Abstract
Circadian regulation of gene expression plays a major role in health and disease. The precise role of the circadian system remains to be clarified, but it is known that circadian proteins generate physiological rhythms in organisms by regulating clock-controlled target genes. The estrogen receptor beta (ERβ) is, together with ERα, a member of the nuclear receptor superfamily and a key mediator of estrogen action. Interestingly, recent studies show that disturbed circadian rhythmicity in humans can increase the risk of reproductive malfunctions, suggesting a link between the circadian system and ER-mediated transcription pathways. Here, we identify a novel level of regulation of estrogen signaling where ERβ, but not ERα, is controlled by circadian clock proteins. We show that ERβ mRNA levels fluctuate in different peripheral tissues following a robust circadian pattern, with a peak at the light-dark transition, which is maintained under free-running conditions. Interestingly, this oscillation is abolished in clock-deficient BMAL1 knockout mice. Circadian control of ERβ expression is exerted through a conserved E-box element in the ERβ promoter region that recruits circadian regulatory factors. Furthermore, using small interfering RNA-mediated knockdown assays, we show that the expression levels of the circadian regulatory factors directly influence estrogen signaling by regulating the intracellular levels of endogenous ERβ.
Circadian rhythms of physiology and behavior are observed in almost all living organisms, ranging from bacteria to eukaryotes. These rhythms are driven by autonomous self-sustained cellular clocks and have a period of about 24 h (26, 29, 43). In mammals, the main circadian pacemaker is localized in the suprachiasmatic nucleus of the ventral hypothalamus (13). This master clock in the suprachiasmatic nucleus is entrained primarily by light but can be reset by exposure to other stimuli, for example, melatonin and certain steroid hormones (28, 40); the master clock coordinates the rhythms of multiple local clocks in peripheral tissues and cells through both neural and hormonal signals (19, 38). On the other hand, apart from being governed by the master clock, the peripheral clocks are also adjusted by nonphotic time cues, for example, feeding time, hormones, growth factors, and metabolites such as glucose and others (41). Moreover, circadian rhythms have been identified even in cells in culture, which can be synchronized by treatment with high concentrations of serum (2), hormones, and growth factors (3).
The core molecular oscillator of the circadian clock consists of an evolutionarily conserved feedback loop that includes both a positive and a negative set of components. The positive loop of the feedback system includes the CLOCK and BMAL1 transcription factors. These proteins are members of the basic helix-loop-helix (bHLH)-Per-ARNT-Sim (PAS; ARNT is aryl hydrocarbon receptor [AHR]nuclear translocator) family of transcription factors which includes key regulatory proteins like the hypoxia-inducible factor HIF-1 and endothelial PAS, which regulate the cellular response to low oxygen tension, as well as the master regulator of the cellular response to xenobiotic insult, the AHR. In addition, the bHLH-PAS family of transcription factors includes the members of the ARNT subfamily which are key heterodimeric partners to bHLH-PAS transcription factors such as AHR, HIF-1, and CLOCK. In fact, previous studies have shown that CLOCK needs to form heterodimeric complexes with BMAL1 (also known as ARNT-3) to acquire DNA binding activity and thus the ability to induce the transcription of clock target genes by binding to E-box motifs (CACGTG) in their promoters. Among these CLOCK-BMAL1 target genes, there are the negative circadian regulators PERIOD (PER1 and PER2) and CRY (CRY1 and CRY2). PER and CRY form multimeric complexes, translocate to the nucleus, and inhibit transcription of their own and other clock-controlled genes (CCGs) by interacting with CLOCK-BMAL1 heterodimers, thereby inhibiting their activity. In addition, during this process, posttranslational modifications such as phosphorylation, ubiquitination, and sumoylation are involved in regulating the subcellular location, stability, and activity of circadian proteins. Therefore, this transcriptional/translational feedback system generates and maintains circadian rhythms (1, 13, 25, 42). Other factors such as the nuclear orphan receptors Rev-erbs and retinoid orphan receptors as well as DEC (differentially expressed in chondrocytes) factors are also playing an important role in the rhythmic activity of the molecular pacemaker (10, 20).
Many autonomic body functions are known to be regulated by this internal clock system, such as the sleep-wake cycle, core body temperature, hormonal secretion rhythms, feeding time, and heart rate (12). Importantly, disturbances of normal circadian rhythms (e.g., shift work) can lead to increased risk of illnesses (9) such as breast cancer (18, 34) and various other health disorders (8). There is growing evidence that disturbed circadian rhythmicity is also associated with reproductive malfunctions (5), such as increased risks of spontaneous abortion and irregular menstruation, which have been observed in female shift workers (23). However, the mechanisms by which the circadian system influences these processes are still unclear. One possibility is dysregulation of certain CCGs, which are under the direct regulation of clock-regulatory proteins.
Estrogen receptor β (ERβ) (NR3A2) was discovered in 1996 and belongs to the nuclear receptor (NR) superfamily, a large family of structurally related ligand-inducible transcription factors (15, 24, 39). Together with the ERα (NR3A1) isoform, ERβ mediates the biological action of estrogens and plays important roles in many biological processes in both females and males, including not only growth, differentiation, and functioning of reproductive system but also many other functions in several tissues including the respiratory system, the central nervous system, immune system, and skeletal muscle (32) (16).
Ligand binding is an essential regulatory step in the activation process of ERs. Upon the binding of ligand, activated ERs interact with estrogen-responsive elements (EREs) located in regulatory regions of estrogen target genes and initiate transcription (33, 39). Furthermore, as for other NRs, ER-mediated gene transcription involves recruitment of auxiliary coregulatory proteins, namely, corepressors mediating the repression of unliganded receptors, as well as coactivators potentiating NR activity either in a ligand-independent or a ligand-dependent manner (44). Although both ER isoforms are widely distributed throughout the body, they have distinct expression patterns and levels in different tissues and cell types (27). This suggests that the intracellular levels of ER isoforms are significant factors in ER-mediated transcription, and mechanisms controlling the expression of the ERs are key to understanding estradiol (E2) signaling. A number of studies have addressed molecular mechanisms regulating the transcription of the ERα gene, involving participation of several distinct promoters (14, 45, 48). However, the mechanisms regulating the expression of the ERβ gene are currently unclear.
In this study, we identify an evolutionarily conserved E box in the 5′ promoter region of ERβ. This E box recruits circadian-regulatory proteins to the ERβ promoter, leading to a circadian oscillation of ERβ expression in both mouse tissues and synchronized cultured cells. Furthermore, our experiments using small interfering RNA (siRNA) suggest that the expression levels of circadian proteins influence ERE transactivation in HC11 cells. Taken together, our studies demonstrate that expression of ERβ is under circadian regulation and that this regulation is important in determining the cellular response to estrogens.
MATERIALS AND METHODS
Plasmids.
A fragment of the mouse ERβ (mERβ) promoter from −1652 to +281 was obtained by PCR amplification from genomic DNA using Taq Polymerase (Roche Diagnostics, Indianapolis, IN) with primers 5′-CGACGCGTTTCTATCTCACAAATGGAGAGAACC −3′ and 5′-CGCTCGAGCTAAATGCAGACACGTACTTTCCTC −3′ incorporating MluI and XholI restriction sites (underlined), respectively. The cleaved PCR product was cloned into the pGL3 vector. The sequence of the promoter fragment was verified by DNA sequencing. The pGL3 human ERβ (hERβ) construct was kindly provided by Silke Kietz. The mutations in E-box sites were created using a Quikchange site-directed mutagenesis kit (Qiagen); the E box (CACGTG) was mutated to GGATCC (22). pCMV CLOCK (where CMV is cytomegalovirus) and pCDNA BMAL1 were gifts from Y. Fujii-Kuriyama (Tohuko University, Sendai, Japan). pHSG396 PER1 and pBCIISK+ PER2 were gifts from Hajime Tei (Institute of Medical Science, University of Tokyo).
