Abstract
Fission yeast Cid14, a component of the TRAMP (Cid14/Trf4-Air1-Mtr4 polyadenylation) complex, polyadenylates nuclear RNA and stimulates degradation by the exosome for RNA quality control. Here, we analyze patterns of global gene expression in cells lacking the Cid14 or the Dis3/Rpr44 subunit of the nuclear exosome. We found that transcripts from many genes induced during meiosis, including key regulators, accumulated in the absence of Cid14 or Dis3. Moreover, our data suggest that additional substrates include transcripts involved in heterochromatin assembly. Mutant cells lacking Cid14 and/or Dis3 accumulate transcripts corresponding to naturally silenced repeat elements within heterochromatic domains, reflecting defects in centromeric gene silencing and derepression of subtelomeric gene expression. We also uncover roles for Cid14 and Dis3 in maintaining the genomic integrity of ribosomal DNA. Our data indicate that polyadenylation-assisted nuclear RNA turnover functions in eliminating a variety of RNA targets to control diverse processes, such as heterochromatic gene silencing, meiotic differentiation, and maintenance of genomic integrity.
Polyadenylation is important for the maturation of mRNAs (26), while our recent work has also uncovered unexpected links between polyadenylation and chromosome replication and segregation (33, 35, 37). In addition to its function in the maturation of mRNAs, nuclear polyadenylation is important for diverse cellular activities, such as RNA interference (RNAi)-mediated heterochromatin assembly and quality control of noncoding RNAs (33, 35, 37). These functions involve Cid12 and Cid14, members of a widespread family of noncanonical poly(A) polymerases found in eukaryotes from yeasts to humans (30). Besides its function in checkpoint control, Cid12 is required for faithful chromosome segregation and RNAi-mediated heterochromatin assembly at centromeres (19, 37). RNAi silencing is triggered by double-stranded RNA, which is processed by the RNase III-like RNase Dicer into small interfering RNA (siRNA) molecules of around 21 nucleotides. These siRNAs become incorporated into the RNA-induced transcriptional silencing (RITS) complex, directing the complex to homologous RNA targets (6). In worms, plants, and fungi, RNAi also requires RNA-directed RNA polymerases (RDRs), which are involved in the siRNA- and template-directed production of double-stranded RNA (1, 17, 29). Motamedi et al. (19) identified Cid12 in an RDR complex (RDRC) which also contains Rdp1 (the fission yeast RDR homolog) and Hrr1 (an RNA helicase). RDRC physically interacts with the RITS complex in a manner that requires Dicer and the histone methyltransferase Clr4. In cells lacking Cid12, RITS complexes are devoid of siRNA and fail to localize to centromeric DNA repeats to initiate heterochromatin assembly.
In Saccharomyces cerevisiae, the Cid1-like proteins Trf4 and Trf5 share an essential function that involves the polyadenylation of nuclear RNAs as part of a pathway of exosome-mediated RNA turnover (10, 12, 31, 38). We have identified Cid14 as a Trf4/5 functional ortholog in the distantly related fission yeast, Schizosaccharomyces pombe (35). Unlike trf4 trf5 double mutants, cells lacking Cid14 are viable, but they suffer an increased frequency of chromosome missegregation. We have identified a minor population of polyadenylated rRNAs which accumulate in an exosome mutant in a manner that is largely dependent on Cid14, in line with a role for Cid14 in rRNA degradation (35). We report here a range of RNAs as additional substrates, including the centromeric transcripts involved in heterochromatin assembly. Cells lacking Cid14 or mutant cells defective in the Dis3/Rpr44 component of the exosome accumulate polyadenylated transcripts corresponding to naturally silenced repeat elements within heterochromatic domains, with consequent defects in centromeric gene silencing. Based on these data, we propose a role for Cid14 in stimulating the degradation of these transcripts to ensure heterochromatic function, which is distinct from the function of Cid12 in RNAi-mediated heterochromatin assembly (19, 37) and from previously suggested models of Cid14 function (3).
MATERIALS AND METHODS
Fission yeast strains and methods.
The strains used were described previously (32, 37). The original gar2-GFP (28) and dis3-54 mutants (23) were gifts from M. Yamamoto and M. Yanagida, respectively. The conditions for the growth, maintenance, and genetic manipulation of fission yeast were as described previously (18). Cells were grown at 30°C, which was also used as the permissive temperature for the dis3-54 mutant and in the microarray study. Where necessary, partially induced gene expression from the nmt1 promoter was performed in the presence of 15 μM thiamine.
RNA isolation and RT-PCR.
Total RNA was isolated by hot phenol extraction and purified by using RNeasy (Qiagen). An amount of 0.5 μg of total S. pombe RNA was reverse transcribed (SuperScript, Invitrogen) using random primers and then PCR amplified with the primer pairs RT-PCR1 and RT-PCR2 (19) and primer pairs specific for cdc2 (normalizer gene) (CCAGCTAGTGAACGGTGTAA and TCCGCAAAGGGACACCAAAT) with 30 and 20 PCR cycles, respectively. For strand-specific reverse transcription-PCR (RT-PCR), RNA was reverse transcribed using strand-specific primer and then PCR amplified with both primer pairs. tlh dh transcripts were amplified with OKR40 (TCGTCTTGTAGCAGCATGTGA) and OKR41 (GAGATGAACGTATCTCTATCGAC) (8). The amplified products were subjected to electrophoresis in 1% agarose gels, followed by staining with ethidium bromide and quantification by densitometry.
