Abstract
Transport of specific mRNAs to defined regions within the cell cytoplasm is a fundamental mechanism for regulating cell and developmental polarity. In the Xenopus oocyte, Vg1 RNA is transported to the vegetal cytoplasm, where localized expression of the encoded protein is critical for embryonic polarity. The Vg1 localization pathway is directed by interactions between key motifs within Vg1 RNA and protein factors recognizing those RNA sequences. We have investigated how RNA-protein interactions could be modulated to trigger distinct steps in the localization pathway and found that the Vg1 RNP is remodeled during cytoplasmic RNA transport. Our results implicate two RNA-binding proteins with key roles in Vg1 RNA localization, PTB/hnRNP I and Vg1RBP/vera, in this process. We show that PTB/hnRNP I is required for remodeling of the interaction between Vg1 RNA and Vg1RBP/vera. Critically, mutations that block this remodeling event also eliminate vegetal localization of the RNA, suggesting that RNP remodeling is required for localization.
RNA localization is a widespread mechanism for generating cell polarity through the spatial restriction of gene expression to a defined subcellular region. Localized RNAs in somatic cells are thought to aid in distinct functions, such as motility and structure, while those RNAs localized in germ cells play roles in establishing early developmental axes and act in germ line specification (reviewed in references 4, 7, 15, 18, 19, 27, and 29). Transport of specific RNAs to defined regions within the cell cytoplasm is initiated by RNA-protein interactions that direct the recognition of the RNA and assembly of a ribonucleoprotein (RNP) transport complex. While RNA localization plays a key role in many cellular functions, the molecular mechanisms directing formation of a localization-competent RNP are not yet understood.
Among vertebrates, Vg1 mRNA is a prominent example of a localized mRNA that plays a role in embryonic patterning (reviewed in references 18 and 29). Vg1 mRNA is localized to the vegetal hemisphere cytoplasm in oocytes of the frog Xenopus laevis, and restricted expression of the peptide growth factor encoded by Vg1 RNA is critical for correct patterning of the embryo (2, 37). The Vg1 RNA localization pathway initiates in the nucleus, where recognition of Vg1 RNA by RNA-binding proteins with roles in localization first occurs (20). Upon export of the early RNP complex from the nucleus to the cytoplasm, additional factors, including molecular motors, are assembled onto the Vg1 RNP (1, 20, 40). Although Vg1 RNA is transcribed from the earliest stages of oogenesis, the RNA remains uniformly distributed within the oocyte cytoplasm until mid-oogenesis, when it is transported to the vegetal cortex (28). The molecular events that trigger the active transport step of the localization pathway are not yet known but may require remodeling of the early RNP complex to facilitate assembly of a transport-competent RNP.
Both cis-acting sequences within localized RNAs and trans-acting factors that interact with those sequences play important roles in the localization process. Localized RNAs contain sequences directing their localization, which are usually found within their 3′ untranslated regions (UTR) (reviewed in reference 35). Transport of Vg1 RNA to the vegetal cortical cytoplasm during Xenopus oogenesis relies on a localization element (LE) found within its 3′ UTR (32). The Vg1 LE (VLE) contains clusters of short sequence motifs implicated in localization (3, 9, 12, 22, 23). Two of these motifs, termed E2 and VM1 sites, are bound by two RNA-binding proteins, Vg1RBP/vera and PTB/hnRNP I, respectively (5, 8, 9, 12). PTB/hnRNP I and Vg1RBP/vera are RNA-binding proteins with diverse roles in posttranscriptional regulation of RNA biogenesis in multiple systems. PTB/hnRNP I is an hnRNP family member with roles in alternative splicing, polyadenylation, mRNA stability, internal ribosome entry site-mediated translation initiation, and mRNA localization (reviewed in reference 10). Vg1RBP/vera is a member of a family of RNA-binding proteins also implicated in multiple posttranscriptional processes, such as mRNA localization, mRNA stability, and translational regulation; other family members include chick ZBP-1, the mammalian IMPs 1 to 3, and CRD-BP (reviewed in reference 39). Both PTB/hnRNP I and Vg1RBP/vera first associate with Vg1 RNA in the oocyte nucleus (20) and are colocalized with the RNA in the vegetal cortical cytoplasm (5, 41). Mutations in E2 and VM1 sites within the VLE block binding of Vg1RBP/vera and PTB/hnRNP I, respectively (5, 8, 9, 12), and eliminate localization, supporting essential roles for these RNA-binding proteins in vegetal localization. However, how such RNA-binding proteins function in RNA localization pathways remains unknown.
In cells, RNAs are present as RNP complexes, a collection of RNA and proteins that define the biogenesis and expression of the RNA. As RNAs mature from transcription to destruction, protein factors are added, removed, modified, or rearranged to control the various steps in RNA metabolism. RNP remodeling during events such as transcription, nuclear export, and degradation have been well documented (reviewed in references 24 and 30), but little is known about remodeling during RNA localization. In Saccharomyces cerevisiae, rearrangement of the ASH1 mRNA localization complex is required for anchoring of the RNA at its destination (14). Certain RNA helicases are required for proper localization and translational regulation of oskar mRNA (33, 34), hinting at a role for remodeling during RNA localization in the Drosophila oocyte. During localization of Vg1 RNA, Vg1RBP/vera and PTB/hnRNP I interact with each other and with Vg1 RNA in both the nucleus and cytoplasm but the interactions are distinct in each compartment, suggesting a remodeling step in the Xenopus vegetal localization pathway (20). Although these results have provided tantalizing clues that RNP remodeling may affect localization, a role for RNP rearrangements during vegetal localization has yet to be shown.
To investigate whether RNP remodeling could promote RNA localization, we have investigated the RNA-protein interactions occurring at distinct time points in the RNA localization pathway. We show that interactions between Vg1RBP/vera and VLE RNA sequences are remodeled during localization. Vg1RBP/vera initially interacts only indirectly with VLE RNA but is bound directly to the RNA later during localization in the cytoplasm. Moreover, we find that PTB/hnRNP I is required for remodeling of the Vg1RBP/vera-VLE interaction, as mutations within PTB/hnRNP I binding sites that block binding of PTB/hnRNP I to VLE RNA also prevent direct interaction between VLE RNA and Vg1RBP/vera, despite recruitment of Vg1RBP/vera to the RNP. Vegetal localization is blocked by these mutations, suggesting that this remodeling event is critical for localization.
MATERIALS AND METHODS
Mutagenesis and cloning.
To introduce point mutations into E2 and VM1 motifs within the minimal VLE, primers containing mutations in either VM1 (23) or E2 (8) sites (in VM1, UUUCU→AUACA, and in E2, UUCAC→UUUGC) were used to amplify fragments from pSP73-2x135 (12) by PCR. The resulting fragments were cloned into pSP73 (Promega), and the mutations were verified by DNA sequencing.
Synthesis of RNA transcripts.