Transient transfection and luciferase assays.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (FBS; Invitrogen) in 12-well plates 24 h before transfection and maintained at 37°C. All transient transfection assays were performed using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Cells were transfected with 200 ng of ERβ-luciferase reporter, 50 ng of pCMV5-β-galactosidase, 50 ng of pCMV CLOCK, 50 ng of pCDNA BMAL1, and 250 ng or 500 ng of pHSG396 PER1 or pBCIISK+ PER2 expression plasmids as indicated in Fig. 4. After 48 h, cells were harvested, and luciferase and β-galactosidase activities were determined using a luciferase assay kit (BioThema, Dalarö, Sweden) and a Galacto Light Plus kit (Tropix), respectively.
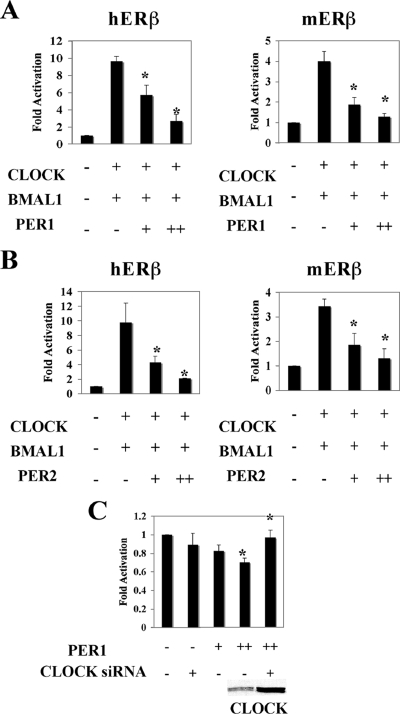
FIG. 4.
PER1 and PER2 inhibit ERβ transactivation through CLOCK-BMAL1. (A and B) Cotransfection of PER1 (A) or PER2 (B) represses expression of the mERβ- and hERβ-regulated luciferase reporter gene constructs activated by CLOCK-BMAL1 in HeLa cells. + and ++ correspond to 250 and 500 ng of PER1 and PER2 expression plasmids, respectively. (C) CLOCK siRNA blocks the ability of PER1 to repress basal activity of pGL3 mERβ in HC11 cells. CLOCK expression levels in HC11 cells were assessed by Western blotting. Results were compared to basic luciferase activity of pGL3-ERβ alone, which was arbitrarily set to be 1, and expressed as means ± standard deviations of three independent experiments performed in triplicate.*, P < 0.05 (Student's t test).
Real-time PCR.
Total RNA was purified using TRIzol reagent (Invitrogen, Carlsbad, CA). Two micrograms of total RNA from each sample was reverse transcribed into cDNA with a SuperScript First-Strand Synthesis System (Invitrogen) according to manufacturer's instructions. mRNA transcripts were quantified by a Sybr green reverse transcription-PCR kit (Applied Biosystems). The sequences of the primers are as follows: ERβ, 5′-TCAGGCACATCAGTAACAAGG-3′ (forward) and 5′-CTCTGTCGAGCAGCACTCAG-3′ (reverse); PER1, 5′-GAAAGAAACCTCTGGCTGTTCCT-3′ (forward) and 5′-GGAATGTTGCAGCTCTCCAAA-3′ (reverse); ERα, 5′-GTGCCTGGCTGGAGATTCTG-3′ (forward) and 5′-GAGCTTCCCCGGGTGTTC-3′ (reverse); Rev-erbα, 5′-CATGGTGCTACTGTGTAAGGTGTGT-3′ (forward) and 5′-CACAGGCGTGCACTCCATAG-3′ (reverse); 36B4, 5′-ACCTCCTTCTTCCAGGCTTT-3′ (forward) and 5′-CCCACCTTGTCTCCAGTCTTT-3′ (reverse).
Real-time PCR assays were conducted using an Applied Biosystems 7500 fast real-time PCR system. The two-step amplification protocol consisted of a 2-min incubation step at 50°C and 10 min at 95°C, followed by target amplification via 40 cycles of 15 s at 95°C and 1 min at 60°C. All real-time PCRs were performed in duplicate, and results were normalized to the expression level of the 36B4 gene (21). In a time course study, the gene expression level at time zero was set to be 1, and results from other time points were related to it. In the RNA interference (RNAi) study, gene expression in the CLOCK siRNA-treated group was related to that in the scrambled siRNA-treated group.
Chromatin immunoprecipitation (ChIP) assay.
HC11 cells were maintained in RPMI 1640 medium with 10% FBS until 80% confluence. Cells were then fixed in 1% formaldehyde for 15 min at room temperature and quenched for 5 min by adding glycine to a final concentration of 0.125 M. The cells were washed twice with phosphate-buffered saline and harvested by centrifugation at 200 × g for 5 min.
The cell pellets were suspended in 600 μl of lysis buffer (50 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% Na-deoxycholate, 150 mM NaCl, and protease inhibitor). ChIP assays were performed essentially following the protocol described in Brunnberg et al. (6). Immunoprecipitation was done with the following antibodies: anti-mouse immunoglobulin G (Sigma), anti-CLOCK, anti-BMAL1 (Abcam), anti-PER1 (Alpha Diagnostic), and anti-acetyl histone H3 (Ac-H3) and anti-Ac-H4 (Upstate Biotechnology). Indicated fragments (see Fig. 5A) were amplified by PCR, and PCR products were separated on 2% agarose gel, purified, and verified by sequencing. The putative CLOCK-BMAL1 target region in the ERβ promoter was amplified with following primer set: forward, 5′-TGACTGTGAAGTGGCTGGAG-3′; reverse, 5′-GCTGGAGAAGCTGCAAAGAC-3′. For the positive control, the promoter region of PER1 containing the proximal E-box enhancer was amplified with the following primer set: forward, 5′-ATCCTCCCTGAAAAGGGGTA-3′; reverse, 5′-GGATCTCTTCCTGGCATCTG-3′. For negative control, the −1997 to −1763 region of the ERβ promoter was amplified with the primer set 5′-AAATCCTTCCACCTCCTTGG-3′ (forward) and 5′-CCCTGTGTCCTTTCCTGTGT-3′ (reverse). For real-time PCR amplification, the primer set 5′-ACATCCAGTGGATCTGGTTGC-3′ and 5′-CAAACCGGGAGCCAGAGA-3′ was used for detecting the CLOCK-BMAL1 target region of the ERβ promoter. Results were normalized to input and to the time zero sample.
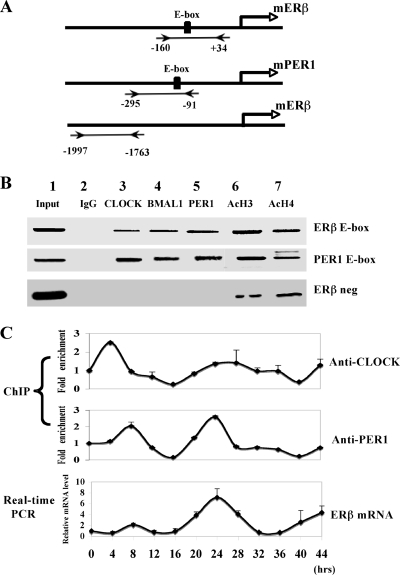
FIG. 5.
