Abstract
The distal end of mouse chromosome 7 (Chr 7) contains a large cluster of imprinted genes. In this region two cis-acting imprinting centers, IC1 (H19 DMR) and IC2 (KvDMR1), define proximal and distal subdomains, respectively. To assess the functional independence of IC1 in the context of Chr 7, we developed a recombinase-mediated chromosome truncation strategy in embryonic stem cells and generated a terminal deletion allele, DelTel7, with a breakpoint in between the two subdomains. We obtained germ line transmission of the truncated Chr 7 and viable paternal heterozygotes, confirming the absence of developmentally required paternally expressed genes distal of Ins2. Conversely, maternal transmission of DelTel7 causes a midgestational lethality, consistent with loss of maternally expressed genes in the IC2 subdomain. Expression and DNA methylation analyses on DelTel7 heterozygotes demonstrate the independent imprinting of IC1 in absence of the entire IC2 subdomain. The evolutionarily conserved linkage between the subdomains is therefore not required for IC1 imprinting on Chr 7. Importantly, the developmental phenotype of maternal heterozygotes is rescued fully by a paternally inherited deletion of IC2. Thus, all the imprinted genes located in the region and required for normal development are silenced by an IC2-dependent mechanism on the paternal allele.
The 1-Mb imprinted domain on distal chromosome 7 (Chr 7) shares syntenic homology to the Beckwith-Wiedemann syndrome (BWS) region on human Chr 11p15.5 (69). Located less than 3 Mb from the telomere (Tel7q), this region contains two imprinting centers, IC1 and IC2. These conserved imprinting control elements are cis-acting sequences which carry opposite germ line DNA methylation marks and regulate the monoallelic expression of different flanking genes (9). In the proximal part of the Chr 7 domain, IC1 is located 2 kb upstream of the H19 promoter (Fig. 1A) (66). This sequence acquires a paternal DNA methylation imprint established during spermatogenesis and maintained throughout development (4, 24, 67). The methylated IC1 is required to initiate silencing of the paternal H19 allele (65), whereas the maternal IC1 controls the paternally expressed genes Igf2 and Ins2 via a methylation-sensitive insulator (6, 35). Distally, IC2 is located in intron 10 of Kcnq1 (Fig. 1A). This sequence is marked by a maternal DNA methylation imprint acquired during oogenesis (19). The unmethylated paternal IC2 is associated with the production of the Kcnq1ot1 noncoding RNA (ncRNA) and the silencing in cis of several genes in this subdomain (26). Consequently, these protein-coding genes are imprinted and expressed preferentially from the maternal Chr 7 homologue (62). These maternally expressed genes (MEGs) include transcripts expressed in the placenta and required for embryonic development, such as Ascl2 (31, 32), Cdkn1c (36), and Phlda2 (58). As in the case of IC1, IC2 appears to carry different allele-specific functions, such as promoter, enhancer, and CTCF binding insulator activities (25, 39).
FIG. 1.
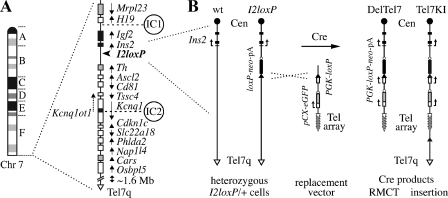
Structure of the imprinted domain on distal Chr 7 and strategy for RMCT. (A) Ideogram of Chr 7 (left) and imprinted gene organization (right) in the distal band 7F5. Genes expressed preferentially from the maternal (white) or paternal (black) homologue, as well as known biallelically expressed transcripts (gray), are identified, with arrows showing their transcriptional orientation. Several nonimprinted genes and novel annotated transcripts also map telomeric to the imprinted domain, in a ∼1.6-Mb region of distal Chr 7. The map also shows the positions of the known imprinting centers (IC1 and IC2) and that of the targeted loxP site insertion allele I2loxP used in this study (arrowhead). (B) Close-up maps of the Ins2 region in I2loxP/+ ES cells and two novel alleles. Shown are structures of the wt Ins2 gene and of the Ins2I2loxP allele (I2loxP), as found in G418-sensitive heterozygous I2loxP/+ ES cells (left). The I2loxP allele is the targeted insertion of a loxP site followed by a promoterless neo-pA cassette (loxP-neo-pA) upstream of Ins2. Deletion of the Chr 7 sequences distal of I2loxP by RMCT was achieved by coelectroporation of a Cre recombinase expression vector together with a linear replacement vector (middle) carrying a terminal array of telomere repeats (Tel array), a pCX-eGFP reporter (56), and a PGK promoter-loxP construct (PGK-loxP). Cre recombination in trans between I2loxP on Chr 7 and the vector regenerates an active PGK-loxP-neo-pA-selectable marker and yields the DelTel7 or Tel7KI alleles in G418-resistant derivatives of I2loxP/+ cells (right). In DelTel7, Chr 7 is truncated by RMCT and sequences distal of the I2loxP breakpoint are replaced by the vector from the loxP site to the array of TTAGGG telomeric repeats. Tel7KI is an insertion (pop-in) of the circular replacement vector at I2loxP. Tel7q, Chr 7 distal telomere; Cen, centromere.
Transgenic and knockout studies have suggested that the imprinting centers IC1 and IC2 function independently of each other, thereby defining two imprinted subdomains on distal Chr 7, despite the evolutionarily conserved linkage between these cis-acting sequences (2, 8, 9, 18, 26). However, the possibility of functional interactions between the two linked subdomains containing these ICs, as suggested by BWS cases (41, 62), or the presence of other shared elements required by both ICs have not been studied in the context of Chr 7. To address these issues and assess directly the ability of IC1 to function as an independent imprinting center in the context of distal Chr 7, we generated a truncated Chr 7 variant in which the entire IC2 subdomain is deleted. For the engineering of this terminal deletion, we developed a new strategy for chromosome truncation in embryonic stem (ES) cells, termed recombinase-mediated chromosome truncation (RMCT). The specificity of this approach is conferred by two consecutive steps: (i) targeting of a loxP site at the desired chromosomal location of the breakpoint; (ii) Cre-mediated recombination in trans between the genomic loxP site and a second loxP site provided by an incoming linear vector carrying a terminal array of telomere repeats, known to act as telomere seeds in telomerase-positive cells (33). Following RMCT, the vector with the telomere repeats replaces the chromosomal fragment distal of the breakpoint, thus generating a site-specific terminal deletion, or truncation of the desired chromosome.
In this report, we used RMCT to engineer a terminal deletion on Chr 7 in ES cells, the DelTel7 allele, with a breakpoint immediately distal of the IC1 subdomain. We present the construction and characterization of this allele in ES cells, the germ line transmission of the truncated Chr 7 variant, and the generation of the first mouse line with an engineered telomere. For the first time, we were able to analyze the independent imprinting of IC1 on Chr 7, and we demonstrate that the H19-Igf2-Ins2 proximal subdomain is able to acquire and maintain appropriate epigenetic identity in the absence of the entire IC2 subdomain. The DelTel7 deficiency also allowed us to characterize the phenotype associated with loss of the entire IC2 subdomain and to demonstrate the central role played by IC2 in the epigenetic silencing of all the developmentally required genes on distal Chr 7. Our work also establishes RMCT as a new avenue for chromosome engineering in the mouse and for the generation of mouse lines with specifically tailored telomeres.
MATERIALS AND METHODS
Plasmid constructions.
The positive selectable marker neo was chosen to construct complementary bipartite cassettes that can be partially collapsed or recycled using the Cre-loxP recombinase system. In the first cassette, PGK-loxP-neo-pA-loxP, the neo-pA is flanked by loxP sites. In the second cassette, it is the PGK promoter alone which can be deleted by Cre (loxP-PGK-loxP-neo-pA). A PGK-loxP-neo-pA construct, common to both cassettes, was first obtained by subcloning the 0.5-kb EcoRI-PstI (blunt) PGK promoter fragment of plasmid pPGKβ-geobpA (27) into the EcoRI-SmaI sites of plasmid pBS65, which contains a single loxP site inserted into pSP65 (51). pBS65 is similar to pBS64 (60) but with the loxP in the opposite orientation. In pPGK-loxP, the loxP site is oriented as follows: EcoRI-PGK pro(PstI/SmaI)-BamHI-loxP (5′-ATAACTTCGTATAATGTATGCTATACGAAGTTAT-3′)-PstI-HindIII. A 1.0-kb blunt EcoRI-SpeI neo-pA fragment from pneobpA, a subclone of neo-bpA from pMC1neopA (Stratagene) into pBlueScript KS(−) (Nagy lab), was inserted into the blunt PstI site of pPGK-loxP to obtain pPGK-loxP-neo-pA. This cassette was subcloned back into pBS65 as a blunt 1.5-kb EcoRI-HindIII fragment in two different ways: (i) in the blunt EcoRI-SmaI sites, 5′ of the loxP site, to obtain pPGK-loxP-neo-pA-loxP, or (ii) into the blunt PstI site, 3′ of the loxP site, to obtain ploxP-PGK-loxP-neo-pA. These two constructs, which provided the complementary components for positive selection of RMCT, were shown to confer G418 resistance in ES cells.
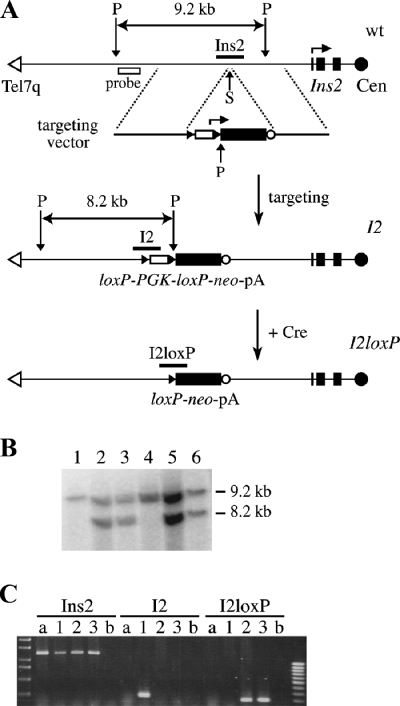
The targeting vector to generate the I2 insertional allele was based on the 5.0-kb EcoRI Ins2 genomic clone P11, isolated from a 129/Sv genomic library and obtained from Jacques Jami (17). A flanking SpeI site from the pSK+ vector was first destroyed by filling with Klenow. The loxP-PGK-loxP-neo-pA cassette was then inserted in this genomic clone as a blunt EcoRI-XhoI fragment into the blunt SpeI site, located 2.6 kb upstream of the transcription start site of Ins2, such that both genes were in the same transcriptional orientation. The resulting targeting vector (pI2TV), shown in its NotI-linearized form in Fig. 2A, has 5′ and 3′ arms of homology of 2.9 and 2.1 kb, respectively.
