Abstract
Regulation of chromatin in eukaryotic transcription requires histone-modifying enzymes, nucleosome remodeling complexes, and histone chaperones. Specific regulation of histone incorporation/eviction by histone chaperones on the promoter (e.g., region specific) is still poorly understood. In the present study, we show that direct and functional interaction of histone chaperone and DNA-binding transcription factor leads to promoter region-specific histone incorporation and inhibition of histone acetylation. We report here that the DNA-binding transcription factor Krüppel-like factor 5 (KLF5) interacts with the novel histone chaperone acidic nuclear phosphoprotein 32B (ANP32B), leading to transcriptional repression of a KLF5-downstream gene. We further show that recruitment of ANP32B onto the promoter region requires KLF5 and results in promoter region-specific histone incorporation and inhibition of histone acetylation by ANP32B. Extracellular stimulus (e.g., phorbol ester) regulates this mechanism in the cell. Collectively, we have identified a novel histone chaperone, ANP32B, and through analysis of the actions of this factor show a new mechanism of promoter region-specific transcriptional regulation at the chromatin level as mediated by the functional interaction between histone chaperone and DNA-binding transcription factor.
Eukaryotic transcription at the chromatin/nucleosome level is regulated by posttranslational chemical modification of histones, chromatin remodeling, and incorporation/eviction of histones including histone variants (11, 13, 17, 20, 22, 40, 52, 53). Regulation of chromatin transcription is mediated by three classes of factors including histone chaperones (a.k.a. ATP-independent nucleosome assembly factors), chemical modification enzymes (e.g., acetylases/deacetylases etc.), and ATP-dependent nucleosome remodeling factors (4, 22, 25, 41, 46). Recent studies have further suggested that the histone chaperones are important key regulators of chromatin/nucleosome structural regulation in many if not all higher-order DNA-associated processes ranging from transcription to DNA replication/repair (1, 2, 8, 12, 16, 23, 35, 38, 50). Of these three classes of chromatin/nucleosome-modulating factors, the mechanisms and actions of the histone chaperones, which are the central regulators of nucleosome assembly/disassembly (histone incorporation/eviction) (4, 5, 17, 52), are the least well understood. Understanding the precise role of histone chaperones in regulation of transcription at the nucleosomal level will therefore be of critical importance to further advancing our understanding of the underlying mechanisms of chromatin transcription.
One important question in chromatin transcription which remains poorly understood is the mechanism underlying region-specific (e.g., promoter-specific) nucleosomal regulation. As gene-specific transcription is likely dictated by the DNA-binding transcription factor whose actions are critically dictated through its cognate binding sequence on the promoter, investigations focused on understanding the role of the DNA-binding transcription factor in regulation of transcription at the nucleosomal level will likely be pivotal in helping our understanding of region-specific nucleosomal regulation in transcription.
To this aim, we have previously investigated the structural and functional basis of mechanisms of action of histone chaperones as well as addressing the role of DNA-binding transcription factor in chromatin transcription using the Sp/KLF (for Sp1- and Krüppel-like factor) family (27, 29-32, 44-47). The Sp/KLF family of transcription factors has received recent attention due to roles in various biological events, among which KLF5 (Krüppel-like factor 5) is involved in the cardiovascular remodeling response to stress and acts at the cellular level on cell proliferation and cell cycle regulation (46, 47). This family has been shown to functionally interact with all three classes of chromatin-related cofactors, which is a unique property for DNA-binding factors aside from histones (18, 27, 29, 44, 45, 47). Previously, we have addressed the pathways and mechanisms of transcriptional regulation mediated by Sp/KLF family members and chromatin-related cofactors, which include differential regulation through interaction and acetylation of Sp/KLF by acetylase (29, 44), inhibition of acetylation and DNA accessibility of Sp/KLF by histone chaperone (29, 45), and inhibition of acetylation and DNA accessibility of KLF5 by direct binding of deacetylase (27). Given our understanding of the actions and regulation of this family of factors on chromatin-related cofactors, they serve as an excellent target for further investigations on the role of DNA-binding transcription factor in transcription at the chromatin level, especially in regard to functional interaction with histone chaperones.
In the present study, we identify the novel histone chaperone acidic nuclear phosphoprotein 32B (ANP32B) as a factor interacting with KLF5 and through the analysis of their functional interaction show promoter region-specific histone incorporation and inhibition of histone acetylation as mediated by histone chaperone activity of ANP32B and KLF5-dependent recruitment of ANP32B onto the promoter.
MATERIALS AND METHODS
Cell culture.
HeLa S3 cells were cultured and nuclear extract was prepared as previously described (29). HeLa cells were cultured as previously described (27).
Isolation and identification of factors interacting with KLF5 ZF/DBD.
Isolation of interactors with KLF5-ZF/DBD was done as previously described with modification by use of HeLa S3 nuclear extract (29). Protein identification was done using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry, and identifications were searched against the nonredundant National Center for Biotechnology Information database as previously described (29).
Construction of ANP32B expression vectors.
Full-length human ANP32B (NM_006401.2) cDNA was amplified from a human aorta cDNA library (Clontech) by PCR and inserted into the expression vectors pcDNA3 (Invitrogen), pBICEP (SIGMA), pGEX4T-3 (GE Healthcare), and pET30b (Novagen).
Preparation of recombinant proteins.
Glutathione S-transferase (GST)-tagged proteins (GST, GST-KLF5-ZF/DBD, and GST-ANP32B) were expressed and affinity purified as previously described (29). GST-KLF5 full length and activation domain were obtained from inclusion bodies and affinity purified. Hexahistidine-tagged proteins (6HIS-KLF5-ZF/DBD, 6HIS-ANP32B, and 6HIS-TAF-Iβ) were expressed and affinity purified as previously described (29). Affinity-purified 6HIS-ANP32B was further purified using Q-Sepharose (GE Healthcare).
Protein-protein interaction assay.
The protein-protein interaction assay for experiments to investigate the binding of KLF5 and its deletion mutants to ANP32B was done as previously described (29). Commercially available recombinant proteins were used for p53, MyoD, and NF-κB (Santa Cruz). Briefly, GST fusion proteins, glutathione-Sepharose 4B resin, and hexahistidine-tagged proteins were incubated in a buffer containing 20 mM HEPES (pH 7.6 at 4°C), 20% glycerol, 0.2 mM EDTA, 0.1% Triton, 100 mM NaCl, and 100 μM ZnSO4 at 4°C for 2 h, and then the mixtures were washed two times in the same buffer. Bound proteins were analyzed by Western blotting using anti-His probe (G-18) antibody (Santa Cruz).
Preparation of anti-ANP32B antibody.
Rabbits were immunized with 100 μg of full-length purified recombinant His6-human ANP32B for five times at 1-week intervals, after which serum was extracted. Antibody specificity was confirmed by Western blotting (see Fig. 1H).
