Abstract
Genome-wide DNA methylation patterns are frequently deregulated in cancer. There is considerable interest in targeting the methylation machinery in tumor cells using nucleoside analogs of cytosine, such as 5-aza-2′-deoxycytidine (5-azadC). 5-azadC exerts its antitumor effects by reactivation of aberrantly hypermethylated growth regulatory genes and cytoxicity resulting from DNA damage. We sought to better characterize the DNA damage response of tumor cells to 5-azadC and the role of DNA methyltransferases 1 and 3B (DNMT1 and DNMT3B, respectively) in modulating this process. We demonstrate that 5-azadC treatment results in growth inhibition and G2 arrest—hallmarks of a DNA damage response. 5-azadC treatment led to formation of DNA double-strand breaks, as monitored by formation of γ-H2AX foci and comet assay, in an ATM (ataxia-telangiectasia mutated)-dependent manner, and this damage was repaired following drug removal. Further analysis revealed activation of key strand break repair proteins including ATM, ATR (ATM-Rad3-related), checkpoint kinase 1 (CHK1), BRCA1, NBS1, and RAD51 by Western blotting and immunofluorescence. Significantly, DNMT1-deficient cells demonstrated profound defects in these responses, including complete lack of γ-H2AX induction and blunted p53 and CHK1 activation, while DNMT3B-deficient cells generally showed mild defects. We identified a novel interaction between DNMT1 and checkpoint kinase CHK1 and showed that the defective damage response in DNMT1-deficient cells is at least in part due to altered CHK1 subcellular localization. This study therefore greatly enhances our understanding of the mechanisms underlying 5-azadC cytotoxicity and reveals novel functions for DNMT1 as a component of the cellular response to DNA damage, which may help optimize patient responses to this agent in the future.
DNA methylation is an essential epigenetic modification required for normal mammalian development, gene regulation, genomic imprinting, and chromatin structure (9). Methylation occurs at the C-5 position of cytosine within the CpG dinucleotide and is carried out by a family of DNA methyltransferases (DNMTs), DNMT1, DNMT3A, and DNMT3B (32). DNMT1 acts primarily as a maintenance methyltransferase by copying existing methylation patterns following DNA replication (53). DNMT3A and DNMT3B exhibit de novo methyltransferase activity and are required for establishing new methylation patterns during embryonic development (72). Deregulation of the DNA methylation machinery has been identified in several disease states, particularly cancer (78), and leads to global DNA hypomethylation of repetitive DNA, which has been linked to genomic instability (21) and, concomitantly, hypermethylation at the promoter regions of certain genes (tumor suppressors), leading to their aberrant silencing (43).
The process of cytosine methylation is reversible and may be altered by biochemical and biological manipulation, making it an attractive target for therapeutic intervention. Demethylation and consequent reactivation of tumor suppressor genes are rational approaches being used in the treatment of cancer (38, 39, 61). Currently available nucleoside-based DNMT inhibitors 5-azacytidine (5-azaC), 5-aza-2′-deoxycytidine (5-azadC), and zebularine (ZEB) are analogues of cytosine that are believed to have a similar mechanism of inhibition (16, 96). 5-azaC, 5-azadC, and ZEB are metabolically activated in vivo and readily incorporated into DNA during replication, and as a result of the chemistry of the methyltransferase reaction, the DNMT becomes covalently linked to DNA, in effect creating genome-wide protein-DNA cross-links (16, 57, 61, 103). This results in depletion of soluble DNMT protein levels, leading to replication-dependent global demethylation and gene reactivation (15, 100). Both 5-azaC and 5-azadC are used clinically for the treatment of myelodysplastic syndrome (MDS) and other leukemias, where the drugs have the approval of the Food and Drug Administration (38). Clinical trials combining aza drugs with other agents, such as histone deacetylase inhibitors and interleukin-2, have also recently been reported (33, 34). Promising results were observed with hematologic malignancies, particularly MDS. In phase II and III clinical trials, 5-azaC treatment yields a better response rate than supportive care alone (91). Similarly encouraging results were obtained with clinical trials using 5-azadC (38).
Although there is considerable literature on the possible antitumor mode of action of aza drugs, their exact in vivo mechanism remains unclear. One model for their effects involves the reactivation of aberrantly silenced growth-regulatory genes accompanied by cell cycle arrest and/or apoptosis (36). A second model for their antitumor activity is related to formation of covalent DNMT-DNA adducts in aza-containing DNA, leading to DNA damage and cytotoxicity (44). While the former model has been extensively studied, the latter has not. 5-azadC treatment is perceived as DNA damage and leads to activation of the G1 checkpoint regulator p53 (49). Furthermore, 5-azadC treatment results in inhibition of cell proliferation due to p53-dependent activation of p21Waf1/Cip1 (49, 104). 5-azaC also induces apoptosis, either in a p53-dependent (88) or p53-independent manner (66). Aside from the role of p53 and p21Waf1/Cip1, the cellular events activated by 5-azaC, 5-azadC, and ZEB that occur downstream of DNA methylation inhibition in DNMT-aza-DNA-containing cells are largely unknown, as is the means by which the cellular DNA repair machinery recognizes and responds to this form of damage. It is likely that cytotoxicity is, at least in part, mediating the antitumor effect of aza drugs since clinical responses do not necessarily correlate with gene-specific or global demethylation (46, 64, 87). Given the interest in using these drugs as part of an “epigenetic”-based chemotherapy regimen and their likely increased use in the clinic, it is essential to better understand how aza-DNA-DNMT adducts mediate their cytotoxic effects and how this contributes to the overall therapeutic outcome.
One of the first steps in the response of cells to DNA damage is activation of the phosphatidylinositol 3-kinase (PI3K) family of high-molecular-weight protein kinases, ATM (ataxia-telangiectasia mutated) and ATR (ATM-Rad3-related) (90). ATM is present in an inactive state and, in response to DNA damage, undergoes auto-phosphorylation at serine 1981 (4). Activated ATM then phosphorylates many downstream effectors, such as p53, NBS1, BRCA1, and SMC1 (90). Phosphorylation of ATM also results in activation of cell cycle checkpoints via the transducing kinases checkpoint kinase 1 (CHK1) and CHK2, which are substrates of ATR and ATM, respectively (6). There also appears to be active cross talk between ATM and ATR as ATM function is required for recruitment of ATR to sites of damage (1). The G1/S checkpoint response is mediated primarily through a pathway involving ATM/CHK2-p53/MDM2-p21Waf1/Cip1 (68). ATM directly phosphorylates p53 at serine 15, contributing to its stabilization and increased activity as a transcription factor (12), which in turn leads to activation of a key transcriptional target of p53, the cyclin-dependent kinase (CDK) inhibitor p21Waf1/Cip1, causing G1 arrest (58, 59). The G2 checkpoint primarily targets the cyclin B/CDK1 kinase and is mediated by ATM/ATR, CHK2/CHK1, and/or p38 kinase, which inhibit the CDC25 family of phosphatases required for CDK1 activation (11, 68, 101). The recruitment and/or retention of DNA repair factors at sites of damage is mediated in part by ATM-dependent phosphorylation of the histone variant H2AX at serine 139 (phospho-Ser139 H2AX, or γ-H2AX), an early event known to occur in response to DNA double-strand breaks (81). H2AX phosphorylation is rapid and extensive, covering megabase chromatin domains adjacent to the break that are easily visualized as discrete foci by immunofluorescence microscopy (74, 82). Phosphorylation of ATM is observed upon radiation-induced DNA double-strand breaks and through other signal transduction events and leads to activation of the G2/M checkpoint and the double-strand break response pathway (4, 90).
Our lack of knowledge of the mechanism of action of 5-azaC and 5-azadC, in particular the contribution of the cytotoxic DNMT-aza-DNA adduct to drug efficacy, prevents them from being used as effectively as possible. We therefore set out to better characterize the cytotoxic properties of these drugs and to determine the relative contribution of DNMT1 and DNMT3B to this process, since these DNMTs mediate, alone or in combination, most of the DNA methylation in cancer cells, and they are often aberrantly expressed (76, 80). Our results indicate that 5-azadC treatment results in growth inhibition, G2 arrest, and reduced clonogenic survival. In addition, cells lacking functional DNMT1 are markedly deficient in cell cycle arrest and growth inhibition while DNMT3B-deficient cells generally show an intermediate effect. 5-azadC treatment also leads to robust induction of γ-H2AX, DNA fragmentation, and activation of DNA repair proteins in both the ATM and ATR pathways, such as CHK1 and CHK2, NBS1, BRCA1, and RAD51, and their relocalization to a focal staining pattern characteristic of cells with damaged DNA. Interestingly, DNMT1 hypomorphic mutant cells (1KO) were profoundly deficient in their ability to induce γ-H2AX and also exhibited altered induction of other DNA repair proteins. Further demonstration of the particularly critical role for DNMT1 was the finding that DNMT1 strongly colocalized with γ-H2AX in 5-azadC-treated cells, demonstrating for the first time the presence of DNMT1 at sites of DNA damage in 5-azadC-treated cells. We have also identified a novel interaction between DNMT1 and CHK1. The defective damage response of 1KO cells is, at least in part, due to altered subcellular localization of CHK1 in the absence of functional DNMT1. Furthermore, other clinically relevant DNMT inhibitors also cause induction of γ-H2AX staining and DNA fragmentation in a manner roughly proportional to their ability to reactivate aberrantly silenced genes. These studies therefore greatly enhance our understanding of how human tumor cells respond to 5-azadC and also reveal an important new role for DNMT1 as a component of the DNA damage response system.
MATERIALS AND METHODS
Cell lines, transient transfection, and quantitative reverse transcriptase PCR (RT-PCR).
HeLa and parental HCT116 cells were purchased from the American Type Culture Collection. Isogenic HCT116 cell lines with knockouts of DNMT1 (1KO, a hypomorph) and DNMT3B (3BKO) were provided by Bert Vogelstein. Cells were cultured in McCoy's 5A medium (Cellgro Mediatech Inc.) supplemented with 10% heat-inactivated fetal bovine serum and 2 mM l-glutamine (Invitrogen) at 37°C in a 5% CO2 incubator. The isogenic cell lines YZ5 and EBS are simian virus 40-transformed fibroblasts derived from an ataxia telangectasia patient; EBS cells are ATM deficient, and YZ5 cells stably express full-length wild-type ATM (105). EBS and YZ5 (Coriell Cell Repositories) were grown in Dulbecco's modified Eagle medium (Cellgro Mediatech Inc.) supplemented with 10% fetal bovine serum and 100 μg/ml G418. For immunofluorescence staining experiments, cells were transfected using the TransIT LT1 transfection reagent (Mirus) according to the manufacturer's protocol. Forty-eight hours later, transfected cells were trypsinized and replated at a 1:3 dilution in wells containing glass coverslips and then treated with 10 μM 5-azadC for 48 h.
For quantitative RT-PCR, 2 μg of RNA was reverse transcribed using Superscript III reverse transcriptase according to the manufacturer's protocol (Invitrogen). One microliter of cDNA was used per PCR. Real-time PCR was performed using the Bio-Rad MiniOpticon Real-Time PCR System (Bio-Rad Laboratories). Each 20-μl reaction mixture consisted of 1× Sybr Green PCR Master Mix (Applied Biosystems) and 200 nM concentrations of forward and reverse primers for either WIF1 (WIF1F, 5′-CAACCGTCAATGTCCCTCTGCT-3′; WIF1R, 5′-TCCACTTCAAATGCTGCCACC-3′) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (described previously in reference 97). The cycling conditions were as follows: 1 cycle at 95°C for 10 min and either 40 (WIF1) or 30 (GAPDH) cycles at 95°C for 30 s, followed by 65°C (WIF1) or 60°C (GAPDH) for 30 s. The threshold cycle (CT) values were determined using the Opticon Monitor 3 software (Bio-Rad Laboratories). WIF1 expression was normalized to GAPDH expression (2[Ct(GAPDH) − Ct(WIF1)]), and relative expression of treated versus untreated cells was determined by the ratio of normalized expression values for the treated cells to the normalized expression value of the calibrator (untreated cells).
Chemicals and plasmids.
5-azaC, 5-azadC, ZEB, bleomycin, hydroxyurea, doxorubicin, and wortmannin were purchased from Sigma. Plasmids expressing green fluorescent protein (GFP)-tagged DNMT1 and Dnmt3b1 have been described elsewhere (28, 77). The FLAG-CHK1 plasmid was provided by Yolanda Sanchez. General chemicals were purchased from Sigma. Protease inhibitors were purchased from Roche. 5-aza, 5-azadC, and ZEB were dissolved in 1× phosphate-buffered saline (PBS; pH 7.0). Cells were treated with 0.1 to 10 μM 5-azadC or 5-azaC and 50 to 500 μM ZEB for the times and doses described in Results. For recovery experiments, cells were treated with 5-azadC for 48 h (fresh drug was added every 24 h), after which the medium was changed. Cells were fixed or harvested and analyzed every 24 h for up to 4 days after drug removal. Cells were treated with 10 μg/ml bleomycin for 4 h, the medium was changed, and the cells were analyzed 20 h later. Wortmannin was used at a final concentration of 10 μM for 24 h of treatment. Hydroxyurea, doxorubicin, and bleomycin treatments for cell viability assays are described in Results.
Growth inhibition.
Cells were plated, allowed to grow overnight and treated with 10 μM 5-azadC for 48 h (fresh drug was added every 24 h), and then the medium was changed. Cell growth was then monitored for 4 days by counting live cells following staining with trypan blue. Growth inhibition was calculated as the ratio of the number of viable treated cells versus untreated cells and plotted against the day posttreatment with 5-azadC as follows: [100 − (number of cells in treated sample/number of cells in untreated sample)] × 100.
Clonogenic assay.
Cells were plated at a density of 200 to 300 cells per dish and treated with 5-azadC (10−4 μM, 10−3 μM, 10−2 μM, 10−1 μM, 1 μM, and 10 μM) or vehicle only (control) medium for 48 h, with fresh medium added after 24 h. After completion of the treatments all cells received fresh growth medium. The cells were allowed to grow for the next 10 to 12 days to allow colony formation; then the medium was removed, and the colonies were stained and fixed with 0.2% methylene blue in 50% methanol and counted. Each experimental treatment was performed in quadruplicate.
Cell cycle analysis.
