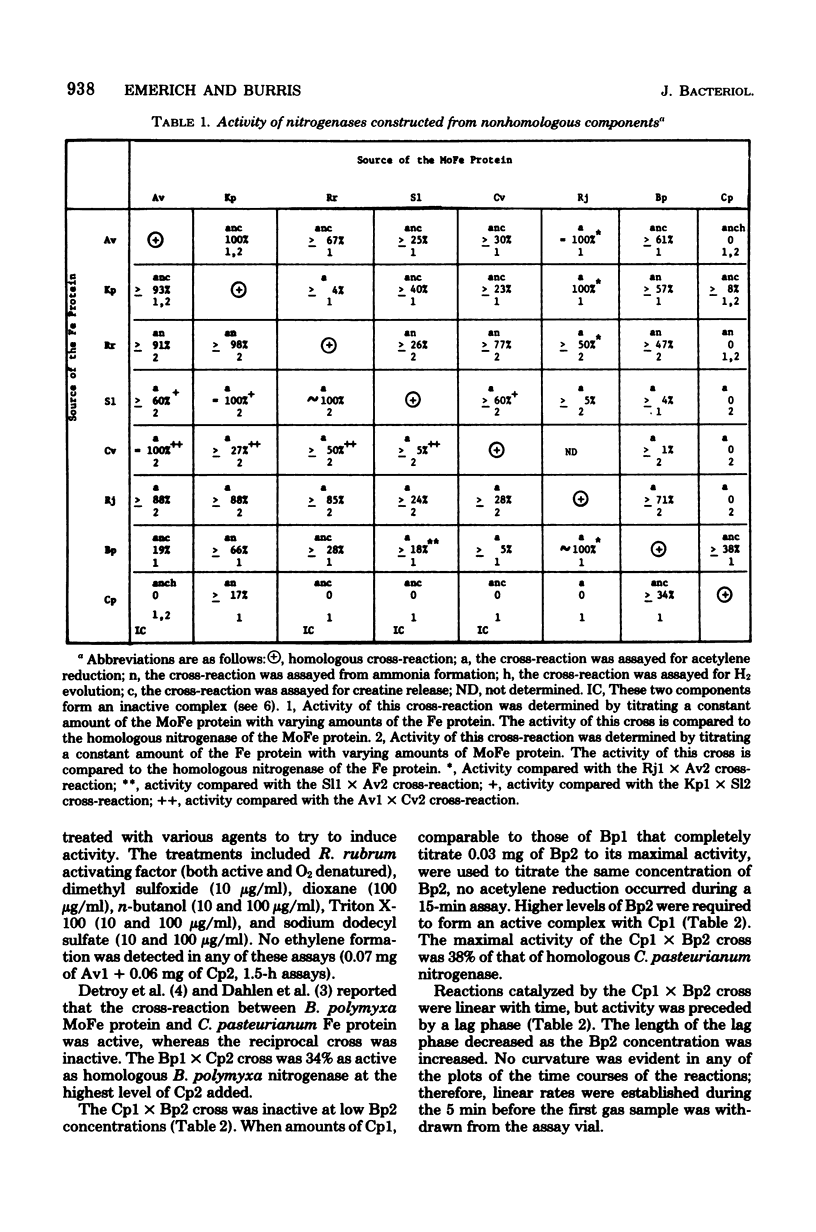
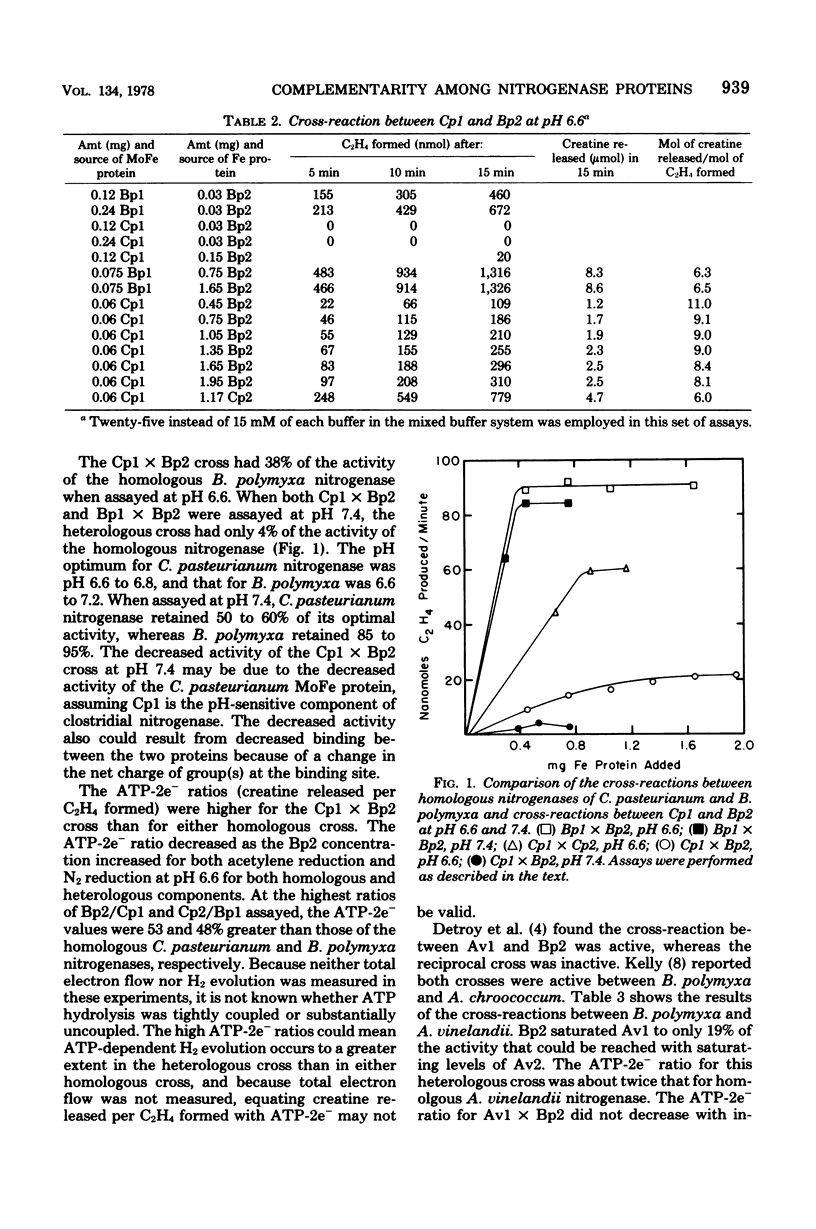
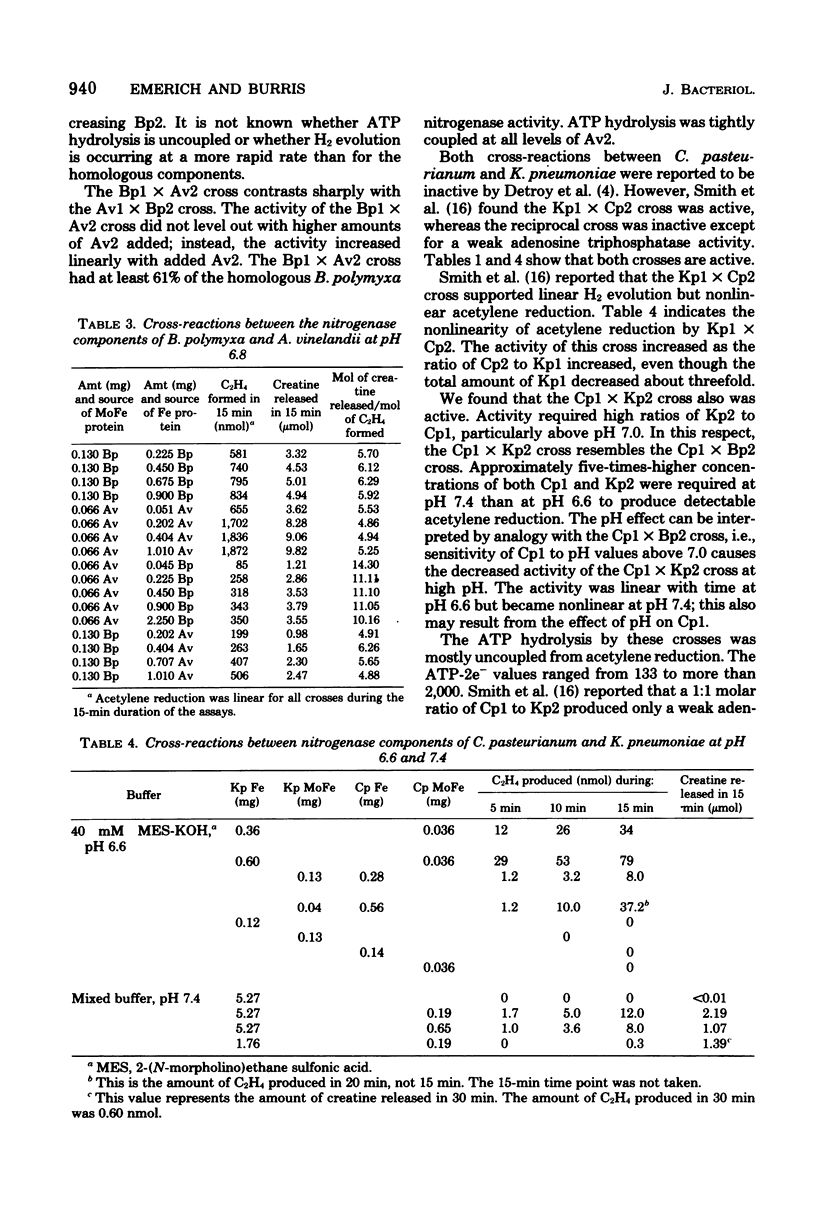
Abstract
The nitrogenase proteins from eight organisms have been highly purified, and a survey of their cross-reactions shows that the nitrogenase proteins from a wide variety of organisms can interact with one another. An active cross-reaction is the complementary functioning of the MoFe protein and the Fe protein from different organisms. Of 64 possible combinations of component proteins, 8 yielded homologous nitrogenases (components from the same organism); 45 of the 56 possible heterologous crosses generated active hybrid nitrogenases; 4 heterologous crosses yielded no measurable nitrogenase activity but did form inactive tight-binding complexes; 6 crosses did not give measurable activity; and 1 cross was not made. All these crosses were assayed for acetylene reduction, and some also were assayed for ammonia formation, hydrogen evolution, and ATP hydrolysis activity. The activity generated by combining two complementary heterologous nitrogenase components depended on pH, component ratio, and protein concentration, the same factors that determine the activity of homologous nitrogenases. However, several crosses showed an unusual dependency on component ratio and protein concentration, and some cross-reactions showed interesting ATP hydrolysis activity.