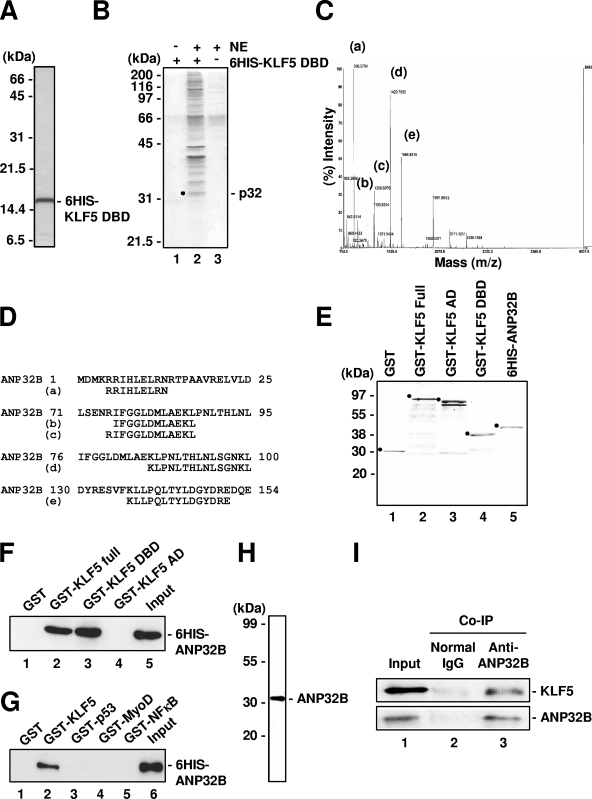
FIG. 1.
Isolation of ANP32B as an interactor with KLF5. (A) Silver-stained gel of 6HIS-KLF5-ZF/DBD. (B) Isolation of factors associating with KLF5-ZF/DBD. Lane 3 is HeLa S3 nuclear extract (NE). p32, 32-kDa band. (C) MALDI-TOF mass spectra obtained from tryptic peptides of p32. Fragment peaks assigned to ANP32B are labeled. (D) Partial peptide sequences of human ANP32B. Numbering is from the initiation methionine of ANP32B. The peptide sequences of peaks a to e obtained by peptide mass fingerprinting are shown. (E) Coomassie blue-stained gel of the recombinant proteins used for protein-protein interaction studies. (F) In vitro binding of KLF5 full-length protein and ANP32B. Bound proteins were detected by anti-His antibody. (G) In vitro binding of KLF5 ZF/DBD and ANP32B. Bound proteins were detected by anti-His antibody. (H) Specific detection of target protein by anti-ANP32B antibody. HeLa whole-cell extract was analyzed by Western blotting using prepared rabbit polyclonal anti-ANP32B. (I) Cellular binding of ANP32B and KLF5. Bound proteins were detected by anti-KLF5 (KM1785) and anti-ANP32B (rabbit polyclonal) antibodies.
Coimmunoprecipitation assay.
For experiments on the effect of phorbol 12-myristate 13-acetate (PMA) stimulation on the interaction of ANP32B and KLF5, 4 × 106 HeLa cells were transfected with 6 μg of Bicep-ANP32B and pCAG-KLF5 using Lipofectamine 2000 (Invitrogen). Cells were treated with Dulbecco modified Eagle medium (DMEM) (10% fetal calf serum [FCS]) for 24 h prior to serum starvation for an additional 24 h and then stimulated by 200 nM of PMA. Cells were extracted in FLAG lysis buffer containing 100 μM ZnSO4, and then extracts were incubated with 1 μg of anti-KLF5 polyclonal antibody or normal rabbit immunoglobulin G (IgG) (Santa Cruz) prior to incubation with protein G-Sepharose. One wash was done with FLAG lysis buffer containing 100 μM ZnSO4, and then three washes were done with buffer containing 20 mM HEPES (pH 7.6 at 4°C), 20% glycerol, 150 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40, 100 μM ZnSO4, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 1 μg/ml pepstatin A.
Experiments on endogenous ANP32B and KLF5 were performed as experiments on the effect of PMA stimulation on the interaction of ANP32B and KLF5 with slight modification. Briefly, transfection, serum starvation, and PMA stimulation were not done; extracts were incubated with 1 μg of anti-ANP32B antibody (Abcam) or normal rabbit IgG; and bound proteins were eluted using 0.5 M glycine (pH 2.5). Precipitate was analyzed by Western blotting using anti-FLAG-horseradish peroxidase (Sigma), rabbit polyclonal anti-ANP32B, and anti-KLF5 (KM1785) antibodies.
Gel shift DNA-binding assay.
Gel shift DNA-binding assays were performed as previously described (27) with some modification. Briefly, recombinant proteins and 2.5 pmol of 5′-fluorescein isothiocyanate-labeled probes were incubated at room temperature for 30 min in BA150 (10 mM Tris HCl [pH 7.5], 10% glycerol, 150 mM NaCl, and 0.1% NP-40) containing 6 μM ZnSO4. Gels were analyzed using Typhoon 3700 (GE Healthcare).
Cotransfection reporter assay.
The cotransfection reporter assay was done as previously described (3) with slight modification. Briefly, HeLa cells were transfected with the platelet-derived growth factor A (PDGF-A) chain (−900)-Luc promoter reporter (0.1 μg) and effector expression vectors. The total effector DNA amount in transfection reaction mixtures was corrected to 1 μg by addition of empty vector.
Histone binding assay.
Core histones were purified from HeLa S3 cells as previously described (43). GST fusion proteins were immobilized onto glutathione-Sepharose 4B resin and incubated with 5 μg of core histones or calf thymus linker histones (Sigma) in BA150 (10 mM Tris HCl [pH 7.5], 10% glycerol, 150 mM NaCl, and 0.1% NP-40) at 4°C and then washed three times with BA150. Precipitated proteins were analyzed by Coomassie blue staining or Western blotting using anti-His antibody.
Plasmid supercoiling assay.
Nucleosome assembly reactions were performed as previously described (30) with slight modification. Briefly, after preincubation for 30 min at 37°C, reaction mixtures and relaxed DNA were incubated at 37°C for 2 h.
MNase digestion assay.
Nucleosome assembly reactions were done with twofold-increased reaction volumes and factors compared with those in the plasmid supercoiling assay, and then the reaction mixtures were incubated with micrococcal nuclease (MNase) (Takara) at each concentration in the presence of 6 mM CaCl2 for 7 min at room temperature followed by DNA extraction.
RNA interference analysis.
The target sequences of small interfering RNA (siRNA) oligonucleotide probes were as follows: ANP32B, 5′-GAAGAGGAGUUUGAUGAAGAAGA-3′ (34); KLF5, 5′-AACCCGGAUCUGGAGAAGCGA-3′ (7); and secreted alkaline phosphatase (SEAP), 5′-AGGGCAACUUCCAGACCAUU-3′ (3). siRNAs were constructed as previously described (3).
Effect of RNA interference of ANP32B on expression levels of PDGF-A chain.
Four micrograms of siRNA was transfected into 2 × 105 HeLa cells using Lipofectamine 2000. After 48 h of incubation using DMEM (10% FCS), cells were harvested and total RNA was obtained with the RNeasy preparation kit (Qiagen) and then reverse transcribed. cDNA was analyzed by quantitative PCR using gene-specific primer sets and Quantum RNA 18S internal standard primer set I (Ambion) as previously described (29). The relative intensity of each gene was calculated in reference to internal 18S rRNA. The gene-specific primer pairs were as follows: ANP32B, 5′-TGGATGGTGTGGATGAAGAGGA-3′ and 5′-TGTTTCTGCAGGTCATCTGGGG-3′; KLF5, 5′-GGCTTGGCGCCCGTGTGCTTCC-3′ and 5′-GGTTGCACAAAAGTTTATAC-3′; and PDGF-A chain, 5′-CAGCATCCGGGACCTCCAGCGACTC-3′ and 5′-TCGTAAATGACCGTCCTGGTCTTGC-3′.
