Abstract
Whereas DNA methylation is essential for genomic imprinting, the importance of histone methylation in the allelic expression of imprinted genes is unclear. Imprinting control regions (ICRs), however, are marked by histone H3-K9 methylation on their DNA-methylated allele. In the placenta, the paternal silencing along the Kcnq1 domain on distal chromosome 7 also correlates with the presence of H3-K9 methylation, but imprinted repression at these genes is maintained independently of DNA methylation. To explore which histone methyltransferase (HMT) could mediate the allelic H3-K9 methylation on distal chromosome 7, and at ICRs, we generated mouse conceptuses deficient for the SET domain protein G9a. We found that in the embryo and placenta, the differential DNA methylation at ICRs and imprinted genes is maintained in the absence of G9a. Accordingly, in embryos, imprinted gene expression was unchanged at the domains analyzed, in spite of a global loss of H3-K9 dimethylation (H3K9me2). In contrast, the placenta-specific imprinting of genes on distal chromosome 7 is impaired in the absence of G9a, and this correlates with reduced levels of H3K9me2 and H3K9me3. These findings provide the first evidence for the involvement of an HMT and suggest that histone methylation contributes to imprinted gene repression in the trophoblast.
More than 80 mammalian genes undergo parent-of-origin-dependent expression. Most of these are clustered in domains, which are broadly conserved between mice and humans (33). The allelic expression along imprinted domains is regulated by “imprinting control regions” (ICRs) (7, 9, 54). DNA methylation is essential for the mechanism of imprinting (34), and all known ICRs are marked by DNA methylation on their maternally, or their paternally, inherited allele. The germ line establishment of these methylation imprints requires the DNA methyltransferase Dnmt3a (1, 20) and the related protein Dnmt3L (1-3, 15, 20). The somatic maintenance of imprints requires the maintenance methyltransferase Dnmt1 (17, 28). In the embryo, and after birth, ICRs are marked by parental allele-specific histone methylation as well. Specifically, together with other histone modifications, ICRs are consistently enriched in histone H3-lysine-9 methylation on their DNA-methylated allele (6, 40, 53, 56, 58). It is unknown which histone methyltransferase (HMT) mediates this H3-lysine-9 methylation and to what extent this epigenetic modification is involved in the maintenance of the allelic chromatin organization at ICRs.
During embryonic development, ICRs bring about parental allele-specific gene expression (7, 26). At some imprinted gene clusters this process involves the establishment of allele-specific histone modifications. Imprinted expression along the Kcnq1 domain on mouse distal chromosome 7 is mediated by a noncoding RNA (31) transcribed from the ICR, and chromatin on the domain's repressed paternal chromosome is enriched in H3-lysine-9 dimethylation (H3K9me2) and H3-lysine-27 trimethylation (H3K27me3). This was observed most extensively in the placenta, in which the majority of the genes in this >800-kb domain are paternally repressed (25, 53). Genetic and biochemical studies have suggested that the Polycomb repressive complex PRC2 regulates H3K27me3 along the Kcnq1 domain (30, 53). It is unknown, however, which HMT could mediate the H3K9me2 on the repressed paternal chromosome. Several SET domain proteins have been found to specifically transfer methyl groups onto lysine-9 of histone H3 (18). Some of these HMTs bring about H3K9me2 preferentially, whereas others mediate H3K9me3. The HMT G9a was shown to be essential for genome-wide levels of H3K9me2, and fluorescence studies suggest that it mediates H3K9me2 at regions other than at the pericentric heterochromatin (39, 41, 48). The G9a protein forms a functional heterodimer with a closely related protein called Glp (G9a-like protein), also called EuHMTase1 in humans (35). Also this SET domain protein is essential for H3K9me2 at euchromatic regions (49).
Given its substrate specificity and its global effects, G9a could potentially regulate the allelic H3K9 methylation at the imprinted Kcnq1 domain and that observed at ICRs. To test this hypothesis, a gene trap approach was used to generate G9a-deficient mouse conceptuses. This allowed us to perform studies on placentas and embryos, rather than on cells in culture, which can sometimes give rise to aberrant epigenetic effects on imprinted genes (5, 59). Our in vivo approach did not provide evidence for G9a to be essential in the allelic regulation of DNA methylation at the different ICRs analyzed, although moderate reductions in H3K9 methylation were observed. Interestingly, however, we found that the absence of G9a has pronounced effects on the paternal repression along the Kcnq1 domain in the placenta. In particular, G9a deficiency affected genes that are imprinted in the trophoblast only and which are not dependent on DNA methylation for the somatic maintenance of their allelic silencing. This provides the first in vivo evidence for the involvement of a SET domain protein in genomic imprinting and emphasizes the relative importance of histone methylation in placenta-specific imprinting.
MATERIALS AND METHODS
G9a-deficient conceptuses.
The G9a gene was trapped by insertion of a β-galactosidase-neomycin phosphotransferase (β-geo) construct comprising a splice acceptor (47). We used a G9a-trapped embryonic stem (ES) cell line, ES62, to generate a transgenic line, which was maintained in a heterozygous state and was crossed for four generations to C57BL/6 mice. Concomitantly, a second line was derived by back-crossing to a (Mus spretus) congenic mouse line, SDP711. G9a−/− embryos and placentas were obtained by intercrossing these two lines. Genotyping was performed by PCR against the β-geo insert and the endogenous G9a gene. At the imprinted loci analyzed, C57BL/6 and M. spretus genotypes were distinguished by PCR (see Table S3 in the supplemental material). For histological examination, placentas were fixed with paraformaldehyde overnight and then dehydrated and embedded in paraffin. Serial sections (5 μm) were stained with hematoxylin and eosin using routine procedures. Cell death was assessed on sections by a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling test with an in situ fluorescein cell death detection kit (Roche).
Analysis of gene expression.
Total RNA was extracted using TRIzol reagent (Invitrogen). First-strand cDNA was generated with SuperScript II (Invitrogen) using random primers. All reverse transcription-PCR (RT-PCR) amplifications (see Table S3 in the supplemental material) were performed in the presence of [32P]dCTP (1% of total dCTP) during all cycles for single-strand conformation polymorphism (SSCP) analysis or only during a last cycle of reamplification (hot-stop PCR) (52). Relative band intensities were determined using ImageQuantTL imaging software (Amersham Biosciences). Allelic ratios were compared between wild-type (WT) and G9a−/− placentas by using the Student t test.
Microarray analysis.