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggins D. R., Kelly M., Postgate J. R. Resolution of nitrogenase of Mycobacterium flavum 30l into two components and cross reaction with nitrogenase components from other bacteria. Eur J Biochem. 1971 May 11;20(1):140–143. doi: 10.1111/j.1432-1033.1971.tb01371.x. [DOI] [PubMed] [Google Scholar]
- Dahlen J. V., Parejko R. A., Wilson P. W. Complementary functioning of two components from nitrogen-fixing bacteria. J Bacteriol. 1969 Apr;98(1):325–326. doi: 10.1128/jb.98.1.325-326.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detroy R. W., Witz D. F., Parejko R. A., Wilson P. W. Reduction of N2 by complementary functioning of two components from nitrogen-fixing bacteria. Proc Natl Acad Sci U S A. 1968 Oct;61(2):537–541. doi: 10.1073/pnas.61.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleton P., Elsden S. R., Gough N. The estimation of creatine and of diacetyl. Biochem J. 1943;37(5):526–529. doi: 10.1042/bj0370526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerich D. W., Burris R. H. Interactions of heterologous nitrogenase components that generate catalytically inactive complexes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4369–4373. doi: 10.1073/pnas.73.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOA J. A micro biuret method for protein determination; determination of total protein in cerebrospinal fluid. Scand J Clin Lab Invest. 1953;5(3):218–222. doi: 10.3109/00365515309094189. [DOI] [PubMed] [Google Scholar]