Chromatin immunoprecipitation (ChIP) assay. (i) Transfection and cell treatment.
For experiments on RNA interference of ANP32B, 80 μg of siRNA was transfected into 2 × 106 HeLa cells. Cells were harvested after 48 h of incubation using DMEM (10% FCS). For experiments on localization of ANP32B, 24 μg of pBICEP-ANP32B was transfected into 2 × 106 HeLa cells. Cells were harvested after 48 h of incubation using DMEM (10% FCS). For experiments on RNA interference of both ANP32B and KLF5, 80 μg of siANP32B and/or siKLF5 was transfected into 6 × 106 HeLa cells. The total siRNA amount in transfection reaction mixtures was corrected to 160 μg by addition of siSEAP. Cells were harvested after 24 h of incubation using DMEM (10% FCS). For experiments on the effect of RNA interference of KLF5 on localization of ANP32B, 24 μg of pBICEP-ANP32B and 80 μg of siRNA were transfected into 2 × 106 HeLa cells. Cells were harvested after 48 h of incubation using DMEM (10% FCS). For experiments on the effect of PMA stimulation, 2 × 106 HeLa cells were treated with DMEM (10% FCS) for 24 h and then stimulated by 200 nM of PMA after treatment with DMEM for 24 h. For experiments on the effect of PMA stimulation on localization of ANP32B, 24 μg of pBICEP-ANP32B was transfected into 4 × 106 HeLa cells and then cells were treated with DMEM (10% FCS) for 24 h and further stimulated with 200 nM of PMA after treatment with DMEM for 24 h.
(ii) Anti-KLF5, FLAG-ANP32B, or acetylated histone H3 and H4 ChIP assays.
Anti-KLF5 and anti-FLAG ChIP assays were performed as previously described (26) with slight modification. For anti-KLF5 ChIP analysis, fixed proteins were incubated with 2 μg of rabbit polyclonal anti-KLF5 (48) or normal rabbit IgG (Santa Cruz). For anti-acetylated histone H3 and H4 ChIP analyses, fixed proteins were incubated with 2 μg of rabbit polyclonal anti-acetylated histone H3 (K9 and K14) (Upstate Biotechnology), rabbit polyclonal anti-acetylated histone H4 (K5, K8, and K12) (Upstate Biotechnology), or normal rabbit IgG (Santa Cruz) in the presence of 50 mM sodium butyrate. For anti-FLAG ChIP analysis, fixed proteins were incubated with 2 μg of mouse monoclonal anti-FLAG (Sigma) or normal mouse IgG (Santa Cruz) antibodies. Proteins and antibodies were adsorbed to protein G Sepharose (GE Healthcare) and then washed. For anti-KLF5 and FLAG-ANP32B ChIP analyses, washing was done with wash buffer (10 mM Tris HCl [pH 8.0], 0.1% sodium dodecyl sulfate, 1% Triton, 1 mM EDTA, and 150 mM NaCl), LiCl buffer (10 mM Tris HCl [pH 8.0], 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 250 mM LiCl), and TE-200 (10 mM Tris HCl [pH 8.0], 1 mM EDTA, and 200 mM NaCl). For anti-acetylated histone H3 and H4 ChIP analyses, washing was done using the same buffer containing 50 mM sodium butyrate.
(iii) Anti-histone H2B and H4 ChIP assays.
Antihistone ChIP assays were performed as previously described (33) with modification of nucleus isolation according to previously described methods for nucleosome mapping with nuclease (21, 24). Cells were harvested and treated in cell lysis buffer (10 mM Tris HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM spermine, 0.5 mM spermidine, and 50 mM sodium butyrate) and centrifuged at 8,000 rpm for 4 min at 4°C, and then precipitate was harvested as nuclei. Nuclei were suspended in nuclear buffer (10 mM Tris HCl [pH 7.5], 50 mM NaCl, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mM EGTA, 0.15 mM spermine, 0.5 mM spermidine, 1 mM β-mercaptoethanol, and 50 mM sodium butyrate). Nuclei were incubated with CaCl2 at a final concentration of 2 mM and 20 units of MNase (Takara) at 37°C for 5 min, and then reactions were stopped by addition of EDTA to a concentration of 10 mM. Soluble chromatin was harvested as previously described (33) and incubated with 8 μg of rabbit polyclonal anti-H4 (H-97; Santa Cruz), whose epitope corresponds to amino acids residues 7 to 103 of the N terminus of human histone H4; rabbit polyclonal anti-H2B (Abcam), whose epitope is derived from a peptide region containing from amino acid residue 100 to the C terminus of human histone H2B; or normal rabbit IgG (Santa Cruz) and then adsorbed to protein G Sepharose (GE Healthcare).
(iv) Analysis of immunoprecipitated DNA.
Immunoprecipitated DNA was analyzed by quantitative PCR using a Light Cycler (Roche) and QuantiTect Sybr green PCR Kit (Qiagen). The region-specific primers pairs were as follows: 5′-(−3,000-bp) region, 5′-GCGAAAGTTTCTTCCCCTCT-3′ and 5′-GAGTCAGGTGCCCCAAAATAG-3′; KLF5 site (promoter region), 5′-TGGCACTGGAGGGTGGGCAA-3′ and 5′-GAAGCGCTAGGGGTTCGT-3′; and 3′ region, 5′-GTGTTACATTCCTGAACCTACTATGTACGGTGC-3′ and 5′-CTGCTTCACCGAGTGCTACAATACTTGCTTTGATG-3′.
Phorbol ester-induced expression of KLF5 and ANP32B.
Cell lysate from HeLa cells stimulated with 200 nM of PMA following 24 h of treatment with DMEM was analyzed by Western blotting using anti-KLF5 (KM1785) or anti-ANP32B antibody. The relative intensity of KLF5 or ANP32B protein in reference to Coomassie blue staining was calculated by using National Institutes of Health Image software. Quantitative PCR using Quantum RNA 18S internal standard primer set I (Ambion) and calculation of the relative intensity of PDGF-A chain mRNA were done as previously described (29).
RESULTS
Isolation of ANP32B as a KLF5 interactor.
To elucidate novel mechanisms of transcriptional regulation mediated by the action of DNA-binding transcription factor, we affinity purified KLF5-interacting proteins from HeLa S3 nuclear extract using the zinc-finger DNA-binding domain (ZF/DBD) region of KLF5 (Fig. 1A). More than 10 major bands were seen when KLF5-ZF/DBD and nuclear extract were used (Fig. 1B, lane 2) and not with either nuclear extract or KLF5-ZF/DBD alone (Fig. 1B, lanes 1 and 3). One band with an apparent molecular mass of 32 kDa was easily separable from other nearby bands. MALDI-TOF (mass spectrometry)-peptide mass fingerprinting with a database search and further confirmation of the amino acid sequence by post-source decay peptide sequencing showed this protein to be ANP32B (UniGene Hs.494604, accession no. NM_006401.2) (Fig. 1C and 1D). ANP32B, whose biochemical activity is unknown, is a member of a conserved superfamily of nuclear proteins whose members regulate cell growth and cell differentiation in a tissue-specific manner (28, 36).
