Abstract
The repression of translation in environmentally stressed eukaryotic cells causes the sequestration of translation initiation factors and the 40S ribosomal subunit into discrete cytoplasmic foci called stress granules (SGs). Most components of the preinitiation complex, such as eIF3, eIF4A, eIF4E, eIF4G, and poly(A)-binding protein, congregate into SGs under stress conditions. However, the molecular basis of translation factor sequestration into SGs has not been clearly elucidated. Here, we report that proline-rich transcript in brain (PRTB) protein interacts with eIF4G and participates in SG formation. PRTB was recruited to SG under sodium arsenite and heat stress conditions. When overexpressed, PRTB inhibited global translation and formed SGs containing TIA-1, eIF4G, and eIF3. Knockdown of PRTB reduced the SG formation induced by sodium arsenite. These results suggest that PRTB not only is a component of SG formed by cellular stresses but also plays an important role in SG formation via an interaction with the scaffold protein eIF4G, which is associated with many translation factors and mRNAs.
The translation rate of eukaryotic mRNAs is modulated by environmental conditions, with that of many mRNAs being inhibited when cells are stressed by heat, high osmolarity, oxidative chemicals, etc. (15). The translation inhibition causes the sequestration of translation initiation factors and the 40S ribosomal subunit into discrete cytoplasmic foci, called stress granules (SGs), that are formed in environmentally stressed eukaryotic cells (1, 2, 14-17, 19, 26, 30). The SGs contain most of the components of the 48S preinitiation complex (i.e., small [but not large] ribosomal subunits, eukaryotic initiation factor 4G [eIF4G], eIF3, eIF4E, eIF2, and eIF2B), other RNA-binding proteins, such as T-cell-restricted intracellular antigen-1 (TIA-1) and T-cell-restricted intracellular antigen-related protein (TIAR), and untranslated mRNAs (1, 2, 15-17). As a consequence, mRNA translation generally is inhibited under stress conditions (6, 31).
eIF4G plays a pivotal role in the initiation of translation, since it recruits many translation factors [poly(A)-binding protein (PABP) (12), eIF4E (20, 23), eIF4A (13), and eIF3 (13)] and the translation modulator Mnk1 (a Ser/Thr kinase) to the 40S ribosomal subunit via protein-protein interactions (28, 37). Moreover, the signaling molecule TRAF2 has been shown to bind to eIF4GI, one of the two functional homologues of eIF4G, and to block proinflammatory signaling via the sequestration of TRAF2 at the SGs under stress conditions (18, 25). This indicates that eIF4G plays important roles in the regulation of cellular activities such as translation and signal transduction.
The proline-rich transcript of the brain (PRTB) protein, which is a 17-kDa protein, originally was isolated in a gene trap screen as a transcript expressed in the developing mouse inner ear (38). PRTB also is known as DAZAP2 [deleted-in-azoospermia (DAZ)-associated protein 2], which was identified as a protein interacting with the protein named DAZ (35), a germ-cell-specific RNA-binding protein. Phylogenetic analysis and structure prediction revealed that PRTB is highly conserved in vertebrates from zebra fish to humans and contains several potential Src homology 2 (SH2)-/SH3-binding sites throughout the protein, as well as a polyproline region at the C terminus (32). PRTB expression is reduced in untreated patients with multiple myelomas at both the mRNA and protein levels (32). In other words, the PRTB level is inversely correlated with the pathogenesis of multiple myelomas. Human PRTB is the orthologue of mouse PRTB that is expressed in various tissues during embryonic development and in the brain of adult mice (38). Sox6, which is a component of the BMP pathway (4), was shown to interact with PRTB protein during mouse cardiac differentiation (4). PRTB mRNA is upregulated in mouse osteoblasts during adhesion (33) and is highly expressed during the differentiation of mouse MC3T3-E1 osteoblasts (33). The level of rat PRTB mRNA increases when cells are exposed to ammonia and hypoosmotic conditions (36). The above-mentioned changes in the level of PRTB suggest that PRTB plays important roles in the physiological responses to various environmental conditions. However, the biological function of PRTB per se remains to be elucidated.
It is known that translation initiation factors, including eIF4G, eIF4E, and eIF3, are recruited in SGs under stress conditions, but the underlying molecular mechanisms remain elusive. Our investigation into the translational regulation of mRNAs under various conditions using the yeast two-hybrid (Y2H) system found that PRTB interacts with eIF4Gs (7), and this was confirmed by coimmunoprecipitation assays and immunocytochemistry. PRTB was recruited to SGs and colocalized with eIF4GI under stress conditions. Interestingly, the overexpression of PRTB induced the formation of SGs and resulted in the inhibition of general translation. Conversely, short interfering RNA (siRNA) treatment against PRTB reduced the level of SG formation after sodium arsenite (SA) treatment. These results indicate that PRTB has an important role in SG formation, possibly via an interaction with the scaffold protein eIF4G, which is associated with many translation factors and mRNAs.
MATERIALS AND METHODS
Plasmid construction.
To construct the pBCT-eIF4GII (amino acids [aa] 1 to 145) plasmid used in the Y2H screening, pSK(−)-eIF4GII (18) was digested with restriction endonucleases SmaI and BamHI, followed by being filled in with a Klenow fragment (New England Biolabs). The expression level of bait protein was controlled using pBCT plasmid, in which the 2μ region of pGBT9 (Clontech) is exchanged for CEN6/ARSH4 of p414GALL (ATCC) through AatII and NsiI sites. The two primers used for pBCT construction were 5′-CGCGGACGTCTTCATCACGTGCTATAAAAATAATT-3′ and 5′-CGCGATGCATTAGGACGGATCGCTTGCCTGTAAC-3′. The EcoRI Klenow fragment of pBCT was ligated with the SmaI/BamHI Klenow fragment of eIF4GII (aa 1 to 145). To construct the plasmid expressing green-fluorescent-protein (GFP)-fused eIF4GI (aa 41 to 168) protein, the nucleotide sequence encoding the open reading frame of eIF4GI (aa 41 to 168) was amplified from pSK-eIF4GI (18) by PCR with two primers, 5′-GGCCAAGCTTCGAACACGCCTTCTCAGC-3′ and 5′-TCCCCGCGGTCATGGCTGGTTCATCAAAAC-3′. The amplified fragments and pEGFP-C1 (Clontech) were digested with HindIII and KspI and were ligated to generate pEGFP-eIF4GI (aa 41 to 168). To make pEBG-eIF4GI (aa 41 to 168), the XhoI-BamHI fragment of pEGFP-eIF4GI (aa 41 to 168) was inserted into pEBG treated with BamHI and Klenow fragment.