Endogenous circadian proteins are recruited to ERβ promoter and drive the rhythmic expression of ERβ in HC11 cells. (A) Schematic illustration of the promoter of mERβ and mouse PER1 (mPER1). Regions amplified by PCR after ChIP assay are indicated. (B) ChIP assay in HC11 cells performed with CLOCK, BMAL1, and PER1 antibodies. Specific regions of the ERβ promoter or PER1 promoter are amplified. The accuracy of ERβ sequence was verified by sequencing as described in Materials and Methods. Presence of Ac-H3 and Ac-H4 on the ERβ promoter was monitored with H3/H4 antibodies. Aliquots of chromatin obtained before immunoprecipitation were analyzed as input. Data are representative of at least three independent experiments. ERβ neg, negative control (−1997 to −1763 region of the ERβ promoter). (C) HC11 cells were synchronized by treatment with 50% horse serum and collected at different time points as shown. ChIP assays and real-time PCR analysis were performed with PER1 and CLOCK antibodies to monitor recruitment at different circadian time points. Relative ERβ mRNA levels at corresponding time points were monitored to compare mRNA expression at different circadian time points in synchronized HC11 cells. Results were compared with the activity of controls (samples at time point 0), which was arbitrarily set to be 1, and are means ± standard errors of the mean (n = 3). ANOVA analyses were used to compare each data set; variations were significant with a P of <0.001.
Serum shock.
HC11 cells were maintained in RPMI 1640 medium supplemented with 10% FBS until confluent and kept in serum-free medium for 2 h before synchronization. At time point zero, the medium was changed to 50% horse serum (Invitrogen)-rich medium, and after 2 h, the medium was replaced with serum-free RPMI 1640 medium. At indicated time points (see Fig. 5C), cells were fixed with formaldehyde for ChIP assays or harvested in TRIzol reagent (Gibco-BRL) for RNA preparation.
RNAi.
The sequences of the CLOCK siRNA oligonucleotides were GGGAUGUAGCACUAAUAAAdTdT and UUUAUUAGUGCCUACAUCCCdTdT (where dT is deoxyribosylthymine); the sequences of the PER1 siRNA oligonucleotides were CGCUCGCCCUGGCCAAUAAdTdT and UUAUUGGCCAGGGCGAGCGdGdG (synthesized by Qiagen). The sequences of the scrambled siRNA oligonucleotides were GGACAAUAGUAACAGUGUAdTdT and UACACUGUUACUAUUGUCCdTdT (synthesized by DNA Technology A/S). At about 50% confluence, HC11 cells or HC11 cells with a stably transfected 3×ERE-TATA-luciferase reporter construct (HC11-3×ERE) were transfected with Lipofectamine and plus reagent (Invitrogen) in phenol red-free, serum-free RPMI 1640 medium. Four hours after transfection, the medium was exchanged with phenol red-free medium supplemented with 5% dextran-coated, charcoal-treated FBS. After 24 h, for real-time PCR and Western blotting, cells were harvested, and RNA and whole-cell extracts were prepared; for reporter assays, cells were treated with indicated ligands (see Fig. 8) for another 24 h before luciferase activity was determined.
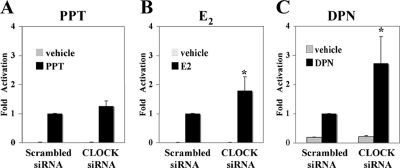
FIG. 8.
siRNA-mediated CLOCK knockdown in HC11-3×ERE luciferase cells influences ERE-dependent transcriptional activity. After CLOCK siRNA transfection, HC11-3×ERE cells were incubated for 24 h and treated with 10 nM PPT (A), 10 nM E2 (B), or 100 nM DPN (C) for another 24 h before luciferase measurement. E2- and DPN (ERβ selective ligand)-induced transcriptional activity but not PPT (ERα selective ligand)-induced transcriptional activity was upregulated upon cotransfection of siRNA toward CLOCK. Activity of scrambled siRNA-transfected groups was arbitrarily set to be 1; data are expressed as means standard deviations of three independent experiments performed in triplicate (*, P < 0.05; Student's t test).
Western blot analysis.
The protein lysate of HC11 cells was prepared in radioimmunoprecipitation assay buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, 0.1 mg/ml phenylmethylsulfonyl fluoride) including 1× protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). About 20 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose transfer membranes (Whatman GmbH, Germany). Blots were probed with phosphate-buffered saline containing 5% milk and the following antibodies and dilutions: anti-CLOCK antibody (Abcam Inc., Cambridge, MA) at 1:2,000, anti-PER1 antibody (Alpha Diagnostic) at 1:1,000, anti-β-actin antibody (Sigma-Aldrich, Stockholm, Sweden) at 1:20,000, anti-ERβ antibody (Abcam Inc., Cambridge, MA) at 1:1,000, and anti-ERα antibody (Santa Cruz Biotechnology) at 1:1,000. Protein-antibody complexes were detected by an ECL chemiluminescence system (Amersham Biosciences, Buckinghamshire, United Kingdom).
Animals.
All animal experiments were conducted in compliance with CNRS ethical guidelines on animal handling or the Rhône-Alpes Ethical Committee for Animal Experiments. Male wild-type (WT) mice and BMAL1 knockout (KO) mice (kindly provided by C. A. Bradfield, Wisconsin) in the C57BL/6j background were used at 8 to 12 weeks of age, before they developed arthropathy (7). Heterozygous animals were crossed to generate KO mice and WT littermates. Animals were exposed to a light-dark (LD) cycle of 12 h of light and 12 h of dark (12 h:12 h), housed in a temperature- and humidity-controlled environment, and fed ad libitum; under these conditions zeitgeber time zero (ZT0) is light-on time. For the constant darkness (dark-dark [DD] cycle) experiment, lights were kept off from ZT0 for 24 h. For the experiment shown in Fig. 2, male WT mice were housed under LD conditions with 12 h:12 h cycles for 15 days and then exposed to either LD conditions or DD conditions for 3 days(see Fig. 2A). Lung and quadriceps muscle were dissected at the indicated time points and snap frozen in liquid N2 and stored at −80°C until use.
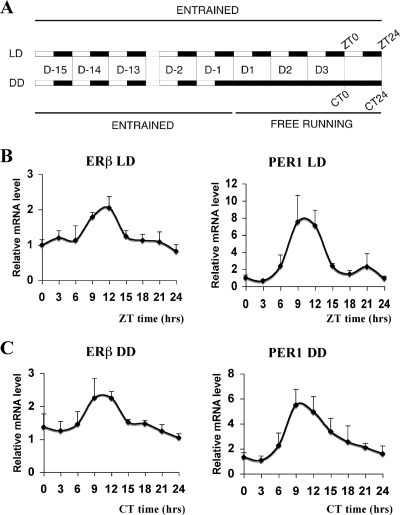
FIG. 2.