FIG. 2.
Targeted loxP site insertion distal of the IC1 subdomain on distal Chr 7. The breakpoint for truncation on Chr 7 was established by targeting of a loxP site 2.6 kb upstream of the Ins2 gene. (A) The heterozygous I2loxP/+ ES cells used for RMCT were derived by two successive steps in R1 ES cells, detailed in these representations of the Ins2 locus oriented from the telomere (Tel7q) on the left to the centromere (Cen). First, the wt Ins2 gene was modified by the targeted insertion of a loxP-PGK-loxP-neo-pA cassette in the SpeI site (S) upstream of Ins2 to generate the Ins2I2 (or I2) allele. Targeted clones were identified by Southern blot analysis of ES cell genomic DNA digested with PstI (P) and hybridized to a 5′-flanking probe (B). In addition to the wt 9.2-kb band seen in R1 ES cells (lane 1) and an untargeted clone (lane 4), I2/+ cells have a new 8.2-kb band indicative of targeting of the Ins2 locus (lanes 2, 3, 5, and 6). The structure of the 3′ end was also confirmed with a 3′ internal probe (see Fig. 3B, below). Second, the loxP-flanked PGK promoter of the I2 allele was deleted by transient production of Cre recombinase from an electroporated expression vector. Individual colonies were picked and expanded in duplicates, and G418-sensitive clones were identified and kept for further analysis. The Cre-mediated deletion of the PGK promoter, to form the I2loxP allele, was confirmed by Southern blot analysis (see Fig. 3B, below) and by genomic PCR (C). PCRs specific to the wt (Ins2), targeted (I2), and excised (I2loxP) alleles, using a common forward primer, were used to monitor the presence of the I2 allele in parental I2/+ ES cells (lane 1) and its modification to I2loxP in G418s Cre-electroporated clones (lanes 2 and 3). Lane a, R1 ES cell DNA; lane b, water control.
The vector used for RMCT contains an array of telomere repeats, a ubiquitous enhanced green fluorescent protein (eGFP) reporter, and a PGK promoter-loxP unit (PGK-loxP) designed to activate the promoterless loxP-neo-pA cassette of the I2loxP allele, following site-specific Cre-mediated recombination in trans (Fig. 1). The 1.6-kb BamHI-BglII fragment from plasmid pSX-neo-1.6-T2AG3 (33), carrying two 0.8-kb tandem arrays of T2AG3 repeats, was cloned into BamHI-digested pCAGGS-eGFP (pCX-eGFP) (56). In pCX-eGFP-Telb, the unique NotI site of the insert is adjacent to the PstI site and poly(A) sequence of pCX-eGFP, such that the arrays are oriented toward the eGFP cassette. The PGK-loxP unit was generated by in vitro Cre treatment of pM2TV, carrying the PGK-loxP-neo-pA-loxP cassette inserted in 3′-flanking sequences from the Ascl2 locus (L. Lefebvre, unpublished data), to form pM2TV-Cre. A 2.4-kb EagI-NsiI fragment of pM2TV-Cre was subcloned into pCX-eGFP-Telb digested with NotI and PstI to form the vector pCX-eGFP-Telb-PGK-loxP. This telomere-seeding replacement vector is represented in its NotI-linearized form in Fig. 1B.
Generation of I2loxP/+ embryonic stem cells.
The mouse ES cell lines are all derived from the R1 line (54). Maintenance, electroporation, selection, and aggregation of ES cells followed standard procedures (53). The I2loxP/+ ES cells used for RMCT were generated by two steps: targeting of the I2 allele and Cre excision of the PGK promoter. First, R1 ES cells were electroporated with ∼20 μg of the I2 targeting vector pI2TV, linearized with NotI. Individual colonies of neo-expressing cells were recovered after 8 to 10 days of selection with G418 (G9516; Sigma) at 150 μg/ml, picked, and expanded for analysis. A confirmed positive I2/+ targeted clone was expanded and electroporated with ∼20 μg of the circular Cre expression vector pCX-nlsCre-puro (A. Nagy, unpublished data). At 24 h postelectroporation, cells were maintained under puromycin (P8833; Sigma) selection (1.5 μg/ml) for 48 h to enrich for cells transiently expressing Cre. Clones in which the loxP-flanked PGK promoter of the I2 allele has been deleted were identified by G418 sensitivity. The structure of the resulting allele, called I2loxP, was confirmed by analysis of genomic DNA (PCR and Southern blotting). A confirmed heterozygous I2loxP/+ ES cell clone was expanded for RMCT. The Ins2I2 and Ins2I2loxP insertions have the official allele names Ins2tm1Nagy and Ins2tm1.1Nagy, respectively.
Modification of distal Chr 7 by RMCT in ES cells.
The heterozygous I2loxP/+ cells used here for RMCT are G418-sensitive cell carrying a promoterless loxP-neo-pA insertion 5′ of Ins2. This silent neo marker can be activated by the Cre-catalyzed delivery of a PGK promoter from a PGK-loxP cassette such as the one cloned into our replacement vector (34). For RMCT, ∼6 × 106 heterozygous Ins2I2loxP/+ ES cells were electroporated with ∼30 μg of NotI-linearized pCX-eGFP-Telb-PGK-loxP and a Cre expression vector as described above. Following 2 days of puromycin selection, positive clones were recovered after 6 to 8 days of G418 selection (150 μg/ml). G418-resistant colonies were picked and expanded as described previously (53). We never observed G418-resistant colonies from Ins2I2loxP/+ ES cells electroporated with pCX-nlsCre-puro alone. This observed null rate of spontaneous reversion, together with the absence of neo sequences in the electroporated vectors, contributed to a high rate of site-specific insertion (100%) in the few G418-resistant clones obtained (∼50 colonies per electroporation).
Mice.
Outbred ICR mice (Harlan) were used for aggregations with ES cells to generate chimeras and to maintain the DelTel7 line. Six recipient females were used per ES cell line aggregated; 18 to 19 embryos were transferred to each pseudopregnant female. The IC2KO mice (KvDMR1 deletion) were originally generated in a 129/SvJae (129S4) ES cell line and maintained on the C57BL/6J background (26). For the phenotypic rescue experiments, we used +/IC2KO heterozygous males maintained on C57BL/6J or ICR backgrounds. Similar results were obtained with each background, and the data were pooled (see Table 2, below). The Ascl2 (Mash2) knockout allele (Ascl2tm1Alj) has been published and was maintained on an ICR background (32). For allele-specific studies, we used reciprocal crosses between +/DelTel7 heterozygotes and 129S1/SvImJ × CAST/EiJ F1 mice, (129CAST)F1. To distinguish reciprocal heterozygotes, all the crosses and genotypes presented in this study give the maternal allele first (maternal/paternal), such that +/DelTel7 and DelTel7/+ embryos have inherited the DelTel7 allele paternally and maternally, respectively.
TABLE 2.
The midgestational lethality of DelTel7/+ embryos is rescued by a paternally inherited deletion of IC2
| Exptl group | Parental genotypes
|
Stage | No. of progeny (%)
|
|||
|---|---|---|---|---|---|---|
| Female | Male | +/+ | Heterozygotes | Δ/Δb | ||
| A | +/+ | +/DelTel7 | P21e | 79 (54) | 68 (46) | |
| B | +/DelTel7 | +/+ | P21 | 86 (100) | 0 (0) | |
| E11.5 | 15 (68) | 7 (32)a | ||||
| E10.5 | 15 (48) | 16 (52)a | ||||
| E9.5 | 19 (50) | 19 (50) | ||||
| C | +/DelTel7 | +/DelTel7 | E9.5 | 5 (50) | 5 (50) | 0 (0) |
| E3.5 | 7 (27) | 12 (46) | 7 (27) | |||
| E3.5c | 7 (29) | 12 (50) | 5 (21)d | |||
| D | +/Ascl2KO | +/IC2KO | P21 | 8 (26) | 0 (0) (Ascl2KO/+) | 14 (45) (Ascl2KO/IC2KO) |
| 9 (29) (+/IC2KO) | ||||||
| E | +/DelTel7 | +/IC2KO | P21 | 39 (40) | 0 (0) (Δ/+) | 36 (37) (Δ/IC2KO) |
| 22 (23) (+/IC2KO) | ||||||
Viable DelTel7/+ embryos were recovered prior to E10.0, but these maternal heterozygotes were never seen at birth and were dead when recovered at E10.5 or E11.5.
Δ, DelTel7.
Genotyped from ICM outgrowths recovered after 6 days in culture (see Fig. 4B).
These samples could not be genotyped, since no material was recovered.
P21, postnatal day 21.
DNA FISH and SKY analysis.