To obtain the specific cDNA sequence of PRTB, cDNA corresponding to full-length (aa 1 to 168) human PRTB was amplified from a human testis library (Clontech) by PCR using the following two primers: 5′-CGGAATTCTCCCGGGGATGAACAGCAAAGGTCAA-3′ and 5′-CGGCATCCTCACCAGATGGTGTAGCCA-3′. The amplified cDNA fragment and plasmid pGAD424 (Clontech) were digested with SmaI and BamHI and then ligated together to construct pGAD424-PRTB. The sequence of PRTB was confirmed by sequencing. To construct pACT2-PRTB, the EcoRI/SalI fragment of pGAD424-PRTB was inserted into plasmid pACT2 through the EcoRI/XhoI site. To make the in-frame fusion protein construct, the plasmid was digested with XmaI and filled in with a Klenow fragment consecutively and then was religated. Two deletion variants of PRTB, pACT2-PRTB (aa 1 to 66) and pACT2-PRTB (aa 67 to 168), were generated by a yeast homologous recombination technique. To construct the eukaryotic expression plasmid for the PRTB gene, the EcoRI/BamHI fragment of the PRTB cDNA was inserted into the same restriction sites of pEGFP-C1. To construct pEGFP-PRTB (aa 1 to 66) and pEGFP-PRTB (aa 67 to 168), the EcoRI/BamHI fragments of pACT2-PRTB (aa 1 to 66) and pACT2-PRTB (aa 67 to 168), respectively, were inserted into plasmid pEGFP-C1 through the same restriction sites. To make the in-frame fusion protein construct, the plasmids were filled in with a Klenow fragment after digestion with HindIII and then were religated.
Antibodies and chemicals.
Antibodies against eIF4GI and eIF4GII were raised in rabbits (18). Anti-FLAG and antiactin antibodies were purchased from Sigma. Antibodies against GFP and TIA-1 and goat antibodies against eIF4GI, HuR, and eIF3b were purchased from Santa Cruz Biotechnology. Antibody against Hsp27 was purchased from Stressgen, and anti-S6 antibody was purchased from Cell Signaling Technology. Anti-L7a antibody was purchased from Ilamm. Glutathione S-transferase (GST) antibody was provided by Sung Ho Ryu (Postech). SA was purchased from Sigma.
Polyclonal antibody against human PRTB.
Antiserum against the human PRTB was raised by subcutaneous immunization of a rabbit with keyhole limpet hemocyanin-conjugated synthetic peptides corresponding to two amino acid sequences (aa 2 to 16 and aa 36 to 50) of human PRTB. Animals were bled after three booster injections. Antibody against PRTB was purified by using an Affigel-15 resin (Bio-Rad) conjugated with PRTB peptides.
Y2H screening.
Y2H screening was performed using the yeast strain PBN204 (MATα ura3-52 his3-200 ade2-101 trp-901 leu2-3,112 gal4Δ gal80Δ ura3::KANMX6-pGAL1-URA3 pGAL1-lacZ ade2::pGAL2-ADE2). pBCT-eIF4GII (aa 1 to 145) was cotransformed with the human testis Matchmaker cDNA library (Clontech) according to the manufacturer's protocol, with some modifications. The yeast cells containing eIF4GII-interacting baits were selected after being cultured on leucine, tryptophan, and uracil-free plates for 7 days. The colonies from these plates subsequently were assayed for β-galactosidase expression with a β-galactosidase filter assay (21). The prey plasmids were isolated and then retransformed into yeast cells with the bait or a negative control vector. The prey genes in real positive prey vectors, which showed positivity only with bait vector, were sequenced.
Cell culture and transfection.
COS7, HeLa, and 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with antibiotics (penicillin, 100 U/ml; streptomycin, 100 μg/ml) and 10% fetal bovine serum (HyClone) at 37°C. Transient transfection was performed as described elsewhere (3).
Pull-down assays and immunoprecipitation.
293T cells transfected with pEBG/pEBG-eIF4GI (aa 41 to 168) DNAs were lysed by being soaked in Triton lysis buffer (0.5% Triton X-100, 25 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 0.1 mM EDTA, 2 mM sodium orthovanadate, 1 μg aprotinin/ml, 1 μg/ml antipain, 1 μg/ml bestatin, 1 μg/ml pepstatin A, and 1 mM phenylmethylsulfonyl fluoride). Lysates were clarified by centrifugation at 14,000 × g at 4°C for 15 min and then were incubated with 20 μl glutathione-Sepharose 4B at 4°C for 3 h with or without 5 μg/ml RNase A (Sigma). Precipitated proteins were washed three times with the lysis buffer, resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE), and then transferred to a nitrocellulose membrane. Western blotting was performed with an antibody against PRTB to confirm the interaction.
Immunoprecipitation of FLAG-fused PRTB (FLAG-PRTB) was performed as described previously (18). Briefly, 293T cells were transfected with pFLAG or pFLAG-PRTB and lysed using the 0.5% Triton lysis buffer described above. The lysates underwent centrifugation at 14,000 × g at 4°C for 15 min. Anti-FLAG monoclonal antibody (4 μg) was incubated with 20 μl of protein G-agarose overnight in 1 ml phosphate-buffered saline (PBS) at 4°C. Lysates were treated with 10 μl of protein G-agarose at 4°C for 3 h and then incubated with antibodies conjugated with protein G-agarose at 4°C for 6 h. Precipitates were washed four times with lysis buffer and analyzed by SDS-6 to 15% PAGE, followed by Western blotting with antibodies against eIF4GI and eIF4GII.
Establishment of cell line expressing GFP and GFP-PRTB.
Plasmids (1 μg each) expressing GFP (pEGFP-C1) and GFP-PRTB (pEGFP-PRTB) were transfected into COS7 cells by electroporation. Beginning 48 h posttransfection, cells were maintained in DMEM containing G418 (600 μg/ml; Calbiochem). After 3 weeks of selection, G418-resistant cell colonies were picked and cultivated independently for further analysis.
Fluorescence microscopy.
Cells were grown on coverslips coated with 0.2% gelatin for 48 h and then washed three times with PBS. The cells were fixed with 3.5% (wt/vol) paraformaldehyde (Sigma) at room temperature (RT) for 12 min. After being washed three times with PBS, the cells were permeabilized with 0.1% Triton X-100 at RT for 2 min and then washed three times with PBS. The samples were soaked in blocking solution (PBS containing 1% bovine serum albumin) for 30 min at RT and then incubated with primary antibodies for 1 h at RT. After being washed with PBS, the samples were treated with Hoechst 33258 (Sigma) for 2 min at RT and then washed three times with PBS. Samples were treated with secondary antibodies conjugated with rhodamine tetramethyl isocyanate, fluorescein isothiocyanate (FITC), and/or Cy5 (Jackson ImmunoResearch Laboratories) for 1 h at RT. Finally, the coverslips were washed three times with PBS, placed on a glass slide, and then sealed with transparent nail polish. The fluorescent images were captured with a Zeiss Axioskop 2 plus microscope (Jena, Germany) and an Olympus FV1000 confocal microscope (Japan). Data were processed using standard software (Photoshop; Adobe, Mountain View, CA). The microscope-based observations of proteins were performed as described elsewhere (3).
Metabolic labeling.
GFP- and GFP-PRTB-expressing stable COS7 cell lines, which were more than 80% confluent on 60-mm culture dishes, were washed twice with PBS and then were incubated in methionine-free DMEM (BMS) for 1 h. Cells were incubated for a further 15 or 30 min after supplementation with [35S]methionine ([35S]Met) (500 μCi/ml; NEN Life Science Products), washed twice with ice-cold PBS, harvested, and then lysed as described above. The cell lysates were spun down, and the protein concentrations in cell lysates were measured using the Bradford assay. For the quantification of [35S]Met-labeled proteins, equal amounts (25 μg) of proteins were subjected to SDS-10% PAGE and analyzed by autoradiography. Alternatively, newly synthesized proteins labeled with [35S]Met were precipitated in 10% trichloroacetic acid (TCA) (wt/vol), and the precipitated proteins were dissolved in water and then subjected to liquid scintillation analysis (Packard).