We next investigated whether ANP32B directly and specifically interacts with KLF5-ZF/DBD by using deletion mutants (Fig. 1E) and other DNA-binding proteins (29) (Fig. 1F and G). A GST pull-down assay showed that ANP32B directly binds GST-KLF5 full length and GST-ZF/DBD (Fig. 1F, lanes 2 and 3) but not the activation domain, which excludes the ZF/DBD (Fig. 1F, lane 4). Further analysis showed that ANP32B binds KLF5 (Fig. 1G, lane 2) but not p53, MyoD, or NF-κB (Fig. 1G, lanes 3 to 5). Therefore, ANP32B directly and specifically binds KLF5 through its ZF/DBD. Further, a coimmunoprecipitation assay using anti-ANP32B antibody, whose specificity was confirmed by Western blotting (Fig. 1H), showed that endogenous ANP32B and KLF5 proteins interact in the cell (Fig. 1I, lane 3). Thus, KLF5 and ANP32B specifically and directly bind in the cell.
Functional effects of interaction between ANP32B and KLF5.
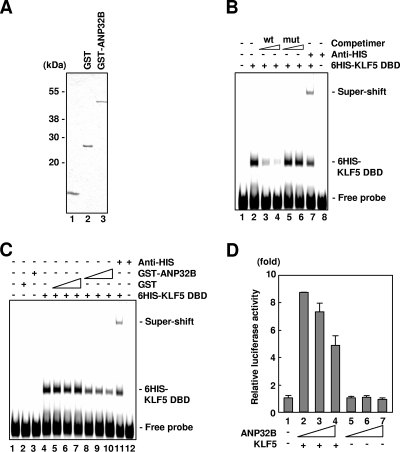
To next address the functional implications of interaction of ANP32B with KLF5, we first examined the effect of ANP32B on the DNA-binding activity of KLF5 by gel shift analysis using KLF5 and ANP32B recombinant proteins (Fig. 2A). Under conditions in which KLF5-ZF/DBD showed sequence-specific DNA binding (Fig. 2B) and ANP32B did not bind DNA (Fig. 2C, lanes 1 to 4), addition of GST-ANP32B inhibited DNA-binding activity of KLF5-ZF/DBD (Fig. 2C, lanes 5 to 7 versus 8 to 10). This indicates that ANP32B possesses the ability to negatively regulate DNA-binding activity of KLF5 or that KLF5 association with ANP32B and DNA is mutually exclusive.
FIG. 2.
Negative regulation of KLF5 activities by ANP32B. (A) Coomassie blue-stained gel of the recombinant proteins used for DNA-protein binding studies. (B) Sequence-specific DNA binding of KLF5-ZF/DBD. wt and mut represent wild-type and mutant oligonucleotide competimers, respectively (lanes 3 and 4 and lanes 5 and 6, respectively) (1 and 4 pmol from left to right). Two picomoles of 6HIS-KLF5-ZF/DBD was used. The 6HIS-KLF5-ZF/DBD protein was supershifted by 1 μg of anti-His antibody (lanes 7 and 8). (C) Effect of ANP32B on DNA-binding activity of KLF5-ZF/DBD. Thirty nanograms of 6HIS-KLF5-ZF/DBD was used. The 6HIS-KLF5-ZF/DBD protein was supershifted by 1 μg of anti-His probe antibody (lanes 7 and 8). The amounts of recombinant proteins were as follows; 1.2, 2.4, and 4.8 μg of GST (lanes 5, 6, and 7, respectively) and GST-ANP32B (lanes 8, 9, and 10, respectively); 4.8 μg of GST (lane 2) and GST-ANP32B (lane 3); and the presence (lanes 4 to 11) or absence (lanes 1 to 3 and 12) of 30 ng of 6HIS-KLF5-ZF/DBD. (D) Effect of ANP32B on KLF5 transactivation. Effectors were as follows; lanes 2, 3, and 4 were 0.5 μg of pCAG-KLF5, respectively, and lanes 3 and 4 and lanes 6 and 7 were 0.25 μg and 0.5 μg of pcDNA4-ANP32B, respectively. Error bars denote standard errors.
Cotransfection reporter assays were further done to examine the effect of ANP32B on KLF5-dependent transcriptional activation (Fig. 2D). Under conditions in which KLF5 showed transactivation of the PDGF-A chain gene promoter (Fig. 2D, lane 2), which is an endogenous target gene of KLF5 (3, 42), and under which ANP32B did not show activation of this promoter (Fig. 2D, lanes 5 to 7), cotransfection of ANP32B and KLF5 showed ANP32B dose-dependent repression of KLF5-mediated transactivation (Fig. 2D, lanes 2 to 4). Therefore, ANP32B inhibits both DNA-binding and transactivational activities of KLF5.
Characterization of the novel histone chaperone ANP32B.
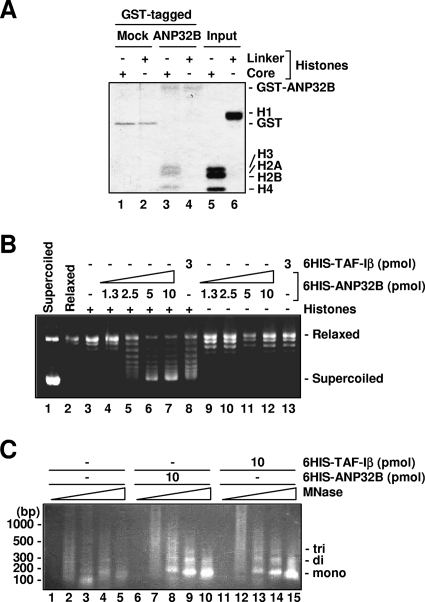
ANP32B is a member of a conserved superfamily which includes pp32/ANP32A, a factor that binds histones and inhibits their acetylation (19, 40). As the biochemical functions of ANP32B are unknown, we first examined whether ANP32B binds histones (Fig. 3A). A GST pull-down assay showed ANP32B to directly bind core histones, with a preference for core histones rather than linker histones (Fig. 3A, lanes 3 and 4).
FIG. 3.
Characterization of the novel histone chaperone ANP32B. (A) In vitro binding of ANP32B and core histones. Bound proteins were detected by Coomassie blue staining. (B) In vitro nucleosome assembly activity of ANP32B. Thirty-five nanograms of 6HIS-ANP32B and 45 ng of 6HIS-TAF-Iβ are equivalent to 1 pmol. Reaction mixtures were incubated in the presence (+) or in the absence (−) of histones with recombinant proteins, whose amounts are indicated. 6HIS-TAF-Iβ was used as a positive control for in vitro nucleosome assembly activity. (C) ANP32B-dependent nucleosome formation in vitro. Reactions were done with histones in either the presence or the absence of indicated factors. Each sample was treated with MNase (0, 6.4, 32, 160, and 800 units/ml from left to right). 6HIS-TAF-Iβ was used as a positive control for in vitro nucleosome assembly activity.
Recent studies have shown that histone-binding nuclear proteins may possess histone chaperone activity, which is an activity to assemble/disassemble nucleosomes in an ATP-independent manner. To test whether ANP32B harbors histone chaperone activity, a plasmid supercoiling assay was done (9) (Fig. 3B). In the absence of ATP, ANP32B assembled nucleosomes in a dose-dependent manner as shown by histone-dependent introduction of negative supercoils into plasmid DNA (Fig. 3B, lanes 4 to 7 and 9 to 12). Histone chaperone TAF-Iβ was used as a positive control (Fig. 3B, lanes 8 and 13). Given that the plasmid supercoiling assay detects only DNA topological changes, an MNase digestion assay was done which confirmed nucleosomal protection of DNA in approximately 150-bp repeats consistent with the size of the nucleosome (ANP32B, Fig. 3C, lanes 6 to 10; 6HIS-TAF-Iβ, Fig. 3C, lanes 11 to 15) (51). Thus, ANP32B is a newly identified histone chaperone.
