Abstract
In Drosophila, dosage compensation—the equalization of most X-linked gene products between XY males and XX females—is mediated by the MSL complex that preferentially associates with numerous sites on the X chromosome in somatic cells of males, but not of females. The complex consists of a noncoding RNA and a core of five protein subunits that includes a histone acetyltransferase (MOF) and an ATP-dependent DEXH box RNA/DNA helicase (MLE). Both of these enzymatic activities are necessary for the spreading of the complex to its sites of action along the X chromosome. MLE is related to the ATPases present in complexes that remodel chromatin by altering the positioning or the architectural relationship between nucleosomes and DNA. In contrast to MLE, none of these enzymatic subunits has been shown to possess double-stranded nucleic acid-unwinding activity. We investigated the function of MLE in the process of dosage compensation by generating mutations that separate ATPase activity from duplex unwinding. We show that the ATPase activity is sufficient for MLE's role in transcriptional enhancement, while the helicase activity is necessary for the spreading of the complex along the X chromosome.
It is widely recognized that the association of DNA, histones, and nonhistone proteins in chromatin leads to a compacted structure that is unfavorable to transcription. This association must be modified in order to allow the transcriptional machinery to access the promoter regions of genes and to carry out RNA synthesis. There are two general means by which such modifications can be achieved: the ATP-dependent remodeling of the DNA-nucleosome association and the covalent modification of histones. Large multiprotein complexes use the ATPase activity of one of their subunits to slide nucleosomes along the DNA molecule or to alter the torsional stress of the DNA as it wraps around nucleosomes. Other complexes bring histone acetyltransferases, methylases, kinases, or ubiquitinases to the nucleosomes in order to modify specific residues, predominantly but not exclusively in the free N-terminal tails. These modifications alter the charge difference between the histone and DNA molecules, reducing their affinity for one another, or serve as signals for the docking of transcription activators and coactivators (reviewed in references 13, 16, and 29). The MSL complex of Drosophila consists of a core of five protein subunits (encoded by male-specific lethal 1, 2, and 3 [msl1, msl2, and msl3], males absent on the first [mof], and maleless [mle]), as well as one of two noncoding RNAs (RNA on the X1 and X2 [roX1 and roX2]). The complex preferentially associates with numerous sites on the X chromosome in somatic cells of males, but not of females. It is responsible for an enhancement of the transcriptional rates of a substantial number of X-linked genes, thereby mediating a compensatory effect for the difference in the dosages of these genes between males and females. The presence of the MSL complex on the male X chromosome is correlated with a significant increase of a specific histone isoform, histone H4 acetylated at Lys16 (H4K16ac). This acetylation, which is the result of the activity of MOF, a histone acetyltransferase of the MYST family, is believed to favor enhanced levels of transcription by lessening internucleosomal interactions or DNA-nucleosomal association and thereby facilitating nucleosomal eviction by the transcription elongation complex (see reference 18 for a review).
The MLE subunit of the MSL complex is an ATP-dependent DEXH box RNA/DNA helicase that prefers double-stranded RNA (dsRNA) or RNA/DNA hybrid substrates with a short 3′ overhang (14). MLE is related to the ATPases present in complexes that remodel chromatin by altering the positioning of or the architectural relationship between nucleosomes and DNA. These ATPases belong to four major subfamilies defined by the first member to be identified: SWI2/SNF2, ISWI, CHD-1, and Ino80 (see reference 8 for a review). In contrast to MLE, none of these enzymatic subunits has been shown to possess double-stranded nucleic acid-unwinding activity (the helicase activity exhibited by the Saccharomyces cerevisiae INO80 complex appears to be due to two subunits that are similar to the bacterial RuvB helicase and that appear to be functionally coupled to the Ino80 ATPase [26]). In view of this catalytic distinction, we wished to determine whether MLE has been subsumed by the MSL complex just for its ATPase function or for its ATP-dependent helicase activity. To this end, we generated stable lines of Drosophila Schneider line 2 (S2) cultured cells that express different mutant MLE proteins under the control of an inducible promoter. We partially purified these proteins and tested them for ATPase and helicase activities. We used an experimental system that reproduces dosage compensation on a plasmid to assess the abilities of complexes formed in the cell lines that expressed mutant MLE proteins to enhance the transcriptional activity of a targeted reporter gene. We also generated transgenic flies to determine the abilities of MSL complexes containing mutant MLE proteins to spread along the X chromosome. Here, we report that the ATPase activity is sufficient for MLE's role in the transcriptional enhancement of a targeted gene while the helicase activity is necessary for the spreading of the complex along the X chromosome.
MATERIALS AND METHODS
Recombinant Flag-mle vectors.
The sequences of the oligonucleotides used for the cloning strategies are presented in Fig. S3 in the supplemental material.
pBS vectors.
Site-directed mutagenesis was performed using the in vitro Quickchange system (Stratagene). The smallest restriction fragment containing the mutation was subcloned in the pFLC-1 plasmid carrying the full mle cDNA. A Flag-pBS recombinant plasmid was generated by annealing two oligonucleotides encoding the Flag epitope with KpnI and SmaI sites at the 5′ end and EcoRV at the 3′ end, respectively, and cloning the resulting dsDNA fragment in a pBS-KS plasmid. An 800-bp fragment starting from the first codon following the ATG and including a new EcoRV site was amplified by PCR and subcloned in the Flag-pBS vector to obtain the Flag-pBS/Ntmle vector. Lastly, HindIII-BamHI fragments from each mle/pFLC-1 recombinant vector were cloned in Flag-pBS/Ntmle.
pMK33 vectors.