To prepare fluorescent transcripts for microinjection, VLE RNA was transcribed from linearized pSP73-2x135 or the various VM1 and E2 site mutants in reaction mixtures containing transcription buffer (40 mM Tris-HCl [pH 7.5], 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol, 40 units RNasin RNase inhibitor [Promega]), 0.5 mM each of CTP and ATP, 0.45 mM UTP, 1 mM diguanosine triphosphate, 0.1 mM GTP, 1 μCi of [α-32P]UTP (800 Ci/mmol; DuPont/NEN), and 50 μM Alexa Fluor 546-14-UTP (Molecular Probes). The RNAs were purified and resuspended to a concentration of 50 nM. For RNA-binding assays, RNAs were transcribed in reaction mixtures containing 1× transcription buffer (Promega), 0.5 mM each of CTP and ATP, 50 μM GTP, 0.5 mM diguanosine triphosphate, and 50 μCi of [α-32P]UTP (800 Ci/mmol; DuPont/NEN). RNA transcripts were resuspended at 1 ng/μl for in vitro UV cross-linking and 250 nM for microinjection. FLAG-tagged versions of Vg1RBP/vera, PTB/hnRNP I (20), and XStau (40) were transcribed using an mMessage mMachine kit (Ambion) according to the manufacturer's instructions. Prior to microinjection, Vg1RBP/vera-FLAG RNA was resuspended to 750 nM, PTB/hnRNP I-FLAG RNA was resuspended to 250 nM, and XStau-FLAG RNA was resuspended to 500 nM. Sequence-specific competitor RNAs were synthesized from linearized pSP73-2x135 (12) and wild-type and mutant 3×VM1 (5) by using a MEGAscript kit (Ambion) according to the manufacturer's protocol. Escherichia coli rRNA used as a nonspecific competitor was a generous gift from A. Dahlberg.
Oocyte culture.
Oocytes were obtained surgically from Xenopus laevis females (Nasco) and defoliculated by incubation in 2 mg/ml collagenase (Sigma-Aldrich). The oocytes were washed with MBSH buffer [88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.82 mM MgSO4·7H2O, 0.33 mM Ca(NO3)2·4H2O, 0.41 mM CaCl2·6H2O, 10 mM HEPES (pH 7.6)], and stage III oocytes (11) were manually sorted. After microinjection, oocytes were cultured in oocyte culture medium (OCM; 50% L15 medium, 15 mM HEPES [pH 7.6], 1 mM glutamine, 1 mg/ml insulin, 100 mg/ml gentamicin, 50 U/ml nystatin, 50 U/ml penicillin, 50 mg/ml streptomycin, and 5 to 10% frog serum containing vitellogenin), as described in reference 36.
In vivo RNA localization assay.
Albino stage III oocytes were microinjected with ∼3 nl of fluorescently labeled RNA and cultured for up to 4 days (36), followed by fixation in MEMFA (16) and storage in 100% methanol at −20°C. For microscopy, oocytes were cleared in 2:1 benzyl benzoate-benzyl alcohol. Oocytes were scored for localization with a Leica MZFL II fluorescence dissecting microscope. Confocal images were obtained using a Leica TCS SP2 AOBS spectral confocal microscope.
In vivo RNA-binding assays.
Stage III oocytes were microinjected with ∼3 nl of RNA transcripts encoding FLAG-tagged proteins (Vg1RBP/vera-FLAG, PTB/hnRNP I-FLAG, or XStau-FLAG). Following a 16-h incubation in OCM, the oocytes were subsequently injected with ∼3 nl of radiolabeled probe RNA and either processed immediately (1 h) or cultured overnight (16 h) in OCM. The oocytes were homogenized in YSS buffer (50 mM Tris [pH 8.0], 75 mM NaCl, 1 mM MgCl2, 0.05% Igepal [Sigma], 1 U/ml RNasin [Promega], 0.1 μg/ml leupeptin, 0.1 μg/ml antipain, 0.1 μg/ml trypsin inhibitor, 0.4 mM Pefabloc SC [Sigma], 1.0 mM dithiothreitol, and 100 mM sucrose) at a concentration of 0.5 oocyte per μl. Following centrifugation at 10,000 × g for 10 min, the lysate supernatant was cross-linked by a 15-min incubation in 0.1% formaldehyde, followed by quenching with 0.25 M glycine for 5 min. For cross-link immunoprecipitation (IP) analysis for assessing direct RNA-protein interactions, the lysate was cross-linked by UV irradiation for 10 min in a Stratalinker (Stratagene). IPs were performed by rocking the 25- to 40-oocyte equivalents of lysate with 10 μl anti-FLAG beads (Sigma) in a total volume of 500 μl of YSS for 2 h at room temperature. After treatment with RNase A (1 mg/ml; Sigma) for 15 min at 37°C, the cross-linked proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography. For RNA IP analysis, anti-FLAG IP was performed as described above, except that 10 oocyte equivalents of lysate were used and samples were not subjected to UV irradiation. RNA was isolated from the immunoprecipitates by addition of 200 μl RNA elution buffer (10 mM Tris [pH 7.5], 0.8 M ammonium acetate, 10 mM EDTA, 0.1% SDS, 50 μg/ml yeast tRNA) and incubation at 70°C for 15 min. After phenol-chloroform extraction and ethanol precipitation, the isolated RNA was separated on a denaturing polyacrylamide gel and visualized by autoradiography.
In vitro RNA-binding assays.
Preparation of oocyte S10 and S100 lysates was performed as described in reference 20, 31, and fractionation of oocyte lysate by heparin agarose chromatography was performed as described in reference 5. In vitro binding was performed as described in Lewis et al. (23), with 10-μl reaction mixtures containing ∼5 μg of oocyte lysate or ∼4 ng partially purified Vg1RBP/vera or PTB/hnRNP I, 600 ng of unlabeled competitor RNA, and 1 ng 32P-labeled VLE RNA. After incubation for 10 min, followed by UV cross-linking for 10 min using a Stratalinker (Stratagene) and treatment with RNase A (1 mg/ml; Sigma) for 15 min at 37°C, cross-linked proteins were separated by SDS-PAGE and visualized by autoradiography or phosphorimage analysis. Quantitation was carried out using a Typhoon 9410 variable mode imager.
RESULTS
In order to dissect distinct steps in the RNA localization pathway, we first established the time course of localization from the oocyte nucleus to the vegetal cortical cytoplasm. For these experiments, we used a vegetal LE (VLE) that is composed of a duplication of the first 135 nucleotides of the VLE residing in the 3′ UTR of Vg1 RNA (12). This VLE faithfully directs vegetal localization yet has reduced sequence complexity relative to other sequences known to carry out vegetal localization by this pathway (3, 12, 22, 32). Fluorescently labeled VLE transcripts were microinjected into nuclei of stage III Xenopus oocytes and cultured to allow localization of the injected RNA. Oocytes were fixed at various time points, and localization was assessed by confocal microscopy. As shown in Fig. 1A, the injected RNA remained in the oocyte nucleus at early time points (1 to 4 h) and was undergoing localization in the cytoplasm by 16 h postinjection (Fig. 1B). Transport was complete after 3 to 6 days, by which time the injected RNA was tightly localized at the vegetal cortex (Fig. 1C). To gain molecular insight into the localization pathway, we analyzed direct interactions between the VLE and RNA-binding proteins by UV cross-linking. Oocytes were microinjected with radiolabeled VLE RNA and cultured as described above. Oocyte lysates were prepared at 1 h and 16 h postinjection to distinguish RNA-protein interactions that may occur before or during transport in the cytoplasm. As shown in Fig. 1D, several RNA-binding proteins bind to VLE RNA both before (1 h) and during (16 h) localization. Notably, however, the 69-kDa protein is bound to VLE RNA only at times when VLE RNA is undergoing localization in the cytoplasm. Several Xenopus RNA-binding proteins have been implicated in vegetal localization (5, 6, 8, 17, 21, 42), and of these, Vg1RBP/vera is of the appropriate molecular mass to represent the 69-kDa protein (8, 17). IP analysis confirmed the identity of the 69-kDa protein as Vg1RBP/vera (Fig. 2 and data not shown).