Effects of loss of function of ANP32B on KLF5-mediated chromatin transcription.
As ANP32B possesses dual activities to stimulate nucleosome assembly and to inhibit DNA-binding activity of KLF5, we reasoned that ANP32B could regulate KLF5-mediated transcription by inhibiting binding of KLF5 to its cognate binding sites and by inducing incorporation of histones.
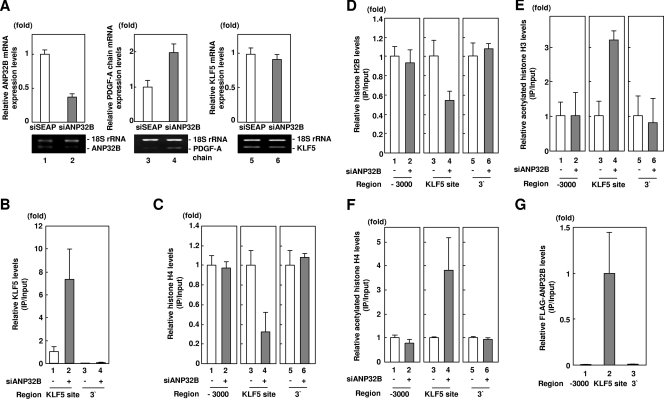
To investigate this, we first examined the effect of RNA interference of ANP32B on endogenous PDGF-A chain expression, as ANP32B represses KLF5-mediated transcription of this gene (Fig. 2D). RNA interference of ANP32B reduced levels of ANP32B (Fig. 4A, lanes 1 and 2) as previously described (34), which resulted in an increase in PDGF-A chain mRNA expression (Fig. 4A, lanes 3 and 4), while KLF5 expression was not affected (Fig. 4A, lanes 5 and 6). This indicates that ANP32B negatively regulates transcription of the KLF5-downstream gene PDGF-A chain.
FIG. 4.
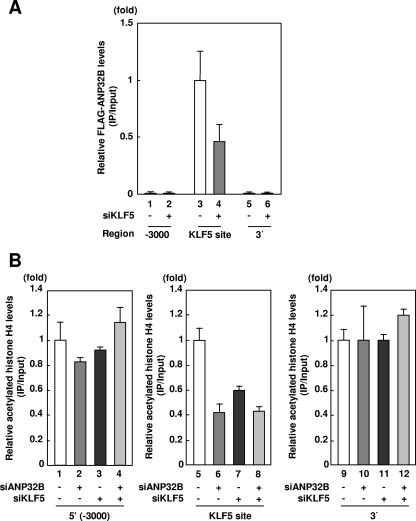
Effect of loss of function of ANP32B on KLF5-mediated chromatin transcription. (A) Effect of RNA interference of ANP32B on expression levels of ANP32B, PDGF-A chain, and KLF5 levels. SEAP was used as a negative control for RNA interference. Relative mRNA expression levels were calculated in reference to the expression levels of 18S rRNA. Error bars denote standard errors. (B) Effect of RNA interference of ANP32B on KLF5 levels over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (C) Effect of RNA interference of ANP32B on histone H4 levels over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (D) Effect of RNA interference of ANP32B on histone H2B levels over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (E) Effect of RNA interference of ANP32B on levels of acetylated histone H3 over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard error. (F) Effect of RNA interference of ANP32B on levels of acetylated histone H4 over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (G) Localization of ANP32B over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors.
Next, we investigated whether ANP32B negatively regulates binding of KLF5 to the PDGF-A chain gene promoter (Fig. 4B). Under conditions in which KLF5 showed specific binding to the KLF5 binding site of the PDGF-A chain gene promoter but not the 3′ end of the gene used as a negative control (Fig. 4B, lanes 3 and 4), ChIP assay showed that RNA interference of ANP32B stimulated binding of KLF5 to the KLF5 binding site on the PDGF-A chain gene promoter (Fig. 4B, lanes 1 and 2). This indicates that endogenous ANP32B negatively regulates promoter access of KLF5, leading to transcriptional repression.
We further examined whether ANP32B participates in histone incorporation over the PDGF-A chain gene (Fig. 4C and D). Anti-histone H4 ChIP analysis showed that RNA interference of ANP32B showed decreased histone H4 levels over the KLF5 binding site on the PDGF-A chain gene promoter (Fig. 4C, lanes 3 and 4) while not affecting histone H4 levels over the 5′-(−3,000 bp) or the 3′ regions of the gene (Fig. 4C, lanes 1, 2, 5, and 6). Anti-histone H2B ChIP analysis showed the same effect on histone H2B levels over the PDGF-A chain gene promoter (Fig. 4D, lanes 1 to 6). These findings indicate that ANP32B participates in incorporation of both histones H2A/H2B and histones H3/H4.
As ANP32B is a member of a conserved superfamily which includes pp32/ANP32A, a factor that binds histones and inhibits their acetylation (40), we reasoned that ANP32B inhibits acetylation of histones leading to transcriptional repression. To address the relationship between histone acetylation, histone eviction, and gene activation, we focused upon regulation of acetylation of K9 and K14 of H3 and K5, K8, and K12 of histone H4, which have been reported to be coupled with transcriptional activation and inhibition as well as recognition by INHAT (inhibition of histone acetyltransferase) subunits (37, 39). To address whether ANP32B participates in inhibition of histone acetylation leading to transcriptional activation, we examined the effect of RNA interference of ANP32B on acetylated histone levels (Fig. 4E and F). Anti-acetylated histone H3 and H4 ChIP assays showed that RNA interference of ANP32B increased levels of acetylated histones H3 and H4 over the KLF5 binding site on the PDGF-A chain gene promoter (Fig. 4E and F, lanes 3 and 4) while not affecting levels of acetylated histone H3 and H4 over the 5′-(−3,000 bp) or the 3′ regions of the gene (Fig. 4E and F, lanes 1, 2, 5, and 6). This indicates that ANP32B participates in inhibition of histone acetylation in a promoter region-specific manner.
To confirm that ANP32B mediates histone incorporation over the KLF5 binding sites, we examined whether ANP32B localizes to the PDGF-A chain gene promoter (Fig. 4G). The ChIP assay showed that relative levels of FLAG-ANP32B over the 5′-(−3,000 bp) or the 3′ regions of the gene were markedly lower than that over the KLF5 sites of the promoter (Fig. 4G, lanes 1 to 3). This indicates that ANP32B localizes to the PDGF-A chain gene in a promoter-specific manner.
These findings collectively suggest that ANP32B represses KLF5-mediated transcription by dual pathways of inhibition of DNA-binding activity of KLF5 by protein-protein interaction and inhibition of promoter access of KLF5 by histone incorporation over its cognate binding sites.
Recruitment of ANP32B by KLF5.
As KLF5 and ANP32B directly interact and ANP32B localizes on the PDGF-A chain gene promoter in a region-specific manner, we reasoned that KLF5 recruits ANP32B onto the promoter region (Fig. 5A). To address whether KLF5 possesses the ability to recruit ANP32B onto the promoter region, we examined effects of RNA interference of KLF5 on ANP32B levels over the promoter region. Under the conditions in which the ChIP assay showed promoter region-specific localization of ANP32B, RNA interference of KLF5 decreased ANP32B levels over the promoter region (Fig. 5A, lanes 3 and 4). This indicates that ANP32B is recruited onto the promoter region of the PDGF-A chain gene in a KLF5-dependent manner.