Cell cycle profiles were determined by analyzing DNA content using propidium iodide (PI) staining and flow cytometry. Cells were treated with various concentrations of 5-azadC (0.1 to 10 μM) for 24, 48, and 72 h. Cells were harvested by trypsinization, washed with 1× PBS, and fixed with ice-cold 70% ethanol overnight. Fixed cells were washed once with 1× PBS and resuspended at 1 × 106 cells/ml in PI staining solution (0.1 mg/ml PI, 0.1 mg/ml RNase, and 0.1% Triton X-100 in PBS) and incubated in the dark at room temperature for 15 min before analysis. Cell cycle profiles were determined using fluorescence-activated cell sorting (FACS) with a Becton Dickinson FACSort. For each sample, 3 × 104 events were recorded. Data were analyzed by ModFit cell cycle analysis software (Verity) to determine the percentage of cells in each phase. For quantitative assessment of G2 arrest, a modification of an assay outlined previously was used (101). Briefly, cells were treated with 10 μM 5-azadC for 48 h; then demecolcine (Sigma-Aldrich) was added to the medium (0.4 μg/ml final concentration), and cells were incubated for an additional 24 h. Cells were harvested and fixed in ice-cold 70% ethanol. After fixation, cells were washed twice in 1× PBS, resuspended in 1 ml of 1× PBS containing 0.25% Triton X-100, and incubated on ice for 5 min. Cells were collected by centrifugation, and the cell pellet was resuspended in 100 μl of PBS containing 1% bovine serum albumin and 1 μg of anti-histone H3 phospho-Ser10 (H3 phosphorylated on serine 10) antibody (06-570; Upstate Biotechnology). Cells were incubated with this antibody for 3 h at room temperature, washed, and subsequently incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody. After a 30-min incubation at room temperature, cells were washed with PBS, resuspended in PBS containing 25 μg/ml PI and 100 μg/ml RNase A, and incubated for an additional 30 min at room temperature. Both FITC and PI fluorescence were simultaneously measured by flow cytometry.
Cell viability and apoptosis.
To assess cell viability following drug treatment, a CellTiter 96 AQueous One Solution kit (Promega) was used, and assays were performed as per the manufacturer's instructions. Cells were plated in 96-well plates, and drug treatments were typically initiated 24 h later. For all drugs tested, parallel cell-free reactions were set up, and the readings from these wells were used for background correction. For all dose-response experiments measurements were performed in quadruplicate. For time course experiments, parallel dishes were set up for each time point for the drug-treated and control cells, and the results were obtained from eight replicates per treatment per cell line. The UV treatments were performed with cells seeded in 96-well plates in a Bio-Rad GS Gene Linker UV chamber (10 mJ/cm2) without removing the growth medium from the plates. The dishes containing the control cells were removed from the incubator and kept outside for the same length of time as the cells receiving the UV treatments. Upon treatment completion, the growth medium was replaced for both control and UV-treated samples. Apoptosis was measured with the Annexin V-PI kit according to the manufacturer's protocol (Trevigen). Briefly, cells were harvested, washed with cold 1× PBS, and resuspended at 1 × 106 cells/ml in 100 μl of annexin V-FITC/PI-containing binding buffer for 15 min in the dark. Apoptotic cells were determined by FACS using a Becton Dickinson FACSort. Data were analyzed using Cellquest software.
Analysis of DNA damage.
A single-cell gel electrophoresis system (Comet assay kit; Trevigen) was used to quantitate DNA damage. Briefly, cells were harvested by trypsinization and mixed with 1% low-melting-point agarose. A total of 75 μl (500 to 1,000 cells) of the cell suspension was spread on a precoated glass slide and placed at 4°C for 30 min. Cells were lysed by submerging the slides in ice cold lysis solution (10 mM Tris, 100 mM EDTA, 2.5 M NaCl, with 1% Triton X-100 and 10% dimethyl sulfoxide added just prior to use) at 4°C for at least 1 h. After lysis, the slides were placed in freshly made alkaline buffer (300 mM NaOH and 1 mM EDTA, pH >13.0) for 45 min at room temperature. The samples were subjected to electrophoresis in 0.5× Tris-borate-EDTA buffer at 0.5 V/cm for 25 min. After electrophoresis, the slides were rinsed gently with 0.4 M Tris-HCl (pH 7.5), and the DNA was stained with Sybr Green (Molecular Probes). Fluorescently stained nucleoids were scored visually using an epifluorescence microscope (Zeiss Axioplan 3). Images were captured at a magnification of ×20. The percentage of cells having a comet-like appearance was calculated by analyzing at least 100 cells from each treatment. The ratio of comet-positive cells in the 5-azadC-treated versus the mock-treated (1× PBS) sample was plotted against time posttreatment with 5-azadC. The tail moment, a measure of the amount of DNA damage, was calculated using CometScore from TriTek Corporation (a minimum of 50 cells was used for each condition).
Immunofluorescence staining.
Cells were grown on 22-mm2 glass coverslips in six-well plates and treated as described in Results. Treated and mock-treated cells were fixed with ice-cold methanol for 15 min at −20°C or 2% paraformaldehyde in 1× PBS, pH 7.0 (RAD51 and ATR only). Cells were washed with 1× PBS (pH 7.0), incubated with diluted primary antibody (in 1× PBS with 0.1% Tween-20 [PBST]) for 1 h at room temperature, washed three times with PBST, and subsequently incubated with appropriate fluorophore-conjugated secondary antibody in PBST. Cells were washed and counterstained for nuclear DNA using Hoechst 33342 (1 μg/ml) for 10 min before being mounted onto glass slides using Fluoromount G (Southern Biotech). Images were captured using a Zeiss Axioplan 3 upright fluorescent microscope and deconvolved using Openlab software.
Antibodies.
Antibodies for Western blotting and immunofluorescence by source include the following: phospho-Ser139 H2AX (γ-H2AX), phospho-Ser1981 ATM, p53, CHK2, BRCA1, and phospho-Ser343 NBS1 from Upstate; phospho-Ser1981 ATM from Rockland; phospho-Ser15 p53, CHK2 phosphorylated at threonine 68 (phospho-Thr68), and phospho-Ser345 CHK1 from Cell Signaling; phospho-Ser317 CHK1, RAD51, p21Waf1/Cip1, ATR, and DNA-dependent protein kinase (DNA-PK) from Calbiochem; DNMT3A (P16), Ku70, CHK1 (G-4), PCNA, and RNA polymerase II (Pol II) from Santa Cruz; anti-FLAG (M2) from Sigma; Mre11 from GeneTex; and GAPDH from Abcam. Epitope affinity-purified rabbit anti-DNMT3B antibody (number 79/80) (29), rabbit polyclonal anti-DNMT1 (74 or N1020) (77), and anti-ATM (pAb 354) (5) have been described previously. Generally, antibodies were used at a 1:50 dilution for immunofluorescence and a 1:500 to 1:1,000 dilution for Western blotting (except GAPDH, which was used at a 1:8,000 dilution). FITC-, tetramethyl rhodamine isocyanate-, and Texas Red-conjugated anti-mouse, anti-rabbit, and anti-goat secondary antibodies were from Southern Biotech. Horseradish peroxidase-conjugated secondary antibodies for Western blotting were used at a 1:2,000 dilution (Santa Cruz).
Immunoprecipitation and Western blotting.
Typically, immunoprecipitations were performed with 1 to 2 mg of HeLa high-salt nuclear extracts or total cell lysates derived from one 150-mm dish of HeLa cells at 80 to 90% confluence per reaction. The immunoprecipitation controls were done with species-matched normal immunoglobulin G (IgG) purchased from Pierce (rabbit and mouse) or Santa Cruz (goat). Immunoprecipitations and nuclear extract preparations were performed as previously described (28). Briefly, for immunoprecipitation, the nuclear extract was supplemented with immunoprecipitation dilution buffer to a final concentration of 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40 plus protease inhibitors. For each reaction, 200 to 300 μl of nuclear protein (1 to 2 mg) was mixed with immunoprecipitation dilution buffer and precleared with 20 μl of protein A/G agarose (Santa Cruz Biotechnology) for 30 min at 4°C with rotation, followed by centrifugation and collection of supernatants. For DNMT1 immunoprecipitations, each tube received 20 μl of DNMT1 (antibody 74) (79) or 10 μl of CHK1 (G-4; Santa Cruz) antibody, and reaction mixtures were incubated overnight at 4°C with rotation. Immune complexes were collected the next day by incubation with 20 μl of protein A/G beads for 2 h at 4°C with rotation. Washes were performed five times in 500 μl of wash buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, protease inhibitors) at 4°C for 5 min each. For whole-cell extracts, cells were washed with cold 1× PBS, and 0.5 to 1 ml of radioimmunoprecipitation assay buffer was added to each plate. Cells were scraped, collected, and incubated on ice for 30 min. Cells were then centrifuged at 12,000 rpm for 15 min. The supernatant was collected, and the protein concentration was determined using the Bio-Rad protein assay reagent. An equal amount of protein (60 μg) from each sample was loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, transferred to polyvinylidene difluoride membrane, and used for Western blotting by standard methods.
Subcellular fractionation.
Fractionation was performed as described previously (60). Briefly, cells were scraped in 1× PBS, collected by brief centrifugation, and counted. Equal numbers of viable cells were suspended in 100 μl of solution A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, protease and phosphatase inhibitors) per 2 × 106 cells. Triton X-100 was added to a final concentration of 0.1%, and the cells were incubated on ice for 5 min. The cytoplasmic fraction was harvested by centrifugation at 1,300 × g for 4 min. The nuclear pellet was washed once with solution A and lysed in 100 μl of solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, protease and phosphatase inhibitors) per 2 × 106 cells by incubating at 4°C with rotation for 15 min. The soluble nuclear fraction was harvested by centrifugation at 1,700 × g for 4 min. The chromatin pellet (P3) was then washed once in solution B, centrifuged at 10,000 × g for 1 min, and resuspended in 100 μl of sample buffer per 2 × 106 cells. The P3 fraction was homogenized by sonication three times for 30 s each time.
RESULTS
Effects of 5-azadC on DNMT protein levels and cell growth.
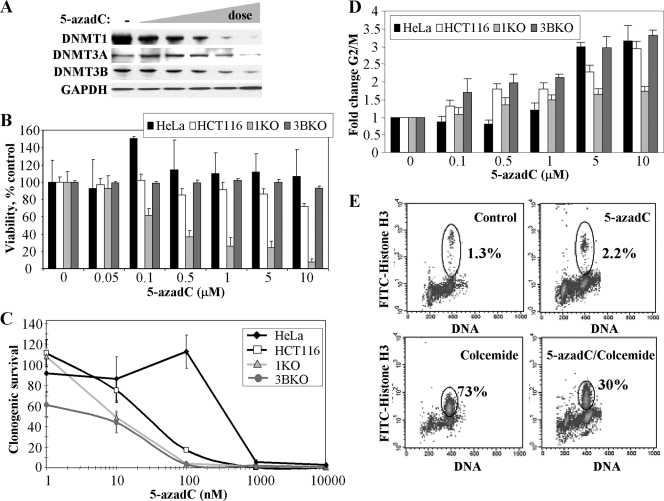
The effects of 5-azadC on DNMT protein levels (30, 71, 100) and cell growth (8, 49, 62, 88) have been examined by others in several different cell lines and experimental systems. As a starting point for defining the role of individual DNMTs in mediating the cytotoxic effects of 5-azadC, we determined its effects on soluble DNMT protein levels in HeLa cells. As has been reported (30, 100), treatment of cells with an increasing concentration of 5-azadC (0.1 to 10 μM) resulted in dose-dependent depletion of DNMTs from soluble cell extracts derived from treated HeLa cells. DNMT1 was affected to the largest extent, followed by DNMT3A and DNMT3B, which showed depletion only at the highest 5-azadC doses (Fig. 1A). Similar results were obtained with HCT116 cells (data not shown).
FIG. 1.
Responses of HeLa and HCT116 cells to the DNA methylation inhibitor 5-azadC and the role of DNMT1 and DNMT3B. (A) Effect of increasing concentrations of 5-azadC on DNMT1, DNMT3A, and DNMT3B protein levels. HeLa cells were mock treated (−) or treated with 0.1, 0.5, 1.0, 5.0, and 10.0 μM 5-azadC for 48 h (indicated by the wedge); then soluble protein extracts were prepared and subjected to SDS-PAGE followed by Western blotting with the antibodies indicated at the left. GAPDH served as a loading control. (B) Effect of 5-azadC on cell viability. Cells were treated with indicted doses of 5-azadC for 48 h (fresh drug added every 24 h); the medium was changed, and then cell viability was determined using the MTT assay. (C) 5-azadC treatment results in decreased clonogenic survival. Cell lines were treated with 5-azadC for 48 h, followed by replacement of the medium and continued growth for 10 to 12 days. Results are presented as the average of quadruplicate measurements, and the bar is the standard deviation. (D) Effect of increasing doses of 5-azadC on the cell cycle summarized as the relative increase in the number of cells (n-fold) arrested in G2/M for each of the cell lines following drug treatment. Experiments were repeated three times and averaged. (E) HeLa cells treated with 10 μM 5-azadC for 48 h preferentially arrest in G2 as determined by DNA content (PI staining shown on the x axis) and staining cells with an antibody for the M phase specific marker phospho-Ser10 histone H3 (y axis) followed by flow cytometry.
We next examined the effects of 5-azadC on HeLa cell viability, clonogenic survival, and cell cycle dynamics. HeLa cells were treated with 0.05 to 10 μM 5-azadC for 48 h (fresh drug added every 24 h). An MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] cell viability assay indicated that HeLa cells were relatively resistant to the effects of 5-azadC within the treatment period (Fig. 1B). In contrast, however, long-term survival of HeLa cells was markedly reduced by doses of 1 μM 5-azadC and above, as monitored using a clonogenic survival assay (Fig. 1C). Despite the relatively minor effect of 5-azadC treatment on short-term HeLa cell viability, identical drug treatment conditions revealed that 5-azadC caused HeLa cells to arrest in the G2/M phase of the cell cycle (Fig. 1D). Since PI staining and flow cytometry analysis of cell cycle profiles cannot distinguish between G2 or M phase arrest, we investigated this issue in greater detail by staining with an M phase-specific marker, phospho-Ser10 histone H3 (101). HeLa cells were treated with 5-azadC for 48 h, after which either they were harvested or colcemide was added to the culture medium. After an additional 24-h incubation, cells were harvested and double stained for DNA and phospho-Ser10 histone H3 and analyzed by flow cytometry. Treatment with 10 μM 5-azadC for 48 h caused nearly 50% of the cells to arrest in G2/M, but only about 2% of these cells were in M phase (Fig. 1E). Colcemide treatment alone led to M phase arrest in nearly 75% of the cells while only 30% of the cells treated with 5-azadC plus colcemide progressed to M phase. These results indicate that cells are arrested in G2 in response to 5-azadC treatment, a finding observed with many other traditional DNA damaging chemotherapeutic agents that do not act directly through DNA methylation-based pathways. G2 arrest with minimal cell death also suggests that HeLa cells may be attempting to repair 5-azadC treatment-induced DNA damage (discussed later).
