAG-SUMO1 (+F-Sumo). The resulting whole-cell lysates were subjected to GST pull-downs with either GST alone or GST-Pin1 and analyzed by immunoblotting with anti-HA and anti-FLAG antibodies. α, anti; WT, wild type.
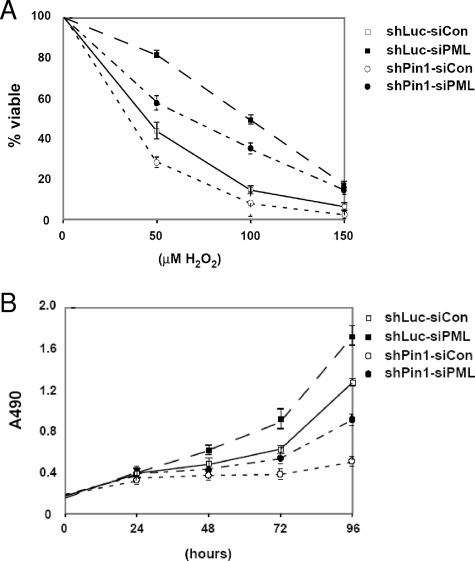
In order to determine the physiological significance of the interaction between PML and Pin1, we investigated the effects of altering this interaction on pathways in which PML has been previously shown to be involved. As PML is known to be involved in inducing cell death as a result of many cellular insults (reviewed in references 1, 17, and 42), we established MDA-MB-231 Luc shRNA control or MDA-MB-231 Pin1 shRNA cell lines and used them to evaluate whether decreases in Pin1 levels have effects on the ability of the cells to respond to hydrogen peroxide-induced death. We examined whether a change in PML levels, due to a change in Pin1 levels, affects the ability of hydrogen peroxide to induce the death of MDA-MB-231 cells. We treated MDA-MB-231-Luc shRNA control or MDA-MB-231-Pin1 shRNA cells with hydrogen peroxide, assayed the amount of cell death by trypan blue staining, and found that when Pin1 was knocked down by shRNA, the cells showed greater sensitivity to hydrogen peroxide (Fig. 7A, open symbols). To test whether this change resulted from different PML levels in the two cell lines and not to off-target effects of the Pin1 knockdown, we knocked down PML levels using siRNA and performed the same cell assay measuring cell death. In both MDA-MB-231 Luc shRNA control and MDA-MB-231 Pin1 shRNA cells, when PML is decreased by siRNA, less cell death is observed (Fig. 7A, compare solid symbols to open symbols). These results indicate that the levels of PML are important in the cellular response to hydrogen peroxide. Furthermore, the amount of Pin1 in the cell, which affects the amount of PML, is involved in the control of the cellular response to hydrogen peroxide.
FIG. 7.
Knockdown of PML reduces cellular sensitivity to hydrogen peroxide and increases cellular proliferation. (A) Changes in PML levels affect the response of cells to treatment with hydrogen peroxide. MDA-MB-231 control and Pin1 shRNA cells were treated with control oligonucleotide or siRNA directed against PML. Cells were then treated with hydrogen peroxide, and cell death was analyzed by trypan blue staining. Each cell line was analyzed in triplicate. The percentage of cell death was calculated by comparing the number of blue cells to the total number of cells counted. Each cell death percentage was adjusted relative to the basal rate of cell death of each set of cells so that the results shown indicate the percentage of cell death due to hydrogen peroxide. (B) Changes in PML levels contribute to the rate of cellular proliferation. MDA-MB-231 control and Pin1 shRNA cells were treated with control oligonucleotide or siRNA directed against PML. Cells were then analyzed at the time points indicated using an MTS [3,4-(5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy phenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt] assay to obtain a value indicative of the number of cells present. Each time point was determined in triplicate. Con, control.
Finally, since PML has previously been shown to be involved in the cell cycle (11), we hypothesized that decreases in PML due to Pin1 may contribute to an increase in the rate of proliferation. To test this, we used the same cell lines and analyzed their rates of proliferation. When we compared the growth of the MDA-MB-231 Luc control cells to that of the MDA-MB-231 Pin1 shRNA cells, we observed a decrease in the rate of proliferation when Pin1 was knocked down (Fig. 7B, open symbols). To determine if PML levels played a role in the decreased proliferation, we performed knockdown of PML by siRNA in both cell lines and compared the resulting changes in proliferation. In support of our hypothesis, when the levels of PML were knocked down in both cell lines, the rate of proliferation increased (Fig. 7B, compare solid symbols to open symbols). These results indicate that the level of PML, as regulated by Pin1, is an important determinant in the rate of proliferation. Taken together, our results imply that PML protein levels are involved in regulating both the cellular response to hydrogen peroxide and the rate of proliferation.