PCR poly(A) test.
Polyadenylation tests were performed as described previously (5) using the 3′ rapid amplification of cDNA ends system (Invitrogen) and then PCR amplified with the primer pairs RT-PCR2 and adapter primer.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described previously (37). To immunoprecipitate hemagglutinin (HA)-tagged Cid12 and Cid14 proteins, the mouse anti-HA monoclonal antibody HA-11 (Babco, Berkeley, CA) was used and then PCR amplified with primer pairs specific for the centromeric dh repeat (GAAAACACATCGTTGTCTTCAGAG and CGTCTTGTAGCTGCATGTGAA) and fbp1 (AATGACAATTCCCCACTAGCC and ACTTCAGCTAGGATTCACCTGG).
Microarray experiments and data evaluation.
We used DNA microarrays displaying probes for >99% of all known and predicted genes of S. pombe spotted in duplicate onto glass slides, performing hybridization and initial data processing and normalization as previously described (13). Two independent biological experiments with RNA preparation and subsequent hybridization were performed for the cid14 and dis3 mutants. Dye swaps were performed for repeated labeling reactions to exclude dye-specific artifacts. The data were analyzed by using GeneSpring (Agilent) and Excel (Microsoft). Hierarchical clustering was performed in GeneSpring using the Pearson correlation, with genes containing no data in ≥50% of the conditions being discarded. The significance of overlaps between different gene lists was calculated in GeneSpring using a standard Fisher's exact test, and the P values were adjusted with a Bonferroni multiple testing correction. An average mutant/wild-type ratio was calculated for each gene. Cut-off values of 1.5-fold were used for comparisons between mutants due to the overall subtle changes in gene expression. The results for selected gene loci were confirmed by real-time PCR analysis as described previously (35). Gene annotations were downloaded from Schizosaccharomyces pombe GeneDB (http://www.genedb.org/genedb/pombe/). The entire processed data set will be available at http://www.sanger.ac.uk/PostGenomics/S_pombe. The primer pairs used in the real-time PCR analysis were the following: c212.08c, CCGAATGGCAAGATGGTAAT and AGGAACTTGCAAGCCAGAAA; c750.01, TCCATTGACGCAAATGAAGA and GCCAACTCTTCCACACGACT; c750.07, AAAGTGGAACGCAGTGTGTG and AGGCGAAGAACCACCTAATG; c1348.02, ATCTCCTGCAATTTGGGTGT and GCAACATTGGAGATGCACAG; pB2B2.01, CAGGAGCCTCAAAGAGGTTG and ATCTGGTTTACCGCCAACAG; pB2B2.06c, GCCCTTAATCCCGGTTATGT and CTGGCGAGGAGGTAGCATTA; c23c4.07, CGTCTGTGTCAACCTCGAAC and CCTCGCACAATTTGCTGTAA; c70.09c, CTGTTGGCTCCTTCAAGCTC and GCAGGTCTGCGAAAATAACC; ssm4, ACAGCTAAAGACCGCAAGGA and CAATGGGCTTGGAATTGAGT; and c17A5.18c, CCCTCCAACTTCGTTTTCAA and AACACGTTAGCAGCCCTTGT.
Pulsed-field gel electrophoresis and PCR analysis of rDNA.
DNA plugs were prepared as previously described (36). Pulsed-field gel electrophoresis was carried out with an 0.8% chromosomal grade agarose gel in 1× TAE buffer (40 mM Tris-acetate, 2 mM EDTA) using a CHEF III apparatus (Bio-Rad, Hercules, CA). The settings were as follows: 2 V/cm; switch time, 30 min; angle, 106°; 14°C, 48 h. For PCR analysis of ribosomal DNA (rDNA), 105 cells were subjected to colony PCR analysis using primer pairs specific for rDNA (GAAGATGGGCGATGGTTGATGAAACGGAAGTG and ACAAATCTTGGGAACAAAGGCTTAATCTCAGCAG) and cdc2 (normalizer gene) (CCAGCTAGTGAACGGTGTAA and CATGCGTTTCCAACGAGGAA) with 25 and 35 PCR cycles, respectively. The amplified products were subjected to electrophoresis in 1.5% agarose gels, followed by staining with ethidium bromide and quantification by densitometry.
Microscopy.
Visualization of green fluorescent protein (GFP)-tagged protein in living cells, embedded in 0.6% low-melting-point agarose, was performed at room temperature as previously described (25). Images were acquired by using a Zeiss Axioplan 2 microscope equipped with a Plan Apochromat 100× objective, an Axiocam cooled, charge-coupled-device camera, and Axiovision software (Carl Zeiss, Welwyn Garden City, United Kingdom) and were assembled using Adobe PhotoShop.
