Abstract
In Saccharomyces cerevisiae, the double-stranded-RNA-specific RNase III (Rnt1p) is required for the processing of pre-rRNA and coprecipitates with transcriptionally active rRNA gene repeats. Here we show that Rnt1p physically interacts with RNA polymerase I (RNAPI) and its deletion decreases the transcription of the rRNA gene and increases the number of rRNA genes with an open chromatin structure. In contrast, depletion of ribosomal proteins or factors that impair RNAPI termination did not increase the number of open rRNA gene repeats, suggesting that changes in the ratio of open and closed rRNA gene chromatin is not due to a nonspecific response to ribosome depletion or impaired termination. The results demonstrate that defects in pre-rRNA processing can influence the chromatin structure of the rRNA gene arrays and reveal links among the rRNA gene chromatin, transcription, and processing.
In Saccharomyces cerevisiae, ribosomes are formed of 78 different ribosomal proteins assembled around four rRNAs. The synthesis of these essential protein factories involves three RNA polymerases and several processing factors (70). The ribosomal protein genes are transcribed by RNA polymerase II, while the rRNAs are transcribed by RNA polymerase I (RNAPI), with the exception of the 5S rRNA that is produced by RNA polymerase III. RNAPI produces the 18S, 5.8S, and 25S rRNAs as a single transcript containing two external transcribed spacers (ETS1 and -2) and two internal transcribed spacers (ITS1 and -2). Once transcribed, the rRNA precursor is processed to form the ribosome small subunit containing the 18S rRNA and the ribosome large subunit (LSU) containing the 5.8S and 25S rRNAs (69).
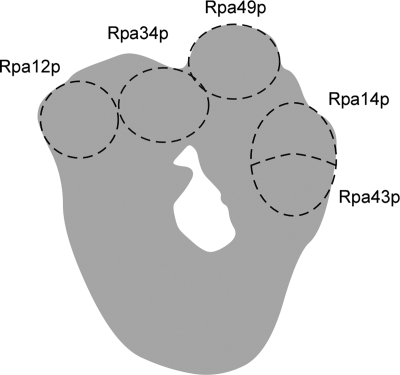
RNAPI (Fig. 1) is composed of five subunits common to all RNA polymerases, two subunits shared with RNA polymerase III, and seven unique subunits (53). All of the shared subunits are essential for yeast growth, as are the two largest unique subunits. The five remaining unique subunits are dispensable; however, the loss of Rpa12p causes temperature sensitivity and impairs RNAPI termination (21, 52). The last four RNAPI-specific subunits form two distinct complexes; one contains the Rpa34p and Rpa49p subunits, and the other contains the Rpa14p and Rpa43p subunits. The Rpa34p-Rpa49p complex is located near Rpa12p (Fig. 1) and is thought to play a role in RNAPI elongation (21, 29). In contrast, it was suggested that the Rpa14p-Rpa43p complex participates in the recruitment of RNAPI to its promoter (51). Both of these RNAPI-associated complexes are functionally interdependent since the double deletion of the nonessential components within each complex (e.g., Rpa34p and Rpa14p) causes synthetic lethality (21).
FIG. 1.
Structure of RNAPI. Schematic representation illustrating the presumed positions of five unshared RNAPI subunits and their spatial relationship to each other. The model is based on electron microscopy and immunolocalization data published by Schultz and coworkers (4, 11).
The rRNA genes are present in multiple copies of two distinct types; one is permissive to transcription, and the other is transcriptionally refractive (26). In higher eukaryotes, rRNA synthesis is regulated by three different mechanisms, (i) designation of the total number of active genes, (ii) modulation of the transcription initiation rate, and (iii) regulation of the RNAPI elongation rate (14, 26, 47, 62). In S. cerevisiae, rRNA synthesis appears to be regulated by varying the proportions of active and inactive rRNA gene copies in response to growth conditions and by modulating the transcription initiation rate of active genes, which is controlled by the enhancer DNA element (3, 15, 53). More recently, it has been proposed that the overall initiation rate, but not the number of active genes, determines the rRNA transcription rate during exponential growth (20). However, changes in the number of active rRNA gene copies were also shown to contribute, albeit modestly, to the overall transcription rate (18).
At the chromatin level, canonical nucleosomes are not present in the coding regions of rRNA genes that are either transcribing or permissive to transcription. In contrast, nucleosomes are present on inactive rRNA genes (44, 61). These two forms of chromatin have been observed in a number of organisms, ranging from yeast to mammals, by a technique that is based on psoralen photo-cross-linking (58, 65). Furthermore, electron micrographs of psoralen-cross-linked rRNA gene chromatin and biochemical studies monitoring DNA accessibility to nuclease-like DNase I, micrococcal nuclease, and restriction enzymes (8, 13, 42, 43, 48, 49, 60) have confirmed the absence of canonical nucleosomes from the coding regions of active rRNA genes in all of the organisms investigated. Recently, it was shown that nucleosomes may associate with active rRNA gene repeats in a genetically engineered yeast strain harboring an artificially reduced number of rRNA gene repeats (33).
After transcription by RNAPI, the 3′ end of the primary rRNA transcript is cleaved by the double-stranded RNA (dsRNA)- specific RNase Rnt1p at site B0 (2, 36) to produce the first biochemically detected rRNA precursor (35S) (69). The 35S rRNA precursor is subjected to a series of further processing events performed by a large ribonucleoprotein complex that includes more than 100 proteins and several snoRNAs (25, 69). The activity of this large RNA-processing complex is affected by mutations that impair RNAPI initiation (59), while mutations of factors involved in pre-rRNA processing impair transcription initiation (22). Other protein factors required for the transcription of the ribosomal protein genes were also implicated in the transcription and processing of pre-rRNA, underscoring the tight regulation of ribosome synthesis (57).
Although it was recently shown by electron microscopy analysis that a cotranscriptional cleavage separating the small subunit from the LSU RNA precursors could occur in as many as 50% of the pre-rRNA transcripts (50), the earliest processing event linked to transcription that is detectable by biochemical techniques is performed by the endoribonuclease Rnt1p (2). Rnt1p is recruited to the nucleolus only in the presence of an actively transcribed rRNA gene (7) and coprecipitates with active rRNA gene repeats (28). In addition, the deletion of Rnt1p affects the accuracy of RNAPI termination (52). This phenotype is similar to that obtained upon the deletion of RNAPI subunit Rpa12p, which is believed to be responsible for transcription termination (52).
In order to better define the link between rRNA gene transcription and pre-rRNA processing, we have searched for proteins that connect Rnt1p to rRNA gene transcription. A two-hybrid screen was performed with Rnt1p as bait and a limited set of RNAPI-associated subunits as prey. The results show that Rnt1p interacts with RNAPI unique subunits Rpa12p and Rpa34p. Rnt1p interacts with these subunits independently with different sets of amino acid residues. Rnt1p was immunoprecipitated with the RNAPI-specific complex containing subunits Rpa34p and Rpa49p, confirming the association of Rnt1p with RNAPI. In vivo labeling showed that in the absence of Rnt1p, rRNA transcription is reduced and unprocessed pre-rRNA is degraded. Psoralen photo-cross-linking indicated that the deletion or inactivation of RNT1 increases the number of rRNA genes with open chromatin. Expression of a catalytically impaired RNT1 allele failed to restore the expression and ratio of an open versus a closed rRNA gene, suggesting that transcription of the rRNA gene is linked to RNA processing by Rnt1p.