The SmaI-BamHI fragment from each recombinant Flag-mle vector was subcloned in the pMK33/pMtHy vector (12).
pCasperhs83 vectors.
Two oligonucleotides containing EcoRI and SacII sites at the 5′ end and BamHI and EcoRI sites at the 3′ end were annealed, and the dsDNA fragment was cloned into the EcoRI site of the pCasperhs83 vector. The SacII-BamHI fragment from each recombinant Flag tag-mle vector was subcloned in this modified pCasperhs83 vector (9).
Transfections and selection of stable lines.
Drosophila S2 cells were transfected with each recombinant mle-pMK33/pMtHy vector using the Qiagen Effectene protocol. Stably transfected cells were selected with increased amounts of hygromycin B (Cellgro). Stably transfected cells grown to 3 × 106 cells/ml were transferred to 500-ml spinner flasks and cultured at 25°C with constant stirring (80 rpm) until a doubling of the cell density with a viability greater than 90% was reached. Copper sulfate was added (200 μM) for 24 h to induce production of recombinant MLE protein.
Purification of the Flag-tagged MLE recombinant proteins.
Preparation of nuclear extracts was performed as described previously (28), using the salt extraction protocol. Extracts were mixed with anti-Flag M2-agarose beads (Sigma) equilibrated with nucleus extraction buffer in high salt (350 mM NaCl), gently rocked at 4°C for 1 h, and then loaded onto a 5-ml column. The beads were washed with 5 volumes of nucleus extraction buffer with 350 mM NaCl, followed by 5 volumes of low-salt extraction buffer (150 mM NaCl). Bound Flag-MLE proteins were eluted with 200 μg/ml of Flag peptide and 20% glycerol in low-salt extraction buffer. Aliquots were quickly frozen in liquid nitrogen and kept at −80°C. The purity of each protein preparation was checked by 7.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using a silver-staining protocol.
Helicase and ATPase assays.
ATP-dependent RNA helicase activity was measured as described previously (14), using the same dsRNA substrate and 5 ng of each Flag-tagged recombinant MLE protein per assay. ATPase activity was measured as described previously (27) at pH 7.6 in 20-μl reaction mixtures containing 5 ng of each Flag-tagged recombinant MLE protein per assay. The reaction mixtures were spotted onto a polyethyleneimine thin-layer chromatography plate (Sigma). ATP and Pi were separated by chromatography in 1 M formic acid/0.5 M LiCl for 45 min and then located by autoradiography. All enzyme reaction products were quantitated with a phosphorimager.
Dosage compensation.
The plasmid model of dosage compensation (summarized in Fig. S2 in the supplemental material) was used as previously described (30). The Flag-MLE stable S2 lines were split to 30 to 60% confluence prior to transfection, which was carried out following the Qiagen Effectene protocol with 15 ng supercoiled roX-bearing firefly luciferase plasmid (roX-FF) or Nesprin-bearing firefly luciferase (N-FF) plasmid (where Nesprin is 1 kb from the human Nesprin introm), 1 ng tTA activator plasmid, 5.4 ng Renilla luciferase plasmid, and 1.2 μg pBluescript (Stratagene) carrier; 10.8 μl enhancer and 19.2 μl Effectene reagent were used per 5 × 106 cells. Just prior to transfection, 200 μM of copper sulfate was added to the medium. The next day, the cells were split to a final concentration of 0.3 × 106 cells/ml, maintaining the same copper sulfate concentration. Three to five days after transfection, the cells were collected for luciferase assay and protein isolation. Luciferase activity was determined using the Dual Luciferase Reporter Assay system (Promega). The firefly luciferase activity was normalized to Renilla luciferase activity for each sample. At least three independent experiments were performed.
Chromatin immunoprecipitation.
Cells were grown to a density of 3 × 106 cells/ml in a 15-ml volume with copper sulfate, and chromatin was immunoprecipitated according to the Upstate EZ ChIP protocol with the following modifications. The final concentration of formaldehyde was 0.3%, and sonication was performed with 25 sets of 4-second pulses. Immunoprecipitates were eluted from the agarose beads with 50 μl of sample buffer. Samples were incubated at 65°C for 5 h to reverse the cross-links. Aliquots of each sample were loaded on a 4 to 20% SDS-polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and probed with MSL1, Flag (Sigma), and H3 (Abcam) antisera. Negative controls were carried out with uncoated beads or with beads coated with RFX5 antiserum (a gift from J. Boss), as well as stock S2 (Flag-minus) cells.
RNA interference knockdown.
The day before transfection, 0.9 × 106 S2 cells were transferred to a six-well culture dish 1 h prior to pretreatment with dsRNA. To knock down wild-type (endogenous) mle RNA while leaving mutant mle(ΔV) RNA intact, we added 30 μg/well of 5′ untranslated region (UTR) dsRNA (Ambion's MEGAscript protocol) plus 10 μg/well of RNA duplex covering the 7-amino-acid ΔV deletion (Integrated DNA Technologies, Inc.). Green fluorescent protein (GFP) dsRNA (20 μg/well) was used as an RNA interference control. Cells were transfected 18 to 22 h later as described under “Dosage compensation” above. The primers used are listed in Fig. S3 in the supplemental material.
Transgenic lines.