RESULTS
Cid14 and Dis3 are required for centromeric gene silencing.
Cells lacking Cid12 accumulate transcripts corresponding to naturally silenced repeat elements within heterochromatic domains (37). To further explore the function of Cid1-like proteins, we extended this analysis to cells carrying deletions of other cid genes. We found that these transcripts also accumulated in cid14Δ mutants, at a lower level than in cid12Δ mutants, but not in cells carrying deletions of other cid genes (cid1, cid11, cid13, and cid16) (Fig. 1A, B). Furthermore, these transcripts predominantly derived from the reverse strand, as determined by strand-specific RT-PCR (Fig. 1C). It is known that the reverse strand is transcribed in wild-type cells but is rapidly converted into siRNA, whereas the forward strand is not transcribed in detectable quantities and is silenced at the transcriptional level (32). Thus, our results indicate that Cid14 plays an important role in posttranscriptional silencing (of the reverse strand) rather than in transcriptional silencing (of the forward strand). Consistent with this interpretation, the integrity of heterochromatin is largely unaffected by the deletion of cid14, as judged by Swi6 binding that acts to repress transcription (35) and by ChIP analysis using antibody against methylated H3-K9 and Chp1 (3).
FIG. 1.
Fission yeast Cid14 and Dis3 are required for centromeric gene silencing. (A) Schematic representation of a portion of centromere III indicating the locations of fragments amplified. (B) Total RNA from cultures of the indicated strains grown at 30°C was reverse transcribed using random primers and then PCR amplified with primer pairs specific for the dh repeat and cdc2 (normalizer gene). A non-reverse-transcribed negative control was included (− RT). (C) Reverse transcription using strand-specific primer was performed as described for panel B before PCR amplification. A non-reverse-transcribed negative control was included. RT-PCR products were separated on a 1% agarose gel. Relative increases in the levels of the PCR products are indicated beneath each lane. (D) Strains indicated, all with an otr1R-inserted ade6+ allele as shown, were spotted onto a yeast extract agar plate and photographed after 3 days of incubation.
The presence of Swi6 at the outer repeats in a dis3/RPR44 exosome mutant at the restrictive temperature supports the above finding (20, 21). In keeping with the genetic interaction between cid14 and dis3 (35), similar results were observed in the dis3-54 mutant grown at the permissive temperature of 30°C. The Dis3-54 mutant protein is partially inactivated at this temperature, as judged by the 30% reduction in the level of accumulated polyadenylated rRNA shown in the results of our previous study (35). As shown in Fig. 1C, we found that these reverse transcripts also accumulated in the dis3-54 mutant and were marginally further increased by the deletion of cid14. Together, these results suggest that Cid14 has an overlapping function with Dis3 in centromeric gene silencing. In accordance with these results, cid14Δ and dis3-54 mutants were defective in silencing the reporter gene ade6+ that is located at the outermost centromeric repeat region otr1R and produced pink colonies on low-adenine plates (Fig. 1D). Deletion of cid14 in the dis3-54 mutant further increased the silencing defects and produced white colonies to an extent similar to that seen for the cid12Δ mutant, whereas wild-type cells efficiently silenced otr1R::ade6+ expression and produced dark red colonies.
Role of polyadenylation in Cid14 function.
The results described above suggest that a polyadenylation-assisted degradation mechanism is required for centromeric gene silencing. In line with its predicted polyadenylation activity, the silencing function was dependent on the conserved aspartate residues 298 and 230 in the nucleotide transferase motif (GS X10 DXD) of Cid14 that are known to be essential for the polyadenylation activity of this protein family (Fig. 2A). When expressed in a cid14Δ strain, the Cid14 DADA mutant protein, unlike the wild-type protein, was unable to suppress the silencing defects in these cells. We conclude that the polyadenylation function of the Cid14 poly(A) polymerase is essential for its silencing function.
FIG. 2.
Role of polyadenylation in Cid14 function. (A) Total RNAs from cultures of cid14Δ cells containing the indicated plasmids (with or without a mutation in the nucleotide transferase motif GS X10 DXD of Cid14) were subjected to RT-PCR assays as described for Fig. 1. − RT, non-reverse-transcribed negative control. (B) Schematic diagram of the genetic organization of the cen3 locus and the primers for the rapid amplification of cDNA ends-polyadenylation technique assay to detect polyadenylated transcripts. Total RNA from cultures of the indicated strains was reverse transcribed with the oligo(dT) adapter primer (AP) and then PCR amplified with adapter primer and RT-PCR2 specific for the reverse transcript. The relative increase in the level of the PCR product is indicated beneath each lane.