FIG. 1.
Time course of RNA-protein interactions during vegetal RNA localization in Xenopus oocytes. (A to C) Alexa Fluor 546-labeled VLE RNA transcripts were injected into the nuclei of stage III Xenopus oocytes. Oocytes were fixed at the indicated time points postinjection (4 h [A], 16 h [B], and 4 days [C]) and viewed by confocal microscopy. The injected RNA (red) is evident in the nucleus in panel A, is asymmetrically distributed within the vegetal hemisphere cytoplasm in panel B, and is restricted to the vegetal cortex in panel C. The vegetal hemisphere is oriented toward the bottom (A to C), and the scale bars are 100 μm. (D) Radiolabeled VLE RNA transcripts were injected into the nuclei of stage III Xenopus oocytes. Oocyte lysates were prepared at either 1 h (lane 1) or 16 h (lane 2) postinjection and cross-linked by UV irradiation. After RNase treatment, labeled proteins were resolved by SDS-PAGE and visualized by autoradiography. The position of Vg1RBP/vera is indicated at the right, and molecular mass standards are on the left.
FIG. 2.
Interaction between Vg1RBP/vera and VLE RNA is remodeled during localization. (A) The experimental strategy for identifying proteins directly bound to RNA versus RNAs contained in RNP complexes is diagrammed. Stage III Xenopus oocytes are microinjected with RNA transcripts encoding FLAG-tagged proteins. After overnight culture to allow protein expression, 32P-labeled VLE RNA transcripts are injected into the oocyte nuclei. Oocyte lysates are prepared and split into two aliquots. One aliquot is analyzed by “cross-link IP,” in which cross-linking by UV irradiation is followed by anti-FLAG IP and RNase treatment. The RNA-bound proteins, labeled by covalent attachment of RNA oligonucleotides, are resolved by SDS-PAGE and visualized by autoradiography. The other aliquot is analyzed by “RNA IP,” in which anti-FLAG IP is carried out directly, and RNAs associated with the FLAG-tagged proteins are detected by autoradiography following PAGE. (B) Cross-link IP analysis. 32P-labeled VLE RNA was injected into nuclei of oocytes expressing Vg1RBP/vera-FLAG (lanes 3 and 4), PTB/hnRNP I-FLAG (lanes 7 and 8), or control oocytes without expression of FLAG-tagged proteins (lanes 1, 2, 5, and 6). Oocyte lysates were prepared either 1 h (lanes 1, 3, 5, and 7) or 16 h (lanes 2, 4, 6, and 8) postinjection and subjected to cross-link IP as detailed for panel A (above). Autoradiograms of cross-linked proteins resolved by SDS-PAGE are shown, and the positions of Vg1RBP/vera-FLAG (lanes 1 to 4) and PTB/hnRNP I-FLAG (lanes 4 to 8) are indicated to the right. (C) RNA IP analysis. 32P-labeled VLE RNA was injected into nuclei of oocytes expressing Vg1RBP/vera-FLAG (lanes 1 to 3), PTB/hnRNP I-FLAG (lanes 4 to 6), XStau-FLAG (lanes 7 to 9), or control oocytes without expression of FLAG-tagged proteins (lanes 10 to 12). Oocyte lysates were prepared either 1 h (lanes 2, 5, 8, and 11) or 16 h (lanes 3, 6, 9, and 12) postinjection and subjected to RNA IP as detailed for panel A (above). Autoradiograms of isolated VLE RNA resolved by PAGE are shown, and the position of VLE RNA is indicated at the left. For each panel, samples were run on the same gel, except for lanes 7 to 9 (XStau) in panel C, which were run together on a separate gel. Lane order was changed for clarity of presentation.
The lack of Vg1RBP/vera binding to VLE RNA in the nucleus was puzzling, as Vg1RBP/vera, along with PTB/hnRNP I, has been shown to associate with endogenous Vg1 RNA in the nucleus (20). A potential explanation for this apparent discrepancy may lie in the nature of the interactions revealed by these earlier experiments. The UV cross-linking experiments whose results are shown in Fig. 1D identify direct RNA-protein interactions, while associations determined by co-IP, as described in Kress et al. (20), are potentially indirect. To test this explicitly, we developed a protocol to compare direct versus potentially indirect interactions in RNP complexes isolated from oocytes. As diagrammed in Fig. 2A, RNAs encoding FLAG-tagged versions of either Vg1RBP/vera or PTB/hnRNP I were injected into stage III oocytes. After overnight culture to allow expression of the FLAG-tagged proteins, radiolabeled VLE RNA was injected into the oocyte nuclei, and oocyte lysates were prepared either after 1 h to assess early interactions or after 16 h to capture interactions that occur in the cytoplasm during localization. To analyze the RNA-protein interactions occurring at these time points, the oocyte lysates were split. In one portion, protein-RNA cross-links were induced by UV irradiation, followed by IP with anti-FLAG antibodies and RNase treatment. Proteins in direct contact with the VLE RNA were detected after SDS-PAGE and autoradiography (Fig. 2B) by virtue of covalent attachment of radiolabeled oligoribonucleotides. The other portion of the oocyte lysate was subjected to anti-FLAG IP without cross-linking to isolate RNA associated, either directly or indirectly, with the FLAG-tagged proteins (Fig. 2C).
As shown in Fig. 2B, PTB/hnRNP I-FLAG (lane 7), but not Vg1RBP/vera-FLAG (lane 3), was cross-linked to VLE RNA at the 1-h time point, whereas both Vg1RBP/vera-FLAG (lane 4) and PTB/hnRNP I-FLAG (lane 8) bound directly to VLE RNA at the 16-h time point, when VLE RNA was undergoing localization in the cytoplasm. These results are in agreement with the results obtained with endogenous proteins (Fig. 1D). By contrast, when RNP complexes containing FLAG-tagged Vg1RBP/vera were captured by IP without cross-linking, VLE RNA associated with Vg1RBP/vera-FLAG at both the 1-h (Fig. 2C, lane 2) and 16-h (lane 3) time points, as did PTB/hnRNP I-FLAG (lanes 5 and 6). As controls, we performed anti-FLAG IPs from oocytes that had not been injected with RNAs encoding FLAG-tagged proteins (lanes 11 and 12) and from oocytes expressing XStau-FLAG (lanes 9 and 10), a double-stranded-RNA-binding protein that associates with the Vg1 RNP in the cytoplasm (20). As expected, VLE RNA coimmunoprecipitated with XStau at the 16-h time point only (lane 9), when the RNA was in the cytoplasm, and was not recovered by anti-FLAG IP from uninjected oocytes (lanes 11 and 12). These results suggest that Vg1RBP/vera is recruited to the VLE RNP in the nucleus but does not contact the RNA directly at this early step (compare Fig. 2B, lane 3, and C, lane 2). Only later, when VLE RNA is undergoing localization in the cytoplasm, does Vg1RBP/vera associate directly with VLE RNA (Fig. 2B, lane 4). Thus, it is possible that rearrangement of the RNP complex to facilitate direct contact between Vg1RBP/vera and VLE RNA is required for vegetal transport.