MATERIALS AND METHODS
Strains, plasmids, and media.
All of the yeast strains used in this study are described in Table 1. Yeast cells were grown and manipulated by standard procedures (27, 56). The plasmids used in this study are listed in Table 2. Temperature shifts from 26°C to 37°C were initiated by adding 1 volume of medium heated to 48°C. The new cultures were incubated at 37°C. TetO7 promoter-dependent depletion was achieved by adding 10 μg/ml doxycycline (Sigma-Aldrich, Oakville, Ontario, Canada) to the growth medium.
TABLE 1.
S. cerevisiae strains used in this work
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-1 can1-100 | 63 |
| Δrnt1 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade2-1 can1-100 rnt1Δ::TRP1 | 10 |
| PJ69-4A | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 32 |
| LLY112 | MATa/MATα ura3Δ/ura3Δ his3Δ/HIS3 leu2Δ/leu2Δ lys2Δ/lys2Δ met15Δ/MET15 ade2Δ/ADE2 | Personal communication |
| RNT1::TAP | MATα ura3Δ his3Δ leu2Δ lys2Δ RNT1::TAP::URA3 | This work |
| RNT1 untagged | MATα ura3Δ his3Δ leu2Δ lys2Δ | This work |
| HI227 | MATaleu2 trp1 ura3-52 lys2Δ his3Δ200 prb1-1122 pep4-3 prc1-407 | 1 |
| Δrnt1::HIS3 | MATaleu2 trp1 ura3-52 lys2Δ his3Δ200 prb1-1122 pep4-3 prc1-407 Δrnt1::HIS3 | 1 |
| R1158 | MATahis3Δ1 leu2Δ0 met15Δ0 URA3::CMV-tTA | 30 |
| TH_3372 | MATahis3Δ1 leu2Δ0 met15Δ0 URA3::CMV-tTA Kanr::TetO7-RPL15A | Open Biosystems (46) |
| TH_7410 | MATahis3Δ1 leu2Δ0 met15Δ0 URA3::CMV-tTA Kanr::TetO7-UTP7 | Open Biosystems (46) |
| TH_5563 | MATahis3Δ1 leu2Δ0 met15Δ0 URA3::CMV-tTA Kanr::TetO7-UTP5 | Open Biosystems (46) |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 5 |
| rpa12Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rpa12Δ::KANMX | 71 |
TABLE 2.
Plasmid constructs used in this study
| Name | Description | Source or reference |
|---|---|---|
| pRS315/GFP | GFP under RNT1 promoter control in pRS315 | 7 |
| pRS315/GFP/RNT1 | RNT1 coding sequence (1-471 aa) cloned in frame in pRS315/GFP | 7 |
| pRS315/GFP/RNT1-D247/R | RNT1 coding sequence (1-471 aa) variant with D247R mutation cloned in frame in pRS315/GFP | 7 |
| pRS315/GFP/RNT1-463st | RNT1 coding sequence (1-463 aa) cloned in frame in pRS315/GFP | 7 |
| pRS315/RNT1-I338/T | RNT1 coding sequence (1-471 aa) temp sensitive variant with I338/T mutation cloned under RNT1 promoter control in pRS315 | 23 |
| pGBDU-C3 | 32 | |
| BD/RNT1 | RNT1 coding sequence (1-471 aa) cloned in frame in pGBDU-C3 | 38 |
| BD/NT2 | RNT1 coding sequence (1-191 aa) cloned in frame in pGBDU-C3 | 38 |
| BD/DS1 | RNT1 coding sequence (344-471 aa) cloned in frame in pGBDU-C3 | 38 |
| BD/DS4 | RNT1 coding sequence (428-471 aa) cloned in frame in pGBDU-C3 | 38 |
| BD/RNT1-D247/R | RNT1 coding sequence (1-471 aa) variant with D247R mutation cloned in frame in pGBDU-C3 | 7 |
| BD/RNT1-K421/A | RNT1 coding sequence (1-471 aa) variant with K421A mutation cloned in frame in pGBDU-C3 | 66 |
| BD/RNT1-KK463/AA | RNT1 coding sequence (1-471 aa) variant with K463A and K464A mutations cloned in frame in pGBDU-C3 | 67 |
| BD/RNT1-463st | RNT1 coding sequence (1-463 aa) cloned in frame in pGBDU-C3 | This work |
| AD/RNT1-463st | RNT1 coding sequence (1-463 aa) cloned in frame in pGAD | 7 |
Characterization of protein interaction.
Two-hybrid assays were carried out as described previously (67). In order to immunoprecipitate Rnt1p, the TAP tag was fused to RNT1 in strain LLY112 via a PCR-based one-step in vivo tagging strategy with integration vector pBS1539 (55). One-liter yeast cultures were grown in YEPD to an optical density at 600 nm (OD600) of 0.7 to 0.8 and were frozen and dissociated with dry ice in a coffee grinder (Krups, Medford, MA). Tagged complexes were purified from the extract on immunoglobulin G (IgG) beads as described previously (35, 55). The purified proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 10% acrylamide gel. Western blot assays were performed with four different antibodies. Blots were incubated with primary antibodies against Rnt1p (1:7,500) (67), Rpa34p (1:20,000), and Rpa49p (1:20,000) (6) for 2 h at room temperature or for 16 h at 4°C. The secondary antibody (anti-rabbit IgG, 1:80,000; Sigma-Aldrich Canada) was incubated with the blots for 1 h at room temperature. The primary antibody against phosphoglycerate kinase (1:500; Molecular Probes, Eugene, OR) was incubated under the same conditions as the other three but was visualized with an anti-mouse IgG (1:15,000, Sigma-Aldrich Canada) secondary antibody.
Northern blot analysis.
Total RNA (10 μg) was separated by electrophoresis on 1% denaturing agarose gels. The rRNA species were visualized as described earlier (2), with probes I (5′ ACTATCTTAAAAGAAGAAGCAACAAGCAG), II (5′ ATGAAAACTCCACAGTG), and III (5′ TTTCGCTGGGTTCTTCATC), which were generated by end labeling of DNA oligonucleotides. Probe IV was generated by random labeling (19) of a PCR fragment corresponding to the sequence downstream of the 25S rRNA 3′ end (+53 to +208).
RNase protection assay.
A probe complementary to the 3′ end of the 25S rRNA and the 3′ ETS was produced by T7 transcription (2). DNase RQ1 (Promega, Madison, WI)-treated total RNA (10 μg) was incubated at 42°C for 16 h with 0.5 fmol of probe in 80% formamide hybridization buffer (45). The hybridization mixture was digested with 100 U of RNase T1 (Roche Diagnostics, Laval, Quebec, Canada) for 1 h at 30°C, extracted with phenol-chloroform, ethanol precipitated, and loaded on 6% denaturing polyacrylamide gels.
Pulse-labeling of cellular RNA.