FIG. 5.
Effects of loss of function of KLF5 on recruitment of ANP32B. (A) Effect of RNA interference of KLF5 on ANP32B levels over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (B) Effect of RNA interference of ANP32B and KLF5 on levels of histone H4 over the PDGF-A chain gene promoter. SEAP was used as a negative control for RNA interference. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors.
We further examined whether KLF5-dependent recruitment of ANP32B mediates histone incorporation over the promoter region of the PDGF-A chain gene (Fig. 5B). Under the conditions in which RNA interference of ANP32B decreased promoter region-specific histone levels, anti-histone H4 ChIP analysis showed that RNA interference of KLF5 decreased histone H4 levels in a promoter region-specific manner. RNA interference of both ANP32B and KLF5 showed a promoter region-specific decrease in histone levels (Fig. 5B). These findings indicate that recruitment of ANP32B onto the promoter region of the PDGF-A chain and histone incorporation by ANP32B over the promoter region of the PDGF-A chain require KLF5.
Dynamics of ANP32B, KLF5, and histones over the PDGF-A chain gene in response to transactivational stimulus.
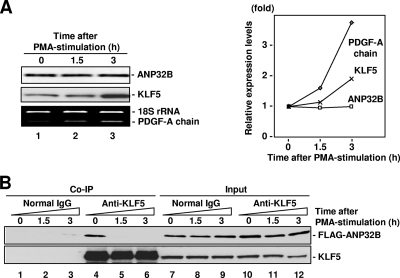
To examine the cellular implications of functional interaction of KLF5 and ANP32B in chromatin transcription, we investigated how ANP32B, KLF5, and histones act over the PDGF-A chain gene promoter in response to a KLF5-activating stimulus. For this purpose, we examined effects of phorbol ester (as represented by PMA) on PDGF-A chain gene expression as mediated by KLF5. PMA is a model agonist of inducible extracellular pathophysiological stimulation known to transcriptionally activate KLF5, which in turn upregulates its endogenous downstream gene, PDGF-A chain (3, 42).
We first assessed expression levels of ANP32B and KLF5 proteins and PDGF-A chain mRNA under PMA stimulation (Fig. 6A). ANP32B protein expression was not affected under the condition in which KLF5 protein expression and PDGF-A chain mRNA expression were induced by PMA stimulation as previously described (29). ANP32B is thus a constitutive factor under this condition. Next, we assessed whether interaction of KLF5 and ANP32B is regulated by PMA stimulation by using a coimmunoprecipitation assay (Fig. 6B). As KLF5 is an inducible factor, experiments were done with cotransfection of ANP32B and KLF5 under the condition in which expression levels of KLF5 and ANP32B were not affected by PMA stimulation (Fig. 6B, lanes 7 to 12). The coimmunoprecipitation assay showed that interaction of KLF5 and ANP32B was attenuated by PMA stimulation (Fig. 6B, lanes 4 to 6). Thus, while ANP32B is not an inducible protein, its interaction with KLF5 is attenuated by PMA stimulation.
FIG. 6.
Dynamics of ANP32B and KLF5 in response to transactivational stimulus. (A) Expression levels of ANP32B and KLF5 proteins and PDGF-A chain mRNA after phorbol ester (PMA) stimulation. Cell lysate was analyzed by Western blotting or Coomassie blue staining. (B) Cellular binding of ANP32B and KLF5 under PMA stimulation. Bound proteins were detected by anti-KLF5 (KM1785) and anti-FLAG-horseradish peroxidase antibodies.
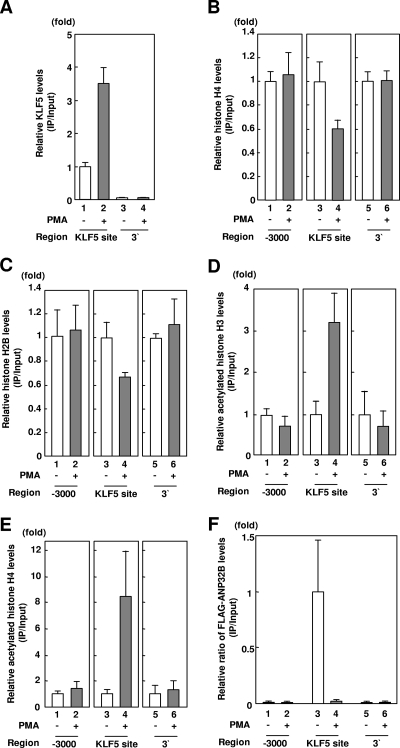
Next, we assessed whether PMA stimulation affects KLF5, in addition to histone H2B and H4 levels over the PDGF-A chain gene promoter in a manner coupled with transcriptional activation by PMA stimulation (Fig. 7A and B). Under conditions in which KLF5 showed specific binding to its cognate binding site on the PDGF-A chain gene promoter (−71 bp to −55 bp) but not the 3′ end of the gene used as a negative control (Fig. 7A, lanes 3 and 4), KLF5 levels over its cognate binding site increased under PMA stimulation (Fig. 7A, lanes 1 and 2). This indicates that PMA stimulation induces binding of KLF5 to the PDGF-A chain gene promoter, resulting in transcriptional activation.
FIG. 7.
Dynamics of KLF5, histones, and ANP32B over the PDGF-A chain gene promoter in response to transactivational stimulus. (A) Effect of PMA stimulation on KLF5 levels over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (B) Effect of PMA stimulation on histone H4 levels over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (C) Effect of PMA stimulation on histone H2B levels over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (D) Effect of PMA stimulation on levels of acetylated histone H3 over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (E) Effect of PMA stimulation on levels of acetylated histone H4 over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors. (F) Effect of PMA stimulation on ANP32B levels over the PDGF-A chain gene promoter. The amount of immunoprecipitated DNA was determined by quantitative PCR with indicated primer pairs. Error bars denote standard errors.
Further, the anti-histone H4 ChIP assay showed that histone H4 levels over the KLF5 binding site on the PDGF-A chain gene promoter decreased under transcriptional activation mediated by PMA stimulation (Fig. 7B, lanes 3 and 4) while not affecting histone H4 levels over the 5′-(−3,000 bp) and 3′ regions of the gene (Fig. 7B, lanes 1, 2, 5, and 6). Anti-histone H2B ChIP analysis showed that PMA stimulation also decreases histone H2B levels over the PDGF-A chain gene (Fig. 7C). These findings indicate that transcriptional activation as induced by PMA stimulation is coupled with promoter region-specific histone eviction.
To address the functional implications of regulation of histone acetylation and transcriptional activation by PMA stimulation, we examined the effect of PMA stimulation on levels of acetylated histones H3 and H4 (Fig. 7D). ChIP assays of anti-acetylated histones H3 and H4 showed that PMA stimulation increased acetylated histone H3 and H4 levels over the promoter region of the PDGF-A chain gene (Fig. 7D, lanes 3 and 4) while not affecting histone H4 levels over the 5′-(−3,000 bp) or the 3′ regions of the gene (Fig. 7D, lanes 1, 2, 5, and 6). These findings indicate that transcriptional activation mediated by PMA stimulation is coupled with promoter access of KLF5 as well as promoter region-specific histone eviction and histone acetylation.