Circadian expression of ERβ in mouse lung. (A) Schematic presentation of the experimental setup. WT male mice were subjected to a LD 12 h:12 h cycle for 15 days and then exposed to either to LD or DD conditions for 3 days. (B) mRNA levels of ERβ and PER1 in lung under LD conditions were then quantified by real-time PCR at nine circadian times (ZT0, ZT3, ZT6, ZT9, ZT12, ZT15, ZT18, ZT21, ZT24). (C) mRNA levels of ERβ and PER1 in lung for DD were quantified by real-time PCR at nine circadian times (CT0, CT3, CT6, CT9, CT12, CT15, CT18, CT21, and CT24). The expression level at ZT0 or CT0 was arbitrarily set to be 1. Results are shown as the means ± standard deviations of values for four animals at each time point. ANOVA analyses were used to compare each set of data; variations were significant at a P of <0.05.
Statistical analysis.
The relative ERβ expression levels among different time groups were compared by one-way analysis of variance (ANOVA). Transfection experiments shown in Fig. 3 were assessed by two-way ANOVA. Other experiments were assessed by a Student's t test. The statistical analysis was performed using R (www.r-project.org), and P values are indicated in figure legends.
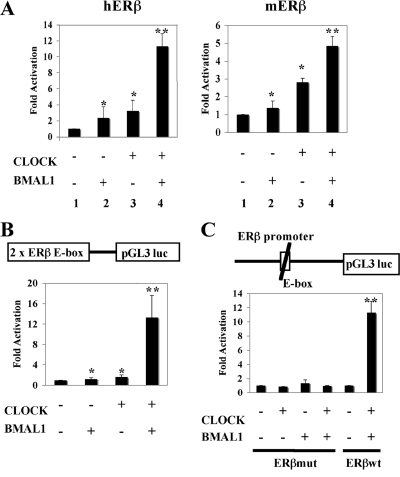
FIG. 3.
CLOCK-BMAL1 heterodimer activates the promoter activity of ERβ gene via the E box. (A) Cotransfection of CLOCK and BMAL1 activates expression of a luciferase reporter construct flanked by the ERβ promoter region of human and mouse in HeLa cells. (B) The minimal 2×ERβ E box (schematically presented) luciferase construct (pGL3 luc) is transcriptionally activated in the presence of both CLOCK and BMAL1 in HeLa cells. (C) Mutation of the ERβ E-box promoter construct impairs the ability of CLOCK and BMAL1 to activate transcription of the luciferase reporter gene construct. All results presented here are compared to reporter luciferase activity of controls (pGL3-hERβ, pGL3-mERβ, pGL3-2×ERβ E box, or pGL3-ERβmut alone), which was arbitrarily set to be 1, and expressed as means ± standard deviations of three independent experiments performed in triplicate. Data were analyzed by using two-way ANOVA. *, the contribution of CLOCK or BMAL1 is significant with a P value of <0.01; **, statistically significant interaction between CLOCK and BMAL1 with a P value of <0.01.
RESULTS
ERβ mRNA levels oscillate with a circadian pattern in mouse lung.
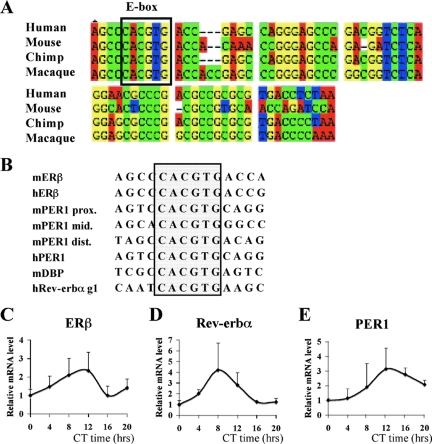
It is well known that E2 signaling pathways can be regulated at multiple levels, such as the intracellular concentration of either ER isoform. Considerable information is available regarding the promoter organization of ERα. However, little is known about the regulation of ERβ expression. Therefore, we decided to screen the 5′ region of the ERβ promoter from different species to identify evolutionarily conserved regulatory elements. We found a conserved E-box motif and flanking sequences in the proximal promoter region of ERβ (Fig. 1A). Interestingly, this region has considerable sequence identity to regulatory sequences of genes that are under circadian regulation (Fig. 1B). Therefore, we decided to test if ERβ expression is also under circadian control. For this purpose, we analyzed the expression of ERβ in mouse lung, since ERβ is the predominant ER isoform in this tissue. In the first 24 h when mice were kept in constant darkness, quantitative real-time PCR analysis of ERβ mRNA showed a circadian oscillation pattern, with a robust peak at circadian time CT12 (CT prefixes are used for DD times) (Fig. 1C). At the same time, we assessed the expression patterns of two well-known clock genes, Rev-erbα (Fig. 1D) and the negative circadian regulator PER1 gene (Fig. 1E). Taken together, these experiments suggest that ERβ can display a circadian oscillatory expression pattern and perhaps a circadian output. If ERβ expression is under the control of an endogenous oscillator, then the oscillation should remain in the absence of external synchronizer. To test this hypothesis, we kept the mice for 3 days in either LD or DD (Fig. 2A) conditions and measured ERβ expression at different circadian time points. Interestingly, we observed a rhythmic expression of ERβ under LD conditions with a peak at ZT12 (Fig. 2B), and this oscillation was maintained after 3 days in constant darkness (Fig. 2C). These experiments demonstrate that ERβ is a CCG in lung and suggest that ERβ might be, as Rev-erbα and PER1 (49), a direct target of the CLOCK-BMAL1 heterodimer in lung.
FIG. 1.
Conserved CLOCK-BMAL1 binding site of the E box in the ERβ promoter and the circadian expression of ERβ in mouse lung. (A) Sequence comparison of the proximal ERβ promoter regions from human, mouse, chimp and macaque. The E-box element is framed. (B) Alignment of the E boxes found in hERβ and mERβ promoters with E boxes found in various circadian promoters. The E-box element is framed. (C to E) mRNA levels of ERβ, Rev-erbα, and PER1 in WT male mouse lung were quantified by real-time PCR at six circadian times (CT0, CT4, CT8, CT12, CT16, and CT20). The expression level at CT0 was arbitrarily set to be 1. Results are shown as the means ± standard deviations of four animals at each time point. ANOVA analyses were used to compare each set of data; variations were significant at a P value of <0.05.
CLOCK-BMAL1 activates ERβ transcription via E-box enhancer.
Important gene regulatory sequences are usually conserved across different species. Alignment of the human, chimp, macaque, and mouse ERβ gene sequences revealed a highly conserved E-box (CACGTG) motif in the proximal promoter region (Fig. 1A). Since an E-box motif is the identified binding site for the circadian regulator CLOCK-BMAL1 heterodimer, we decided to determine whether the CLOCK-BMAL1 heterodimer could regulate the activity of ERβ promoter. To test this possibility we performed transient cotransfection assays in HeLa cells. We cotransfected a luciferase reporter gene fused to the human or mouse ERβ promoter including the E-box element (Fig. 3A) together with expression plasmids for CLOCK and BMAL1. Following transfection, cells were grown for 48 h and lysed, and luciferase activity was determined as described in Materials and Methods. Interestingly, while CLOCK or BMAL1 alone induced only a weak luciferase signal from the ERβ promoter (Fig. 3A, bars 2 and 3), coexpression of both CLOCK and BMAL1 strongly increased the luciferase signal from both hERβ and mERβ promoters 11- or 5-fold, respectively (Fig. 3A, bars 4, respectively), suggesting that the CLOCK-BMAL1 heterodimer is involved in the regulation of ERβ expression. Moreover, similar results were observed in HC11 cells (data not shown). To verify that the E-box motif in the ERβ promoter is necessary to mediate the effects of the CLOCK-BMAL1 heterodimer, we constructed a reporter plasmid carrying two copies of the ERβ-E box (2×ERβ-E box) (Fig. 3B) as well as E-box mutants of the ERβ promoter reporter plasmid (Fig. 3C) and transfected them together with CLOCK and BMAL1 into HeLa cells. The transcription of 2×ERβ-E box was dramatically activated (12-fold) in the presence of both CLOCK and BMAL1 (Fig. 3B), whereas the mutated version of the E-box motif, where CACGTG was mutated to GGATCC in the context of the natural promoter, failed to mediate transcriptional activation by CLOCK-BMAL1 (Fig. 3C, ERβmut). As shown previously, the original E-box motif of the ERβ gene efficiently mediated the CLOCK-BMAL1 transcriptional response (Fig. 3C, ERβwt). These experiments show that the ERβ promoter contains a functional element conferring responsiveness to the CLOCK and BMAL1 core clock-positive components.