Yeast cells were grown in YC medium to an OD600 of 0.5 at 26°C. The RNA was labeled in vivo by incubation of 20-ml culture aliquots with 50 μCi/ml [32P]orthophosphate (Amersham Biosciences, Baie d'Urfé, Quebec, Canada) in phosphate-depleted YC medium (67). The RNA was extracted, separated on a 1% denaturing agarose gel, and visualized by a PhosphorImager (Storm 860; Amersham Biosciences). For the pulse-chase experiment, 5-ml aliquots of a culture grown in YC medium lacking uracil to an OD600 of 0.6 were pulse-labeled for 2 min with 30 μCi/ml [3H]uracil (Amersham Biosciences). The chase was performed by the addition of excess cold uracil (240 μg/ml) to the culture (64).
Nuclear extraction and psoralen photo-cross-linking.
Cell disruption and nucleus preparation were performed as described previously (12). Yeast cells (∼1.6 × 109) were collected, washed with ice-cold phosphate-buffered saline, suspended in 1.5 ml of nuclear isolation buffer (50 mM morpholinepropanesulfonic acid [MOPS, pH 8.0], 150 mM potassium acetate, 2 mM MgCl2, 17% glycerol, 0.5 mM spermine, 0.15 mM spermidine), and transferred to 15-ml polypropylene tubes containing 1.5 ml of glass beads (425 to 600 μm; Sigma-Aldrich Canada). Cross-linking of nuclei was performed in 24-well multiwell plates (Falcon, uncoated). A psoralen (4,5′,8-trimethylpsoralen; Sigma-Aldrich Canada) stock solution (400 μg/ml) was added at a volume equal to 0.025× the nuclear suspension volume. After 5 min on ice in the dark, the nuclear suspension was irradiated on ice for 10 min as described previously (13). The irradiation step was repeated twice. Southern blot assays were performed on cross-linked DNA extracted from nuclei, digested with EcoRI, and separated on 1% native agarose gels with a probe corresponding to a 140-bp sequence located in the middle of the 25S rRNA (14).
RESULTS
RNAPI subunits Rpa12p and Rpa34p interact independently with the C-terminal domain of Rnt1p in vivo.
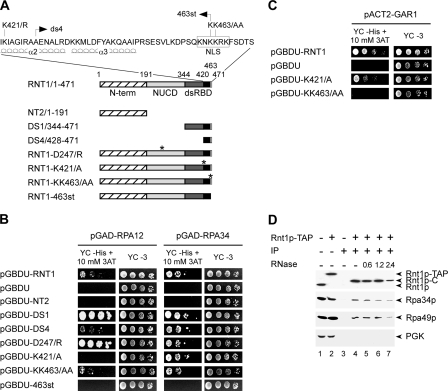
Several lines of evidence suggest that Rnt1p-dependent processing of the 25S pre-rRNA 3′ end is coupled to transcription (28, 36, 52, 54), suggesting a physical interaction between Rnt1p and RNAPI. To test this hypothesis, we assayed the interaction of Rnt1p with the RNAPI-specific subunits (Rpa12p, Rpa14p, Rpa34p, Rpa43p, Rpa49p, Rpa135p, and Rpa190p) by a two-hybrid interaction assay. RNAPI subunits were fused to the Gal4 activation domain (AD) (31), and Rnt1p was fused to the DNA-binding domain (BD) of Gal4 (pGBDU-RNT1). Plasmids carrying the different RNAPI subunits were transformed with pGBDU-RNT1 into a yeast strain harboring three marker genes (32). Interactions were scored by monitoring yeast growth on medium lacking histidine. Of the seven subunits screened, two (Rpa12p and Rpa34p) induced specific growth on the selective medium in the presence of Rnt1p (Fig. 2).
FIG. 2.
Rnt1p interacts with RNAPI-specific subunits. (A) Schematic representation of the Rnt1p truncations and point mutations used in this study. The hatched boxes represent the Rnt1p N-terminal (N-term) domain. The Rnt1p nuclease domain (NUCD) is light gray, and its dsRBD is dark gray. The portion of the dsRBD in black represents the Rnt1p protein-binding site. The white box indicates the Rnt1p nuclear localization signal (NLS). The positions of the different point mutations are indicated by asterisks. The sequence of the C-terminal 51 aa of Rnt1p and the positions of the different mutations in this region are shown at the top, as well as the extents of α-helices 2 and 3 of the dsRBD. (B) The interaction between RNAPI subunits Rpa12p and Rpa34p with the different domains of Rnt1p was assessed by analyzing the abilities of plasmid pairs to support the growth of yeast strain PJ69-4A on medium lacking histidine and supplemented with 10 mM 3-aminotriazole (YC −His + 10 mM 3AT) compared to growth on nonselective medium (YC −3). (C) The two-hybrid assay was carried out as described for panel B, with Gar1p as bait, in order to evaluate the specificity of Rnt1p interaction with the RNAPI subunits. (D) RNAPI subunits Rpa34p and Rpa49p coimmunoprecipitate with TAP-tagged Rnt1p (Rnt1p-TAP). Immunoprecipitations with cell extracts expressing either tagged or untagged Rnt1p were performed, and the Rnt1p-bound proteins (IP) were released by enzymatic cleavage of the Rnt1p tag (Rnt1p-C). The released proteins were visualized by Western blot analysis with protein-specific antibodies. Antibodies against phosphoglycerate kinase (PGK) were used as a control. The Rnt1p complex was treated with different concentrations (units) of a mixture of RNases V1, T1, and T2 in order to evaluate the contribution of RNA to the formation of the protein complex.
Gal4 AD fusions with Rpa12p (pGAD-RPA12) and Rpa34p (pGAD-RPA34) were tested for interaction with the different domains of Rnt1p (Fig. 2A). As shown in Fig. 2B, both AD-Rpa12p and AD-Rpa34p interacted with full-length BD-Rnt1p. However, the interaction of Rnt1p with Rpa34p was somewhat stronger than that with Rpa12p. In order to better define the Rpa12p and Rpa34p binding sites, a series of Rnt1p truncation mutants were examined for interaction with both subunits. Deletion of the C terminus (BD-NT2/1-191) disrupted the interaction with both subunits, while deletion of both the N-terminal domain and the nuclease domain (BD-DS1/344-471) did not (Fig. 2A and B). The last 44 amino acids (aa) of Rnt1p (BD-DS4/428-471), which include 4 aa of α-helix 2 (α2), a conserved element of the αβββα dsRNA-binding motif (34), and all of α-helix 3 (α3), an element unique to Rnt1p (41, 72), interacted with an efficiency similar to that of BD-DS1 with Rpa34p (Fig. 2B). In contrast, BD-DS4 interacted with Rpa12p much less efficiently than BD-DS1. This difference in binding efficiency suggests that Rpa34p binds to Rnt1p primarily through an interaction with the amino acid residues downstream of the canonical dsRNA BD (dsRBD) (Fig. 2A), while Rpa12p requires additional elements of the dsRBD present in BD-DS1. As shown in Fig. 2B, the introduction of a stop codon at position 463 of Rnt1p eliminates the terminal 8 aa and blocks interaction with both subunits. Point mutations that change the positively charged residue lysine (K) 421 to the hydrophobic residue alanine (A) disrupt the binding to Rpa12p but not to Rpa34p. As shown in Fig. 2C, this mutation does not significantly affect the binding previously documented for Gar1p (67) (a component of the H/ACA pseudouridylation complex involved in the modification and cleavage of the 18S pre-rRNA). Therefore, although the C terminus of Rnt1p constitutes the principal protein-binding site, the interaction of each protein uses a distinct set of amino acid residues. A point mutation in Rnt1p disrupting its catalytic activity (BD-D247/R) without affecting its interaction with the RNA (37) did not disrupt the interaction of Rnt1p with either of the subunits of RNAPI. In fact, the disruption of the Rnt1p catalytic activity enhanced the binding of Rnt1p with Rpa12p without affecting the interaction with Rpa34p (Fig. 2B). We conclude that Rnt1p binding to RNAPI is independent of its capacity to cleave pre-rRNA.