Flag-tagged recombinant mle/pCasperhs83 constructs were purified by using a Qiaprep Spin Miniprep kit (Qiagen) and used for germ line transformation of a w1118 mutant strain by Genetic Services Inc. The G1 progeny were mated to w1118; T(2;3)apXa/CyO P{ActGFP.w−}CC2;TM6 Sb Tb flies in order to determine the chromosome of insertion. Males, w1118/Y; pr mle1/Bc; [hsp83-Flag-mletg w+]/TM6 Sb Tb (where mletg indicates a transgene), were then mated with y w; pr mle1/pr mle1; H83msl2/H83msl2 females to monitor the rescue of homozygous mle1 male lethality and to visualize the localization of MSL complexes carrying the Flag-MLE recombinant proteins on female polytene chromosomes.
Cytoimmunofluorescence.
Salivary gland polytene chromosome squashes were performed on y w1118/w1118; pr mle1/pr mle1; [H83msl2]/[hsp83-Flag-mletg w+] females. Glands were dissected from third-instar larvae, and chromosomes were prepared for immunofluorescence as previously described (25).
Quantitative RT-PCR of roX RNAs and gene transcripts.
Total RNA was extracted from 12 third-instar larvae using the Trizol Plus RNA purification kit (Invitrogen) and subsequently treated with Turbo DNase (Ambion). Quantitative reverse transcription (RT)-PCR was performed in triplicate on at least three different preparations with the respective gene-specific primer pairs (see Fig. S4 in the supplemental material) using a Bio-Rad iCycler. Enrichment was determined by raising 2 to a power equal to the cycle difference between experimental and control RNA samples.
RESULTS
The helicase and ATPase activities of MLE can be separated.
A comparison of the amino acid sequence of the MLE protein with those of other known helicases (see Fig. S1 in the supplemental material) revealed a series of putative functional domains (Fig. 1A). The mutation used by Lee et al. (14) is a substitution of lysine for glutamate at position 413 [MLE(K413E)] in the ATP-binding site (domain I) that eliminates both ATPase and helicase activities. In order to inactivate the helicase activity without affecting the ATPase activity, we engineered mutations at positions 539 and 541 in domain III [MLE(S539A,T541A)] and at position 764 in domain VI [MLE(Q764H)], hypothesizing that they would have no effect or partially reduce the ATPase activity, respectively, while eliminating helicase function. We also engineered mutations at positions 717 and 723 [MLE(T717A,S723A)], as well as a deletion starting at position 721 in domain V [ΔETSITID, abbreviated henceforth as MLE(ΔV)]. This domain forms a distinct loop in SWI2/SNF2; its deletion does not alter the ATPase activity of the protein but cripples the ability of the SWI/SNF complex to remodel chromatin (27). Even though the amino acid sequence of this domain is very poorly conserved in the remodeling-complex ATPases belonging to the other subfamilies, a loop is predicted to form in the general region of the MLE protein where the domain should occur, suggesting that a deletion of this region may have an effect similar to the SWI2/SNF2 deletion.
FIG. 1.
Purification and assays of mutant MLE proteins. (A) Schematic of the MLE protein structure. The domains that are involved in the ATP-dependent helicase activity are numbered I to VI. The mutations analyzed are indicated. (B) Silver staining of 146-kDa purified Flag-tagged recombinant MLE proteins from S2 cells run on a 7.5% SDS-polyacrylamide gel. KE, Flag-MLE(K413E); MLE, wild type Flag-MLE; ΔV, Flag-MLE(ΔV); STA, Flag-MLE(S539A,T541A); QH, Flag-MLE(Q764H); TSA, Flag-MLE(T717A,S723A). The faster bands in the ΔV lane may represent degradation products. (C) Activity assays of mutant MLE proteins performed as described in Materials and Methods. The abbreviations are the same as in panel B; Mock, extract from untransfected (stock) S2 cells treated with Flag-agarose beads. The brackets indicate the windows used in the phosphorimager quantification of the assays. (Top) ATPase activity measured as a function of inorganic phosphate (Pi) released from [γ-32P]ATP. (Bottom) Helicase activity measured as a function of single-stranded RNA released from the dsRNA/DNA substrate. The linearity of the assay was established with different amounts of wild-type MLE protein. (D) Quantitative representation of ATPase and helicase assay results; each bar is an average of three assays. The error bars represent standard deviations of the mean. The boxes indicate the two mutants that separate ATPase from helicase activities. In the quantitations provided under the histograms, the ATPase and helicase activities of MLE are set at 100%.
Drosophila S2 cells were transfected with vectors carrying sequences encoding Flag-tagged mutant MLE proteins under the control of a metallothionein promoter. Stable cell lines were established by hygromycin selection. Mutant proteins were partially purified (Fig. 1B) and tested for ATPase and helicase activities (Fig. 1C and D). The amino acid substitutions in domain V (T717A and S723A) and in domain VI (Q764H) had a very small effect and were not pursued further. The MLE protein carrying a deletion of domain V had a wild-type level of ATPase activity but completely lacked helicase activity. The mutation in domain III (S539A, T541A) exhibited reduced but still substantial ATPase activity with no helicase function.
The ATPase activity of MLE is required for the transcriptional enhancement necessary to achieve dosage compensation.