ES cells and embryonic fibroblasts were harvested following a 4-hour treatment with Colcemid (0.05 μg/ml). Subsequently, cells were exposed to 0.075 M KCl hypotonic treatment for 20 min at 37°C and then fixed in three changes of methanol-acetic acid (3:1 ratio). The suspension of fixed cells was dropped onto slides to obtain metaphase spreads. The structure of distal Chr 7 in mutant ES cell lines was analyzed by DNA fluorescence in situ hybridization (FISH) using probes generated from P1-derived artificial chromosome (PAC) clones spanning the H19 (29G9) and Th (231E13) genes (13). PAC DNA was directly labeled with Spectrum Green (H19) or Spectrum Orange (Th) using a nick translation kit (Vysis, Downers Grove, IL). PAC DNA labeling, consequent hybridization to ES cells metaphase spreads, and posthybridization washes were carried out according to the nick translation kit protocol (Vysis, Downers Grove, IL). For spectral karyotyping (SKY) analysis, metaphase spreads were obtained from primary embryonic fibroblasts derived from embryonic day 14.5 (E14.5) embryos heterozygous for the DelTel7 allele and their wild-type littermates. The SKY mouse probe from Applied Spectral Imaging (ASI; Carlsbad, CA) was hybridized to the prepared slides following the manufacturer's instructions. The SKY metaphase images were captured using an SD 200 spectral bio-imaging system (ASI Ltd., MigdalHaemek, Israel) attached to an Axioplan 2 microscope (Zeiss, Canada). Captured metaphase images were analyzed using the SKYView software version 1.6.2 (ASI, Carlsbad, CA). Ten SKY metaphase spreads were karyotyped according to spectral and inverted 4′,6′-diamidino-2-phenylindole images. Description of chromosomal changes followed the ISCN 2005 guidelines and standard mouse chromosome nomenclature.
Genotyping.
PCR genotyping was performed on purified ES cell genomic DNA or on crude lysates from yolk sacs or ear punches (53). All primer sequences are given in Table S1 in the supplemental material. Note that the Δ5′ PCR is specific to DelTel7 and Tel7KI, whereas the Δ3′ PCR is also shared with I2 (see Fig. 3, below). In reciprocal crosses, the Mus mus castaneus (CAST) Chr 7 was identified by a single nucleotide polymorphism (SNP) in exon 3 of Ascl2. For this assay, primers in2F1 and 726R were used to amplify a 511-bp PCR product which was then digested with HpaII; the CAST allele yields 198-bp and 313-bp fragments, whereas Mus domesticus alleles have an extra HpaII recognition site and give products of 96, 198, and 217 bp. For the phenotypic rescue experiments, +/DelTel7 females were bred to +/IC2KO males and progeny were genotyped using the DelTel7 PCR (Δ5′) (see Fig. 3A, below) and reactions specific for the IC2KO allele (primers KvDMR1 F90+ R322) or IC2 wild-type (wt) allele (primers KvDMR1 F239+ R240).
FIG. 3.
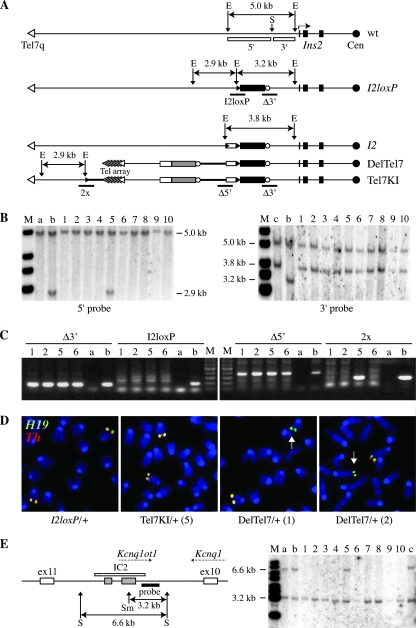
The maternal allele of Chr 7 is truncated in ES cells carrying the DelTel7 allele. (A) Diagrams of the Ins2 region on Chr 7, oriented from the distal telomere (Tel7q) to the centromere (Cen), for the wt locus and the parental I2loxP and I2 alleles, as well as the DelTel7 and Tel7KI alleles obtained by Cre recombination. The positions of the genomic probes 5′ and 3′ on either side of the insertion site of the I2loxP allele (SpeI site [S]) are shown below the wild-type allele (white boxes). The EcoRI (E) fragments hybridizing to these probes are shown above each allele, except for the 5′ 2.9-kb fragment of the I2 allele. The sequences hybridizing to the 5′ probe are deleted in DelTel7. The positions of diagnostic PCRs (I2loxP, Δ3′, Δ5′, and 2x), amplifying key junctions, are given below the diagrams. (B) Southern blot analysis of ES cell genomic DNA digested with EcoRI and hybridized to the 5′ and 3′ genomic probes. Control lanes show genomic DNA from R1 ES cells (a), the parental I2loxP/+ ES cell line (b), and I2/+ ES cells (c). Lanes 1 to 10: 10 of 24 G418-resistant ES cell clones were analyzed. (C) Representative results of PCR analyses on four G418-resistant ES cell clones (1, 2, 5, and 6 from panel B), using the four junctional PCRs (Δ3′, I2loxP, Δ5′, and 2x) mapped in panel A. For each PCR, controls included genomic DNA from wild-type ES cells (a) and from mutations containing the expected junctions (b) for Δ3′ and I2loxP, I2loxP/+ ES cell DNA, for Δ5′, Del7AI/+ DNA, and for 2x, Dup7AI/+ DNA. The Del7AI and Dup7AI alleles carry the same junctions as DelTel7 and Tel7KI for the Δ5′ and 2x reactions (L. Lefebvre, unpublished data). For primer sequences, see Table S1 in the supplemental material. (D) DNA FISH on ES cell metaphase chromosomes, using genomic PAC clones from Chr 7 regions proximal (H19, green) or distal (Th, red) to the I2loxP insertion site as probes. Arrows point to Chr 7 variants negative for the Th probe. (E) DNA methylation status at IC2 (KvDMR1) as determined by Southern blot analysis of ES cell genomic DNA. The diagram illustrates intron 10 of Kcnq1 (left), showing the position of IC2 (white rectangle), associated with the production of the antisense noncoding RNA Kcnq1ot1 from the unmethylated paternal allele. The IC2 genomic probe (black rectangle) hybridizes next to the CpG-rich sequences (gray boxes) of IC2, which are methylated on the maternal allele (62). The expected methylated (6.6-kb) and unmethylated (3.2-kb) DNA fragments recognized by this probe in genomic DNA digested with SpeI (S) and the methylation-sensitive enzyme SmaI (Sm) are shown below the diagram. The 10 samples analyzed in panel B were digested with SpeI and SmaI, and the blot was hybridized with the IC2 probe (right panel). Controls included genomic DNA samples from R1 ES cells (lane a), an E10.5 embryo (lane b), and the parental I2loxP/+ ES cells (lane c).
Southern blot analyses.
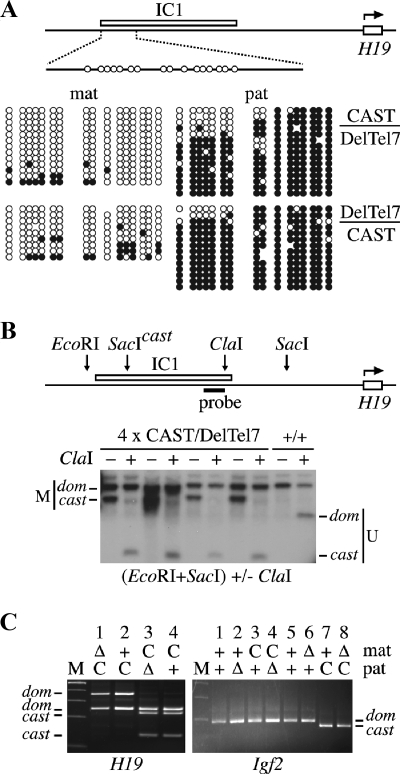
Preparation of genomic DNA from ES cell clones and Southern blot analysis were performed as described previously (44). Positive targeted I2/+ ES cell clones were identified by Southern blotting of PstI-digested ES cell genomic DNA and hybridization with a 5′-flanking probe, a 1.3-kb AccI fragment purified from an Ins2 cosmid clone (obtained from A.-K. Hadjantonakis) (Fig. 2). The structure of the 3′ end was also confirmed using the 3′ probe on genomic DNA digested with EcoRI (see Fig. 3B, below) and several other restriction enzymes (L. Lefebvre, data not shown). The analysis of RMCT clones was performed on EcoRI-digested genomic DNA. The 5′ probe is the 3.0-kb EcoRI-SpeI fragment from the Ins2 5′-flanking region immediately upstream of the loxP site insertion in I2loxP (at the SpeI site). The 3′ probe is the 1.6-kb HindIII-EcoRI fragment located in between the loxP insertion site and exon 1 of Ins2. These probes were purified from the Ins2 genomic clones P11 and P13 (17). Methylation at IC2 was analyzed with SpeI-SmaI digests of ES cell genomic DNA probed with the Kcnq1 intronic probe sc34 (62). This probe detects bands of 6.6 and 3.3 kb for methylated and unmethylated IC2 molecules, respectively (see Fig. 3E, below). For the IC1 methylation analysis (see Fig. 6B, below), genomic DNA was prepared from E14.5 CAST/DelTel7 embryos and digested with SacI-EcoRI with and without the methylation-sensitive enzyme ClaI. Maternal CAST and paternal M. domesticus alleles were distinguished by a SacI restriction fragment length polymorphism within IC1 (see Fig. 6B, below) (66). The probe is the 0.7-kb HpaII-ClaI fragment of IC1, purified from a genomic clone of the H19 upstream sequences (3.8-kb EcoRI fragment, cloned into pBluescript II KS; obtained from Mika Tanaka).
FIG. 6.