Rnt1p coimmunoprecipitates with RNAPI-specific subunits.
In order to confirm the interaction between Rnt1p and RNAPI, we immunoprecipitated a TAP-tagged version of Rnt1p and monitored the coprecipitation of RNAPI subunits with specific antibodies. The TAP-tagged protein was generated from a chromosomal copy of the RNT1 gene fused to the calmodulin BD, a TEV protease cleavage site, and the IgG BD of protein A from Staphylococcus aureus (55). The expression of the tagged protein was confirmed by Western blot assay with antibodies raised against Rnt1p (Fig. 2D, lane 2). As expected, Rnt1p-TAP migrated more slowly than the untagged Rnt1p version (Fig. 2D, lane 1) due to the presence of the TAP tag. The tagged RNT1 allele was able to complement the deletion of RNT1, and no effect on either RNA processing or cell growth was observed (data not shown). As shown in Fig. 2D, Rnt1p-TAP coprecipitated with IgG beads and was released upon cleavage by the TEV protease (Fig. 2D, lanes 4 to 7). Western blot analysis with antibodies against Rpa34p and Rpa49p revealed that these two interacting subunits specific for RNAPI precipitate with Rnt1p-TAP (Fig. 2D, lanes 4 to 7) and not in the absence of the tag (Fig. 2D, lane 3). The interaction of Rnt1p with Rpa49p could not be reproduced by the two-hybrid system, suggesting that this interaction might be either too weak or indirect. Since Rpa34p and Rpa49p form a stable complex (21), we presume that the observed coimmunoprecipitation of Rpa49p with Rnt1p, shown in Fig. 2D and previously noted in genome-wide protein coprecipitation assays (35), is mediated by Rpa34p. These data confirm the results of the two-hybrid assay and indicate the formation of a stable Rnt1p/RNAPI complex.
In order to determine whether or not Rnt1p interaction with Rpa34p is RNA dependent, we examined the effects of RNase treatment on the Rnt1p/Rpa34p-Rpa49p complex. Incubation of the immunoprecipitated Rnt1p/Rpa34p-Rpa49p complex with a cocktail of RNases V1, T1, and T2 completely degraded all of the RNA species, as detected by gel analysis (data not shown). As shown in Fig. 2D, lanes 5 to 7, treatment with RNases had no effect on Rnt1p/Rpa34p-Rpa49p complex formation. Therefore, we suggest that the interaction between Rnt1p and the Rpa34p-Rpa49p complex is not likely to be via RNA, although we cannot rule out the possibility that an amount of RNA undetectable by gel electrophoresis remains present inside the protein complex.
Deletion of Rnt1p reduces synthesis and induces the degradation of pre-rRNA.
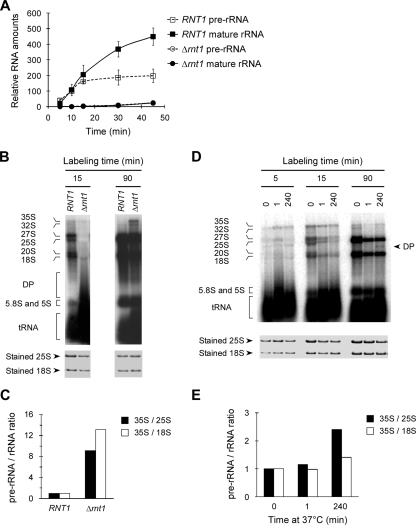
Microarray analysis (23) indicated that deletion or inactivation of Rnt1p leads to the accumulation of polyadenylated fragments of the 25S rRNA 3′ end. Northern blot assays with a probe covering the 3′ end of the 25S rRNA also confirmed the presence of these truncated species (data not shown). On the other hand, in vivo labeling of newly synthesized rRNA indicated that in the absence of Rnt1p, the overall rate of rRNA synthesis is reduced (Fig. 3A). As shown in Fig. 3B, after a 15-min labeling period, pre-rRNA precursors were visible in wild-type (RNT1) cells but the majority of the rRNA was either mature or in the form of processing intermediates. In contrast, very little RNA was observed in Δrnt1 cells at the same time point. At 90 min postincubation with [32P]PO4, most of the pre-rRNA is processed in wild-type cells while a significant amount of the 35S pre-rRNA precursor remains visible in Δrnt1 cells. It is important to note that after 90 min of labeling, the 35S rRNA continued to accumulate in Δrnt1 cells but not in wild-type cells (Fig. 3B and C), indicating a defect in the maturation of the pre-rRNA.
FIG. 3.
Deletion of RNT1 inhibits rRNA synthesis. (A) The rate of rRNA synthesis was determined by following the incorporation of [32P]PO4 in both pre-rRNA and mature rRNA species. Labeled RNAs separated on denaturing agarose gels were quantified and plotted using the amount of total stained RNA as a reference point. The data presented are an average of three experiments. (B) Pulse-labeling of rRNA upon the deletion of RNT1. Wild-type and Δrnt1 cells were incubated for either 15 or 90 min with [32P]PO4 at 26°C. The RNA was extracted, and equal counts were loaded on an agarose gel and visualized by autoradiography. To evaluate the RNA quality, equal amounts of total RNA were loaded onto agarose gel and visualized by direct staining. The positions of the different RNA species, as well as of the degradation products (DP), are indicated on the left. (C) Chart illustrating the ratio of rRNAs quantified after 90 min of labeling with a PhosphorImager. The expression level of 35S rRNA relative to that of mature rRNA was calculated and normalized to that of wild-type RNA. (D) Pulse-labeling of rRNA upon the inactivation of Rnt1p. Cells expressing a temperature-sensitive allele of Rnt1p (rnt1-ts) were grown for 0, 1, or 240 min at 37°C and then incubated with [32P]PO4 for 5, 15, or 90 min. The RNA was extracted and visualized as described for panel B. DP indicates a degradation product observed upon the inactivation of Rnt1p. (E) Chart illustrating the relative expression levels of the rRNA species detected after 90 min of labeling. The RNA values were normalized with the RNA extracted from rnt1-ts cells growing at the permissive temperature as a reference.