Polysome analysis.
Cells from two 150-mm culture dishes containing GFP- and GFP-PRTB-overexpressing COS7 cell lines were treated with 25 μg/ml cycloheximide (Sigma) for 5 min at 37°C and harvested by scraping. The cytoplasmic extracts were prepared as described elsewhere (29). Cytoplasmic extracts of equal optical densities at 260 nm from GFP- and GFP-PRTB-overexpressing cells were layered over 0.5 to 1.5 M linear sucrose gradients and centrifuged at 40,000 rpm in a Beckman SW-41 Ti rotor for 2 h at 4°C. Gradients were fractionated using an ISCO tube piercer (Brandel) and a liquid chromatography system equipped with an absorbance monitor (254 nm) and a fraction collector (Bio-Rad).
Knockdown of PRTB using siRNA.
Duplex siRNAs targeting PRTB and a control siRNA were purchased from Bioneer Inc. (Korea). The siRNA sequence targeting PRTB was 5′-CAC CAU GUC AGC CGC AUU UdTdT-3′ (sense). The control siRNA sequence was 5′-CCU ACG CCA CCA AUU UCG UdTdT-3′. In order to transfect siRNA into cells, 100 nM of siRNA mixed with 6 μl of Lipofectamine 2000 (Invitrogen) and diluted in OptiMEM (Invitrogen) was added to each well of a 6-well plate and then incubated for 48 h before further analysis.
In vitro transcription and translation assays.
In vitro transcription and translation reactions were carried out as previously described (3), with some modifications. Briefly, plasmid DNA of pRL-CMV (Promega) linearized by HpaI was used for transcription with T7 RNA polymerase (Roche) at 37°C for 90 min. Translation reactions were carried out in micrococcal nuclease-treated rabbit reticulocyte lysates (Promega), as recommended by the manufacturer, in the presence of purified control GST protein (200, 400, and 800 ng) or GST-PRTB protein (200, 400, and 800 ng). Translation reactions were carried out at 30°C for 30 min. Translational efficiencies were monitored by Renilla luciferase activity as described previously (18). Luciferase activity in each reaction was normalized to the amount of protein measured by Bradford assays.
RESULTS
PRTB interacts with the translation factor eIF4G.
We recently showed that the translation factor eIF4GI is involved in the regulation of the tumor necrosis factor signaling molecule TRAF2 under stress conditions via a protein-protein interaction (18). In order to further understand the role of translation factors in the cellular responses to various cellular conditions, we performed Y2H analysis using the N-terminal region of eIF4GII (aa 1 to 145) as bait. Even though eIF4G is known to interact with other translation factors, such as eIF4A, eIF4E, eIF3, and PABP, as well as the signaling molecules Mnk1 and TRAF2, no protein has been reported to interact with the N-terminal region of eIF4GII. The PRTB protein was shown to interact with eIF4GII by Y2H screening (data not shown). A Y2H analysis with PRTB and eIF4GI, which is a homologue of eIF4GII, showed that PRTB also interacted with the N-terminal region of eIF4GI (aa 1 to 370), which is equivalent to the N-terminal region of eIF4GII used in the primary Y2H screening (Fig. 1A). These results suggest that both eIF4GI and eIF4GII interact with PRTB.
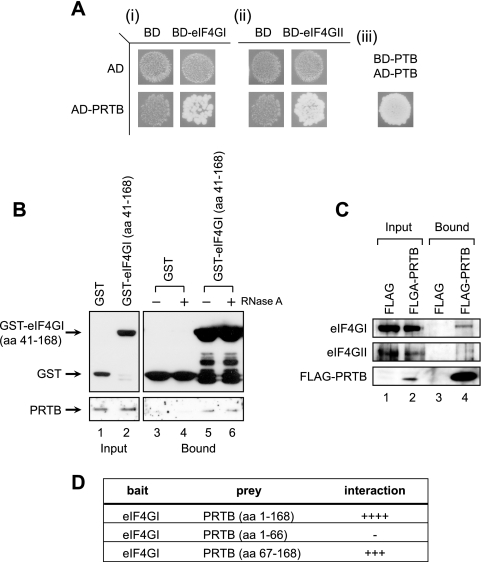
FIG. 1.
PRTB interacts with the N-terminal region of eIF4G. (A) Protein-protein interactions between PRTB and eIF4G proteins (eIF4GI and eIF4GII) were monitored using the Y2H system with a prey vector containing the PRTB gene encoding aa 12 to 168 and a bait vector containing either the eIF4GI gene encoding aa 1 to 370 (i) or the eIF4GII gene encoding aa 1 to 145 (ii). Yeast cells transfected with both bait and prey vectors were cultivated on a synthetic dextrose medium deficient in leucine, tryptophan, and uracil. Yeasts containing bait and prey vectors producing proteins interacting with each other grew on this medium (lower right images of i and ii), but the bait or prey with control pairs did not grow (the other images in i and ii). Bait and prey vectors containing the polypyrimidine-tract-binding protein (PTB) gene, which makes a homopolymer, were used as a positive control (iii) (27). BD, GAL4 DNA binding domain; AD, GAL4 activation domain. GST-pull down assays (B) and coimmunoprecipitation assays (C) were performed to verify the protein-protein interactions in eukaryotic cells. (B) 293T cells were transfected with a plasmid expressing GST (lanes 1, 3, and 4) or a plasmid expressing GST-tagged eIF4GI (aa 41 to 168) (lanes 2, 5, and 6). Input images (lanes 1 and 2) show proteins in 5% of the cell extracts before a pull-down assay was performed. Cell lysates treated with 5 μg/ml of RNase A (lanes 4 and 6) or left untreated (lanes 3 and 5) were pulled down using glutathione-Sepharose 4B, and coprecipitated PRTB was visualized by Western blotting with an antibody against PRTB. (C) 293T cells were transfected with a plasmid expressing FLAG (lanes 1 and 3) or a plasmid expressing FLAG-tagged PRTB (FLAG-PRTB) (lanes 2 and 4). Cell lysates were subjected to immunoprecipitation using a monoclonal anti-FLAG antibody, and coprecipitated eIF4GI and eIF4GII were visualized by Western blotting with antibodies against eIF4GI, eIF4GII, and FLAG (FLAG-PRTB). The input images show proteins in the 1% of lysates used in immunoprecipitation. (D) Y2H analysis of the eIF4G-binding region in PRTB. The numbers in parentheses indicate the PRTB amino acids included in the prey vector. A bait vector containing the portion of the eIF4GI gene encoding aa 41 to 370 was used. Positive and negative results in the Y2H analyses are indicated by plus and minus signs, respectively. The number of pluses gives an arbitrary indication of the strength of interaction.