Vg1RBP/vera has been shown to bind directly to RNA sequence elements termed E2 motifs in vitro (8), and mutation or deletion of E2 motifs disrupts VLE localization in vivo (3, 8, 9, 22, 23). However, our results (Fig. 1D and 2B) suggest that E2 motifs alone are not sufficient to mediate direct VLE RNA binding by Vg1RBP/vera in oocytes. The VLE contains two copies of the E2 motif, yet Vg1RBP/vera is not bound directly to the RNA at early time points in the localization pathway. A possible explanation for this result is that other factors may act either to block VLE RNA binding by Vg1RBP/vera in the nucleus or to facilitate RNA binding in the cytoplasm. A candidate for such a factor is PTB/hnRNP I, as Vg1RBP/vera and PTB/hnRNP I have been shown to interact with one another in both the nucleus and the cytoplasm, and the interactions differ in the two compartments such that the interaction is RNA dependent in the cytoplasm but not in the nucleus (20).
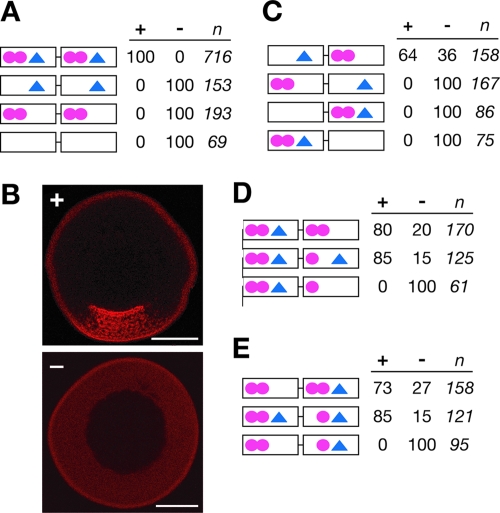
As a first test of whether PTB/hnRNP I might modulate interactions between Vg1RBP/vera and VLE RNA, we systematically mutated both the PTB/hnRNP I binding sites (VM1 motifs) and the E2 motifs within the VLE. As shown in Fig. 3A, the VLE (top), consisting of the duplicated 135-nucleotide Vg1 sequence, contains four VM1 sites and two E2 motifs. The VM1 and E2 sites were mutated by engineering specific point mutations that had previously been shown to block binding of PTB/hnRNP I and Vg1RBP/vera to their respective sites in vitro (5, 8, 22, 23). To test for potentially synergistic effects on localization in vivo, fluorescently labeled wild-type and mutant RNA transcripts were injected into nuclei of stage III oocytes and cultured to allow transport of the injected RNA. The oocytes were fixed and viewed by confocal microscopy, and localization was scored by comparison to the localization of the wild-type VLE, which was set at 100%. Oocytes scored as positive for localization exhibited strong accumulation of the injected RNA in the vegetal cytoplasm (Fig. 3B, top), whereas oocytes showing no detectable asymmetry in RNA distribution were scored as negative for localization (Fig. 3B, bottom). As expected, mutation of all four VM1 sites or both E2 sites disrupted VLE RNA localization in vivo, as did mutation of all VM1 and E2 sites together (Fig. 3A). The positioning of the VM1 and E2 sites relative to one another dramatically affected localization (Fig. 3C). A minimum of one E2 site and a pair of VM1 sites was necessary to support vegetal localization, but placement of the sites closer to or farther apart from one another disrupted localization. Synergistic effects were also apparent when different combinations of VM1 and E2 sites were mutated. For example, mutation of the downstream E2 site (Fig. 3D, top) had only a modest effect on localization, as did mutation of the downstream VM1 site (Fig. 3D, middle). However, when both of these sites were mutated together, localization was abolished (Fig. 3D, bottom). Similar effects were observed with other combinations of VM1 and E2 mutations (Fig. 3E and data not shown). As shown in Fig. 3E, mutation of either the upstream E2 site or the first downstream VM1 site resulted in minimal reduction in localization, while mutation of both of those sites eliminated localization. These results indicate that the spacing and positioning of VM1 and E2 sites relative to one another are critical for proper VLE function during localization, consistent with the possibility that PTB/hnRNP I may influence interactions between Vg1RBP/vera and VLE RNA.
FIG. 3.
E2 and VM1 motifs exhibit spacing and synergistic effects within the VLE. (A) Schematics of wild-type VLE and mutant RNAs, with VM1 motifs (YYUCU, where Y = U or C) shown as circles and E2 motifs (WYCAC, where W = A or C and Y = U or C) represented as triangles. Introduction of mutations into the VM1 sites (YYUCU→AYACA) or E2 sites (WYCAC→UUUGC) is indicated by removal of the site(s) from the diagrams. The results of in vivo localization assays (percent localization) are indicated at the right: + represents normal vegetal localization (set to 100% for the wild type), and − indicates no detectable localization. The number of oocytes (n) injected for each transcript is indicated at the far right. (B) Xenopus stage III oocytes were injected with Alexa 546-labeled RNA transcripts and scored for localization as indicated above for panel A. An example of normal vegetal localization (+) is shown on the top, and an example with no detectable localization (−) is at the bottom. Localization of the injected RNA (red) is evident by accumulation of the RNA in the vegetal cytoplasm, which is at the bottom of the image. The scale bars represent 100 μm. (C to E) Schematics of VLE RNA transcripts containing mutations in E2 motifs (triangles) or VM1 sites (circles), with localization analyses as detailed for panel A, above.
