Abstract
Methylation of histone H3 on lysine 9 is critical for diverse biological processes including transcriptional repression, heterochromatin formation, and X inactivation. The biological effects of histone methylation are thought to be mediated by effector proteins that recognize and bind to specific patterns of methylation. Using an unbiased in vitro biochemical approach, we have identified ICBP90, a transcription and cell cycle regulator, as a novel methyl K9 H3-specific binding protein. ICBP90 and its murine homologue Np95 are enriched in pericentric heterochromatin of interphase nuclei, and this localization is dependent on H3K9 methylation. Specific binding of ICBP90 to methyl K9 H3 depends on two functional domains, a PHD (plant homeodomain) finger that defines the binding specificity and an SRA (SET- and RING-associated) domain that promotes binding activity. Furthermore, we present evidence that ICBP90 is required for proper heterochromatin formation in mammalian cells.
Covalent modifications of the histone tails regulate virtually all aspects of chromatin biology. In addition to affecting histone-histone and histone-DNA interactions, posttranslational marks on the histone tails exert their modulatory role by generating docking sites for downstream effectors. Such molecules, often possessing enzymatic activities, serve as readers of histone modifications and participate in vital cellular processes, including transcription, replication, chromosome segregation, recombination, and DNA repair (15, 30, 58).
One of the most extensively studied histone tail modifications is methylation of histone H3 on lysine 9 (2). Histone H3 K9 methylation has been shown to be critical for regulation of gene expression, and it is enriched in transcriptionally inactive regions of the genome. It is considered a molecular mark of heterochromatin, the cytologically defined, gene-poor, and highly compacted regions of the chromatin. Interplay between H3 K9 methylation and DNA methylation has also been proposed in various models of heterochromatin formation and maintenance (32, 43, 51, 65). Furthermore, H3 K9 methylation is implicated in gene silencing phenomena such as X chromosome inactivation in female mammals and DNA elimination in the microscopic protozoon Tetrahymena (4, 60).
Several mammalian proteins, including SUV39H1, SUV39H2, G9a, ESET/SETDB1, and EuHTMase1, have been shown to have methyltransferase activity toward K9 of H3 (46, 47, 54, 56, 59, 72). Though they target the same histone residue, important differences exist among the above enzymes regarding their chemistry and distribution and consequently their biological roles. The reversibility of H3 K9 methylation has been an object of speculation for many years. Evidence for the removal of this covalent mark was obtained recently with the identification of specific histone demethylases (10, 17, 34, 40, 66, 67, 71). Although reversible, methylation appears to be much more stable compared to other histone modifications. Therefore, it is considered to play a major role in the establishment and maintenance of cellular memory.
In addition to the enzymes that write and erase this modification, identification of proteins that read the H3 K9 methyl mark is equally important in understanding its biology. Identification of heterochromatin protein 1 (HP1) as a protein that recognizes and interacts with methyl K9 H3 via its chromodomain provided a mechanistic link between H3 K9 methylation and heterochromatin formation, as well as related phenomena such as position effect variegation (3, 27, 35, 37, 44, 45).
To identify novel effectors of H3-K9 methylation, we undertook an unbiased in vitro biochemical approach using pull-down experiments. We identified ICBP90 as a protein that specifically binds to the histone H3 N-terminal tail when methylated on K9. Consistent with its specific binding of the K9-methylated H3 tail in vitro, ICBP90 and its murine homologue Np95 localize preferentially to pericentric heterochromatin in mouse cells in an H3K9me3-dependent fashion. Experiments addressing the biological function of ICBP90 in cultured mammalian cells suggest that ICBP90 is required for proper higher-order chromatin organization. Furthermore, ICBP90 possesses E3 ligase activity toward H3 both in vitro and in cells and its E3 ligase activity appears to be involved in heterochromatin formation and/or maintenance.
MATERIALS AND METHODS
Pull-down assays.
Nuclear extracts were prepared from HeLa cells by the protocol of Dignam et al. (11) and precleared with streptavidin-coated agarose beads. Biotinylated histone tail peptides were synthesized and purified by Genemed Synthesis Inc. For pull-down assays, the histone tail peptides were immobilized on streptavidin-coated beads, and after washing to remove the unbound peptide, they were incubated with HeLa cell nuclear extracts diluted once with binding buffer (20 mM HEPES [pH 7.9], 150 mM KCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10% glycerol, 0.1% NP-40, proteinase inhibitors) for 3 h at 4°C. Unbound proteins were removed by washing the beads with washing buffer (20 mM HEPES [pH 7.9], 150 mM KCl, 1 mM DTT, 1 mM PMSF, 0.1% NP-40, proteinase inhibitors). The proteins that remained bound to the peptides were boiled in sodium dodecyl sulfate (SDS) loading buffer and analyzed by polyacrylamide gel electrophoresis (PAGE). Mass spectrometry (53) was used to identify the proteins that preferentially bound to methyl K9 H3 versus unmodified H3. For competition assays, 2.5, 10, or 40 μg of nonbiotinylated peptide (H3 or H3K9me3) was included in the pull-down assay mixture with 0.25 μg of biotinylated H3K9me3. These peptides (residues 1 to 18 or 1 to 20; W. M. Keck Biotechnology Resource Center), as well as the H4 and H4K20me peptides, were a kind gift from M. Bedford. Radiolabeled proteins were generated with the TNT coupled reticulocyte lysate system (Promega).
Plasmid constructs and antibodies.
Human ICBP90 cDNA (full length and delta RING) cloned in vector pcDNA3.1 was a kind gift from M. Unoki (64). Mutant RING and deletion constructs were generated by site-directed mutagenic PCR and cloned into vector pSG5 with an N-terminal MS2-Flag. The JMJD2A construct has been previously described (34). For generation of the short hairpin RNA (shRNA) stable cell lines, oligonucleotides targeting ICBP90 were annealed and cloned into the pSuper vector (OligoEngine) by following the manufacturer's protocol. The following antibodies were obtained commercially: anti-H3K9me3 (Upstate), anti-H3 (Novus), anti-ICBP90 (BD Transduction), anti-HP1α and -β (Chemicon), anti-HA (anti-hemagglutinin; Cell Signaling), anti-β-actin, and anti-Flag (Sigma). Anti-Np95 was a kind gift from I. M. Bonapace (9). Polyclonal rabbit anti-JMJD2A antibody has been described previously (74). Flag-conjugated beads were purchased from Sigma.
Cell culture and transfection.
Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics. Stable cell lines were maintained in Dulbecco's modified Eagle's medium containing 5 μg/ml puromycin. Transfections were performed with Lipofectamine 2000 (Invitrogen) by following the manufacturer's protocol. For small interfering RNA (siRNA) treatment, SMART-pool against Np95 was purchased from Dharmacon.
Preparation of whole cell extracts and immunoprecipitation.
Cells were harvested 2 days posttransfection and lysed with IP lysis buffer (150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10 mM Tris [pH 7.6], 0.5% NP-40, 10% glycerol, 0.2 mM PMSF, leupeptin, pepstatin, aprotinin) for 30 min at 4°C with gentle shaking. Lysates were passed through a syringe with a needle and centrifuged for 15 min at maximum speed. The supernatant was used in an immunoprecipitation reaction with 0.5 to 1 μl of antibody and 5 μl of protein A/G+ agarose beads (Santa Cruz) for 2.5 h with rotation at 4°C, followed by five 10-min washes with IP lysis buffer. Samples were boiled in SDS loading buffer and analyzed by Western blotting.