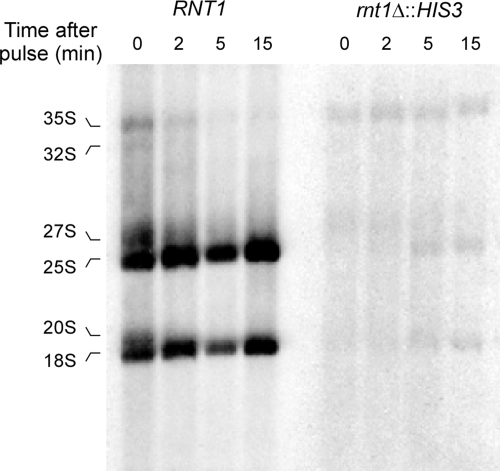
Deletion of RNT1 impairs the processing and degradation of many snRNAs (1, 9), snoRNAs (24), and mRNAs (23, 39), causing major changes in cell metabolism (7). Moreover, the reduced growth rate of Δrnt1 cells may indirectly cause a general reduction in transcription (2). Therefore, we wanted to examine pre-rRNA transcription and processing shortly after the inactivation of the enzyme to avoid indirect effects arising from prolonged growth in the absence of Rnt1p. Cells carrying a temperature-sensitive allele of Rnt1p (rnt1-ts) were grown at the permissive temperature and then shifted to the restrictive temperature for 1 or 240 min and incubated with [32P]PO4. As shown in Fig. 3D, after 5 min of incubation with [32P]PO4, very little rRNA was detected in RNA extracted from cells incubated at either the permissive or the restrictive temperature. Instead, the precursor 35S rRNA was observed under all conditions and RNA extracted from cells shifted to the restrictive temperature for 240 min exhibited a reduced amount of 27S rRNA. After 15 min of labeling, defects in the accumulation of both the 18S and 25S rRNAs, compared to the 35S rRNA, were observed even when the cells were incubated for only 1 min at the restrictive temperature prior to labeling. After 90 min of labeling, a band migrating faster than the 25S rRNA was detected, indicating an aberrant rRNA species, and an increased accumulation of 35S over the mature rRNAs was observed in the cells shifted to the restrictive temperature for 240 min (Fig. 3E). Pulse-chase analysis of pre-rRNA processing in the wild-type (RNT1) and Δrnt1 (Δrnt1::HIS3) strains grown at 26°C (Fig. 4) also indicates that the deletion of RNT1 reduces the synthesis and delays the processing of the 35S rRNA into mature rRNA. We conclude that the inactivation of Rnt1p delays both the synthesis and the processing of pre-rRNA transcripts.
FIG. 4.
Deletion of RNT1 delays rRNA processing. Wild-type (H1227) and Δrnt1::HIS3 cells were pulse-labeled for 2 min with [3H]uracil, followed by a chase with excess unlabeled uracil. Aliquots of the cultures were collected at the indicated time points starting from the beginning of the chase. RNA was extracted and separated on denaturing agarose gels and transferred to nylon membranes. The observed rRNA species are indicated on the left.
Deletion or inactivation of RNT1 increases the number of rRNA genes with open chromatin.
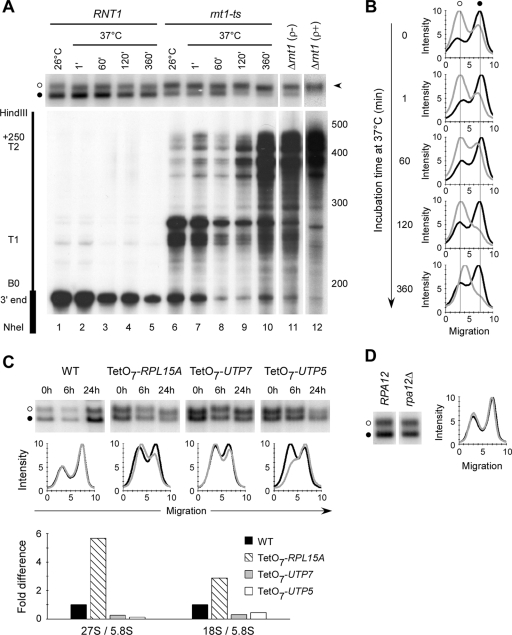
In vivo labeling suggests that deletion of RNT1 impairs both the transcription and the processing of rRNA. In order to further investigate the impact of Rnt1p on the regulation of rRNA gene transcription, we used psoralen photo-cross-linking to monitor the ratio of open to closed forms of chromatin in the rRNA gene repeats. An open form of chromatin is indicative of genes that are transcribed or poised for transcription by RNAPI, while a closed form is indicative of inactive genes (65). Yeast cells either carrying a deletion in RNT1 (Δrnt1) or expressing a temperature-sensitive allele (rnt1-ts) were harvested, and nuclei were prepared and photo-cross-linked with psoralen. The principle of psoralen cross-linking is that the preactive and active (nonnucleosomal or open) forms of the rRNA gene bind more psoralen than the inactive (nucleosomal or closed) rRNA gene. Consequently, psoralen cross-linked rRNA gene fragments from open genes have a slower migration rate on electrophoretic gels than do fragments from closed genes (14, 65). As shown in Fig. 5A, wild-type (RNT1) cells (lane 1) contained about 30% open and 70% closed forms of the rRNA gene, indicating that the majority of the rRNA genes under these growth conditions (at 26°C in synthetic medium) are inactive. Interestingly, the rRNA genes extracted from Δrnt1 cells were mostly in the open form (Fig. 5A, lanes 11 and 12), while the unrelated RNA polymerase II-transcribed genes (e.g., GLT1 or POL2) were not (data not shown).
FIG. 5.
Loss of Rnt1p activity increases the number of rRNA genes with an open chromatin conformation. (A) Cells carrying a wild-type copy of RNT1 or a temperature-sensitive allele (rnt1-ts) were shifted to 37°C, and aliquots were collected at different time points for RNA extraction or nucleus preparation and psoralen photo-cross-linking. Southern blot analysis of the cross-linked DNA (top) was performed with a probe corresponding to the 25S rRNA. The open and closed rRNA gene forms are indicated by open and black circles, respectively. The arrowhead on the right indicates the open form of the rRNA genes. The bottom part shows an RNase protection assay performed to assess the impact of Rnt1p inactivation on the processing of the 25S rRNA 3′ end (B0) as a control. The values on the right are molecular sizes in nucleotides. (B) The psoralen-cross-linked rRNA gene shown in panel A was quantified with a PhosphorImager, and the results are presented as migration profiles. The profiles of RNT1 DNA are in black, while those of rnt1-ts DNA are in gray. (C) Ribosomal protein Rpl15Ap, 5′-end processing factor Utp7p, and Utp5p, a member of the t-Utp complex, were expressed from tetracycline (TetO7)-controllable promoters, and the rRNA genes forms were examined after inhibition of the transcription of each gene. Psoralen cross-linking and Southern blot assays (top) were prepared as described for panel A, and the migration profiles (middle) of the cross-linked DNA were prepared as described for panel B. The migration profiles of DNA extracted after 0 and 24 h of transcription inhibition are shown as black and gray lines, respectively. The amounts of 27S pre-rRNA and 18S rRNA were examined by Northern blot analysis after 24 h of transcription inhibition, and the RNA amount was quantified and plotted relative to the amount of 5.8S rRNA (bottom). WT, wild type. (D) Psoralen cross-linking was performed with nuclei extracted from wild-type cells (RPA12) or cells lacking RPA12 (rpa12Δ). The Southern blot assay (left side) was prepared as described for panel A, and the migration profile (right side) was prepared as described for panel B. The profile of RPA12 DNA is in black, and that of rpa12Δ is in gray.