To determine the effects of the mutant proteins on the function of the dosage compensation MSL complex, we used an experimental model reproducing dosage compensation on a plasmid transfected and transiently expressed in cultured cells (30). Plasmids containing a reporter gene (firefly luciferase) with (roX-FF) or without (N-FF) a 1,087-bp fragment from the roX2 genomic sequence that nucleates the MSL complex were transfected into Drosophila S2 cells, together with another plasmid (R) containing a different reporter gene (Renilla luciferase) as a control for levels of transfection. The firefly luciferase gene was under the control of the tetracycline resistance operator promoter (tetO) that is induced by a synthetic transcriptional activator (tTA). A plasmid containing the gene for the activator under the control of the constitutive alpha-tubulin promoter (ptTA) was cotransfected into S2 cells. The Renilla luciferase gene was under the control of a constitutive Copia promoter. In the presence of the roX2 sequence, there was an almost perfect twofold enhancement of transcription of the firefly luciferase gene. The roX-FF plasmid is highly enriched in histone H4 acetylated at lysine 16. This mark, as well as the transcriptional enhancement, was abrogated by knockdown of the MSL complex with RNA interference or by cotransfection with a plasmid expressing Sex-lethal (30) (the SXL protein prevents the translation of MSL2 and therefore formation of the complex [3, 11]).
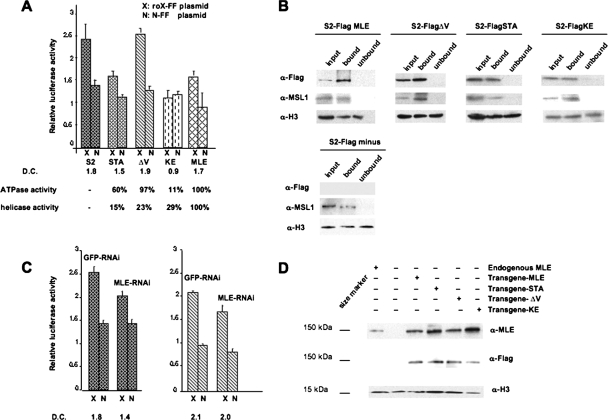
Stable S2 cell lines expressing mutant or wild-type mle transgenes were induced with copper sulfate and then transfected with the plasmid system described above. This assay is equivalent to a dominant-negative assay in that the mle transgene in each stable line is overexpressed and its product is expected to be incorporated into the majority of assembled MSL complexes. Dosage compensation was monitored as the ratio of the relative firefly luciferase activity of the roX-FF plasmid to that of the N-FF plasmid (Fig. 2A). As expected, this ratio was similar in transfected cells expressing the wild-type mle transgene (1.73 ± 0.09) and in stock S2 cells (1.86 ± 0.06). In the S2 cells expressing the MLE protein with the K413E mutation, this ratio was 0.94 ± 0.15, indicating that the MSL complex formed in these cells is incapable of mediating a transcriptional enhancement of the firefly gene. roX-FF plasmids transfected into cells expressing MLE with the domain V deletion exhibited normal levels of dosage compensation (1.94 ± 0.21), and cells expressing MLE with the S539A, T541A mutation in domain III had an intermediate level of compensation (1.53 ± 0.02). These results indicate that the function of the MSL complex depends on the level of ATPase activity, but not on the helicase activity of its MLE subunit.
FIG. 2.
Effects of mutant MLE proteins on dosage compensation. (A) S2 cells were transiently transfected with the roX-FF plasmid (X) or the control N-FF plasmid (N) and cotransfected with a plasmid that expresses Renilla luciferase as an internal control. The heights of the columns represent the firefly luciferase activity relative to the Renilla luciferase activity. There is a significant enhancement in relative firefly luciferase activity produced by the roX-bearing plasmids in stock S2 cells and in the stably transformed S2 cell lines expressing the wild-type Flag-MLE protein (MLE) or the Flag-MLE(ΔV) protein (ΔV), evidencing the occurrence of dosage compensation of the reporter gene. This enhancement is abrogated by expression of Flag-MLE(K413E) protein (KE) and is decreased to an intermediate value by the Flag-MLE(S539A,T541A) protein (STA). The error bars represent standard deviations of the mean. (B) The Flag-tagged mutant MLE proteins coimmunoprecipitate with MSL1. Shown are Western blots of immunoprecipitated proteins separated by SDS-polyacrylamide gel electrophoresis and probed with anti-FLAG, anti-MSL1, and anti-H3 sera. The input lanes represent 4% of the chromatin extract. The stable cell lines providing the extracts are indicated at the top of each panel. (C) Partial knockdown of wild-type endogenous MLE using RNA interference with dsRNAs homologous to the sequence deleted in the ΔV deletion mutant and to the 5′ UTR that is replaced by the Flag sequence in this mutant. The ratio of relative firefly luciferase levels in X and N cells represents the level of dosage compensation of the firefly gene. (Left) RNA interference treatment of stock S2 cells reduces this ratio by approximately 33% (1.8 versus 1.4). (Right) In contrast, this treatment has no effect on the level of compensation of Flag-MLE(ΔV)-expressing cells (2.1 versus 2.0), indicating that the Flag-MLE protein from the transgene is able to participate in MSL formation and function. (D) Expression of Flag-tagged MLE proteins in transgenic flies. Western blots of crude lysates from adult fly heads in a mle-null background (mle1) reacted with anti-MLE, anti-Flag, and anti-H3 sera. The level of histone H3 was used as a loading control. Similar results were obtained with three independent transgenic lines used for immunofluorescence and rescue experiments. The abbreviations are the same as in Fig. 1.
