Abstract
Parvin-β is a focal adhesion protein downregulated in human breast cancer cells. Loss of Parvin-β contributes to increased integrin-linked kinase activity, cell-matrix adhesion, and invasion through the extracellular matrix in vitro. The effect of ectopic Parvin-β expression on the transcriptional profile of MDA-MB-231 breast cancer cells, which normally do not express Parvin-β, was evaluated. Particular emphasis was placed upon propagating MDA-MB-231 breast cancer cells in three-dimensional culture matrices. Interestingly, Parvin-β reexpression in MDA-MB-231 cells increased the mRNA expression, serine 82 phosphorylation (mediated by CDK9), and activity of the nuclear hormone receptor peroxisome proliferator-activated receptor gamma (PPARγ), and there was a concomitant increase in lipogenic gene expression as a downstream effector of PPARγ. Importantly, Parvin-β suppressed breast cancer growth in vivo, with associated decreased proliferation. These data suggest that Parvin-β might influence breast cancer progression.
The integrin-linked kinase (ILK) is a multifunctional adaptor protein and serine/threonine kinase that binds the cytoplasmic domains of β1- and β3-integrin receptor subunits (18, 19, 68). ILK is a key constituent of the molecular bridge between cell surface integrins and the cortical actin cytoskeleton, namely, focal adhesion complexes (32, 49, 57). In addition to a structural role, integrin-extracellular matrix (ECM) engagement or stimulation with growth factors activates ILK kinase activity in a phosphatidylinositol 3′ kinase-dependent manner, resulting in phosphorylation of downstream substrates, such as AKT Ser473 and glycogen synthase kinase 3β Ser9 (13). ILK also provides integrins with a connection to certain receptor tyrosine kinases via the adaptor proteins PINCH1/2 and NCK2 (64, 72). Overexpression of ILK in cell lines results in anchorage-independent growth, E-cadherin loss, increased invasiveness, and tumorigenicity in nude mice (13, 19). Moreover, increased ILK expression and activity in mouse mammary tumor virus ILK transgenic mice leads to mammary hyperplasias and breast cancers (67). These data suggest that ILK activity must be regulated carefully for effective tumor suppression in vivo and raise the possibility that modulators of ILK function or kinase activity could be deregulated during epithelial oncogenesis.
Parvin-α, -β, and -γ comprise a small family of widely expressed ILK-binding proteins with tandem calponin homology domains (30, 48, 51, 57, 70). The two best-characterized members, Parvin-α and -β, interact directly with the ILK kinase domain in a mutually exclusive manner (73) and modulate both its kinase activity and connections to the actin cytoskeleton (51), although the molecular mechanisms underlying these actions are only now beginning to emerge (43, 66, 73). Less is known about Parvin-γ function.
Data from several studies suggest that Parvin-α and Parvin-β may have divergent actions in the regulation of ILK signaling and cytoskeletal dynamics. For example, Parvin-α was reported to facilitate ILK-mediated phosphorylation of AKT Ser473, with subsequent protection from apoptosis (13, 73), whereas Parvin-β overexpression in HeLa cells promoted apoptosis (73). Parvin-β also inhibited ILK kinase activity and reduced AKT Ser473 and glucogen synthase kinase 3β Ser9 phosphorylation in response to epidermal growth factor stimulation, as previously reported by us (43), consistent with negative regulation of ILK signaling. In contrast to Parvin-α, Parvin-β directly bound to the actin-binding protein α-actinin and was required for proper focal adhesion formation, lamellipodium maturation, and cell spreading (69, 70). In addition to its regulation of ILK, Parvin-β was also found to activate αPIX (ARHGEF6), a GTPase exchange factor for RAC and CDC42 (41, 55). Hence, Parvin-β is also implicated in RAC- and CDC42-mediated rearrangements of the actin cytoskeleton following adhesion to the ECM.
The PARVA gene is located on human chromosome 11p15, whereas PARVB and PARVG are juxtaposed on human chromosome 22q13.31 within an approximately 1-Mb region that undergoes frequent loss of heterozygosity in sporadic breast cancers (7, 27) and mismatch repair-proficient colorectal cancers (6, 27). Mutational analysis of PARVG in sporadic breast and colorectal tumors revealed several germ line polymorphisms but no evidence of somatic mutations (8). However, we demonstrated that Parvin-β mRNA and protein levels are reduced in primary breast tumors compared with adjacent normal breast tissue and also in certain breast cancer cell lines (43). Restoration of Parvin-β expression in metastatic MDA-MB-231 breast cancer cells resulted in reduced colony-forming ability in semisolid medium, increased adhesion to type I collagen, and impaired invasion through ECM, without affecting proliferation (43).
To identify the signaling pathways regulated by Parvin-β in breast cancer cells and gene expression alterations that may mediate the perturbed in vitro behavior of Parvin-β transfectants, we propagated control and Parvin-β-expressing populations on type I collagen-coated plastic (two dimensional [2D]), within a fibrillar type I collagen gel (three-dimensional [3D] collagen), or within basement membrane-containing Matrigel (3D Matrigel) and then subjected them to expression profiling. A compelling rationale for interrogation of breast cancer cells in 3D is that it enables elucidation of the impact of extracellular matrix composition on epithelial biology (24, 28, 54, 66) and evaluation of the influence of physical stiffness on underlying biological processes mediated by Parvin-β (53). We describe herein novel findings through the demonstration that Parvin-β reexpression in MDA-MB-231 cells: (i) increased levels of transcription factors associated with epithelial differentiation (inhibitor of DNA binding 2 [ID2] and Krüppel-like factor 4 [KLF4]) and (ii) increased cyclin-dependent kinase 9 (CDK9)-mediated Ser82 phosphorylation, and transcriptional activity of the nuclear hormone receptor peroxisome proliferator-activated receptor gamma 1 (PPARγ1). There was also a selected increase in expression of PPARγ target genes involved in lipid biosynthesis, lipid droplet formation, and cholesterol efflux in Parvin-β transfectants, particularly in 3D Matrigel culture. This may be due to significantly higher levels of the PPARγ coactivator 1α (PGC-1α) in 3D compared with 2D conditions. Finally, ectopic Parvin-β expression inhibited growth of MDA-MB-231 xenografts in vivo. These novel findings may start to explain how Parvin-β loss may influence breast cancer progression.
MATERIALS AND METHODS
Cell culture in 2D and 3D and drug treatments.
MDA-MB-231 cells stably transfected with pcDNA3.1/Myc-His (clone B) plasmid or the plasmid containing the full-length coding sequence of the long isoform of Parvin-β [Parvin-β(l)] were maintained at 37°C in a 5% CO2 atmosphere on bovine type I collagen-coated plates (Purecol; Inamed, Fremont, CA) in Dulbecco's minimal essential (DME) medium supplemented with 10% fetal bovine serum (FBS; Sigma Chemical Co., St Louis, MO). For embedded growth in Matrigel, six-well plates were coated with 1 ml/well of 80% growth factor-reduced Matrigel (BD Biosciences, Bedford, MA) diluted in serum-free DME medium and allowed to set at 37°C. Cells (3 × 105) were mixed with 1 ml of 40% growth factor-reduced Matrigel (also diluted in DME medium), spread on top of the solidified 80% Matrigel layer, and allowed to set prior to addition of FBS-supplemented DME medium (29). For 3D growth in collagen, cells were combined with 1.5 mg/ml bovine type I collagen as described previously (50) and allowed to set at 37°C before addition of medium. For layered growth in 3D, six-well plates were coated with 80% growth factor-reduced Matrigel and allowed to set and 2 ml of FBS-supplemented medium containing 3 × 105 cells and 2% growth factor-reduced Matrigel was placed on top. Culture medium was replenished every 2 days. Phase-contrast images were generated using a Nikon Eclipse TS100 microscope. Dimethyl sulfoxide was used to reconstitute the CDK9 inhibitor 5,6-dichloro-1-β-ribofuranosyl-benzimidazole (DRB; Sigma) and the PPARγ agonist rosiglitazone (Cayman Chemical Co., Ann Arbor, MI). U0126 (Sigma) and SP600125 (A.G. Scientific Inc., San Diego, CA) were reconstituted as recommended, and each was used at 10 μM.
Gene expression profiling and bioinformatics analysis.
MDA-MB-231 vector control (231_VC) and MDA-MB-231 Parvin-β (231_PARVβ) cells were propagated on type I collagen in monolayer (2D), in type I collagen gels (3D collagen), or in growth factor-reduced Matrigel gels (3D Matrigel) for 7 days. Cells were directly lysed in Trizol reagent (Invitrogen, Carlsbad, CA), and total RNA was isolated according to the manufacturer's instructions for expression profiling. Briefly, 50 ng of total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase (Invitrogen) primed by a poly(T) oligomer that incorporated the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript. The resulting cRNA (200 ng) was used as template for randomly primed cDNA synthesis and a second round of in vitro transcription, which incorporated biotinylated CTP and UTP. The cRNA products were fragmented to 200 bp or less, heated at 99°C for 5 min, and hybridized for 16 h at 45°C to U133Plus 2.0 oligonucleotide microarrays (Affymetrix Inc., Santa Clara, CA). Microarrays were subsequently washed at low (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}]) and high (100 mM morpholineethanesulfonic acid [MES], 0.1 M NaCl) stringency and stained with streptavidin-phycoerythrin. The fluorescence signal was amplified by addition of biotinylated antistreptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to acquire the fluorescent signal after excitation (570 nm).
Affymetrix microarray suite 5.0 (MAS5) was used to quantitate mRNA expression levels; default values provided by Affymetrix were applied to all analysis parameters. The number of probe pairs meeting the default discrimination threshold (tau = 0.015) was used to assign a call of absent, present, or marginal for each assayed gene, and a P value was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for nonspecific signal by subtracting the mismatch probe value) was calculated using the one-step Tukey biweight estimate. Global scaling was applied to allow comparison of gene signals across multiple microarrays. All signal values from one microarray were then multiplied by the appropriate scaling factor. The MAS5 algorithm was used to determine gene expression ratios (flagged as I [increased], D [decreased], MI [marginally increased], MD [marginally decreased], or NC [no change]) and P values for the change. Ratios of mRNA expression levels between 231_VC and 231_PARVβ cells were calculated by determining the inverse log2 of the signal log ratio for each differentially expressed gene. Array results from 2D and 3D experiments were compared, and Venn diagrams were generated using Genespring GX software (Agilent Technologies, Palo Alto, CA). Comparisons of the 3D microarray results to the myoepithelial and luminal epithelial gene expression signatures derived by Grigoriadis and colleagues (16) were performed with Genespring GX. Pathway analysis and functional grouping of genes were carried out using Ingenuity Systems (Redwood City, CA) software.
RT-PCR.