Deletion of RNT1 inhibits yeast growth, reducing the cell doubling time in culture from 3.6 to 12.4 h (at 26°C in synthetic medium) (7) and causes the cells to easily lose their mitochondria (data not shown). Thus, we examined the impact of mitochondrial loss and short-term depletion of Rnt1p on the two forms of rRNA gene chromatin. As shown in Fig. 5A, lanes 11 and 12, the rRNA gene chromatin phenotype is not altered by the presence or absence of mitochondria. To ensure that the changes in chromatin structure do not arise from unspecific long-term metabolic adaptation due to the slow growth of Δrnt1 cells, we followed the ratio of open versus closed rRNA gene chromatin after inactivation of the temperature-sensitive allele of Rnt1p (rnt1-ts). As shown in Fig. 5A and B, changes in the growth temperature did not affect the ratio of open to closed rRNA gene fractions of the wild type (RNT1), even after 6 h (Fig. 5A, lanes 1 to 5). Although there is already an increase in the number of open rRNA gene repeats extracted from rnt1-ts cells when they are grown at the permissive temperature (26°C) (Fig. 5A, lane 6), more rRNA gene repeats were open after 2 h at the restrictive temperature (37°C) (lane 9). An increase in the processing defects at B0 in this mutant strain is also observed at almost the same time. The results indicate that the inactivation of Rnt1p induces the opening of additional rRNA gene units, which suggests that the reduced rate of rRNA synthesis results from defects in later steps like transcription initiation or elongation.
Deletion of RNT1 could indirectly induce the opening of the rRNA gene chromatin by reducing ribosome stability, perturbing general rRNA processing, or by decreasing rRNA gene transcription. To examine these possibilities, we expressed three genes, with different documented effects on ribosome synthesis, from repressible promoters and monitored the impact of depleting each protein on rRNA gene chromatin. As shown in Fig. 5C, the conditional depletion of the ribosomal protein (Rpl15Ap), which decreases the stability of the ribosome LSU (40), did not increase the number of open rRNA genes despite a substantial reduction in the amount of LSU RNA. Similarly, depletion of 5′-end pre-rRNA processing factor Utp7p (17) did not induce a notable change in the ratio of the two forms of rRNA gene chromatin, even when the amount of 18S rRNA was reduced threefold. In contrast, depletion of Utp5p, a component of the t-Utp complex required for optimal rRNA gene transcription (22), reduced rRNA synthesis and the amount of the open rRNA gene form (Fig. 5C).
To determine whether the effect of RNT1 deletion on the rRNA gene chromatin is due to transcriptional readthrough, we deleted RPA12, the RNAPI subunit required for transcription termination (52), and monitored the ratio of the two rRNA gene forms. As shown in Fig. 5D, deletion of RPA12 did not alter the ratio of open to closed rRNA gene repeats, indicating that transcription readthrough alone does not alter the chromatin structure of the rRNA gene locus. We conclude that the observed changes in rRNA gene chromatin are specific for cells lacking Rnt1p and not an indirect response due to general perturbation of ribosome synthesis.
Mutations that disrupt the Rnt1p/RNAPI complex alter the rRNA gene chromatin conformation and reduce the efficiency of pre-rRNA processing.
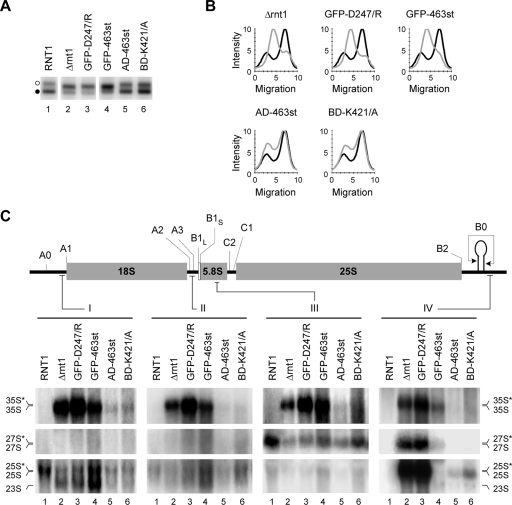
To better define the mechanism that induces the opening of rRNA gene chromatin, we introduced mutations that block Rnt1p catalytic activity or its interaction with RNAPI and monitored their impact on the state of the rRNA genes and pre-rRNA processing. As shown in Fig. 6A and B, mutations that impair Rnt1p catalysis (GFP [green fluorescent protein]-D247/R) or inhibit the nuclear localization and interaction with RNAPI (GFP-463st) increased the number of open rRNA gene repeats (Fig. 6A, lanes 3 and 4) and blocked pre-rRNA processing (Fig. 6C, lanes 3 and 4), as observed in Δrnt1 cells. Cells expressing a GFP-Rnt1p fusion exhibit a slight accumulation of 35S pre-rRNA that could only be detected after prolonged exposure, but the processing pattern was very similar to that of wild-type cells (data not shown). Interestingly, mutations that inhibit the interactions between Rnt1p and either one (BD-K421/A) or two (AD-463st) RNAPI subunits also altered the ratio of the two forms of rRNA gene (Fig. 6A, lanes 5 and 6) and reduced pre-rRNA efficiency (Fig. 6C, lanes 5 and 6), albeit moderately. These results suggest that the formation of a complex of RNAPI and Rnt1p is required for efficient pre-rRNA processing and maintaining the number of closed rRNA genes.
FIG. 6.
Mutations that disrupt the Rnt1p/RNAPI complex increase the number of open rRNA genes and reduce the efficiency of pre-rRNA processing. (A) The ratios of open to closed rRNA genes were examined in cells expressing different mutations that impair Rnt1p catalytic activity (GFP-D247/R), block the localization of Rnt1p in the nucleolus (GFP-463st), or disrupt the interaction with RNAPI (AD-463st and BD-K421/A). The open and closed rRNA genes are indicated by open and black circles, respectively, on the left. The profiles corresponding to the samples shown in panel A are presented in panel B. Wild-type profiles are in black, while those of the different mutants are in gray. (C) Northern blot analyses of rRNAs extracted from cells expressing the different Rnt1p mutants examined in panel A. The pre-rRNA processing intermediates were visualized with probes against sequences in the 5′ ETS, ITS1, 5.8S rRNA, or 3′ ETS (I to IV). Schematic representations of the primary rRNA transcript and probe positions are shown at the top. The positions of the different processing intermediates are indicated at the sides, with asterisks denoting 3′-extended species detected with probe IV.
DISCUSSION
This study describes a new link between rRNA gene transcription and rRNA processing. Two-hybrid assay and immunoprecipitation have indicated that the 25S rRNA processing enzyme Rnt1p binds at least two different subunits of RNAPI (Fig. 2). Deletion of Rnt1p inhibited rRNA synthesis and altered the chromatin conformation of the rRNA gene locus (Fig. 3, 4, 5, and 6). Reduced rRNA gene transcription in Δrnt1 cells is underscored by its increased sensitivity to transcription inhibitors (unpublished data). Deletion of Rnt1p increased the number of rRNA genes having an open chromatin conformation, indicating an increase in transcriptionally poised rRNA genes (Fig. 5 and 6). The opening of the rRNA genes was specifically induced by defects in Rnt1p cleavage activity or binding to RNAPI but not in response to a generic perturbation of ribosome synthesis. Together, the results suggest that the opening of the rRNA gene chromatin, transcription, and pre-rRNA processing are interdependent processes linked, at least in part, by physical interactions between Rnt1p and RNAPI.