Normal imprinting at IC1 in DelTel7 heterozygotes. (A) Diagram of the upstream region of H19, showing the position of the IC1 (H19 DMR) (66) and the structure of a 473-bp sequence containing the 16 CpG sites analyzed by bisulfite sequencing. Individual embryos were recovered at E9.5 from reciprocal crosses between +/DelTel7 and (129CAST)F1 mice. PCR genotyping on yolk sac lysates was used to identify DelTel7 heterozygous embryos carrying the CAST alleles for distal Chr 7. Genomic DNA samples from a CAST/DelTel7 and a DelTel7/CAST embryo were used for bisulfite sequencing analysis of DNA methylation patterns at IC1. Unlike IC2, IC1 and H19 are not deleted in DelTel7 (Fig. 1A). Both parental alleles were analyzed, and SNPs in the sequenced products were used to identify the parental origin of each DNA strand analyzed. Methylated CpG sites are represented by filled circles, and unmethylated sites are shown as open circles. Omitted sites indicate sequencing ambiguities. (B) Southern blot analysis at IC1 for genomic DNA samples from E14.5 CAST/DelTel7 embryos and a wild-type (+/+) littermate. Digestions were with EcoRI and SacI, without (-) or with (+) the methylation-sensitive enzyme ClaI. A CAST SacI polymorphism was used to distinguish maternal and paternal alleles. The diagram shows the positions of the enzyme cut sites relative to IC1 in the H19 5′-flanking region and that of the probe used. Methylated (M) and unmethylated (U) bands are identified for the M. domesticus (dom) and M. castaneus (cast) alleles. (C) Allele-specific expression analysis of H19 and Igf2 in placental RNA samples at E9.5. For each sample, the maternal and paternal alleles are shown as follows: C, wild-type M. castaneus allele; Δ, DelTel7; +, wild-type M. domesticus allele. Note that both Δ and wild-type alleles have M. domesticus variants of H19 and Igf2 on distal Chr 7.
DNA bisulfite modification and sequencing.
Genomic DNA samples isolated from reciprocal CAST/DelTel7 and DelTel7/CAST E9.5 embryos were subjected to bisulfite modification, PCR amplification, subcloning, and sequencing as described elsewhere (10). For IC1 (H19 DMR) (see Fig. 6A, below), nested PCRs encompassing known M. domesticus/M. castaneus SNPs were carried out as described previously (see Table S1 in the supplemental material for primer modifications), with primers BMsp2t1 and BHha1t3 followed by BMsp2t2.2 and BHha2t4.2 (12). For IC2 (KvDMR1) (see Fig. 5A, below), the assays were performed as described with primers Kcnq1ot1 OF plus Kcnq1ot1 OR followed by Kcnq1ot1 IF plus Kcnq1ot1 IR (19). The parental alleles are distinguished by an A50C transversion, identified by sequencing CAST genomic DNA (M. domesticus A50/ M. castaneus C50). PCR products were cloned into the pGEM-T vector (Promega) and transformed into TOP10 cells (Invitrogen). Primer sequences are available in Table S1 of the supplemental material. For each DNA sample, we performed two independent bisulfite treatments; each bisulfite-treated DNA was amplified in two independent PCRs. We sequenced three to five cloned stands per PCR and analyzed the data using BiQ Analyzer (7).
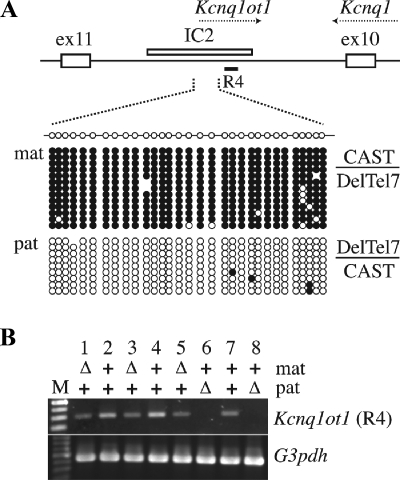
FIG. 5.
Normal imprinting at IC2 in reciprocal DelTel7 heterozygotes. (A) Diagram of the intron 10 region of Kcnq1, showing the position of IC2 (KvDMR1, white rectangle) (26), the structure of a 335-bp sequence containing the 31 CpG sites analyzed by bisulfite sequencing, and the region 4 fragment (R4) amplified by RT-PCR in panel B. Reciprocal DelTel7 heterozygous embryos were recovered at E9.5 from crosses between +/DelTel7 and (129CAST)F1 mice. Genomic DNA samples were analyzed by sodium bisulfite mutagenesis and sequencing. The entire IC2 subdomain is deleted in the DelTel7 allele such that mutant embryos are hemizygous for the wild-type IC2 CAST allele. The results show normal differential methylation of the paternal (pat; unmethylated) and maternal (mat; methylated) CAST alleles of IC2 in these embryos. Methylated CpG sites are represented by filled circles, and unmethylated sites are indicated by open circles. Omitted sites indicate sequencing ambiguities. (B) Allele-specific expression of Kcnq1ot1 in DelTel7 embryos. The expression of Kcnq1ot1 was assessed by RT-PCR (region 4 [R4] in panel A) (68) on embryonic RNA samples collected at E9.5 from reciprocal crosses between +/DelTel7 and (129CAST)F1 mice. For each embryo, the maternally (mat) and paternally (pat) inherited alleles are shown. Δ, Deltel7; +, wild type. Embryos with a wild-type CAST allele are numbers 1, 2, and 5 (pat CAST allele); numbers 6 and 7 have the mat CAST allele. RT-PCR for the housekeeping gene G3pdh was used as a positive control.
Allele-specific expression analysis.
Random-primed cDNA (SuperScript II) was generated from E9.5 placental and embryonic RNA samples (purified using TRIzol), collected from reciprocal crosses between +/DelTel7 and (129CAST)F1 mice. The H19 reverse transcription-PCR (RT-PCR) (see Fig. 6C, below) was carried out as described previously (66), with primers RT1 and RT2. The 640-bp product was digested with SmaI and Cac8I, which yielded fragments of 352, 244, and 44 bp for the 129 allele and 244, 222, 130, and 44 bp for the CAST allele. The 44-bp band is not visible in Fig. 5C, below. For Igf2, RT-PCR (see Fig. 6C, below) was carried out with primers Igf2F and Igf2R2. The 207-bp product was digested with Tsp5091, yielding multiple bands with the largest being 183 bp and 170 bp for the 129 and CAST alleles, respectively. For Kcnq1ot1 (see Fig. 5B, below), cDNA amplification was carried out with primers Lit1r4F and Lit1r4R as described elsewhere (68), yielding a 300-bp product. Primer sequences are available in Table S1 of the supplemental material.
RESULTS
Engineering of a 2.6-Mb terminal deletion by RMCT in ES cells.
The imprinted domain on distal Chr 7 is close to the Tel7q telomere, spanning ∼2.75 to 1.6 Mb from Mrgprd, the most distal gene identified on the current Chr 7 sequence (Ensembl release 46, August 2007) (Fig. 1A). Telomeric to the imprinted domain itself, the last 1.6-Mb region of Chr 7 contains several known and novel protein-coding genes. No imprinting effects have been described in this region. Because of this telomeric position, we considered the possibility of using the Cre-loxP system to engineer a terminal deletion leaving the proximal IC1 subdomain intact but deleting everything distal of Ins2, the most telomeric transcriptional unit known to be regulated by IC1 (15, 29). To establish the breakpoint for this truncation on Chr 7, first we derived ES cells carrying a promoterless loxP-neo-pA insertion targeted upstream of Ins2 (Fig. 1B). This allele, called Ins2I2loxP or I2loxP, was obtained by insertion of a loxP-PGK-loxP-neo-pA cassette at the Ins2 locus by homologous recombination in R1 ES cells to generate the Ins2I2 allele (or I2), followed by Cre-mediated deletion of the loxP-flanked PGK promoter (Fig. 2). ES cells carrying the I2loxP allele do not express the neo marker, are sensitive to G418, and provide a strong selection for efficient Cre-mediated site-specific integrations (28, 34). We constructed a replacement vector carrying a telomere seed (a cloned array of T2AG3 repeats) and a PGK promoter-loxP cassette suitable to activate the silent neo marker at I2loxP (Fig. 1B). Cre-mediated recombination between this linear vector and I2loxP can produce two new alleles: the desired truncation of Chr 7 by RMCT (DelTel7 allele) and a simple insertion of the vector (Tel7KI allele) (Fig. 1B).
We electroporated ES cells heterozygous for the I2loxP allele with the linearized replacement vector plus a Cre recombinase-expressing construct and selected for activation of the promoterless neo marker. From ∼50 G418-resistant clones recovered, 24 colonies were picked and expanded for Southern blot analysis. These ES cell clones all showed evidence of site-specific recombination at I2loxP. This was demonstrated by loss of the proximal 3.2-kb EcoRI band of I2loxP and gain of a new 3.8-kb band, indicating delivery of the PGK promoter at I2loxP (Fig. 3A and B, 3′ probe). In 22 clones, the distal 2.9-kb I2loxP band hybridizing to the 5′ probe was deleted, as expected for the structure of DelTel7 (Fig. 3B, 5′ probe). Two clones, such as clone 5 in Fig. 3B, showed a structure consistent with a simple insertion of the vector (Tel7KI allele), with retention of the 2.9-kb I2loxP band and no loss of distal Chr 7 sequences. Such an insertion at I2loxP might have occurred because of residual circular vector or via an intermediate array of multimerized vectors recircularized and inserted by Cre. The structures of both of these new alleles were also confirmed by PCRs amplifying novels junctions (Fig. 3A and C).
To provide cytogenetic evidence for the Chr 7 truncation, we performed DNA FISH on metaphase spreads from ES cells, using genomic PAC clones from regions proximal (H19) and distal (Th) to the I2loxP breakpoint as probes. Parental cells (I2loxP/+) as well as those with the simple insertion (Tel7KI/+, clone 5) gave signals for both the H19 and the Th probes, at the distal end of each Chr 7 homologue (Fig. 3D). Conversely, for clones 1 and 2 a single Chr 7 homologue was positive for both probes, whereas the other only gave a signal for the proximal H19 probe (Fig. 3D), as expected for cells heterozygous for the DelTel7 allele.