Analysis of mRNA expression by reverse transcription-PCR (RT-PCR) was performed as previously described (47). For oligonucleotide sequences of the PCR primer pairs used, see the supplemental materials and methods. Normal human colon was obtained from the Cooperative Human Tissue Network at the hospital of the University of Pennsylvania, and total RNA was isolated from the purified epithelium as described previously (27).
qPCR.
mRNA expression levels were determined by SYBR green (Applied Biosystems, Warrington, United Kingdom) quantitative real-time RT-PCRs (qPCRs) with 231_VC and 231_PARVβ cells as described previously (43). Optimal PCR conditions were determined by performing primer matrix reactions and generating standard curves for each primer pair used. Assays for both the gene of interest and the β-actin internal control were performed in triplicate in an ABI PRISM 7000 sequence detection system, and results were analyzed as previously described (43). Two separate RNA preparations from each growth condition were evaluated and data combined. Thus, means and standard deviations were calculated from a total of six independent PCRs on cDNA synthesized from two separate total RNA preparations. For the primer pairs used see the supplementary materials and methods. TaqMan gene expression assays were used to determine expression levels of total PPARγ, PPARγ transcript variant 2 (PPARγ2), PGC-1α, and ABCA1 and were in accordance with the manufacturer's recommendations (Applied Biosystems, Foster City, CA). Student's t test for means was applied for evaluation of expression level differences using SPSS (version 12) statistical software (SPSS Inc., Chicago, IL). Frozen primary human breast tumors were obtained from the Cooperative Human Tissue Network at the hospital of the University of Pennsylvania, and cDNA was synthesized as previously described (27).
Western blot analysis.
Cells were isolated from Matrigel using cell recovery solution (BD Biosciences) according to the manufacturer's instructions. Whole-cell, nuclear, and cytoplasmic extracts were prepared as described previously (43). For analysis of PPARγ expression, 30 to 60 μg of whole-cell lysate (15 to 20 μg of nuclear extract) was used, whereas 15 to 25 μg of whole-cell lysate was used for analysis of other proteins. Polyvinylidene difluoride membranes were incubated with primary antibody overnight (4°C), followed by the appropriate horseradish peroxidase-conjugated secondary antibody (Amersham, GE Healthcare UK, Little Chalfont, United Kingdom) for 1 h at room temperature. Either β-actin (Sigma), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Ambion, Austin, TX), or lamin A/C (Cell Signaling Technologies) was used as a loading control. The primary antibodies used were as follows: PPARγ, mouse monoclonal antibody, clone E-8 (Santa Cruz Biotechnology, Santa Cruz, CA); phospho-PPARγ (pPPARγ; Ser82), rabbit polyclonal antibody (Upstate, Charlottesville, VA); green fluorescent protein, rabbit polyclonal antibody (Clontech); CDK9, rabbit polyclonal antibody (Santa Cruz Biotechnology); CDK7, rabbit polyclonal antibody (Santa Cruz Biotechnology); total mitogen-activated protein kinase (MAPK), rabbit polyclonal antibody (Upstate); and phospho-MAPK (pMAPK; T202, Y204), rabbit polyclonal antibody; phospho-Jun N-terminal protein kinase (pJNK; T183, Y185), rabbit polyclonal antibody; total JNK, rabbit polyclonal antibody; phospho-c-JUN (T183, Y185), rabbit polyclonal antibody, total c-JUN, rabbit polyclonal antibody; lamin A/C, rabbit polyclonal antibody; and hemagglutinin, mouse monoclonal antibody, clone 6E2 (all from Cell Signaling Technology Inc., Danvers, MA). Antibodies recognizing Parvin-α, Parvin-β, Parvin-γ, KLF4, and PGC-1α are described elsewhere (35, 43, 71).
Dual luciferase assays.
Dual luciferase assays (Promega, Madison, WI) were conducted as previously described (26). Briefly, cells were seeded into 24-well plates, and each well was transiently transfected the following morning with the PPARγ response element (PPRE) firefly luciferase reporter vector (1 μg) and either 2 ng of the pRL-CMV Renilla control vector or 20 ng of the pRL-SV40 Renilla control vector using 2 μl/well of FuGene6 reagent (Roche). The respective firefly and Renilla luciferase activities of cell lysates (20 to 40 μl) were acquired sequentially using an Orion microplate luminometer (Berthold Detection Systems, Pforzheim, Germany).
Transient transfection of CDK9 and CDK7 siRNA.
Cells were seeded into Vitrogen-coated six-well plates and transiently transfected at 50% to 70% confluence with Lipofectamine 2000 reagent (Invitrogen) mixed with prevalidated small interfering RNAs (siRNAs) to either human CDK9 or CDK7 or scrambled negative control siRNA no. 5 (all from Ambion) at a final concentration of 40 nM. Cells were propagated for 2 to 3 days before harvesting without a medium change.
Generation of firefly luciferase- and tdTomato-expressing MDA-MB-231 derivatives.
The pBabe-puro_luciferase expression vector (a generous gift from Wafik El-Deiry) was used to produce retroviral supernatants as described previously (14). Supernatant (400 μl) was used to transduce MDA-MB-231 derivatives by the spin infection method as previously described. Six separate populations of luciferase-transduced 231_VC and 231_PARVβ cells were then assayed for firefly luciferase activity in duplicate, and activity was normalized to total protein levels. One population from each cell line was selected for expansion in puromycin (5 μg/ml). tdTomato cDNA in pRSET-B (a generous gift from Roger Tsien) was PCR amplified and subcloned (BamHI/EcoRI) into the pBabe-BlaS expression vector to produce pBabe-BlaS_tdTomato (45, 58). Luciferase-transduced 231_VC and 231_PARVβ cells were transduced further with pBabe-BlaS_tdTomato as described above and selected in Blasticidin S (20 μg/ml; Sigma Chemical Co.). Percentages of tdTomato expression and fluorescence intensity levels were measured by fluorescence microscopy and fluorometry, respectively. Fluorescence intensity (excitation filter λ, 530/25 nm; emission filter λ, 580/35 nm; Synergy HT; Biotek Instruments Inc., Winooski, VT) was normalized to protein levels and expressed as mean intensity/μg protein ± standard deviation (n = 5).
Subcutaneous injections and in vivo optical imaging.
MDA-MB-231 cells (1 × 106) were resuspended in serum-free DME medium containing 30% Matrigel and injected subcutaneously into the flanks (four injection sites per mouse) of anesthetized nonlethally irradiated immunodeficient mice (NCRNU-M; Taconic, Hudson, NY) using a 27.5-gauge needle. Mice were maintained with the approval of and according to the guidelines produced by the University of Pennsylvania Institutional Animal Care and Use and Committee and Small Animal Imaging Facility. In vivo bioluminescent imaging (BLI) was performed using an IVIS 100 series optical imaging system (Xenogen Corporation, Hopkinton, MA) as described previously (14). Briefly, following intraperitoneal injection of d-luciferin, three or four anesthetized mice were placed 25 cm from the camera, and luminescence was measured every 1 min (30-s exposure, medium bin) until the maximum luminescence intensity was reached (approximately 15 min postinjection). For quantitation, counts were converted to flux (photons of light/s) keeping the same-size field for each time point. A mixed-effects generalized linear regression model using a random effect for tumor (that is, accounting for the repeated measurements of each tumor) was used to compare 231_VC and 231_PARVβ growth rates over time (SAS version 9.1; SAS Institute, Cary, NC). A P value of <0.05 was taken to represent a significant difference in growth rates between the two cell lines. In vivo fluorescence images of tdTomato were acquired using Maestro instrumentation (Cambridge Research and Instrumentation, Woburn, MA) with DsRed filter settings. Anesthetized mice were scanned (550 to 700 nm) using a 5-nm step, and background fluorescence was eliminated by employing the spectral unmixing capability.
Immunohistochemistry.
Tumors were fixed in 4% paraformaldehyde for 8 h. Sections (5 μm) were subjected to antigen retrieval in a citrate buffer. Immunohistochemistry was performed as previously described (25). The primary antibodies used were antipancytokeratin (Sigma Chemical Co.), antivimentin (Novus Biologicals, Littleton, CO), anti-Ki-67 (Vector Laboratories, Burlingame, CA), anti-cyclin D1 (Lab Vision, Fremont, CA), and anti-CD34 (BD Biosciences). Slides were imaged using a Nikon TE2000 Eclipse microscope with a QiCam (Q Imaging, Surrey, Canada) camera and IPLab imaging software (BD Bioscience Bioimaging, Rockville, MD). The Ki-67 proliferation index was obtained by choosing five random fields from each of the two 231_VC and 231_PARVb tumors stained (total n = 10) and calculating (following blinded counting) the average number of Ki-67-positive nuclei.
Microarray data accession number.
Microarray data were deposited in the Gene Expression Omnibus database with accession no. GSE9747.
RESULTS
Gene expression profiling of 231_VC and 231_PARVβ cells.
Different growth conditions were exploited for comparison of gene expression levels between 231_VC and 231_PARVβ cells. The effects of varying ECM composition and physical stiffness/tension (3D type I collagen gel versus 3D basement membrane gel [3D Matrigel]) as well as varying external force/stiffness (2D type I collagen versus 3D collagen gel) on Parvin-β-regulated gene expression were studied by gene chip analysis (see Fig. S1 in the supplemental material). Our particular focus was to reveal those changes in 3D culture that might help unravel Parvin-β's biological functions and then use this as a platform for in vivo studies.
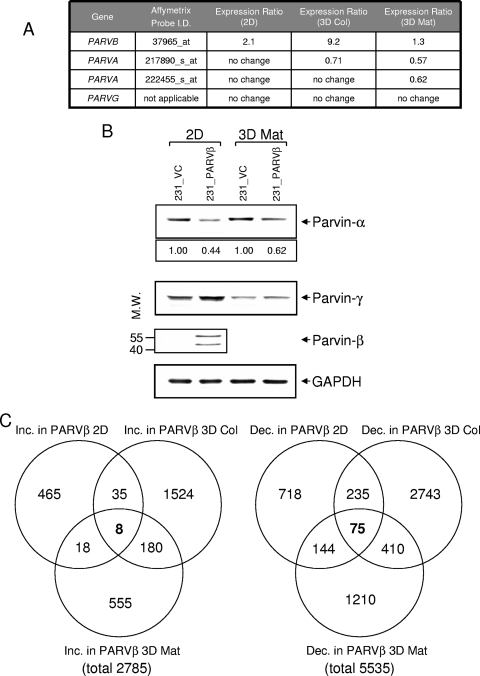
As expected, Parvin-β mRNA itself was higher in 231_PARVβ cells than control cells in each of the three growth conditions (Fig. 1A), and this was corroborated by Western blotting in 2D (Fig. 1B). Interestingly, endogenous Parvin-α mRNA levels were decreased in 231_PARVβ cells cultured in both 3D conditions (Fig. 1B), whereas Parvin-α protein levels were reduced by approximately 50% in both 2D and 3D Matrigel growth conditions (Fig. 1B). Parvin-γ mRNA and protein levels were not changed. The number of genes with differential expression between the two cell lines cultured under each of the three conditions is summarized using Venn diagrams (Fig. 1C). From a global perspective, more genes were repressed in 231_PARVβ cells (5,535) than were activated (2,785) (see Tables S1 and S2 in the supplemental material). This provided the basis for interrogation of specific families of genes that might relate to Parvin-β mediated function(s).
FIG. 1.