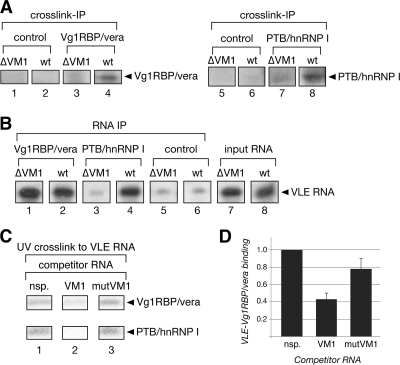
To test explicitly whether PTB/hnRNP I binding to the VLE could affect RNA binding by Vg1RBP/vera, we tested in vitro binding of Vg1RBP/vera to RNA transcripts that are unable to bind PTB/hnRNP I. For this, we used VLE transcripts in which all four VM1 sites are mutated (VLE-ΔVM1), but the Vg1RBP/vera binding sites (E2 motifs) are intact. Radiolabeled RNA transcripts were combined with partially purified preparations of Vg1RBP/vera and PTB/hnRNP I, and RNA-protein interactions were assessed by UV cross-linking (Fig. 4A). As expected, binding of PTB/hnRNP I to VLE-ΔVM1 RNA was dramatically reduced (Fig. 4A, lanes 1 and 2) compared to that for wild-type VLE RNA (lanes 3 and 4). Binding of Vg1RBP/vera to the VM1 mutant RNA (VLE-ΔVM1) was reduced as well (Fig. 4A, lanes 1 and 2), indicating that Vg1RBP/vera-RNA interactions are disrupted by mutations that do not impinge on the Vg1RBP/vera binding site E2. To assess whether disruption of PTB/hnRNP I binding to VLE RNA similarly blocks RNA binding by Vg1RBP/vera in vivo, we next analyzed RNP complexes formed in oocytes. Stage III oocytes were injected with radiolabeled VM1 mutant (VLE-ΔVM1) or wild-type VLE RNAs and cultured for 16 h to promote localization of the injected RNAs. RNA-protein interactions were examined by UV cross-linking in lysates prepared from the injected oocytes. As shown in Fig. 4B, Vg1RBP/vera bound poorly to VM1 mutant RNA (lane 1), yet binding to the wild-type VLE RNA (lane 2) was robust. While it was not possible to discern PTB/hnRNP I binding in these crude oocyte lysates, as PTB/hnRNP I is not resolved from the p54/p56 RNA-binding proteins (25) under these conditions, the results observed for Vg1RBP/vera in Fig. 4B are identical to those obtained in vitro (Fig. 4A). The inability of Vg1RBP/vera to bind to VLE RNA containing intact E2 sites and mutated VM1 sites (Fig. 4A and B) supports a model in which PTB/hnRNP I may modulate interactions between Vg1RBP/vera and VLE RNA. Importantly, mutation of the VM1 sites blocks localization in vivo (Fig. 3A and 4C), and this could be due, at least in part, to disruption of Vg1RBP/vera-VLE RNA interactions.
FIG. 4.
VM1 site mutations eliminate direct binding of both PTB/hnRNP I and Vg1RBP/vera to VLE RNA. (A) UV cross-linking analysis was performed using radiolabeled VM1 mutant (ΔVM1; lanes 1 and 2) and wild-type (lanes 3 and 4) VLE RNA to test the ability to be directly bound by partially purified preparations of Vg1RBP/vera (top panels) or PTB/hnRNP I (bottom panels). Specificity of in vitro binding was assessed by challenging the binding reactions with unlabeled specific (sp.; lanes 1 and 3) or nonspecific (nsp.; lanes 2 and 4) competitor RNAs. (B) Radiolabeled VM1 mutant (ΔVM1; lane 1) and wild-type (wt; lane 2) VLE RNA transcripts were injected into the nuclei of stage III oocytes. After culture for 16 h, oocyte lysates were prepared, cross-linked by UV irradiation, and treated with RNase. Proteins interacting with the injected RNA were resolved by SDS-PAGE and autoradiography. The position of Vg1RBP/vera is indicated at the right, and molecular mass markers are shown on the left. (C) Stage III oocytes were injected with Alexa 546-labeled VM1 mutant (ΔVM1; left) and wild-type (wt; right) VLE RNA transcripts. VM1 mutant VLE RNA (left) shows no detectable localization after culture, whereas vegetal localization of the injected RNA (red) is evident in oocytes injected with wild-type VLE RNA (right). The oocytes are oriented with the vegetal hemisphere toward the bottom. Scale bar, 100 μm.
Our results have shown that interactions between Vg1RBP/vera and VLE RNA are modulated during localization (Fig. 1D), as are interactions between Vg1RBP/vera and PTB/hnRNP I (20). Thus, the observed defects in RNA binding to VM1 mutant VLE RNA by Vg1RBP/vera may reflect a specific RNP remodeling event that is necessary for the transition from early to later steps in the RNA localization pathway. To test whether the RNP complexes formed with VM1 mutant (ΔVM1) VLE RNA are biochemically similar to those detected early in the normal localization pathway (Fig. 2B and C), we again used the experimental approach diagrammed in Fig. 2A to analyze direct versus indirect RNA-protein interactions using wild-type and ΔVM1 VLE RNAs. As shown in Fig. 5A, both Vg1RBP/vera (lane 4) and PTB/hnRNP I (lane 8) bound directly to the wild-type VLE, but neither Vg1RBP/vera (lane 3) nor PTB/hnRNP I (lane 7) bound to the ΔVM1 VLE. These results are in agreement with the in vitro and in vivo results shown in Fig. 4A and B. To test whether the ΔVM1 VLE RNA was present in RNP complexes containing FLAG-tagged Vg1RBP/vera or PTB/hnRNP I, RNA was isolated from anti-FLAG immunoprecipitates without UV cross-linking (Fig. 5B). In contrast to the UV cross-linking analyses (Fig. 4A and B and 5A), ΔVM1 VLE RNA was recovered by anti-FLAG IP from oocyte lysates containing FLAG-tagged Vg1RBP/vera (Fig. 5B, lane 1). Wild-type VLE RNA was recovered from RNP complexes containing Vg1RBP/vera (lane 2) and from PTB/hnRNP I RNP complexes (lane 4), but PTB/hnRNP I was not associated with VM1 mutant (ΔVM1) VLE RNA (lane 3). These results indicate that VLE RNA lacking VM1 sites (ΔVM1) does not interact either directly or indirectly with PTB/hnRNP I and that while Vg1RBP/vera associates with VM1 mutant VLE RNA (Fig. 5B, lane 1), it does so only indirectly (Fig. 5A, lane 3). Thus, PTB/hnRNP I does not appear to be necessary for recruitment of Vg1RBP/vera to VLE RNA, as Vg1RBP/vera is able to associate, albeit indirectly, with VM1 mutant (ΔVM1) VLE RNA. Moreover, the interaction of Vg1RBP/vera with ΔVM1 VLE RNA is indistinguishable from the Vg1RBP/vera-VLE RNA interaction observed at the earliest time points in the localization pathway (Fig. 2B, lane 3, and C, lane 2), suggesting that PTB/hnRNP I promotes remodeling of interactions between Vg1RBP/vera and VLE RNA during transport in the cytoplasm.
FIG. 5.