The interaction between PRTB and the eIF4G proteins was further confirmed biochemically. We raised a polyclonal antibody that specifically interacts with PRTB proteins in 293T and HeLa cells (see Fig. S1 in the supplemental material). To confirm the interaction between PRTB and eIF4GI, a plasmid expressing the N-terminal region of eIF4GI (aa 41 to 168) fused with GST (GST-eIF4GI [aa 41 to 168]) was transfected into 293T cells, and a GST pull-down experiment was performed in the presence or absence of RNase A (Fig. 1B). Endogenous PRTB was coprecipitated with GST-eIF4GI (aa 41 to 168) in the presence or absence of RNase A (Fig. 1B, lanes 5 and 6) but not with GST (Fig. 1B, lanes 3 and 4). PRTB also was coprecipitated with the N terminus of eIF4GII (aa 1 to 130), which is fused with GST irrespective of RNase A treatment (data not shown). These results indicate that PRTB interacts with the N-terminal regions of eIF4GI and eIF4GII independently of cellular RNAs. In addition, a plasmid expressing the human PRTB fused with FLAG (FLAG-PRTB) was introduced into 293T cells, and an immunoprecipitation assay was performed with an anti-FLAG antibody (Fig. 1C). Endogenous eIF4GI and eIF4GII were coprecipitated with FLAG-PRTB (Fig. 1C, lane 4) but not with FLAG (Fig. 1C, lane 3). This indicates that full-length PRTB can interact with full-length eIF4G proteins.
Y2H analysis also was performed using several deleted prey clones of PRTB to determine the region of PRTB required for the interaction with eIF4GI (Fig. 1D). The interaction of eIF4GI (aa 41 to 370) with the C-terminal region of PRTB (aa 67 to 168), which is well conserved among vertebrates, was almost as strong as the interaction with full-length PRTB (aa 1 to 168). The N-terminal third of PRTB (aa 1 to 66) did not interact with eIF4GI. Taken together, these results indicate that the N-terminal region of eIF4GI most likely interacts with the C-terminal region of PRTB.
PRTB migrates to SGs under cellular stress conditions.
The eIF4G-PRTB interaction was further investigated in vivo by analyzing the subcellular localization of PRTB and eIF4GI using polyclonal antibodies against PRTB and eIF4GI (Fig. 2A). We analyzed the subcellular distribution of PRTB under normal, SA, and heat treatment (44°C) conditions. This approach was taken because the localization pattern of eIF4GI previously has been shown to change under stress conditions (1, 5, 19). Under normal conditions, PRTB was enriched in the nucleus in a punctate pattern but was absent from the nucleoli (Fig. 2A, image a). PRTB also was observed in the cytoplasm with punctate patterns (Fig. 2A, image a). Using a goat antibody against eIF4GI, the eIF4GI protein also was shown to localize to the cytoplasm and nucleus (Fig. 2A, image b). However, most of the cytoplasmic PRTB and eIF4GI proteins were localized to distinct regions of the cells (Fig. 2A, image c). Interestingly, when cells were treated with 400 μM of SA for 30 min, some of the cytoplasmic PRTB proteins were redistributed to distinct speckles, whereas the nuclear pattern of PRTB remained unchanged (Fig. 2A, image d). Some eIF4GI proteins also migrated to the cytoplasmic speckles (Fig. 2A, image e). A significant number of the PRTB and eIF4GI proteins in the cytoplasmic speckles were colocalized, as shown by yellow dots in the merged image; however, some of the PRTB and eIF4GI proteins remained in distinct regions, as shown by red and green signals, respectively (Fig. 2A, image f). Similarly, after heat treatment of cells at 44°C for 1 h, many of the cytoplasmic PRTB and eIF4GI proteins were colocalized in the cytoplasmic speckles (Fig. 2A, images g to i). These results suggest that PRTB and eIF4GI proteins migrated to distinct regions in the cytoplasm under SA and heat stress conditions.
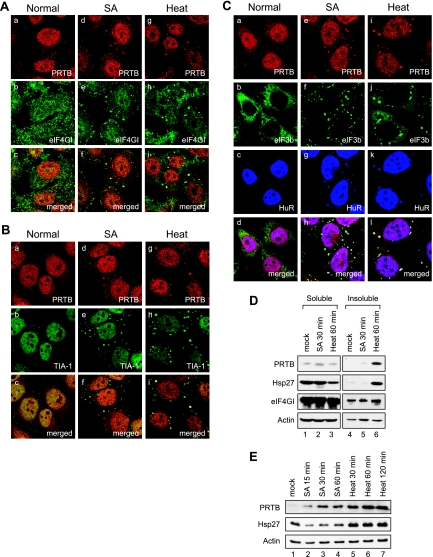
FIG. 2.
PRTB migrates to SGs under stress conditions. (A) HeLa cells were mock treated (images a to c), treated with 400 μM of SA for 30 min (images d to f), or treated with heat at 44°C for 1 h (images g to i). After the treatments, cells were subjected to immunocytochemistry analyses using an antibody against PRTB and a goat polyclonal antibody against eIF4GI. Donkey antibodies against rabbit immunoglobulin G (IgG) conjugated with rhodamine and against goat IgG conjugated with FITC were used as secondary antibodies. The localization patterns of PRTB and eIF4GI are shown in red (images a, d, and g) and green (images b, e, and h), respectively. Images c, f, and i are merged images of a and b, d and e, and g and h, respectively. (B) The same sets of samples shown in panel A were visualized using a rabbit antibody against PRTB and a goat antibody against TIA-1. (C) Distribution patterns of PRTB, eIF3b, and HuR before (Normal) and after SA or heat treatments were visualized using anti-PRTB, anti-eIF3b, and anti-HuR antibodies, respectively. Donkey antibodies against rabbit IgG conjugated with rhodamine, against goat IgG conjugated with FITC, and against mouse IgG conjugated with Cy5 were used as secondary antibodies. PRTB, eIF3b, and HuR proteins are shown in red (images a, e, and i), green (images b, f, and j), and blue (images c, g, and k), respectively. Merged images are shown in white (images d, h, and l). (D) Solubility of proteins under normal and stress conditions. HeLa cells were mock treated (lanes 1 and 4), treated with 400 μM of SA for 30 min (lanes 2 and 5), or treated with heat for 1 h (lanes 3 and 6). The HeLa cell extracts were fractionated as described elsewhere (5). The soluble (lanes 1 to 3) and insoluble (lanes 4 to 6) fractions were analyzed by Western blotting with anti-PRTB, anti-Hsp27, anti-eIF4GI, and anti-actin antibodies. (E) Protein levels of PRTB at various time points under stress conditions. HeLa cells were treated with 400 μM of SA for 15, 30, or 60 min (lanes 2 to 4) or with heat (44°C) for 30, 60, or 120 min (lanes 5 to 7). Cell extracts were analyzed by Western blotting with antibodies against PRTB, Hsp27, and actin.