We also wanted to provide molecular evidence for the expected hemizygosity on distal Chr 7 in heterozygous DelTel7 cells, but this could not be achieved by demonstration of loss of heterozygosity at genetic markers in our ES cells. The R1 ES cells were derived from an F1 embryo between two different 129 substrains (42), but they are homozygous for SNPs distal of Ins2 on Chr 7. However, for the allele-specific studies at IC1 (see below), we eventually recovered informative embryos by crossing +/DelTel7 animals with mice carrying the M. mus castaneus variant of distal Chr 7. The CAST/DelTel7 progeny from such crosses were used to confirm the hemizygosity at two markers (Ascl2 and Fgf3) distal of I2loxP, providing further molecular evidence for the truncation of Chr 7 in the DelTel7 allele (R. Ho and L. Lefebvre, unpublished data). Furthermore, differential DNA methylation at IC2 did provide an epigenetic polymorphism to assess directly the hemizygosity in DelTel7 cells and to determine which parental homologue of Chr 7 was deleted in these cells (19). We performed a Southern blot analysis on ES cell genomic DNA digested with a methylation-sensitive enzyme cutting within the differentially methylated CpG-rich region of IC2 (Fig. 3E). Both parental epigenotypes were detected at IC2 in heterozygous I2loxP/+ and Tel7KI/+ ES cells (clone 5), confirming that the Chr 7 sequences distal of I2loxP have not been deleted in this interstitial insertion allele (Fig. 3E, lane 5). On the other hand, all of the ES clones carrying the DelTel7 allele show an unmethylated paternal allele but have lost the methylated maternal allele of IC2 (Fig. 3E). These results confirm that the original I2loxP allele was thus targeted on the maternal homologue of Chr 7 in the heterozygous cells used here and that the DelTel7 cells are missing the maternal end of Chr 7 (DelTel7/+, not +/DelTel7 heterozygotes).
Germ line transmission and stable maintenance of the truncated Chr 7 from viable heterozygous males.
The DelTel7/+ ES cells are viable in culture, with no apparent growth defect. The truncated Chr 7 variant appears stable in undifferentiated ES cells, as demonstrated by maintenance of the heterozygous genotype after several passages without selection for the engineered chromosome (L. Lefebvre, unpublished data). We then asked whether transmitting germ line chimeras could be obtained from these ES cells, which carry the truncated Chr 7 and the engineered Tel7q. Three independent DelTel7/+ ES cell clones were aggregated to wild-type diploid embryos to generate chimeras, using Tel7KI/+ ES cells as a control. For the DelTel7 clones, nearly all medium- and strong-contribution chimeras died perinatally (Table 1). This was not observed in control chimeras, several of which gave germ line transmission of the Tel7KI allele. Despite this lethality, some DelTel7 chimeras survived to weaning, and germ line transmission of the DelTel7 allele was obtained from one chimeric male.
TABLE 1.
Survival of chimeras between DelTel7/+ ES cells, or control Tel7KI/+ ES cells, and diploid embryosa
| ES cell line | No. of pups born | No. found dead or lost | No. of chimeras (no. of males) at relative levelb
|
||
|---|---|---|---|---|---|
| Strong | Medium | Weak | |||
| Tel7KI/+ #5 | 16 | 0 | 4 (4) | 3 (3) | 9 (3) |
| DelTel7/+ #1 | 12 | 4 | 0 | 0 | 8 (5) |
| DelTel7/+ #2 | 29 | 24 | 0 | 0 | 5 (4) |
| DelTel7/+ #3 | 15 | 4 | 0 | 1 (1) | 10 (7) |
| DelTel7/+ (all three cell lines) | 56 | 32 | 0 | 1 (1) | 23 (16) |
For each cell line aggregated and transferred to ICR recipient females, the table presents the total number of pups recorded at delivery and the number of pups found dead perinatally or lost before weaning.
Relative level of chimerism in surviving 6-week-old mice. Chimerism was estimated on the basis of the percentage of the coat contributed by Agouti R1 ES cells: weak, 0 to 25%; medium, 26 to 74%; strong, >75% chimerism. The numbers of male chimeras are given in parentheses.
The paternal heterozygous progeny (+/DelTel7) from the transmitting chimera are viable and fertile. We were thus able to follow the inheritance of the deletion allele from mature carrier males. The DelTel7 allele was recovered at the expected frequency from heterozygous males at weaning (46%) (Table 2, group A). We have now maintained the DelTel7 mouse line by paternal transmission for nine generations, with no apparent abnormal phenotype. The integrity and stability of the truncated Chr 7 was assessed by SKY analysis of eighth-generation primary embryonic fibroblasts. As in cells from a wild-type littermate, nonclonal aneuploidy and/or chromosomal breaks were documented in ∼10% of the fibroblasts analyzed, but none of the aberrations observed involved Chr 7 (see Fig. S1 in the supplemental material). The truncated Chr 7 of the DelTel7 allele is therefore stable in mice, compatible with germ line transmission in males, and is a silent mutation when paternally inherited.
The maternally inherited DelTel7 allele causes embryonic lethality at midgestation.
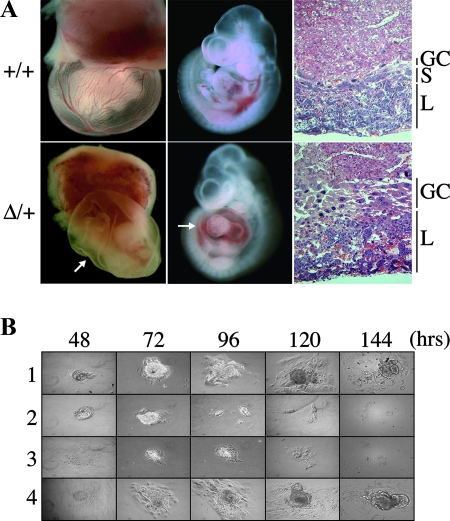
Heterozygous females inheriting the DelTel7 allele paternally are also viable and fertile. However, when litters from +/DelTel7 females were genotyped, no DelTel7/+ maternal heterozygotes were recovered at weaning (Table 2, group B). To analyze this imprinted phenotype further, we dissected litters obtained from crosses between heterozygous females and wild-type males at different developmental stages. We found that maternal transmission of DelTel7 is embryonic lethal at approximately E10 (Table 2, group B). Viable DelTel7/+ embryos were recovered prior to E10.0, but later stages were compromised by a defective development of the placenta, characterized by a lack of spongiotrophoblast and a thicker giant cell layer (Fig. 4A). Additional abnormalities were also observed at E10.5, notably, the absence of yolk sac blood and pericardial effusion, possibly as secondary consequences of placental vasculature defects (Fig. 4A).
FIG. 4.
Embryonic phenotypes cause by the DelTel7 deficiency. (A) Imprinted midgestational lethality phenotype in DelTel7/+ (Δ/+) maternal heterozygotes. Healthy and normal DelTel7/+ embryos were recovered at E9.5 (not shown). At E10.5 mutant conceptuses are characterized by an abnormal placenta and lack of blood in the yolk sac, as well as a greatly enlarged pericardium (arrows). Hematoxylin and eosin-stained histological sections of E10.5 placentas (left) show abnormal development of trophoblast lineages in DelTel7/+ heterozygotes, including the lack of spongiotrophoblast (S) and an increased giant cell (GC) population. L, labyrinth layer. (B) Preimplantation phenotype in DelTel7/DelTel7 homozygotes. The growth of attached blastocysts, cultured in the absence of leukemia inhibitory factor, was followed for 6 days, after which ICM outgrowths were collected and genotyped (Table 2, group C). Whereas wild-type (lane 1, 29%) and heterozygous (lane 4, 50%) embryos showed normal proliferation in this assay, no homozygous mutants were recovered, and 21% of the blastocysts failed to grow (lanes 2 and 3).
We also assessed the phenotype of mutant homozygous embryos by analyzing the progeny of heterozygous crosses. At E9.5, these crosses only yielded viable wild-type and heterozygous embryos, suggesting an early embryonic lethality in DelTel7 homozygotes (Table 2, group C). We looked at the genotypes of preimplantation-stage embryos by collecting blastocysts at E3.5. At that early stage, DelTel7/DelTel7 homozygous embryos are viable and indistinguishable from wild-type and heterozygous littermates (Table 2, group C). By culturing these blastocysts for 6 days in vitro, we found that embryos of all genotypes except homozygous mutant blastocysts showed normal inner cell mass growth (Fig. 4B).
The embryonic phenotype of DelTel7/+ embryos is rescued fully by a deletion of IC2 on paternal Chr 7.
The unmethylated paternal IC2 is associated with the silencing in cis of eight genes which are consequently transcribed preferentially from the maternal homologue (MEGs). A paternally inherited deletion of IC2 causes the loss of the Kcnq1ot1 ncRNA and the biallelic expression of these eight genes (MEGs) normally silenced on paternal Chr 7 (26, 45). Since this entire gene cluster is deleted in the DelTel7 allele and since the paternal DelTel7 heterozygotes are viable, we hypothesized that the absence of these IC2-regulated genes from the maternal DelTel7 allele is responsible for the imprinted lethality of DelTel7/+ embryos. To address this possibility, we asked whether expression of these MEGs from a paternal homologue with the IC2KO can rescue the embryonic lethality caused by DelTel7. A similar phenotypic rescue experiment has previously been performed for the placental overgrowth phenotype caused by loss of the IC2-regulated Phlda2 gene (59). Ascl2 is another MEG from the IC2 subdomain that is required for placental development; maternal transmission of the Ascl2 knockout allele causes a midgestational lethality (31, 32). By crossing heterozygous Ascl2+/KO females with males carrying a small 2.8-kb deletion of IC2 (KvDMR1 KO allele, or IC2KO) (26), we first showed that the IC2KO allele can rescue the Ascl2 deficiency (Table 2, group D). We then performed crosses between +/DelTel7 females and IC2KO heterozygous males. Whereas DelTel7/+ embryos die at midgestation, as described above, DelTel7/IC2KO mice are viable and were recovered at the expected frequency at weaning (Table 2, group E). Both male and female DelTel7/IC2KO mice are indistinguishable from their wild-type littermates and are fertile. Thus, the lethality observed upon maternal transmission of DelTel7 is a true imprinted embryonic phenotype and is not due to some incompatibility between the truncated allele and transmission through oogenesis itself.
The hemizygous IC2 shows stable imprinting in reciprocal DelTel7 heterozygotes.