PER represses the CLOCK-BMAL1-induced ERβ transcription.
Previous studies have shown that the circadian feedback mechanism is composed of two limbs: the positive drivers of circadian expression are bHLH-PAS proteins BMAL1 and CLOCK while the repressors of CLOCK-BMAL1 transcriptional activity are PER and CRY (25). Inhibition by PER represents a negative feedback since PER itself is regulated by the CLOCK-BMAL1 complex. We therefore tested if CLOCK-BMAL1-dependent transactivation of the ERβ promoter could be antagonized by the negative limb components. For this purpose we introduced expression plasmids of CLOCK and BMAL1 together with PER into HeLa cells. ERβ promoter-regulated transcription of the luciferase reporter construct was strongly repressed by both PER1 (Fig. 4A) and PER2 (Fig. 4B). Similar results were observed in HC11 cells (data not shown). This suggests that ERβ expression is regulated by both the positive and negative circadian regulators through the E-box motif. In addition, introduction by transient transfection of PER1 into HC11 cells, a mouse mammary epithelial cell line that expresses endogenous circadian proteins (data not shown) and ERs (11, 30), reduced the basal expression of the ERβ promoter-regulated luciferase reporter construct in a dose-dependent manner (Fig. 4C). Moreover, the negative effect of PER1 on ERβ transcription in HC11 cells was abolished in the presence of siRNA against CLOCK (Fig. 4C), suggesting that the repression by PER1 is CLOCK-BMAL1 dependent.
Endogenous circadian proteins are recruited to the ERβ promoter and drive the rhythmic expression of ERβ in mammary epithelial HC11 cells.
The transient transfection experiments presented above suggest that the circadian system regulates ERβ expression through the conserved E-box region of the ERβ promoter. We first evaluated HC11 cells as a suitable system to assess circadian rhythms and tested the presence of the circadian regulators CLOCK, BMAL1, CRY1, and PER1. Using Western blot experiments (data not shown), we could show that all these factors are present in HC11 cells, suggesting that they represent a suitable cellular system to study circadian cycling. Using ChIP assays, we then used ERβ-expressing HC11 cells as a model system to test whether the circadian regulators accomplish this through direct interaction with the promoter. We utilized antibodies against CLOCK, BMAL1, and PER1 for immunoprecipitation; we also included antibodies against Ac-H3 and Ac-H4 to detect transcriptionally active open chromatin and mouse immunoglobulin G as a negative control. In addition, the promoter region of PER1 was included as a positive control since PER1 is a key core gene (13), and the −1997 to −1763 promoter region of ERβ was included as a negative control (Fig. 5A). Interestingly, both the positive circadian proteins CLOCK (Fig. 5B, lane 3) and BMAL1 (Fig. 5B, lane 4) and the negative circadian protein PER1 (Fig. 5B, lane 5) were efficiently recruited to the ERβ promoter in nonsynchronized HC11 cells. Ac-H3 and Ac-H4 (Fig. 5B, lanes 6 and 7) showed strong association with this promoter region as well, indicating ongoing transcription. Furthermore, these recruitments were also observed on the PER1 promoter (Fig. 5B), whereas no obvious recruitment was observed to the −1997 to −1763 region of the ERβ promoter, demonstrating the specificity of the analysis (Fig. 5B, ERβ neg). In summary, these experiments together with our transient transfection studies indicate that ERβ expression is under circadian control and is modulated by both the positive and the negative limbs of the circadian clock pacemaker. In addition, we show that the target sequence necessary to impose circadian control of ERβ overlaps with the conserved E-box element present in the ERβ promoter.
Previous studies have shown that exposure of cultured cells to a serum shock synchronizes their clocks (2). We applied this treatment in combination with ChIP assays to analyze if the recruitment of the circadian transcription factors to the ERβ promoter in HC11 cells would follow a circadian rhythmic pattern. In addition, we monitored ERβ gene expression to assess if ERβ mRNA levels oscillate in consonance with the presence of different circadian factors. For this purpose ERβ expression was monitored by real-time PCR analysis. Confluent HC11 cells were treated with medium containing 50% horse serum for 2 h (time points 0 to 2 h) and were subsequently cultured in serum-free medium. Samples were collected at different time points, as indicated on Fig. 5C, and ChIP assays were performed. In parallel experiments, cells were collected, the mRNA was prepared, and real-time PCR analysis was performed. Interestingly, the binding of PER1 in the ERβ promoter (Fig. 5C, middle panel) fluctuated in parallel with the rhythmic expression of ERβ mRNA (Fig. 5C, bottom panel), with the peak at 24 h after serum shock; however, we observed no obvious rhythm in the recruitment of CLOCK (Fig. 5C, top panel). Thus, these observations suggest that recruitments of both positive and negative circadian regulators are important in ERβ regulation; however, these regulators confer different effects on the circadian expression of ERβ.
ERβ expression is increased by siRNA-mediated depletion of CLOCK or PER1.
The experiments presented above provide clear evidence that expression of the ERβ gene is under circadian control. However, these experiments do not provide insights as to which component of the circadian clock is the main driver of ERβ cycling expression. In an effort to study the functional consequences of this regulation on E2 transcriptional signaling and to identify the main regulator of ERβ cycling, we used RNAi technology to target individual components of the circadian clock and to assess the effects on the intracellular levels of ERβ. Following introduction of the siRNA against CLOCK or scrambled siRNA as a negative control into HC11 cells, we measured the intracellular concentrations of CLOCK, PER1, and actin (Fig. 6A) and ERβ (Fig. 6B) and ERα (Fig. 6C). Western blotting experiments showed that cells treated with CLOCK siRNA showed a reduced level of PER1 (Fig. 6A), as expected. Remarkably, real-time PCR and Western blotting experiments demonstrated that ERβ expression was dramatically increased both at the mRNA (Fig. 6B, graph) and protein (Fig. 6B, blot) levels, whereas, in contrast, the expression of ERα did not change significantly (Fig. 6C). Furthermore, when we knocked down PER1 in this cell line (Fig. 6D, top panel), ERβ expression was also upregulated (Fig. 6D), whereas no change was observed for ERα expression (Fig. 6D). These experiments show that only ERβ and not ERα is under the control of the circadian system in HC11 cells. Furthermore, the observations suggest that the negative circadian regulator PER could be the main driver of the circadian expression of ERβ.
FIG. 6.