Next, we attempted to determine the polyadenylation states of the reverse transcripts detected in cid14Δ mutant cells by using a polyadenylation assay (see Materials and Methods). As shown in Fig. 2B, using primers corresponding to the reverse transcripts, we obtained a major band of 0.9-kb products, in addition to the 1.7-kb transcripts spanning dh and dg repeats at cen3, in dis3 and/or cid14 mutants but much-lower levels of these bands in the wild-type strain, consistent with the normally rapid turnover of centromeric transcripts by the RNAi machinery. A similar result has been obtained by using RNA from mutants defective in the RNAi machinery (32), indicating that these transcripts are normally produced in the cells. We conclude that centromeric transcripts are polyadenylated even in cid14 mutants. At present, the enzyme responsible for the polyadenylation of these transcripts is not known. We have previously shown that the polyadenylation of the pre-siRNA transcripts is independent of Cid12 (37). Given the involvement of RNA polymerase II in generating centromeric transcripts (5, 11) and the fact that the other known Cid1-like poly(A) polymerases are cytoplasmic (our unpublished data and reference 27), the polyadenylation of these RNAs is likely to depend on the canonical poly(A) polymerase. The dependence of these products on the dis3 mutation implied that these molecules are destined for exosome-mediated degradation. Consistent with a role of Cid14 in stimulating exosome-mediated degradation, the accumulation of both transcripts in the dis3-54 mutant was increased further by the deletion of cid14 (Fig. 2B). In conclusion, despite the requirement of its catalytic activity for the silencing function of Cid14, we did not detect any Cid14-dependent polyadenylated intermediate even in the dis3-54 genetic background. This could reflect the relatively low abundance of these intermediates given that, as shown in the results of the reporter assay (Fig. 1D), centromeric gene silencing is still maintained to some degree in the dis3-54 mutant.
Cid14 functions off chromatin.
To test whether Cid14 is associated with chromatin at the site of transcription, we performed ChIP experiments. As shown in Fig. 3A, although Cid12 was preferentially enriched at the centromeres, we observed no enrichment of Cid14 at the centromeres, suggesting that any modification of centromeric transcripts by Cid14 occurs off chromatin. This is consistent with the nucleolar localization of Cid14 (35). As shown in Fig. 3B, the Cid14 signals were separated from both the large and small Swi6 foci representing centromeric heterochromatin and telomeric regions, respectively.
FIG. 3.
Cid14 functions off chromatin. (A) ChIP analysis of Cid14-HA in wild-type cells using the HA-11 antibody. The relative enrichment (n-fold) of dh centromeric repeats is indicated beneath each lane. As a positive control, the Cid12-HA strain was used. WCE, whole-cell extracts. (B) Merged images of fluorescence micrographs showing Swi6-GFP (green) and Cid14-red fluorescent protein (RFP) (red) localization in living cells. Arrowheads indicate the localization of centromeric (open) and telomeric (filled) heterochromatin. Bar, 10 μm.
Gene expression patterns in cid14Δ and dis3 mutants.
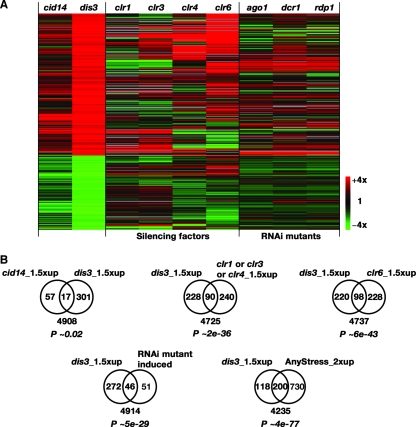
To understand further the role of Cid14 in gene silencing, we used microarrays to compare the RNA expression profiles of cid14Δ cells with those of dis3-54 cells at the permissive temperature of 30°C and with those of mutants defective in silencing or RNAi (7). A global comparison between these mutants is presented in Fig. 4A. The dis3 mutation affected similar genes as the cid14 mutation, although these genes were less strongly modulated in the latter. This could reflect differential effects on exosome activity in the two mutants. Consistent with this view, the exosome retains some basal activity in the absence of the TRAMP complex (12). Consistent with the genetic interaction between cid14 and dis3 (35), the induced genes significantly overlapped between the two mutants (P ∼ 0.02). Cid14 and Dis3 mainly function to reduce transcript levels, since most of the affected genes were upregulated in the mutant strains. Seventy-four and 318 genes were upregulated ≥1.5-fold in the cid14 and dis3 mutants, respectively. Consistent with a function in gene silencing for Dis3, the expression profile is similar to those of mutants defective in silencing and RNAi (Fig. 4B). The expression profile of a clr6 mutant, defective in a histone deacetylase, is most similar to those of cid14/dis3 mutants (Fig. 4A). As seen in the expression profiles of these silencing mutants, many of the genes that are regulated by Dis3 were also induced by stress (Fig. 4B) (7). Although a direct role of Clr6 and Clr3 in the regulation of stress-inducible genes has been suggested, we cannot rule out that some of the effects that occurred in the dis3 cells were indirect and caused by stress in sick mutant cells.
FIG. 4.