Expression profiling of MDA-MB-231 stable transfectants cultured in 2D and 3D conditions (A) Expression ratios of Parvin-α and Parvin-β mRNAs in 231_PARVβ cells versus 231_VC cells grown on type I collagen-coated plastic (2D), in a 3D fibrillar type I collagen gel (3D Col), or embedded in 40% Matrigel (3D Mat). (B) Western blot analysis of Parvin-α, Parvin-γ, and Parvin-β expression in 231_VC and 231_PARVβ cells grown in 2D or 3D Mat conditions. Numbers indicate relative Parvin-α expression following normalization to GAPDH (bottom panel). The lower Parvin-β band may result from translation initiation from a downstream in-frame ATG. Molecular weights (M.W.) are in thousands. (C) Venn diagrams depicting numbers of genes significantly increased (Inc.) or decreased (Dec.) in 231_PARVβ cells compared with 231_VC cells following gene expression profiling using Affymetrix U133_Plus2 gene chips (see Materials and Methods). The numbers of genes increased or decreased in each growth condition are shown. For a complete description of the genes altered, see Table S1 in the supplemental material. The total numbers of genes significantly upregulated and downregulated in 231_PARVβ cells were 2,785 and 5,535, respectively.
Deregulation of genes involved in epithelial differentiation and lipid metabolism in 231_PARVβ cells.
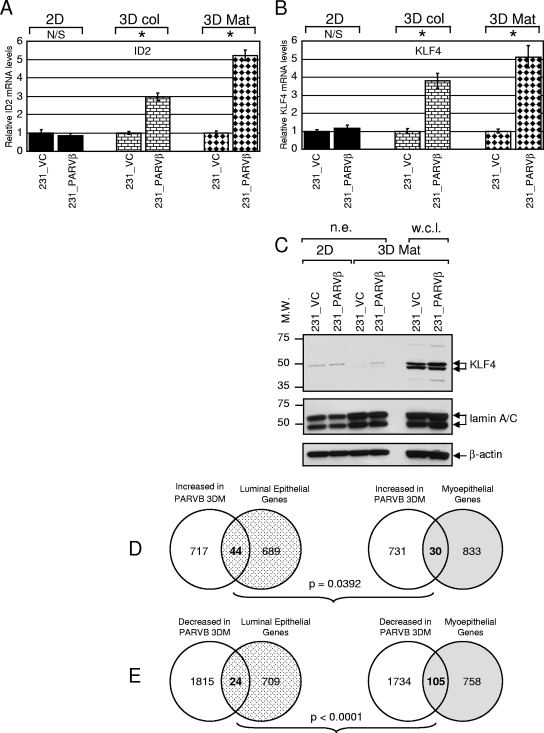
Pathway analysis and functional grouping of array results (see Materials and Methods) revealed that genes relating to epithelial cell differentiation and lipid and steroid metabolism were particularly affected in 231_PARVβ cells in 3D compared to control cells (Table 1; see Table S1 in the supplemental material). mRNA expression levels were validated by qPCR in two separate sets of 231_VC/231_PARVβ cDNA pairs prepared from two independent 2D, 3D collagen, and 3D Matrigel cultures, and the results were averaged. ID2 and KLF4, two transcription factors in control of epithelial differentiation (12, 23, 42, 46, 60), were approximately fivefold upregulated in 3D Matrigel cultures and three- to fourfold upregulated in 3D collagen cultures of 231_PARVβ cells (Fig. 2A and B). KLF4 protein levels were also higher in 231_PARVβ cells grown in 3D Matrigel (Fig. 2C). To assess potential Parvin-β-mediated effects on differentiation in MDA-MB-231 cells in a global fashion, we directly compared our array results from 3D Matrigel culture conditions with gene expression signatures from purified human luminal (differentiated) mammary epithelial cells and myoepithelial (undifferentiated) cells (16), obtained using the same approach as the present study (Affymetrix U133_Plus_2 gene chip). A four-way binary comparison (that is, genes increased or decreased in 231_PARVβ cells versus luminal epithelium- or myoepithelium-enriched genes) revealed that a significantly higher number of luminal epithelium-enriched genes than myoepithelium-enriched genes were upregulated in the Parvin-β transfectants propagated in 3D Matrigel (Fig. 2D; P = 0.04), whereas significantly more myoepithelium-enriched genes than luminal genes were downregulated in the 231_PARVβ transfectants (Fig. 2E; P < 0.01). Together, these expression data suggest that the presence of Parvin-β may drive breast cancer cells toward an earlier differentiated state, although this was not manifest in obvious morphological changes that are associated with terminal differentiation (see Fig. S1 in the supplemental material). This may place the changes in ID2 and KLF4 in context.
TABLE 1.
Altered gene expression in 231_PARVβ cells following 2D or 3D culturea
| Category | Gene | Protein encoded | Affymetrix probe ID | Ratio for:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 2D
|
3D col
|
3D Mat
|
|||||||
| Array | qPCR | Array | qPCR | Array | qPCR | ||||
| Transcription factors | ID2 | Inhibitor of DNA binding 2 | 201565_s_at | NC | 0.9 | NC | 3.0 | 8.6 | 5.2 |
| KLF4 | Krüppel-like factor 4 | 221841_s_at | 0.8 | 1.2 | NC | 3.8 | 2.3 | 5.1 | |
| Nuclear hormone receptors | PPARG | Peroxisome proliferator-activated receptor gamma (total) | 208510_s_at | 2.1 | 1.6 | NC | 2.0 | 4.0 | 3.5 |
| PPARG | Peroxisome proliferator-activated receptor gamma 2 | NA | NA | 1.6 | NA | 2.9 | NA | 1.8 | |
| Lipid metabolism | SLC25A1 | Citrate transporter | 210010_s_at | NC | ND | 1.7 | ND | 1.9 | ND |
| ACLY | ATP-citrate lyase | 210337_s_at | NC | ND | 0.8 | ND | 1.4 | ND | |
| FASN | Fatty acid synthase | 212218_s_at | 0.8 | 0.7 | 1.2 | 1.3 | 2.0 | 4.7 | |
| SCD | Stearoyl-CoA desaturase | 200831_s_at | NC | 0.9 | 0.4 | 1.2 | 1.7 | 1.6 | |
| SCD | Stearoyl-CoA desaturase | 200832_s_at | 0.5 | NC | 1.5 | ||||
| SCD | Stearoyl-CoA desaturase | 223839_s_at | 0.5 | 1.9 | 0.7 | ||||
| SCD | Stearoyl-CoA desaturase | 211162_x_at | 0.3 | 0.3 | 1.6 | ||||
| ECH1 | Enoyl-CoA hydratase | 200789_at | NC | ND | NC | ND | 3.2 | ND | |
| Lipid droplet formation | ADFP | Adipophilin/adipocyte differentiation-related protein | 209122_at | NC | 1.5 | NC | 2.7 | 2.5 | 3.5 |
| CAV2 | Caveolin 2 | 203324_s_at | 1.2 | ND | NC | ND | 1.3 | ND | |
| VIM | Vimentin | 201426_s_at | NC | ND | NC | ND | 1.3 | ND | |
| Cholesterol efflux | NR1H3 | Liver X receptor alpha | 203920_at | NC | ND | NC | ND | 2.6 | ND |
| ABCA1 | ATP-binding cassette, subfamily A member 1 (ABC1 subfamily) | 203505_at | NC | 0.4 | NC | 15.5 | 2.1 | 5.4 | |
| ABCA1 | ATP-binding cassette, subfamily A member 1 (ABC1 subfamily) | 203504_s_at | 0.5 | NC | NC | ||||
| SNTB1 | Syntrophin, beta 1 | 208608_s_at | NC | ND | NC | ND | 16.0 | NC | |
| Steroid metabolism | AKR1C1 | Aldo-keto reductase family 1, member C1 | 204151_x_at | 0.4 | 0.4 | NC | 9.2 | 3.0 | 7.4 |
| AKR1C1 | Aldo-keto reductase family 1, member C1 | 216594_x_at | 0.5 | NC | 2.1 | ||||
| AKR1C2 | Aldo-keto reductase family 1, member C2 | 209699_x_at | NC | 0.9 | NC | 9.5 | 3.1 | 6.4 | |
| AKR1C2 | Aldo-keto reductase family 1, member C2 | 211653_x_at | NC | NC | 2.5 | ||||
| AKR1B1 | Aldo-keto reductase family 1, member B1 | 201272_at | NC | ND | 2.1 | ND | 1.5 | ND | |
| AKR1D1 | Aldo-keto reductase family 1, member D1 | 207102_at | NC | ND | 8.0 | ND | NC | ND | |
Cells were cultured in either 2D on type I collagen (2D), 3D embedded in type I collagen (3D col), or 3D embedded in Matrigel (3D Mat), followed by transcriptomic analysis using Affymetrix U133_Plus2 gene chips. Genes are listed according to functional category. The gene expression ratios (231_PARVβ/231_VC) from the microarray (array) and from qPCR validation are shown. Genes listed in boldface were validated by qPCR. ID, identification; NA, not applicable; NC, no change; ND, not done.
FIG. 2.
Parvin-β induces transcription factors linked to epithelial differentiation. Levels of ID2 mRNA (A) and KLF4 mRNA (B) expression were evaluated by SYBR green qPCR analysis with 231_VC and 231_PARVβ cells cultured in 2D, 3D collagen (3D col), or 3D Matrigel (3D Mat). Graphs show the mean relative gene expression levels and standard deviations, and expression in 231_VC cells was set to 1 in each condition. N/S, not significant; *, P < 0.05. (C) Western blot analysis of KLF4 expression in 231_VC and 231_PARVβ cells. Both nuclear extracts (n.e.; 15 μg, 2D and 3D Mat conditions) and whole-cell lysates (w.c.l.; 80 μg, 3D Mat only) were examined (top). The membrane was then reprobed for lamin A/C (middle), and β-actin (bottom), as loading controls. Molecular weights (M.W.) are in thousands. (D and E) Comparison of gene expression alterations in 231_PARVβ cells cultured in 3D with gene expression signatures of purified human luminal and myoepithelial mammary epithelial cells (16) (D) Significantly more luminal epithelial transcripts (n = 44) were upregulated in 231_PARVβ 3D cultures than were myoepithelial transcripts (n = 30; P = 0.0392). (E) Significantly more myoepithelial transcripts were downregulated (n = 105) in 231_PARVβ 3D cultures than were luminal epithelial transcripts (n = 24; P < 0.0001).