Role of individual DNMTs in mediating the growth effects of 5-azadC on human tumor cells.
We next sought to extend our work with HeLa cells to an experimental system in which we could directly examine the effects of individual DNMTs in mediating the growth-suppressive properties of 5-azadC. We chose to use the well-studied HCT116 colorectal cancer cell line because of the availability of isogenic variants in which the DNMT1 or the DNMT3B genes have been disrupted by homologous recombination (76). Recent studies have revealed that the DNMT1-deficient HCT116 line is actually a hypomorph, expressing significantly reduced levels of a DNMT1 protein lacking its PCNA binding domain (22). Previous studies employed murine embryonic stem cell lines carrying deletions in DNMT genes (44, 71); however, in this study we specifically sought to determine the effects of 5-azadC on human tumor cells, the target of a chemotherapeutic regimen containing 5-azadC. HCT116 parental, HCT116 DNMT1 knockout (1KO), and HCT116 DNMT3B knockout (3BKO) cells were treated with 0.05 to 10 μM 5-azadC for 48 h (fresh drug added every 24 h), and cell viability was assessed. Interestingly, while the parental HCT116 and 3BKO lines behaved similarly to HeLa cells, showing relatively minor reductions in cell viability; the HCT116 1KO cells displayed significantly reduced viability (Fig. 1B). The long-term survival of all three HCT116 lines was reduced with increasing doses of 5-azadC, and overall all lines were more sensitive to 5-azadC than HeLa cells. In contrast to previous experiments utilizing complete genetic knockouts of Dnmt1 in murine embryonic stem (ES) cells (44), however, we found that cells deficient (or hypomorphic) in DNMT1 or DNMT3B were significantly more sensitive to 5-azadC in a clonogenic survival assay (Fig. 1C). Cell cycle analysis showed that HCT116 parental and 3BKO cells arrested preferentially in G2, like HeLa cells; however, 1KO cells were impaired in their ability to arrest in G2 upon 5-azadC treatment (Fig. 1D). These results suggest that 1KO cells are markedly defective in their responses to 5-azadC treatment and that their reduced capacity to arrest in G2 and potentially repair DNA damage may, in turn, contribute to the reduced viability of these cells we have observed. Given that reduced short- and long-term viability and G2 arrest are characteristic responses of cells treated with traditional DNA damage-inducing chemotherapeutic agents, we next sought to examine in more detail the nature of the cellular DNA damage response to 5-azadC treatment.
5-azadC treatment induces formation of γ-H2AX foci, a marker of DNA double-strand breaks, in an ATM- and DNMT1-dependent manner.
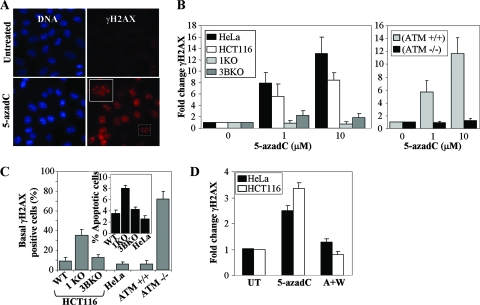
The data presented in the preceding sections clearly implicate DNMT1 in mediating much of the survival and cell cycle-related effects of 5-azadC. Interestingly, lack of a major target of 5-azadC, DNMT1, actually resulted in greater cell death and reduced G2 arrest (Fig. 1), suggesting that reduced DNMT1 levels and/or the presence of the truncated hypomorphic form of DNMT1 may result in a defective DNA damage response. The type of DNA damage induced in 5-azadC-treated cells and how cells respond to this form of damage have not been well characterized. In order to investigate this in greater detail, we analyzed whether 5-azadC treatment caused induction of a well-known marker of DNA double-strand breaks, γ-H2AX. H2AX is rapidly phosphorylated by ATM and other PI3K family members (ATR and DNA-PK) in response to DNA double-strand breaks due to naturally occurring endogenous processes, such as V(D)J recombination, or exposure to DNA damaging agents from the environment (75). Treatment of HCT116 cells (Fig. 2A) with 10 μM 5-azadC for 48 h resulted in a large increase in cells positive for γ-H2AX, as monitored by immunofluorescence staining (cells were scored positive if they contained five or more γ-H2AX foci). 5-azadC-treated cells exhibited intense γ-H2AX staining in highly localized foci, a characteristic of many agents known to induce DNA double-strand breaks (26, 82). HeLa and parental HCT116 cells demonstrated a dose-dependent increase in γ-H2AX staining, with foci readily detectable following a 48-h exposure to 1 and 10 μM 5-azadC (Fig. 2B). Of notable significance, cells deficient in DNMT1 or ATM were completely devoid of a γ-H2AX response upon 5-azadC treatment (Fig. 2B). DNMT3B-deficient cells displayed an intermediate reduction in γ-H2AX staining when treated with 5-azadC (Fig. 2B). The lack of γ-H2AX staining we observed in 5-azadC-treated ATM-deficient cells is in keeping with previously published studies on the role of ATM in phosphorylating H2AX in response to DNA damaging agents (95). Interestingly, the basal level of γ-H2AX staining in untreated cells (not exposed to drug and grown under standard laboratory conditions) varied significantly, with DNMT1- and ATM-deficient cells showing fourfold or higher levels of staining compared to the respective line's wild-type for each of these proteins (Fig. 2C). In keeping with this result, 1KO cells also undergo apoptosis under standard growth conditions at approximately twice the frequency of all other cell lines (Fig. 2C, inset graph), suggesting that 1KO cells may have a chronically elevated level of DNA damage.
FIG. 2.
Treatment of cells with 5-azadC results in induction of γ-H2AX, a marker of DNA double-strand breaks, that is dependent on DNMT1 and the PI3K ATM. (A) Representative immunofluorescence staining result for γ-H2AX (red) and DNA (blue) in untreated and 10 μM 5-azadC-treated HCT116 cells. A magnified view of the boxed cell is shown in the upper left corner of the 5-azadC-treated cells. (B) Dose-dependent increase in γ-H2AX-positive HeLa, HCT116 parental, 1KO, and 3BKO cells treated with 5-azadC (left graph). Induction of γ-H2AX staining in isogenic YZ5 and EBS cell lines following 5-azadC treatment (right graph). Results are plotted as the average relative change in cells showing five or more γ-H2AX foci compared to untreated cells. Data are graphed as the average increase (n-fold) in level of staining, relative to the respective parental cell lines (set at 1.0). (C) Quantitation of the percentage of γ-H2AX-positive cells and level of apoptosis (inset graph) in each cell line in the absence of any drug treatment (basal levels) showing that cell lines with reduced or absent induction of γ-H2AX staining upon 5-azadC treatment (1KO and EBS) exhibit elevated basal levels of γ-H2AX expression and apoptosis. (D) PI3Ks are required for H2AX phosphorylation in response to 5-azadC treatment. HeLa and parental HCT116 cells were mock treated (UT), treated with 10 μM 5-azadC alone for 24 h (5-azadC), or treated with both 10 μM 5-azadC and 10 μM wortmannin (a general PI3K inhibitor) for 24 h (A+W); then, cells were fixed and stained with a γ-H2AX antibody, and positive cells were quantitated (cells with five or more γ-H2AX foci). All values are an average of at least three experiments, and the error bar is standard deviation from the mean.
The ATM protein kinase is considered the master regulator of the DNA damage response to double-strand breaks, phosphorylating, among many other proteins, p53, H2AX, NBS1, BRCA1, and CHK2 to activate the G1/S, S, and G2 checkpoints as well as to load ATR at sites of DNA damage for the later phases of the damage response (1, 70, 90, 101). ATM and the related PI3K-like proteins ATR and DNA-PK regulate many aspects of DNA damage recognition and repair with partially overlapping substrates and differential activation kinetics (90), and these kinases are inhibited to various degrees by wortmannin (DNA-PK > ATM ≫ ATR) (85). The ATM-defective EBS cell line was completely deficient in induction of γ-H2AX upon 5-azadC treatment, strongly implicating ATM as the primary mediator of this process (Fig. 2B). To gain additional support for the involvement of PI3Ks in general, we treated HeLa and parental HCT116 cells with 10 μM 5-azadC or 10 μM 5-azadC plus 10 μM wortmannin (added at the same time) and stained for γ-H2AX. Wortmannin completely suppressed γ-H2AX induction in response to 5-azadC treatment in both cell lines (Fig. 2D), lending further support to the involvement of the PI3Ks in the cellular DNA damage response to 5-azadC. Taken together, these results reveal that 5-azadC treatment leads to DNA double-strand breaks, either directly or indirectly, and that DNMT1 plays a critical role in this process. It is also noteworthy that cell lines deficient in γ-H2AX staining in response to 5-azadC treatment (DNMT1 KO and ATM-deficient cells) also exhibit elevated basal levels of γ-H2AX and apoptosis, suggesting that these same proteins have roles in suppressing spontaneously occurring DNA strand breaks. Indeed, a previous study noted that genetic disruption of DNMTs in the HCT116 system leads to increased genomic instability (48).
5-azadC treatment induces DNA double-strand breaks that are recognized and repaired differentially in DNMT-proficient versus -deficient cells.
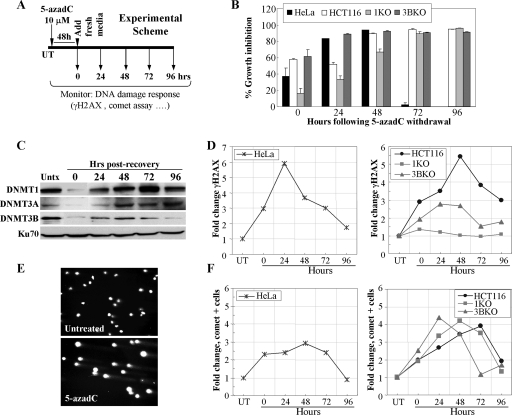
Our previous studies clearly showed that treatment of HeLa and HCT116 cells with 5-azadC results in G2 arrest and induction of DNA double-strand breaks. We therefore wondered if cells may be accumulating DNA damage and then repairing it. To address this, we treated HeLa and HCT116 cells as shown in the experimental scheme in Fig. 3A. Cells were treated with 10 μM 5-azadC for 48 h (fresh drug added every 24 h), and the medium was changed. In this way, all cells in the population should have gone through at least one round of DNA replication in the presence of 5-azadC. Cells were then allowed to grow for an additional 96 h without drug, during which time growth inhibition and DNMT protein levels were measured. Data obtained from cell counting following trypan blue staining of cells revealed that HeLa cells show a peak of growth inhibition in this treatment scheme at 48 h following 5-azadC removal, while HCT116 cells and the KO lines peak at 72 h after drug removal (Fig. 3B). HCT116 1KO cells displayed reduced growth inhibition at early time points (0 to 48 h after drug removal) but caught up to the other HCT116 lines by 72 h (Fig. 3B). Levels of all DNMT proteins were heavily depleted after the end of the drug treatment but recovered to posttreatment levels by 48 h. DNMT1 protein levels rebounded by 48 h, increased further at 72 h to a level above the untreated cells, and then returned to posttreatment levels again by 96 h (Fig. 3C). We next analyzed γ-H2AX staining by immunofluorescence at 24-h intervals after 5-azadC removal. Both HeLa and HCT116 parental lines showed a five- to sixfold increase in γ-H2AX staining that peaked 24 to 48 h following withdrawal of 5-azadC from the medium (Fig. 3D). This level then steadily declined over the remainder of the experiment to nearly the levels of untreated cells at 96 h, suggesting that cells are accumulating DNA damage and then repairing it. The relatively low level of apoptosis, particularly in HCT116 cells (data not shown), and the 48-h treatment time (at least one cell doubling in the presence of drug) support the notion of repair rather than cell death and repopulation of the culture by cells that had not incorporated 5-azadC. Interestingly, similar to the results presented in Fig. 2, loss of normal DNMT1 function resulted in a profound deficiency in γ-H2AX induction following 5-azadC treatment, whereas cells deficient in DNMT3B showed an intermediate effect (Fig. 3D, right panel).
FIG. 3.
5-azadC treatment results in formation of DNA strand breaks that are repaired upon drug withdrawal. (A) Experimental scheme used for the assays in this figure. Cells were treated with 5-azadC for 48 h with fresh drug added each day. The medium was then changed, and the cells were allowed to recover for 4 days. Numbering in all graphs is relative to the day post-drug withdrawal, which starts at zero hours. (B) Growth inhibition of all cell lines after removal of 5-azadC. Results are presented as the average percent growth inhibition ([100 − (number of cells in treated sample/number of cells in untreated sample)] × 100) for quadruplicate experiments. (C) Soluble DNMT protein levels before and during drug treatment and recovery as monitored by Western blotting. Ku70 served as a loading control. Hrs, hours. (D) Time course (or recovery) of γ-H2AX staining in HeLa cells following withdrawal of 5-azadC (left). Time course of HCT116 parental, 1KO, and 3BKO cells following 5-azadC withdrawal from the growth medium (right). (E) Representative comet assay result showing formation of DNA strand breaks (formation of a “comet tail”) in 5-azadC-treated HCT116 cells. The photograph is taken at a magnification of ×20. (F) Time course of comet tail formation in HeLa cells (left) or HCT116, 1KO, and 3BKO cells (right) following withdrawal of 5-azadC from the medium. Results are presented as the relative change (n-fold) in cells displaying a comet tail compared to untreated cells. Experiments were repeated at least twice, and the results shown are representative.