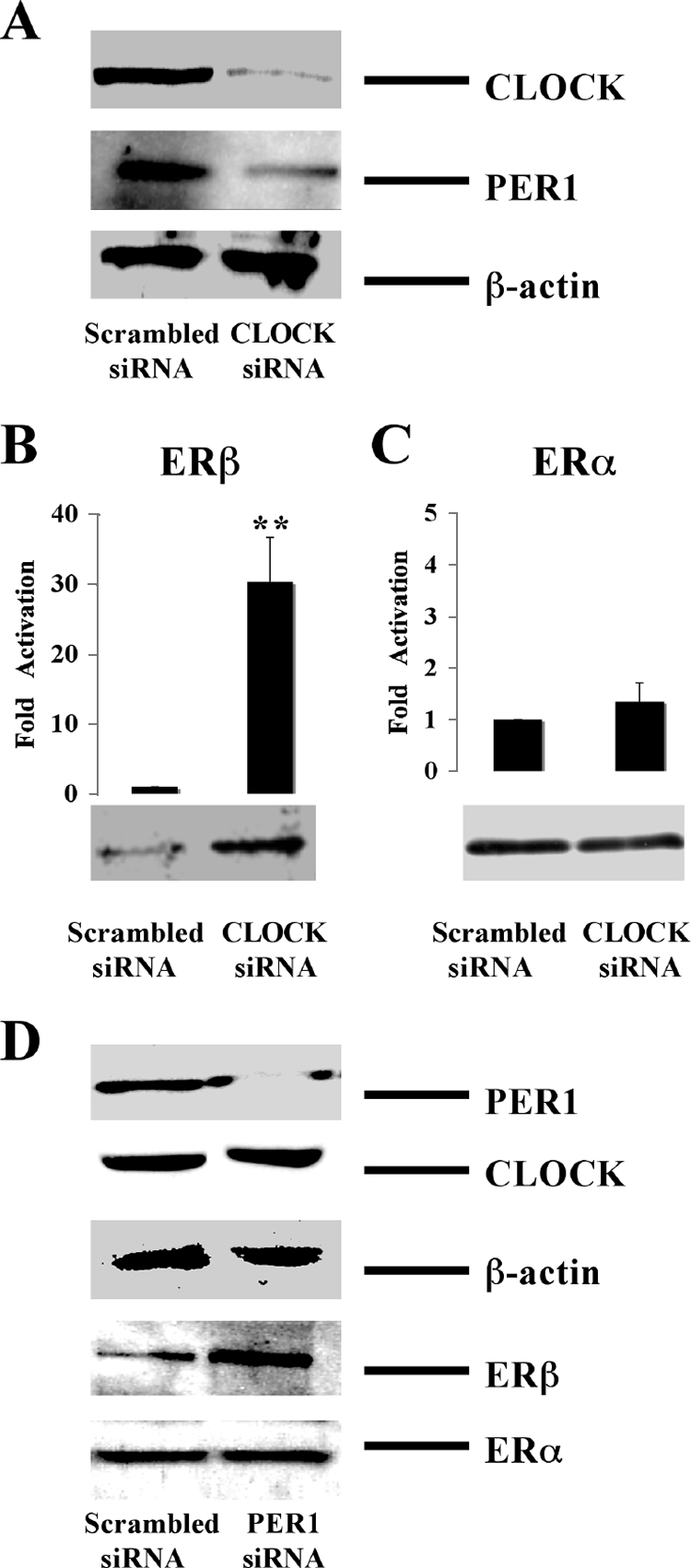
Effect of CLOCK and PER1 siRNA on ER expression. (A) siRNA against CLOCK was introduced in HC11 cells, and protein levels of CLOCK, PER1, and β-actin were assessed by Western blotting. (B and C) siRNA was introduced into HC11 cells, and ERα and ERβ mRNA and protein levels were monitored by real-time PCR and Western blotting. mRNA levels of scrambled siRNA-treated groups were arbitrarily set to be 1. (D) siRNA against PER was introduced in HC11 cells, and protein levels of PER1, CLOCK, β-actin, ERβ, and ERα were assessed by Western blotting. All Western blotting data were representative of three independent experiments. Real-time PCR data were expressed as means ± standard deviations of three independent experiments (**, P < 0.01; Student's t test).
Cycling of ERβ expression is abolished in BMAL1 KO mice.
To test genetically that ERβ is a circadian clock target gene in vivo, we compared the ERβ expression profile in skeletal muscles (where ERα and ERβ are coexpressed) from WT and BMAL1 KO mice. In the WT mouse muscle, the expression of ERβ mRNA showed a peak at ZT12 (Fig. 7A), corresponding to the ERβ expression pattern in lung presented in Fig. 1C. The ERβ expression pattern differed significantly between WT and KO animals; in KO animals, the ZT12 ERβ expression peak was missing. Interestingly, ERβ expression in KO mice was higher than in WT mice (Fig. 7A). Therefore, this experiment again confirmed that the rhythmic expression of ERβ in vivo is driven by the circadian system, mainly by the negative proteins, just as in HC11 cells, and suggested that the inactivation of BMAL1 blocks the access of the negative limb proteins to the ERβ promoter.
FIG. 7.
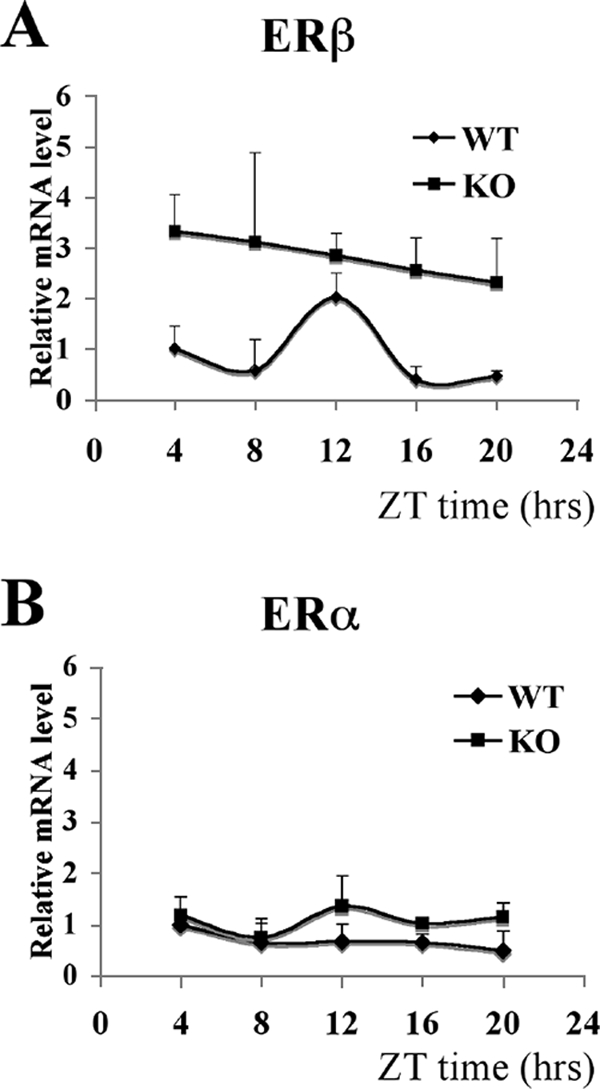
ER expression in WT and KO mouse muscle. (A) mRNA levels of ERβ in WT and KO male mouse muscle were quantified by real-time PCR at five circadian times (ZT4, ZT8, ZT12, ZT16, and ZT20). ERβ expression level in WT muscle at ZT4 was arbitrarily set to be 1. Results are shown as the means ± standard deviations of data for three animals at each time point. ANOVA analyses were used to compare each set of data. For WT mice, P = 0.015; for KO mice, P = 0.76. (B) mRNA levels of ERα in WT and KO male mouse muscle were quantified by real-time PCR at five circadian times (ZT4, ZT8, ZT12, ZT16, and ZT20). The ERα expression level at ZT4 was arbitrarily set to be 1. Results are shown as the means ± standard deviations of three animals at each time point. ANOVA analyses were used to compare each set of data. For WT mice, P = 0.24; for KO mice, P = 0.44.