In vitro ubiquitination assays.
Purified histones and recombinant H3 were obtained commercially (Upstate), and HeLa nucleosome preparations were a kind gift from Y. Zhang. Recombinant ICBP90 protein was generated with the Bac-to-Bac baculovirus expression system (Invitrogen) as described by Feng et al. (13). Immunoprecipitated or recombinant ICBP90 was used in an in vitro ubiquitination reaction with E1, E2, ubiquitin, and ATP purchased from Boston Biochem. Standard ubiquitination reaction mixtures contained 600 ng of recombinant ICBP90 protein, 10 μg of ubiquitin, 100 ng of E1, 300 ng of E2, 3 mM Mg-ATP, and 2 μg of histones as a substrate in ubiquitination reaction buffer (25 mM HEPES [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.05% Triton X-100, 0.5 mM DTT). The mixture was incubated at 37°C for 1 h, and the reaction was stopped by adding an equal volume of 2× SDS loading buffer. The reaction was resolved by SDS-PAGE, followed by Western blotting.
Preparation of core histones from mammalian cells.
Cells were lysed with IP lysis buffer and centrifuged at maximum speed. Two hundred microliters of water was added to the pellet, which was vortexed vigorously. Two hundred microliters of 25% trichloroacetic acid was added, and the samples were left on ice for 15 min. After centrifugation, the supernatant was discarded and 1 ml of ice-cold acetone was added to the pellet. The samples were vortexed, left at −20°C for 20 min, and centrifuged again. The supernatant was discarded, and the samples were air dried. Following the addition of 1× SDS loading buffer, the samples were boiled for 5 min, vortexed, and centrifuged. The supernatant was used for SDS-PAGE and Western blot analysis.
Immunocytochemistry.
Cells were grown on glass coverslips. Staining was performed at room temperature. Cells were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 10 min, and subsequently washed with PBS. Following permeabilization with 0.5% Triton X-100 in PBS for 10 min, cells were washed with PBS and incubated in blocking solution (5% bovine serum albumin [BSA] in PBS) for 30 min. Primary antibody staining was performed in 1% BSA in PBS for 1 h, followed by three washes with 1% BSA in PBS. Cells were then incubated with Alexa Fluor-conjugated secondary antibodies (Molecular Probes) for 1 h and washed with 1% BSA in PBS. DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI). Following washing with 1% BSA in PBS and one final wash with water, the coverslips were mounted on the slides and sealed with nail polish. Images were captured with a Zeiss Axioskop2 plus microscope and a Zeiss Axiocam MRc camera with a 63× oil objective. Alternatively, slides were visualized with an Applied Precision deconvolution microscope (DeltaVision). Images were processed with Adobe Photoshop (adjustment of brightness and contrast). Antibodies were used at the following dilutions: anti-ICBP90, 1:3,000; anti-Np95, 1:500; anti-H3K9me3, 1:3,000; anti-Flag, 1:5,000; anti-HP1α, β 1:5,000; anti-JMJD2A, 1:1,000.
RESULTS
ICBP90 selectively binds to di- and trimethyl K9 histone H3 peptides.
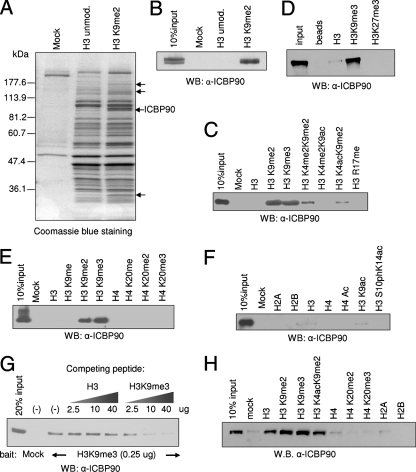
To identify proteins that selectively bind to methylated H3 K9, we performed in vitro pull-down assays. Similar approaches have previously identified effectors of other histone marks (69, 73). Biotinylated synthetic histone H3 peptides (amino acids 1 to 27), unmodified or dimethylated on K9, were immobilized on streptavidin-coated agarose beads and incubated with HeLa cell nuclear extracts. After washing, the proteins that remained bound to the peptides were dissolved in SDS loading buffer, resolved by SDS-PAGE, and visualized by Coomassie staining. Several proteins (Fig. 1A, arrows) bound preferentially to the dimethyl K9 versus the control H3 peptide. These bands were excised, digested with trypsin, and subjected to mass spectrometric analysis. The analysis identified HP1α and HP1γ (low-molecular-weight bands indicated by the arrows in Fig. 1A) among the proteins enriched in the H3K9me2 pull-down, providing a positive control for the assay. A polypeptide migrating at ∼90 kDa, enriched in the dimethyl K9 H3 pull-down assays, was identified as ICBP90 (Fig. 1A), a 793-amino-acid protein previously implicated in transcriptional regulation and cell cycle control. The identities of all of the other proteins in Fig. 1A will be described elsewhere. The specific binding of ICBP90 to the immobilized H3K9me2 peptide was confirmed by Western blotting with a specific anti-ICBP90 antibody (Fig. 1B).
FIG. 1.
ICBP90 interacts specifically with di- and trimethyl K9 H3 in vitro. (A) Pull-down assays were performed with HeLa cell nuclear extracts and unmodified (unmod.) or dimethyl K9 H3 peptides. Bound polypeptides were analyzed by SDS-PAGE and stained with Coomassie. Arrows indicate polypeptides enriched in the H3K9me2 pull-down. A polypeptide of ∼90 kDa enriched in the dimethyl K9 reaction was identified by mass spectrometry as ICBP90. (B) Pull-down assays were performed as described for panel A and analyzed by Western blotting (WB) with anti-ICBP90 antibody. (C, D, E, and F) Western blotting with anti-ICBP90 following pull-down of HeLa cell nuclear extracts with various histone tail peptides, as indicated. (G) Competition assays using pull-down of HeLa cell nuclear extracts with immobilized trimethyl K9 H3 and increasing amounts of free unmodified or trimethyl K9 H3 peptides. Bound polypeptides were analyzed by Western blotting with anti-ICBP90. (H) Pull-down of baculovirus-expressed ICBP90 with various histone tail peptides. Excess BSA was included as a blocking medium.
Pull-down assays with various immobilized synthetic histone peptides revealed that the binding of ICBP90 is highly selective for H3K9me2 and H3K9me3 (Fig. 1C, D, E, and F), whereas additional acetylation or methylation on lysine 4 reduces the affinity of the protein for the methyl K9 H3 tail (Fig. 1C). Furthermore, ICBP90 did not bind the H3K27me3 peptide (Fig. 1D), despite the similarity between the flanking amino acid sequences of H3K9 and H3K27. No interaction was detected with mono-, di-, or trimethyl K20 histone H4 tail peptides either, suggesting that the observed interaction is highly selective for methyl K9 H3 and not a general interaction with the methyl group (Fig. 1E). The specificity was also demonstrated in a competition assay where the binding of ICBP90 in HeLa cell nuclear extracts to immobilized H3K9me3 peptide was challenged with increasing amounts of free unmodified or trimethyl K9 H3 peptide. Western blot analysis showed that only the H3K9me3 peptide was able to compete for the binding of ICBP90 (Fig. 1G).