To further investigate the nature of the DNA damage induced by 5-azadC and how cells respond to it, we employed the comet assay. The comet assay is a sensitive fluorescent microscopic method to examine DNA damage and repair at the individual cell level (92). Treatment of HeLa and HCT116 cells with 10 μM 5-azadC resulted in DNA fragmentation characteristic of DNA strand breaks, as monitored by formation of a comet-like tail following single-cell gel electrophoresis (Fig. 3E). Using the same experimental scheme shown in Fig. 3A, we found that DNA fragmentation (or comet-positive cells) in HeLa and HCT116 cells increased three- to fourfold, and like γ-H2AX staining, comet-positive cells decreased to nearly background levels after 96 h (Fig. 3F). The peak of comet-positive cells differed by approximately 24 h between HeLa and HCT116 cells. Treated HCT116 parental, 1KO, and 3BKO cultures accumulated nearly equal levels of cells with DNA damage, but there was a marked difference in the kinetics (Fig. 3F). 1KO and 3BKO cells accumulated DNA damage more rapidly (24 to 48 h earlier) than the parental HCT116 cells. Interestingly, however, most of the DNA damage appeared to be repaired by the end of the 96-h experiment, as shown by the return of comet-positive cells to nearly background levels. Taken together, the results of γ-H2AX staining and the comet assay suggest that 5-azadC treatment results in formation of DNA double-strand breaks and that cells are capable of repairing this damage. Furthermore, loss of DNMT1 function, in particular, results in a profound change in the DNA damage response, with drastically reduced induction of γ-H2AX and more rapid accumulation of DNA damage (as measured by the comet assay) following 5-azadC treatment. The comet assay also demonstrates that the lack of γ-H2AX response in the 1KO cells is not due to decreased DNA strand breaks in the absence of fully functional DNMT1 protein. The observation that DNMT1 or DNMT3B depletion in HCT116 cells results in minor changes in doubling time (<30%) (data not shown) supports the notion that the effects we observe are not a result of differential incorporation of 5-azadC due to differences in the time each cell type spends in S phase. These results also imply that the nature of the 5-azadC-DNMT1 adducts differs from the 5-azadC-DNMT3B adducts in the structure of the lesion, quantity, or perhaps location within the genome. 5-azadC-DNMT1 adducts and/or a fully functional and mobile DNMT1 enzyme are therefore critical for induction not only of most of the cell cycle and growth inhibitory effects but also of the cell's response to these strand breaks in the form of H2AX phosphorylation.
5-azadC treatment induces a DNA damage response that is altered in DNMT-deficient cells, particularly in 1KO cells.
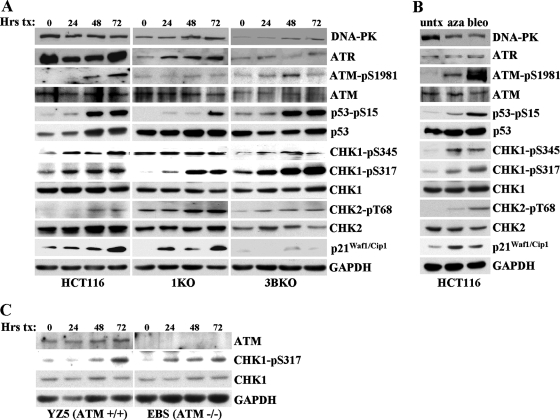
Our previous studies strongly implicated ATM in the response to 5-azadC-mediated DNA damage. ATM is primarily activated by DNA double-strand breaks caused by ionizing radiation or other radiomimetic agents, whereas ATR is involved in the damage response to replicative stress or other forms of damage that result in formation of single-stranded DNA. Both ATM and ATR phosphorylate a large number of downstream substrates, such as the CHK2 and CHK1 kinases, respectively, which initiate the DNA damage response cascade (90). The exact structural nature of the DNA damage produced in 5-azadC-treated cells is unknown; however, we initiated a more detailed analysis of the cells' response to this damage, which may provide clues as to how cells respond to and repair it. For this purpose, we treated HCT116 parental, 1KO, and 3BKO cells with 10 μM 5-azadC for 72 h and monitored the changes in both the absolute levels of key DNA damage signaling molecules and induction of their active, phosphorylated forms, whenever possible, by quantitative Western blotting. Equal microgram amounts of protein from all cell lines and time points were loaded on SDS-PAGE gels, transferred, probed with a given antibody in a single experiment, and exposed to the same piece of film during Western blotting to allow for comparison of absolute levels of each protein between different cell lines. Upon 5-azadC treatment, we detected a time-dependent increase in the active, phosphorylated forms of ATM (at serine 1981), p53 (at serine 15), and CHK1 (at serines 317 and 345) in HCT116 parental cells (Fig. 4A, left). We also detected increases in the total levels of p53 and p21Waf1/Cip1 in 5-azadC-treated HCT116 cells, consistent with previous studies in HCT116 and other cell lines (49, 66, 88, 104). The active form of CHK2 (phospho-Thr68) was only weakly induced by 5-azadC treatment. GAPDH levels served as a loading control. Interestingly, cells deficient in DNMT1 or DNMT3B exhibited a number of distinct differences in their DNA damage response compared to parental HCT116 cells. For example, basal and 5-azadC-induced levels of phospho-Thr68 CHK2 increased, relative to parental HCT116, in both the 1KO and 3BKO cells. The 1KO cells stood apart from parental and 3BKO cells in a number of respects. 1KO cells displayed reduced levels and/or delayed activation of phospho-Ser15 p53, phospho-Ser317 CHK1, and phospho-Ser1981 ATM but were constitutively activated for the phospho-Ser345 form of CHK1 (Fig. 4A, middle). The 3BKO cells behaved similarly to the parental HCT116 cells with regard to these markers. One response of 3BKO cells to 5-azadC that was particularly notable was the nearly complete lack of p21Waf1/Cip1 induction despite an apparently normal p53 response. Total levels of the other PI3Ks, DNA-PK and ATR, were unchanged upon 5-azadC treatment except for ATR levels in 1KO cells and DNA-PK levels in 3BKO cells (Fig. 4A). As a control for these studies, we treated HCT116 cells with bleomycin, an agent known to induce DNA double-strand breaks and mimic many of the effects of irradiation (89). For the case of bleomycin-treated cells, all of the phosphorylated forms of the proteins we studied were robustly induced, and total levels of p53 and p21Waf1/Cip1 also increased, as would be expected (Fig. 4B). CHK1, although generally considered an ATR target, can be phosphorylated by ATM (65). We therefore monitored CHK1 phosphorylation in 5-azadC-treated ATM-negative EBS cells. Quantitative Western blotting revealed that CHK1 was indeed phosphorylated in ATM-negative cells treated with 5-azadC, further supporting a role for ATR in the 5-azadC-mediated DNA damage response (Fig. 4C). Taken together, this analysis reveals that 5-azadC-induced DNA damage activates components of both the ATM and ATR pathways, suggesting that it involves the direct or indirect generation of DNA strand breaks and that other forms of damage or replicative stress are also created. Importantly, this study also demonstrates that DNMT1 (the loss of which blunts both p53 and alters CHK1 activation) and, to a lesser extent, DNMT3B (the loss of which enhances the CHK2 response and severely impairs p21Waf1/Cip1 induction) or the adducts they form with 5-azadC play important roles in initiating the DNA damage response to this drug. Differential activation of proteins like ATM, CHK1, and CHK2 may also help explain the marked differences in induction of γ-H2AX staining we observed in 5-azadC-treated cells.
FIG. 4.
DNA damage response to 5-azadC, as monitored by quantitative Western blotting, and the role of DNMT1 and DNMT3B. (A) The HCT116 cell lines indicated at the bottom of the Western panel were treated with 10 μM 5-azadC for 0, 24, 48, and 72 h (with fresh drug added every 24 h), and whole-cell extracts were prepared. Equal microgram amounts of extract were loaded onto SDS-PAGE gels, transferred to polyvinylidene difluoride membrane, and probed with the antibodies indicated at the right. Note that all cell lines and treatments were run on gels, probed with antibody, and exposed to film at the same time to allow for accurate and quantitative comparison between cell lines for a given antibody. GAPDH served as a loading control. (B) HCT116 cells untreated or treated with 10 μM 5-azadC (aza) for 48 h or 10 μg/ml bleomycin (bleo) for 4 h. (C) ATM-deficient cells show induction of the active phosphorylated form of CHK1 (phospho-Ser317) in response to 5-azadC treatment (10 μM), demonstrating involvement of the ATR pathway in response to 5-azadC-mediated DNA damage. Equal microgram amounts of whole-cell extract from EBS and YZ5 cells treated with 5-azadC for 0, 24, 48, and 72 h were used as described in panel A. pS, phospho-serine; pT, phospho-threonine. Hrs tx, hours of treatment; untx, untreated.
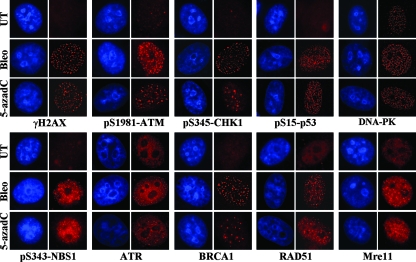
Most of the proteins involved in DNA double-strand break repair, such as γ-H2AX and the phosphorylated forms of ATM and p53, rapidly accumulate in discreet foci, where it is believed that each focus corresponds to a double-strand break (25, 27, 82). To gain additional insight into the proteins involved in sensing and repairing 5-azadC-induced DNA damage, we used immunofluorescence microscopy to monitor localization of key players in the DNA damage response. Two main pathways contribute to DNA double-strand break repair, homologous recombination and nonhomologous end joining, and each pathway makes use of both shared and unique protein components (70). HeLa cells were treated with 10 μM 5-azadC for 48 h and then fixed and stained. As a control for focus formation and/or protein relocalization, we also treated HeLa cells with bleomycin for 4 h and then washed out the drug and fixed them 20 h later. Results clearly show accumulation of γ-H2AX and the phosphorylated forms of ATM, CHK1, p53, and NBS1, as well as ATR, BRCA1, RAD51, and Mre11 at discreet foci in 5-azadC-treated cells (Fig. 5, red staining). We did not detect substantial relocalization of DNA-PK (Fig. 5) or Ku80 (data not shown) under the same conditions. Nearly identical results were obtained in bleomycin-treated cells. These results therefore provide further support that 5-azadC treatment of cells leads to the formation of DNA double-strand breaks and for the involvement of both ATM- and ATR-related pathways in responding to the damage. Our data also suggest a role for the homologous recombination system in repairing 5-azadC-mediated DNA damage since we observed formation of RAD51-containing foci, although the involvement of nonhomologous end joining as well cannot be excluded (99).
FIG. 5.
5-azadC treatment induces characteristic relocalization of DNA damage response proteins. Immunofluorescence staining of HeLa cells that were mock treated (UT), treated with 10 μM 5-azadC for 48 h, or treated with 10 μg/ml bleomycin (bleo) for 4 h. Cells were then fixed and stained with the indicated antibodies (red staining) and for DNA (blue staining), and representative images were collected on an upright fluorescence microscope at a magnification of ×100. Note the formation of foci in drug-treated cells and the induction of phosphorylated (active) forms of select proteins.
DNMT1 colocalizes with sites of DNA damage in 5-azadC-treated cells.
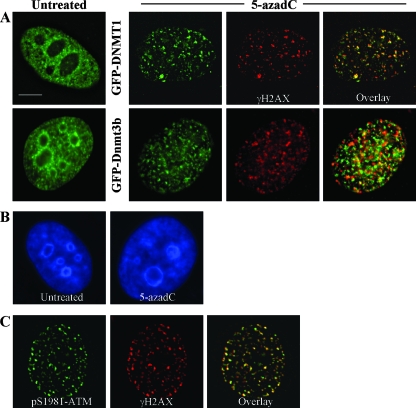
During the course of these studies we noted that, upon treatment of HeLa (Fig. 6A) and HCT116 cells (data not shown) with 5-azadC, there was a dramatic relocalization of GFP-tagged DNMTs within the nucleus. Localization of DNMT1 and Dnmt3b1 changed from their normal nucleoplasmic staining with concentration in DAPI (4′,6′-diamidino-2-phenylindole)-dense heterochromatic regions (3, 28, 56, 77) to one where the DNMTs were aggregated into many small and large foci and little if any of the protein remained in heterochromatic regions (Fig. 6A, compare images with green staining). This was not due to a generalized disruption in DAPI-dense heterochromatin regions (Fig. 6B), suggesting that DNMT relocalization is not a result of nonspecific effects of 5-azadC on nuclear morphology. Similar relocalization in the presence of 5-azadC and dependence on an active catalytic domain has been observed for DNMT1 (86).
FIG. 6.
DNMTs relocalize in the presence of 5-azadC, and DNMT1 colocalizes with γ-H2AX DNA damage foci in 5-azadC-treated cells. (A) HeLa cells were transfected with GFP-DNMT1 or GFP-Dnmt3b1. Cells were then mock treated (untreated) or were treated with 10 μM 5-azadC for 48 h, fixed, and then examined for localization of GFP signal. Both GFP-DNMTs localize diffusely throughout the nucleoplasm and are also concentrated in DAPI-dense regions corresponding to heterochromatin in untreated cells (particularly Dnmt3b; green staining), as has been shown previously by us along with other investigators. Upon 5-azadC treatment, DNMT1 and Dnmt3b1 relocalize, becoming aggregated into more discrete foci and absent from DAPI-dense heterochromatin regions. 5-azadC-treated cells were also stained for γ-H2AX (red staining), showing the formation of characteristic foci in cells with damaged DNA. Overlaid images (far right) of the red and green signals show that DNMT1 is highly colocalized with γ-H2AX (yellow signal). (B) DNA staining of untreated and 5-azadC-treated HeLa cells to demonstrate that there is no gross disruption of heterochromatin or nuclear morphology in 5-azadC-treated cells. (C) Colocalization of activated ATM (phosphorylated at serine 1981) and γ-H2AX in 5-azadC-treated HeLa cells. Bar, 5 μm.
The punctate pattern of staining for the DNMTs in the presence of 5-azadC, particularly that of DNMT1, was reminiscent of the focal patterns observed with γ-H2AX and other DNA repair proteins in 5-azadC- and bleomycin-treated cells (Fig. 5). To examine this potential correlation more directly, we stained GFP-DNMT transfected cells that had been treated with 10 μM 5-azadC with the γ-H2AX antibody. Interestingly, there was a very high degree of colocalization between GFP-DNMT1 and γ-H2AX in 5-azadC-treated cells. GFP-Dnmt3b1, while relocalizing from its normal staining pattern into nucleoplasmic foci due to 5-azadC treatment, exhibited much weaker colocalization with γ-H2AX (Fig. 6A, 5-azadC frames). We used double staining of 5-azadC-treated cells with γ-H2AX and phospho-Ser1981 ATM as a positive control, and this revealed nearly complete colocalization of these two DNA damage markers, as would be expected (Fig. 6C). These results therefore demonstrate the presence of DNMT1 at sites of DNA damage that likely correspond to DNA double-strand breaks. The particularly high degree of colocalization between DNMT1 and γ-H2AX, which was comparable to the colocalization between γ-H2AX and activated ATM, further supports our previous data that DNMT1 is a major mediator of the effects of 5-azadC on cell growth and induction of the DNA damage response.