DISCUSSION
There has been a recent realization that PML is involved in many cellular processes and is not just a passive translocation partner with RARα. It is thus important to understand how this versatile protein is regulated both at steady-state levels and in response to various extracellular stimuli and, in turn, how this regulation may affect formation of PML NBs. Our work indicates that PML interacts with Pin1, which leads to degradation of PML protein. We observed no change in PML mRNA levels due to Pin1 knockdown (Fig. 4E). We show that PML interaction with Pin1 can be detected both in vitro and in mammalian cells. We believe that the small amount of interaction observed is primarily due to two factors. First, both PML and Pin1 bind many different proteins in the cell, and any specific complexes that contain both PML and Pin1 are likely only a small fraction of protein complexes that contain either protein alone. This is supported by our immunofluorescence microscopy studies that indicate only fractional colocalization between the proteins (Fig. 1C). Second, the interaction between PML and Pin1 is likely transient, where Pin1 binds to PML, enzymatically induces a conformational change in PML, and then dissociates to allow other proteins to interact with PML. Such transient interactions have been reported to occur between Pin1 and other target proteins such as c-Myc and SRC-3 (54, 55). While we cannot decisively conclude that there is a direct interaction between the two proteins from out data, there are several lines of evidence that strongly suggest a direct interaction. First, our data show that the interaction is dependent on phosphorylation of PML and is mediated through the WW domain of Pin1, which is known to bind pS/T-P motifs in other protein targets. Furthermore, we have generated a mutant of PML with S-to-A substitutions at four pS/T-P motifs that no longer binds to Pin1. Taken together, these data support the idea that it is Pin1 binding to one or more of these four sites on PML rather than another cellular protein as well as that the phosphorylation of these Ser residues is important in the interaction rather than a different modification on another location of PML. Lastly, we know that the C-terminal region of PML is important for the Pin1 interaction, specifically with regard to the four S/T-P motifs. However, there also appears to be some contribution of the N-terminal region of PML that remains to be explored in detail. We hypothesize that the folding of PML is disrupted when half of the protein is deleted, and as a result the stable binding of PML to proteins such as Pin1 is disrupted. Predictions based on consensus motifs for candidate kinases suggest that possibilities include mitogen-activated protein kinases (including ERK1 and ERK2), GSK-3β, CDK2, and protein kinase A. The possibility of the involvement of a mitogen-activated protein kinase is strengthened by the fact that S527 has previously been shown to be phosphorylated in vitro by ERK2 (16). We believe that identification of the relevant kinases is an important future objective in our quest to elucidate the signaling cascades that may influence the interaction between Pin1 and PML in order to regulate PML protein levels.
Our data further suggest that the result of the interaction between PML and Pin1 is a decrease in the steady-state level of PML. Increasing levels of Pin1 result in decreased PML levels, whereas decreased Pin1 levels result in increased PML levels. While we cannot conclude that this reciprocal relationship between PML and Pin1 is a direct consequence of the binding of PML to Pin1, our results with mutants of Pin1 and PML strongly support this conclusion. When mutated, neither the Pin1 mutants unable to bind to PML nor a catalytically inactive Pin1 mutant was able to promote PML degradation. Finally, the HA-PML4 4X mutant that cannot bind Pin1 is more stable than wild-type HA-PML4. While we are unsure of the mechanisms by which PML is degraded in response to Pin1 binding, previous work by others has shown that Pin1 works with many different E3 ligases to target proteins for degradation, such as the SCF ubiquitin ligases which target c-Myc (44), cyclin E1 (41) and SRC-3 (47); Hdm2 which targets Che-1 (6); and the p65 subunit of NF-κB which is bound by SOCS-1 (36). We have begun to evaluate potential effects of these and other E3 ligases in facilitating the Pin1-dependent degradation of PML.
Work by other investigators has suggested that phosphorylation and sumoylation of PML are involved in both PML regulation and localization (16, 31). It has long been speculated that PML sumoylation is linked to regulation of its steady-state level. Our work is the first to demonstrate that sumoylation of PML can protect it from Pin1-mediated degradation. We show that overexpression of SUMO1 leads to decreased interaction between PML and Pin1 and that a sumoylation-defective mutant of PML shows more binding to Pin1 than wild-type PML. We were not able to separate the effects of PML sumoylation on stability and localization, so we are unable to confirm the relationship between sumoylation and PML activity in a cell culture system.