To address whether the physical interaction of ICBP90 with the histone H3 tail is direct, baculovirus-expressed ICBP90 protein was used in pull-down reactions in the presence of excess BSA. The recombinant protein also interacted preferentially with di- and trimethyl K9 H3 peptides, suggesting that the interaction is direct (Fig. 1H).
ICBP90 and Np95 localize in heterochromatic regions in interphase nuclei.
Since ICBP90 specifically bound to the H3K9me3 peptides in vitro, we wanted to determine if the same binding activity could be observed in cultured cells. In mammalian cells, H3K9 trimethylation is enriched in heterochromatic regions. In mouse cells, pericentric heterochromatin can be visualized by its intense DAPI staining. In interphase nuclei, these regions cluster into bright, DAPI-dense foci. We transfected human ICBP90 into NIH 3T3 cells and examined if transfected ICBP90 localized to the DAPI-dense foci. Immunostaining of NIH 3T3 cells for transfected human ICBP90 revealed predominantly heterochromatic localization of the protein (see Fig. 4D). Thus, in agreement with the in vitro binding data, ICBP90 localizes to pericentromeric heterochromatin, which is highly enriched in H3K9me3.
FIG. 4.
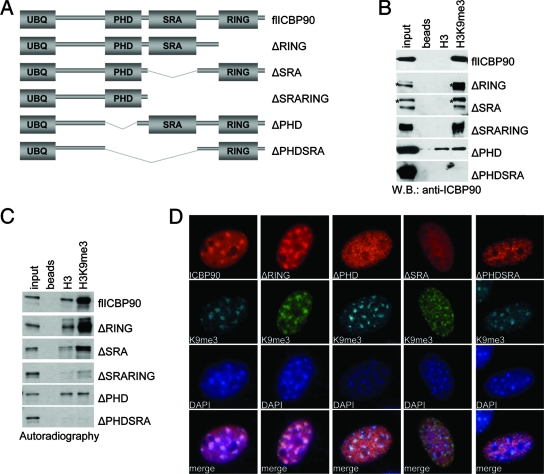
The PHD and the SRA domain of ICBP90 are required for trimethyl K9 H3 binding specificity and heterochromatic localization. (A) Schematic representation of the ICBP90 domain architecture as predicted by SMART (http://smart.embl-heidelberg.de/) and of the ICBP90 deletion constructs used in panels B, C, and D. (B) Pull-down assays with whole extracts of cells transfected with different ICBP90 deletion constructs (indicated on the right). Western blotting (W.B.) was done with anti-ICBP90 antibody. An asterisk indicates endogenous (full-length) protein. (C) Autoradiography following SDS-PAGE analysis of pull-down assays using different in vitro-translated, 35S-labeled ICBP90 deletion constructs (indicated on the right), with unmodified or trimethyl K9 H3 peptides. Input is 5%. (D) Immunolocalization of full-length ICBP90 (red) or ICBP90 deletion constructs (red) transfected into NIH 3T3 cells. Cells were also stained with anti-H3K9me3 (green) and DAPI (blue). Staining was performed 2 days posttransfection.
We also analyzed the localization of endogenous murine Np95 by immunostaining of NIH 3T3 cells with an anti-Np95-specific antibody. This analysis revealed enrichment of Np95 protein in pericentric heterochromatin (see Fig. 3C), consistent with previous studies on the subcellular localization of Np95 (9, 48).
FIG. 3.
Overexpression of JMJD2A demethylase results in diffuse nuclear ICBP90 and Np95 staining. (A) NIH 3T3 cells were transfected with Flag-tagged JMJD2A and stained with anti-Flag and anti-H3K9me3 antibodies. Overexpression of the demethylase results in reduced H3K9me3 in transfected nuclei. (B) NIH 3T3 cells were transfected with Flag-JMJD2A and stained with anti-JMJD2A and anti-HP1α antibodies. Transfected cells display reduced levels and diffuse localization of HP1α. (C) NIH 3T3 cells were transfected with Flag-JMJD2A and stained with anti-Flag and anti-Np95 antibodies. Transfected cells display reduced levels and diffuse localization of Np95. (D) NIH 3T3 cells were cotransfected with ICBP90 and JMJD2A. Two days posttransfection, the cells were stained with anti-ICBP90 (red), anti-trimethyl K9 H3 (green), and DAPI (blue). An arrow indicates a nucleus with reduced trimethyl K9 H3 signal, as a result of JMJD2A overexpression. An arrowhead indicates a cell with normal H3K9me3 levels, transfected with ICBP90. Brightness and contrast were adjusted with Adobe Photoshop.
The above results indicate that ICBP90 and Np95 exhibit similar localization with HP1, particularly the alpha and beta isoforms (3, 35), and thus support ICBP90 and Np95 as a family of novel methyl K9 H3 binding proteins. However, a notable distinction exists between the localization of HP1 and that of Np95 and ICBP90 during mitosis. More specifically, HP1 dissociates from the mitotic chromosomes as a result of H3 serine 10 phosphorylation by mitotic kinases (14, 19). Immunostaining of murine NIH 3T3 cells for Np95 indicates that the protein remains associated with the mitotic chromosomes (9, 63; data not shown). Similar observations have also been previously reported for green fluorescent protein-fused ICBP90 (29). We were not able to detect chromosome-bound endogenous ICBP90 by immunocytochemistry, perhaps due to epitope masking or modification of ICBP90 during mitosis, which may interfere with antibody recognition.
Methylated H3 K9 signals heterochromatic localization of ICBP90 and Np95.
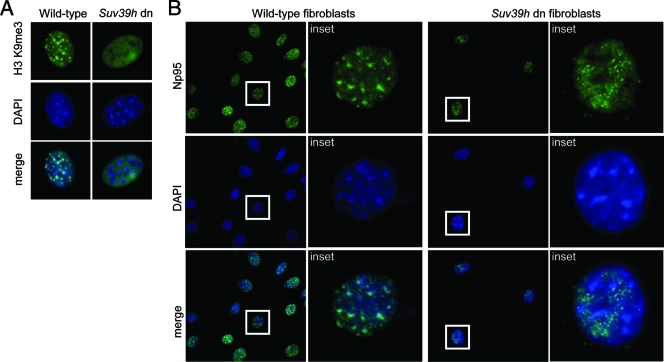
To examine whether heterochromatic localization of ICBP90 and Np95 depends on the H3K9me3 modification, we first used Suv39h double-null mouse embryonic fibroblasts (MEFs). These cells display a significant reduction of H3 K9 methylation at pericentric heterochromatin, whereas the broad H3 K9 methylation in other regions is not severely impaired (50) (Fig. 2A). Heterochromatic enrichment of Np95 is disrupted in Suv39h double-null MEFs. In these cells, the protein exhibits a granular staining pattern throughout the nucleoplasm, excluding the nucleoli (Fig. 2B). A similar effect has been described for HP1 (35, 38).
FIG. 2.