Binding of PTB/hnRNP I to VLE RNA is required for direct interaction between Vg1RBP/vera and VLE RNA. (A) Cross-link IP analysis (as detailed in Fig. 2A) was carried out using oocytes expressing Vg1RBP/vera-FLAG (lanes 3 and 4), PTB/hnRNP I-FLAG (lanes 7 and 8), or control oocytes without expression of FLAG-tagged proteins (lanes 1, 2, 5, and 6). 32P-labeled VM1 mutant (ΔVM1; lanes 1, 3, 5, and 7) or wild-type (wt; lanes 2, 4, 6, and 8) VLE RNA transcripts were injected into the oocyte nuclei, and lysates were prepared after 16 h of culture. After cross-linking by UV irradiation, RNase treatment, and anti-FLAG IP, proteins directly bound to the injected RNA transcripts were resolved by SDS-PAGE and detected by autoradiography. The positions of Vg1RBP/vera-FLAG (lanes 1 to 4) and PTB/hnRNP I-FLAG (lanes 4 to 8) are indicated to the right. (B) RNA IP analysis (as diagrammed in Fig. 2A) was carried out using oocytes expressing Vg1RBP/vera-FLAG (lanes 1 and 2), PTB/hnRNP I-FLAG (lanes 3 and 4), or control oocytes without expression of FLAG-tagged proteins (lanes 5 and 6). 32P-labeled VM1 mutant (ΔVM1; lanes 1, 3, 5, and 7) or wild-type (wt; lanes 2, 4, 6, and 8) VLE RNA transcripts were injected into the oocyte nuclei, and lysates were prepared either immediately (input RNA; lanes 7 and 8) or after 16 h of culture (lanes 1 to 6). RNA was isolated after anti-FLAG IP and resolved by PAGE; the position of input VLE RNA is indicated at the right. (C) UV cross-linking analysis was performed using radiolabeled VLE RNA and stage III oocyte lysates. In vitro binding reactions included a 500-fold molar excess of nonspecific RNA (nsp.; lane 1) or a 500-fold binding site excess of wild-type VM1 RNA (VM1; lane 2) or RNAs containing mutated VM1 sites (mutVM1; lane 3). VLE-bound Vg1RBP/vera is at the top, and VLE-bound PTB/hnRNP I is at the bottom. (D) Quantitation of UV cross-linking results (as shown in panel C). Binding of Vg1RBP/vera to VLE RNA was quantitated by phosphorimage analysis, and the level of VLE-bound Vg1RPB/vera obtained in the presence of a 500- to 1,000-fold molar excess of nonspecific RNA (nsp.) was set to 1. The results of three experiments are graphed, and the levels of VLE-bound Vg1RBP/vera averaged 0.44 (standard deviation = 0.07) in the presence of 500- to 1,000-fold binding site excesses of unlabeled wild-type VM1 RNA (VM1) and averaged 0.78 (standard deviation = 0.12) in the presence of mutant VM1 RNA (mutVM1). For panels A to C, samples were run on the same gel, but lane order was changed for presentation in the figure.
Our results (Fig. 4A and B and 5A and B) for VLE RNAs lacking PTB/hnRNP I binding sites (ΔVM1) suggest that interaction between PTB/hnRNP I and VLE RNA is required for Vg1RBP/vera to directly bind VLE RNA. However, the possibility that the VM1 site mutations could cause secondary effects, unrelated to their abilities to bind PTB/hnRNP I, potentially affecting Vg1RBP/vera-VLE RNA interaction, remained. To address this issue, we sought to investigate Vg1RBP/vera interaction with wild-type VLE RNA under conditions where PTB/hnRNP I activity could be reduced. To reduce PTB/hnRNP I binding activity, we included a molar excess of unlabeled RNA transcripts consisting of three copies of the PTB/hnRNP I binding site (VM1) in in vitro binding reactions prior to UV cross-linking to radiolabeled VLE RNA. The 3×VM1 RNA contains only VM1 sites (no E2 sites) and has been shown to specifically bind PTB/hnRNP I, but not to Vg1RBP/vera (5, 23). As controls, we also included either nonspecific competitor RNA or RNA transcripts with three mutated VM1 sites, which cannot bind PTB/hnRNP I (5). As expected, binding of PTB/hnRNP I to VLE RNA (Fig. 5C, bottom) is eliminated in the presence of excess VM1 RNA (lane 2) and is unaffected by the excess mutant VM1 RNA (lane 3). Importantly, reduction of PTB/hnRNP I binding activity by inclusion of excess VM1 RNA also affected binding of Vg1RBP/vera to VLE RNA (Fig. 5C, top). Binding of Vg1RBP/vera to VLE RNA was reduced in the presence of excess wild-type (lane 2) but not mutant (lane 3) VM1 RNA. As shown in Fig. 5D, incubation with excess VM1 RNA reduced the binding of Vg1RBP/vera to VLE RNA more than twofold relative to the level observed in the presence of nonspecific competitor RNA, while only modest effects were detected in the presence of mutant VM1 RNA. As VM1 RNA interacts specifically with PTB/hnRNP I and does not itself bind to Vg1RBP/vera (5), the observed effects on binding of Vg1RBP/vera to VLE RNA are likely to be exerted through reduction in binding of PTB/hnRNP I to VLE RNA. These results suggest that binding of PTB/hnRNP I to VLE RNA is required to facilitate direct interactions between Vg1RBP/vera and VLE RNA.
DISCUSSION
RNA localization promotes cell polarity by spatially restricting protein expression. Targeting of mRNA molecules to discrete regions within the cell cytoplasm proceeds through multistep pathways (7, 35), but the molecular mechanisms directing transitions between steps in RNA localization pathways have remained unresolved. Remodeling of RNP complexes is an attractive mechanism for regulating such transitions, and we have found that the Vg1 RNP is remodeled in the cytoplasm during vegetal localization. At an early step in the Vg1 localization pathway, Vg1RBP/vera is recruited to the Vg1 RNP but does not bind VLE RNA directly. It is only later, when VLE RNA is undergoing transport in the cytoplasm, that Vg1RBP/vera associates directly with VLE RNA. In probing the mechanism of this remodeling event, we have uncovered a requirement for the RNA-binding protein PTB/hnRNP I. Moreover, mutations that prevent recruitment of PTB/hnRNP I to the Vg1 RNP block both remodeling of the VLE-Vg1RBP/vera interaction and RNA transport, suggesting a functional requirement for RNP remodeling during RNA localization.