Intriguingly, our data indicate that MDA-MB-231 breast cancer cells exploit the interaction between PML and Pin1 to promote malignant characteristics. Pin1 has been reported to be up-regulated in many breast cancers (50), and one mechanism proposed involves activation of the Her2/Neu/ErbB2 receptor (34). Since MDA-MB-231 cells express the Her2/Neu/ErbB2 receptor, it seems likely that these breast cancer cells also stably overexpress Pin1, which we show correlates with decreased steady-state PML expression. Due to PML's ability to promote apoptosis by both intrinsic and extrinsic pathways (17, 42), it acts to protect cells against transformation. We show that knocking down Pin1 in the breast cancer cells results in increased PML and sensitizes them to hydrogen peroxide-induced death. Knockdown of the increased PML level due to the decrease in Pin1 can reverse these effects, suggesting that by maintaining high levels of Pin1, these cells are less susceptible to oxidative stress-induced death. Next, PML has also been characterized as a growth suppressor because adenoviral infection of MCF-7 and SK-BR-3 breast cancer cells with PML leads to cell cycle arrest at the G1 phase (26). Correlating with this, our data indicate that when Pin1 levels are decreased and PML is increased, there is less proliferation that can be rescued by a transient decrease in PML levels. These data strongly suggest that the decreased cell proliferation that we observed in MDA-MB-231 cells when Pin1 is knocked down is at least partially due to an increase in PML expression.
In addition to the regulation of PML by Pin1 in MDA-MD-231 cells reported herein, other studies have suggested a role for PML in cancer phenotypes since PML levels are lower in many oncogenic samples relative to corresponding normal tissues (14). Recent work showing that casein kinase 2 can phosphorylate PML and lead to its degradation highlights the relationship between PML levels and transformation in that it provides additional evidence for an inverse correlation between PML protein levels and casein kinase 2 activity in human lung cancer-derived cell lines (37). This appears to be one important mechanism in the regulation of PML that bears directly on its role as a tumor suppressor. Our work with the breast cancer-derived cell line MDA-MB-231 indicates that regulation of PML by Pin1 represents another mechanism by which PML is deregulated. The reduction of PML protein levels in these cells may be an important step in the series of events that lead to their transformation, and this idea may hold true for other cancers with decreased cellular levels of PML.
Our research reveals a previously uncharacterized mechanism of PML regulation. This mechanism allows control of cellular PML levels and thus governs the role of PML in its functions inside and outside of the NBs. A better understanding of the mechanisms that conspire to regulate PML may lead to identification of novel targets for therapeutic development. In this regard, it will be important to identify the additional cellular signals and pathways that modulate the association between PML and Pin1.
Acknowledgments
We thank D. Samols and M. Snider for their comments on the manuscript. We thank Ron Evans and Hsiu-Ming Shih for reagents.
H.-Y. Kao is a recipient of the James T. Pardee-Carl A. Gerstacker Assistant of Cancer Research Faculty Chair in Cancer Research at CWRU Cancer Center and an American Cancer Society Research Scholar (RSG-04-052-01-GMC). This research was supported by the Aging Cancer Research Program at the Case Comprehensive Cancer Center (P20 CA103736), the Confocal Microscopy Core Facility of the Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland (P30 CA43703-12), an NIH grant to A.R.M. (CA082845), and an NIH grant to K.-S.C. (CA055577). E.L.R. is supported by a Case Comprehensive Cancer Center NIH/NCI Research Oncology Training Grant (T32 CA059366-11). K.J.S. is supported by an NIH cellular and molecular biology training grant (T32 GM08056).