Heterochromatic localization of ICBP90 and Np95 depends on H3K9 methylation. (A) Immunostaining of Suv39h wild-type (left) and double-null (dn; right) MEFs with anti-trimethyl K9 H3 (green) and DAPI (blue). Representative interphase nuclei are shown. (B) Suv39h wild-type and double-null MEFs stained with anti-Np95 (green) and DAPI (blue). Np95 displays diffuse nuclear localization in Suv39h double-null fibroblasts.
We then analyzed the effect of acute reduction of H3K9 methylation on the localization of Np95 to heterochromatin by overexpressing an H3K9 demethylase, JMJD2A/JHDM3A, in NIH 3T3 cells (Fig. 3A). In addition to the reduction of H3K9me3, overexpression of JMJD2A in these cells resulted in diffuse HP1α and -β localization (Fig. 3B; also see reference 34). Similarly, cells transfected with JMJD2A alone also demonstrated reduced intensity and a diffuse localization pattern of Np95 staining in the nucleus (Fig. 3C).
We then cotransfected NIH 3T3 cells with JMJD2A and ICBP90 to determine if acute reduction of H3K9me3 affected the localization of ICBP90 to heterochromatin. Because the antibodies against ICBP90 and the Flag tag (for detection of transfected JMJD2A) were both mouse monoclonal antibodies, we indirectly determined cells overexpressing JMJD2A by immunostaining against H3K9me3. While ICBP90 exhibited heterochromatic enrichment in cells with normal levels of H3K9me3 (Fig. 3D, arrowhead), it displayed diffuse localization in cells with significantly reduced H3K9me3, presumably due to JMJD2A overexpression (Fig. 3D, arrow). We conclude that in these cell culture models, H3K9me3 is required for proper localization of ICBP90 and Np95 to heterochromatin, consistent with the in vitro binding data.
The PHD (plant homeodomain) and the SRA domain of ICBP90 are required for specific binding to methyl K9 H3.
ICBP90 contains a ubiquitin-like domain in the amino terminus, a PHD, a SET- and RING-associated (SRA) domain, and a carboxy-terminal RING finger motif (Fig. 4A). Since ICBP90 lacks a conserved chromodomain, which is known to interact with methyl K9 H3, we were interested in identifying the structural domain(s) that mediates its interaction with the K9-methylated H3 tail. To this end, different ICBP90 deletion constructs were expressed in mammalian cells and their binding specificity was tested in pull-down assays with unmodified or H3K9me3 peptides, followed by Western analysis (Fig. 4B). Deletion of the RING domain had no significant effect on the binding of ICBP90 to the H3K9me3 peptide. Endogenous ICBP90 (marked by asterisks) was also detected, serving as an internal control for comparison of the efficacy of binding between full-length ICBP90 and the deletion mutant forms. Deletion of the SRA domain resulted in reduced binding of the H3K9me3 peptide. Previous studies have implicated the SRA domain as a chromatin- and methylcytosine-binding domain (9, 31, 64, 68). Our data indicate that the SRA domain also contributes to the binding of ICBP90 to H3K9me3. Interestingly, deletion of the PHD resulted in complete loss of binding specificity for the H3K9me3 peptide. The resulting molecule bound equally well, but with reduced affinity, to both unmodified and H3K9me3 peptides. Furthermore, deletion of both the PHD and the SRA domain completely abolished the binding of ICBP90 to either H3 peptide. The same set of ICBP90 mutants were also expressed by in vitro transcription and translation reactions, and their binding to control and H3K9me3 peptides was tested (Fig. 4C). The results were essentially identical to the data in Fig. 4B, with the exception of a weak binding to the unmodified H3 peptide observed for full-length ICBP90 and most of the deletion mutants. Together, these results define the PHD as a major determinant for binding of ICBP90 to H3K9me3, with the SRA domain contributing to the overall H3 binding affinity.
The PHD and the SRA domain are required for localization of ICBP90 to pericentric heterochromatin.
We then examined the subcellular localization of the above ICBP90 deletion mutants. Constructs encoding full-length ICBP90 and ICBP90 with the PHD deleted, with the SRA domain deleted, with the PHD and the SRA domain deleted, and with the RING domain deleted were transfected into NIH 3T3 cells. All of the proteins were Flag tagged, and their localization was analyzed by immunocytochemistry with an anti-Flag antibody (Fig. 4D). The immunostaining analysis revealed that, similar to the full-length protein, ICBP90 with the RING domain deleted localized predominantly to the DAPI-dense chromocenters in interphase nuclei. In contrast, ICBP90 protein lacking the SRA domain, the PHD, or both the SRA domain and the PHD did not exhibit enrichment in pericentromeric heterochromatin. Instead, all of the above domain deletions resulted in diffuse localization of ICBP90 throughout the nucleoplasm. These results suggest that the PHD and the SRA domain of ICBP90 are required for proper heterochromatic localization of the protein.
Downregulation of ICBP90 and Np95 disrupts the distribution of H3K9me3 and HP1α, respectively, in interphase nuclei.
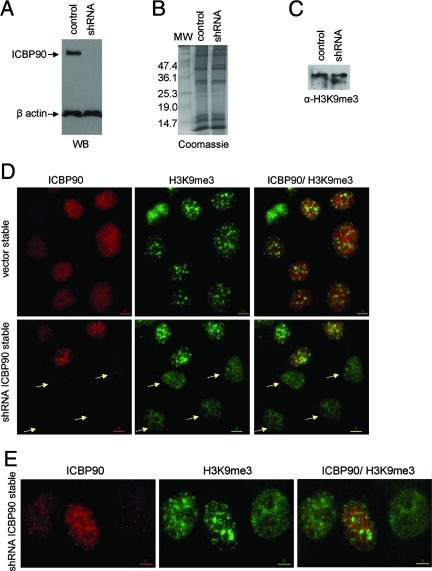
To address the biological significance of the interaction between ICBP90 and the methylated histone H3 tail, we stably transfected HeLa cells with a construct encoding shRNA against ICBP90. As a control, we used cells stably transfected with the empty vector (pSuper). We note here that in HeLa cells, endogenous ICBP90 exhibits a punctate pattern throughout the nucleus (see Fig. 8A) that is different from the focal enrichment in the heterochromatic regions of NIH 3T3 nuclei. In spite of that, we used HeLa cells for this assay because they provide a relatively easy system for stable transfection. Efficient downregulation of ICBP90 protein was confirmed by Western blot analysis and immunocytochemistry (Fig. 5A and D). Since there are several paradigms of proteins that recognize a histone modification and also affect the levels of this modification in the cell (1, 55, 69), we asked whether downregulation of ICBP90 had any effect on the global H3 K9 methylation levels. We prepared crude core histones from the control and ICBP90 shRNA stable cells. Coomassie blue staining showed similar levels of core histones from the two samples (Fig. 5B). Western blot analysis with an H3K9me3-specific antibody did not reveal any change in the levels of H3K9me3 between control cells and cells stably expressing shRNA against ICBP90 (Fig. 5C). However, analysis of cells stably transfected with shRNA against ICBP90 revealed that, in contrast to the focal heterochromatic enrichment of H3K9me3 in control cells and shRNA cells with normal levels of ICBP90 protein, shRNA cells with low levels of ICBP90 displayed a diffuse granular pattern of H3K9me3 staining (Fig. 5D and E). Note that the cell in the upper part of Fig. 5D is in mitosis; therefore, as mentioned above, endogenous ICBP90 is hardly detectable by immunostaining.
FIG. 8.