Per genotype, three total RNA samples were pooled and then quantified on an Agilent Bioanalyser. Three μg of pooled RNA sample was used to synthesize double-stranded cDNA using the Superscript cDNA synthesis kit (Invitrogen). This was then used as input for an in vitro transcription reaction with biotin-labeled dUTPs, using the BioArray high-yield RNA transcript labeling kit (Enzo). The biotinylated target cRNA was fragmented with 5× fragmentation buffer (Affymetrix) and quantified prior to hybridization using the Agilent Bioanalyser. Per array hybridization mixture, 20 μg of labeled cRNA was used. Hybridization, washing, and staining were all performed on an Affymetrix Fluidics Station 450 using standard Affymetrix protocols (Affymetrix GeneChip Expression Analysis Technical Manual [http://www.affymetrix.com]). An Affymetrix Scanner 3000 was used to generate the raw array image data. The above process was repeated twice, using biologically replicate G9a−/− and WT placentas. The first replicate experiment was performed using two 430A and two 430B mouse expression set arrays (Affymetrix), while two mouse genome 430 2.0 arrays (also from Affymetrix) were employed in the second experiment. The raw array images were transformed into CEL files and analyzed at the probe level, using the Affymetrix GCOS v1.2 statistical algorithms description document (http://www.affymetrix.com/support/). The GCOS comparative gene expression analysis feature used was similar to that used by Schulz et al. (45) to carry out three comparative probe-level analyses (G9a−/− replicate 1 on 430A versus WT replicate 1 on 430A, G9a−/− replicate 1 on 430B versus WT replicate 1 on 430B, and G9a−/− replicate 2 on 430 2.0 versus WT replicate 2 on 430 2.0), quantifying absolute and differential gene expression levels measured by the respective two arrays. For each probe set, GCOS computes an absolute expression level and associated detection P value (dtcP) per array and a signal log2 ratio (SLR) and an associated change P value (chgP) per each comparison of two arrays. The SLR expresses the observed direction and degree of change in expression measured by the probe set between the two arrays, while the change P value is a measure of confidence in any observed difference in expression. In this study, a change in expression was considered statistically significant whenever the chgP was ≤0.003 or the chgP was ≥0.997 in both biological replicate experiments. Absolute expression in either G9a−/− or WT placentas was judged statistically significant whenever dtcP was ≤0.06 in both biological replicate experiments. The respective P value thresholds are the Affymetrix default values. For imprinted genes, we compiled a list of 82 known imprinted loci, drawing from the MRC (Mammalian Genetics Unit at Harwell, United Kingdom [http://www.mgu.har.mrc.ac.uk/research/imprinting/]) and Otago University (33) imprinting resources. We then aligned the target sequences of all probe sets on the Affymetrix 430 2.0 array to the mouse genome (NCBI build 36) using BLAT (21) and the UCSC genome browser (mm8 mouse genome) (22).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays on native chromatin were performed as described elsewhere (53). We used different antisera against H3K9me2 (Upstate 07-212 as antibody A and Upstate 07-441 as antibody B), H3K9me3 (Upstate 07-442 as antibody A and Abcam 1186 as antibody B), and H3K4me2 (Upstate 07-030). As a negative control (mock precipitation), we used a rabbit antiserum directed against chicken immunoglobulin G (IgG; C2288; Sigma). Precipitation levels were determined by real-time PCR, using a SYBR Green PCR kit (Qiagen). Each PCR was run in triplicate, and results are presented as the average value of the precipitated material corrected for the average value of the corresponding mock precipitation. ChIP was also performed on placentas at 9.5 days postcoitum (dpc) after cross-linking with 1% formaldehyde (10 min at 20°C) using antisera against G9a (Upstate 07-551) and RNA polymerase II (Abcam 5131). As a negative control, a rabbit antiserum to chicken IgG (C2288; Sigma) was used.
Analysis of DNA methylation.
A 200-ng aliquot of genomic DNA was digested in a volume of 20 μl with appropriate restriction enzymes. Aliquots were taken for PCR in the presence of [32P]dCTP (1% of total dCTP). PCR products were denatured and analyzed by SSCP gel electrophoresis. Primer sequences are provided in Table S3 in the supplemental material. Bisulfite sequencing on embryo and placental genomic DNA was performed as described before (2).
Microarray data.
Data from the microarray analysis of the G9a HMT were deposited in the GEO repository and are accessible at https://atlas.genetics.kcl.ac.uk/.
RESULTS
Effects of G9a deficiency on the embryo and placenta.
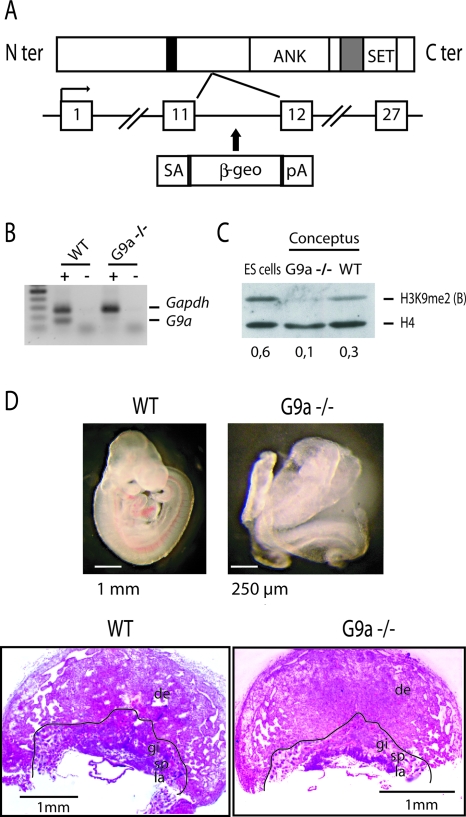
To explore the role of G9a, we derived embryos and placentas deficient for this HMT. This was achieved as part of a gene trap targeting approach on ES cells, using a promoterless β-geo construct containing a splice acceptor and a polyadenylation signal. Insertion of this construct into a gene's intron leads to a chimeric splice product and, consequently, the production of a lacZ fusion protein that lacks the protein sequence encoded by exons of the trapped gene that are 3′ of the gene-trap insertion. Sequence analyses of insertions into mouse genes encoding nuclear proteins (47) identified one ES line in which the construct had inserted in intron 11 of the G9a gene (Fig. 1A). Heterozygous mice were derived by making chimeric animals using the targeted ES cells, followed by germ line transmission. Heterozygous mice were intercrossed to generate G9a−/− conceptuses in which the site of gene-trap insertion was confirmed by PCR amplification and DNA sequencing. No transcription was detected from the 3′-half of the G9a gene (Fig. 1B). The G9a-β-geo fusion protein lacks the ankyrin repeats and, most importantly, the catalytic SET domain of the wild-type protein and so is likely to be functional null. Accordingly, Western blotting showed that in the G9a−/− conceptuses there was a strong reduction of global H3K9me2 (Fig. 1C).