Relationship between the transcriptomes in cid14, dis3, and silencing mutants. (A) Hierarchical cluster analysis with columns representing different mutants and rows representing genes. The mRNA levels relative to the levels in wild-type cells are color coded as indicated at the bottom right, with missing data in gray. The 472 genes whose levels of expression were up- or downregulated 1.5-fold in the dis3 mutant are shown for comparison. (B) Numbers of genes that were upregulated ≥1.5-fold in the indicated mutants are presented in Venn diagrams. The numbers of genes not included in the gene lists are indicated below the diagrams. The P values shown indicate the probabilities that the observed overlaps occurred by chance.
Cid14 and Dis3 could indirectly affect the silencing of centromeric transcripts if these proteins were to regulate the RNAi machinery. However, transcripts encoding components of the RNAi machinery were not downregulated in cid14 or dis3 mutants, and some of these transcripts were even somewhat increased in the mutants (Table 1). These data are consistent with a model of a polyadenylation-assisted degradation mechanism directly targeting heterochromatic transcripts to promote silencing.
TABLE 1.
The expression of genes encoding components of the RNAi machinery is not significantly downregulated by the absence of Cid14 or Dis3a
| Gene | Relative transcript level (fold) in:
|
GeneDB description | |
|---|---|---|---|
| cid14Δ mutant | dis3-54 mutant | ||
| ago1 | 0.9 | 1.5 | RITS complex subunit |
| tas3 | 0.9 | 1.0 | RITS complex subunit |
| chp1 | 1.2 | 1.5 | RITS complex subunit |
| swi6 | 1.1 | 0.9 | Chromodomain protein |
| rdp1 | 1.2 | 1.0 | RDRC complex subunit |
| cid12 | 1.0 | 1.5 | RDRC complex subunit |
| hrr1 | 1.0 | 1.4 | RDRC complex subunit |
| dcr1 | 1.1 | 1.5 | Dicer |
| clr3 | 1.0 | 1.0 | Histone deacetylase |
| clr4 | 1.0 | 1.3 | Histone H3 methyltransferase |
| clr6 | 1.2 | 1.2 | Histone deacetylase |
Two independent biological experiments with RNA preparation and subsequent hybridization were performed for the cid14 and dis3 mutants. Dye swaps were performed for each strain to exclude dye-specific artifacts. The average increase in expression was calculated for each gene.
Subtelomeric genes are repressed by Cid14 and Dis3.
In line with the data described above, many of the genes whose mRNA levels increased in the cid14 or dis3 mutants were located at subtelomeric regions of chromosomes I and II (Fig. 5A, B). The absolute signal intensities for these genes are low in wild-type cells as expected for silenced genes. Increased transcription of subtelomeric regions was confirmed by real-time PCR analysis of selected gene loci (Fig. 5B and data not shown) and further supported by RT-PCR analysis of transcripts from tlh1, an RecQ helicase with a cenH-like repeat in its coding sequence, which located at the ends of chromosomes I and II (14, 15). As shown in Fig. 5C, an accumulation of forward transcripts was detected in the cid14 and dis3 mutants. Unlike the results for the centromere loci, only forward and no reverse transcripts were detected in the cid12 mutants, which reflects the functional redundancy of RNAi factors with Taz1, a telomeric DNA binding protein (8).
FIG. 5.
Subtelomeric genes are repressed by Cid14 and Dis3. (A) Horizontal bars represent the three S. pombe chromosomes. Genes whose levels of expression were upregulated ≥1.5-fold in the indicated mutants are represented by vertical bars above or below the chromosome depending on their direction of transcription. (B) Mutant versus wild-type expression ratios of subtelomeric genes of chromosomes I and II, left (L) and right (R) arms, were plotted for the indicated mutants. Arrowheads and lines indicate 1.5-fold-expression threshold. Asterisks indicate gene loci confirmed by real-time PCR analysis. (C) Schematic representation of the tlh1 gene indicating the location of fragments amplified by RT-PCR. Total RNA from cultures of the indicated strains was reverse transcribed using strand-specific primer for the tlh1 dh repeat and cdc2 (normalizer gene) before PCR amplification. A non-reverse-transcribed negative control was included (− RT). The relative increase in the level of the PCR product is indicated beneath each lane.
Meiotic RNAs accumulate in dis3-54 mutant.
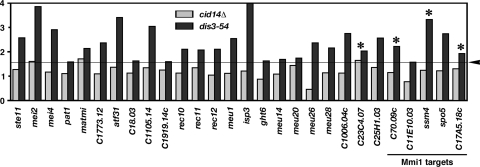
We have previously shown that mutations of cid14 or dis3 lead to the accumulation of mei2 mRNA, which encodes an RNA binding protein that is essential for the initiation of meiosis (34), suggesting that polyadenylation-assisted degradation plays a role in meiotic entry (35). In line with these data, Harigaya et al. (9) recently described a function of the exosome, regulated by an Mmi1-dependent mechanism, in the repression of meiotic genes during vegetative growth. As shown in Fig. 6, the microarray data revealed that many genes induced during meiosis, including key regulators (16), were accumulated in the dis3-54 mutant. This is not an indirect effect of starvation or nitrogen limitation as many of these genes are meiosis specific and include several targets of Mmi1 confirmed by real-time PCR analysis (Fig. 6 and data not shown) (9). Intriguingly, the dis3 mutation seemed to affect a larger number of genes than the mmi1 mutation, raising the possibility that the exosome also regulates meiotic entry through an Mmi-independent mechanism.