Other clinically relevant nucleoside-based DNMT inhibitors induce DNA damage.
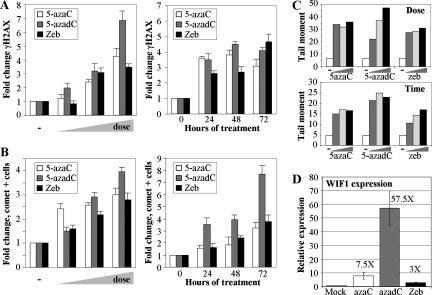
While 5-azadC and 5-azaC have been used in the laboratory and clinic for some time, renewed interest in their clinical use, particularly for the treatment of MDS and other leukemias, has increased in recent years (47, 64, 102). ZEB, a related analog, also inhibits DNMTs and induces expression of aberrantly hypermethylated genes, although it is considerably less potent. ZEB is, however, more stable in aqueous solution than 5-azaC and 5-azadC; therefore, there is interest in developing it for clinical use (14, 15, 96). While the demethylating properties of these agents have been directly compared (96), their capacity to induce DNA damage has not. We compared the ability of 5-azaC, 5-azadC, and ZEB to induce the well-known marker of damaged DNA, γ-H2AX, by immunofluorescence microscopy. Treatment of HCT116 cells with increasing doses of each drug (0.1, 1.0, and 10 μM for 5-azaC and 5-azadC and 50, 250, and 500 μM ZEB) for 48 h (fresh drug added every 24 h) resulted in a dose-dependent increase in γ-H2AX-positive cells, with 5-azadC being the most effective of the three drugs (Fig. 7A, left graph). The time course of γ-H2AX induction was also monitored over 3 days after the addition of 10 μM 5-azaC, 10 μM 5-azadC, and 250 μM ZEB. Treatment of HCT116 cells with each of the three agents caused a time-dependent increase in γ-H2AX staining that peaked between 48 and 72 h for 5-azaC and 5-azadC while ZEB reached its highest level at 72 h of treatment (Fig. 7A, right graph).
FIG. 7.
Other clinically relevant nucleoside inhibitors of DNA methylation induce DNA damage in a manner related to their ability to demethylate genes. (A) Direct comparison of the ability of 5-azaC, 5-azadC, and ZEB to induce formation of γ-H2AX foci in HCT116 cells in a dose-dependent (left) and time-dependent (right) manner. Doses (indicated by the wedge) of 5-azaC and 5-azadC used were 0.1, 1.0, and 10 μM, while 50, 250, and 500 μM ZEB was used since it is less potent at inducing demethylation of aberrantly methylated genes. (B) Time- and dose-dependent induction of strand breaks in HCT116 cells treated with the same three drugs as monitored by comet assay. Values are the average of three independent determinations. (C) Calculation of the tail moment, a measure of the degree of DNA damage, from the data in panel B. The upper panel shows the tail moments for the dose-response experiment, and the lower panel shows the tail moments for the time course. −, mock-treated cells or zero hours. (D) Quantitative RT-PCR analysis for expression of the WIF1 gene, which is densely hypermethylated in parental HCT116 cells. RNA prepared from HCT116 cells treated with 10 μM 5-azaC, 10 μM 5-azadC, or 250 μM ZEB was reverse transcribed and PCR amplified with primers for WIF1 and GAPDH. WIF1 expression was normalized to GAPDH expression as described in Materials and Methods, and relative expression of treated versus untreated cells was determined by the ratio of normalized expression values for the treated cells to the normalized expression value of the calibrator (untreated cells) All drug treatments are significantly elevated over values of the mock-treated cells (paired t test, P < 0.04).
To more directly compare the DNA damage caused by the three drugs, we measured DNA strand break formation with the comet assay. As was seen for the γ-H2AX staining, 5-azaC, 5-azadC, and ZEB treatment resulted in a dose-dependent increase in DNA strand breaks, with 5-azadC being the most effective at the highest dose (Fig. 7B, left graph). The time course experiment also indicated that the number of cells with strand breaks accumulated progressively with time, and the largest number of comet tail-positive cells was observed after 5-azadC treatment (Fig. 7B, right graph). Experiments shown in Fig. 7B quantitated the numbers of cells showing comet tails, indicative of DNA fragmentation. To compare the amount of DNA fragmentation (or DNA damage) in individual cells caused by each drug, we calculated the tail moment. These results are summarized in Fig. 7C and reveal that, in general, 5-azadC induced the greatest amount of DNA damage at the highest dose and that 5-azaC and ZEB induced slightly less damage and were roughly comparable. The amount of γ-H2AX- and comet-positive cells also roughly correlated with the efficacy of each drug to reverse aberrant promoter hypermethylation of the WIF1 gene, which we have shown to be densely hypermethylated in HCT116 cells (2). Quantitative RT-PCR analysis of WIF1 expression revealed that 10 μM 5-azadC most efficiently induced reexpression (∼58-fold), followed by 10 μM 5-azaC (7.5-fold) and 250 μM ZEB (3-fold) in HCT116 cells (Fig. 7D). Taken together, these results indicate that all three nucleoside-based DNMT inhibitors result in induction of DNA strand breaks and that their capacity to do so roughly correlates with their ability to demethylate aberrantly hypermethylated genes. Therefore, DNA damage-mediated cytotoxicity, as well as reexpression of aberrantly silenced growth regulatory genes, may contribute to the clinical efficacy of all of these agents.
Defective DNA damage responses in HCT116 1KO cells are not limited to DNA methylation inhibitors.
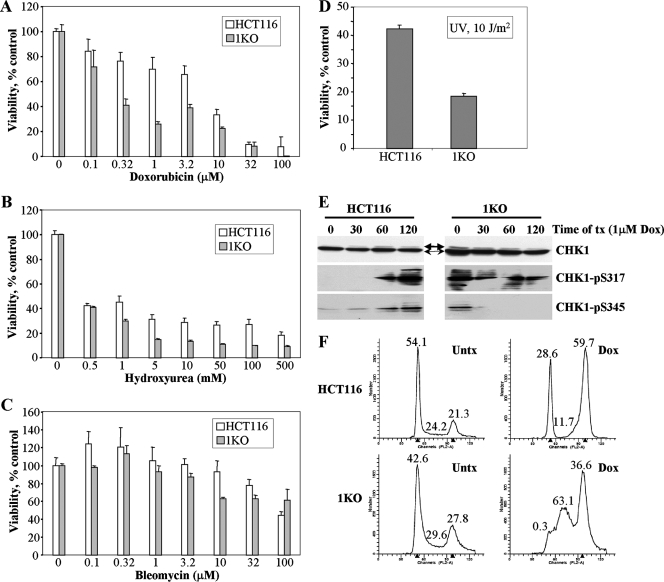
Our previous experiments indicated that HCT116 1KO cells were defective in their DNA damage response in a number of ways, including altered induction of the activated forms of γ-H2AX, CHK1, ATM, and p53, as well as reduced G2 arrest and increased cell death following 5-azadC treatment. Our observations that HCT116 cells lacking a fully functional DNMT1, a major target of 5-azadC, were actually affected to a greater extent than parental or 3BKO cells, coupled with the defects just mentioned, led us to wonder if there was a more generalized defect in the DNA damage response in 1KO cells. To test this idea, we treated HCT116 parental and 1KO cells with four other well-characterized DNA damaging agents, doxorubicin, hydroxyurea, bleomycin, and exposure to UV light and measured cell viability. Following a 24-h drug exposure, 1KO cells showed significantly reduced cell viability, relative to parental HCT116 cells, at doses of 0.3 to 10 μM doxorubicin, 1 to 500 mM hydroxyurea, and 10 to 32 μM bleomycin (Fig. 8A to C). In addition, 1KO cells demonstrated significantly reduced viability following exposure to UV light (Fig. 8D). Since our previous results indicated that activation of CHK1 was markedly altered in 1KO cells after 5-azadC treatment, we used Western blotting of extracts from doxorubicin-treated cells to monitor activation of CHK1 (phosphorylation at both serines 317 and 345). This revealed that both phospho-Ser317 and phospho-Ser345 CHK1 were activated in parental HCT116 cells (Fig. 8E), as would be expected and consistent with the 5-azadC results presented earlier (Fig. 4A). In contrast, 1KO cells demonstrated constitutively activated phospho-Ser345 CHK1 that actually decreased following doxorubicin treatment, while levels of phospho-Ser317 were elevated, and no induction upon doxorubicin treatment was observed (Fig. 8E). We then compared the cell cycle dynamics of HCT116 and 1KO cells following treatment with 1 μM doxorubicin for 24 h. This experiment revealed that parental HCT116 cells arrested predominantly in G2 and G1 in response to doxorubicin treatment, consistent with published results (Fig. 8F) (20). 1KO cells, however, were markedly deficient in both G1 and G2 arrest after the identical doxorubicin treatment conditions and appeared to accumulate primarily in S phase (Fig. 8F). Taken together, these results show that the defective DNA damage response of 1KO cells is not restricted to agents that impact DNA methylation. Rather, 1KO cells are significantly more sensitive to other agents that directly or indirectly lead to DNA double-strand breaks. This increased sensitivity may, at least in part, be mediated by aberrant activation of CHK1 and subsequent defects in cell cycle checkpoints since CHK1 is critical for G2 arrest in response to doxorubicin and other DNA damaging agents (37).
FIG. 8.
Altered cellular responses of HCT116 1KO cells when exposed to other genotoxic agents. (A to D) MTT cell viability assays in parental HCT116 and 1KO cells after exposure to increasing doses of doxorubicin, hydroxyurea, bleomycin, and UV light. All values, presented as the percent viability relative to mock-treated cells, are the average of quadruplicate experiments, and the error bar is the standard deviation. Doxorubicin, hydroxyurea, and bleomycin treatments were for 24 h. (E) CHK1 activation following treatment of parental and 1KO HCT116 cells with 1 μM doxorubicin for the indicated times (in minutes), monitored by quantitative Western blotting. Equal amounts of whole-cell extract were used at each time point, and Western blotting was performed with the antibodies indicated at the right. Thin and thick arrows denote unmodified and phosphorylated forms of CHK1, respectively. (F) Representative cell cycle profiles of HCT116 and 1KO cells exposed to 1 μM doxorubicin for 24 h. The percentage of cells in each phase of the cell cycle is given above the G1, S, and G2/M peaks. Parental HCT116 cells arrest predominantly in G2/M and also in G1, while the G2/M and G1 checkpoints appear to be defective in 1KO cells after doxorubicin treatment. Untx, untreated; tx, treatment; Dox, doxorubicin.
DNMT1 interacts with CHK1 and influences its subcellular localization following DNA damage.
Aberrant DNA damage responses and G2 arrest following treatment of HCT116 cells deficient in DNMT1 function with 5-azadC and other DNA damaging agents, coupled with recent findings showing that HCT116 cells completely null for DNMT1 undergo mitotic catastrophe with minimal losses of DNA methylation (13), led us to test the hypothesis that DNMT1 interacts with components of the DNA damage response or repair machinery. Interestingly, loss of CHK1 function also induces mitotic catastrophe in certain experimental settings (69). To address this possibility, we performed immunoprecipitations using HeLa nuclear extract and an antibody directed against CHK1. Subsequent Western blotting of the immunoprecipitated material with a DNMT1 antibody clearly showed that DNMT1 coimmunoprecipitated with CHK1 (Fig. 9A, top panel). Positive controls included immunoprecipitations with antibodies directed against PCNA and p53, both of which have been shown to interact with DNMT1 (17, 24), while preimmune IgG served as a negative control (Fig. 9A). In addition, the DNMT1 antibody used was clearly capable of immunoprecipitating endogenous DNMT1 from HeLa cells (Fig. 9B). In a reciprocal reaction, DNMT1 antibody immunoprecipitated CHK1 but not CHK2, from HeLa nuclear extract, showing that while these two kinases have some overlapping functions and interact with some of the same proteins, only CHK1 interacts with DNMT1 under our experimental conditions (Fig. 9A, bottom panel). Our data showing that endogenous DNMT1 and CHK1 interact in HeLa cells without exposure to a DNA damaging agent suggested that the interaction was constitutive; however, low levels of DNA damage are likely always present in cell cultures. To determine if DNA damage alters the interaction between DNMT1 and CHK1, we first transfected HeLa cells with GFP-tagged DNMT1 and FLAG-tagged CHK1 in order to facilitate detection of interactions, followed by treatment with 1 μM doxorubicin for 24 h. Whole-cell extract was prepared, and immunoprecipitations were performed with an antibody directed against DNMT1. Subsequent Western blotting with a FLAG antibody revealed that FLAG-CHK1 does indeed bind to GFP-DNMT1 in untreated cells, consistent with immunoprecipitations using HeLa nuclear extract, and that upon treatment with doxorubicin, the interaction between DNMT1 and CHK1 was markedly increased (Fig. 9C, upper panel). This enhancement was not attributable to differences in expression of ectopic DNMT1 and CHK1 (Fig. 9C, lower panels). These results therefore indicate that DNMT1 interacts with CHK1 but not CHK2 in HeLa cells, revealing a direct connection between DNMT1 and the DNA damage response system, and this interaction is enhanced by DNA damage.
FIG. 9.