In control experiments, the expression of ERα, which is coexpressed with ERβ in skeletal muscle (4), was monitored. ERα expression in both WT and KO mouse muscle remained at stable levels throughout the whole experiment without significant changes at any time point (Fig. 7B). This result provides genetic evidence that ERβ is specifically regulated by the circadian clock in vivo.
CLOCK modulates E2 signaling through ERβ.
Since both experiments with HC11 cells and BMAL1 KO animals suggest a subtype-specific circadian regulation of ER expression, diurnal variation of ER-controlled transcriptional activity could also be subtype specific. To test this possibility, we studied the effect of the circadian system on ERE-dependent transcription, using an HC11 cell line with a stably transfected 3×ERE-TATA-luciferase reporter construct (11). Cells were transfected with CLOCK siRNA and treated with different ER subtype-specific ligands, i.e., the ERα agonist propyl pyrazole triol (PPT) (47) and the ERβ agonist diarylpropionitrile (DPN) (31) or the panagonist E2. Following CLOCK depletion, no significant difference in luciferase activity levels compared to the control was observed in PPT-treated cells (Fig. 8A), supporting the previous observation that ERα is not under circadian control. On the other hand, the luciferase activity was increased 1.8-fold in E2-treated cells (Fig. 8B) and 2.8-fold in DPN-treated cells (Fig. 8C) following CLOCK depletion, supporting the notion that ERβ is under circadian control. Moreover, this observation demonstrates that E2 signaling is modulated by CLOCK protein intracellular levels through specific clock regulation of ERβ.
DISCUSSION
Previous studies on circadian rhythms have linked the endogenous clock system to various biological functions and diseases. For instance, ERβ-mediated and circadian-controlled physiological processes and diseases have been identified in both reproductive disorders and nonreproductive systems. Examples are mood disorders, which might arise following uncoupled phase of sleep and other biological rhythms (46). Interestingly, ERβ appears to be important in both the pathogenesis and therapy of depression (37). This finding, among others, suggests connections between the circadian system and ERβ-mediated physiological processes. Circadian proteins may affect these physiological functions by regulating the expression of ERβ. Therefore, when the normal circadian rhythm in the organism is disrupted, this would lead to alterations in the ERβ-dependent transcription pathway, which might increase the risk of the development of various diseases. However, more studies are needed to fully understand the precise mechanisms involved.
Our study shows that circadian proteins are strongly recruited to the E-box-containing region in the ERβ promoter, and from this element the circadian factors drive the rhythmic expression of ERβ in HC11 cells. In transient transfections we show that the CLOCK-BMAL1 complex binds to an E-box element in the ERβ promoter. The CLOCK-BMAL1 complex can subsequently serve as a platform for PER and CRY, which repress transcription. Maintaining circadian oscillation is thus a complex interplay between positive and negative regulatory steps. To identify which of the positive or the negative components of the circadian cycle are the critical components regulating ERβ expression, siRNA was used. Our experiments show that knocking down either CLOCK or PER1 expression leads to elevated expression of ERβ. This result suggests that ERβ circadian cycling is primarily maintained through PER1 and that CLOCK-BMAL1 serves as a docking point for PER1. Positive regulation of ERβ is likely to be maintained by additional transcription factors not necessarily involved in the cycling behavior of ERβ. In addition, our in vivo results demonstrate that expression of ERβ displays a circadian oscillation pattern in both lung and skeletal muscle of WT mice, and this oscillation is abolished in BMAL1 KO mice, indicating that ERβ is a direct target of the circadian system. Previous studies have identified clock-regulated NRs including Rev-erbα (49), ERRα (21) and peroxisome proliferator-activated receptor α (36). Here, our finding that another important NR, ERβ, is under circadian control again indicates a robust cross talk between bHLH-PAS circadian transcription factors and nuclear hormone receptors. Our experiments with HC11-3×ERE-luciferase cells show that depleting endogenous CLOCK using an siRNA approach induces the intracellular level of ERβ. Furthermore, ER-regulated transcription is modulated upon CLOCK depletion. The ERE transcriptional response is not affected when CLOCK-depleted HC11 cells are treated with the ERα-specific agonist PPT, in consonance with our finding that ERα is not under circadian regulation. In sharp contrast, however, 3×ERE-luciferase activity in CLOCK-depleted HC11 cells is significantly increased in the presence of the ERβ-specific agonist DPN. Moreover, CLOCK depletion leads to an increased ERE-dependent transcription upon E2 treatment. These results provide evidence of a novel regulatory pathway through which the circadian transcription factors influence estrogen signaling. Therefore, in living organisms, by regulating ERβ expression, the circadian clock may influence expression of ERβ target genes, a process which could have significant impact in various physiological contexts.
Although both ERs are expressed in the body, there are considerable differences in their tissue distribution. For example, ERβ is primarily expressed in lung, ovary, prostate, gastrointestinal tract, bladder, and central nervous system, whereas ERα is mainly expressed in liver, uterus, and kidney (27). In this study, we observed a robust cyclic expression of ERβ but not of ERα mRNA in mouse skeletal muscle, where both ERs are present (4, 50), in line with our finding that the circadian regulation of ER is subtype specific. Moreover, our RNAi experiments with HC11 cells indicate that this selective regulation may affect estrogen signaling, depending on the expression ratio of ERα to ERβ, which varies from tissue to tissue. In ERβ-dominant tissues, estrogen responses might fluctuate between day and night whereas estrogen actions in ERα-dominant tissues would be expected to show little or no diurnal variation. In tissues where the two receptors are present at comparable levels, more complicated estrogen response scenarios might be expected. The effect of estrogen on these tissues might differ from day to night due to the changed expression ratio of ERα to ERβ. Therefore, administration of compounds or drugs that have estrogenic or antiestrogenic function could lead to distinct effects, depending on the tissue type and time point selected for administering the drug. Furthermore, this suggests that circadian variations need to be taken into account when biological responses to compounds that affect estrogenic signaling are considered.