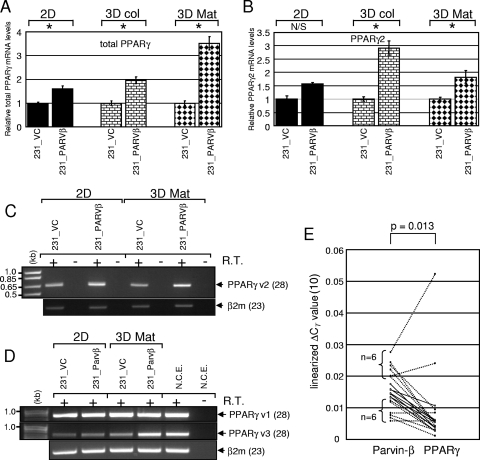
Interestingly, total levels of PPARγ mRNA, encoding a pleiotrophic nuclear hormone receptor implicated in breast cancer progression (9, 31, 34, 65), were increased in both 3D cultures (2.0-fold for 3D collagen and 3.5-fold for 3D Matrigel) of 231_PARVβ cells, and the increases were greater than that for 2D cultures (1.6-fold) (Fig. 3A; Table 1). The human PPARG gene contains three major promoters that together generate up to four discrete transcript variants, which may ultimately produce two protein isoforms (see Fig. S2 and S3 in the supplemental material), namely, epithelium-enriched PPARγ1 (synthesized from mRNAs transcribed from promoters 1 and 3) and adipocyte-enriched PPARγ2 (generated from promoter 2) (9, 34, 65). Thus, expression of each PPARγ transcript variant was investigated individually. Variant 2 was expressed at higher levels in 231_PARVβ cells cultured in both 2D and 3D Matrigel (Fig. 3B and C); however, its relative expression in MDA-MB-231 cells was significantly lower than that of variants 1 and 3 (data not shown), as expected (9). Whereas expression of PPARγ variant 1 appeared unchanged between control and 231_PARVβ cells, importantly, variant 3 mRNA was augmented in 231_PARVβ cells grown in 3D Matrigel (Fig. 3D). Of note, differential expression of variant 3 was not evident in 2D cultures (Fig. 3D).
FIG. 3.
Parvin-β induces PPARγ mRNA expression. Levels of total PPARγ mRNA (A) and PPARγ2 mRNA (B) expression in 231_VC and 231_PARVβ cells cultured in 2D, 3D collagen (3D col), or 3D Matrigel (3D Mat) were evaluated by TaqMan qPCR analysis. Graphs show the mean relative gene expression levels (normalized to β-actin mRNA) and standard deviations, and expression in 231_VC cells was set to 1 in each condition. N/S, not significant; *, P < 0.05. (C) Semiquantitative RT-PCR analysis of PPARγ2 mRNA expression (28 cycles) in 231_VC and 231_PARVβ cells grown in 2D or 3D Matrigel. β2-Microglobulin expression (23 cycles) was measured as an internal control. (D) Semiquantitative RT-PCR analysis (28 cycles) of PPARγ variant 1 (top) and variant 3 (middle) levels in cells cultured in either 2D or 3D Matrigel conditions. RNA extracted from purified normal human colonic epithelial cells (N.C.E.) was used as a positive control for both transcripts. β2-Microglobulin expression (23 cycles) was measured as an internal control (bottom). (E) Relationship between Parvin-β and PPARγ mRNA expression levels in primary breast tumors. Parvin-β and total PPARγ mRNA levels in 19 primary breast ductal adenocarcinomas were determined by SYBR green and TaqMan qPCR, respectively, and normalized to β-actin mRNA levels. The linearized ΔCT values for Parvin-β and PPARγ are represented on the y axis. Dotted lines are used to link the Parvin-β and PPARγ data points of the six specimens with the highest Parvin-β mRNA expression and the six specimens with the lowest. Solid lines are used for the seven remaining tumors. Significance of the correlation (P = 0.013) was calculated by simple linear regression (r = 0.56).
Since Parvin-β reexpression in breast cancer cells resulted in accumulation of PPARγ mRNA, we compared Parvin-β and PPARγ mRNA expression levels in sporadic primary breast tumors by qPCR. Analysis of 19 primary ductal adenocarcinomas revealed a statistically significant correlation (P = 0.013; correlation coefficient [r] = 0.56) between Parvin-β mRNA and total PPARγ mRNA levels (Fig. 3E).
Increased PPARγ Ser82 phosphorylation, transcriptional activity, and lipogenic gene expression in 231_PARVβ cells.
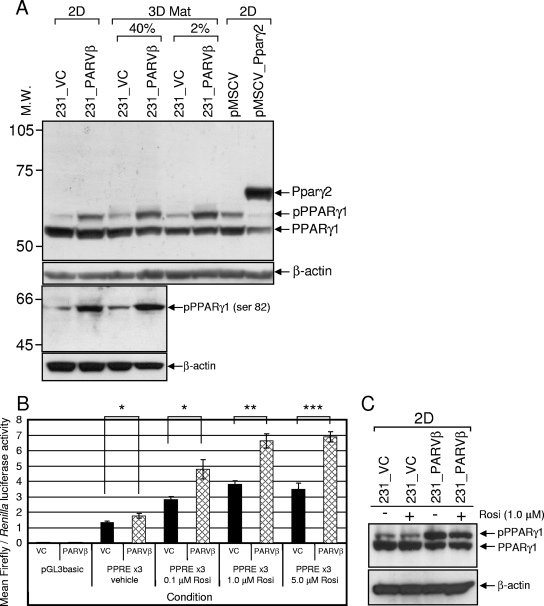
Since PPARγ mRNA was upregulated in 231_PARVβ cells, PPARγ protein expression was also assessed. Since posttranslational modification of PPARγ had been shown to regulate its function (1, 4, 5, 21), we also examined PPARγ phosphorylation and transcriptional activity. As expected, only the PPARγ1 isoform was detected in MDA-MB-231 cells (Fig. 4A), and levels of unphosphorylated PPARγ1 in 231_VC and 231_PARVβ cells did not appear appreciably different. However, the abundance of a slower-migrating pPPARγ1 species was increased in 231_PARVβ cells (22), regardless of the culture conditions (Fig. 4A and data not shown). Serine 82 of PPARγ1 (Ser112 in PPARγ2) was identified as the target residue for enhanced phosphorylation in 231_PARVβ cells through use of a pPPARγ (Ser82/112)-specific antibody (Fig. 4A, bottom). The transcriptional activity of PPARγ was evaluated using a PPRE-driven reporter gene assay. Basal PPARγ transcriptional activity was increased in 231_PARVβ cells compared to control cells (Fig. 4B). Incubation with the synthetic PPARγ agonist rosiglitazone increased reporter gene activity in both cell lines in a dose-dependent manner (33); however, rosiglitazone-stimulated reporter activity in 231_PARVβ cells was approximately 1.7-fold higher than in control cells at each concentration used (Fig. 4B). Increased PPARγ1 activity in rosiglitazone-stimulated 231_PARVβ cells was not due to modulation of pPPARγ1 abundance or phosphorylation by rosiglitazone (Fig. 4C).
FIG. 4.
Expression and activity of PPARγ protein in 231_VC and 231_PARVβ cells. (A, top two panels) Western blot analysis of PPARγ1 and pPPARγ1 expression in 231_VC and 231_PARVβ cells cultured in 2D or 3D Matrigel (3D Mat; embedded in 40% growth factor-reduced Matrigel [40%] or layered on 80% Matrigel in medium containing 2% Matrigel [2%]) using anti-total PPARγ (E-8) monoclonal antibody. MDA-MB-231 cells transduced with pMSCV encoding murine Pparγ2 or pMSCV alone served as a control to distinguish pPPARγ1 from PPARγ2. β-Actin was used as a loading control. Bottom two panels) A pPPARγ (Ser82/112)-specific antibody was used in Western blot analysis of whole-cell lysates from 2D or 3D Matrigel cultures and shows higher pPPARγ in 231_PARVβ cells than in 231_VC cells. β-Actin was used as a loading control. Molecular weights (M.W.) are in thousands. (B) Transcriptional activity of PPARγ1 in 231_VC (VC) and 231_PARVβ (PARVβ) cells cultured in 2D. Cells were cotransfected with the PPREx3 vector and the Renilla control vector as described in Materials and Methods. Cells were then treated with dimethyl sulfoxide alone (vehicle) or increasing concentrations of the selective PPARγ agonist rosiglitazone (Rosi). The empty firefly luciferase vector (pGL3basic) was used as a negative control. The graph shows the mean normalized luciferase activities and standard errors from four independent transfections. The experiment was conducted three times. *, P < 0.05; **, P < 0.005; ***, P < 0.001. (C) Rosiglitazone treatment did not augment PPARγ1 phosphorylation in 231_VC or 231_PARVβ cells grown in either 2D or 3D Matrigel (data not shown) conditions. Cells were treated with rosiglitazone (1.0 μM) for 48 h, followed by Western blot analysis of whole-cell lysates. β-Actin was used as a loading control (bottom).
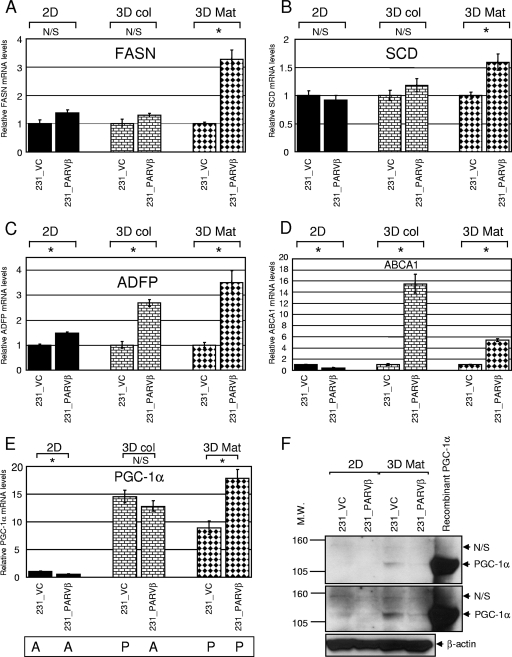
PPARγ activates a transcriptional cascade promoting both lipogenesis and adipogenesis (9, 37, 40, 63). Since PPARγ-dependent transcriptional activity was higher in 231_PARVβ cells compared to control cells, as measured by a reporter gene assay (Fig. 4B), the array results were interrogated for evidence of differential expression of potential PPARγ targets. Importantly, five genes encoding enzymes with direct roles in the lipid biosynthetic pathway (citrate transporter gene [SLC25A1], ATP-citrate lyase gene [ACLY], fatty acid synthase gene [FASN], stearoyl-coenzyme A [CoA] desaturase gene [SCD], and enoyl-CoA hydratase 1 gene [ECH1]) were found to be coordinately upregulated in 231_PARVβ cells, but only when cultured in 3D Matrigel and to a lesser extent in 3D collagen (Table 1). Increased FASN and SCD mRNA levels in 231_PARVβ cells were confirmed by qPCR (Fig. 5A and B; Table 1). In addition, mRNA levels of adipocyte differentiation-related protein, vimentin, and caveolin 2, each of which plays a structural role in intracellular lipid droplet formation (20, 38, 42), were also upregulated after 3D culture in Matrigel (Fig. 5C; Table 1). Finally, PPARγ was shown to orchestrate activation of a cholesterol efflux pathway in macrophage foam cells through PPARγ-mediated upregulation of liver X receptor-α (LXRα), which in turn induced expression of the cholesterol transporter ABCA1 (10, 44). Both LXRα and ABCA1 mRNAs were increased in 231_PARVβ cells, and again only when cultured in the 3D conditions (Fig. 5D; Table 1). Taken together, these data indicate that only the 3D Matrigel culture system was permissive for consistent and robust activation of putative or known key PPARγ-regulated genes and cellular processes.