FIG. 1.
Targeting of G9a. (A) Schematic presentation of G9a with its ankyrin (ANK) and SET domains. Black and gray boxes indicate the nuclear localization signal and the pre-SET domains, respectively. The β-geo gene trap (with a splice acceptor [SA] and a polyadenylation site [pA]) was inserted in intron 11. This creates a truncated transcript lacking the SET domain. (B) Lack of G9a expression in G9a−/− placentas. Reverse transcription was performed with (+) or without (-) reverse transcriptase followed by duplex PCR of G9a and Gapdh in WT and G9a−/− placentas and in ES cells. (C) Western analysis of H3K9me2; the control antiserum is against histone H4. The ratios between the H3K9me2 and H4 signals are indicated underneath. (D) Histology of WT and G9a−/− conceptuses. The upper panel shows 9.5-dpc WT (left) and G9a−/− (right) embryos. For the 9.5-dpc WT and G9a−/− placentas in the lower panel, the labyrinthine layer (la), spongiotrophoblast (sp), maternal decidua (de), and trophoblast giant cells (gi) are indicated.
G9a−/− embryos were viable and present at the expected frequency up to 10 dpc. In agreement with an earlier study (48), at later stages we observed embryonic death and resorption. Development of the embryos was grossly abnormal at 8.5 to 9.5 dpc. The ectoderm showed a consistent nonclosure of the neural groove, and the G9a−/− embryos were about half the size of WT embryos (Fig. 1D). The placenta, in contrast, did not show gross developmental abnormalities, with a normal morphology of the maternal decidua and the three embryonic layers: labyrinthine trophoblast, spongiotrophoblast, and the giant cell layer (Fig. 1D). However, size measurements on several G9a−/− versus WT placentas showed an ∼10% reduction in placental diameter. To assess trophoblastic differentiation and cell death, we counted the polyploid giant cells on sequential sections of G9a−/− and WT placentas. There was an 18% reduction in the number of giant cells in the G9a−/− placentas, and these showed a twofold increase in cell death compared to WT (see Fig. S1 in the supplemental material). Giant cell reduction was higher than expected given the size reduction of these placentas, indicating that G9a deficiency had a moderate effect on trophoblastic differentiation.
G9a deficiency causes loss of imprinting in the placenta but not the embryo.
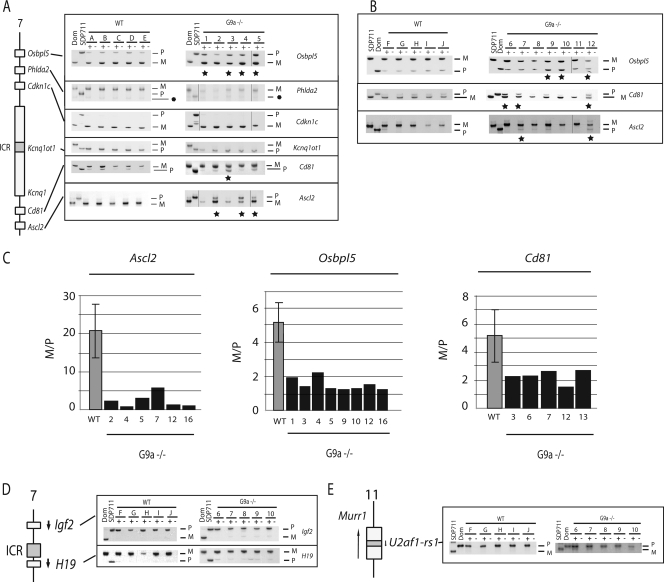
To be able to distinguish the parental chromosomes in our gene expression studies, we crossed the G9a line onto a congenic mouse line (SDP711) in which distal chromosome 7 and proximal chromosome 11 were derived from M. spretus on an otherwise C57BL/6J (Mus musculus) background. The original line (C57BL6/J background) and the newly derived G9a heterozygous line (congenic SDP711 background) were intercrossed to generate G9a−/− placentas and embryos. Single nucleotide polymorphisms were used to distinguish the maternal and paternal transcripts of imprinted genes. This was done by RT-PCR followed by electrophoretic detection of SSCPs or by hot-stop PCR (52, 53). WT and G9a−/− placentas and embryos (data not shown) were compared at 9.5 dpc. The carefully dissected embryonic portions of WT placentas showed maternal expression of Osbpl5, Phlda2, Cdkn1c, Cd81, and Ascl2 at the Kcnq1 domain. Expression of the noncoding RNA Kcnq1ot1, which is transcribed from the domain's ICR (called KvDMR1) (46), was from the paternal chromosome exclusively (Fig. 2A).
FIG. 2.
Altered imprinted gene expression in G9a−/− placentas. (A) RT-PCR in the presence (+) or absence (-) of reverse transcriptase on (C57BL6 × SDP711)F1 placentas at 9.5 dpc (WT and G9a−/−). In all panels, the first two lanes show control amplifications from C57BL/6 (Dom) and SDP711 WT placentas, respectively. Maternal (M) and paternal (P) specific bands are indicated. Asterisks indicate placentas in which the M/P ratio was different from that observed in the cohorts of WT placentas. In the Phlda2 analysis, the black dot indicates a secondary, maternal-specific band. (B) Loss of imprinting (asterisks) at Osbpl5, Cd81, and Ascl2 was also observed in (SDP711 × C57BL6)F1 placentas. (C) M/P band intensity ratios. In each of the panels, the average ratio for all WT placentas analyzed is shown to the left. Examples of G9a−/− placentas that were below this range for Ascl2, Osbpl5, or CD81 are shown to the right. In the G9a−/− placentas, M/P ratios were significantly lower than in WT placentas for Ascl2 (mean, 2.3 versus 5.2; P < 0.0001) and Osbpl5 (mean, 2.3 versus 5.2; P < 0.00001). For Cd81, the means were not different between the WT and G9a−/− mice (5.2 versus 8.4; P = 0.014). (D) Unaltered allelic expression of the Igf2 and H19 genes in G9a−/− placentas. (E) Unaltered paternal expression of the U2af1-rs1 gene.