Under stress conditions, translationally inactive mRNAs, which are associated with the 40S ribosomal subunit and most translation initiation factors, including eIF4G and eIF3, accumulate in SGs. The sequestration of mRNAs, 40S ribosomal subunits, and translation factors into SGs reduces the translation of many mRNAs. In order to determine the identity of the cytoplasmic speckles formed by SA and heat stresses in cells in which PRTB and eIF4GI exist, we performed immunocytochemistry with pairs of antibodies against PRTB and TIA-1, a well-known SG marker (Fig. 2B). TIA-1 proteins were localized mostly in the nucleus as green signals under normal condition (Fig. 2B, image b). However, a portion of them migrated to the SGs in the cytoplasm after SA and heat treatments (Fig. 2B, images e and h). Under these conditions, most of the cytoplasmic PRTB and TIA-1 proteins were colocalized at the cytoplasmic speckles, the SGs (Fig. 2B, images h and i). In order to rule out the possibility that these results were caused by phenomena specific to HeLa cells, we performed the same set of immunocytochemistry tests with COS7 cells. PRTB also migrated to the cytoplasmic speckles and colocalized with TIA-1 by SA and heat treatments (data not shown). The migration of PRTB to SGs was further investigated by immunocytochemistry with antibodies against eIF3b and HuR, which also are well known to migrate to SGs during SA and heat stresses (8, 15, 16). Although PRTB, eIF3b, and HuR were distributed separately in cells under normal conditions (Fig. 2C, images a to c), these proteins colocalized under stress conditions, as shown by white speckles in the cytoplasm (Fig. 2C, images h and l). These data suggest that PRTB migrates to SGs under stress conditions.
The migration of PRTB to SGs was further investigated by measuring the solubility of PRTB under normal and stress conditions (Fig. 2D). Under normal conditions, most of the PRTB proteins existed in the soluble fraction of HeLa cell extracts (Fig. 2D, lanes 1 and 4). After SA treatment for 30 min, some of the PRTB proteins were accumulated in the insoluble fraction, in which components of SGs are known to be enriched, although some of the PRTB proteins still remained in the soluble fraction (Fig. 2D, lanes 2 and 5). Upon heat treatment for 1 h, however, the levels of PRTB in the insoluble fraction were markedly increased (Fig. 2D, lane 6), whereas the levels of PRTB in the soluble fraction were slightly reduced (Fig. 2D, lane 3). Hsp27 and eIF4GI proteins also were accumulated in the insoluble fraction by heat treatment (Fig. 2D, lane 6), as shown previously (5, 18).
Interestingly, the total protein levels of PRTB seem to be increased under stress conditions (Fig. 2D). Therefore, we measured total protein levels of PRTB at various time points after SA and heat treatments (Fig. 2E). The levels of Hsp27 protein also were monitored as a control (Fig. 2E), because this protein is known to be induced by heat stress (5). Total protein levels of PRTB began to be elevated by 15 min of SA treatment and continued to rise until 60 min (Fig. 2E). Even more marked increases in the PRTB levels were observed in cells heat treated for 30 min (Fig. 2E, lane 6), and the high level of PRTB remained for 120 min (Fig. 2E). As expected, the levels of Hsp27 protein also were increased under heat stress conditions (Fig. 2E, lanes 5 to 7). On the other hand, the Hsp27 levels were slightly reduced with SA treatment (Fig. 2E, lanes 2 to 4). Taken together, these data indicate that the PRTB levels are markedly increased under stress conditions.
Overexpression of PRTB protein induces SG formation.
In order to investigate further the function of PRTB under stress conditions, we overexpressed PRTB using GFP-fused PRTB. At first, the subcellular localization of PRTB was analyzed using GFP-PRTB (Fig. 3). The control GFP was distributed uniformly in the cytoplasm and nucleus (Fig. 3A, image b), as reported by Gerdes and Kaether (9). On the other hand, GFP-PRTB showed a punctate distribution in the cytoplasm and nucleus of COS7 cells (Fig. 3A, image e). Interestingly, GFP-PRTB speckles were observed only in the cytoplasm in some cells, only in the nucleus in some cells, and in both the cytoplasm and nucleus in the remaining cells. The proportions of cytoplasmic and nuclear speckles varied by the cell line tested, with the former being higher in COS7 cells and the latter being higher in HeLa cells (data not shown). This explains the apparent discrepancy between the nuclear speckle pattern reported by Hamilton et al. (11) and the cytoplasmic speckle pattern reported by Shi et al. (32), which we attribute to their use of different cell lines. In addition, the portion of PRTB that localizes in cytoplasmic speckles was proportional to the expression level of PRTB, since the number of cells containing GFP-PRTB in cytoplasmic speckles was increased in a dose-dependent manner by the amount of transfected GFP-PRTB plasmid (data not shown).
FIG. 3.
Overexpressed PRTB proteins are localized in SGs. (A) COS7 cells were subjected to immunocytochemistry using polyclonal antibodies against eIF4GI (images a to f) and eIF3b (images g to i) 48 h after transfection with pEGFP (images a to c) and pEGFP-PRTB (images d to i). Antibodies against rabbit immunoglobulin G (IgG) and goat IgG conjugated with rhodamine were used as secondary antibodies. GFP and GFP-fused PRTB appear green (images b, e, and h), and eIF4GI and eIF3b proteins appear red (images a, d, and g). Images c, f, and i are merged images of images a and b, d and e, and g and h, respectively. (B) COS7 cells expressing GFP and GFP-PRTB were subjected to immunocytochemistry using anti-TIA-1 antibody. TIA-1 protein appears red (images a and d), since rhodamine-conjugated secondary antibody against goat IgG (upper two rows) was used in the immunocytochemistry. GFP and GFP-PRTB appear green (images b and e). Images c and f show the merged images of a and b and d and e, respectively.
eIF4GI was recruited to the cytoplasmic speckles by GFP-PRTB overexpression and colocalized with PRTB at the cytoplasmic speckles; it also colocalized in dispersed patterns, as indicated by the yellow speckles in the merged images in image f of Fig. 3A. To further investigate the functions of PRTB, stable cell lines expressing either GFP or GFP-PRTB were generated using the plasmid pEGFP-PRTB and COS7 cells. The same patterns of distribution of GFP, GFP-PRTB, and eIF4GI were observed in these stable cell lines (data not shown). In order to rule out adverse effects of GFP fusion on the subcellular localization of PRTB, we performed similar experiments using FLAG-PRTB in COS7 cells. eIF4GI colocalized with FLAG-PRTB with the same cytoplasmic speckle pattern as that for GFP-PRTB (data not shown). This indicates that the cytoplasmic speckle is not caused by the fusion of GFP but is caused by the overexpression of PRTB per se. The effect of PRTB on the localization of other initiation factors also was examined. We investigated the subcellular localization of eIF3b to monitor the behavior of the eIF3 complex, which revealed that eIF3b also was colocalized with PRTB at the cytoplasmic speckles in the cells overexpressing PRTB (Fig. 3A, images g to i).
We next tested whether the cytoplasmic speckles induced by the overexpression of PRTB are SGs by using immunocytochemistry with an antibody against TIA-1, since the expression level of PRTB was increased by SA and heat stresses (Fig. 2E). Most of the TIA-1 proteins reside in the nucleus (Fig. 3B, image a). On the other hand, some TIA-1 proteins migrated to the cytoplasm and were concentrated in particular regions of PRTB-overexpressed cells (Fig. 3B, images d and f). The cytoplasmic TIA-1 was colocalized exactly with GFP-PRTB at the cytoplasmic speckles, as indicated by the yellow speckles in image f of Fig. 3B. The colocalization of TIA-1 with GFP-PRTB was not changed even in the SA-treated condition (data not shown), which revealed that the cytoplasmic speckles induced by the overexpression of PRTB are SGs. Taken together, these results suggest that overexpressed PRTB proteins induce SGs, which recruit translation initiation factors such as eIF4G, eIF3, and TIA-1 into distinct regions in the cytoplasm.