FIG. 5.
PPARγ-regulated pathways involved in lipogenesis, lipid droplet formation, and cholesterol efflux are induced in 231_PARVβ cells cultured in 3D conditions only. Levels of fatty acid synthase (FASN) mRNA (A), stearoyl-CoA desaturase (SCD) mRNA (B), adipocyte differentiation-related protein (ADFP) mRNA (C), and cholesterol transporter (ABCA1) mRNA (D) in 231_VC and 231_PARVβ cells cultured in 2D, 3D collagen (3D col), and 3D Matrigel (3D Mat) were evaluated by TaqMan qPCR analysis. (E) Evaluation of PGC-1α mRNA expression in 231_VC and 231_PARVβ cells by TaqMan qPCR. Graphs show the mean relative gene expression levels (normalized to β-actin mRNA) and standard deviations, and expression in 231_VC cells was set to 1 in each condition. N/S, not significant; *, P < 0.05. “A” (absent) and “P” (present) refer to the call tags given (PGC-1α probe set 219195_at) by the Affymetrix MAS5.0 algorithm (see Materials and Methods). (F) Western blot analysis of PGC-1α protein expression in nuclear extracts (25 μg) of 231_VC and 231_PARVβ cells grown in 2D and 3D Mat conditions. Lysate from 293T cells engineered to overexpress PGC-1α was used as a positive control (35). Two different exposures are shown, and β-actin was used as a loading control (bottom). Molecular weights (M.W.) are in thousands.
We next postulated that there may be differences in expression of PPARγ transcriptional coactivators or corepressors between the 2D and 3D systems. Interrogation of the microarray data suggested that the PPARγ coactivator PGC-1α was induced when MDA-MB-231 cells were cultured in either 3D growth condition (data not shown). Indeed, qPCR analysis confirmed that PGC-1α mRNA levels were approximately 10- to 20-fold upregulated in both 231_VC and 231_PARVβ cells grown in either 3D system (type I collagen or Matrigel) compared to the 2D system (Fig. 5E). PGC-1α protein levels were also shown to be induced by 3D Matrigel culture (Fig. 5F).
Assessment of the role of CDK9 in Parvin-β-induced phosphorylation of PPARγ.
Considering that hyperphosphorylation of PPARγ1 was observed in 231_PARVβ cells and that MAPK and JNK had been demonstrated to phosphorylate PPARγ1/2 on Ser82/112 (1, 4, 5, 21), MAPK and JNK pathway activation was evaluated as natural candidates initially for mediating the phosphorylation. Both pMAPK and pJNK levels were suppressed in cells cultured in 3D compared with 2D culture, and no differences in pMAPK and pJNK levels between control and 231_PARVβ cells were found, suggesting that MAPK and/or JNK may not be responsible for the increased pPPARγ1 in 231_PARVβ cells (see Fig. S5 in the supplemental material). Furthermore, pharmacologically mediated inhibition of MAPK or JNK was unrevealing (see Fig. S4 in the supplemental material). These data indicated that a kinase(s) other than MAPK or JNK was responsible for phosphorylating PPARγ in the 231_PARVβ cells.
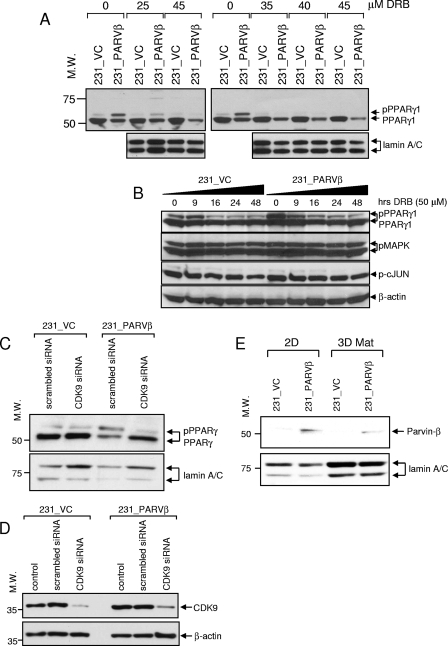
When bound to cyclin T partners, CDK9 comprises part of the basal machinery for RNA polymerase II-directed gene transcription (36). Mouse Cdk9 was shown recently to phosphorylate Pparγ2 on Ser112 during adipogenic differentiation of 3T3-L1 cells (22), a modification that was associated with enhanced Pparγ2 transcriptional activity. We evaluated initially whether a small-molecule CDK9 inhibitor, DRB, could abrogate PPARγ1 phosphorylation in breast cancer cells (39). A DRB dose-response experiment revealed that treatment with 25 μM DRB for 24 h attenuated PPARγ1 phosphorylation by approximately 50%, whereas higher concentrations (35 to 45 μM) appeared to completely inhibit phosphorylation in 231_PARVβ cells (Fig. 6A). The inhibitory effect of DRB (50 μM) on pPPARγ levels was observed by 16 h in control cells and by 9 h in Parvin-β-transfected cells, and pPPARγ levels continued to decline over time (Fig. 6B). Importantly, DRB treatment did not alter pMAPK or phospho-c-JUN levels in either cell line (Fig. 6B). We next perturbed CDK9 function using a genetic approach. Transfection with CDK9-specific siRNA was used to abrogate endogenous CDK9 function. CDK9 protein was reduced to negligible levels in both cell lines following transfection with CDK9-specific siRNA but not with a scrambled (control) siRNA (Fig. 6D). Diminished CDK9 expression abrogated PPARγ1 phosphorylation in 231_PARVβ cells, which was associated with a simultaneous increase in levels of the unphosphorylated protein (Fig. 6C), thereby establishing that Parvin-β mediates phosphorylation of PPARγ via CDK9. Since the data suggested that Parvin-β could influence CDK9 function, we hypothesized that Parvin-β protein may be present in the nucleus. Indeed, subcellular fractionation revealed that both CDK9 (Fig. 6C), and Parvin-β (Fig. 6E) were contained in the nuclei of MDA-MB-231 cells, previously not described for Parvin-β.
FIG. 6.
CDK9 mediates increased PPARγ1 Ser82 phosphorylation in 231_PARVβ cells. (A, top) Cells cultured in 2D were treated with the indicated concentrations of DRB for 24 h, and nuclear extracts were evaluated for total PPARγ expression by Western blotting. pPPARγ1 levels were reduced by approximately 50% following treatment with 25 μM DRB. Lamin A/C was used as a loading control (bottom). (B) Cells were incubated in DRB (50 μM) for the indicated times followed by Western blot analysis for total PPARγ expression. Reduced pPPARγ1 levels were observed at 16 h of treatment in 231_VC cells and at 9 h of treatment in 231_PARVβ cells (top). DRB treatment did not affect pMAPK or pJNK levels (middle). β-Actin was used as a loading control (bottom). (C) 231_VC and 231_PARVβ cells were transfected with siRNA specific to CDK9 or with scrambled (control) siRNA, followed by Western blotting of nuclear extracts for PPARγ levels (top). Lamin A/C was used as an internal control (bottom). (D) Knockdown of CDK9 in 231_VC and 231_PARVβ cells transfected with CDK9-specific siRNA, scrambled siRNA, or vehicle alone (control) was confirmed by Western blotting for CDK9 expression (top). β-Actin was used as a loading control. (E) Nuclear extracts were prepared from 231_VC and 231_PARVβ cells cultured in 2D or 3D Matrigel (3D Mat) and analyzed for Parvin-β expression (top). Lamin A/C was used as an internal control (bottom). For panels A and C to E, molecular weights (M.W.) are in thousands.
Parvin-β negatively regulates breast cancer cell growth in vivo.
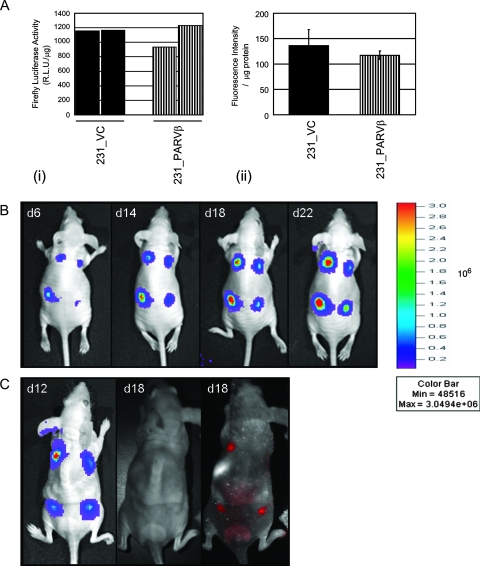
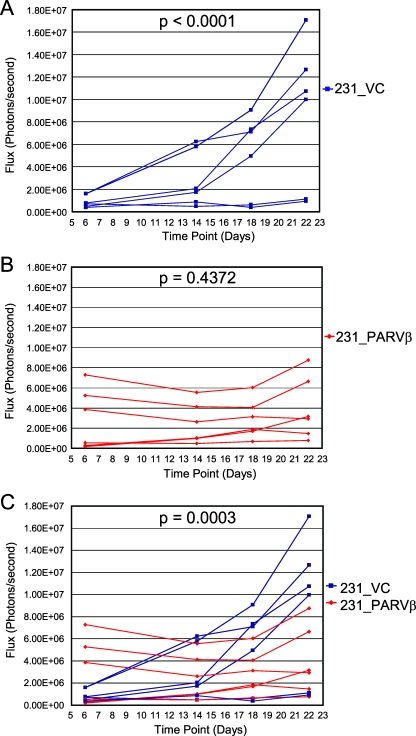
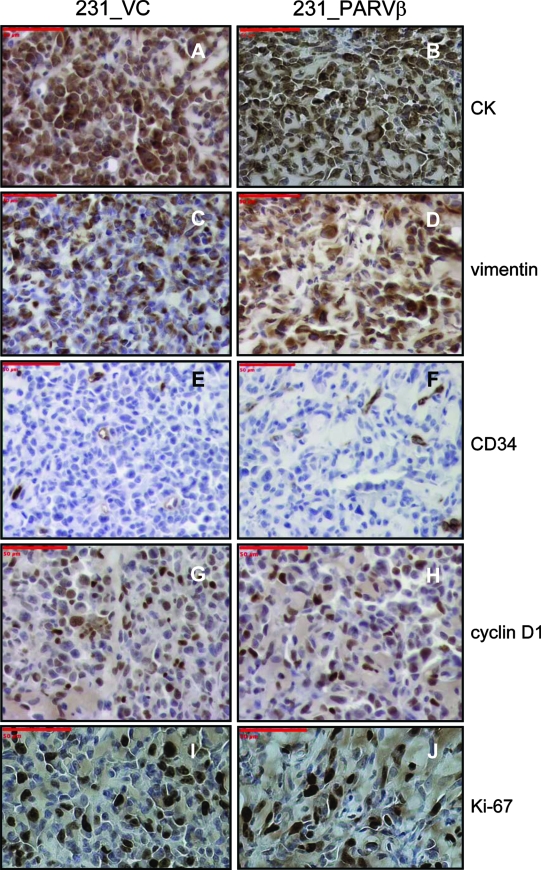
We had previously reported that Parvin-β reexpression reduced MDA-MB-231 colony formation in soft agar and invasiveness in vitro. Therefore, we tested whether Parvin-β could alter the behavior of MDA-MB-231 xenografts in irradiated immunodeficient mice in vivo. The MDA-MB-231 transfectants were transduced initially with both firefly luciferase and tdTomato for in vivo BLI and fluorescence imaging of tumor behavior, respectively (Fig. 7A; see Fig. S5 in the supplemental material). In the first cohort, three animals were administered two 231_VC/231_PARVβ pairs each (total n = 12 injection sites) and tumor growth was measured at four time points over a 22-day period using BLI (Fig. 7B and 8). Quantitation of emitted photons showed that there was a significant increase in luciferase activity, and therefore volume, of the 231_VC tumors over time (Fig. 8A; P < 0.0001), whereas the 231_PARVβ tumors did not significantly increase in size (Fig. 8B; P = 0.4372). Moreover, the growth rate of the group of 231_VC tumors was significantly faster than that of the group of 231_PARVβ tumors (P = 0.0003). A second cohort of mice (n = 3) were followed for an 18-day period, where both BLI and fluorescence imaging were performed for qualitative comparison of the two imaging modalities. The relative fluorescence signal intensity from the tdTomato protein showed close agreement with the relative luciferase activity for each tumor within each animal and provided independent corroboration of the BLI results demonstrating slower growth of the 231_PARVβ xenografts (Fig. 7C). The tumors were excised and subjected to histologic and immunohistochemical analyses (Fig. 9). The tumors displayed the expected poorly differentiated state of MDA-MB-231 cells, with expression of both epithelial (pancytokeratin) and mesenchymal (vimentin) markers. However, Ki-67 staining (Fig. 9I and J) revealed that proliferation was suppressed (P = 0.03) in the 231_PARVβ-derived tumors, in agreement with the reduced tumor growth quantified by BLI and tdTomato-mediated fluorescence signal intensity (Fig. 7B and C and 8).