It has been proposed that Rnt1p cleaves the primary rRNA transcript cotranscriptionally because of a failure to detect the unprocessed precursor in wild-type cells (54), perturbation of transcriptional termination upon the deletion of RNT1 (52), and the coimmunoprecipitation of Rnt1p with actively transcribed rRNA genes (28). The results presented here support this proposition and provide evidence for a physical interaction between Rnt1p and at least two subunits of RNAPI (Rpa12p and Rpa34p). The Rpa12p subunit is not shared with other RNA polymerases and is required for transcription termination (52, 68). Thus, an interaction between Rnt1p and Rpa12p may link transcription termination and processing. Indeed, the deletion of RNT1 or RPA12 results in transcription readthrough in vitro (52). However, while Rnt1p might be required for optimal transcription termination (52), Rpa12p itself is not required for pre-rRNA processing in vivo (unpublished data). Rnt1p also interacts with RNAPI-specific subunit Rpa34p, which is located near Rpa12p and forms a stable complex with Rpa49p. Deletions of any of these three subunits are not lethal, unless they occur in combination with the deletion of RPA14, whose product is another nonessential subunit residing on the opposite side of the RNAPI holoenzyme (21). Individual deletion of RPA12, RPA34, or RPA49 did not affect pre-rRNA processing, while the deletion of RPA14 delayed the processing of the 35S rRNA (unpublished data). Interestingly, deletions of RNT1 and RPA49 are synthetically lethal (16), suggesting that Rnt1p interacts both physically and functionally with the RNAPI subunit.
Ribosome biogenesis is a complex process that requires coordinated transcription, maturation, and assembly of many RNA and protein components (22, 57, 59). In this study, we provide a new link between early pre-rRNA processing events and the predisposition of rRNA genes to transcription. Normally, the number of rRNA genes predisposed to transcription remains constant during exponential growth and is decreased during cell starvation (15). Surprisingly, deletion of pre-rRNA processing factor Rnt1p induced the opening of most rRNA genes, suggesting that almost all rRNA gene repeats could be opened under special conditions. The increase in open rRNA genes did not lead to an increase in rRNA gene transcription, suggesting either that there are additional defects preventing transcription in Δrnt1 cells or that the transcription is controlled by a number of events in addition to the opening of the rRNA gene chromatin. Deletion of Rnt1p shifts the transcription profile but does not significantly reduce the transcription elongation rate, as judged by transcription run-on (52; Nick J. Proudfoot, personal communication), suggesting that the defect in RNA synthesis observed in vivo is due to defects in transcription steps other than elongation.
What is the signal that triggers chromatin changes in the rRNA gene locus? The available data suggest that the opening of rRNA genes is triggered by a failure in the processing of the 3′ end of the primary rRNA transcript. Perturbation of transcription initiation, ribosome stability, or processing of the pre-rRNA 5′ end failed to induce the chromatin opening of rRNA gene repeats (Fig. 5). In contrast, a point mutation that only disrupts the catalytic activity of Rnt1p fully reproduced the phenotype caused by RNT1 deletion and induced the chromatin opening of most rRNA genes (Fig. 6). These data suggest that the overall reduction in the number of ribosomes is not sufficient to promote changes in the number of transcriptionally competent rRNA genes. On the other hand, it appears that opening of rRNA gene chromatin is not enough to activate transcription in response to a reduction in the amount of mature rRNA. We suggest that new copies of rRNA genes are opened as a reaction to a continuous failure of the cells to produce the correct pre-rRNA 3′ end from a given set of active rRNA genes.
Acknowledgments
We thank R. J. Wellinger for critical reading of the manuscript and for yeast strain LLY112, T. R. Hughes for yeast strain R1158, B. Séraphin for plasmid pBS1539, and T. Ito for plasmids pGAD-RPA12 and pGAD-RPA34. We also thank M. Riva for providing antibodies against Rpa34p and Rpa49p. We are indebted to Nick J. Proudfoot for the communication of unpublished data and helpful discussion.
This work was supported by grant 216854-04 from the Natural Sciences and Engineering Research Council of Canada (NSERC). Support for the RNA group core was provided by CIHR. S.A. is a Chercheur Boursier Senior of the Fonds de la Recherche en Santé du Québec. M.T. and A.C. were supported by NSERC grant 326873-06.
Footnotes
Published ahead of print on 8 November 2007.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 173738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela, S., H. Igel, and M. Ares, Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85115-124. [DOI] [PubMed] [Google Scholar]
- 3.Banditt, M., T. Koller, and J. M. Sogo. 1999. Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 194953-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischler, N., L. Brino, C. Carles, M. Riva, H. Tschochner, V. Mallouh, and P. Schultz. 2002. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 214136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14115-132. [DOI] [PubMed] [Google Scholar]
- 6.Buhler, J. M., J. Huet, K. E. Davies, A. Sentenac, and P. Fromageot. 1980. Immunological studies of yeast nuclear RNA polymerases at the subunit level. J. Biol. Chem. 2559949-9954. [PubMed] [Google Scholar]
- 7.Catala, M., B. Lamontagne, S. Larose, G. Ghazal, and S. Abou Elela. 2004. Cell cycle-dependent nuclear localization of yeast RNase III is required for efficient cell division. Mol. Biol. Cell 153015-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalli, G., D. Bachmann, and F. Thoma. 1996. Inactivation of topoisomerases affects transcription-dependent chromatin transitions in rDNA but not in a gene transcribed by RNA polymerase II. EMBO J. 15590-597. [PMC free article] [PubMed] [Google Scholar]
- 9.Chanfreau, G., S. Abou Elela, M. Ares, Jr., and C. Guthrie. 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 112741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 173726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chédin, S., M. Riva, P. Schultz, A. Sentenac, and C. Carles. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 123857-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conconi, A., V. A. Bespalov, and M. J. Smerdon. 2002. Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc. Natl. Acad. Sci. USA 99649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conconi, A., M. Paquette, D. Fahy, V. A. Bespalov, and M. J. Smerdon. 2005. Repair-independent chromatin assembly onto active ribosomal genes in yeast after UV irradiation. Mol. Cell. Biol. 259773-9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57753-761. [DOI] [PubMed] [Google Scholar]
- 15.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 212331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davierwala, A. P., J. Haynes, Z. Li, R. L. Brost, M. D. Robinson, L. Yu, S. Mnaimneh, H. Ding, H. Zhu, Y. Chen, X. Cheng, G. W. Brown, C. Boone, B. J. Andrews, and T. R. Hughes. 2005. The synthetic genetic interaction spectrum of essential genes. Nat. Genet. 371147-1152. [DOI] [PubMed] [Google Scholar]
- 17.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahy, D., A. Conconi, and M. J. Smerdon. 2005. Rapid changes in transcription and chromatin structure of ribosomal genes in yeast during growth phase transitions. Exp. Cell Res. 305365-373. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1326-13. [DOI] [PubMed] [Google Scholar]
- 20.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 231558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gadal, O., S. Mariotte-Labarre, S. Chédin, E. Quemeneur, C. Carles, A. Sentenac, and P. Thuriaux. 1997. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol. 171787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 182506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge, D., B. Lamontagne, and S. Abou Elela. 2005. RNase III-mediated silencing of a glucose-dependent repressor in yeast. Curr. Biol. 15140-145. [DOI] [PubMed] [Google Scholar]