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Bernardi, R., and P. P. Pandolfi. 2003. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 229048-9057. [DOI] [PubMed] [Google Scholar]
- 2.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 225259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buschbeck, M., I. Uribesalgo, A. Ledl, A. Gutierrez, S. Minucci, S. Muller, and L. Di Croce. 2007. PML4 induces differentiation by Myc destabilization. Oncogene 263415-3422. [DOI] [PubMed] [Google Scholar]
- 4.Chelbi-Alix, M. K., L. Pelicano, F. Quignon, M. H. Koken, L. Venturini, M. Stadler, J. Pavlovic, L. Degos, and H. de The. 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 92027-2033. [PubMed] [Google Scholar]
- 5.Ching, R. W., G. Dellaire, C. H. Eskiw, and D. P. Bazett-Jones. 2005. PML bodies: a meeting place for genomic loci? J. Cell Sci. 118847-854. [DOI] [PubMed] [Google Scholar]
- 6.De Nicola, F., T. Bruno, S. Iezzi, M. Di Padova, A. Floridi, C. Passananti, G. Del Sal, and M. Fanciulli. 2007. The prolyl isomerase Pin1 affects Che-1 stability in response to apoptotic DNA damage. J. Biol. Chem. 28219685-19691. [DOI] [PubMed] [Google Scholar]
- 7.de Stanchina, E., E. Querido, M. Narita, R. V. Davuluri, P. P. Pandolfi, G. Ferbeyre, and S. W. Lowe. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13523-535. [DOI] [PubMed] [Google Scholar]
- 8.de The, H., C. Lavau, A. Marchio, C. Chomienne, L. Degos, and A. Dejean. 1991. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66675-684. [DOI] [PubMed] [Google Scholar]
- 9.Doucas, V., and R. M. Evans. 1996. The PML nuclear compartment and cancer. Biochim. Biophys. Acta 1288M25-M29. [DOI] [PubMed] [Google Scholar]
- 10.Dyck, J. A., G. G. Maul, W. H. Miller, Jr., J. D. Chen, A. Kakizuka, and R. M. Evans. 1994. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76333-343. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 1124581-4588. [DOI] [PubMed] [Google Scholar]
- 12.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 142015-2027. [PMC free article] [PubMed] [Google Scholar]
- 13.Galat, A. 2003. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity—targets—functions. Curr. Top. Med. Chem. 31315-1347. [DOI] [PubMed] [Google Scholar]
- 14.Gurrieri, C., P. Capodieci, R. Bernardi, P. P. Scaglioni, K. Nafa, L. J. Rush, D. A. Verbel, C. Cordon-Cardo, and P. P. Pandolfi. 2004. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J. Natl. Cancer Inst. 96269-279. [DOI] [PubMed] [Google Scholar]
- 15.Hamdane, M., C. Smet, A. V. Sambo, A. Leroy, J. M. Wieruszeski, P. Delobel, C. A. Maurage, A. Ghestem, R. Wintjens, S. Begard, N. Sergeant, A. Delacourte, D. Horvath, I. Landrieu, G. Lippens, and L. Buee. 2002. Pin1: a therapeutic target in Alzheimer neurodegeneration. J. Mol. Neurosci. 19275-287. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa, F., and M. L. Privalsky. 2004. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell 5389-401. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann, T. G., and H. Will. 2003. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 101290-1299. [DOI] [PubMed] [Google Scholar]
- 18.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss, 3rd, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 207223-7233. [DOI] [PubMed] [Google Scholar]
- 20.Joseph, J. D., E. S. Yeh, K. I. Swenson, A. R. Means, and Winkler. 2003. The peptidyl-prolyl isomerase Pin1. Prog. Cell Cycle Res. 5477-487. [PubMed] [Google Scholar]
- 21.Kakizuka, A., W. H. Miller, Jr., K. Umesono, R. P. Warrell, Jr., S. R. Frankel, V. V. Murty, E. Dmitrovsky, and R. M. Evans. 1991. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66663-674. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., S. Akira, and J. C. Reed. 2003. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol. Cell. Biol. 236174-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 1931361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11871-876. [PubMed] [Google Scholar]
- 25.Lavoie, S. B., A. L. Albert, and M. Vincent. 2003. Unexpected roles of the peptidyl-prolyl cis/trans isomerase Pin1. Med. Sci. (Paris) 191251-1258. [In French.] [DOI] [PubMed] [Google Scholar]
- 26.Le, X. F., S. Vallian, Z. M. Mu, M. C. Hung, and K. S. Chang. 1998. Recombinant PML adenovirus suppresses growth and tumorigenicity of human breast cancer cells by inducing G1 cell cycle arrest and apoptosis. Oncogene 161839-1849. [DOI] [PubMed] [Google Scholar]
- 27.Lu, P. J., X. Z. Zhou, M. Shen, and K. P. Lu. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 2831325-1328. [DOI] [PubMed] [Google Scholar]
- 28.Mann, K. K., and W. H. Miller, Jr. 2004. Death by arsenic: implications of PML sumoylation. Cancer Cell 5307-309. [DOI] [PubMed] [Google Scholar]
- 29.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129278-287. [DOI] [PubMed] [Google Scholar]