- Kelly M. Comparisons and cross reactions of nitrogenase from Klebsiella pneumoniae, Azotobacter chroococcum and Bacillus polymyxa. Biochim Biophys Acta. 1969;191(3):527–540. doi: 10.1016/0005-2744(69)90346-5. [DOI] [PubMed] [Google Scholar]
- Klucas R. V., Koch B., Russell S. A., Evans H. J. Purification and Some Properties of the Nitrogenase From Soybean (Glycine max Merr.) Nodules. Plant Physiol. 1968 Dec;43(12):1906–1912. doi: 10.1104/pp.43.12.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Ludden P. W., Burris R. H. Activating factor for the iron protein of nitrogenase from Rhodospirillum rubrum. Science. 1976 Oct 22;194(4263):424–426. doi: 10.1126/science.824729. [DOI] [PubMed] [Google Scholar]
- Murphy P. M., Koch B. L. Compatibility of the components of nitrogenase from soybean bacteroids and free-living nitrogen-fixing bacteria. Biochim Biophys Acta. 1971 Nov 2;253(1):295–297. doi: 10.1016/0005-2728(71)90257-x. [DOI] [PubMed] [Google Scholar]
- Okon Y., Houchins J. P., Albrecht S. L., Burris R. H. Growth of Spirillum lipoferum at constant partial pressures of oxygen, and the properties of its nitrogenase in cell-free extracts. J Gen Microbiol. 1977 Jan;98(1):87–93. doi: 10.1099/00221287-98-1-87. [DOI] [PubMed] [Google Scholar]
- Rivera-Ortiz J. M., Burris R. H. Interactions among substrates and inhibitors of nitrogenase. J Bacteriol. 1975 Aug;123(2):537–545. doi: 10.1128/jb.123.2.537-545.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V. K., Brill W. J. Nitrogenase. IV. Simple method of purification to homogeneity of nitrogenase components from Azotobacter vinelandii. Biochim Biophys Acta. 1973 May 30;305(2):445–454. doi: 10.1016/0005-2728(73)90190-4. [DOI] [PubMed] [Google Scholar]