The possibility existed that, whereas the overexpressed MLE(K413E) protein is readily incorporated into the MSL complex, MLE(ΔV) or MLE(S539A,T541A) is not, and the levels of dosage compensation exhibited in cells expressing these mutations are the result of MSL complexes assembled with the endogenous wild-type MLE protein. We addressed this question by determining the presence of the mutant protein in MSL complexes using immunoprecipitation and by attempting to deplete the cells of the wild-type protein using RNA interference. Immunoprecipitation was carried out with MSL1 antiserum on nuclear extracts of cells treated with copper sulfate to induce expression of the different transgenes. Because the association of MLE with the MSL complex is relatively weak (28), we used formaldehyde cross-linking followed by sonication to stabilize this association in the immunoprecipitates. The results indicate that the vast majority of MSL complexes formed contain the Flag-MLE proteins produced by the transgenes (Fig. 2B). We attempted to achieve RNA interference of the endogenous MLE gene product without affecting the synthesis of Flag-MLE by synthesizing dsRNA homologous to the 5′ UTR uniquely present in the endogenous mle transcript. This approach resulted in a small but insufficient reduction of MLE levels. As an alternate strategy, applicable only to the MLE(ΔV)-expressing cell line, we synthesized dsRNA complementary to the domain V deletion (Fig. 2C). Treatment of stock S2 cells with both types of dsRNAs led to a modest decrease in MLE, reflected in a dosage compensation level of 1.41 ± 0.01 versus 1.77 ± 0.08. An identical treatment of the MLE(ΔV)-expressing cell line had no effect on dosage compensation (2.02 ± 0.08 versus 2.13 ± 0.10 in cells treated with GFP-specific dsRNA).
The helicase activity of MLE is required for the spreading of the MSL complex along the X chromosome.
In the roX-FF plasmid, the MSL complex targets the roX2 sequence that is immediately adjacent to the firefly gene. The enhancement of transcription, i.e., the dosage compensation of this gene, is not dependent on the helicase activity of MLE. Could this activity be required for the spreading of the complex to its affinity sites along the X chromosome and, ultimately, to all of its sites of action? In order to answer this question, transgenic lines carrying wild-type or mutant Flag-tagged MLE proteins were generated by germ line transformation. Expression of the transgenes was determined by Western analysis (Fig. 2D). The transgenes were introduced into the genomes of females that carried an msl2 cDNA insertion that allows synthesis of MSL2 and the assembly of functional MSL complexes (3, 11). These females were made homozygous for a null allele of mle, causing the formation of incomplete MSL complexes that bind only to the high-affinity sites. The extent of spreading beyond these sites by MSL complexes assembled with the Flag-MLE proteins can be determined by immunofluorescence on salivary gland polytene chromosome spreads.
In females that express the msl2 cDNA transgene but that lack MLE protein because they are homozygous for an mle-null allele, the partial complex detected by the presence of MSL1 is found only at the high-affinity sites on the paired X chromosomes (Fig. 3A). Expression of a wild-type Flag-MLE protein in these females leads to the presence of an MSL complex that has a distribution comparable to its normal distribution in males (Fig. 3B and C). Although the number of sites accessed by the complex is greater in the presence of the Flag-MLE(S539A,T541A) protein than the Flag-MLE(ΔV) protein, both mutant proteins clearly fail to induce normal spreading of the MSL complex (Fig. 3D and E).
FIG. 3.
Analysis of binding to the X chromosome of MSL complexes formed in the presence of Flag-tagged mutant MLE proteins. Indirect immunofluorescence was performed on salivary gland polytene chromosomes from y w1118/w; pr mle1/pr mle1; H83msl2/hsp83-Flag-mletg w+ females, where mletg represents one of the transgenes. (A) Control female homozygous for H83msl2 and mle1. (B) Female in which mletg is mle+. (C) Wild-type males. (D) Female in which mletg is mleS539A,T541A. (E) Female in which mletg is mleΔV. The MLE antiserum (green) recognizes the product of the transgenes that is the only MLE epitope present in panels B, D, and E; the MSL1 antiserum (red) detects the presence of the MSL complex. wt, wild type.
Spreading of the complex beyond the high-affinity sites to its numerous sites of action along the X chromosome is a prerequisite for normal dosage compensation and male viability. As expected, expression of the Flag-MLE(S539A,T541A) or the Flag-MLE(ΔV) protein failed to rescue males that lacked endogenous MLE (Table 1). Previously published results reported that, in spite of a lack of in vitro helicase activity, the MLE(S539A,T541A) mutation allowed normal spreading of the complex and full male viability (24). It is possible that the difference between these and our own results may be ascribable to a difference between genomic versus cDNA constructs.
TABLE 1.
Effects of mutant MLE proteins on the viability of mletg mutant males
| Transgene | No. of lines tested | Genotypea | No. (%) of females | No. (%) of males |
|---|---|---|---|---|
| [mleS539A,T541A] | 3 | 1 | 107 (31) | 98 (28) |
| 2 | 46 (13) | 0 | ||
| 3 | 41 (12) | 37 (11) | ||
| 4 | 20 (6) | 0 | ||
| [mleΔV] | 3 | 1 | 102 (21) | 98 (20) |
| 2 | 63 (13) | 0 | ||
| 3 | 90 (19) | 74 (15) | ||
| 4 | 53 (11) | 0 | ||
| [mle] | 2 | 1 | 55 (21) | 57 (22) |
| 2 | 32 (13) | 29 (11) | ||
| 3 | 23 (9) | 20 (8) | ||
| 4 | 20 (8) | 21 (8) |
w; mle1/CyO; [hsp83-Flag-mletg w+]/TM6, Sb Tb virgins were crossed with males carrying the same genotype (where mletg indicates a transgene). Genotypes were as follows: 1, w; mle1/CyO; [hsp83-Flag-mletg w+]/TM6, Sb Tb; 2, w; mle1/mle1; [hsp83-Flag-mletg w+]/TM6, Sb Tb; 3, w; mle1/CyO; [hsp83-Flag-mletg w+]/[hsp83-Flag-mletg w+]; and 4; w; mle1/mle1; [hsp83-Flag-mletg w+]/[hsp83-Flag-mletg w+].