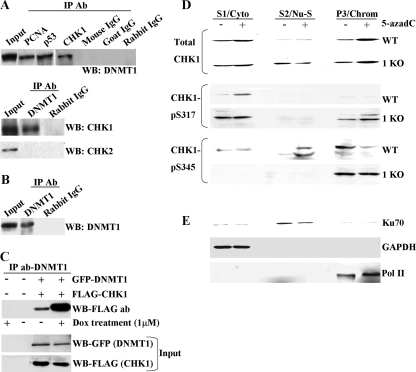
DNMT1 interacts with CHK1 and influences its subcellular distribution after DNA damage. (A) Demonstration that DNMT1 interacts with CHK1 in reciprocal coimmunoprecipitations. In the top panel, a CHK1 antibody (Ab) was used in the immunoprecipitation (IP), followed by Western blotting (WB) with a DNMT1 antibody. Immunoprecipitation with PCNA and p53 antibodies served as positive controls since they are known to interact with DNMT1, while preimmune IgG served as a negative control. In the bottom panel DNMT1 was used as the immunoprecipitating antibody followed by Western blotting with either CHK1 or CHK2 antibody. DNMT1 does not interact with CHK2 under these conditions. (B) Validation that the DNMT1 antibody used in panel A does indeed immunoprecipitate endogenous DNMT1 from HeLa nuclear extract. Untreated HeLa nuclear extract was used for panels A and B. (C) The DNMT1-CHK1 interaction is enhanced after DNA damage. HeLa cells were mock transfected (−) or transfected with GFP-DNMT1 and FLAG-CHK1. After 24 h, doxorubicin (Dox) was added to the cultures as indicated (+), and whole-cell extract was prepared 24 h later and used in immunoprecipitations with a DNMT1 antibody. CHK1 was detected in Western blotting with an antibody directed against FLAG (upper panel). Expression levels of DNMT1 and CHK1 were equal between untreated and drug-treated cultures (input). (D) Subcellular fractionation of untreated and 5-azadC-treated parental (WT) and 1KO HCT116 cells. Each cell line was mock treated (−) or treated with 10 μM 5-azadC for 48 h (+), followed by isolation of the cytoplasmic (S1/Cyto), soluble nuclear (S2/Nu-S), and chromatin-enriched (P/Chrom) fractions. Each protein fraction was used for Western blotting with the antibodies shown at the left. (E) Validation of the fractionation protocol for HCT116 cells by showing the expected localization of Ku70, GAPDH, and Pol II in the nuclear, cytoplasmic, and chromatin-enriched fractions, respectively. Similar results were obtained with the 1KO cells (data not shown).
We next sought to better understand the interplay between CHK1 and DNMT1 and to learn why 1KO cells are defective in CHK1 activation and G2 arrest after exposure to DNA damaging agents. It has been well established that CHK1 is associated with chromatin in untreated cells and that, upon DNA damage, the differentially phosphorylated forms of CHK1 redistribute to the cytoplasm and/or the soluble fraction of the nucleus, and this redistribution is essential for CHK1's checkpoint functions (42, 93). Since DNMT1 is also associated with chromatin and interacts with CHK1, we were interested in determining if normal DNMT1 function contributed to CHK1 relocalization following DNA damage. We therefore performed a subcellular fractionation on untreated and 10 μM 5-azadC-treated (48 h) HCT116 parental and 1KO cells to isolate cytoplasmic, soluble nuclear, and chromatin-enriched fractions according to protocols established for monitoring CHK1 relocalization (60). For each of the fractions in each cell line, we monitored localization of total CHK1 and CHK1 phosphorylated at serines 317 and 345. For both parental HCT116 and 1KO cells, 5-azadC treatment resulted in an increase in the total CHK1 levels bound to chromatin and increased CHK1 in the cytoplasmic fraction in parental cells. CHK1 levels in the soluble nuclear fraction were markedly increased in both mock-treated and 5-azadC-treated 1KO cells and did not change with drug treatment (Fig. 9D, upper panel). Levels of phospho-Ser317 CHK1 were increased in 5-azadC-treated parental HCT116 cells, and this form of CHK1 relocalized to the cytoplasm, consistent with previous reports using UV light (93). In contrast, there was a slight increase in phospho-Ser317 CHK1 with 5-azadC treatment in the chromatin-associated fraction and a constitutively elevated level of phospho-Ser317 CHK1 in the cytoplasm of 1KO cells (Fig. 9D). Western blotting for phospho-Ser345 CHK1 revealed that there was a low basal level of CHK1 phosphorylated at serine 345 in mock-treated HCT116 cells. After DNA damage, this form of CHK1 became reduced in the chromatin-associated fraction and moved to the soluble nuclear phase, consistent with other studies (42). The pattern of phospho-Ser345 CHK1 in 5-azadC-treated 1KO cells was very different. While basal levels of this form of CHK1 were nearly equal in mock-treated parental and 1KO cells in the chromatin-associated fraction, there was no detectable relocalization of phospho-Ser345 CHK1 to the soluble nuclear or cytoplasmic fractions in 5-azadC-treated 1KO cells (Fig. 9D). As controls for the fractionation procedure, we used antibodies against Ku70, GAPDH, and Pol II, which are known to localize predominantly in the soluble nuclear, cytoplasmic, and chromatin-associated fractions, respectively, consistent with our results and demonstrating that the procedure was successful (Fig. 9E). Taken together, these results demonstrate a novel interaction between DNMT1 and CHK1 and indicate that at least some of the defects in the DNA damage response we have observed in cells lacking functional DNMT1 may be due to an inability to properly relocalize the activated forms of CHK1 to the cellular compartment where they exert their function. This appears particularly true of phospho-Ser345 CHK1, which was completely unable to be released from the chromatin fraction in 1KO cells following DNA damage.
DISCUSSION
In the present study we have conducted a detailed investigation into the cytotoxic action of the antitumor DNA demethylating drug 5-azadC. 5-azadC has been extensively studied, and its ability to inhibit cellular DNMTs and mediate reexpression of aberrantly hypermethylated growth-regulatory genes is well known (16, 36). Due to its inhibitory mechanism of action and its incorporation into newly synthesized DNA, 5-azadC results in covalent trapping of DNMTs, essentially causing genome-wide protein-DNA cross-links. This very likely contributes to 5-azadC's ability to inhibit tumor cell growth even at low doses but has been less well studied. We have shown, using human tumor cell lines, that 5-azadC treatment causes depletion of DNMT protein from the soluble nuclear phase, growth inhibition, G2 arrest, and a reduction in clonogenic survival. These results are in keeping with several previous studies of the mechanism of action of 5-azadC or 5-azaC (8, 30, 49, 62, 88, 100, 104). In addition, we have investigated how cells respond to 5-azadC in terms of the DNA damage machinery. We demonstrate that both the ATM and ATR pathways are activated in response to 5-azadC-mediated DNA damage, as shown by use of ATM-deficient cells, activation of both CHK1 and CHK2 kinases, and by use of the PI3K inhibitor wortmannin to blunt this activation. Induction of γ-H2AX by 5-azadC treatment, along with formation of DNA strand breaks measured by comet assay, supports the notion that 5-azadC directly or indirectly leads to formation of DNA double-strand breaks. We also present evidence, using immunofluorescence microscopy, for relocalization of DNA damage response proteins of the ATM and ATR pathways in 5-azadC-treated cells, as well as components of the homologous recombination-based repair machinery, into foci characteristic of cells with damaged DNA. Significantly, γ-H2AX foci, which are thought to represent sites of DNA strand breaks and are an assembly point for the DNA repair machinery (70, 82), colocalize highly with GFP-tagged DNMT1. Other clinically relevant nucleoside-based DNA demethylating drugs also mediate formation of γ-H2AX foci and DNA strand breaks. Finally, we show for the first time that cells deficient in normal DNMT1 function are defective in their response to 5-azadC and DNA damaging agents in general. This led us to discover a novel interaction between DNMT1 and CHK1 and a role for this interaction in regulating association of CHK1 with chromatin in response to DNA damage.
During the methylation reaction of cytosine, the DNMT forms a covalent thioester linkage with the C-6 position of cytosine, increasing electron flow to C-5 with subsequent attack on the methyl donor S-adenosyl-l-methionine. Proton abstraction from C-5, followed by β-elimination, allows for reformation of the 5-6 double bond and DNMT release. Presence of the more electronegative nitrogen at the 5 position in 5-azadC permits the normal methylation reaction to begin; however, it is halted prior to enzyme release, resulting in a relatively stable covalent DNA-DNMT linkage (16, 84). DNA damage mediated by 5-azadC could result from chemical instability of the drug or from an enzymatic repair process. Decay of 5-azadC-DNA-DNMT adducts is thought to yield predominantly two end products: a ring-open 1-β-d-ribofuranosyl-3-guanylurea species and inactivated DNMT formylated at the active site cysteine (7, 16, 41). Spontaneous decay of 5-azadC in aqueous solution is also thought to yield the same 1-β-d-ribofuranosyl-3-guanylurea species; however, given that the vast majority of mutations in 5-azadC-treated cells occur at CpG sites despite its random incorporation into the genome during replication, spontaneous decay is unlikely to play a major role in 5-azadC's DNA damage-inducing functions (7, 41). Furthermore, alkali-labile sites (indicative of strand breaks) in DNA from 5-azadC-treated leukemia cells did not increase linearly with drug incorporation, suggesting that, rather than simple chemical breakdown of incorporated 5-azadC molecules giving rise to strand breaks, an active and saturable repair process is involved (55). The fate of 5-azadC-DNA-DNMT adducts in vivo has not been determined. It is conceivable that the formylated DNMT is actively targeted for degradation (30), although our finding that DNMT1 strongly colocalizes with γ-H2AX foci in 5-azadC-treated cells suggests that the 5-azadC-DNA-DNMT adducts persist even if the level of soluble DNMT proteins decreases. Thus, it will be of great importance to identify the breakdown products of 5-azadC-DNA-DNMT-containing DNA in vivo to better understand how these adducts are causing DNA damage.
The DNA damage recognition and repair machinery in mammalian cells is extensive and mounts a rapid and efficient response to the myriad forms of damage that occur, depending on the environmental or endogenous insult. Exactly how cells repair the damage induced by 5-azadC-DNMT covalently linked DNA lesions is unknown although our data strongly support the involvement of double-strand break repair pathways. Alternatively, a DNA glycosylase may remove the damaged 5-azadC base degradation product once the inactivated methyltransferase has been removed from DNA. Whether loss of covalently bound DNMT occurs spontaneously or also requires an active removal process in vivo mediated by the nucleotide excision repair machinery, which is often required for removal of bulky adducts from DNA, remains unclear (19). Our data showing induction of γ-H2AX, which colocalizes with DNMT1 in 5-azadC-treated cells, and activation of many proteins in the double-strand break repair pathway (ATM and ATR for example) strongly suggest not only that the 5-azadC-DNA-DNMT adduct leads to DNA strand breaks but also that the cell is responding to and repairing this damage using proteins in both the ATM and ATR arms of the double-strand break repair system. Another key factor in determining which DNA repair pathways respond in 5-azadC-treated cells is where in the genome it is incorporated. For example, 5-azadC incorporated at replication foci, where DNMT1 is also localized, may result in defects in the structure of the replication fork, a finding supported by formation of foci containing ATR and activation of CHK1 in 5-azadC-treated cells in our study. Alternatively, the active removal of the 5-azadC-DNA-DNMT adduct by an unknown mechanism may directly lead to strand breaks. The expression level of a particular DNMT may also influence the DNA damage response to 5-azadC. Interestingly, DNMT1 and DNMT3A are highly overexpressed in chronic myelogenous leukemia (CML) bone marrow samples (52), and CML is one of the most 5-azadC-responsive tumor types known. It will therefore be critical to gain a better understanding of these issues as part of future studies so that human cancer patients that will respond best to 5-azadC can be identified and also to potentially exploit this knowledge to further enhance the antitumor potential of 5-azadC.
Accumulating evidence suggests that DNMT1, DNMT3A, and DNMT3B methylate the genome with some degree of redundancy but that there is functional specialization as well (31, 45, 54). For example, studies using ICF (immunodeficiency, centromeric instability, and facial anomalies) syndrome cells have demonstrated the particularly prominent role for DNMT3B in methylating pericentromeric satellite repeats (23). We examined the role of DNMT1 and DNMT3B, two DNMTs responsible for the majority of cellular DNA methylation (76) that are frequently deregulated in human tumor cells (80), in mediating the response to 5-azadC. Interestingly, cells deficient in DNMT1 exhibit consistent and markedly different responses to 5-azadC. For example, 1KO cells show delayed growth inhibition and reduced G2 arrest and clonogenic survival in response to 5-azadC treatment. DNMT 3BKO cells generally showed either an intermediate response or responded similarly to parental HCT116 cells with regard to most of these properties. Perhaps the most notable difference in 3BKO cells was the lack of p21Waf1/Cip1 induction, a finding which may merit further investigation. Results obtained using the 1KO cells are somewhat paradoxical in the sense that removal of one of the main targets of 5-azadC, DNMT1, actually resulted in more severe reductions in short- and long-term viability and reduced G2 arrest, suggesting a fundamental difference between human tumor cells and other cell systems used or a novel function for DNMT1 as a component of the DNA damage signaling machinery revealed through the use of hypomorphic 1KO cells (22). For example, a prior study in ES cells using complete knockout of Dnmt1 showed that reducing Dnmt1 levels also reduced the cytotoxic effects of 5-azadC (44, 71). Another recent study showed that Dnmt3a and Dnmt3b played a greater role in mediating the cytotoxic effect of 5-azadC on the growth of murine ES cells (71). Difference in species or the use of transformed versus normal cells could account for some of the divergent results; however, the particularly unique defect in DNMT1 identified in 1KO cells may be the most significant contributor to the differences, and this will be discussed further below. We focused our studies on human tumor cells because they are the intended target of a chemotherapeutic regimen utilizing 5-azadC.
We identified a number of other significant differences between parental and 1KO HCT116 cells while examining the DNA damage response to 5-azadC. For example, 1KO cells were completely deficient in induction of γ-H2AX foci and displayed reduced and/or delayed induction of the phosphorylated forms of p53, ATM, and CHK1 following 5-azadC treatment. Differences in the damage responses of 1KO cells to genotoxic agents were not limited to DNA methylation inhibitors. 1KO cells were significantly more sensitive to doxorubicin, hydroxyurea, bleomycin, and UV light, and activation of CHK1 and G2 arrest in 1KO cells treated with doxorubicin was also defective, indicating that 1KO hypomorph cells have a generalized defect in the damage response. These results suggest that DNMT1-aza-DNA adducts are distinct from DNMT3B-aza-DNA adducts in terms of either location throughout the genome, quantity, or ability of each lesion to activate a DNA damage response. For example, localization of DNMT1 to replication foci is mediated, at least in part, by its interaction with PCNA (17, 53); thus, this localization may make DNMT1-aza-DNA adducts more readily recognized by the repair machinery or cause defects in replication fork structure. The latter of these possibilities is supported by our data demonstrating activation of ATR-dependent signaling in 5-azadC-treated cells, since ATR senses stalled or collapsed replication forks (51), and by a recent publication using DNMT1 knock-down approaches (98). The prominent role of DNMT1 in mediating many of the effects of 5-azadC may therefore be attributable to its function as a maintenance DNMT responsible for copying methylation patterns throughout the genome.