To address possible epigenetic defects caused by the DelTel7 allele, we first analyzed imprinting at IC2 in reciprocal DelTel7 heterozygotes recovered at E9.5. This particular developmental stage was chosen since it precedes the lethality observed in maternal heterozygotes. DNA methylation patterns at IC2 were analyzed by sodium bisulfite sequencing of a 335-bp region of IC2 encompassing 31 CpG dinucleotides (Fig. 5A). Paternal and maternal hemizygotes showed a single epigenotype and maintenance of normal parental DNA methylation imprints at IC2 (Fig. 5A). Similarly, the Kcnq1ot1 ncRNA, transcribed from the paternal IC2 in wild-type embryos, was detected in all the embryonic samples analyzed, except those in which the DelTel7 deletion was paternally inherited (Fig. 5B, embryos 6 and 8).
Thus, the absence of DNA methylation at IC2 and expression of Kcnq1ot1 from the paternal allele is maintained normally in DelTel/+ embryos. As demonstrated by the rescue experiments described above, the presence of this unmethylated paternal IC2, via the long-range silencing it mediates, is responsible for the developmental phenotype of conceptuses of DelTel7/+ genotype.
Normal epigenetic imprinting at IC1 in the absence of the entire IC2 subdomain.
We designed the DelTel7 allele with a breakpoint distal of the IC1 subdomain (Fig. 1). Our analysis of the structure of DelTel7 in ES cells by Southern blot analysis and DNA FISH confirmed that the IC1 region is intact on the truncated Chr 7 of DelTel7 (Fig. 3B and D). This offers the possibility to look at the epigenetic function of IC1 independently of the IC2 subdomain on Chr 7. For allele-specific analyses at IC1, we collected heterozygous E9.5 embryos carrying the DelTel7 allele and a wild-type Chr 7 with M. mus castaneus alleles of distal Chr 7 genes. We first studied DNA methylation at 31 CpG sites within IC1 by sodium bisulfite sequencing. The 335-bp region of IC1 studied also encompasses strain-specific sequence polymorphisms, allowing the determination of the parental origin of each DNA strand amplified (67). This analysis showed that IC1 carries normal parental imprints with hypermethylation of the paternal allele in reciprocal DelTel7 heterozygotes (Fig. 6A).
These results were confirmed by Southern blot analysis of genomic DNA isolated from four CAST/DelTel7 embryos (Fig. 6B). In each heterozygote, only the maternal CAST allele is unmethylated at a ClaI site within IC1. The DNA methylation imprint at IC1 is inherited from the heterozygous males and maintained normally on the truncated paternal DelTel7 chromosome in these embryos.
We also analyzed the allele-specific expression of two IC1-regulated transcripts, H19 and Igf2, in embryonic RNA purified from reciprocal DelTel7 heterozygotes. The allele-specific RT-PCR results confirmed the normal imprinted expression of both genes in DelTel7 heterozygotes: irrespective of the genotype, H19 is only expressed from the maternal allele, and Igf2 is expressed only from the paternal allele (Fig. 6C). Together with the results from the DNA methylation studies, this confirms the independent function of IC1 on distal Chr 7 and the absence of essential interactions between the two subdomains for the initiation and maintenance of imprinted gene expression regulated by IC1.
DISCUSSION
The imprinted domain on distal Chr 7 provides a model system for the study of the mechanism, function, and evolution of genomic imprinting in mammals. Since the identification of Igf2 and H19 as the first linked imprinted genes (5, 14), several other loci transcribed exclusively or preferentially from one of the two parental homologues have been identified in this region. At least 10 protein-coding genes are known to be regulated by imprinting on distal Chr 7 and under the control of two imprinting centers carrying opposite germ line DNA methylation imprints. The developmental importance of some of these imprinted genes was first suggested by the analysis of embryos carrying an abnormal dosage of parental alleles of distal Chr 7 genes. Both maternal and paternal duplications for distal Chr 7 (partial disomies) cause developmental phenotypes (61). In humans, paternal uniparental disomy for the region of syntenic homology at 11p15.5 and the presence of androgenetic cell mosaicism in the placenta both cause BWS (38, 46). Individual imprinted genes implicated in these phenotypes have also been studied by gene targeting in the mouse. Here we have introduced a new approach for the analysis of telomeric gene clusters in the mouse genome and apply it to the analysis of the mechanism and function of imprinting on distal Chr 7. This work makes important contributions to three separate areas: (i) the development of RMCT as a new tool for functional genome analysis in the mouse; (ii) the epigenetic properties and roles of the imprinting centers IC1 and IC2 in the distal Chr 7 domain; and (iii) the developmental function of distal Chr 7 imprinted genes.
We have described here a new approach for chromosome engineering in ES cells, termed RMCT. This strategy was inspired by previous studies demonstrating that cloned arrays of (T2AG3)n telomeric repeats could be used as telomere seeds for chromosome fragmentation in mammalian cell lines (3, 21, 22, 33, 37). In RMCT, we refined such approaches by delivering the telomeric array to a defined chromosomal location in mouse ES cells, using the Cre-loxP site-specific recombination system. More importantly, we have shown here that the engineered telomere is stable in mouse ES cells and that the modified cells are germ line competent and can lead to the establishment of a mutant mouse line with the DelTel7 truncation. RMCT thus offers new opportunities for functional genome analysis, from the introduction of specific chromosomal truncations to the marking of specific telomeres and the analysis of telomere function and biology in vivo. Since it is based on the Cre-loxP system, RMCT could be applied to available genome-wide resources for chromosome engineering, such as the MICER clones, which are based on a bipartite Hprt selectable marker system similar to the one described here with the PGK-loxP-neo-pA cassette (1). Universal RMCT vectors compatible with these clones are currently under development.
The seeding of a new telomere at a distal location, following the introduction of a double-strand break by the I-SceI endonuclease or the random integration of a linear telomere seed vector, has previously been reported in ES cells (57, 64). In the I-SceI system, the gradual addition of telomere repeats by telomerase could be followed over 150 cell divisions in culture (64). We have not documented this dynamic process for the DelTel7 allele, but after several passages of the DelTel7/+ ES cells, the average telomeric length was similar for the wild-type and truncated Chr 7 (L. Lefebvre, L. Chavez, and P. Lansdorp, unpublished data). This observation, together with the mitotic stability of the DelTel7 allele, suggests that the end of the truncated chromosome is elongated and maintained in a stable structure after the Cre-mediated replacement reaction. We did however observe a high level of lethality in the chimeras obtained from the heterozygous DelTel7 ES cells. Although we cannot rule out the possibility that the engineered telomere is involved in this phenotype, it is more likely the deletion itself that is responsible for the observed lethality. Indeed, we have shown that the DelTel7 truncation and thus the original I2loxP targeted allele are both on the maternal Chr 7 homologue. The DelTel7/+ ES cells are thus expected to be deficient for all the MEGs in the IC2 subdomain. The cumulative loss of these gene products from distal Chr 7, or only of Cdkn1c, has previously been shown to cause cell autonomous defects and perinatal lethality in high-contribution chimeras, as described here for the DelTel7/+ ES cells (11, 49). Furthermore, if the engineered telomere somehow contributed to differentiation abnormalities in our chimeras, this effect would have to be erased upon germ line transmission, since we obtained viable and fertile paternal heterozygotes from surviving chimeras and Mendelian transmission of DelTel7 from heterozygous males. The embryonic lethality observed upon maternal transmission is also in support of a cell autonomous defect implicating loss of imprinted gene products in DelTel7/+ ES cells.
The presence of a telomere unbuffered by subtelomeric repeats is also known to affect the expression of nearby genes by stochastic epigenetic silencing. This telomere position effect (TPE) is a conserved phenomenon observed in several eukaryotic model organisms, including the mouse (57). Since RMCT can be used to deliver essentially any construct in the context of a de novo telomere seed, to specific chromosome sites, it offers new opportunities to study TPE and telomere biology in the mouse. Two transcriptional units are inserted next to the telomere repeats in the DelTel7 allele: a ubiquitous eGFP cassette as well as the PGK-neo marker regenerated upon Cre recombination. We observed that transcription from both of these promoters is regulated by stochastic TPE in ES cells (L. Lefebvre, unpublished results). Whether or not this effect extends to the distal Chr 7 endogenous genes, notably the nearby Ins2 locus, remains to be determined. In the case of Igf2, which is located ∼20 kb proximal of the introduced telomere, our RT-PCR and phenotypic data suggest that it is not subject to TPE on the expressed paternal allele in +/DelTel7 embryos.
In the context of imprinting, our characterization of the truncated Chr 7 allele DelTel7 allowed us to study the consequences of deleting ∼90% of an evolutionarily conserved imprinted domain (19). We showed that +/DelTel7 paternal heterozygous mice are viable and fertile (Fig. 7C). From these results, we conclude that there is no haploinsufficiency effect associated with the terminal deletion of ∼2.6 Mb on paternal Chr 7. In addition to the imprinted domain, this region also contains ∼20 currently known genes, including a cluster of Fgf genes (Fgf3, Fgf4, and Fgf15), Fadd, and Ccnd1, coding for cyclin D1. Gene knockout experiments reported for these genes are in support of this absence of haploinsufficiency effects in heterozygotes (20, 23, 48, 70, 71). Furthermore, the proliferation defects of DelTel7 homozygous blastocysts is consistent with the phenotype described previously for Fgf4-deficient embryos (a nonimprinted gene from distal Chr 7 deleted in the DelTel7 allele) (23).
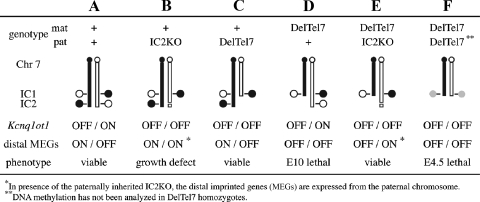
FIG. 7.