Moreover, to confirm that ANP32B participates in regulation of promoter region-specific histone eviction and histone acetylation as mediated by PMA stimulation, we examined the effect of PMA stimulation on localization of ANP32B over the PDGF-A chain gene promoter (Fig. 7E). Anti-ANP32B ChIP analysis showed that PMA stimulation decreased ANP32B levels over the promoter region (Fig. 7E, lanes 3 and 4), while not affecting ANP32B levels over the 5′-(−3,000 bp) and 3′ regions of the gene (Fig. 7E, lanes 1, 2, 5, and 6). This indicates that ANP32B localizes over the PDGF-A chain promoter during transcriptional repression while not during transcriptional activation.
These findings collectively indicate that a transactivating stimulus regulates interaction of ANP32B and KLF5 from an associative to a dissociative state, thus leading to transcriptional activation in a manner correlated with region-specific histone eviction. Thus, transcriptional activation of the PDGF-A chain gene is mediated by histone eviction over the KLF5 binding site and binding of KLF5 to its cognate binding site.
DISCUSSION
New mechanism of transcriptional regulation mediated by the novel histone chaperone ANP32B and DNA-binding transcription factor KLF5.
We identified ANP32B as an interactor with DNA-binding transcription factor KLF5 (Fig. 1). ANP32B directly binds core histones and assembles nucleosomes in vitro (Fig. 3) and participates in histone incorporation in vivo (Fig. 4). Therefore, ANP32B is a novel bona fide histone chaperone. ANP32B is a member of the ANP32 family (family of proteins with acidic- and leucine-rich stretches), whose members, while often functioning as oncogenic regulators (i.e., ANP32A/pp32 are tumor suppressors whereas ANP32C/pp32R1 and ANP32D/pp32R2 are tumorigenic factors) (19), harbor divergent biochemical activities that range from control of histone acetylation, histone-dependent plasmid supercoiling, and mRNA stability to specialized forms of apoptosis among others for those that have been documented (14, 36).
We showed that ANP32B harbors multiple activities which include nucleosome assembly in vitro (Fig. 3), histone incorporation in vivo (Fig. 4), inhibition of histone acetylation (Fig. 4), inhibition of DNA-binding activity of KLF5 by protein-protein interaction (Fig. 2), and repression of transcription of PDGF-A chain, which is a downstream gene of KLF5 (Fig. 2 and 4).
Importantly, functional interaction of histone ANP32B and KLF5 resulted in promoter region-specific histone incorporation and inhibition of histone acetylation on the KLF5-downstream PDGF-A chain gene which was coupled with transcriptional repression (Fig. 4 and 5). An extracellular stimulus (e.g., phorbol ester) further regulated this mechanism in the cell, which supports the biological relevance of this interaction (Fig. 6 and 7). RNA interference of KLF5 resulted in decreased ANP32B and histone levels over the PDGF-A chain gene promoter (Fig. 5), which indicates that recruitment of ANP32B onto the PDGF-A chain gene promoter leading to histone incorporation by ANP32B requires KLF5. PMA stimulation showed an increase in KLF5 levels and a decrease in ANP32B and histone levels (Fig. 7), which indicates that recruitment of ANP32B or KLF5 onto the PDGF-A chain gene promoter results in eviction of ANP32B or KLF5. Further, as ANP32B inhibited the DNA-binding activity of KLF5 (Fig. 2), it would seem that, while KLF5 and ANP32B cooperatively interact during recruitment, occupancy on the promoter is mutually exclusive.
In perspective, the coupling of transcriptional regulation and histone eviction/incorporation has been best studied on the Saccharomyces cerevisiae PHO5 promoter. The transcription factors Pho2 and Pho4 activate PHO5 by promoter access (10, 49). A recent study showed that the histone chaperone Spt6 incorporates histones during transcriptional repression of PHO5 and that histone incorporation by Spt6 and promoter access to Pho2/Pho4 are independent (2). Importantly, as protein-protein interaction and/or coregulation of Spt6 and Pho2/Pho4 has not been addressed (as Spt6 and Pho2/Pho4 are not regulated by protein-protein interaction and/or coregulation), regulation of histone incorporation/eviction and DNA-binding activity of transcription factors are mutually independent in the Spt6-Pho2/Pho4 system. In contrast to the Spt6-Pho2/Pho4 system, histone incorporation/eviction and DNA-binding activity of transcription factor are coupled in regulation of transcription as mediated by ANP32B and KLF5.
A recent study showed that the histone chaperone JDP2, an interactor with the transcription factor Jun (6, 15), inhibits DNA-binding activity of Jun, assembles nucleosomes, and represses Jun-mediated transcription (16). We have shown that the histone chaperone TAF-Iβ inhibits KLF5-mediated transcription by protein-protein interaction with KLF5 ZF/DBD (29). These histone chaperones, JDP-2 and TAF-I, directly bind transcription factors and negatively regulate transcription; however, involvement of these histone chaperones in histone incorporation in a promoter region-specific manner was not addressed.
Collectively, we have described cooperative regulation of promoter region-specific histone incorporation in transcriptional repression by DNA-binding transcription factor and histone chaperone.
Acknowledgments
This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology; the New Energy and Industrial Technology Development Organization; the Ministry of Health, Labor, and Welfare; the Japan Science and Technology Corporation; the Takeda Medical Research Foundation; the Japan Heart Foundation; and the Japan Foundation for Applied Enzymology.