- 24.Ghazal, G., D. Ge, J. Gervais-Bird, J. Gagnon, and S. Abou Elela. 2005. Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals. Mol. Cell. Biol. 252981-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granneman, S., and S. J. Baserga. 2005. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr. Opin. Cell Biol. 17281-286. [DOI] [PubMed] [Google Scholar]
- 26.Grummt, I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 171691-1702. [DOI] [PubMed] [Google Scholar]
- 27.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- 28.Henras, A. K., E. Bertrand, and G. Chanfreau. 2004. A cotranscriptional model for 3′-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA 101572-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huet, J., J. M. Buhler, A. Sentenac, and P. Fromageot. 1975. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl. Acad. Sci. USA 723034-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102109-126. [DOI] [PubMed] [Google Scholar]
- 31.Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 971143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, H. S., J. Kawauchi, P. Braglia, C. M. Alen, N. A. Kent, and N. J. Proudfoot. 2007. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 14123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kharrat, A., M. J. Macias, T. J. Gibson, M. Nilges, and A. Pastore. 1995. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 143572-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13225-239. [DOI] [PubMed] [Google Scholar]
- 36.Kufel, J., B. Dichtl, and D. Tollervey. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamontagne, B., and S. Abou Elela. 2004. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 2792231-2241. [DOI] [PubMed] [Google Scholar]
- 38.Lamontagne, B., A. Tremblay, and S. Abou Elela. 2000. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 201104-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larose, S., N. Laterreur, G. Ghazal, J. Gagnon, R. J. Wellinger, and S. Abou Elela. 2007. RNase III-dependent regulation of yeast telomerase. J. Biol. Chem. 2824373-4381. [DOI] [PubMed] [Google Scholar]
- 40.Lee, J. C., B. Henry, and Y. C. Yeh. 1983. Binding of proteins from the large ribosomal subunits to 5.8 S rRNA of Saccharomyces cerevisiae. J. Biol. Chem. 258854-858. [PubMed] [Google Scholar]
- 41.Leulliot, N., S. Quevillon-Cheruel, M. Graille, H. Van Tilbeurgh, T. C. Leeper, K. S. Godin, T. E. Edwards, S. T. Sigurdsson, N. Rozenkrants, R. J. Nagel, M. Ares, and G. Varani. 2004. A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNase III. EMBO J. 232468-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucchini, R., U. Pauli, R. Braun, T. Koller, and J. M. Sogo. 1987. Structure of the extrachromosomal ribosomal RNA chromatin of Physarum polycephalum. J. Mol. Biol. 196829-843. [DOI] [PubMed] [Google Scholar]
- 43.Lucchini, R., and J. M. Sogo. 1992. Different chromatin structures along the spacers flanking active and inactive Xenopus rRNA genes. Mol. Cell. Biol. 124288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucchini, R., and J. M. Sogo. 1998. The dynamic structure of ribosomal RNA gene chromatin, p. 255-276. In M. R. Paule (ed.), Transcription of ribosomal genes by eukaryotic RNA polymerase I. Springer-Verlag and R. G. Landes Company, New York, NY.
- 45.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 127035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 11831-44. [DOI] [PubMed] [Google Scholar]
- 47.Moss, T. 2004. At the crossroads of growth control: making ribosomal RNA. Curr. Opin. Genet. Dev. 14210-217. [DOI] [PubMed] [Google Scholar]
- 48.Muller, M., R. Lucchini, and J. M. Sogo. 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5767-777. [DOI] [PubMed] [Google Scholar]
- 49.Ness, P. J., P. Labhart, E. Banz, T. Koller, and R. W. Parish. 1983. Chromatin structure along the ribosomal DNA of Dictyostelium. Regional differences and changes accompanying cell differentiation. J. Mol. Biol. 166361-381. [DOI] [PubMed] [Google Scholar]
- 50.Osheim, Y. N., S. L. French, K. M. Keck, E. A. Champion, K. Spasov, F. Dragon, S. J. Baserga, and A. L. Beyer. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16943-954. [DOI] [PubMed] [Google Scholar]
- 51.Peyroche, G., E. Levillain, M. Siaut, I. Callebaut, P. Schultz, A. Sentenac, M. Riva, and C. Carles. 2002. The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc. Natl. Acad. Sci. USA 9914670-14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prescott, E. M., Y. N. Osheim, H. S. Jones, C. M. Alen, J. G. Roan, R. H. Reeder, A. L. Beyer, and N. J. Proudfoot. 2004. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl. Acad. Sci. USA 1016068-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reeder, R. H. 1999. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 62293-327. [DOI] [PubMed] [Google Scholar]
- 54.Reeder, R. H., P. Guevara, and J. G. Roan. 1999. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol. 197369-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 171030-1032. [DOI] [PubMed] [Google Scholar]
- 56.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Rudra, D., J. Mallick, Y. Zhao, and J. R. Warner. 2007. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 274815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz, C., E. Gorab, M. F. Ruiz, J. M. Sogo, and J. L. Diez. 2007. Chromatin structure of ribosomal genes in Chironomus thummi (Diptera: Chironomidae): tissue specificity and behaviour under drug treatment. Chromosome Res. 15429-438. [DOI] [PubMed] [Google Scholar]
- 59.Schneider, D. A., A. Michel, M. L. Sikes, L. Vu, J. A. Dodd, S. Salgia, Y. N. Osheim, A. L. Beyer, and M. Nomura. 2007. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell 26217-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sogo, J. M., P. J. Ness, R. M. Widmer, R. W. Parish, and T. Koller. 1984. Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J. Mol. Biol. 178897-919. [DOI] [PubMed] [Google Scholar]
- 61.Sogo, J. M., and F. Thoma. 2004. The structure of rDNA chromatin, p. 73-87. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academics/Plenum Publishers, New York, NY.
- 62.Stefanovsky, V., F. Langlois, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21629-639. [DOI] [PubMed] [Google Scholar]
- 63.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 64.Tollervey, D., H. Lehtonen, M. Carmo-Fonseca, and E. C. Hurt. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toussaint, M., G. Levasseur, M. Tremblay, M. Paquette, and A. Conconi. 2005. Psoralen photocrosslinking, a tool to study the chromatin structure of RNA polymerase I-transcribed ribosomal genes. Biochem. Cell Biol. 83449-459. [DOI] [PubMed] [Google Scholar]
- 66.Tremblay, A. 2002. Étude de la fonction de la RNase III eucaryote et identification de ses partenaires cellulaires dans un criblage double-hybrides. M.S. thesis. Université de Sherbrooke, Sherbrooke, Ontario, Canada.
- 67.Tremblay, A., B. Lamontagne, M. Catala, Y. Yam, S. Larose, L. Good, and S. Abou Elela. 2002. A physical interaction between Gar1p and Rnt1p is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol. Cell. Biol. 224792-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Mullem, V., E. Landrieux, J. Vandenhaute, and P. Thuriaux. 2002. Rpa12p, a conserved RNA polymerase I subunit with two functional domains. Mol. Microbiol. 431105-1113. [DOI] [PubMed] [Google Scholar]
- 69.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33261-311. [DOI] [PubMed] [Google Scholar]
- 70.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24437-440. [DOI] [PubMed] [Google Scholar]
- 71.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 72.Wu, H., A. Henras, G. Chanfreau, and J. Feigon. 2004. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. USA 1018307-8312. [DOI] [PMC free article] [PubMed] [Google Scholar]