Cdkn1c and Phlda2, located in the central portion of the domain, faithfully maintained their paternal repression in the absence of G9a. Furthermore, the Kcnq1ot1 noncoding RNA remained expressed from the paternal allele only. However, altered imprinted expression was detected at the proximal and distal portions of the domain (Fig. 2A; see also Fig. S2 in the supplemental material). Loss of imprinting was defined as detection of an allelic ratio between the maternal and the paternal allele (maternal/paternal ratio [M/P]), which was below that observed in the cohort of all WT placentas (Fig. 2C; see also Table S1 in the supplemental material). In several of the G9a−/− placentas, there was clear derepression of the paternal alleles of the Osbpl5, Ascl2, and Cd81 genes. However, Osbpl5, Ascl2, and Cd81 did not show loss of paternal repression in concert. In one placenta there was loss of imprinting at Ascl2 only, whereas in three others, there was loss of imprinting at both Ascl2 and Ospbl5 (Fig. 2A; see also Fig. S1A in the supplemental material). Morphologically, these placentas appeared comparable to the other G9a−/− placentas.
To verify that the partial loss of imprinting was not linked to the parental backgrounds used, we performed the crosses between the G9a heterozygous mouse lines in the reciprocal orientation. This resulted in the same phenotype, with frequent relaxation of imprinting at Osbpl5, Cd81, and Ascl2, which in one of the placentas was observed at all three genes (Fig. 2B). As in the initial cross, no loss of imprinting was observed at Cdkn1c and Kcnq1ot1 (data not shown).
Since we had detected a moderate reduction in the number of giant cells in the G9a−/− placentas, the observed loss of imprinting could have been related to a less-advanced trophoblastic development. We excluded this possibility by analyzing WT placentas 1 day earlier in development, at 8.5 dpc. This showed that Osbpl5, Cd81, and Ascl2 were expressed from the maternal chromosome at this earlier developmental stage as well (see Fig. S3 in the supplemental material).
Ascl2, Obpl5, and Cd81 are imprinted in the trophobast only. The Cdkn1c, Phlda2 and Kcnq1ot1 genes, in contrast, are also imprinted in the embryo (38, 53). We therefore studied these genes in embryos as well and found that their allelic expression is not altered in the absence of G9a (data not shown). Together, these findings indicate that, in the absence of G9a, there is normal paternal repression at these genes in the central part of the Kcnq1 domain, but that the establishment or the maintenance of silencing is affected at the distal and proximal genes, which are imprinted in the trophoblast only.
At the neighboring Igf2-H19 domain (54), no allelic changes in gene expression were observed. H19 remained expressed from the maternal chromosome exclusively, and the Igf2 gene from the paternal chromosome only, in all G9a−/− placentas and embryos analyzed (Fig. 2D and data not shown). As an additional imprinted region, we studied the U2af1-rs1 gene on proximal chromosome 11. This imprinted gene is DNA methylated on its repressed maternal allele (16). No loss of imprinting was observed in placentas (Fig. 2E) or embryos (data not shown). U2af1-rs1 remained paternally expressed only.
To globally assess levels of gene expression, we performed microarray (Affymetrix) analyses on WT versus G9a−/− placentas. Embryos were not included in this study, given the gross developmental abnormalities induced by the absence of G9a. 39.000 transcripts were analyzed in two independent experiments on pools of G9a−/− and WT placentas at 9.5 dpc.
G9a itself was readily expressed in WT placenta and, as expected, not in G9a−/− placentas. Sixty genes were altered fourfold or more in their expression levels. Of these, 27 showed a >10-fold change in G9a−/− placentas (see Table S2 in the supplemental material). Strongly upregulated genes included the Mage-a1-6 genes and Cdkn1a (p21). The latter is a negative cell cycle regulator, which had been proposed earlier to be controlled by G9a (8). Its upregulation could explain the moderate reduction in size of the G9a−/− placentas. The derepression of the Mage-a gene family extends recent in vitro studies on the involvement of G9a and Glp in the silencing of the Mage-a2 gene (23, 48, 49). Of the 65 imprinted genes that were included in the microarray study, 9 showed a significant change in their expression levels, of two- to fourfold. Significantly altered expression was not detected for Ascl2, Cd81, or Osbpl5 though, agreeing with our finding that the relaxation of imprinting is partial and not detected in all the placentas (see Table S2 in the supplemental material). This finding was confirmed by real-time PCR amplification for four of the G9a−/− placentas (see Fig. S1B in the supplemental material).
G9a recruitment regulates H3-K9 methylation.
The loss of imprinting at Ascl2, Cd81, and Osbpl5 in the G9a−/− placentas suggested that these genes could be marked by H3-K9 methylation on their repressed paternal alleles. To address this question in more detail, we performed ChIP on nonfixed chromatin extracted from 25 WT placentas at 9.5 dpc, using two antisera directed against H3K9me2 and two against H3K9me3 (Fig. 3). Enrichment of both H3K9me2 and H3K9me3 was detected on the repressed alleles of Ascl2 and Cd81. Also, at the Cdkn1c gene and at KvDMR1, there was H3K9me2 and H3K9me3 enrichment on the repressed allele. In contrast to Ascl2 and Cd81, these regions have DNA methylation on their repressed parental allele as well (25, 46). Although only little chromatin was precipitated, levels of H3K9me2 were higher than background. For H3K9me3, the highest levels of precipitation were detected at the KvDMR1. These data indicate that in WT placenta, Ascl2, Cd81, Cdkn1c, and KvDMR1 are enriched both in H3K9me2 and H3K9me3 on their repressed alleles.
FIG. 3.
ChIP on wild-type placenta. (A) Chromatin purified from 9.5-dpc WT placentas was precipitated with two different antisera directed against H3K9me2 (indicated by A and B in parentheses) and two antisera against H3K9me3 (indicated by A and B in parentheses). As an internal control, additional precipitations were performed with an antiserum against H3K4me2, which is present on the opposite parental allele. PCR was performed on antibody-bound (B) and unbound (U) fractions, followed by SSCP-based discrimination of the maternal (M) and paternal (P) alleles. In all panels, the first four lanes show amplifications from C57BL/6 (Dom), SDP711, and (C57BL/6 × SDP711)F1 (F1) genomic DNAs and from the used input chromatin used for the ChIP (input). Black dots indicate an allelic ratio which is higher than 2 (after correction for the ratio in the input chromatin). (B) Real-time PCR quantification of bound fractions corresponding to H3K9me2 and H3K9me3 from the precipitations shown in panel A. Precipitation was defined as enrichment over that of a mock precipitation with an unrelated IgG antiserum. In terms of the percentage of input chromatin that was precipitated, this corresponds to 2% for H3Kme2 at Cdkn1c and 10% for H3K9me3 at KvDMR1.