FIG. 7.
231_VC and 231_PARVβ cells were engineered to express both firefly luciferase and tdTomato. (A, i) Luciferase activity was measured in firefly luciferase-transduced cells in duplicate and normalized to protein levels. (ii) tdTomato fluorescence intensity was measured using a fluorometer and normalized to protein levels. Results are expressed as mean fluorescence intensities/μg protein ± standard deviations (n = 5). (B) Luciferase activities of subcutaneously implanted tumors (left, two 231_VC tumors; right, two 231_PARVβ tumors) from a representative mouse at four time points following injection. Images were acquired using the IVIS system (see Materials and Methods). The color bar to the right represents luminescence intensity in flux (light photons/s). Greyscale and color images were superimposed for visualization of tumor location. 231_PARVβ tumors demonstrate reduced growth rate compared with 231_VC tumors. (C) Qualitative correlation of bioluminescent and fluorescent signals from a representative mouse 12 days and 18 days following subcutaneous implantation, respectively (left, two 231_VC tumors; right, two 231_PARVβ tumors). One 231_PARVβ tumor (front right flank) shows marked growth retardation compared to its 231_VC control (front left flank).
FIG. 8.
Growth rates of 231_VC- and 231_PARVβ-derived tumors in vivo. The growth of tumors from three mice (231_VC, n = 6; 231_PARVβ, n = 6) was followed longitudinally for a period of 22 days using in vivo BLI. (A) Tumors derived from 231_VC cells increased in size over time (P < 0.0001). (B) Tumors derived from 231_PARVβ cells did not increase in size over time (P = 0.4372). (C) Statistical comparison of the curves from 231_VC and 231_PARVβ cells (see Materials and Methods) demonstrated that the rate of change in tumor size was significantly greater in 231_VC tumors than in 231_PARVβ tumors (P = 0.0003).
FIG. 9.
Analysis of tumors for histology and immunohistochemistry. Four tumors (two of 231_VC and two of 231_PARVβ) were excised from the mouse shown in Fig. 7B and analyzed by immunohistochemistry for a marker of epithelial intermediate filaments (pancytokeratin [CK]; A and B), a marker of mesenchymal intermediate filaments (vimentin; C and D), a marker of mouse vascular endothelial cells (CD34; E and F), and markers of proliferation (cyclin D1 [G and H] and Ki-67 [I and J]) and counterstained with hematoxylin. One representative image is shown for each marker (original magnification, ×200). Scale bars, 50 μm. The number of Ki-67-positive nuclei was significantly lower for 231_PARVβ-derived tumors than for 231_VC-derived tumors (P = 0.03, Student's t test; data not shown).
DISCUSSION
Parvin-β gene transcription can be initiated from either of two alternative promoters, thereby resulting in translation of two protein isoforms (30, 43, 57). Transcription from the proximal promoter (promoter 1A) produces the shorter Parvin-β [Parvin-β(s)] isoform (364 amino acids), whereas initiation from the distal promoter (promoter 1) leads to synthesis of the longer Parvin-β [Parvin-β(l)] isoform (397 amino acids), also referred to as Parvin-β3 or CLINT (43). Parvin-β(s) contains 37 unique N-terminal amino acids, whereas Parvin-β(l) has a 70-amino-acid N-terminal extension unique to this isoform. Differences in function or expression pattern between the two isoforms have not been reported to date. MDA-MB-231 breast cancer cells do not express detectable levels of either Parvin-β protein isoform (Fig. 1B), and thus cell populations with stable expression of Parvin-β(l) were generated (43).
Global gene expression profiling revealed major differences between Parvin-β-regulated transcriptional profiles of cells grown on type I collagen (2D) and those grown embedded in either type I collagen (3D collagen) or basement membrane proteins (3D Matrigel). It is well documented that 3D growth more closely mimics both the normal tissue microenvironment and tumor microenvironment than 2D culture (29, 53, 66), in terms of both chemical properties (allowing proper cell-ECM interactions) and physical properties (appropriate tissue stiffness). We wished to interrogate how differences between 2D and 3D systems, as well as ECM composition, affected Parvin-β-mediated functions. For example, entire classes of genes were deregulated in 231_PARVβ cells compared to control cells and were restricted to 3D growth conditions only (see Tables S1 and S2 in the supplemental material). Examples include metabolic genes and those encoding chromatin-modifying factors, DNA repair proteins, and replication factors (see Table S1 in the supplemental material). Additionally, coordinated regulation of multiple genes involved in a specific cellular process or signaling pathway was also observed, again particularly when cells were propagated in 3D. First, evidence for increased expression of genes involved in epithelial differentiation in the Parvin-β-transfected cells was obtained. ID2, encoding a basic helix-loop-helix transcriptional regulator (3), was upregulated in 231_PARVβ cells in embedded 3D culture (Fig. 2A). ID2 plays a critical role in differentiation of mammary epithelial cells in vitro (26, 30) and is also required for mammary epithelial cell proliferation and differentiation during pregnancy in vivo (46, 42). Likewise, KLF4, a zinc finger transcription factor expressed in many differentiated epithelial cell types, regulates cell cycle exit and induction of differentiation programs in both columnar and squamous epithelia (12, 15) and was specifically upregulated in 3D culture (Fig. 2B and C). Comparison of transcriptional profiles in a global fashion showed that Parvin-β was more likely to upregulate luminal epithelium-expressed genes than those found in myoepithelial cells (Fig. 2D) and was more likely to downregulate myoepithelium-enriched transcripts than luminal epithelial ones. Thus, it is conceivable that Parvin-β is involved in “early” differentiation of breast cancer cells in 3D but is not sufficient by itself to trigger “terminal” differentiation, as another gene(s) is needed to orchestrate expression of the full repertoire of differentiation-linked genes and a differentiated morphology.
Increased mRNA expression, Ser82 phosphorylation, and activity of PPARγ were observed in 231_PARVβ cells and serve as a key novel component of our findings. How exactly might Parvin-β regulate PPARγ? The first effect may be transcriptional in nature. PPARγ3 was specifically increased in 231_PARVβ cells cultured in 3D Matrigel. Alignment of the proximal sequence of PPARG promoter 3 from several species revealed conserved consensus δEF1/TCF8 (56), AP-1, C/EBP β/δ (2), Sp1 (62), and RREB-1 (61) DNA binding sites (see Fig. S3 in the supplemental material). Indeed, the proximal and overlapping putative C/EBP β/δ and Sp1 binding sites were almost totally conserved among humans, chimpanzees, rhesus monkeys, mice, and rats, as were the adjacent putative δEF1/TCF8 and AP-1 binding sites. Accordingly, qPCR analysis of C/EBP β mRNA levels revealed a twofold increase in 231_PARVβ cells cultured in 3D Matrigel only (data not shown). Similarly, mRNA levels of ras-responsive element binding protein 1 (RREB-1), a dual transcriptional activator/repressor (61), were found to be decreased in 231_PARVβ cells cultured in 3D Matrigel by array analysis (see Table S1 in the supplemental material). Therefore, C/EBP β and RREB-1 are candidate transcriptional regulators of PPARG promoter 3 in 3D cultures and require further evaluation. Surprisingly, despite an increase in mRNA levels, overall PPARγ protein levels were not higher in 231_PARVβ cells, suggesting that MDA-MB-231 cells exhibit tight regulation of PPARγ protein levels. Our data demonstrated that Parvin-β's effect(s) on PPARγ is posttranslational in nature. CDK9 was a particularly attractive candidate kinase, and pharmacologic and siRNA strategies were employed to perturb CDK9 function. DRB (25 μM) reduced pPARG1 levels by about 50% (Fig. 6A). Since the concentration of DRB required to inhibit CDK9 activity by 50% in vitro (IC50) was reported to be 3 μM (37) and the in vivo (using intact cells) IC50 was predicted to be in the vicinity of 30 μM, these results supported the notion of CDK9 involvement in regulation of PPARγ1 phosphorylation. Indeed, siRNA-mediated knockdown of CDK9 reduced pPPARγ1 levels in 231_PARVβ cells (Fig. 6C), thus demonstrating clearly a role for CDK9 in Parvin-β-mediated PPARγ Ser82 phosphorylation. Ectopically expressed Parvin-β was also resident in the nuclei of MDA-MB-231 cells, therefore suggesting potential regulation of CDK9 activity.
It was intriguing that potential PPARγ target genes such as those involved in lipid biosynthesis, lipid droplet formation, and cholesterol efflux were essentially activated only in 231_PARVβ cells maintained in 3D Matrigel culture, and to a lesser extent in 3D collagen (Table 1). PPARγ transcriptional activity is controlled not only by level of expression, availability of ligand, and phosphorylation but also by the availability of transcriptional cofactors (17, 31). Levels of PGC-1α, a coactivator for PPARγ, were substantially elevated in both 3D growth conditions (Fig. 5E and F), whereas expression of other PPARγ coactivators, such as NCOA1/SRC-1 (steroid receptor coactivator 1), and PGC-1β was unchanged (data not shown). In this model, induction of PGC-1α expression by 3D culture may then permit ligand-independent transactivation of PPARγ target genes. This induction may contribute to the apparently enhanced ability of Ser82-phosphorylated PPARγ in the Parvin-β transfectants to activate endogenous target genes when the cell-basement membrane interaction is maintained. We extended our 3D studies to in vivo xenotransplantation experiments with nude athymic mice. Indeed, Parvin-β transfectants exhibited suppressed tumor growth, as determined by two different optical imaging modalities, and a reduced proliferation index. Perhaps the suppression of proliferation in vivo is related to the induction of a specific early differentiation program observed in 3D cultures.