Model for genotype-phenotype correlation on distal Chr 7. The analyses of mutant mice carrying the deletion alleles IC2KO and DelTel7 reveal new genetic effects on distal Chr 7. The columns (A to F) represent mice of different genotypes on Chr 7, drawn schematically with a focus on the epigenetic status of IC1 and IC2 on the maternal (mat; black) and paternal (pat; white) homologues. The allelic expression levels of the ncRNA Kcnq1ot1 and several protein-coding genes under its regulation (distal MEGs) are shown for each genotype, as is the observed phenotype. Epigenetic imprinting at IC1 has not been analyzed in DelTel7/DelTel7 preimplantation embryos (F; shaded circles), but normal imprinting of IC1 is observed in animals A to E. Our results with the DelTel7 allele (C and D) confirm the independent epigenetic function of IC1 on Chr 7 in the absence of the entire distal domain. DelTel7 also gives a new imprinted embryonic phenotype upon maternal transmission (D). This phenotype is fully rescued by a paternal IC2KO (E), demonstrating that all of the developmentally required genes normally expressed from the maternal Chr 7 (as in animals A to C) are under IC2-mediated silencing on the paternal Chr 7.
Our results also provide genetic evidence that telomeric to Ins2, distal Chr 7 does not harbor any additional paternally expressed genes required for normal development. However, our analysis cannot rule out the presence of imprinted genes involved in more subtle adult phenotypes (as suggested for imprinted genes such as Mest [43], Htr2a [40], and Zac1 [63]). The ncRNA Kcnq1ot1, transcribed from the unmethylated paternal IC2, is therefore the only known PEG in this region.
We describe here a new imprinted phenotype, the embryonic lethality of DelTel7/+ heterozygotes (Fig. 7D). This phenotype could formally be a maternal effect, attributed to a specific incompatibility of the truncated chromosome with transmission through oogenesis. However, since the lethality of DelTel7 heterozygotes can be rescued by a paternally inherited IC2 deletion (Fig. 7E), our results confirm that it is the loss of maternally expressed genes (Fig. 7E, distal MEGs OFF) that is responsible for this phenotype. This conclusion is further supported by the fact that similar imprinted phenotypes, characterized by placental defects, have been described previously in embryos with a paternal disomy for distal Chr 7 or a maternally inherited Ascl2 deletion (31, 50).
The mechanism whereby the unmethylated IC2 leads to the silencing in cis of several proximal and distal genes is unknown, but recent studies suggest a role for transcriptional elongation and/or production of the ncRNA in this process (47; M. Higgins, unpublished results). The association of silencing with the production of a long ncRNA, recruitment of Polycomb group complexes, and acquisition of repressive histone modifications all point to a parallel between IC2 and X inactivation center-mediated silencing (26, 68). Consistent with a central role for IC2 on distal Chr 7, our genetic data show that all of the developmentally essential genes controlled by genomic imprinting and normally silenced on the paternal chromosome are under IC2 regulation. In the rescued DelTel7/IC2KO mice (Fig. 7E), the monoallelic expression of distal genes is reversed, being provided from the paternal Chr 7 with the IC2KO allele (26), a situation fully compatible with normal development.
Our methylation and expression studies at IC2 in DelTel7 heterozygotes showed that the expected parental epigenotype is maintained stably at the hemizygous IC2. These results do not address the erasure and establishment of imprints in the germ line of heterozygous embryos, since the IC2 allele analyzed here was inherited by the wild-type parent in our crosses. However, our genetic data from the rescue experiment show that wild-type mice are viable and recovered at the expected frequency from crosses between +/DelTel7 females and +/IC2KO males (Table 2, group E). This suggests that the maternal IC2 allele is methylated normally when inherited from +/DelTel7 females; otherwise, Kcnq1ot1 expression would silence the distal MEGs and cause embryonic lethality, as seen in the DelTel7 maternal heterozygotes (Fig. 7A and D).
Analysis of BWS patients has shown that epigenetic defects at IC2 or rearrangements between IC1 and IC2 can lead to loss of imprinting at IGF2 (41, 62). We show here that IC1 acts as an independent imprinting center on distal mouse Chr 7 and that Igf2 is imprinted normally in the absence of the entire distal IC2 subdomain. Based on our results in the mouse, we propose that IC1-independent biallelic IGF2 expression is caused by linkage with new cis-acting enhancers (in the case of translocations) or compensatory activation of nonimprinted promoters as a consequence of loss of CDKN1C expression (in the case of loss of DNA methylation at IC2). IGF2 is known to be biallelically expressed in certain normal tissues, such as adult liver and bone marrow (52), and compensatory interactions between IGF2 and CDKN1C have been documented in embryonic fibroblasts (30). The ability of IC1 to function as an independent imprinting center on Chr 7, despite the evolutionarily conserved linkage between the two subdomains, suggests that if a unique regulatory mechanism led to the emergence of long-range imprinting in the domain in ancestral mammals, the strict requirement for IC1-IC2 linkage has been lost in the mouse, at least as far as IC1 function is concerned. A similar evolutionary relaxation of constraint imposed by linkage has recently been described for Hox gene clusters, which show conservation of clustering and colinearity in several species (55).
Nearly 50% of sporadic BWS cases involve loss of imprinting at KCNQ1OT1 (62). Although the mechanism by which this occurs remains unknown, biallelic KCNQ1OT1 expression is known to cause the down-regulation of CDKN1C in BWS patients and, therefore, loss of maternally expressed genes (16). The terminal deletion presented here models this loss of distal maternal genes in the mouse and has the potential to offer new insights into the long-range regulation of Kcnq1ot1-mediated epigenetic silencing on Chr 7 and the function of the distal genes in imprinted developmental phenotypes. The rescue of the maternal deletion also provides genetic evidence demonstrating that inhibition of IC2-mediated silencing in trans could overcome the complex developmental phenotype caused by the simultaneous loss of several imprinted gene products.
Supplementary Material
Acknowledgments
We thank Titia de Lange for the cloned array of telomeric repeats, Jörn Walter and Jacquetta Trasler for help with the IC2 bisulfite sequencing protocols, Marisa Bartolomei for sharing results before publication, and Aaron Bogutz for help with blastocyst collections. We thank Wendy Robinson and Meaghan Jones for comments on the manuscript, and Vincenzo Pirrotta for bringing the Hox gene data to our attention.
All animal experiments were performed under certificate A03-0289 from the UBC Animal Care Committee and complied with the national CCAC guidelines to the care and use of experimental animals.
This work was supported in part by the CIHR operating grants MOP-64193 (to L.L.) and FRN-13687 (to A.N.). L.L. is a Michael Smith Foundation for Health Research Scholar and holds a Canada Research Chair.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, D. J., P. J. Biggs, T. Cox, R. Davies, L. van der Weyden, J. Jonkers, J. Smith, B. Plumb, R. Taylor, I. Nishijima, Y. Yu, J. Rogers, and A. Bradley. 2004. Mutagenic insertion and chromosome engineering resource (MICER). Nat. Genet. 36867-871. [DOI] [PubMed] [Google Scholar]
- 2.Ainscough, J. F., T. Koide, M. Tada, S. Barton, and M. A. Surani. 1997. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development 1243621-3632. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, M. A., V. J. Buckle, E. P. Evans, A. C. Porter, D. Rout, A. G. Smith, and W. R. Brown. 1993. Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 2127-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 71663-1673. [DOI] [PubMed] [Google Scholar]
- 5.Bartolomei, M. S., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351153-155. [DOI] [PubMed] [Google Scholar]
- 6.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 7.Bock, C. 2005. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 214067-4068. [DOI] [PubMed] [Google Scholar]
- 8.Caspary, T., M. A. Cleary, C. C. Baker, X. J. Guan, and S. M. Tilghman. 1998. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol. 183466-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerrato, F., A. Sparago, I. Di Matteo, X. Zou, W. Dean, H. Sasaki, P. Smith, R. Genesio, M. Bruggemann, W. Reik, and A. Riccio. 2005. The two-domain hypothesis in Beckwith-Wiedemann syndrome: autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Hum. Mol. Genet. 14503-511. [DOI] [PubMed] [Google Scholar]
- 10.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 222990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary, M. A., C. D. van Raamsdonk, J. Levorse, B. Zheng, A. Bradley, and S. M. Tilghman. 2001. Disruption of an imprinted gene cluster by a targeted chromosomal translocation in mice. Nat. Genet. 2978-82. [DOI] [PubMed] [Google Scholar]
- 12.Davis, T. L., J. M. Trasler, S. B. Moss, G. J. Yang, and M. S. Bartolomei. 1999. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 5818-28. [DOI] [PubMed] [Google Scholar]
- 13.Day, C. D., N. J. Smilinich, G. V. Fitzpatrick, P. J. deJong, T. B. Shows, and M. J. Higgins. 1999. The imprinted domain in mouse distal chromosome 7: reagents for mutagenesis and sequencing. Mamm. Genome 10182-185. [DOI] [PubMed] [Google Scholar]
- 14.DeChiara, T. M., E. J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64849-859. [DOI] [PubMed] [Google Scholar]
- 15.Deltour, L., X. Montagutelli, J. L. Guenet, J. Jami, and A. Paldi. 1995. Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev. Biol. 168686-688. [DOI] [PubMed] [Google Scholar]