Next, we analyzed by ChIP a small number of available G9a−/− placentas (3) versus WT placentas (6). In the absence of G9a, no allelic enrichment of H3K9me2 and H3K9me3 was observed at Ascl2, Cd81, or Cdkn1c, and precipitation levels were considerably reduced compared to WT placentas (Fig. 4A; see also Table S1 in the supplemental material). This points to a reduction in H3K9me2/me3 at these imprinted genes and extends the recent finding that G9a regulates specific gene loci and controls local levels of both H3K9me2 and H3K9me3 (23). The KvDMR1, in contrast, retained high levels of H3K9me2 but showed decreased precipitation of H3K9me3. Major satellite DNA at pericentric heterochromatin retained high levels of H3K9me3 in the absence of G9a (Fig. 4B). At the ICR upstream of the H19 gene, precipitation levels of both H3K9me2 and H3K9me3 were lower than at the KvDMR1, and only the latter modification was reduced in the absence of G9a.
FIG. 4.
Kcnq1 domain genes have reduced H3K9me2 and H3K9me3 in the absence of G9a. (A) ChIP was performed concomitantly on limited numbers of G9a−/− and WT placentas (at 9.5 dpc), using the same reagents. Chromatin was precipitated with antisera against H3K9me2 and H3K9me3. PCR was performed on antibody-bound (B) and unbound (U) fractions, followed by SSCP-based discrimination of the maternal (M) and paternal (P) alleles (for the precise location of primers, see Table S3 in the supplemental material). In all panels, the first four lanes show amplifications from C57BL/6 (Dom), SDP711, and (C57BL/6 × SDP711)F1 (F1) genomic DNAs and from the used input chromatin (input). Black dots indicate allelic ratios which are >2 (after correction for the ratio in the input chromatin). (B) Levels of precipitation are presented as the enrichment over that of a mock precipitation with an unrelated IgG antiserum. In terms of percentage of input chromatin that was precipitated, this corresponds to 6% for H3Kme2 at Cdkn1c and 32% for H3K9me3 at the KvDMR1. (C) ChIP on fixed placental chromatin (WT). The first three lanes show control amplifications from C57BL/6 (Dom), SD7P711, and input chromatin, respectively. The allelic enrichment is marked by black dots when it was >2. Association of G9a was detected on the repressed paternal allele of Ascl2; RNA Pol II is associated with the active maternal allele.
Given the reduction in histone methylation at the Kcnq1 domain genes in the G9a−/− placentas, we explored whether G9a could be bound to these genes in WT placenta. Cross-linked chromatin was precipitated with an antibody directed against the N-terminal domain of G9a. At Ascl2, G9a was preferentially precipitated on the repressed paternal allele of Ascl2 (Fig. 4C). As a control we used an antiserum against the serine-5 phosphorylated form of RNA polymerase II (Pol II), which was detected predominantly on the active maternal allele of the Ascl2 gene. Thus, histone modifications (including H3-K9 methylation) on the silenced paternal alleles could prevent binding of Pol II. Under the experimental conditions used, however, we had little precipitation above background at Cdkn1c and Cd81, and so we could not determine whether at these genes the paternal allele also binds G9a (data not shown).
Maintenance of imprinted DNA methylation.
H3-K9 trimethylation is consistently associated with the DNA-methylated allele of ICRs (6, 40, 53, 56, 58; this study). It is enriched on the repressed allele of several imprinted gene promoters as well, including H19 and Cdkn1c. Furthermore, part of the cellular G9a is associated with Dnmt1, at replication foci (10), suggesting a link between the maintenance of H3-K9 and DNA methylation (29, 43). Indeed, the HMTs Suv39h1 and h2 are required for directing DNA methylation to the satellite DNA underlying pericentric heterochromatin (24). To determine whether G9a could be required for DNA methylation at ICRs and imprinted genes, we analyzed genomic DNAs extracted from G9a−/− embryos and placentas. Digestion was with HpaII (which cuts unmethylated DNA only), McrBC (which cuts methylated DNA only), or MspI (which cuts both methylated and unmethylated DNA). Digestion profiles were visualized by PCR amplification and were identical between WT and G9a−/− embryos at the KvDMR1 and the H19 ICR, with methylation being detected on the paternal alleles only (Fig. 5A). We also found unaltered paternal DNA methylation at the differentially methylated region 2 (DMR2) of the neighboring Igf2 gene (11) and unaltered maternal DNA methylation at the U2af1-rs1 gene. The Cdkn1c promoter was analyzed by bisulfite sequencing. Both in WT and G9a−/− embryos, methylation was present on the repressed paternal allele only (Fig. 4A). Also in the G9a−/− placentas, levels of allelic DNA methylation were unaltered at the KvDMR1 and the H19 ICRs, the Igf2 DMR2, and at Cdkn1c and U2af1-rs1 (Fig. 5B).
FIG. 5.
Unaltered DNA methylation in G9a−/− conceptuses. (A) Genomic DNAs from WT and G9a−/− embryos were digested with HpaII, McrBc, or MspI. They were PCR amplified subsequently and analyzed by SSCP electrophoresis. In all panels, the first two lanes show control amplifications from nondigested (-) C57BL/6 and SDP711 genomic DNA, respectively. Maternal (M) and paternal (P) specific bands are indicated. Bisulfite sequencing of the Cdkn1c promoter included 28 CpG dinucleotides. Each row of dots represents one individual chromosome. Methylated CpGs are shown as solid circles, and unmethylated CpGs are open circles. Two of the CpGs are polymorphic and are absent in the M. spretus (SDP711) genotype. (B) Unaltered DNA methylation in G9a−/− placenta. The first two lanes show PCR amplifications from undigested C57BL/6 (Dom) and SDP711 DNA, respectively.
DISCUSSION
The main finding from this study is that the HMT G9a contributes to the allelic repression of genes that are imprinted in the trophoblast only. This suggests that histone H3-lysine-9 methylation is one of the factors involved in placenta-specific imprinting. Importantly, no effects were observed on imprinting control regions, which stably maintained their allelic DNA methylation imprints in the absence of G9a, both in the placenta and the embryo.