The lack of spreading along the X chromosome of the Flag-MLE(ΔV)-containing complex is not due to a reduction in the level of roX RNAs.
MLE is required for the stabilization of roX1 RNA in early embryos (20) and has been implicated in the transcription regulation of the roX2 gene (15). These published observations led us to ask whether the lack of spreading by complexes that included either of the two helicase-deficient MLE proteins was caused by misregulation of the levels of roX RNAs. We used quantitative RT-PCR to measure the levels of roX1 and roX2 RNAs in wild-type males and in females that expressed the msl2 cDNA transgene and either a wild-type Flag-mle or the Flag-mle(ΔV) transgene (Fig. 4A). In flies expressing either of these transgenes, the levels of the two roX RNAs were comparable to their levels in wild-type males. Therefore, we conclude that the lack of spreading by complexes that contain helicase-deficient MLE proteins is not caused by an effect of these proteins on the levels of the roX RNAs.
FIG. 4.
Effects of mutant MLE proteins on the levels of roX1 and roX2 RNAs and on the transcription of a gene adjacent to a high-affinity site. (A) Quantitative RT-PCR measurements of roX2 and roX1 RNAs in third-instar female larvae of the following genotypes, (i) y w1118/w; pr mle1/pr mle1; H83msl2/H83msl2, (ii) y w1118/w; pr mle1/pr mle1; H83msl2/hsp83-Flag-mle+ w+, (iii) y w1118/w; pr mle1/pr mle1; H83msl2/hsp83-Flag-mleΔV w+, and (iv) y w1118/y w1118, and (v) y w1118/Y male larvae. The heights of the columns represent the enrichment of females (bars 2 and 3 in relation to bar 1) and of males (bar 5) in relation to females (bar 4). The error bars represent standard deviations of the mean. (B, left) Genomic region including the roX1, yin, and ec genes. The ec gene is not dosage compensated in wild-type males and was not analyzed further. (B, right) Quantitative RT-PCR measurements of yin gene transcripts in female larvae of the following genotypes: (i and ii) same as above, (iii) y w1118/w; pr mle1/pr mle1; H83msl2/hsp83-Flag-mleKE w+, and (iv) y w1118/w; pr mle1/pr mle1; H83msl2/hsp83-Flag-mleΔV w+. The heights of the columns represent the enrichment of females (bars 2, 3, and 4 in relation to bar 1).
An MSL complex containing the Flag-MLE(ΔV) protein can enhance the transcription of a gene adjacent to a high-affinity site.
MSL complexes containing mutant mle alleles that encode proteins with ATPase but no helicase activity cannot spread along polytene chromosomes yet allow the hyperactivation of the firefly luciferase reporter gene on the roX-bearing plasmid. We tested whether the Flag-MLE(ΔV) protein-containing complex would be able to have the same effect on a gene neighboring one of the entry sites where it is located. The yin gene is located near the roX1 high-affinity site (Fig. 4B, left), and the steady-state levels of yin transcripts are the same in wild-type males and females (1.0 ± 0.1 for yin and 1.07 ± 0.15 for Gpdh, an autosomal gene measured as a control), indicating that yin is dosage compensated. In females that express the msl2 cDNA transgene and the Flag-MLE(K413E) protein, the level of the yin transcript is equivalent to that in wild-type females (Fig. 4B, right). The presence of a wild-type mle transgene leads to a doubling in the level of the transcript, as expected, because these females assemble a fully functional and normally distributed MSL complex (Fig. 3B). In females that express the Flag-MLE(ΔV) protein, the level of yin gene transcription is enhanced once again, indicating that the MSL complex that is present at the roX high-affinity site is capable of implementing dosage compensation.
DISCUSSION
Previous studies of dosage compensation made use of mle mutations that yielded a truncated protein (23) or that interfered with ATP binding and therefore produced a full-length protein lacking the ATP-dependent helicase activity (14). This protein can be incorporated into the MSL complex but prevents its spreading along the X chromosome and its function in dosage compensation (6). MLE is related to the ATPases that drive the activities of chromatin-remodeling complexes, such as SWI/SNF, ISWI, NURF, CHRAC, ACF, and NURD, yet to date, it is the only protein that has been shown to have duplex-unwinding activity in vitro (8, 14), raising the question of whether its helicase activity is necessary for the function of the MSL complex. We attempted to separate the ATPase and the helicase activities of MLE by creating several mutations in those domains that, by comparison with other helicases, are thought to mediate the latter. Two of these mutations, MLE(ΔV) and MLE(S539A,T541A), completely abrogate the in vitro unwinding ability of the recombinant proteins they produce while exhibiting wild-type or substantial levels of ATPase activity, respectively. We determined that both proteins mediate an enhancement in the expression of the firefly reporter gene on a plasmid that nucleates the MSL complex. This enhancement was equivalent to the wild-type control in the case of MLE(ΔV) and intermediate in the case of MLE(S539A,T541A). The correlation between the levels of ATPase activity [wild type for the MLE(ΔV) protein and approximately 60% for the MLE(S539A,T541A) protein] and the level of transcriptional enhancement of the firefly gene that they allow led us to conclude that the ATPase activity of MLE, but not its unwinding function, is required for dosage compensation in our plasmid model.