Accumulating evidence indicates that RNA localization pathways initiate in the nucleus, as factors that bind to the RNA and assemble an early RNP influence localization of the RNA later in the cytoplasm (reviewed in reference 13). Indeed, PTB/hnRNP I and Vg1RBP/vera associate in RNP complexes with Vg1 RNA in the Xenopus oocyte nucleus and cytoplasm, although the nuclear and cytoplasmic RNP complexes are distinct (20). Potentially indirect protein-protein contacts appear to mediate the nuclear interaction between PTB/hnRNP I and Vg1RBP/vera, while in the cytoplasm, their interaction appears to be based on association with a shared target RNA (20). These results could suggest a role for PTB/hnRNP I in recruitment of Vg1RBP/vera to the Vg1 RNP, but our results disfavor this idea. Although PTB/hnRNP I appears to be critical for Vg1RBP/vera to gain direct contact with VLE RNA in the cytoplasm, our results show that initial recruitment of Vg1RBP/vera does not require PTB/hnRNP I. Instead, we propose a model (Fig. 6) in which an early step in the localization pathway is recruitment of PTB/hnRNP I to VLE RNA, through direct RNA-protein interactions. Recruitment of Vg1RBP/vera is an early step as well, but association with VLE RNA is indirect, mediated by protein-protein interactions. Initial recruitment of Vg1RBP/vera does not rely on PTB/hnRNP I, as mutations (ΔVM1) that prevent both binding of PTB/hnRNP I to VLE RNA and its association with the Vg1 RNP block direct binding of Vg1RBP/vera to VLE RNA but allow recruitment of Vg1RBP/vera to the Vg1 RNP (Fig. 5A and B). This early step in the localization pathway is likely to occur in the nucleus, as evidenced by comparison of the timing of direct Vg1RBP/vera-VLE RNA interaction with the time course of VLE localization in vivo. Vg1RBP/vera is not bound to VLE RNA at 1 h postinjection (Fig. 1D and 2B), at which point VLE RNA is still in the nucleus (Fig. 1A). At this same time point, PTB/hnRNP I is bound directly to VLE RNA (Fig. 2B), and both Vg1RBP/vera and PTB/hnRNP I are associated with the Vg1 RNP (Fig. 2C). We propose (Fig. 6) that PTB/hnRNP I facilitates remodeling of the Vg1 RNP such that Vg1RBP/vera can contact VLE RNA directly. Support for this proposal comes from our results (Fig. 4 and 5) showing that mutant VLE RNAs (ΔVM1) unable to bind PTB/hnRNP I can still recruit Vg1RBP/vera but that remodeling of the Vg1RBP/vera interaction to allow direct VLE RNA binding is blocked. We suggest that this remodeling event is a necessary step in the localization pathway, as the ΔVM1 VLE RNAs, which fail to remodel the Vg1RBP/vera-VLE RNA interaction, also fail to localize in vivo (Fig. 3, 4, and 6).
FIG. 6.
Model of RNA-protein interactions affecting RNP assembly and remodeling during RNA localization. (A) RNP assembly initiates in the nucleus, and during the early steps in the vegetal RNA localization pathway, PTB/hnRNP I (orange ovals) and potentially other nuclear factors (gray squares) are bound directly to VLE RNA sequences. Vg1RBP/vera (green triangles) is associated only indirectly with VLE RNA, through protein-protein interactions. The RNP is remodeled at later times during the vegetal RNA localization pathway, such that Vg1RBP/vera binds directly to VLE RNA. (B) VLE RNA transcripts containing mutations within VM1 sites cannot bind either directly or indirectly to PTB/hnRNP I. VM1 mutant VLE RNA transcripts bind Vg1RBP/vera (green triangles) indirectly, through protein-protein interactions, but remodeling of the RNP to allow direct interaction between Vg1RBP/vera and VLE RNA is blocked, as is vegetal RNA localization.
One question raised by these results is how PTB/hnRNP I might act to facilitate remodeling of the Vg1 RNP. It has been shown previously that the interaction between Vg1RBP/vera and PTB/hnRNP I is remodeled upon export from the nucleus to the cytoplasm, with the nuclear interaction being potentially direct, mediated by protein-protein contacts (20). These results could point toward a role for PTB/hnRNP I in blocking direct Vg1RBP/vera-RNA interactions in the nucleus during the early steps in the localization pathway. However, PTB/hnRNP I cannot act by simply blocking access of Vg1RBP/vera to VLE RNA early in localization, as VLE RNAs (ΔVM1) lacking PTB/hnRNP I fail to bind Vg1RBP/vera directly (Fig. 4 and 5). Instead, our results indicate that binding of PTB/hnRNP I to VM1 motifs within VLE RNA is required for Vg1RBP/vera to access its RNA-binding sites (E2 motifs). Consistent with this idea, reduction of PTB/hnRNP I binding activity also reduces binding of Vg1RBP/vera to VLE RNA in vitro (Fig. 5C and D). However, binding of PTB/hnRNP I to VLE RNA is not sufficient to permit Vg1RBP/vera-VLE RNA interaction, as evidenced by our analysis of the Vg1 RNP early in the localization pathway (Fig. 1 and 2). At early time points, PTB/hnRNP I is bound to VLE RNA, but Vg1RBP/vera is not directly bound to VLE RNA. It is only later, either during or likely after export of the Vg1 RNP from the nucleus to the cytoplasm, that direct interaction between Vg1RBP/vera and VLE RNA can be detected (Fig. 1 and 2). It is notable that export of PTB/hnRNP I from the nucleus to the cytoplasm is accompanied by its phosphorylation (38). Thus, it is possible that phosphorylation of PTB/hnRNP I may play a role in remodeling the Vg1 RNP by modulating protein-protein interactions within the Vg1 RNP.
A role for PTB/hnRNP I in RNP remodeling does not exclude functions for other nuclear (6) and cytoplasmic (20, 40, 43) components of the Vg1 RNP. For example, Vg1RBP/vera must be recruited to the Vg1 RNP in the nucleus, but PTB/hnRNP I cannot play this role, as recruitment can occur in the absence of PTB/hnRNP I binding (Fig. 4, 5, and 6). It is possible that access of Vg1RBP/vera to VLE RNA is blocked in the nucleus by interactions with other nuclear components of the Vg1 RNP and that PTB/hnRNP I may be required to promote RNP remodeling by displacing this factor after export of the Vg1 RNP to the cytoplasm. PTB/hnRNP I could also promote RNP remodeling by recruiting components of the Vg1 RNP during later steps in the localization pathway. Precedents for such a role can be found for PTB/hnRNP I homologs in other systems. For example, in neurons a PTB/hnRNP I isoform, nPTB, has been shown to act in recruitment of other factors to an RNP complex (26). In the Xenopus oocyte cytoplasm, additional factors are recruited to the Vg1 RNP (20) and could play roles in Vg1 RNP remodeling. Potential functions for other components of the Vg1 RNP in regulating transitions between steps in the RNA localization pathway remain to be explored.
Our results have revealed a function for PTB/hnRNP I in remodeling of the Vg1 RNP during cytoplasmic RNA transport. PTB/hnRNP I is assembled into the Vg1 RNP early in the RNA localization pathway, along with Vg1RBP/vera. At a later step in the localization pathway, the interaction between Vg1RBP/vera and Vg1 RNA is remodeled, resulting in a direct RNA-protein interaction. PTB/hnRNP I is necessary for this remodeling event, and mutations that block direct Vg1RBP/vera-VLE RNA interaction also disrupt vegetal localization in vivo. The identification of PTB/hnRNP I as a factor that promotes a necessary remodeling step in the Vg1 RNA localization pathway provides insight into the molecular pathway of vegetal RNA localization. We suggest that RNP remodeling events similar to those observed here may serve to regulate transitions between critical steps in other RNA localization pathways.
Acknowledgments
This work was supported by Public Health Service grant GM071049 from the National Institute of General Medicine to K.L.M. R.A.L. and J.A.G. were predoctoral trainees, supported in part by grant T32-GM07601 from the National Institute of General Medicine.