A variable degree of paternal derepression was observed at the Ascl2, Cd81, and Ospbl5 genes in G9a−/− placentas. Why are these trophoblast-specific genes susceptible to loss of imprinting, whereas other genes that are imprinted more broadly appear unaffected? One distinction of these placenta-specific genes is that they do not acquire DNA methylation on their repressed paternal promoters during development and remain imprinted in the absence of Dnmt1 (25, 50). Their imprinting maintenance is thus independent of DNA methylation. As a consequence, these genes may rely more heavily on covalent histone modifications, including H3-lysine-9 and -lysine-27 methylation and histone H3 deacetylation (25, 53; this study). The involvement of multiple layers of silencing explains the incomplete penetrance of the loss of imprinting that we observed in the G9a−/− placentas. Many transcriptional repressors act as part of a set of redundant silencing mechanisms. Such a multilayered silencing can cause partial gene derepression in a stochastic manner in case one of the mechanisms is deficient. For instance, studies on the silencing of genes by X chromosome inactivation show that this is controlled by multiple layers of silencing mechanisms (37), each of which reduces the change in each cell of gene reactivation occurring (4). At imprinted genes in the central portion of the Kcnq1 domain, as well as at the H19 and U2af1-rs1 genes, the allelic repression was unaltered in the absence of G9a. These genes, however, use the additional, firm layer of repression put into place by DNA methylation and would therefore not readily lose imprinting due to G9a deficiency. Moreover, the continued paternal repression of some genes of the central part of the Kcnq1 domain (including Cdkn1c) may be controlled by the KvDMR1. Recent studies indicate that, on its unmethylated allele, this intronic ICR binds the CTCF protein and could function as a chromatin boundary, thereby preventing promoter-enhancer interactions that are required for the expression of nearby genes (13, 14, 19, 26). The maternal DNA methylation at the KvDMR1 was not affected in the G9a−/− conceptuses, and G9a deficiency would therefore not have changed its allelic boundary function.
Our data do not exclude that G9a deficiency had stochastically affected the establishment of the allelic repression at the placenta-specific genes. It is technically challenging to determine when precisely during development the repressive H3-lysine-9 methylation becomes established. In case the chromatin repression is an early event, as suggested to be the case for some of the genes in the domain (27), maternally transmitted G9a protein could influence this process, and this might explain some of the differences between individual G9a−/− placentas. Imprinting establishment at the Kcnq1 domain requires the KvDMR1 (13) and, in particular, transcriptional elongation of the noncoding RNA Kcnq1ot1, which is expressed from the ICR (31). Presumably, during early development the full-length Kcnq1ot1 RNA mediates the local recruitment of chromatin-modifying complexes, including PRC2 proteins and G9a, but this remains to be demonstrated.
G9a deficiency led to a two-thirds reduction in global H3K9me2, indicating that this is not the only HMT regulating H3K9me2. That G9a is involved in the allelic repression of placenta-specific genes follows from the strongly reduced H3-lysine-9 methylation levels that we observed in the G9a-deficient placentas. Interestingly, the reduced histone methylation concerned both H3K9me2 and H3K9me3. This was rather unexpected given that, globally, G9a deficiency leads to a major reduction in H3K9me2 but not H3K9me3 (39, 41, 48). However, in a recent study (23) reduction of G9a led to reduced levels of both H3K9me2 and H3K9me3 at specific genes, including the Mage-a2 gene. Also, the embryonic repression of the Oct3/4 gene involves G9a-mediated acquisition of H3K9me3 (12). Most likely, other HMTs contribute to maintaining the allelic H3-lysine-9 methylation at imprinted genes and ICRs as well. Glp, for instance, is present in mammalian cells together with G9a and has an effect on global levels of H3K9me2 as well (49). One other candidate to be tested is SETDB1/ESET, an H3-K9-specific HMT which is associated with a methyl-CpG binding protein and with Dnmt1 (29, 43). This SET domain protein could be important for ICRs and for imprinted genes that acquire allelic DNA methylation during embryonic development (40).
Our finding of unaltered DNA methylation at ICRs extends an earlier study on G9a-deficient embryos which showed unchanged DNA methylation at the ICR controlling the Snrpn imprinted domain on central chromosome 7 (57). In this study, however, loss of Snrpn methylation was observed in G9a-deficient ES cells. One explanation for this discrepancy could be that G9a deficiency affects methylation maintenance more readily in cultured ES cells than in the embryo. Even in WT ES cells, in vitro culture can give rise to DNA methylation changes at ICRs (5, 59). Although we did detect reduced H3K9me2/3, the combined data indicate that G9a is not essential for the in vivo maintenance of DNA methylation at ICRs or at imprinted promoters that acquire their DNA methylation during embryogenesis. Also genes undergoing X chromosome inactivation acquire DNA methylation on their repressed promoters during early development (37), and X inactivation was reported to be unaffected by G9a deficiency (36).
In conclusion, the loss of imprinting in the placenta did not affect the genes along the Kcnq1 domain in concert but, rather, occurred in a stochastic manner. This implies that H3-lysine-9 methylation is not the only epigenetic modification that maintains the paternal silencing along this domain (Fig. 6). Removal of this layer of repression in the placenta induces a less-efficient maintenance of repression, particularly at genes that do not also use CpG methylation as part of their silencing mechanism. One further layer of repression is provided by H3K27me3, similar to that on the inactive X chromosome (30, 37, 53). It remains to be explored whether other mechanisms linked to G9a can explain the paternal silencing of genes at the Kcnq1 domain as well. For instance, recent studies indicate that G9a, via its partner protein Glp, can mediate the local recruitment of transcriptional corepressor molecules, such as CtBP (35, 51). Whatever G9a's precise additional modes of action, our study provides the first in vivo evidence for involvement of an HMT in imprinted gene repression. It highlights the importance of histone methylation rather than DNA methylation in imprinting maintenance in the mouse trophoblast. Intriguingly, several other imprinted mouse loci comprise genes that seem to be imprinted in the placenta only, without the involvement of promoter DNA methylation (55). It should now be interesting to determine whether G9a also plays a contributing role here. From a more general perspective, our data expand earlier work on several imprinted genes, showing that they have lower levels of DNA methylation in the placenta than in the embryo or are not methylated at all in this extraembryonic tissue (25, 44, 50). Concordantly, the maintenance of imprinted gene repression would be less tightly controlled in the trophoblast lineage than in the embryo proper. This could be particularly crucial during early development, given the finding that culture of preimplantation embryos leads to a preferential loss of imprinting in the placenta (32, 42).
FIG. 6.