Our results also suggest that DNMT1 itself plays an active role in the DNA damage/repair response, rather than simply mediating formation of DNA strand breaks in the presence of 5-azadC. For example, DNMT1 KO cells were as deficient for γ-H2AX induction as ATM-deficient cells in response to 5-azadC, and both lines showed elevated basal levels of γ-H2AX staining. The comet assay clearly showed that 1KO cells accumulated strand breaks to a level comparable to parental HCT116 cells. Indeed, a recent study showed that DNMT1-deficient cells display increased genomic instability (48), and DNMT1 has been implicated in playing a direct role in DNA mismatch repair (35, 50). DNMT1 is recruited to sites of DNA damage caused by UV radiation in a manner dependent on its PCNA interaction domain (63), and it interacts and cooperates with p53 to repress transcription (24). While p53 appeared to stimulate DNA methylation by DNMT1, an effect of DNMT1 on p53's DNA damage/repair functions was not reported (24). Interestingly, we found an inverse relationship between γ-H2AX staining and the accumulation of comet-positive cells. For example, 1KO cells showed almost no induction of γ-H2AX staining when treated with 5-azadC, yet they accumulated strand breaks with more rapid kinetics, suggesting that cells lacking DNMT1 (and ATM) have more unstable or fragile genomes. This inverse relationship is in keeping with the role of γ-H2AX not only as a protein involved in recruitment and retention of DNA repair proteins at sites of damage but also as a DNA damage signaling molecule (74, 75, 82). Therefore, the muted γ-H2AX induction in 1KO cells could potentially reflect impaired or delayed recognition of DNA damage in the absence of DNMT1. Taken together, our studies show that, in human tumor cell lines, DNMT1 is required to orchestrate the proper response to 5-azadC-mediated DNA damage.
We show for the first time an interaction between DNMT1 and CHK1 in cells under unperturbed growth conditions, and this interaction is enhanced upon DNA damage. This supports our hypothesis that DNMT1 possesses novel functions as a DNA damage signaling molecule and that the mechanism for this may be mediated through DNMT1's ability to regulate CHK1-chromatin interactions. DNMT1 hypomorphic cells were defective in their ability to activate CHK1, demonstrating blunted phosphorylation of CHK1 Ser317 and constitutively activated phosphorylation CHK1 Ser345 in most cases, and the phospho-Ser345 CHK1 appeared unable to migrate from the chromatin fraction to the soluble phase, a process that is essential for CHK1 function (93). The defect in 1KO cells is actually a small deletion within the N-terminal regulatory domain of DNMT1 yielding a protein lacking its PCNA interaction domain that is expressed at markedly reduced levels (22). Complete knockouts of DNMT1 were recently generated, and this revealed that HCT116 cells completely lacking DNMT1 led to activation of a DNA damage response involving ATM and rapid cell death by mitotic catastrophe (13). Interestingly, depletion of CHK1, which is essential for normal DNA replication in the absence of DNA damage, stabilizing stalled replication forks, and arresting damaged cells in G2/M, or mutation of the CHK1 serine 345 phosphorylation site, also led to mitotic catastrophe. Mutation of CHK1 serine 317 was compatible with cell survival but resulted in enhanced genomic instability (67). We do not at present know exactly how DNMT1 regulates CHK1 activation, nor if the interaction is direct, but several scenarios can be envisaged. DNMT1 may create a particular chromatin state required for CHK1 binding or recruitment of other CHK1 regulators, such as the 9-1-1 (RAD9-HUS1-RAD1) PCNA-like checkpoint complex (73). Since hypomorphic 1KO cells and HCT116 cells with complete DNMT1 knockout lose comparable amounts of genomic DNA methylation (∼20%) with radically different effects on cell viability, this mechanism seems unlikely. Another possible mechanism is that DNMT1 may tether CHK1 to chromatin and or replication forks, and a subsequent modification (e.g., phosphorylation) of DNMT1 or other interacting protein allows for CHK1 release upon DNA damage. Current models suggest that CHK1 activation is regulated in a complex manner; the RAD17-RFC (clamp loader)/9-1-1 complex (clamp) and ATM/ATR independently accumulate at sites of DNA damage, followed by recruitment and activation of CHK1 via the kinase activity of ATR. Interestingly, the 9-1-1 complex is structurally similar to the PCNA replicative sliding clamp. The truncated form of DNMT1 expressed in 1KO hypomorph HCT116 cells lacks its PCNA binding domain and is not efficiently recruited to replication foci or sites of UV-induced DNA damage (63, 94). It is intriguing to speculate that, if a similar interaction exists between DNMT1 and the 9-1-1 complex, this may also be disrupted in the 1KO hypomorphs, resulting in reduced DNMT1-CHK1 interaction at sites of DNA damage culminating in defective CHK1 activation. As we have suggested previously, this novel function of DNMT1 may be revealed only through the use of cells expressing an unusual form of DNMT1 or through the extreme depletion conditions of a complete genetic knockout of DNMT1 (10). Low levels of DNMT1 may be able to sustain proper CHK1 activation and replication fork stability and allow for continued cell division. Under these conditions, however, genomic DNA demethylation and reactivation of tumor suppressor genes may occur and further influence cell growth dependent on cell type, the complement of aberrantly hypermethylated genes, and the integrity of cell cycle checkpoints.
This work also has implications for the clinical use of 5-azadC and related nucleoside-based DNMT inhibitors. 5-azadC and 5-azaC possess significant clinical efficacy in MDS and other leukemias, and in the case of MDS, they represent the only currently available treatment option for many patients due to their advanced age. Total and complete response rates varied between 50 to 80% and 10 to 50%, respectively, depending on the study, dose, number of treatment courses, and method of drug delivery (18, 38, 40, 64, 102). Recent studies have suggested that lower-dose, longer-term treatment of MDS patients with 5-azadC improves response rates, reduces toxic side effects, and maximizes demethylating activity (102). Treatment of human cancer patients with 5-azadC results in global, repetitive element, and gene-specific demethylation (18, 64, 83, 102). Interestingly, however, the correlation between patient response and degree of demethylation has, in general, been limited. For example, a study of 5-azadC in MDS patients revealed that p15Ink4b demethylation correlated with patient response but also that responses were seen in patients without p15Ink4b methylation (18). Microarray gene expression profiling of acute myeloid leukemia and MDS cells before and after 5-azadC treatment revealed that about 80 genes were up-regulated; however, the majority of them were not methylated before or after treatment, suggesting a methylation-independent mechanism (87). In a recent study of 5-azadC for the treatment of CML, hypomethylation correlated with response rate at low doses of 5-azadC but not at higher doses. The DNA methylation status of several frequently hypermethylated growth-regulatory genes, including HoxA5, H19, and the p15Ink4b gene, either did not show demethylation or the demethylation observed did not correlate with clinical response. This study also showed that patients with hypermethylated alleles of the p15Ink4b gene were less likely to respond to 5-azadC treatment (102). In another study of MDS patients, demethylation of the p21Waf1/Cip1 gene was readily evident upon 5-azadC treatment and increased with the number of courses of treatment; however, karyotype normalization preceded DNA demethylation, suggesting that it was not dependent on demethylation (64). The lack of correlation between gene-specific DNA demethylation and patient response suggests that reactivation of other genes not studied are mediating 5-azadC's antitumor effects, that cytotoxic effects due to formation of the DNMT-aza-DNA adduct are important even at the lower doses achieved in vivo, or that other mechanisms come into play, such as activation of an immune response. All of these mechanisms are likely to be important, and one may predominate in a particular cell type or treatment condition. It should also be noted that we observed induction of γ-H2AX foci and DNA strand breakage following treatment with 1 μM 5-azadC and below (Fig. 2 and data not shown) and that even a single unrepaired DNA double-strand break is a lethal event. In addition, DNMT-aza-DNA adducts must be forming in vivo in cancer patients at all doses of 5-azadC in order for the observed global and gene-specific demethylation changes to occur. Therefore, the cellular response to 5-azadC-mediated DNA damage is likely an important contributor to its antitumor activity, and further characterization of its cytotoxic mechanism of action, as well as the interplay between CHK1 and DNMT1 in mediating the DNA damage response, will be important to fully understand its clinical effects in cancer patients.
Acknowledgments
This work was supported by NIH grants K22CA084535, R01CA114229, R01CA116028 (K.D.R.), and R01CA102289 (K.D.B.).
We thank Bert Vogelstein (Johns Hopkins) for providing the DNMT knockout HCT116 cells and Yolanda Sanchez (University of Cincinnati) for the FLAG-CHK1 plasmid.
Footnotes
Published ahead of print on 8 November 2007.
REFERENCES
- 1.Adams, K. E., A. L. Medhurst, D. A. Dart, and N. D. Lakin. 2006. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN complex. Oncogene 253894-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ai, L., Q. Tao, S. Zhong, C. R. Fields, W.-J. Kim, M. W. Lee, Y. Cui, K. D. Brown, and K. D. Robertson. 2006. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis 271341-1348. [DOI] [PubMed] [Google Scholar]
- 3.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 27632282-32287. [DOI] [PubMed] [Google Scholar]
- 4.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. [DOI] [PubMed] [Google Scholar]
- 5.Barlow, C., M. Liyanage, P. B. Moens, M. Tarsounas, K. Nagashima, K. Brown, S. Rottinghaus, S. P. Jackson, D. Tagle, T. Ried, and A. Wynshaw-Boris. 1998. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 1254007-4017. [DOI] [PubMed] [Google Scholar]
- 6.Bartek, J., and J. Lukas. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3421-429. [DOI] [PubMed] [Google Scholar]
- 7.Beisler, J. A. 1978. Isolation, characterization, and properties of a labile hydrolysis product of the antitumor nucleoside, 5-azacytidine. J. Med. Chem. 21204-208. [DOI] [PubMed] [Google Scholar]
- 8.Bender, C. M., M. M. Pao, and P. A. Jones. 1998. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 5895-101. [PubMed] [Google Scholar]
- 9.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 166-21. [DOI] [PubMed] [Google Scholar]
- 10.Brown, K. D., and K. D. Robertson. 2007. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. 39289-290. [DOI] [PubMed] [Google Scholar]
- 11.Bulavin, D. V., Y. Higashimoto, I. J. Popoff, W. A. Gaarde, V. Basrur, O. Potapova, E. Appella, and J. A. J. Fornace. 2001. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature 411102-107. [DOI] [PubMed] [Google Scholar]
- 12.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 2811677-1679. [DOI] [PubMed] [Google Scholar]
- 13.Chen, T., S. Hevi, F. Gay, N. Tsujimoto, T. He, B. Zhang, Y. Ueda, and E. Li. 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39391-396. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, J. C., D. J. Weisenberger, F. A. Gonzales, G. Liang, G.-L. Xu, Y.-G. Hu, V. E. Marquez, and P. A. Jones. 2004. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol. Cell. Biol. 241270-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng, J. C., C. B. Yoo, D. J. Weisenberger, J. Chuang, C. Wozniak, G. Liang, V. E. Marquez, S. Greer, T. F. Orntoft, T. Thykjaer, and P. A. Jones. 2004. Preferential response of cancer cells to zebularine. Cancer Cell 6151-158. [DOI] [PubMed] [Google Scholar]
- 16.Christman, J. K. 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 215483-5495. [DOI] [PubMed] [Google Scholar]
- 17.Chuang, L. S.-H., H.-I. Ian, T.-W. Koh, H.-H. Ng, G. Xu, and B. F. L. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex is a target for p21Waf1. Science 2771996-2000. [DOI] [PubMed] [Google Scholar]
- 18.Daskalakis, M., T. T. Nguyen, C. Nguyen, P. Guldberg, G. Kohler, P. Wijermans, P. A. Jones, and M. Lubbert. 2002. Demethylation of a hypermethylated p15/INK4B gene in patients with myelodysplastic syndrome by 5-aza-2′-deoxycytidine (decitibine) treatment. Blood 1002957-2964. [DOI] [PubMed] [Google Scholar]
- 19.de Boer, J., and H. J. Hoeijmakers. 2000. Nucleotide excision repair and human syndromes. Carcinogenesis 21453-460. [DOI] [PubMed] [Google Scholar]
- 20.Demidenko, Z. N., D. Halicka, J. Kunicki, J. A. McCubrey, Z. Darzynkiewicz, and M. V. Blagosklonny. 2005. Selective killing of adriamycin-resistant (G2 checkpoint-deficient and MRP1-expressing) cancer cells by docetaxel. Cancer Res. 654401-4407. [DOI] [PubMed] [Google Scholar]
- 21.Eden, A., F. Gaudet, A. Waghmare, and R. Jaenisch. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300455. [DOI] [PubMed] [Google Scholar]
- 22.Egger, G., S. Jeong, S. G. Escobar, C. C. Cortez, T. W. H. Li, Y. Saito, C. B. Yoo, P. A. Jones, and G. Liang. 2006. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. USA 10314080-14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrlich, M. 2003. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 10917-28. [DOI] [PubMed] [Google Scholar]
- 24.Esteve, P.-O., H. G. Chin, and S. Pradhan. 2005. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc. Natl. Acad. Sci. USA 1021000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falck, J., J. Coates, and S. P. Jackson. 2005. Conserved modes of recruitment of ATM, ATR, and DNA-PKcs to sites of DNA damage. Nature 434605-611. [DOI] [PubMed] [Google Scholar]
- 26.Furuta, T., H. Takemura, Z.-Y. Liao, G. J. Aune, C. Redon, O. A. Sedelnikova, D. R. Pilch, E. P. Rogakou, A. Celeste, H. T. Chen, A. Nussenzweig, M. I. Aladjem, W. M. Bonner, and Y. Pommier. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 27820303-20312. [DOI] [PubMed] [Google Scholar]
- 27.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25115-119. [DOI] [PubMed] [Google Scholar]