Could the induction of pPPARγ by Parvin-β play a role in mammary epithelial cell differentiation as a consequence? PPARγ1 is expressed highly in differentiated colonic epithelial cells and is implicated in the colonic epithelial differentiation program (15). More recently, the generation of conditional knockout mice revealed a requirement for Pparg in the normal differentiation of airway epithelial cells in the lung (59), where Pparγ1-dependent regulation of both differentiation-linked (for example, moesin) and developmentally regulated (for example, cathepsin B) gene expression was invoked. Prior work also implicated PPARγ1 in the differentiation of breast cancer cells since treatment of several breast cancer cell lines with PPARγ agonists stimulated lipid synthesis and also reduced proliferation and clonogenic growth in soft agar (47). However, no apparent phenotype was observed in the mammary tissues of mice lacking Pparγ expression in mammary epithelium (11), indicating that Pparγ-dependent effects may be subtle or that another factor can compensate in its absence. One unanswered question is whether loss of Pparg expression enhances or suppresses oncogene-induced breast carcinogenesis in vivo (52). A potential clue was obtained recently from the observation that in a large prospective study of patients with diabetes mellitus receiving the glucose-lowering PPARγ agonist pioglitazone (Actos) for at least 2.5 years, a significant reduction (P = 0.034) in the incidence of breast cancer was observed only in the group receiving pioglitazone (n = 3/2,605) compared with the blinded placebo control group (n = 11/2,633) (http://www.proactive-results.com/).
Supplementary Material
Acknowledgments
We thank Don Baldwin, Grace Straszewski, and Donna Wilson (Penn Microarray Core Facility) for the gene expression profiling, Colleen Brensinger for statistical analysis, Jonathan Katz for the KLF4 antibody, Xinghai Li, David Tucker, and Morris Birnbaum for the PGC-1α antibody and protein, and Mitch Lazar for the pMSCV_Pparγ2 construct and helpful reading of the manuscript. We thank members of the Rustgi and Lazar labs for helpful discussions.
This work was supported in part by NIH/NIDDK grant R01-DK056645 (to A.K.R. and C.N.J.), a grant from the National Colorectal Cancer Research Alliance/EIF (to A.K.R.), a grant from the Irving A. Hansen Foundation (to A.K.R.), a Pennsylvania Department of Health Fellowship in Basic Cancer Research from the American Association for Cancer Research (to C.N.J.), a Concept Award (W81XWH-04-1-0658) from the U.S. Department of Defense Breast Cancer Research Program (to C.N.J.), and in part by grants from the National Cancer Institute of Canada (with funds from the Terry Fox Run) and the Canadian Institutes of Health Research (both to G.E.H.). G.E.H. was a CIHR scholar. This study was also supported by the Penn Center for Molecular Studies in Digestive and Liver Diseases and its core facilities (Morphology, Molecular Biology, and Cell Culture) through NIH/NIDDK grant P30 DK50306, the Penn Diabetes and Endocrinology Research Center (NIH/NIDDK grant DK19525), and the Optical/Bioluminescence Core of the Penn Small Animal Imaging Facility (supported in part by NIH grant CA105008).
Footnotes
Published ahead of print on 12 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, M., M. J. Reginato, D. Shao, M. A. Lazar, and V. K. Chatterjee. 1997. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J. Biol. Chem. 2725128-5132. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., H. Isshiki, T. Sugita, O. Tanabe, S. Kinoshita, Y. Nishio, T. Nakajima, T. Hirano, and T. Kishimoto. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 91897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggs, J., E. V. Murphy, and M. A. Israel. 1992. A human Id-like helix-loop-helix protein expressed during early development. Proc. Natl. Acad. Sci. USA 891512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camp, H. S., and S. R. Tafuri. 1997. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J. Biol. Chem. 27210811-10816. [DOI] [PubMed] [Google Scholar]
- 5.Camp, H. S., S. R. Tafuri, and T. Leff. 1999. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma 1 and negatively regulates its transcriptional activity. Endocrinology 140392-397. [DOI] [PubMed] [Google Scholar]
- 6.Castells, A., Y. Ino, D. N. Louis, V. Ramesh, J. F. Gusella, and A. K. Rustgi. 1999. Mapping of a target region of allelic loss to a 0.5-cM interval on chromosome 22q13 in human colorectal cancer. Gastroenterology 117831-837. [DOI] [PubMed] [Google Scholar]
- 7.Castells, A., J. F. Gusella, V. Ramesh, and A. K. Rustgi. 2000. A region of deletion on chromosome 22q13 is common to human breast and colorectal cancers. Cancer Res. 602836-2839. [PubMed] [Google Scholar]
- 8.Castellvi-Bel, S., A. Castells, C. N. Johnstone, V. Pinol, M. Pellise, J. I. Elizalde, N. Romo, A. K. Rustgi, and J. M. Pique. 2003. Evaluation of PARVG located on 22q13 as a candidate tumor suppressor gene for colorectal and breast cancer. Cancer Genet. Cytogenet. 14480-82. [DOI] [PubMed] [Google Scholar]
- 9.Chawla, A., E. J. Schwarz, D. D. Dimaculangan, and M. A. Lazar. 1994. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 135798-800. [DOI] [PubMed] [Google Scholar]
- 10.Chinetti, G., S. Lestavel, V. Bocher, A. T. Remaley, B. Neve, I. P. Torra, E. Teissier, A. Minnich, M. Jaye, N. Duverger, H. B. Brewer, J. C. Fruchart, V. Clavey, and B. Staels. 2001. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 753-58. [DOI] [PubMed] [Google Scholar]
- 11.Cui, Y., K. Miyoshi, E. Claudio, U. K. Siebenlist, F. J. Gonzalez, J. Flaws, K. U. Wagner, and L. Hennighausen. 2002. Loss of the peroxisome proliferation-activated receptor gamma (PPARgamma) does not affect mammary development and propensity for tumor formation but leads to reduced fertility. J. Biol. Chem. 27717830-17835. [DOI] [PubMed] [Google Scholar]
- 12.Dang, D. T., J. Pevsner, and V. W. Yang. 2000. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 321103-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcommenne, M., C. Tan, V. Gray, L. Rue, J. Woodgett, and S. Dedhar. 1998. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc. Natl. Acad. Sci. USA 9511211-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deramaudt, T. B., M. Takaoka, R. Upadhyay, M. J. Bowser, J. Porter, A. Lee, B. Rhoades, C. N. Johnstone, R. Weissleder, S. R. Hingorani, U. Mahmood, and A. K. Rustgi. 2006. N-cadherin and keratinocyte growth factor receptor mediate the functional interplay between Ki-RASG12V and p53V143A in promoting pancreatic cell migration, invasion, and tissue architecture disruption. Mol. Cell. Biol. 264185-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drori, S., G. D. Girnun, L. Tou, J. D. Szwaya, E. Mueller, K. Xia, R. A. Shivdasani, and B. M. Spiegelman. 2005. Hic-5 regulates an epithelial program mediated by PPARgamma. Genes Dev. 19362-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigoriadis, A., A. Mackay, J. S. Reis-Filho, D. Steele, C. Iseli, B. J. Stevenson, C. V. Jongeneel, H. Valgeirsson, K. Fenwick, M. Iravani, M. Leao, A. J. Simpson, R. L. Strausberg, P. S. Jat, A. Ashworth, A. M. Neville, and M. J. O'Hare. 2006. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on mpss and array expression data. Breast Cancer Res. 8R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan, H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 19453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannigan, G., A. A. Troussard, and S. Dedhar. 2005. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat. Rev. Cancer 551-63. [DOI] [PubMed] [Google Scholar]
- 19.Hannigan, G. E., C. Leung-Hagesteijn, L. Fitz-Gibbon, M. G. Coppolino, G. Radeva, J. Filmus, J. C. Bell, and S. Dedhar. 1996. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature 37991-96. [DOI] [PubMed] [Google Scholar]
- 20.Heid, H. W., M. Schnolzer, and T. W. Keenan. 1996. Adipocyte differentiation-related protein is secreted into milk as a constituent of milk lipid globule membrane. Biochem. J. 3201025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, E., J. B. Kim, P. Sarraf, and B. M. Spiegelman. 1996. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science 2742100-2103. [DOI] [PubMed] [Google Scholar]
- 22.Iankova, I., R. K. Petersen, J. S. Annicotte, C. Chavey, J. B. Hansen, I. Kratchmarova, D. Sarruf, M. Benkirane, K. Kristiansen, and L. Fajas. 2006. Peroxisome proliferator-activated receptor gamma recruits the positive transcription elongation factor b complex to activate transcription and promote adipogenesis. Mol. Endocrinol. 201494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itahana, Y., J. Singh, T. Sumida, J. P. Coppe, S. Parrinello, J. L. Bennington, and P. Y. Desprez. 2003. Role of Id-2 in the maintenance of a differentiated and noninvasive phenotype in breast cancer cells. Cancer Res. 637098-7105. [PubMed] [Google Scholar]
- 24.Johnson, K. R., J. L. Leight, and V. M. Weaver. 2007. Demystifying the effects of a three-dimensional microenvironment in tissue morphogenesis. Methods Cell Biol. 83547-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnstone, C. N., N. C. Tebbutt, H. E. Abud, S. J. White, K. L. Stenvers, N. E. Hall, S. H. Cody, R. H. Whitehead, B. Catimel, E. C. Nice, A. W. Burgess, and J. K. Heath. 2000. Characterization of mouse A33 antigen, a definitive marker for basolateral surfaces of intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 279G500-G510. [DOI] [PubMed] [Google Scholar]
- 26.Johnstone, C. N., S. J. White, N. C. Tebbutt, F. J. Clay, M. Ernst, W. H. Biggs, C. S. Viars, S. Czekay, K. C. Arden, and J. K. Heath. 2002. Analysis of the regulation of the A33 antigen gene reveals intestine-specific mechanisms of gene expression. J. Biol. Chem. 27734531-34539. [DOI] [PubMed] [Google Scholar]
- 27.Johnstone, C. N., S. Castellvi-Bel, L. M. Chang, X. Bessa, H. Nakagawa, H. Harada, R. K. Sung, J. M. Pique, A. Castells, and A. K. Rustgi. 2004. ARHGAP8 is a novel member of the RHOGAP family related to ARHGAP1/CDC42GAP/p50RHOGAP: mutation and expression analyses in colorectal and breast cancers. Gene 33659-71. [DOI] [PubMed] [Google Scholar]