ICBP90 promotes H3 ubiquitination when transfected into cells. (A) 293T cells were transfected (transf.) with full-length ICBP90 or ICBP90 with the RING domain deleted (ΔRING) and HA-tagged ubiquitin (HAUb) in the combinations shown at the top. Two days posttransfection, core histones were prepared from the cells and analyzed by Western blotting (WB) with anti-HA antibody to detect ubiquitinated chromatin-associated products. The values on the left are molecular sizes in kilodaltons. (B) Western blotting of the core histone preparations described for panel A with anti-H3 antibody to detect modified forms of H3. The bottom part shows total H3 levels and an exposure time shorter than that in the top part.
FIG. 5.
Downregulation of ICBP90 disrupts the heterochromatic distribution of trimethyl K9 H3. (A) Analysis of whole cell extracts from a stable control (empty pSuper vector) and shRNA against ICBP90 cells by Western blotting (WB) with anti-ICBP90 and anti-β-actin. Coomassie staining (B) and anti-trimethyl K9 H3 Western blotting (C) of crude chromatin preparations from control and shRNA stable cells. (D) Stable control and shRNA against ICBP90 cells stained with anti-ICBP90 (red) and anti-trimethyl K9 H3 (green). The cell in the upper left corner (vector stable) is in mitosis. Mitotic HeLa cells show reduced staining by anti-ICBP90 antibody. (E) Higher magnification of representative cells stably transfected with shRNA against ICBP90.
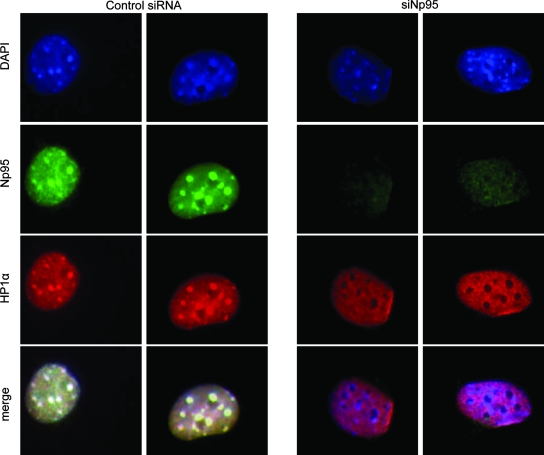
Similar to the effect of shRNA-mediated downregulation of ICBP90, siRNA-mediated downregulation of endogenous Np95 in NIH 3T3 cells resulted in disruption of the heterochromatic localization of HP1α (Fig. 6). It is noteworthy that this effect was observable only after long-term siRNA treatment (at least 5 days). Based on these observations, we propose that ICBP90 and Np95 contribute to the establishment and/or maintenance of the specific organization of H3K9me3 and HP1α in heterochromatic regions of the nucleus.
FIG. 6.
siRNA-mediated downregulation of Np95 disrupts heterochromatic localization of HP1α. NIH 3T3 cells were transfected with siRNA against Np95 and stained with anti-Np95 (green), anti-HP1α (red), and DAPI (blue) at 5 days posttransfection.
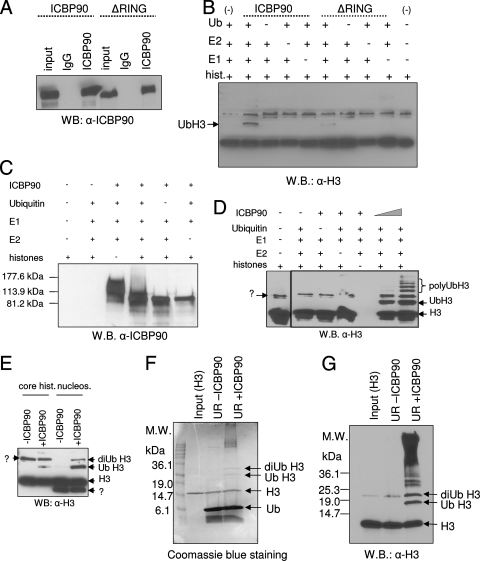
ICBP90 promotes H3 ubiquitination in transfected cells and acts as an E3 ubiquitin ligase for H3 in vitro.
Np95 has been shown to ubiquitinate core histones, especially H3 in vitro (9). ICBP90 has not been reported to have E3 ligase activity for histones but possesses autoubiquitination activity, dependent on its RING domain (29). To test if ICBP90 can ubiquitinate its binding substrate, H3, full-length ICBP90 or ICBP90 with the RING domain deleted was immunoprecipitated from transfected HeLa cells (Fig. 7A) and incubated with core histones in the absence or presence of recombinant E1, E2, and ubiquitin. Immunoprecipitated ICBP90 could ubiquitinate histone H3 in vitro in a RING domain-dependent fashion (Fig. 7B). Similarly, when used in an in vitro ubiquitination reaction, recombinant ICBP90 protein also exhibited strong autoubiquitination activity (Fig. 7C and reference 29), as well as E3 ligase activity for H3. Inclusion of histones in the reaction mixture greatly diminished the autoubiquitination of ICBP90 (Fig. 7C), indicating a possible competition between the substrates (histone versus self). Recombinant ICBP90 protein exhibited E3 ligase activity for H3 in vitro when tested with core histones (Fig. 7D), nucleosomes (Fig. 7E), or recombinant H3 (Fig. 7F and G) as a substrate. Among these forms, nucleosomal H3 appears to be a better substrate for ICBP90 (Fig. 7E).
FIG. 7.
ICBP90 functions as an E3 ubiquitin ligase for H3 in vitro. (A) HeLa cells were transfected with full-length ICBP90 or ICBP90 with the RING domain deleted. Immunoprecipitation reaction mixtures were analyzed by Western blotting (WB) with anti-ICBP90. IgG, immunoglobulin G. (B) The immunoprecipitated proteins from panel A were used in an in vitro ubiquitination reaction with core histones (hist.), ubiquitin (Ub), E1, and E2, as indicated at the top. The reaction products were analyzed by SDS-PAGE, and Western blot analysis was performed with anti-H3 antibody. (C) Recombinant ICBP90 has autoubiquitination activity. Western blotting with anti-ICBP90 was done following the in vitro ubiquitination reaction. Lower autoubiquitination levels are observed in the presence of histones in the reaction mixture. (D and E) In vitro ubiquitination reaction with baculovirus-expressed ICBP90, ubiquitin, E1, E2, and core histones or nucleosomes as the substrate. Results were subject to Western blot analysis with anti-Η3 antibody. (F and G) In vitro ubiquitination reaction with baculovirus-expressed ICBP90, ubiquitin, E1, E2, and recombinant H3 as the substrate. Results were analyzed by SDS-PAGE and Coomassie staining (E) or Western blotting with anti-Η3 antibody (F). UR, ubiquitination reaction; M.W., molecular mass; hist., histones; nucleos., nucleosomes.