Model for involvement of G9a in placenta-specific imprinting. Paternal repression along the Kcnq1 domain is mediated by the KvDMR1 (ICR) and expression of the noncoding RNA Kcnq1ot1 (30). The central portion of the domain is imprinted in both the embryo and placenta. These genes require DNA methylation for the maintenance of their paternal allele-specific repression (25), either directly, at their repressed promoter, or indirectly (14, 19, 26), via a DNA methylation-dependent boundary function of the KvDMR1 involving binding of the CTCF protein (14). These genes maintain imprinting in the absence of G9a. The telomeric and centromeric portions of the Kcnq1 domain comprise genes that are imprinted in the mouse placenta only. These genes do not use DNA methylation as one layer of their allelic repression. Consequently, G9a deficiency and the resulting reduction of H3K9me2/me3 readily gives rise to loss of imprinting at these placental genes.
Supplementary Material
Acknowledgments
We thank Sheila Webb for blastocyst injections, Patricia Cavelier for histology, and Philippe Arnaud for discussion and comments.
R.F. and W.B. thank the 6th Framework NoE EPIGENOME (LSHG-CT-2004-503433) for support and encouragement. R.F. acknowledges grant support from ESF EUROCORES program EuroSTELLS, the ARC program ARECA, the ANR (Programme Blanc), the Cancéropôle Grand Sud-Ouest, and the ACI program of the French Ministry of Education and Science. K.W. received a Peter Baker traveling fellowship, and A.W. acknowledges an ARC Ph.D. Fellowship. R.S. was the recipient of an EMBO long-term fellowship.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arima, T., K. Hata, S. Tanaka, M. Kusumi, E. Li, K. Kato, K. Shiota, H. Sasaki, and N. Wake. 2006. Loss of maternal imprint in Dnmt3Lmat−/− mice leads to a differentiation defect in the extraembryonic tissue. Dev. Biol. 297361-373. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud, P., K. Hata, M. Kaneda, E. Li, H. Sasaki, R. Feil, and G. Kelsey. 2006. Stochastic imprinting in the progeny of Dnmt3L−/− females. Hum. Mol. Genet. 15589-598. [DOI] [PubMed] [Google Scholar]
- 3.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bolman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 2942536-2539. [DOI] [PubMed] [Google Scholar]
- 4.Csankovszki, G., A. Nagy, and R. Jaenisch. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean, W., L. Bowden, A. Aitchison, J. Klose, T. Moore, J. J. Meneses, W. Reik, and R. Feil. 1998. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 1252273-2282. [DOI] [PubMed] [Google Scholar]
- 6.Delaval, K., J. Govin, F. Cerqueira, S. Rousseaux, S. Khochbin, and R. Feil. 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 26720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaval, K., and R. Feil. 2004. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 14188-195. [DOI] [PubMed] [Google Scholar]
- 8.Duan, Z., A. Zarebski, D. Montoya-Durango, H. L. Grimes, and M. Horwitz. 2005. Gfl1 coordinates epigenetic repression of the P21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 2510338-10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, C. A., and A. C. Ferguson-Smith. 2007. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 191-9. [DOI] [PubMed] [Google Scholar]
- 10.Estève, P. O., H. G. Chin, A. Smallwood, G. R. Feehery, O. Gangisetty, A. R. Karpf, M. F. Carey, and S. Pradhan. 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 203089-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, R., J. Walter, N. D. Allen, and W. Reik. 1994. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 1202933-2943. [DOI] [PubMed] [Google Scholar]
- 12.Feldman, N., A. Gerson, J. Fang, E. li, Y. Zhang, Y. Shinkai, H. Cedar, and Y. Bergman. 2006. G9A-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8188-194. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32426-431. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick, G. V., E. M. Pugacheva, J.-Y. Shin, Z. Abdullaev, Y. Yang, K. Khatod, V. V. Lobanenkov, and M. J. Higgins. 2007. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 272636-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo methyltransferases to establish maternal imprints in mice. Development 1291983-1993. [DOI] [PubMed] [Google Scholar]
- 16.Hatada, I., K. Kitagawa, T. Yamaoka, X. Wang, Y. Arai, K. Hashido, S. Ohishi, J. Masuda, J. Ogata, and T. Mukai. 1995. Allele-specific methylation and expression of an imprinted U2af1-rs1 (SP2) gene. Nucleic Acids Res. 2336-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell, C. Y., T. H. Bestor, F. Ding, K. E. Latham, C. Mertineit, J. M. Trasler, and J. R. Chaillet. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104829-838. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein, T. 2006. The epigenetic magic of histone lysine methylation. FEBS J. 2733121-3135. [DOI] [PubMed] [Google Scholar]
- 19.Kanduri, C., G. Fitzpatrick, R. Mukhopadhyay, M. Kanduri, V. Lobanenkov, and R. Ohlsson. 2002. A differentially methylated imprinting control region within the Kcnq1 locus harbors a methylation-sensitive chromatin insulator. J. Biol. Chem. 27718106-18110. [DOI] [PubMed] [Google Scholar]
- 20.Kaneda, M., M. Okano, K. Hata, T. Sado, N. Tsujimoro, E. Li, and H. Sasaki. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429900-903. [DOI] [PubMed] [Google Scholar]
- 21.Kent, J. W. 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent, J. W., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The Human Genome Browser at UCSC. Genome Res. 12996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubicek, S., R. J. O'Sullivan, E. M. August, E. R. Hickey, Q. Zhang, M. L. Teodoro, S. Rea, K. Mechtler, J. A. Kowalski, C. A. Homon, T. A. Kelly, and T. Jenuwein. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25473-481. [DOI] [PubMed] [Google Scholar]
- 24.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 131192-1200. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 361291-1295. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, A., and W. Reik. 2006. How imprinting centres work. Cytogenet. Genome Res. 11381-89. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, A., K. Green, C. Dawson, L. Redrup, K. D. Huyn, J. T. Lee, M. Hemberger, and W. Reik. 2006. Epigenetic dynamics of the Kcnq1 domain in the early embryo. Development 1334203-4210. [DOI] [PubMed] [Google Scholar]