We declare that we have no competing financial interest.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14657-666. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21405-416. [DOI] [PubMed] [Google Scholar]
- 3.Aizawa, K., T. Suzuki, N. Kada, A. Ishihara, K. Kawai-Kowase, T. Matsumura, K. Sasaki, Y. Munemasa, I. Manabe, M. Kurabayashi, Y. Collins, and R. Nagai. 2004. Regulation of platelet-derived growth factor-A chain by Kruppel-like factor 5: new pathway of cooperative activation with nuclear factor-κB. J. Biol. Chem. 27970-76. [DOI] [PubMed] [Google Scholar]
- 4.Akey, C. W., and K. Luger. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 136-14. [DOI] [PubMed] [Google Scholar]
- 5.Annunziato, A. T. 2005. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 28012065-12068. [DOI] [PubMed] [Google Scholar]
- 6.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 173094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanchevalap, S., M. O. Nandan, B. B. McConnell, L. Charrier, D. Merlin, J. P. Katz, and V. W. Yang. 2006. Krüppel-like factor 5 is an important mediator for lipopolysaccharide-induced proinflammatory response in intestinal epithelial cells. Nucleic Acids Res. 341216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chimura, T., T. Kuzuhara, and M. Horikoshi. 2002. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc. Natl. Acad. Sci. USA 999334-9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earnshaw, W. C., B. M. Honda, R. A. Laskey, and J. O. Thomas. 1980. Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell 21373-383. [DOI] [PubMed] [Google Scholar]
- 10.Fascher, K. D., J. Schmitz, and W. Hörz. 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 92523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda, H., N. Sano, S. Muto, M., and Horikoshi. 2006. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief. Funct. Genomics Proteomics 5190-208. [DOI] [PubMed] [Google Scholar]
- 12.Groth, A., D. Ray-Gallet, J. P. Quivy, J. Lukas, J. Bartek, and G. Almouzni. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17301-311. [DOI] [PubMed] [Google Scholar]
- 13.Guermah, M., V. B. Palhan, A. J. Tackett, B. T. Chait, and R. G. Roeder. 2006. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125275-286. [DOI] [PubMed] [Google Scholar]
- 14.Haruki, H., M. Okuwaki, M. Miyagishi, K. Taira, and K. Nagata. 2006. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 80794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin, C., H. Ugai, J. Song, T. Murata, F. Nili, K. Sun, M. Horikoshi, and K. K. Yokoyama. 2001. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 48934-41. [DOI] [PubMed] [Google Scholar]
- 16.Jin, C., K. Kato, T. Chimura, T. Yamasaki, K. Nakade, T. Murata, H. Li, J. Pan, M. Zhao, K. Sun, R. Chiu, T. Ito, K. Nagata, M. Horikoshi, and K. K. Yokoyama. 2006. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat. Struct. Mol. Biol. 13331-338. [DOI] [PubMed] [Google Scholar]
- 17.Jin, J., Y. Cai, B. Li, R. C. Conaway, J. L. Workman, J. W. Conaway, and T. Kusch. 2005. In and out: histone variant exchange in chromatin. Trends Biochem. Sci. 30680-687. [DOI] [PubMed] [Google Scholar]
- 18.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 142441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadkol, S. S., J. R. Brody, J. Pevsner, J. Bai, and G. R. Pasternack. 1999. Modulation of oncogenic potential by alternative gene use in human prostate cancer. Nat. Med. 5275-279. [DOI] [PubMed] [Google Scholar]
- 20.Kaeser, M. D., and B. M. Emerson. 2006. Remodeling plans for cellular specialization: unique styles for every room. Curr. Opin. Genet. Dev. 16508-512. [DOI] [PubMed] [Google Scholar]
- 21.Kanduri, C., C. Holmgren, M. Pilartz, G. Franklin, M. Kanduri, L. Liu, V. Ginjala, E. Ulleras, R. Mattsson, and R. Ohlsson. 2000. The 5′ flank of mouse H19 in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 10449-457. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 23.Kuzuhara, T., and M. Horikoshi. 2004. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 11275-283. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H., H. Kang, R. Liu, X. Chen, and K. Zhao. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 226471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lusser, A., and J. T. Kadonaga. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 251192-1200. [DOI] [PubMed] [Google Scholar]
- 26.Manabe, I., and G. K. Owens. 2001. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J. Clin. Investig. 107823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumura, T., T. Suzuki, K. Aizawa, Y. Munemasa, S. Muto, M. Horikoshi, and R. Nagai. 2005. The deacetylase HDAC1 negatively regulates the cardiovascular transcription factor Kruppel-like factor 5 through direct interaction. J. Biol. Chem. 28012123-12129. [DOI] [PubMed] [Google Scholar]
- 28.Mencinger, M., I. Panagopoulos, J. A. Contreras, F. Mitelman, and P. Aman. 1998. Expression analysis and chromosomal mapping of a novel human gene, APRIL, encoding an acidic protein rich in leucines. Biochim. Biophys. Acta 1395176-180. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto, S., T. Suzuki, S. Muto, K. Aizawa, A. Kimura, Y. Mizuno, T. Nagino, Y. Imai, N. Adachi, M. Horikoshi, and R. Nagai. 2003. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol. Cell. Biol. 238528-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munakata, T., N. Adachi, N. Yokoyama, T. Kuzuhara, and M. Horikoshi. 2000. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5221-233. [DOI] [PubMed] [Google Scholar]
- 31.Muto, S., M. Senda, Y. Akai, L. Sato, T. Suzuki, R. Nagai, R. M. Horikoshi, and T. Senda. 2007. Relationship between the structure of SET/TAF-Iβ/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. USA 1044285-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natsume, R., M. Eitoku, Y. Akai, N. Sano, M. Horikoshi, and T. Senda. 2007. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H. Nature 446338-341. [DOI] [PubMed] [Google Scholar]
- 33.O'Neill, L. P., and B. M. Turner. 2003. Immunoprecipitation of native chromatin: NChIP. Methods 3176-82. [DOI] [PubMed] [Google Scholar]
- 34.Pegoraro, G., A. Marcello, M. P. Myers, and M. Giacca. 2006. Regulation of adeno-associated virus DNA replication by the cellular TAF-I/set complex. J. Virol. 806855-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polo, S. E., D. Roche, and G. Almouzni. 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127481-493. [DOI] [PubMed] [Google Scholar]
- 36.Santa-Coloma, T. A. 2003. Anp32e (Cpd1) and related protein phosphatase 2 inhibitors. Cerebellum 2310-320. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, R., A. J. Bannister, C. Weise, and T. Kouzarides. 2004. Direct binding of INHAT to H3 tails disrupted by modifications. J. Biol. Chem. 27923859-23862. [DOI] [PubMed] [Google Scholar]
- 38.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22415-422. [DOI] [PubMed] [Google Scholar]
- 39.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104119-130. [DOI] [PubMed] [Google Scholar]
- 40.Seo, S. B., T. Macfarlan, P. McNamara, R. Hong, Y. Mukai, S. Heo, and D. Chakravarti. 2002. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem. 27714005-14010. [DOI] [PubMed] [Google Scholar]
- 41.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75243-269. [DOI] [PubMed] [Google Scholar]
- 42.Shindo, T., I. Manabe, Y. Fukushima, K. Tobe, K. Aizawa, S. Miyamoto, K. Kawai-Kowase, N. Moriyama, Y. Imai, H. Kawakami, H. Nishimatsu, T. Ishikawa, T. Suzuki, H. Morita, K. Maemura, M. Sata, Y. Hirata, M. Komukai, H. Kagechika, T. Kadowaki, M. Kurabayashi, and R. Nagai. 2002. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat. Med. 8856-863. [DOI] [PubMed] [Google Scholar]
- 43.Simon, R. H., and G. Felsenfeld. 1979. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 529-41. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, T., S. Muto, S. Miyamoto, K. Aizawa, M. Horikoshi, and R. Nagai. 2003. Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J. Biol. Chem. 27828758-28764. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, T., K. Aizawa, T. Matsumura, and R. Nagai. 2005. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler. Thromb. Vasc. Biol. 251135-1141. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, T., T. Matsumura, and R. Nagai. 2005. Transcriptional regulation at the chromatin level in the cardiovasculature through protein-protein interactions and chemical modifications. Trends Cardiovasc. Med. 15125-129. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki, T., T. Nishi, T. Nagino, K. Sasaki, K. Aizawa, N. Kada, D. Sawaki, Y. Munemasa, T. Matsumura, S. Muto, M. Sata, K. Miyagawa, M. Horikoshi, and R. Nagai. 2007. Functional interaction between the transcription factor Kruppel-like factor 5 and poly (ADP-ribose) polymerase-1 in cardiovascular apoptosis. J. Biol. Chem. 2829895-9901. [DOI] [PubMed] [Google Scholar]
- 49.Svaren, J., and W. Hörz. 1997. Transcription factors vs nucleosomes: regulation of the PH05 promoter in yeast. Trends Biochem. Sci. 2293-97. [DOI] [PubMed] [Google Scholar]
- 50.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402555-560. [DOI] [PubMed] [Google Scholar]
- 51.Wolffe, A. P. 1998. Chromatin (structure and function), 3rd ed. Academic Press, San Diego, CA.
- 52.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 202009-2017. [DOI] [PubMed] [Google Scholar]
- 53.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25619-623. [DOI] [PubMed] [Google Scholar]