In Drosophila males, the complex is believed to assemble at the loci of the two roX genes and then spread to additional sites along the X chromosome for which it has a complete range of affinity levels (4). Approximately 30 to 40 of these sites are defined as “high affinity” because a partial complex can bind to them (19). A complex that is fully assembled and that includes either a full-length but inactive histone acetyltransferase (MOF) or a full-length MLE protein with no ATPase and helicase activities [MLE(K413E)] has a similarly limited distribution (6). These partial or inactive complexes that bind to the high-affinity sites are unable to spread along the X chromosome. In the roX-FF experimental plasmid used to assay the level of dosage compensation of the mutant Flag-MLE proteins, the roX sequence is immediately downstream from the simian virus 40 polyadenylation signal of the firefly gene. This proximity may allow the complex to modify the chromatin of this gene, but it leaves open the possibility that the helicase function of MLE is required for the MSL complex to spread along the X chromosome—a prerequisite for dosage compensation in males. MSL complexes assembled in vivo with MLE(ΔV) or MLE(S539A,T541A) protein fail to spread along the X chromosome, leading to the conclusion that the unwinding activity of MLE is required for this function. Consistent with the effects of partial or inactive complexes described above, the MLE(ΔV) or MLE(S539A,T541A) mutation is lethal in males.
Although mutant complexes fail to spread in vivo beyond the high-affinity sites, they were able to mediate an increase in the transcriptional activity of the reporter gene in the plasmid system. These facts suggested that these complexes should also be able to increase the transcription of genes located near the high-affinity sites. In testing this possibility, we were limited by the fact that only three of these sites have been mapped on the genomic sequence (∼200-bp sequences in the coding regions of the roX1 and roX2 genes [10, 22] and an ∼500-bp site at position 18D10 on the cytological map [21]). We were also limited by the fact that several of the genes that flank these sites are not expressed in the third-instar larval stage used for the recognition of the pertinent genotypes. Finally, some of these genes have different levels of expression in wild-type males and females. The yin gene, adjacent to the roX1 high-affinity site, was found to be dosage compensated, and its level of transcription was significantly increased by the presence of a complex containing the MLE(ΔV) protein. This observation represents an in vivo validation of the results obtained with the plasmid model system.
The nature of the mechanism that translates helicase activity into chromosomal spreading is not resolved. Several observations have documented the probable RNA-dependent association of MLE with the MSL complex. Treatment of salivary gland polytene chromosome preparations with RNase releases MLE from the complex but does not affect the localization of the other subunits (24). The presence of MLE during the partial purification of the MSL complex requires the use of RNA-friendly conditions, and its presence in the complex within S2 cells is very sensitive to a moderately high salt concentration (28). As mentioned in the introduction, in vitro unwinding experiments have established that MLE prefers dsRNA or RNA/DNA hybrid substrates with a short 3′ RNA overhang; it does not bind to single-stranded DNA (14). The roX RNAs that are present in the complex may form double-stranded secondary structures that MLE could unwind, perhaps causing conformational changes in other RNA-binding subunits of the complex, such as MOF and MSL3 (1); these changes could be necessary for the complex to spread.
An alternate possibility is that MLE's function does not involve duplex unwinding; rather, it consists of a remodeling activity similar to that exhibited by such ATPases as SWI2/SNF2, ISWI, and BRG1 that have no demonstrable helicase activity. SWI2/SNF2 within the SWI/SNF complex and recombinant ISWI and BRG1 have been shown to generate superhelical torsion on chromatin templates, leading to increased torsional stress that could result in a rotation of the DNA or in a looping out of the DNA from the surface of the nucleosome (7). The superhelical torsion generated by the SWI/SNF complex appears to be due to its translocase activity (31). The observation that other SWI/SNF family members have also been shown to translocate along tethered DNA molecules (2, 17) suggests that MLE may mediate spreading of the MSL along DNA or RNA/DNA hybrid molecule complexes by a similar mechanism.
Finally, the possibility that MLE may have both unwinding capability and the ability to displace protein complexes in an unwinding-independent manner is suggested by results obtained with two DEXH/D helicases, NPH-II and DED1 (5). These two helicases are able to remove a tryptophan RNA-binding attenuation protein complex or an exon junction complex, respectively, from their cognate binding sites on single-stranded RNA.
Information on the types of enzymatic activities that the MLE protein is capable of performing may be obtained by using assays with single nucleic acid molecules. Once these results are in hand, the challenge will be to determine if the enzymatic activities that have been identified are carried out by MLE when it is present in MSL complexes associated with the X chromosome chromatin.
Supplementary Material
Acknowledgments
We are grateful to Eugene Koonin for his help in identifying putative mutations in domain III and to Mitzi Kuroda for fly stocks. We thank Arri Eisen for his comments on the manuscript. We acknowledge the highly skilled technical help of Huiping Ling.