- Smith B. E., Thorneley R. N., Eady R. R., Mortenson L. E. Nitrogenases from Klebsiella pneumoniae and Clostridium pasteurianum. Kinetic investigations of cross-reactions as a probe of the enzyme mechanism. Biochem J. 1976 Aug 1;157(2):439–447. doi: 10.1042/bj1570439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. V., Telfer A., Evans M. C. Complementary functioning of nitrogenase components from a blue-green alga and a photosynthetic bacterium. J Bacteriol. 1971 Aug;107(2):574–575. doi: 10.1128/jb.107.2.574-575.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneley R. N., Eady R. R. Nitrogenase of Klebsiella pneumoniae: evidence for an adenosine triphosphate-induced association of the iron-sulphur protein. Biochem J. 1973 Jun;133(2):405–408. doi: 10.1042/bj1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso M. Y., Ljones T., Burris R. H. Purification of the nitrogenase proteins from Clostridium pasteurianum. Biochim Biophys Acta. 1972 Jun 23;267(3):600–604. doi: 10.1016/0005-2728(72)90193-4. [DOI] [PubMed] [Google Scholar]
- Wang R., Healey F. P., Myers J. Amperometric measurement of hydrogen evolution in chlamydomonas. Plant Physiol. 1971 Jul;48(1):108–110. doi: 10.1104/pp.48.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]