FIG. 6.
Effects of cid14 and dis3 mutations on selected genomic elements. Mutant versus wild-type expression ratios of genes induced during meiosis that were increased ≥1.5-fold (indicated by arrowhead and line) in the dis3 or cid14 mutants are shown. The Mmi1 target genes, determined by independent microarray experiments (9), are indicated at bottom right. Asterisks indicate gene loci confirmed by real-time PCR analysis.
Cid14 and Dis3 maintain genomic integrity of rDNA.
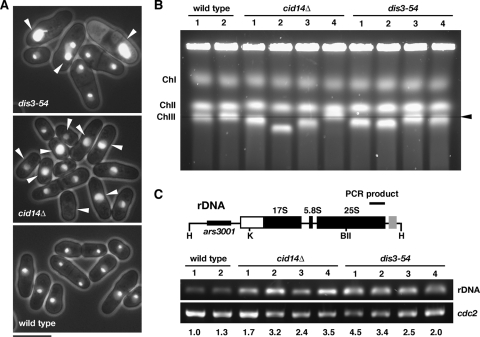
Despite producing the overwhelming majority of RNAs in the cells, rDNA is associated with heterochromatin (4). However, unlike the results for the centromere and telomere loci, no considerable increase in rRNA levels has been observed in cid14 and/or dis3 mutants, and Cid14 and Dis3 seem to exert functions other than silencing in this region (35). Consistent with this interpretation, a recent study has uncovered an important role for the RNAi pathway in maintaining the locus integrity at the tandem rDNA repeats by suppressing homologous recombination (4). The arrangement of rDNA genes strongly influences the organization and localization of the nucleolus (22). Consistent with the role of Cid14 in heterochromatic function, we have previously shown that Cid14 plays a role in the organization of the nucleolus (35). Here, we extend this function to Dis3, using the nucleolar protein Gar2 tagged with GFP to visualize nucleolar structure (28). In wild-type cells, Gar2-GFP occupied roughly half of the nucleus, in a discrete region distinct from the bulk chromosomal DNA, against a background of fainter nuclear signal (Fig. 7A). In contrast, a variety of abnormal morphologies were observed in cid14Δ and dis3-54 cells, even at the permissive temperature (72% and 30% of cell population, respectively). Some cells showed a reduction or complete loss of discrete signals, while others appeared to have increased and more-diffuse Gar2-GFP signals. Dispersion of Gar2-GFP in the nucleoplasm was also observed.
FIG. 7.
Cid14 and Dis3 maintain genomic integrity of rDNA. (A) Fluorescence micrographs showing Gar2-GFP localization in living cells. Arrowheads indicate aberrant nucleolar structures in cells with the indicated mutations. Bar, 10 μm. (B) Pulsed-field gel electrophoresis analyses of chromosomes from cid14 and dis3-54 mutants. Equal numbers of cells were prepared in agarose gel plugs from exponentially growing cultures of the indicated strains (four independent isolates and two wild-type isolates). Pulsed-field gel electrophoresis was carried out as described in Materials and Methods. The position of normal chromosome III is indicated by the arrowhead and line. (C) Schematic representation of the HindIII fragment of rDNA indicating the locations of amplified fragments. Equal numbers of cells from exponentially growing cultures of the indicated strains, as described for panel B, were subjected to PCR analysis as described in Materials and Methods. The relative increase (n-fold) in the level of the PCR product is indicated beneath each lane.
This phenomenon was further explored using pulsed-field gel electrophoresis to assess the integrity of the three S. pombe chromosomes. As shown in Fig. 7B, we consistently observed anomalous migration of chromosome III in these cells (four independent isolates from cid14Δ and three from dis3-54 mutants), which suggests genomic instability in the rDNA reflected in changes in the number of repeats located at the ends of chromosome III. This was further supported by PCR analysis of genomic DNA using whole-cell extracts of cid14Δ and dis3-54 mutants. As shown in Fig. 7C, we consistently observed increased levels of PCR products (increase of repeated DNA) from these cells (four independent isolates), despite the reduction of chromosome III length possibly due to the loss of rDNA repeats. It has been previously shown that mutations in proteins that regulate silencing in Drosophila and S. cerevisiae result in extrachromosomal circular rDNA formation (2, 24). Experiments are under way to determine whether or not the apparent increase in repeated rDNA shown by PCR is due to circular rDNA. Together with the aberrant nucleolar structures observed in cid14Δ and dis3-54 cells, these results suggest that a polyadenylation-assisted degradation mechanism plays a role in the maintenance of the genomic integrity of rDNA.