The C-terminal two-thirds of PRTB protein mediates its recruitment to SGs.
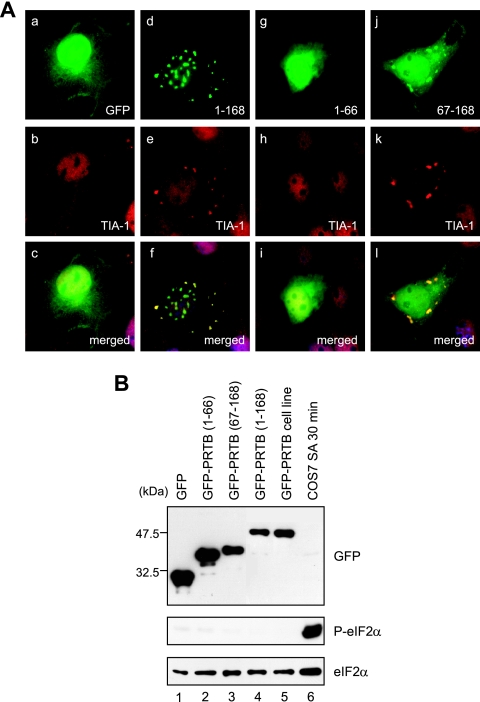
To understand how PRTB is recruited to SGs and induces SG formation, we fused PRTB deletion mutants to GFP and expressed them in COS7 cells in order to analyze their subcellular localization. In PRTB protein, there is no motif or conserved domain, such as an RNA recognition motif, that is known to be required for SG formation, so we divided PRTB into two portions: an N-terminal portion (aa 1 to 66), which does not interact with eIF4GI, and a C-terminal portion (aa 67 to 168), which interacts with eIF4GI. Each of these portions was fused with GFP. In all cases, double labeling with TIA-1 was included to positively identify SGs (Fig. 4A). Overexpression of GFP-PRTB (aa 67 to 168) showed cytoplasmic speckles that colocalize with TIA-1 (Fig. 4A, images j to l) in a manner similar to that of full-length PRTB (GFP-PRTB [aa 1 to 168]) (Fig. 4A, images d to f). In contrast, GFP-PRTB (aa 1 to 66) overexpression showed the same localization pattern (Fig. 4A, image g) as that of GFP alone (Fig. 4A, image a), and TIA-1 in cells overexpressing the N-terminal PRTB did not localize in cytoplasmic speckles (Fig. 4A, image h). When we analyzed the subcellular localization of eIF4GI in the cells overexpressing GFP-fused PRTB deletion mutants, we found that eIF4GI was colocalized with GFP-PRTB (aa 67 to 168), but not with GFP-PRTB (aa 1 to 66), in the cytoplasmic speckles (data not shown). The punctate pattern of full-length GFP-PRTB in the nucleus was not observed for the N- or C-terminal PRTB deletion mutants (Fig. 4A, compare image d to images g and j), which indicates that full-length PRTB is required for the distinct nuclear localization. From these data, we concluded that the C-terminal two-thirds of PRTB is necessary and sufficient for the induction of SGs.
FIG. 4.
C-terminal two-thirds of PRTB has an important role in formation of SGs. (A) COS7 cells were transiently transfected with GFP, GFP-PRTB (aa 1 to 168), or GFP-fused PRTB deletion mutants (aa 1 to 66 and aa 67 to 168). Numbers depict the first and last residues in the constructs. After transfections, cells were subjected to immunocytochemistry using a goat polyclonal antibody against TIA-1. GFP and GFP-PRTB appear green. TIA-1 protein appears red, because a rhodamine-conjugated secondary antibody against goat immunoglobulin G was used in the immunocytochemistry. Images c, f, i, and l are merges of images a and b, d and e, g and h, and j and k, respectively. (B) COS7 cell extracts from the sets of transfections shown in panel A were analyzed by Western blotting with antibodies against GFP, phosphorylated eIF2α (P-eIF2α), and eIF2α. Equal amounts of COS7 cell extracts treated with SA (400 μM for 30 min) were used as a control.
In order to rule out the possibility that the induction of SGs by GFP-PRTB is due to the transfection and that the overexpression of proteins causes the phosphorylation of eIF2α and SG formation, we monitored the level of eIF2α phosphorylation in cells expressing GFP-PRTB and its deletion mutants by Western blotting with antibodies against phosphorylated eIF2α and eIF2α (Fig. 4B). The phosphorylation of eIF2α was strongly induced by SA treatment in COS7 cells for 30 min (Fig. 4B, lane 6). However, overexpression of GFP, GFP-PRTB, and its two deletion mutants did not induce the phosphorylation of eIF2α (Fig. 4B, lanes 1 to 5). These data indicate that the induction of SGs by GFP-PRTB is not attributable to the phosphorylation of eIF2α.
Knockdown of PRTB results in reduction of SG formation.
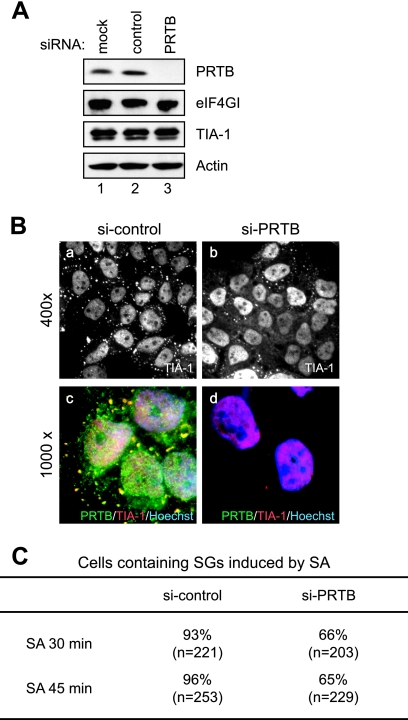
The role of PRTB in SG formation also was investigated by knocking down PRTB expression by using an siRNA against PRTB. Synthetic siRNA against PRTB reduced the expression of PRTB protein (Fig. 5A). The effect of PRTB knockdown on SG formation was monitored immunocytochemically by using an antibody against TIA-1 and HeLa cells treated with SA (Fig. 5B). Treatment with siRNA against PRTB reduced the number of cells showing more than four SGs after SA treatment by about 30% (Fig. 5C). In addition, the average number of SGs in cells treated with siRNA against PRTB also was reduced. The SG formation in the PRTB knockdown cells by SA might be due to other SG inducers, such as TIA-1 (22) and G3BP (34). These data indicate that the knockdown of PRTB partially disrupts SG formation.
FIG. 5.