We thank Jeffrey Laney, Michael McKeown, Timothy Messitt, Catherine Pratt, Adrian Reich, and Tricia Serio for critical reading of the manuscript and for helpful advice and discussions.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Betley, J. N., B. Heinrich, I. Vernos, C. Sardet, F. Prodon, and J. O. Deshler. 2004. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14219-224. [DOI] [PubMed] [Google Scholar]
- 2.Birsoy, B., M. Kofron, K. Schaible, C. Wylie, and J. Heasman. 2006. Vg1 is an essential signaling molecule in Xenopus development. Development 13315-20. [DOI] [PubMed] [Google Scholar]
- 3.Bubunenko, M., T. L. Kress, U. D. Vempati, K. L. Mowry, and M. L. King. 2002. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 24882-92. [DOI] [PubMed] [Google Scholar]
- 4.Condeelis, J., and R. H. Singer. 2005. How and why does beta-actin mRNA target? Biol. Cell 9797-110. [DOI] [PubMed] [Google Scholar]
- 5.Cote, C. A., D. Gautreau, N. A. Terry, and K. L. Mowry. 1999. A Xenopus protein related to hnRNP I has an essential role in cytoplasmic RNA localization. Mol. Cell 4431-437. [DOI] [PubMed] [Google Scholar]
- 6.Czaplinski, K., T. Kocher, M. Schelder, A. Segref, M. Wilm, and I. W. Mattaj. 2005. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev. Cell 8505-515. [DOI] [PubMed] [Google Scholar]
- 7.Czaplinski, K., and R. H. Singer. 2006. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 31687-693. [DOI] [PubMed] [Google Scholar]
- 8.Deshler, J. O., M. I. Highett, T. Abramson, and B. J. Schnapp. 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8489-496. [DOI] [PubMed] [Google Scholar]
- 9.Deshler, J. O., M. I. Highett, and B. J. Schnapp. 1997. Localization of Xenopus Vg1 mRNA by vera protein and the endoplasmic reticulum. Science 2761128-1131. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3195-205. [DOI] [PubMed] [Google Scholar]
- 11.Dumont, J. N. 1972. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 136153-179. [DOI] [PubMed] [Google Scholar]
- 12.Gautreau, D., C. A. Cote, and K. L. Mowry. 1997. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development 1245014-5020. [DOI] [PubMed] [Google Scholar]
- 13.Giorgi, C., and M. J. Moore. 2007. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 18186-193. [DOI] [PubMed] [Google Scholar]
- 14.Gonsalvez, G. B., J. L. Little, and R. M. Long. 2004. ASH1 mRNA anchoring requires reorganization of the Myo4p-She3p-She2p transport complex. J. Biol. Chem. 27946286-46294. [DOI] [PubMed] [Google Scholar]
- 15.Gonsalvez, G. B., C. R. Urbinati, and R. M. Long. 2005. RNA localization in yeast: moving towards a mechanism. Biol. Cell 9775-86. [DOI] [PubMed] [Google Scholar]
- 16.Harland, R. M. 1991. In situ hybridization: an improved whole-mount method for Xenopus embryos, p. 685-695. In B. K. Kay and H. B. Peng (ed.), Xenopus laevis: practical uses in cell and molecular biology, vol. 36. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 17.Havin, L., A. Git, Z. Elisha, F. Oberman, K. Yaniv, S. P. Schwartz, N. Standart, and J. K. Yisraeli. 1998. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 121593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King, M. L., T. J. Messitt, and K. L. Mowry. 2005. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 9719-33. [DOI] [PubMed] [Google Scholar]
- 19.Kloc, M., and L. D. Etkin. 2005. RNA localization mechanisms in oocytes. J. Cell Sci. 118269-282. [DOI] [PubMed] [Google Scholar]
- 20.Kress, T. L., Y. J. Yoon, and K. L. Mowry. 2004. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 165203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroll, T. T., W. M. Zhao, C. Jiang, and P. W. Huber. 2002. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development 1295609-5619. [DOI] [PubMed] [Google Scholar]
- 22.Kwon, S., T. Abramson, T. P. Munro, C. M. John, M. Köhrmann, and B. J. Schnapp. 2002. UUCAC- and vera-dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 12558-564. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, R. A., T. L. Kress, C. A. Cote, D. Gautreau, M. E. Rokop, and K. L. Mowry. 2004. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech. Dev. 121101-109. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, R. A., and K. L. Mowry. 2007. Ribonucleoprotein remodeling during RNA localization. Differentiation 75507-518. [DOI] [PubMed] [Google Scholar]
- 25.Marello, K., J. La Rovere, and J. Sommerville. 1992. Binding of Xenopus oocyte masking proteins to mRNA sequences. Nucleic Acids Res. 205593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 207463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, K. C., and R. S. Zukin. 2006. RNA trafficking and local protein synthesis in dendrites: an overview. J. Neurosci. 267131-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melton, D. A. 1987. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature 32880-82. [DOI] [PubMed] [Google Scholar]
- 29.Minakhina, S., and R. Steward. 2005. Axes formation and RNA localization. Curr. Opin. Genet. Dev. 15416-421. [DOI] [PubMed] [Google Scholar]
- 30.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 3091514-1518. [DOI] [PubMed] [Google Scholar]
- 31.Mowry, K. L. 1996. Complex formation between stage-specific oocyte factors and a Xenopus mRNA localization element. Proc. Natl. Acad. Sci. USA 9314608-14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mowry, K. L., and D. A. Melton. 1992. Vegetal messenger RNA localization directed by a 340-nt sequence element in Xenopus oocytes. Science 255991-994. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, A., R. Amikura, K. Hanyu, and S. Kobayashi. 2001. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 1283233-3242. [DOI] [PubMed] [Google Scholar]
- 34.Palacios, I. M., D. Gatfield, D. St. Johnston, and E. Izaurralde. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427753-757. [DOI] [PubMed] [Google Scholar]
- 35.St. Johnston, D. 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6363-375. [DOI] [PubMed] [Google Scholar]
- 36.Wallace, R. A., and Z. Misulovin. 1978. Long-term growth and differentiation of Xenopus oocytes in a defined medium. Proc. Natl. Acad. Sci. USA 755534-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weeks, D. L., and D. A. Melton. 1987. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-β. Cell 51861-867. [DOI] [PubMed] [Google Scholar]
- 38.Xie, J., J.-A. Lee, T. L. Kress, K. L. Mowry, and D. L. Black. 2003. Protein kinase A phosphorylation modulates transport of polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 1008776-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yisraeli, J. K. 2005. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell 9787-96. [DOI] [PubMed] [Google Scholar]
- 40.Yoon, Y. J., and K. L. Mowry. 2004. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 1313035-3045. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Q., K. Yaniv, F. Oberman, U. Wolke, A. Git, M. Fromer, W. L. Taylor, D. Meyer, N. Standart, E. Raz, and J. K. Yisraeli. 1999. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech. Dev. 88101-106. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, W. M., C. Jiang, T. T. Kroll, and P. W. Huber. 2001. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 202315-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Y., and M. L. King. 1996. RNA transport to the vegetal cortex of Xenopus oocytes. Dev. Biol. 179173-183. [DOI] [PubMed] [Google Scholar]