- 28.Geiman, T. M., U. T. Sankpal, A. K. Robertson, Y. Chen, M. Mazumdar, J. T. Heale, J. A. Schmiesing, W. Kim, K. Yokomori, Y. Zhao, and K. D. Robertson. 2004. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic Acids Res. 322716-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiman, T. M., U. T. Sankpal, A. K. Robertson, Y. Zhao, Y. Zhao, and K. D. Robertson. 2004. DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochem. Biophys. Res. Commun. 318544-555. [DOI] [PubMed] [Google Scholar]
- 30.Ghoshal, K., J. Datta, S. Majumder, S. Bai, H. Kutay, T. Moitiwala, and S. T. Jacob. 2005. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteosomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 254727-4741. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Gius, D., H. Cui, C. M. Bradbury, J. Cook, D. K. Smart, S. Zhao, L. Young, S. A. Brandenburg, Y. Hu, K. S. Bisht, A. S. Ho, D. Mattson, L. Sun, P. J. Munson, E. Y. Chuang, J. B. Mitchell, and A. P. Feinberg. 2004. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 6361-371. [DOI] [PubMed] [Google Scholar]
- 32.Goll, M. G., and T. H. Bestor. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74481-514. [DOI] [PubMed] [Google Scholar]
- 33.Gollob, J. A., C. J. Sciambi, B. L. Peterson, T. Richmond, M. Thoreson, K. Moran, H. K. Dressman, J. Jelinek, and J.-P. Issa. 2006. Phase I trial of sequential low-dose 5-aza-2′-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clin. Cancer Res. 124619-4627. [DOI] [PubMed] [Google Scholar]
- 34.Gore, S. D., S. Baylin, E. Sugar, H. Carraway, C. B. Miller, M. Carducci, M. Grever, O. Galm, T. Dauses, J. E. Karp, M. A. Rudek, M. Zhao, B. D. Smith, J. Manning, A. Jiemjit, G. Dover, A. Mays, J. Zwiebel, A. Murgo, L.-J. Weng, and J. G. Herman. 2006. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 666361-6369. [DOI] [PubMed] [Google Scholar]
- 35.Guo, G., W. Wang, and A. Bradley. 2004. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature 429891-895. [DOI] [PubMed] [Google Scholar]
- 36.Herman, J. G., and S. B. Baylin. 2003. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 3492042-2054. [DOI] [PubMed] [Google Scholar]
- 37.Ho, C. C., W. Y. Siu, J. P. H. Chow, A. Lau, T. Arooz, H. Y. Tong, I. O. L. Ng, and R. Y. C. Poon. 2005. The relative contribution of CHK1 and CHK2 to adriamycin-induced checkpoint. Exp. Cell Res. 3041-15. [DOI] [PubMed] [Google Scholar]
- 38.Issa, J.-P. 2005. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: dose, duration, and patient selection. Nat. Clin. Pract. Oncol. 2S24-S29. [DOI] [PubMed] [Google Scholar]
- 39.Issa, J.-P., and H. Kantarjian. 2005. Azacitidine. Nat. Rev. Drug Discov. Suppl:S6-S7. [DOI] [PubMed]
- 40.Issa, J.-P. J., G. Garcia-Manero, F. J. Giles, R. Mannari, D. Thomas, S. Faderl, E. Bayar, J. Lyons, C. S. Rosenfeld, J. Cortes, and H. M. Kantarjian. 2004. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 1031635-1640. [DOI] [PubMed] [Google Scholar]
- 41.Jackson-Grusby, L., P. W. Laird, S. N. Magge, B. J. Moeller, and R. Jaenisch. 1997. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl. Acad. Sci. USA 944681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang, K., E. Pereira, M. Maxfield, B. Russell, D. M. Goudelock, and Y. Sanchez. 2003. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J. Biol. Chem. 27825207-25217. [DOI] [PubMed] [Google Scholar]
- 43.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3415-428. [DOI] [PubMed] [Google Scholar]
- 44.Juttermann, R., E. Li, and R. Jaenisch. 1994. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. USA 9111797-11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneda, M., M. Okano, K. Hata, T. Sado, N. Tsujimoto, E. Li, and H. Sasaki. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429900-903. [DOI] [PubMed] [Google Scholar]
- 46.Kantarjian, H. M., and J.-P. J. Issa. 2005. Decitabine dosing schedules. Semin. Hematol. 42S17-S22. [DOI] [PubMed] [Google Scholar]
- 47.Kantarjian, H. M., S. O'Brien, J. Cortes, F. J. Giles, S. Faderi, J.-P. Issa, G. Garcia-Manero, M. B. Rios, J. Shan, M. Andreeff, M. Keating, and M. Talpaz. 2003. Results of decitabine (5-aza-2′deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer 98522-528. [DOI] [PubMed] [Google Scholar]
- 48.Karpf, A. R., and S.-I. Matsui. 2005. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 658635-8639. [DOI] [PubMed] [Google Scholar]
- 49.Karpf, A. R., B. C. Moore, T. O. Ririe, and D. A. Jones. 2001. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol. Pharmacol. 59751-757. [PubMed] [Google Scholar]
- 50.Kim, M., B. N. Trinh, T. I. Long, S. Oghamian, and P. W. Laird. 2004. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 325742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 988461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langer, F., J. Dingemann, H. Kriepe, and U. Lehmann. 2005. Up-regulation of DNA methyltransferases DNMT1, 3A, and 3B in myelodysplastic syndrome. Leuk. Res. 29325-329. [DOI] [PubMed] [Google Scholar]
- 53.Leonhardt, H., A. W. Page, H. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71865-873. [DOI] [PubMed] [Google Scholar]
- 54.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Limonta, M., T. Columbo, G. Damia, C. V. Catapano, V. Conter, M. Gervasoni, G. Masera, V. Liso, G. Speechia, G. Giodici, and M. D'Incalci. 1993. Cytotoxic activity and mechanism of action of 5-aza-2′-deoxycytidine in human CML cells. Leuk. Res. 17977-982. [DOI] [PubMed] [Google Scholar]
- 56.Ling, Y., U. T. Sankpal, A. K. Robertson, J. G. McNally, T. Karpova, and K. D. Robertson. 2004. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 32598-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu, K., Y. F. Wang, C. Cantemir, and M. T. Muller. 2003. Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell. Biol. 232709-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 9710389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes. Dev. 151067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 208602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Momparler, R. L. 2005. Pharmacology of 5-aza-2′-deoxycytidine (decitabine). Semin. Hematol. 42S9-S16. [DOI] [PubMed] [Google Scholar]
- 62.Momparler, R. L., J. Samson, L. F. Momparler, and G. E. Rivard. 1984. Cell cycle effects and cellular pharmacology of 5-aza-2′-deoxycytidine. Cancer Chemother. Pharmacol. 13191-194. [DOI] [PubMed] [Google Scholar]
- 63.Mortusewicz, O., L. Schermelleh, J. Walter, M. C. Cardoso, and H. Leonhardt. 2005. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA 1028905-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mund, C., B. Hackanson, C. Stresemann, M. Lubbert, and F. Lyko. 2005. Characterization of DNA demethylation effects induced by 5-aza-2′-deoxycytidine in patients with myelodysplastic syndrome. Cancer Res. 657086-7090. [DOI] [PubMed] [Google Scholar]
- 65.Myers, J. S., and D. Cortez. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2819346-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nieto, M., E. Samper, M. F. Fraga, G. G. de Buitrago, M. Esteller, and M. Serrano. 2004. The absence of p53 is critical for the induction of apoptosis by 5-aza-2′-deoxycytidine. Oncogene 23735-743. [DOI] [PubMed] [Google Scholar]
- 67.Niida, H., Y. Katsuno, B. Banerjee, M. P. Hande, and M. Nakanishi. 2007. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol. Cell. Biol. 272572-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niida, H., and M. Nakanishi. 2006. DNA damage checkpoints in mammals. Mutagenesis 213-9. [DOI] [PubMed] [Google Scholar]
- 69.Niida, H., S. Tsuge, Y. Katsuno, A. Konishi, N. Takeda, and M. Nakanishi. 2005. Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J. Biol. Chem. 28039246-39252. [DOI] [PubMed] [Google Scholar]
- 70.O'Driscoll, M., and P. A. Jeggo. 2006. The role of double-strand break repair—insights from human genetics. Nature Rev. Genet. 745-54. [DOI] [PubMed] [Google Scholar]
- 71.Oka, M., A. M. Meacham, T. Hamazaki, N. Rodic, L.-J. Chang, and N. Terada. 2005. De novo methyltransferases Dnmt3a and Dnmt3b primarily mediate the cytotoxic effect of 5-aza-2′-deoxycytidine. Oncogene 243091-3099. [DOI] [PubMed] [Google Scholar]
- 72.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99247-257. [DOI] [PubMed] [Google Scholar]
- 73.Parrilla-Castellar, E. R., S. J. H. Arlander, and L. Karnitz. 2004. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair 31009-1014. [DOI] [PubMed] [Google Scholar]
- 74.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10886-895. [DOI] [PubMed] [Google Scholar]
- 75.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12162-169. [DOI] [PubMed] [Google Scholar]
- 76.Rhee, I., K. E. Bachman, B. H. Park, K.-W. Jair, R.-W. C. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416552-556. [DOI] [PubMed] [Google Scholar]
- 77.Robertson, A. K., T. M. Geiman, U. T. Sankpal, G. L. Hager, and K. D. Robertson. 2004. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun. 322110-118. [DOI] [PubMed] [Google Scholar]
- 78.Robertson, K. D. 2005. DNA methylation and human disease. Nat. Rev. Genet. 6597-610. [DOI] [PubMed] [Google Scholar]
- 79.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1, and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25338-342. [DOI] [PubMed] [Google Scholar]
- 80.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a, and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 272291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 2735858-5868. [DOI] [PubMed] [Google Scholar]
- 82.Rothkamm, K., and M. Lobrich. 2003. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc. Natl. Acad. Sci. USA 1005057-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Samlowski, W. E., S. A. Leachman, M. Wade, P. Cassidy, P. Porter-Gill, L. Busby, R. Wheeler, K. Boucher, F. Fitzpatrick, D. A. Jones, and A. R. Karpf. 2005. Evaluation of a 7-day continuous intravenous infusion of decitabine: Inhibition of promoter-specific and global genomic DNA methylation. J. Clin. Oncol. 233897-3905. [DOI] [PubMed] [Google Scholar]
- 84.Santi, D. V., A. Norment, and C. E. Garrett. 1984. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl. Acad. Sci. USA 816993-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarkaria, J. N., R. S. Tibbetts, E. C. Busby, A. P. Kennedy, D. E. Hill, and R. T. Abraham. 1998. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 584375-4382. [PubMed] [Google Scholar]
- 86.Schermelleh, L., F. Spada, H. P. Easwaran, K. Zolghadr, J. B. Margot, M. C. Cardoso, and H. Leonhardt. 2005. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods 10751-756. [DOI] [PubMed] [Google Scholar]
- 87.Schmelz, K., N. Sattler, M. Wagner, M. Lubbert, B. Dorken, and I. Tamm. 2005. Induction of gene expression by 5-aza-2′-deoxycytidine in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) but not epithelial cells by DNA-methylation-dependent and -independent mechanisms. Leukemia 19103-111. [DOI] [PubMed] [Google Scholar]
- 88.Schneider-Stock, R., M. Diab-Assef, A. Rohrbeck, C. Foltzer-Jourdainne, C. Boltze, R. Hartig, P. Schonfeld, A. Roessner, and H. Gali-Muhtasib. 2005. 5-Aza-cytidine is a potent inhibitor of DNA methyltransferase 3a and induces apoptosis in HCT-116 colon cancer cells via Gadd45- and p53-dependent mechanisms. J. Pharmacol. Exp. Ther. 312525-536. [DOI] [PubMed] [Google Scholar]
- 89.Shaham, M., Y. Becker, I. Lerer, and R. Voss. 1983. Increased level of bleomycin-induced chromosome breakage in ataxia telangiectasia skin fibroblasts. Cancer Res. 434244-4247. [PubMed] [Google Scholar]
- 90.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3155-168. [DOI] [PubMed] [Google Scholar]
- 91.Silverman, L. R., E. P. Demakos, B. L. Peterson, A. B. Kornblith, J. C. Holland, R. Odchimar-Reissig, R. M. Stone, D. Nelson, B. L. Powell, C. M. De Castro, J. Ellerton, R. A. Larson, C. A. Schiffer, and J. F. Holland. 2002. Randomized controlled trial of azacytidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J. Clin. Oncol. 202429-2440. [DOI] [PubMed] [Google Scholar]
- 92.Singh, N. P., M. T. McCoy, R. R. Tice, and E. L. Schneider. 1988. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175184-191. [DOI] [PubMed] [Google Scholar]
- 93.Smits, V. A. J., P. M. Reaper, and S. P. Jackson. 2006. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr. Biol. 16150-159. [DOI] [PubMed] [Google Scholar]
- 94.Spada, F., A. Hemmer, D. Kuch, U. Rothbauer, L. Schermelleh, E. Kremmer, T. Carell, G. Langst, and H. Leonhardt. 2007. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176656-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stiff, T., M. O'Driscoll, N. Rief, K. Iwabuchi, M. Lobrich, and P. A. Jeggo. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 642390-2396. [DOI] [PubMed] [Google Scholar]
- 96.Stresemann, C., B. Brueckner, T. Musch, H. Stopper, and F. Lyko. 2006. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 662794-2800. [DOI] [PubMed] [Google Scholar]
- 97.Su, N., Y. X. Pan, M. Zhou, R. C. Harvey, S. P. Hunger, and M. S. Kilberg. 18 May 2007. Correlation between asparaginase sensitivity and asparagine synthetase protein content, but not mRNA, in acute lymphoblastic leukemia cell lines. Pediatr. Blood Cancer. [Epub ahead of print.] doi: 10.1002/pbc.21213. [DOI] [PMC free article] [PubMed]
- 98.Unterberger, A., S. D. Andrews, I. C. G. Weaver, and M. Szyf. 2006. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol. Cell. Biol. 267575-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Gent, D. C., J. H. J. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2196-206. [DOI] [PubMed] [Google Scholar]
- 100.Weisenberger, D. J., M. Velicescu, J. C. Cheng, F. A. Gonzales, G. Liang, and P. A. Jones. 2004. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 262-72. [PubMed] [Google Scholar]
- 101.Xu, B., S.-T. Kim, D.-S. Lim, and M. B. Kastan. 2002. Two molecularly distinct G2/M checkpoints are induced by ionizing radiation. Mol. Cell. Biol. 221049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang, A. S., K. D. Doshi, S.-W. Choi, J. B. Mason, R. K. Mannari, V. Gharybian, R. Luna, A. Rashid, L. Shen, M. R. H. Estecio, H. M. Kantarjian, G. Garcia-Manero, and J.-P. J. Issa. 2006. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 665495-5503. [DOI] [PubMed] [Google Scholar]
- 103.Zhou, L., X. Cheng, B. A. Connolly, M. J. Dickman, P. J. Hurd, and D. P. Hornby. 2002. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 321591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu, W.-G., T. Hileman, Y. Ke, P. Wang, S. Lu, W. Duan, Z. Dai, T. Tong, M. A. Villalona-Calero, C. Plass, and G. A. Otterson. 2004. 5-Aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J. Biol. Chem. 27915161-15166. [DOI] [PubMed] [Google Scholar]
- 105.Ziv, Y., A. Bar-Shira, I. Pecker, P. Russell, T. J. Jorgensen, I. Tsarfati, and Y. Shiloh. 1997. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15159-167. [DOI] [PubMed] [Google Scholar]