- 28.Li, E., C. Beard, and R. Jaenisch. 1993. Role of DNA methylation in genomic imprinting. Nature 366362-365. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., T. Rauch, Z. X. Chen, P. E. Szabo, A. D. Riggs, and G. P. Pfeifer. 2006. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 28119489-19500. [DOI] [PubMed] [Google Scholar]
- 30.Mager, J., N. D. Montgomery, F. P. de Villena, and T. Magnuson. 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33502-507. [DOI] [PubMed] [Google Scholar]
- 31.Mancini-Dinardo, D., S. J. Steele, J. M. Levorse, R. S. Ingram, and S. M. Tilghman. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 201268-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann, M. R., S. S. Lee, A. S. Doherty, R. I. Verona, L. D. Nolen, R. M. Schultz, and M. S. Bartolomei. 2004. Selective loss of imprinting in the placenta following preimplantation development in culture. Development 1313727-3735. [DOI] [PubMed] [Google Scholar]
- 33.Morison, I. M., J. P. Ramsay, and H. G. Spencer. 2005. A census of mammalian imprinting. Trends Genet. 21457-465. [DOI] [PubMed] [Google Scholar]
- 34.Neumann, B., and D. P. Barlow. 1996. Multiple roles for DNA methylation in gametic imprinting. Curr. Opin. Genet. Dev. 6159-163. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 2961132-1136. [DOI] [PubMed] [Google Scholar]
- 36.Ohhata, T., M. Tachibana, M. Tada, T. Tada, H. Sasaki, Y. Shinkai, and T. Sado. 2004. X-inactivation is stably maintained in mouse embryos deficient for histone methyl transferase G9a. Genesis 40151-156. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto, I., and E. Heard. 2006. The dynamics of imprinted X inactivation during preimplantation development in mice. Cytogenet. Genome Res. 113318-324. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen, M., K. R. Davies, L. M. Bowden, A. J. Villar, O. Franck, M. Fuermann, W. L. Dean, T. F. Moore, N. Rodrigues, K. E. Davies, R. J. Hu, A. P. Feinberg, E. R. Maher, W. Reik, and J. Walter. 1998. Syntenic organisaton of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5. Hum. Mol. Genet. 71149-1159. [DOI] [PubMed] [Google Scholar]
- 39.Peters, A. H., S. Kubicek, K. Mechter, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 121577-1589. [DOI] [PubMed] [Google Scholar]
- 40.Regda, K., M. A. Sloane, R. Huang, F. M. Pauler, K. E. Warczok, B. Melikant, M. Radolf, J. H. Martens, G. Schotta, T. Jenuwein, and D. P. Barlow. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz, D. F. Hunt, Y. Shinkai, and C. D. Allis. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 121591-1598. [DOI] [PubMed] [Google Scholar]
- 42.Rivera, R. M., P. Stein, J. R. Weaver, J. Mager, R. M. Schultz, and M. S. Bartolomei. 27 September 2007, posting date. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum. Mol. Gen. [Epub ahead of print.] [DOI] [PubMed]
- 43.Sarraf, S. A., and I. Stancheva. 2004. Methyl-CpG binding protein MBD1 couples H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15595-605. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki, H., A. C. Ferguson-Smith, A. S. Shum, S. C. Barton, and M. A. Surani. 1995. Temporal and spatial regulation of H19 imprinting in normal and uniparental mouse embryos. Development 1214195-4202. [DOI] [PubMed] [Google Scholar]
- 45.Schulz, R., T. R. Menheniott, K. Woodfine, A. J. Wood, J. D. Choi, and R. J. Oakey. 2006. Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res. 34e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smilinich, N. J., C. D. Day, G. V. Fitzpatrick, G. M. Caldwell, A. C. Lossie, P. R. Cooper, A. C. Smallwood, J. A. Joyce, P. N. Schofield, W. Reik, R. D. Nicholls, R. Weksberg, D. J. Driscoll, E. R. Maher, T. B. Shows, and M. J. Higgins. 1999. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. USA 968064-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutherland, H. G., G. K. Mumford, K. Newton, L. V. Ford, R. Farrall, G. Dellaire, J. F. Caceres, and W. A. Bickmore. 2001. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 101995-2011. [DOI] [PubMed] [Google Scholar]
- 48.Tachibana, M., K. Sugimoto, M. Nozaki, J. Ueda, T. Ohta, M. Ohki, M. Fukuda, N. Takeda, H. Niida, H. Kato, and Y. Shinkai. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 161779-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tachibana, M., J. Ueda, M. Fukuda, N. Takeda, T. Ohta, H. Iwanari, T. Sakihama, T. Kodama, T. Hamakubo, and Y. Shinkai. 2005. Histone methyltransferase G9a and Glp form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka, M., M. Puchyr, M. Gerstenstein, K. Harpal, R. Jaenisch, J. Rossant, and A. Nagy. 1999. Parental origin-specific expression of Mash2 is established at the time of implantation with its imprinting mechanism highly resistant to genome-wide demethylation. Mech. Dev. 87129-142. [DOI] [PubMed] [Google Scholar]
- 51.Ueda, J., M. Tachibana, T. Ikura, and Y. Shinkai. 2006. Zinc finger protein Wiz links G9a/Glp histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 28120120-20128. [DOI] [PubMed] [Google Scholar]
- 52.Uejima, H., M. P. Lee, H. Cui, and A. P. Feinberg. 2000. Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat. Genet. 25375-376. [DOI] [PubMed] [Google Scholar]
- 53.Umlauf, D., Y. Goto, R. Cao, F. Cerqueira, A. Wagschal, Y. Zhang, and R. Feil. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 361296-1300. [DOI] [PubMed] [Google Scholar]
- 54.Verona, R. I., M. R. Mann, and M. S. Bartolomei. 2003. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19237-259. [DOI] [PubMed] [Google Scholar]
- 55.Wagschal, A., and R. Feil. 2006. Genomic imprinting in the placenta. Cytogen. Genome Res. 11390-98. [DOI] [PubMed] [Google Scholar]
- 56.Wu, M.-Y., T.-F. Tsai, and A. L. Beaudet. 2006. Deficiency of Rbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 202859-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xin, Z., M. Tachibana, M. Guggiari, E. Heard, Y. Shinkai, and J. Wagstaff. 2003. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 27814996-15000. [DOI] [PubMed] [Google Scholar]
- 58.Yang, Y., T. Li, T. H. Vu, G. A. Ulaner, J. F. Hu, and A. R. Hoffman. 2003. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology 1445658-5670. [DOI] [PubMed] [Google Scholar]
- 59.Zvetkova, I., A. Apedaile, B. Ramsahoye, J. Mermoud, L. Crompton, R. John, R. Feil, and N. Brockdorff. 2005. Global hypomethylation of the genome in XX ES cells. Nat. Genet. 371274-1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.