- 30.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 933167-3215. [PubMed] [Google Scholar]
- 31.Muller, S., W. H. Miller, Jr., and A. Dejean. 1998. Trivalent antimonials induce degradation of the PML-RAR oncoprotein and reorganization of the promyelocytic leukemia nuclear bodies in acute promyelocytic leukemia NB4 cells. Blood 924308-4316. [PubMed] [Google Scholar]
- 32.Ranganathan, R., and E. M. Ross. 1997. PDZ domain proteins: scaffolds for signaling complexes. Curr. Biol. 7R770-R773. [DOI] [PubMed] [Google Scholar]
- 33.Reineke, E. L., H. Liu, M. Lam, Y. Liu, and H. Y. Kao. 2007. Aberrant association of promyelocytic leukemia protein-retinoic acid receptor-alpha with coactivators contributes to its ability to regulate gene expression. J. Biol. Chem. 28218584-18596. [DOI] [PubMed] [Google Scholar]
- 34.Ryo, A., Y. C. Liou, G. Wulf, M. Nakamura, S. W. Lee, and K. P. Lu. 2002. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 225281-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryo, A., M. Nakamura, G. Wulf, Y. C. Liou, and K. P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3793-801. [DOI] [PubMed] [Google Scholar]
- 36.Ryo, A., F. Suizu, Y. Yoshida, K. Perrem, Y. C. Liou, G. Wulf, R. Rottapel, S. Yamaoka, and K. P. Lu. 2003. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 121413-1426. [DOI] [PubMed] [Google Scholar]
- 37.Scaglioni, P. P., T. M. Yung, L. F. Cai, H. Erdjument-Bromage, A. J. Kaufman, B. Singh, J. Teruya-Feldstein, P. Tempst, and P. P. Pandolfi. 2006. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 126269-283. [DOI] [PubMed] [Google Scholar]
- 38.Schmid, F. X. 1995. Protein folding. Prolyl isomerases join the fold. Curr. Biol. 5993-994. [DOI] [PubMed] [Google Scholar]
- 39.Shen, M., P. T. Stukenberg, M. W. Kirschner, and K. P. Lu. 1998. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 12706-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadler, M., M. K. Chelbi-Alix, M. H. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 112565-2573. [PubMed] [Google Scholar]
- 41.Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt, and S. I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413316-322. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi, Y., V. Lallemand-Breitenbach, J. Zhu, and H. de The. 2004. PML nuclear bodies and apoptosis. Oncogene 232819-2824. [DOI] [PubMed] [Google Scholar]
- 43.Urist, M., and C. Prives. 2004. The linchpin? Pin1 meets p73. Cancer Cell 5515-517. [DOI] [PubMed] [Google Scholar]
- 44.van Drogen, F., O. Sangfelt, A. Malyukova, L. Matskova, E. Yeh, A. R. Means, and S. I. Reed. 2006. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol. Cell 2337-48. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 2791547-1551. [DOI] [PubMed] [Google Scholar]
- 46.Winkler, K. E., K. I. Swenson, S. Kornbluth, and A. R. Means. 2000. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 2871644-1647. [DOI] [PubMed] [Google Scholar]
- 47.Wu, R. C., Q. Feng, D. M. Lonard, and B. W. O'Malley. 2007. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 1291125-1140. [DOI] [PubMed] [Google Scholar]
- 48.Wu, W. S., S. Vallian, E. Seto, W. M. Yang, D. Edmondson, S. Roth, and K. S. Chang. 2001. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol. 212259-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wulf, G., G. Finn, F. Suizu, and K. P. Lu. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat. Cell Biol. 7435-441. [DOI] [PubMed] [Google Scholar]
- 50.Wulf, G., P. Garg, Y. C. Liou, D. Iglehart, and K. P. Lu. 2004. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 233397-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wulf, G., A. Ryo, Y. C. Liou, and K. P. Lu. 2003. The prolyl isomerase Pin1 in breast development and cancer. Breast Cancer Res. 576-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, Z. X., R. X. Zhao, T. Ding, T. T. Tran, W. Zhang, P. P. Pandolfi, and K. S. Chang. 2004. Promyelocytic leukemia protein 4 induces apoptosis by inhibition of survivin expression. J. Biol. Chem. 2791838-1844. [DOI] [PubMed] [Google Scholar]
- 53.Yaffe, M. B., M. Schutkowski, M. Shen, X. Z. Zhou, P. T. Stukenberg, J. U. Rahfeld, J. Xu, J. Kuang, M. W. Kirschner, G. Fischer, L. C. Cantley, and K. P. Lu. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 2781957-1960. [DOI] [PubMed] [Google Scholar]
- 54.Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W. C. Hahn, P. T. Stukenberg, S. Shenolikar, T. Uchida, C. M. Counter, J. R. Nevins, A. R. Means, and R. Sears. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6308-318. [DOI] [PubMed] [Google Scholar]
- 55.Yi, P., R. C. Wu, J. Sandquist, J. Wong, S. Y. Tsai, M. J. Tsai, A. R. Means, and B. W. O'Malley. 2005. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol. Cell. Biol. 259687-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 952748-2752. [PubMed] [Google Scholar]