We next tested if ICBP90 could ubiquitinate histone H3 in cultured cells. HA-tagged ubiquitin was cotransfected with or without either full-length ICBP90 or ICBP90 with the RING domain deleted into 293T cells similar to a previous report (41). Crude chromatin fractions were analyzed by Western blotting with anti-HA to detect ubiquitinated proteins (Fig. 8A) or with anti-H3 to detect modified forms of H3 (Fig. 8B). Higher levels of ubiquitination in cells transfected with HA-ubiquitin alone are most likely due to increased HA-ubiquitin expression in these cells compared to cells cotransfected with full-length ICBP90 or ICBP90 with the RING domain deleted (Fig. 8A, compare lanes 2 and 3 with lanes 4 and 5). Cotransfection of ICBP90 and HA-ubiquitin altered the pattern of ubiquitinated proteins, suggesting that these proteins were ubiquitinated in an ICBP90-dependent manner (Fig. 8A, lane 4). A band of approximately 25 kDa was detected by both the anti-HA and anti-H3 antibodies. We conclude that this band corresponds to ubiquitinated H3 (Fig. 8A and B, lanes 4). The level of ubiquitinated H3 was substantially lower in cells cotransfected with HA-ubiquitin and ICBP90 with the RING domain deleted (Fig. 8A and B, lanes 5), indicating that the ubiquitination of H3 is dependent on ICBP90 E3 ligase activity. In addition to H3, another chromatin-associated protein (Fig. 8A, arrow and upper question mark) was also abundantly ubiquitinated in an ICBP90-dependent manner. The identity of this protein is unknown. We conclude that ICBP90 can function as an E3 ubiquitin ligase for histone H3 both in vitro and in transfected cells.
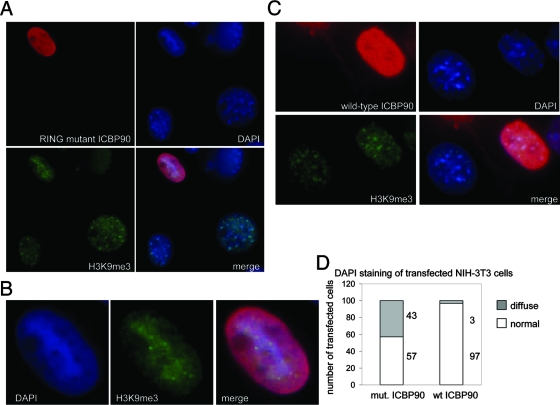
Overexpression of an enzymatically inactive ICBP90 mutant in mouse cells disrupts the higher-order chromatin structure.
To understand whether the observed effect of ICBP90 on heterochromatin depends on its E3 ligase activity, we generated a point mutant (H741R) that is enzymatically inactive due to disruption of the ability of the RING domain to complex with zinc (25, 29). Since all of the other domains of the protein remain intact, we predicted that overexpression of this mutant might exert a deleterious effect toward the endogenous wild-type protein and potentially act as a dominant-negative form. Indeed, NIH 3T3 cells highly overexpressing ICBP90 H741R displayed a phenotype characterized by loss of the distinct DAPI-dense heterochromatic regions (Fig. 9A and B). Instead, the DAPI staining of interphase nuclei became homogeneous. Note here that upon high overexpression, localization of ICBP90 is no longer limited to the heterochromatic regions but can be detected throughout the nucleoplasm. Moderate or lower overexpression of the mutant did not cause any observable change in the DAPI staining of the transfected cells (data not shown). As a control, overexpression of the wild-type ICBP90 protein, at any level, did not disrupt the normal heterochromatic pattern (Fig. 9C and D). Note here that we are looking at cells highly overexpressing either wild-type or RING mutant ICBP90, which is why the proteins are localized throughout the nucleus. These results support the idea that overexpression of a RING mutant ICBP90 form disrupts higher-order organization of heterochromatin in interphase mouse cells.
FIG. 9.
Overexpression of RING mutant ICBP90 results in diffuse DAPI staining of transfected nuclei. (A) NIH 3T3 cells were transfected with ICBP90 carrying a point mutation in the RING finger and stained with anti-ICBP90 (red) and DAPI (blue). (B) Enlarged pictures of transfected cells from panel A. (C) NIH 3T3 cells were transfected with wild-type ICBP90 and stained with anti-ICBP90 (red) and DAPI (blue). Brightness and contrast were adjusted with Adobe Photoshop. (D) Graphic representation of the percentage of cells transfected with RING point mutant (mut.) or wild-type (wt) ICBP90 displaying diffuse DAPI staining. Fifty-seven percent of the cells transfected with the RING mutant (H741R) show normal DAPI staining, and 43% show diffuse DAPI staining. Ninety-seven percent of the cells transfected with wild-type ICBP90 show normal DAPI staining.
DISCUSSION
Identification of ICBP90 as a novel methyl K9 H3 effector.
The critical role of H3 K9 methylation in cellular processes, combined with the small number of known proteins recognizing this mark, led us to screen for new interacting partners for this modification. We report here the identification of ICBP90 as a novel di- and trimethyl K9 H3 binding protein. This property of ICBP90 is added to an already long list of interesting functions of this protein, such as regulation of topoisomerase IIα and Rb levels (20, 21, 22, 28), cell cycle regulation (5, 29), binding to methylated CpG islands, and interaction with HDAC1 (64). It has been previously reported that the normally fluctuating expression of ICBP90 in cycling cells is perturbed in most cancer cell lines (5, 18, 21, 42). Since there is increasing evidence suggesting a role for epigenetic mechanisms in carcinogenesis (12, 62), it would be interesting to determine if the role of ICBP90 in cancer is dependent upon its methyl K9 H3 binding activity.
Our results outline several similarities of ICBP90 and Np95 and HP1, particularly the alpha and beta isoforms, such as recognition of methyl K9 H3, heterochromatic localization, and involvement in the establishment and/or maintenance of a higher-order chromatin organization. Analogously to HP1, ICBP90 and Np95 also appear to participate in a self-propagating circuit by recognizing a histone mark and aiding its stabilization. However, the difference in the mitotic localization of these proteins is intriguing. By remaining attached to the mitotic chromosomes, ICBP90 and Np95 may have a role in protecting the methyl marks throughout a phase of dramatic changes in chromatin organization and be involved in the maintenance of cellular memory.
Recently, Woo and colleagues reported a phenotype characterized by defective heterochromatin formation, upon depletion of VIM1, an ICBP90-related protein in Arabidopsis (68). Furthermore, murine Np95 has also been suggested to have a role in the replication of pericentric heterochromatin (48). It would be interesting, therefore, to examine whether other proteins with a similar domain organization, such as NIRF, have functions in common with ICBP90 and its homologues.
The PHD in the recognition of methylated histone marks.
Recent studies have revealed the PHD as a binding motif for methylated H3 K4 (36, 37, 39, 49, 57, 61, 70). In addition, a PHD motif from the SMCX demethylase was shown to bind H3K9me3 in vitro (26). Our data also support a role for the PHD of ICBP90 in the preferential interaction with H3K9me3, although the minimal PHD of ICBP90 may not be sufficient for this interaction (data not shown). These observations may aid in the discovery of new methyl K9 H3 effectors. The ability of the PHD to read different methylated lysines is reminiscent of the behavior of the other known methyl-lysine-recognizing motif, the chromodomain, which has been shown to interact with both methyl K4 and methyl K9 H3 (6, 7, 16, 52). Thus, variations of the same motif may exert different biological effects. Combination of different modules, such as the SRA domain and PHD of ICBP90, appears to further increase diversity in the reading of chromatin modifications.
ICBP90 may provide a link between histone methylation and DNA methylation.