- 28.Kass, L., J. T. Erler, M. Dembo, and V. M. Weaver. 2007. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. Int. J. Biochem. Cell Biol. 391987-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinman, H. K., and G. R. Martin. 2005. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15378-386. [DOI] [PubMed] [Google Scholar]
- 30.Korenbaum, E., T. M. Olski, and A. A. Noegel. 2001. Genomic organization and expression profile of the parvin family of focal adhesion proteins in mice and humans. Gene 27969-79. [DOI] [PubMed] [Google Scholar]
- 31.Krey, G., H. Keller, A. Mahfoudi, J. Medin, K. Ozato, C. Dreyer, and W. Wahli. 1993. Xenopus peroxisome proliferator activated receptors: genomic organization, response element recognition, heterodimer formation with retinoid X receptor and activation by fatty acids. J. Steroid Biochem. Mol. Biol. 4765-73. [DOI] [PubMed] [Google Scholar]
- 32.Legate, K. R., E. Montanez, O. Kudlacek, and R. Fassler. 2006. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 720-31. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann, J. M., L. B. Moore, T. A. Smith-Oliver, W. O. Wilkison, T. M. Willson, and S. A. Kliewer. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J. Biol. Chem. 27012953-12956. [DOI] [PubMed] [Google Scholar]
- 34.Lehrke, M., and M. A. Lazar. 2005. The many faces of PPARgamma. Cell 123993-999. [DOI] [PubMed] [Google Scholar]
- 35.Li, X., B. Monks, Q. Ge, and M. J. Birnbaum. 2007. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature 4471012-1016. [DOI] [PubMed] [Google Scholar]
- 36.Majello, B., G. Napolitano, A. Giordano, and L. Lania. 1999. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene 184598-4605. [DOI] [PubMed] [Google Scholar]
- 37.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 112633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchesan, D., M. Rutberg, L. Andersson, L. Asp, T. Larsson, J. Boren, B. R. Johansson, and S. O. Olofsson. 2003. A phospholipase D-dependent process forms lipid droplets containing caveolin, adipocyte differentiation-related protein, and vimentin in a cell-free system. J. Biol. Chem. 27827293-27300. [DOI] [PubMed] [Google Scholar]
- 39.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 27012335-12338. [DOI] [PubMed] [Google Scholar]
- 40.Meggio, F., D. Shugar, and L. A. Pinna. 1990. Ribofuranosyl-benzimidazole derivatives as inhibitors of casein kinase-2 and casein kinase-1. Eur. J. Biochem. 18789-94. [DOI] [PubMed] [Google Scholar]
- 41.Mishima, W., A. Suzuki, S. Yamaji, R. Yoshimi, A. Ueda, T. Kaneko, J. Tanaka, Y. Miwa, S. Ohno, and Y. Ishigatsubo. 2004. The first CH domain of affixin activates Cdc42 and Rac1 through alphaPIX, a Cdc42/Rac1-specific guanine nucleotide exchanging factor. Genes Cells 9193-204. [DOI] [PubMed] [Google Scholar]
- 42.Miyoshi, K., B. Meyer, P. Gruss, Y. Cui, J. P. Renou, F. V. Morgan, G. H. Smith, M. Reichenstein, M. Shani, L. Hennighausen, and G. W. Robinson. 2002. Mammary epithelial cells are not able to undergo pregnancy-dependent differentiation in the absence of the helix-loop-helix inhibitor Id2. Mol. Endocrinol. 162892-2901. [DOI] [PubMed] [Google Scholar]
- 43.Mongroo, P. S., C. N. Johnstone, I. Naruszewicz, C. Leung-Hagesteijn, R. K. Sung, L. Carnio, A. K. Rustgi, and G. E. Hannigan. 2004. Beta-parvin inhibits integrin-linked kinase signaling and is downregulated in breast cancer. Oncogene 238959-8970. [DOI] [PubMed] [Google Scholar]
- 44.Moore, K. J., E. D. Rosen, M. L. Fitzgerald, F. Randow, L. P. Andersson, D. Altshuler, D. S. Milstone, R. M. Mortensen, B. M. Spiegelman, and M. W. Freeman. 2001. The role of PPAR-gamma in macrophage differentiation and cholesterol uptake. Nat. Med. 741-47. [DOI] [PubMed] [Google Scholar]
- 45.Morgenstern, J. P., and H. Land. 1990. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 181068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori, S., S. I. Nishikawa, and Y. Yokota. 2000. Lactation defect in mice lacking the helix-loop-helix inhibitor Id2. EMBO J. 195772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller, E., P. Sarraf, P. Tontonoz, R. M. Evans, K. J. Martin, M. Zhang, C. Fletcher, S. Singer, and B. M. Spiegelman. 1998. Terminal differentiation of human breast cancer through PPAR gamma. Mol. Cell 1465-470. [DOI] [PubMed] [Google Scholar]
- 48.Nikolopoulos, S. N., and C. E. Turner. 2000. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J. Cell Biol. 1511435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikolopoulos, S. N., and C. E. Turner. 2001. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 27623499-23505. [DOI] [PubMed] [Google Scholar]
- 50.O'Brien, L. E., W. Yu, K. Tang, T. S. Jou, M. M. Zegers, and K. E. Mostov. 2006. Morphological and biochemical analysis of Rac1 in three-dimensional epithelial cell cultures. Methods Enzymol. 406676-691. [DOI] [PubMed] [Google Scholar]
- 51.Olski, T. M., A. A. Noegel, and E. Korenbaum. 2001. Parvin, a 42 kDa focal adhesion protein, related to the alpha-actinin superfamily. J. Cell Sci. 114525-538. [DOI] [PubMed] [Google Scholar]
- 52.Papadaki, I., E. Mylona, I. Giannopoulou, S. Markaki, A. Keramopoulos, and L. Nakopoulou. 2005. PPARgamma expression in breast cancer: clinical value and correlation with ERbeta. Histopathology 4637-42. [DOI] [PubMed] [Google Scholar]
- 53.Paszek, M. J., N. Zahir, K. R. Johnson, J. N. Lakins, G. I. Rozenberg, A. Gefen, C. A. Reinhart-King, S. S. Margulies, M. Dembo, D. Boettiger, D. A. Hammer, and V. M. Weaver. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell 8241-254. [DOI] [PubMed] [Google Scholar]
- 54.Petersen, O. W., L. Ronnov-Jessen, A. R. Howlett, and M. J. Bissell. 1992. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA 899064-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenberger, G., I. Jantke, A. Gal, and K. Kutsche. 2003. Interaction of alphaPIX (ARHGEF6) with beta-parvin (PARVB) suggests an involvement of alphaPIX in integrin-mediated signaling. Hum. Mol. Genet. 12155-167. [DOI] [PubMed] [Google Scholar]
- 56.Sekido, R., K. Murai, J.-I. Funahashi, Y. Kamachi, A. Fujisawa-Sehara, Y.-I. Nabeshima, and H. Kondoh. 1994. The δ-crystallin enhancer-binding protein δEF1 is a repressor of E2-box-mediated gene activation. Mol. Cell. Biol. 145692-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sepulveda, J. L., and C. Wu. 2006. The parvins. Cell. Mol. Life Sci. 6325-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaner, N. C., P. A. Steinbach, and R. Y. Tsien. 2005. A guide to choosing fluorescent proteins. Nat. Methods 2905-909. [DOI] [PubMed] [Google Scholar]
- 59.Simon, D. M., M. C. Arikan, S. Srisuma, S. Bhattacharya, L. W. Tsai, E. P. Ingenito, F. Gonzalez, S. D. Shapiro, and T. J. Mariani. 2006. Epithelial cell PPAR[gamma] contributes to normal lung maturation. FASEB J. 201507-1509. [DOI] [PubMed] [Google Scholar]
- 60.Stighall, M., C. Manetopoulos, H. Axelson, and G. Landberg. 2005. High ID2 protein expression correlates with a favourable prognosis in patients with primary breast cancer and reduces cellular invasiveness of breast cancer cells. Int. J. Cancer 115403-411. [DOI] [PubMed] [Google Scholar]
- 61.Thiagalingam, A., A. De Bustros, M. Borges, R. Jasti, D. Compton, L. Diamond, M. Mabry, D. W. Ball, S. B. Baylin, and B. D. Nelkin. 1996. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol. Cell. Biol. 165335-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiesen, H.-J., and C. Bach. 1990. Target detection assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 183202-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1995. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor gamma. Curr. Opin. Genet. Dev. 5571-576. [DOI] [PubMed] [Google Scholar]
- 64.Tu, Y., Y. Huang, Y. Zhang, Y. Hua, and C. Wu. 2001. A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J. Cell Biol. 153585-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, X., R. C. Southard, and M. W. Kilgore. 2004. The increased expression of peroxisome proliferator-activated receptor-gamma1 in human breast cancer is mediated by selective promoter usage. Cancer Res. 645592-5596. [DOI] [PubMed] [Google Scholar]
- 66.Weaver, V. M., A. H. Fischer, O. W. Peterson, and M. J. Bissell. 1996. The importance of the microenvironment in breast cancer progression: recapitulation of mammary tumorigenesis using a unique human mammary epithelial cell model and a three-dimensional culture assay. Biochem. Cell Biol. 74833-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White, D. E., R. D. Cardiff, S. Dedhar, and W. J. Muller. 2001. Mammary epithelial-specific expression of the integrin-linked kinase (ILK) results in the induction of mammary gland hyperplasias and tumors in transgenic mice. Oncogene 207064-7072. [DOI] [PubMed] [Google Scholar]
- 68.Wu, C., S. Y. Keightley, C. Leung-Hagesteijn, G. Radeva, M. Coppolino, S. Goicoechea, J. A. McDonald, and S. Dedhar. 1998. Integrin-linked protein kinase regulates fibronectin matrix assembly, E-cadherin expression, and tumorigenicity. J. Biol. Chem. 273528-536. [DOI] [PubMed] [Google Scholar]
- 69.Yamaji, S., A. Suzuki, Y. Sugiyama, Y. Koide, M. Yoshida, H. Kanamori, H. Mohri, S. Ohno, and Y. Ishigatsubo. 2001. A novel integrin-linked kinase-binding protein, affixin, is involved in the early stage of cell-substrate interaction. J. Cell Biol. 1531251-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamaji, S., A. Suzuki, H. Kanamori, W. Mishima, R. Yoshimi, H. Takasaki, M. Takabayashi, K. Fujimaki, S. Fujisawa, S. Ohno, and Y. Ishigatsubo. 2004. Affixin interacts with alpha-actinin and mediates integrin signaling for reorganization of F-actin induced by initial cell-substrate interaction. J. Cell Biol. 165539-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, Y., B. G. Goldstein, H. H. Chao, and J. P. Katz. 2005. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol. Ther. 41216-1221. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Y., K. Chen, L. Guo, and C. Wu. 2002. Characterization of PINCH-2, a new focal adhesion protein that regulates the PINCH-1-ILK interaction, cell spreading, and migration. J. Biol. Chem. 27738328-38338. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, Y., K. Chen, Y. Tu, and C. Wu. 2004. Distinct roles of two structurally closely related focal adhesion proteins, alpha-parvins and beta-parvins, in regulation of cell morphology and survival. J. Biol. Chem. 27941695-41705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.