DISCUSSION
Eukaryotic cells have developed several quality control systems to ensure the fidelity of gene expression. One of these systems in budding yeast involves the addition of poly(A) tails to target RNAs by the TRAMP complex prior to exosome-dependent degradation. Substrates for these poly(A) polymerases include aberrantly modified tRNAs, precursors of snoRNAs and rRNA, and so-called cryptic unstable transcripts (10, 12, 31, 38). We have recently demonstrated that the fission yeast protein Cid14 is the functional ortholog of Trf4/5 that is required for the polyadenylation of rRNAs and proper chromosome segregation (35). In line with these data, Bühler et al. (3) recently confirmed that Cid14 has poly(A) polymerase activity in vitro and resides in a complex similar to TRAMP in budding yeast. These authors link heterochromatic gene silencing to two of the TRAMP components, Cid14 and Mtr4, and suggest that transcripts emerging from heterochromatic domains are polyadenylated by Cid14 to facilitate their degradation and/or processing by the RNAi machinery.
Based on our data, we propose the distinct model that a polyadenylation-assisted degradation mechanism is actually an essential part of heterochromatic silencing. We show that this is not an indirect effect of transcript downregulation, as mRNAs encoding components of the silencing machinery are not significantly affected by the absence of Cid14 or Dis3 (Table 1). To further understand the role of Cid14 in gene silencing, we have determined the polyadenylation states of the centromeric transcripts accumulated in the cid14 and/or dis3 mutants and found that centromeric transcripts in cid14Δ cells contain poly(A) tails (Fig. 2B). Together with the nucleolar localization and ChIP analyses (Fig. 3), these results suggest that Cid14 functions after transcription (Fig. 8A), which differs from the previously proposed model (3) where the main function of Cid14 occurs on chromatin. Our model is also consistent with the previous notion that centromeric transcripts are polyadenylated (15), probably by the canonical poly(A) polymerase. The dependence of these products on the dis3 mutation implied that these molecules are produced at a certain level but rapidly turned over by exosome-mediated degradation. Failure to eliminate these transcripts may interfere with the silencing machinery and compromise the heterochromatin structures (20, 21). To account for the difference in siRNA levels observed in the cid14 and rrp6 mutants (3), we suggest that Cid14 has an additional function in generating siRNA that is not shared by the exosome components. More rigorous evidence is awaited to validate this model.
FIG. 8.
Cid14 and Cid12 contribute to heterochromatic gene silencing through distinct but overlapping functions. (A) In contrast to the function of Cid12 in the RNAi machinery (RITS/RDRC/Dcr1), Cid14 acts to assist the posttranscriptional RNA turnover mechanism to limit transcripts from heterochromatic domains, thus promoting silencing. In addition, Cid14 might have a function in generating siRNA that is not shared by the exosome components as, unlike the level in the cid14 mutant, centromeric siRNA levels are not affected in the exosome mutants (20, 21). (B) Schematic representation of the roles of polyadenylation-assisted nuclear RNA turnover in eliminating a variety of RNA targets to control diverse biological functions.
Our genome-wide data indicate that Cid14 and Dis3 also play a role in the control of meiotic entry. We found that transcripts of many genes induced during meiosis, including key regulators, accumulated in the absence of Cid14 or Dis3 (Fig. 6) (35). The accumulation in exosome mutants of these RNAs required for meiosis suggests that these transcripts are constantly degraded by the exosome in mitotic cells to prevent ectopic meiosis (9). We are in the process of determining whether or not a polyadenylation-assisted degradation mechanism directly targets these transcripts.
Although further studies are required, our data support a function of polyadenylation-assisted nuclear RNA turnover in eliminating a variety of RNA targets to control diverse biological functions (Fig. 8B). It is unclear how the TRAMP complex differentiates various substrates to target them for degradation. Cid14 is associated with ribosomal synthesis factors (3). We speculate that these factors act to recruit the TRAMP complex if not displaced by timely maturation. A similar mechanism of recognition of RNA-protein complexes may apply to the other substrates. Our results also suggest an important role for Cid14 and Dis3 in the maintenance of the genomic integrity of the rDNA (Fig. 7). Experiments are under way to determine whether or not this function applies to repeated DNA in general. The chromosomes of higher eukaryotes contain complex chromatin structures and a high frequency of repeated sequences. It will be interesting to see if this also holds in mammals, in which case defects in polyadenylation-assisted degradation by the exosome could contribute to the chromosomal instability that characterizes most cancer cells.
Acknowledgments
We thank M. Yamamoto and M. Yanagida for yeast strains, T. Z. Win for initiation of this project, and S. Marguerat for critical reading of the manuscript.
This work was supported by Cancer Research UK and grant 96A1-MGPP13-018 from the National Health Research Institute, Taiwan.