The SRA domains of several proteins, including ICBP90, have been shown to interact with methylated DNA (65). Our results also support the idea that the SRA domain may also contribute to histone tail recognition. Mutant ICBP90 lacking the SRA domain maintains specific interaction with H3K9me3, though with reduced affinity compared to the full-length protein. However, ICBP90 with the SRA domain deleted, similar to ICBP90 with the PHD deleted, is not preferentially enriched in pericentromeric heterochromatin. Given that the PHD appears to be required for specific interaction between ICBP90 and H3K9me3, we propose that in vivo heterochromatic localization of ICBP90 depends on interaction with both H3K9me3 and methylated DNA. Interestingly, Bostick and colleagues recently reported that ICBP90 interacts with DNMT1 and is involved in the maintenance of DNA methylation in mammalian cells (6). Therefore, ICBP90 may provide a link between histone methylation and DNA methylation.
Chromatin ubiquitination and heterochromatin formation.
Genetic and biochemical studies of Schizosaccharomyces pombe have revealed a critical role for protein ubiquitination in heterochromatin formation and H3K9 methylation (8, 23, 24). In this unicellular eukaryote, Clr4 is considered the sole enzyme responsible for H3K9 methylation (43). Clr4 was found to associate with Cul4, a member of the cullin family, which acts as a scaffold for the assembly of ubiquitin ligases (33). Ubiquitin ligase-active Cul4 is required for proper Clr4 localization, H3K9 methylation, and heterochromatin formation (33).
The results obtained in this study suggest a role for ubiquitination in heterochromatin formation in mammalian cells. Our present data are not sufficient to establish whether H3 is the physiological in vivo substrate for ICBP90 and its principal biological effector, although the binding specificity for methylated H3 may be accompanied by an in vivo preference for this histone. Finally, we can only speculate about the potential outcome of ICBP90-dependent chromatin ubiquitination. It is possible that the modification might target its substrate for degradation. Alternatively, attachment of a bulky moiety like ubiquitin might affect molecular interactions or change the local chromatin structure. Based on our observations, we favor a model in which ICBP90 binds to H3K9me3 and promotes local chromatin ubiquitination. This event could subsequently signal the recruitment of downstream molecules, such as methyltransferases and deacetylases, which will induce spreading of heterochromatin. Notwithstanding several remaining mechanistic questions, the present studies establish the importance of ICBP90 and implicate its ubiquitin ligase activity in the maintenance of heterochromatin in mammalian cells.
Acknowledgments
We thank Qin Feng for technical assistance, Thomas Jenuwein for Suv39h double-null and wild-type MEFs, Fred Pereira for permission to use laboratory equipment, Motoko Unoki for plasmids, Yi Zhang for nucleosome preparations, Ian Marc Bonapace for anti-Np95 antibody, and Mark Bedford for peptides. We are grateful to Jeffrey Rosen, Sharon Dent, Sophia Tsai, and David Stewart for critical readings of the manuscript and to Michelle Barton and Tae Ho Shin for helpful discussions.
This work was supported by grant DK58679 to J.W.
Footnotes
Published ahead of print on 29 October 2007.
REFERENCES
- 1.Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 181923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109801-806. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410120-124. [DOI] [PubMed] [Google Scholar]
- 4.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 3073-76. [DOI] [PubMed] [Google Scholar]
- 5.Bonapace, I. M., L. Latella, R. Papait, F. Nicassio, A. Sacco, M. Muto, M. Crescenzi, and P. P. Di Fiore. 2002. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 157909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bostick, M., J. K. Kim, P. O. Esteve, A. Clark, S. Pradhan, and S. E. Jacobsen. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 3171760-1764. [DOI] [PubMed] [Google Scholar]
- 7.Brehm, A., K. R. Tufteland, A. Rein, and P. B. Becker. 2004. The many colours of chromodomains. Bioessays 26133-140. [DOI] [PubMed] [Google Scholar]
- 8.Choi, E. S., H. S. Kim, Y. K. Jang, S. H. Hong, and S. D. Park. 2002. Two ubiquitin-conjugating enzymes, Rhp6 and UbcX, regulate heterochromatin silencing in Schizosaccharomyces pombe. Mol. Cell. Biol. 228366-8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citterio, E., R. Papait, F. Nicassio, M. Vecchi, P. Gomiero, R. Mantovani, P. P. Di Fiore, and I. M. Bonapace. 2004. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 242526-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloos, P. A. C., J. Christensen, K. Agger, A. Maiolica, J. Rappsilber, T. Antal, K. H. Hansen, and K. Helin. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442307-311. [DOI] [PubMed] [Google Scholar]
- 11.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducasse, M., and M. A. Brown. 2006. Epigenetic aberrations and cancer. Mol. Cancer 560. doi: 10.1186/1476-4598-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, Q., P. Yi, J. Wong, and B. W. O'Malley. 2006. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 267846-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischle, W., B. S. Tseng, H. L. Dormann, B. M. Ueberheide, B. A. Garcia, J. Shabanowitz, D. F. Hunt, H. Funabiki, and C. D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 4381116-1122. [DOI] [PubMed] [Google Scholar]
- 15.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15172-183. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan, J. F., L.-Z. Mi, M. Chruszcz, M. Cymborowski, K. L. Clines, Y. Kim, W. Minor, F. Rastinejad, and S. Khorasanizadeh. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 4381181-1185. [DOI] [PubMed] [Google Scholar]
- 17.Fodor, B. D., S. Kubicek, M. Yonezawa, R. J. O'Sullivan, R. Sengupta, L. Perez-Burgos, S. Opravil, K. Mechtler, G. Schotta, and T. Jenuwein. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 201557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimori, A., Y. Matsuda, Y. Takemoto, Y. Hashimoto, E. Kubo, R. Araki, R. Fukumura, K. Mita, K. Tatsumi, and M. Muto. 1998. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome 91032-1035. [DOI] [PubMed] [Google Scholar]
- 19.Hirota, T., J. J. Lipp, B. H. Toh, and J. M. Peters. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 4381176-1180. [DOI] [PubMed] [Google Scholar]
- 20.Hopfner, R., M. Mousli, J.-M. Garnier, R. Redon, S. du Manoir, B. Chatton, N. Ghyselinck, P. Oudet, and C. Bronner. 2001. Genomic structure and chromosomal mapping of the gene coding for ICBP90, a protein involved in the regulation of the topoisomerase IIα gene expression. Gene 26615-23. [DOI] [PubMed] [Google Scholar]
- 21.Hopfner, R., M. Mousli, J.-M. Jeltsch, A. Voulgaris, Y. Lutz, C. Marin, J.-P. Bellocq, P. Oudet, and C. Bronner. 2000. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIα expression. Cancer Res. 60121-128. [PubMed] [Google Scholar]
- 22.Hopfner, R., M. Mousli, P. Oudet, and C. Bronner. 2002. Overexpression of ICBP90, a novel CCAAT-binding protein, overcomes cell contact inhibition by forcing topoisomerase IIα expression. Anticancer Res. 223165-3170. [PubMed] [Google Scholar]
- 23.Horn, P. J., J. N. Bastie, and C. L. Peterson. 2005. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 191705-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horn, P. J., and C. L. Peterson. 2006. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 1483-94. [DOI] [PubMed] [Google Scholar]