This research was supported by grant GM15691 from the National Institutes of Health.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407405-409. [DOI] [PubMed] [Google Scholar]
- 2.Amitani, I., R. J. Baskin, and S. C. Kowalczykowski. 2006. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell 23143-148. [DOI] [PubMed] [Google Scholar]
- 3.Bashaw, G. J., and B. S. Baker. 1997. The regulation of the Drosophila Msl-2 gene reveals a function for Sex-Lethal in translational control. Cell 89789-798. [DOI] [PubMed] [Google Scholar]
- 4.Fagegaltier, D., and B. S. Baker. 2004. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2e341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairman, M. E., P. A. Maroney, W. Wang, H. A. Bowers, P. Gollnick, T. W. Nilsen, and E. Jankowsky. 2004. Protein displacement by Dexh/D “RNA helicases” without duplex unwinding. Science 304730-734. [DOI] [PubMed] [Google Scholar]
- 6.Gu, W., X. Wei, A. Pannuti, and J. C. Lucchesi. 2000. Targeting the chromatin-remodeling Msl complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J. 195202-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 1031133-1142. [DOI] [PubMed] [Google Scholar]
- 8.Hogan, C., and P. Varga-Weisz. 2007. The regulation of ATP-dependent nucleosome remodelling factors. Mutat. Res. 61841-51. [DOI] [PubMed] [Google Scholar]
- 9.Horabin, J. I., and P. Schedl. 1993. Sex-Lethal Autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 137734-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kageyama, Y., G. Mengus, G. Gilfillan, H. G. Kennedy, C. Stuckenholz, R. L. Kelley, P. B. Becker, and M. I. Kuroda. 2001. Association and spreading of the Drosophila dosage compensation complex from a discrete Rox1 chromatin entry site. EMBO J. 202236-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley, R. L., J. Wang, L. Bell, and M. I. Kuroda. 1997. Sex Lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387195-199. [DOI] [PubMed] [Google Scholar]
- 12.Koelle, M. R., W. S. Talbot, W. A. Segraves, M. T. Bender, P. Cherbas, and D. S. Hogness. 1991. The Drosophila Ecr gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 6759-77. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 14.Lee, C. G., K. A. Chang, M. I. Kuroda, and J. Hurwitz. 1997. The Ntpase/helicase activities of Drosophila Maleless, an essential factor in dosage compensation. EMBO J. 162671-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, C. G., T. W. Reichman, T. Baik, and M. B. Mathews. 2004. Mle functions as a transcriptional regulator of the Rox2 gene J. Biol. Chem. 27947740-47745. [DOI] [PubMed] [Google Scholar]
- 16.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 17.Lia, G., E. Praly, H. Ferreira, C. Stockdale, Y. C. Tse-Dinh, D. Dunlap, V. Croquette, D. Bensimon, and T. Owen-Hughes. 2006. Direct observation of DNA distortion by the Rsc complex. Mol. Cell 21417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucchesi, J. C., W. G. Kelly, and B. Panning. 2005. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 39615-651. [DOI] [PubMed] [Google Scholar]
- 19.Lyman, L. M., K. Copps, L. Rastelli, R. L. Kelley, and M. I. Kuroda. 1997. Drosophila male-specific Lethal-2 protein: structure/function analysis and dependence on Msl-1 for chromosome association. Genetics 1471743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meller, V. H. 2003. Initiation of dosage compensation in Drosophila embryos depends on expression of the Rox RNAs. Mech. Dev. 120759-767. [DOI] [PubMed] [Google Scholar]
- 21.Oh, H., J. R. Bone, and M. I. Kuroda. 2004. Multiple classes of Msl binding sites target dosage compensation to the X chromosome of Drosophila. Curr. Biol. 14481-487. [DOI] [PubMed] [Google Scholar]
- 22.Park, Y., G. Mengus, X. Bai, Y. Kageyama, V. H. Meller, P. B. Becker, and M. I. Kuroda. 2003. Sequence-specific targeting of Drosophila Rox genes by the Msl dosage compensation complex. Mol. Cell 11977-986. [DOI] [PubMed] [Google Scholar]
- 23.Rastelli, L., and M. I. Kuroda. 1998. An analysis of Maleless and histone H4 acetylation in Drosophila melanogaster Spermatogenesis. Mech. Dev. 71107-117. [DOI] [PubMed] [Google Scholar]
- 24.Richter, L., J. R. Bone, and M. I. Kuroda. 1996. RNA-dependent association of the Drosophila Maleless protein with the male X chromosome. Genes Cells 1325-336. [DOI] [PubMed] [Google Scholar]
- 25.Sass, G. L., A. Pannuti, and J. C. Lucchesi. 2003. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc. Natl. Acad. Sci. USA 1008287-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406541-544. [DOI] [PubMed] [Google Scholar]
- 27.Smith, C. L., and C. L. Peterson. 2005. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 255880-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, E. R., A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook, C. D. Allis, and J. C. Lucchesi. 2000. The Drosophila Msl complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 202009-2017. [DOI] [PubMed] [Google Scholar]
- 30.Yokoyama, R., A. Pannuti, H. Ling, E. R. Smith, and J. C. Lucchesi. 2007. A plasmid model system shows that Drosophila dosage compensation depends on the global acetylation of histone H4 at lysine 16 and is not affected by depletion of common transcription elongation chromatin marks. Mol. Cell. Biol. 277865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, Y., C. L. Smith, A. Saha, S. W. Grill, S. Mihardja, S. B. Smith, B. R. Cairns, C. L. Peterson, and C. Bustamante. 2006. DNA translocation and loop formation mechanism of chromatin remodeling by Swi/Snf and Rsc. Mol. Cell 24559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.