- 16.Diaz-Meyer, N., C. D. Day, K. Khatod, E. R. Maher, W. Cooper, W. Reik, C. Junien, G. Graham, E. Algar, V. M. Der Kaloustian, and M. J. Higgins. 2003. Silencing of CDKN1C (p57KIP2) is associated with hypomethylation at KvDMR1 in Beckwith-Wiedemann syndrome. J. Med. Genet. 40797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duvillie, B., N. Cordonnier, L. Deltour, F. Dandoy-Dron, J. M. Itier, E. Monthioux, J. Jami, R. L. Joshi, and D. Bucchini. 1997. Phenotypic alterations in insulin-deficient mutant mice. Proc. Natl. Acad. Sci. USA 945137-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elson, D. A., and M. S. Bartolomei. 1997. A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol. Cell. Biol. 17309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engemann, S., M. Strodicke, M. Paulsen, O. Franck, R. Reinhardt, N. Lane, W. Reik, and J. Walter. 2000. Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 92691-2706. [DOI] [PubMed] [Google Scholar]
- 20.Fantl, V., G. Stamp, A. Andrews, I. Rosewell, and C. Dickson. 1995. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 92364-2372. [DOI] [PubMed] [Google Scholar]
- 21.Farr, C., J. Fantes, P. Goodfellow, and H. Cooke. 1991. Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 887006-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farr, C. J., M. Stevanovic, E. J. Thomson, P. N. Goodfellow, and H. J. Cooke. 1992. Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nat. Genet. 2275-282. [DOI] [PubMed] [Google Scholar]
- 23.Feldman, B., W. Poueymirou, V. E. Papaioannou, T. M. DeChiara, and M. Goldfarb. 1995. Requirement of FGF-4 for postimplantation mouse development. Science 267246-249. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson-Smith, A. C., H. Sasaki, B. M. Cattanach, and M. A. Surani. 1993. Parental-origin-specific modification of the mouse H19 gene. Nature 362751-755. [DOI] [PubMed] [Google Scholar]
- 25.Fitzpatrick, G. V., E. M. Pugacheva, J. Y. Shin, Z. Abdullaev, Y. Yang, K. Khatod, V. V. Lobanenkov, and M. J. Higgins. 2007. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 272636-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32426-431. [DOI] [PubMed] [Google Scholar]
- 27.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 51513-1523. [DOI] [PubMed] [Google Scholar]
- 28.Fukushige, S., and B. Sauer. 1992. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 897905-7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giddings, S. J., C. D. King, K. W. Harman, J. F. Flood, and L. R. Carnaghi. 1994. Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat. Genet. 6310-313. [DOI] [PubMed] [Google Scholar]
- 30.Grandjean, V., J. Smith, P. N. Schofield, and A. C. Ferguson-Smith. 2000. Increased IGF-II protein affects p57kip2 expression in vivo and in vitro: implications for Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. USA 975279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillemot, F., T. Caspary, S. M. Tilghman, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, D. J. Anderson, A. L. Joyner, J. Rossant, and A. Nagy. 1995. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9235-242. [DOI] [PubMed] [Google Scholar]
- 32.Guillemot, F., A. Nagy, A. Auerbach, J. Rossant, and A. L. Joyner. 1994. Essential role of Mash-2 in extraembryonic development. Nature 371333-336. [DOI] [PubMed] [Google Scholar]
- 33.Hanish, J. P., J. L. Yanowitz, and T. de Lange. 1994. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 918861-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardouin, N., and A. Nagy. 2000. Gene-trap-based target site for cre-mediated transgenic insertion. Genesis 26245-252. [DOI] [PubMed] [Google Scholar]
- 35.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 36.Hatada, I., and T. Mukai. 1995. Genomic imprinting of p57KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat. Genet. 11204-206. [DOI] [PubMed] [Google Scholar]
- 37.Itzhaki, J. E., M. A. Barnett, A. B. MacCarthy, V. J. Buckle, W. R. A. Brown, and A. C. G. Porter. 1992. Targeted breakage of a human chromosome mediated by cloned human telomeric DNA. Nat. Genet. 2283. [DOI] [PubMed] [Google Scholar]
- 38.Kaiser-Rogers, K. A., D. E. McFadden, C. A. Livasy, J. Dansereau, R. Jiang, J. F. Knops, L. Lefebvre, K. W. Rao, and W. P. Robinson. 2006. Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J. Med. Genet. 43187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10853-856. [DOI] [PubMed] [Google Scholar]
- 40.Kato, M. V., Y. Ikawa, Y. Hayashizaki, and H. Shibata. 1998. Paternal imprinting of mouse serotonin receptor 2A gene Htr2 in embryonic eye: a conserved imprinting regulation on the RB/Rb locus. Genomics 47146-148. [DOI] [PubMed] [Google Scholar]
- 41.Lee, M. P., R. J. Hu, L. A. Johnson, and A. P. Feinberg. 1997. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat. Genet. 15181-185. [DOI] [PubMed] [Google Scholar]
- 42.Lefebvre, L., N. Dionne, J. Karaskova, J. A. Squire, and A. Nagy. 2001. Selection for transgene homozygosity in embryonic stem cells results in extensive loss of heterozygosity. Nat. Genet. 27257-258. [DOI] [PubMed] [Google Scholar]
- 43.Lefebvre, L., S. Viville, S. C. Barton, F. Ishino, E. B. Keverne, and M. A. Surani. 1998. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet. 20163-169. [DOI] [PubMed] [Google Scholar]
- 44.Lefebvre, L., S. Viville, S. C. Barton, F. Ishino, and M. A. Surani. 1997. Genomic structure and parent-of-origin-specific methylation of Peg1. Hum. Mol. Genet. 61907-1915. [DOI] [PubMed] [Google Scholar]
- 45.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 361291-1295. [DOI] [PubMed] [Google Scholar]
- 46.Maher, E. R., and W. Reik. 2000. Beckwith-Wiedemann syndrome: imprinting in clusters revisited. J. Clin. Investig. 105247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mancini-Dinardo, D., S. J. Steele, J. M. Levorse, R. S. Ingram, and S. M. Tilghman. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 201268-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mansour, S. L., J. M. Goddard, and M. R. Capecchi. 1993. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development 11713-28. [DOI] [PubMed] [Google Scholar]
- 49.McLaughlin, K. J., H. Kochanowski, D. Solter, G. Schwarzkopf, P. E. Szabo, and J. R. Mann. 1997. Roles of the imprinted gene Igf2 and paternal duplication of distal chromosome 7 in the perinatal abnormalities of androgenetic mouse chimeras. Development 1244897-4904. [DOI] [PubMed] [Google Scholar]
- 50.McLaughlin, K. J., P. Szabo, H. Haegel, and J. R. Mann. 1996. Mouse embryos with paternal duplication of an imprinted chromosome 7 region die at midgestation and lack placental spongiotrophoblast. Development 122265-270. [DOI] [PubMed] [Google Scholar]
- 51.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 127035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morison, I. M., M. R. Eccles, and A. E. Reeve. 2000. Imprinting of insulin-like growth factor 2 is modulated during hematopoiesis. Blood 963023-3028. [PubMed] [Google Scholar]
- 53.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 908424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Negre, B., and A. Ruiz. 2007. HOM-C evolution in Drosophila: is there a need for Hox gene clustering? Trends Genet. 2355-59. [DOI] [PubMed] [Google Scholar]
- 56.Okabe, M., M. Ikawa, K. Kominami, T. Nakanishi, and Y. Nishimune. 1997. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 407313-319. [DOI] [PubMed] [Google Scholar]
- 57.Pedram, M., C. N. Sprung, Q. Gao, A. W. Lo, G. E. Reynolds, and J. P. Murnane. 2006. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol. Cell. Biol. 261865-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian, N., D. Frank, D. O'Keefe, D. Dao, L. Zhao, L. Yuan, Q. Wang, M. Keating, C. Walsh, and B. Tycko. 1997. The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum. Mol. Genet. 62021-2029. [DOI] [PubMed] [Google Scholar]
- 59.Salas, M., R. John, A. Saxena, S. Barton, D. Frank, G. Fitzpatrick, M. J. Higgins, and B. Tycko. 2004. Placental growth retardation due to loss of imprinting of Phlda2. Mech. Dev. 1211199-1210. [DOI] [PubMed] [Google Scholar]
- 60.Sauer, B., M. Whealy, A. Robbins, and L. Enquist. 1987. Site-specific insertion of DNA into a pseudorabies virus vector. Proc. Natl. Acad. Sci. USA 849108-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Searle, A. G., and C. V. Beechey. 1990. Genome imprinting phenomena on mouse chromosome 7. Genet. Res. 56237-244. [DOI] [PubMed] [Google Scholar]
- 62.Smilinich, N. J., C. D. Day, G. V. Fitzpatrick, G. M. Caldwell, A. C. Lossie, P. R. Cooper, A. C. Smallwood, J. A. Joyce, P. N. Schofield, W. Reik, R. D. Nicholls, R. Weksberg, D. J. Driscoll, E. R. Maher, T. B. Shows, and M. J. Higgins. 1999. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. USA 968064-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, R. J., P. Arnaud, G. Konfortova, W. L. Dean, C. V. Beechey, and G. Kelsey. 2002. The mouse Zac1 locus: basis for imprinting and comparison with human ZAC. Gene 292101-112. [DOI] [PubMed] [Google Scholar]
- 64.Sprung, C. N., G. E. Reynolds, M. Jasin, and J. P. Murnane. 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 966781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srivastava, M., S. Hsieh, A. Grinberg, L. Williams-Simons, S. P. Huang, and K. Pfeifer. 2000. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 141186-1195. [PMC free article] [PubMed] [Google Scholar]
- 66.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 123693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tremblay, K. D., J. R. Saam, R. S. Ingram, S. M. Tilghman, and M. S. Bartolomei. 1995. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9407-413. [DOI] [PubMed] [Google Scholar]
- 68.Umlauf, D., Y. Goto, R. Cao, F. Cerqueira, A. Wagschal, Y. Zhang, and R. Feil. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 361296-1300. [DOI] [PubMed] [Google Scholar]
- 69.Verona, R. I., M. R. Mann, and M. S. Bartolomei. 2003. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19237-259. [DOI] [PubMed] [Google Scholar]
- 70.Wright, T. J., R. Ladher, J. McWhirter, C. Murre, G. C. Schoenwolf, and S. L. Mansour. 2004. Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev. Biol. 269264-275. [DOI] [PubMed] [Google Scholar]
- 71.Yeh, W. C., J. L. Pompa, M. E. McCurrach, H. B. Shu, A. J. Elia, A. Shahinian, M. Ng, A. Wakeham, W. Khoo, K. Mitchell, W. S. El-Deiry, S. W. Lowe, D. V. Goeddel, and T. W. Mak. 1998. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 2791954-1958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.