- 25.Huang, J., Q. Huang, X. Zhou, M. M. Shen, A. Yen, S. X. Yu, G. Dong, K. Qu, P. Huang, E. M. Anderson, S. Daniel-Issakani, R. M. L. Buller, D. G. Payan, and H. H. Lu. 2004. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J. Biol. Chem. 27954110-54116. [DOI] [PubMed] [Google Scholar]
- 26.Iwase, S., F. Lan, P. Bayliss, L. de la Torre-Ubieta, M. Huarte, H. H. Qi, J. R. Whetstine, A. Bonni, T. M. Roberts, and Y. Shi. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 1281077-1088. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 2952080-2083. [DOI] [PubMed] [Google Scholar]
- 28.Jeanblanc, M., M. Mousli, R. Hopfner, K. Bathami, N. Martinet, A. Q. Abbady, J. C. Siffert, E. Mathieu, C. D. Muller, and C. Bronner. 2005. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene 247337-7345. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins, Y., V. Markovtsov, W. Lang, P. Sharma, D. Pearsall, J. Warner, C. Franci, B. Huang, J. Huang, G. C. Yam, J. P. Vistan, E. Pali, J. Vialard, M. Janicot, J. B. Lorens, D. G. Payan, and Y. Hitoshi. 2005. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell 165621-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, L. M., M. Bostick, X. Zhang, E. Kraft, I. Henderson, J. Callis, and S. E. Jacobsen. 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, L. M., X. Cao, and S. Jacobsen. 2002. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 121360-1367. [DOI] [PubMed] [Google Scholar]
- 33.Jia, S., R. Kobayashi, and S. I. Grewal. 2005. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 71007-1013. [DOI] [PubMed] [Google Scholar]
- 34.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442312-316. [DOI] [PubMed] [Google Scholar]
- 35.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410116-120. [DOI] [PubMed] [Google Scholar]
- 36.Li, H., S. Ilin, W. Wang, E. M. Duncan, J. Wysocka, C. D. Allis, and D. J. Patel. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 44291-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5296-305. [DOI] [PubMed] [Google Scholar]
- 38.Maison, C., D. Bailly, A. H. F. M. Peters, J.-P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30329-334. [DOI] [PubMed] [Google Scholar]
- 39.Martin, D. G., K. Baetz, X. Shi, K. L. Walter, V. E. MacDonald, M. J. Wlodarski, O. Gozani, P. Hieter, and L. Howe. 2006. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 267871-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. F. M. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437436-439. [DOI] [PubMed] [Google Scholar]
- 41.Minsky, N., and M. Oren. 2004. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell 16631-639. [DOI] [PubMed] [Google Scholar]
- 42.Mousli, M., R. Hopfner, A. Q. Abbady, D. Monte, M. Jeanblanc, P. Oudet, B. Louis, and C. Bronner. 2003. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer 89120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama, J.-I., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. S. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292110-113. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7729-739. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416103-107. [DOI] [PubMed] [Google Scholar]
- 46.O'Carroll, D., H. Scherthan, A. H. F. M. Peters, S. Opravil, A. R. Haynes, G. Laible, S. Rea, M. Schmid, A. Lebersorger, M. Jerratsch, L. Sattler, M. G. Mattei, P. Denny, S. D. M. Brown, D. Schweizer, and T. Jenuwein. 2000. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 209423-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa, H., K.-I. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 2961132-1136. [DOI] [PubMed] [Google Scholar]
- 48.Papait, R., C. Pistore, D. Negri, D. Pecoraro, L. Cantarini, and I. M. Bonapace. 2007. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell 181098-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peña, P. V., F. Davrazou, X. Shi, K. L. Walter, V. V. Verkhusha, O. Gozani, R. Zhao, and T. G. Kutateladze. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107323-337. [DOI] [PubMed] [Google Scholar]
- 51.Peters, A. H., and D. Schubeler. 2005. Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 17230-238. [DOI] [PubMed] [Google Scholar]
- 52.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433434-438. [DOI] [PubMed] [Google Scholar]
- 53.Qin, J., D. Fenyo, Y. Zhao, W. W. Hall, D. M. Chao, C. J. Wilson, R. A. Young, and B. T. Chait. 1997. A strategy for rapid, high-confidence protein identification. Anal. Chem. 693995-4001. [DOI] [PubMed] [Google Scholar]
- 54.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z.-W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406593-599. [DOI] [PubMed] [Google Scholar]
- 55.Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn, and G. Reuter. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 211121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz, D. C., K. Ayyanathan, D. Negorev, G. G. Maul, and F. J. Rauscher III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Michishita, T. Hung, D. Carney, P. Pena, F. Lan, M. R. Kaadige, N. Lacoste, C. Cayrou, F. Davrazou, A. Saha, B. R. Cairns, D. E. Ayer, T. G. Kutateladze, Y. Shi, J. Cote, K. F. Chua, and O. Gozani. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 44296-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 59.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. SET domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 27625309-25317. [DOI] [PubMed] [Google Scholar]
- 60.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110701-711. [DOI] [PubMed] [Google Scholar]
- 61.Taverna, S. D., S. Ilin, R. S. Rogers, J. C. Tanny, H. Lavender, H. Li, L. Baker, J. Boyle, L. P. Blair, B. T. Chait, D. J. Patel, J. D. Aitchison, A. J. Tackett, and C. D. Allis. 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ting, A. H., K. M. McGarvey, and S. B. Baylin. 2006. The cancer epigenome—components and functional correlates. Genes Dev. 203215-3231. [DOI] [PubMed] [Google Scholar]
- 63.Uemura, T., E. Kubo, Y. Kanari, T. Ikemura, K. Tatsumi, and M. Muto. 2000. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct. Funct. 25149-159. [DOI] [PubMed] [Google Scholar]
- 64.Unoki, M., T. Nishidate, and Y. Nakamura. 2004. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 237601-7610. [DOI] [PubMed] [Google Scholar]
- 65.Völkel, P., and P.-O. Angrand. 2007. The control of histone lysine methylation in epigenetic regulation. Biochimie 891-20. [DOI] [PubMed] [Google Scholar]
- 66.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125467-481. [DOI] [PubMed] [Google Scholar]
- 67.Wissmann, M., N. Yin, J. M. Muller, H. Greschik, B. D. Fodor, T. Jenuwein, C. Vogler, R. Schneider, T. Gunther, R. Buettner, E. Metzger, and R. Schule. 2007. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9347-353. [DOI] [PubMed] [Google Scholar]
- 68.Woo, H. R., O. Pontes, C. S. Pikaard, and E. J. Richards. 2007. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121859-872. [DOI] [PubMed] [Google Scholar]
- 70.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 44286-90. [DOI] [PubMed] [Google Scholar]
- 71.Yamane, K., C. Toumazou, Y.-I. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125483-495. [DOI] [PubMed] [Google Scholar]
- 72.Yang, L., L. Xia, D. Y. Wu, H. Wang, H. A. Chansky, W. H. Schubach, D. D. Hickstein, and Y. Zhang. 2002. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21148-152. [DOI] [PubMed] [Google Scholar]
- 73.Zegerman, P., B. Canas, D. Pappin, and T. Kouzarides. 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 27711621-11624. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, D., H.-G. Yoon, and J. Wong. 2005. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 256404-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]