Effect of PRTB knockdown on SG formation. (A) Level of PRTB protein in cells treated with an siRNA against PRTB mRNA. Levels of PRTB, eIF4GI, TIA-1, and actin proteins were analyzed by Western blotting using antibodies against PRTB, eIF4GI, TIA-1, and actin, respectively. HeLa cells were used in the Western blotting before treatment (lane 1) and in treatments with control siRNA (lane 2) and synthetic siRNA against PRTB (lane 3) for 48 h. (B) Effect of siRNA against PRTB on SG formation. HeLa cells were treated with control siRNA (si-control) (images a and c) and siRNA against PRTB (si-PRTB) (images b and d) for 48 h and then were treated with 400 μM of SA for 30 min. SG formation was monitored by immunocytochemistry using anti-TIA-1 antibody (images a and b). Donkey antibodies against rabbit immunoglobulin G conjugated with FITC and against goat immunoglobulin G conjugated with rhodamine were used as secondary antibodies. Merged images of PRTB (green), TIA-1 (red), and Hoechst 33258-stained nuclei (blue) are shown in images c and d. Microscopic magnifications of the pictures are ×400 (images a and b) and ×1,000 (images c and d). (C) Proportions of cells containing SGs. After treatment with SA for 30 and 45 min, HeLa cells were prepared as described for panel B. The numbers of cells containing more than four SGs were counted under a fluorescence microscope. The total numbers of the cells observed are listed in parentheses, and the proportions of SG-containing cells among the observed cells are listed as percentages.
Overexpression of PRTB reduces translational efficiency.
The congregation of translation factors into specific regions (i.e., the SGs) induced by the overexpression of PRTB strongly suggests that PRTB modulates mRNA translation. To further investigate this, we measured the rate of incorporation of [35S]Met into proteins in stable COS7 cell lines overexpressing GFP-PRTB or GFP (as the control). The COS7 cells were labeled with 200 μCi/ml [35S]Met for 15 min (Fig. 6A, lanes 1 and 2) or 30 min (Fig. 6A, lanes 3 and 4), and then proteins were resolved on SDS-10% PAGE. Newly synthesized proteins were observed by autoradiography. As shown in Fig. 6A, the level of newly synthesized protein was significantly lower in GFP-PRTB-overexpressing cells than in GFP-overexpressing cells (Fig. 6A, compare lanes 2 and 4 to lanes 1 and 3, respectively). Coomassie brilliant blue staining of proteins, which reflects the total amount of protein in cells, revealed that both the overall pattern and total amounts of protein were similar in the two cell types (Fig. 6B). The difference in protein incorporation of [35S]Met between GFP-PRTB-overexpressing and GFP-overexpressing cells was reduced after 30 min of incubation with [35S]Met (differences of ∼4.5- and 1.5-fold after 15 and 30 min of incubation, respectively) (Fig. 6C). This indicates that the rate of protein synthesis is lowered in PRTB-overexpressing cells, although it is not halted altogether.
FIG. 6.
Overexpression of PRTB represses translation of mRNAs. Newly synthesized proteins were monitored by metabolic labeling of proteins with [35S]Met. (A) Incorporation of [35S]Met into proteins for 15 min (lanes 1 and 2) and 30 min (lanes 3 and 4) was monitored by autoradiography. Samples (25 μg each) from GFP-overexpressing (lanes 1 and 3) and GFP-PRTB-overexpressing (lanes 2 and 4) cells were analyzed by SDS-10% PAGE. (B) The set of samples used for panel A was subjected to SDS-10% PAGE and stained with Coomassie brilliant blue. (C) Aliquots of each [35S]Met-labeled protein were precipitated by 10% TCA, and the radioactivities in the precipitated proteins were measured by a liquid scintillation counter. The mean counts per minute (CPM) are shown for control cells (open bars) and PRTB-overexpressing cells (solid bars). Each TCA precipitation experiment was performed independently four times. The lines depict standard deviations. (D) Polysome profiles of (i) GFP- and (ii) GFP-PRTB-overexpressing cell lines. Cells were subjected to a sucrose gradient analysis. Absorbance at 254 nm of the sucrose gradient was measured continuously, and the positions of the 40S and 60S ribosomal subunits, 80S monosome, and polysome are indicated.
The effect of PRTB overexpression on translation also was analyzed by comparing the ribosome profiles of the cells using a sucrose density gradient centrifugation method (Fig. 6D). The identities of the observed peaks were confirmed by immunoblotting of the individual peaks with antibodies against ribosomal proteins S6 and L7a (data not shown). The levels of ribosomal subunits participating in translation (the 80S monosome and polysome) were reduced in cells overexpressing PRTB, as indicated by the lower peak heights of the 80S monosome and polysome (Fig. 6D, compare graph i to graph ii). This suggests that PRTB overexpression inhibits translation initiation by delaying the formation of the 80S complex from the 40S and 60S subunits of ribosomes. The lower translation efficiency of the PRTB-overexpressing cells likely causes the slower growth of the PRTB-overexpressing cell line compared to that of the control cell line.
DISCUSSION
Most components of the 48S translation preinitiation complex congregate into SGs under stress conditions. However, little is known about the molecular basis of the sequestration of the translation factors into SGs. Here, we present evidence that PRTB plays an important role in the recruitment of translation factors into SGs. First, PRTB physically interacts with eIF4G, which in turn interacts with eIF3, eIF4A, eIF4E, and PABP; this interaction was confirmed by the Y2H system (Fig. 1A), coimmunoprecipitation (Fig. 1B), and immunocytochemistry (Fig. 2). Second, PRTB migrates to SGs under SA and heat stress conditions, and PRTB protein expression is greatly induced under these conditions (Fig. 2). The identity of the SGs was confirmed by the colocalization of PRTB with eIF4GI, eIF3, and TIA-1. Third, SG was induced by the overexpression of PRTB (Fig. 3). Fourth, knockdown of PRTB gene expression using a PRTB-specific siRNA inhibited SG formation induced by SA treatment (Fig. 5). Fifth, the overexpression of PRTB resulted in the repression of gene expression at the level of translation, as indicated by the delayed incorporation of [35S]Met into proteins (Fig. 6A and C). The level of formation of the 80S ribosomal complex, which is required for the initiation of translation, was reduced in the PRTB-overexpressing cells (Fig. 6D). This reduction in translation probably is due to the sequestration of translation initiation factors, mRNAs, and 40S ribosomal subunits into the SGs induced by PRTB overexpression.
Several proteins have been shown to be involved in the induction of SG formation. For instance, the overexpression of TIA-1 was shown to induce SG formation via prion-like aggregation of the protein (10), and its knockout resulted in an elevated level of translation of mRNAs containing the TIA-1-binding motif (22). SG assembly either is dominantly induced by overexpression of the RasGAP-associated endonuclease G3BP or is inhibited by the expression of a central domain of G3BP (34). Similarly, overexpression of fragile X mental retardation protein (FMRP) resulted in SG formation and repressed the translation of a cotransfected gene (24). All of these SG-inducing proteins are RNA-binding proteins, and they have not been suggested to be directly related to translation initiation factors or the 40S ribosomal subunit, which is sequestered in the SGs. On the other hand, no RNA recognition motif or other known RNA-binding motif exists in PRTB. Here, we have shown that the C-terminal portion of PRTB, which interacts with eIF4GI, is important for SG formation. Therefore, it is likely that PRTB is associated with a stalled preinitiation complex via a protein-protein interaction with the N-terminal region of eIF4G (which is a component of the preinitiation complex) under stress conditions. It is likely that PRTB, translational initiation factors, mRNAs, and mRNA-binding proteins cooperatively function in SG formation. However, the functional relationship among the SG components in SG formation remains to be elucidated.