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Baulcombe, D. 2004. RNA silencing in plants. Nature 431356-363. [DOI] [PubMed] [Google Scholar]
- 2.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73417-435. [DOI] [PubMed] [Google Scholar]
- 3.Bühler, M., W. Haas, S. P. Gygi, and D. Moazed. 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129707-721. [DOI] [PubMed] [Google Scholar]
- 4.Cam, H. P., T. Sugiyama, E. S. Chen, X. Chen, P. C. FitzGerald, and S. I. Grewal. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37809-819. [DOI] [PubMed] [Google Scholar]
- 5.Djupedal, I., M. Portoso, H. Spahr, C. Bonilla, C. M. Gustafsson, R. C. Allshire, and K. Ekwall. 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 192301-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannon, G. J. 2002. RNA interference. Nature 418244-251. [DOI] [PubMed] [Google Scholar]
- 7.Hansen, K. R., G. Burns, J. Mata, T. A. Volpe, R. A. Martienssen, J. Bähler, and G. Thon. 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 25590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, K. R., P. T. Ibarra, and G. Thon. 2006. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 3478-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harigaya, Y., H. Tanaka, S. Yamanaka, K. Tanaka, Y. Watanabe, C. Tsutsumi, Y. Chikashige, Y. Hiraoka, A. Yamashita, and M. Yamamoto. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 44245-50. [DOI] [PubMed] [Google Scholar]
- 10.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 181227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, H., D. B. Goto, R. A. Martienssen, T. Urano, K. Furukawa, and Y. Murakami. 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309467-469. [DOI] [PubMed] [Google Scholar]
- 12.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121713-724. [DOI] [PubMed] [Google Scholar]
- 13.Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici, D. Chen, C. Langford, D. Vetrie, and J. Bähler. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandell, J. G., J. Bähler, T. A. Volpe, R. A. Martienssen, and T. R. Cech. 2005. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol. 6R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandell, J. G., K. J. Goodrich, J. Bähler, and T. R. Cech. 2005. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J. Biol. Chem. 2805249-5257. [DOI] [PubMed] [Google Scholar]
- 16.Mata, J., R. Lyne, G. Burns, and J. Bähler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32143-147. [DOI] [PubMed] [Google Scholar]
- 17.Mello, C. C., and D. Conte, Jr. 2004. Revealing the world of RNA interference. Nature 431338-342. [DOI] [PubMed] [Google Scholar]
- 18.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 19.Motamedi, M. R., A. Verdel, S. U. Colmenares, S. A. Gerber, S. P. Gygi, and D. Moazed. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119789-802. [DOI] [PubMed] [Google Scholar]
- 20.Murakami, H., D. B. Goto, T. Toda, E. S. Chen, S. I. Grewal, R. A. Martienssen, and M. Yanagida. 2007. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS ONE 2e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas, E., T. Yamada, H. P. Cam, P. C. Fitzgerald, R. Kobayashi, and S. I. Grewal. 2007. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 14372-380. [DOI] [PubMed] [Google Scholar]
- 22.Oakes, M., J. P. Aris, J. S. Brockenbrough, H. Wai, L. Vu, and M. Nomura. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 14323-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkura, H., Y. Adachi, N. Kinoshita, O. Niwa, T. Toda, and M. Yanagida. 1988. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 71465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, J. C., and G. H. Karpen. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 925-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pidoux, A. L., S. Uzawa, P. E. Perry, W. Z. Cande, and R. C. Allshire. 2000. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 1134177-4191. [DOI] [PubMed] [Google Scholar]
- 26.Rosonina, E., S. Kaneko, and J. L. Manley. 2006. Terminating the transcript: breaking up is hard to do. Genes Dev. 201050-1056. [DOI] [PubMed] [Google Scholar]
- 27.Saitoh, S., A. Chabes, W. H. McDonald, L. Thelander, J. R. Yates, and P. Russell. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109563-573. [DOI] [PubMed] [Google Scholar]
- 28.Shimada, T., A. Yamashita, and M. Yamamoto. 2003. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 142461-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107465-476. [DOI] [PubMed] [Google Scholar]
- 30.Stevenson, A. L., and C. J. Norbury. 2006. The Cid1 family of non-canonical poly(A) polymerases. Yeast 23991-1000. [DOI] [PubMed] [Google Scholar]
- 31.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2971833-1837. [DOI] [PubMed] [Google Scholar]
- 33.Wang, S. W., K. Asakawa, T. Z. Win, T. Toda, and C. J. Norbury. 2005. Inactivation of the pre-mRNA cleavage and polyadenylation factor Pfs2 in fission yeast causes lethal cell cycle defects. Mol. Cell. Biol. 252288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe, Y., and M. Yamamoto. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78487-498. [DOI] [PubMed] [Google Scholar]
- 35.Win, T. Z., S. Draper, R. L. Read, J. Pearce, C. J. Norbury, and S. W. Wang. 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 261710-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Win, T. Z., A. Goodwin, I. D. Hickson, C. J. Norbury, and S. W. Wang. 2004. Requirement for Schizosaccharomyces pombe Top3 in the maintenance of chromosome integrity. J. Cell Sci. 1174769-4778. [DOI] [PubMed] [Google Scholar]
- 37.Win, T. Z., A. L. Stevenson, and S. W. Wang. 2006. Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control. Mol. Cell. Biol. 264435-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121725-737. [DOI] [PubMed] [Google Scholar]