Interestingly, the expression of the PRTB protein was significantly induced by SA and heat treatments (Fig. 2E). In addition, the overexpression of PRTB led to the recruitment of the translation factors and components of SGs (Fig. 3). Therefore, an increased PRTB level seems to be important for the induction of SG formation. However, it remains to be determined how the PRTB level is regulated under normal and cellular stress conditions.
Based on the above information, we describe a hypothetical mechanism for SG formation. When environmental stresses are applied to cells, translation initiation factors and the 40S ribosomal subunit complex stall on an mRNA, without the formation of the 80S translation complex, either by the phosphorylation of eIF2 (17) or by the depletion of ATP and GTP (16). This RNA-protein complex is further stabilized and grows by associations with RNA-binding proteins such as TIA-1, G3BP, and FMRP (which interact with mRNAs and oligomerize themselves) and/or by the association with PRTB protein that interacts with eIF4G. The increased PRTB level under stress conditions further stimulates SG formation. Either RNA-binding proteins or PRTB protein appear to be associated first with the stalled preinitiation complex, since either the RNA-binding proteins or PRTB induces SG formation when it is overexpressed.
The experiments reported herein provide new clues for understanding the molecular mechanism of SG formation and the sequestration of translation factors and signaling molecules into an SG, which is a key subcellular structure modulating cellular responses to environmental stresses. Further investigations are required to understand the switching mechanism of SG formation and the physiological role(s) of PRTB in the nucleus.
Supplementary Material
Acknowledgments
We thank Sung Ho Ryu (Department of Life Science, Pohang University of Science and Technology) and Kunsoo Rhee (Department of Biological Sciences, Seoul National University) for anti-GST antibody and purified GST-PRTB protein, respectively.
This work was supported in part by the Program for the Training of Graduate Students in Regional Innovation, which was conducted by the Ministry of Commerce, Industry, and Energy of the Korean Government, by grants from MOST and NCRC (R15-2004-033-05001-0), grant FPR05B 1-310 of the 21C Frontier Functional Proteomics Project from KMST, grant R17-2007-089-01001-0 of the NRL from KOSEF, and a grant from POSCO.
Footnotes
Published ahead of print on 5 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 1153227-3234. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Back, S. H., Y. K. Kim, W. J. Kim, S. Cho, H. R. Oh, J. E. Kim, and S. K. Jang. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3Cpro. J. Virol. 762529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen-Barak, O., Z. Yi, N. Hagiwara, K. Monzen, I. Komuro, and M. H. Brilliant. 2003. Sox6 regulation of cardiac myocyte development. Nucleic Acids Res. 315941-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuesta, R., G. Laroia, and R. J. Schneider. 2000. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 141460-1470. [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan, R. F., and J. W. Hershey. 1989. Protein synthesis and protein phosphorylation during heat stress, recovery, and adaptation. J. Cell Biol. 1091467-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340245-246. [DOI] [PubMed] [Google Scholar]
- 8.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 973073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes, H. H., and C. Kaether. 1996. Green fluorescent protein: applications in cell biology. FEBS Lett. 38944-47. [DOI] [PubMed] [Google Scholar]
- 10.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 155383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton, M. H., I. Tcherepanova, J. M. Huibregtse, and D. P. McDonnell. 2001. Nuclear import/export of hRPF1/Nedd4 regulates the ubiquitin-dependent degradation of its nuclear substrates. J. Biol. Chem. 27626324-26331. [DOI] [PubMed] [Google Scholar]
- 12.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 177480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 176940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov, P. A., E. M. Chudinova, and E. S. Nadezhdina. 2003. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290227-233. [DOI] [PubMed] [Google Scholar]
- 15.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30963-969. [DOI] [PubMed] [Google Scholar]
- 16.Kedersha, N., S. Chen, N. Gilks, W. Li, I. J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13195-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1471431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, W. J., S. H. Back, V. Kim, I. Ryu, and S. K. Jang. 2005. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 252450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimball, S. R., R. L. Horetsky, D. Ron, L. S. Jefferson, and H. P. Harding. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284C273—C284. [DOI] [PubMed] [Google Scholar]
- 20.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 27021975-21983. [DOI] [PubMed] [Google Scholar]
- 21.Lim, J., Y. H. Moon, G. An, and S. K. Jang. 2000. Two rice MADS domain proteins interact with OsMADS1. Plant Mol. Biol 44513-527. [DOI] [PubMed] [Google Scholar]
- 22.López de Silanes, I., S. Galban, J. L. Martindale, X. Yang, K. Mazan-Mamczarz, F. E. Indig, G. Falco, M. Zhan, and M. Gorospe. 2005. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 259520-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 154990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazroui, R., M. E. Huot, S. Tremblay, C. Filion, Y. Labelle, and E. W. Khandjian. 2002. Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 113007-3017. [DOI] [PubMed] [Google Scholar]
- 25.McDunn, J. E., and J. P. Cobb. 2005. That which does not kill you makes you stronger: a molecular mechanism for preconditioning. Sci. STKE 2005pe34. [DOI] [PubMed] [Google Scholar]
- 26.Nover, L., K. D. Scharf, and D. Neumann. 1983. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 31648-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez, I., J. G. McAfee, and J. G. Patton. 1997. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 3611881-11890. [DOI] [PubMed] [Google Scholar]
- 28.Pyronnet, S., H. Imataka, A. C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 931065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scharf, K. D., H. Heider, I. Hohfeld, R. Lyck, E. Schmidt, and L. Nover. 1998. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 182240-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scorsone, K. A., R. Panniers, A. G. Rowlands, and E. C. Henshaw. 1987. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. J. Biol. Chem. 26214538-14543. [PubMed] [Google Scholar]
- 32.Shi, Y., S. Luo, J. Peng, C. Huang, D. Tan, and W. Hu. 2004. The structure, expression and function prediction of DAZAP2, a down-regulated gene in multiple myeloma. Genomics Proteomics Bioinformatics 247-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommerfeldt, D. W., J. Zhi, C. T. Rubin, and M. Hadjiargyrou. 2002. Proline-rich transcript of the brain (prtb) is a serum-responsive gene in osteoblasts and upregulated during adhesion. J. Cell Biochem. 84301-308. [DOI] [PubMed] [Google Scholar]
- 34.Tourrière, H., K. Chebli, L. Zekri, B. Courselaud, J. M. Blanchard, E. Bertrand, and J. Tazi. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Tsui, S., T. Dai, S. Roettger, W. Schempp, E. C. Salido, and P. H. Yen. 2000. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics 65266-273. [DOI] [PubMed] [Google Scholar]
- 36.Warskulat, U., S. Kreuels, H. W. Muller, and D. Haussinger. 2001. Identification of osmosensitive and ammonia-regulated genes in rat astrocytes by Northern blotting and differential display reverse transcriptase-polymerase chain reaction. J. Hepatol. 35358-366. [DOI] [PubMed] [Google Scholar]
- 37.Waskiewicz, A. J., J. C. Johnson, B. Penn, M. Mahalingam, S. R. Kimball, and J. A. Cooper. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 191871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, W., and S. L. Mansour. 1999. Expression and genetic analysis of prtb, a gene that encodes a highly conserved proline-rich protein expressed in the brain. Dev. Dyn. 215108-116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.