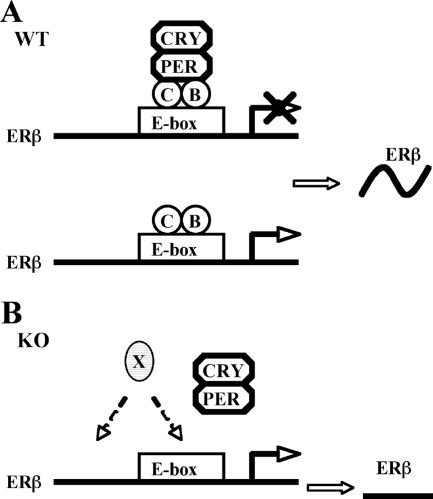
A number of previous studies have revealed that many circadian-regulated genes are strongly repressed in the absence of positive circadian regulators (17, 21, 35, 36). In our experiments we observed that in transient transfection experiments, the activity of the ERβ promoter-luciferase reporter construct is regulated by overexpressed CLOCK-BMAL1 and PER. This observation suggests that both positive and negative circadian transcription factors are involved in regulation of ERβ expression, raising the possibility that ERβ expression could be controlled by CLOCK-BMAL1-mediated transcriptional activation or, alternatively, by PER-dependent transcriptional repression. These two possibilities were tested in HC11 cells. ERβ is expressed in HC11 cells, and our experiments show that the main driver of the ERβ circadian rhythm is not recruitment of the positive CLOCK-BMAL1 complex; rather, the ERβ rhythm is maintained by recruitment of the circadian repressor PER. In addition, we addressed these possibilities using an in vivo strategy. In this study we show that depletion of CLOCK or BMAL1 results in up-regulation of ERβ expression in both cells and animal models. One of the possible mechanisms could be that the CLOCK-BMAL1 heterodimer, binding constantly to the ERβ promoter (as shown in Fig. 5C), would function as a binding platform for the negative circadian regulator PER1. Circadian recruitment and release of the negative regulator PER1 likely drives the rhythmic expression of ERβ (Fig. 9A). In the absence of CLOCK-BMAL1, negative circadian regulators cannot be recruited, and the ER promoter displays high basal activity with no circadian expression pattern (Fig. 9B). However, further studies are required to clarify the details of the mechanism.
FIG. 9.
Schematic model of circadian regulation of ERβ expression. CLOCK-BMAL1 heterodimer mediates the circadian regulation of ERβ by binding to the E-box enhancer in the ERβ promoter, but the negative regulators work as the main driver of the rhythmic expression of ERβ. (A) In WT mice and cells, recruitment of negative circadian regulator PER-CRY causes an inhibition of ERβ expression, and the release of PER-CRY results in an up-regulation of ERβ expression induced by CLOCK-BMAL1 and other unknown activating transcription factors. (B) In BMAL1 KO mice or CLOCK-deficient cells, where CLOCK-BMAL1 heterodimer is not formed, the negative regulators are not recruited to the E-box enhancer, and the expression of ERβ is induced by unknown activating transcription factors (X) and kept at a constant, high level.
In summary, in this study we have demonstrated that ERβ is under circadian regulation. This is likely to have significant consequences for estrogen action in a large number of tissues, particularly in those where ERβ is the dominating ER subtype.
Acknowledgments
This work was supported by the European Union thorough the CASCADE Network of Excellence and the CRESCENDO Integrated Project, by the Swedish Cancer Fund, by the Swedish Research Council, by the CNRS, the MENRT, the Association pour la Recherche sur le Cancer, and by Université de Nice-Sophia-Antipolis.
We thank Gerard Triqueneaux for technical help.
Jan-Åke Gustafsson is a shareholder, research grant recipient, and consultant of KaroBio AB.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Albrecht, U., and G. Eichele. 2003. The mammalian circadian clock. Curr. Opin. Genet. Dev. 13271-277. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre, A., F. Damiola, and U. Schibler. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93929-937. [DOI] [PubMed] [Google Scholar]
- 3.Balsalobre, A., L. Marcacci, and U. Schibler. 2000. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 101291-1294. [DOI] [PubMed] [Google Scholar]
- 4.Barros, R. P., U. F. Machado, M. Warner, and J. A. Gustafsson. 2006. Muscle GLUT4 regulation by estrogen receptors ERβ and ERα. Proc. Natl. Acad. Sci. USA 1031605-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boden, M. J., and D. J. Kennaway. 2006. Circadian rhythms and reproduction. Reproduction 132379-392. [DOI] [PubMed] [Google Scholar]
- 6.Brunnberg, S., K. Pettersson, E. Rydin, J. Matthews, A. Hanberg, and I. Pongratz. 2003. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc. Natl. Acad. Sci. USA 1006517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunger, M. K., J. A. Walisser, R. Sullivan, P. A. Manley, S. M. Moran, V. L. Kalscheur, R. J. Colman, and C. A. Bradfield. 2005. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis 41122-132. [DOI] [PubMed] [Google Scholar]
- 8.Canaple, L., T. Kakizawa, and V. Laudet. 2003. The days and nights of cancer cells. Cancer Res. 637545-7552. [PubMed] [Google Scholar]
- 9.Caruso, C. C., S. L. Lusk, and B. W. Gillespie. 2004. Relationship of work schedules to gastrointestinal diagnoses, symptoms, and medication use in auto factory workers. Am. J. Ind. Med. 46586-598. [DOI] [PubMed] [Google Scholar]
- 10.Emery, P., and S. M. Reppert. 2004. A rhythmic Ror. Neuron 43443-446. [DOI] [PubMed] [Google Scholar]
- 11.Faulds, M. H., H. Olsen, L. A. Helguero, J. A. Gustafsson, and L. A. Haldosen. 2004. Estrogen receptor functional activity changes during differentiation of mammary epithelial cells. Mol. Endocrinol. 18412-421. [DOI] [PubMed] [Google Scholar]
- 12.Gachon, F., E. Nagoshi, S. A. Brown, J. Ripperger, and U. Schibler. 2004. The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113103-112. [DOI] [PubMed] [Google Scholar]
- 13.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 2801564-1569. [DOI] [PubMed] [Google Scholar]
- 14.Grandien, K., M. Backdahl, O. Ljunggren, J. A. Gustafsson, and A. Berkenstam. 1995. Estrogen target tissue determines alternative promoter utilization of the human estrogen receptor gene in osteoblasts and tumor cell lines. Endocrinology 1362223-2229. [DOI] [PubMed] [Google Scholar]
- 15.Gronemeyer, H., J. A. Gustafsson, and V. Laudet. 2004. Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov. 3950-964. [DOI] [PubMed] [Google Scholar]
- 16.Gustafsson, J. A. 2006. ERβ scientific visions translate to clinical uses. Climacteric 9156-160. [DOI] [PubMed] [Google Scholar]
- 17.Hamaguchi, H., K. Fujimoto, T. Kawamoto, M. Noshiro, K. Maemura, N. Takeda, R. Nagai, M. Furukawa, S. Honma, K. Honma, H. Kurihara, and Y. Kato. 2004. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem. J. 38243-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen, J. 2006. Risk of breast cancer after night- and shift work: current evidence and ongoing studies in Denmark. Cancer Causes Control 17531-537. [DOI] [PubMed] [Google Scholar]
- 19.Hirota, T., and Y. Fukada. 2004. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog. Sci. 21359-368. [DOI] [PubMed] [Google Scholar]
- 20.Honma, S., T. Kawamoto, Y. Takagi, K. Fujimoto, F. Sato, M. Noshiro, Y. Kato, and K. Honma. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419841-844. [DOI] [PubMed] [Google Scholar]
- 21.Horard, B., B. Rayet, G. Triqueneaux, V. Laudet, F. Delaunay, and J. M. Vanacker. 2004. Expression of the orphan nuclear receptor ERRα is under circadian regulation in estrogen-responsive tissues. J. Mol. Endocrinol. 3387-97. [DOI] [PubMed] [Google Scholar]
- 22.Jin, X., L. P. Shearman, D. R. Weaver, M. J. Zylka, G. J. de Vries, and S. M. Reppert. 1999. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 9657-68. [DOI] [PubMed] [Google Scholar]
- 23.Knutsson, A. 2003. Health disorders of shift workers. Occup. Med. (London) 53103-108. [DOI] [PubMed] [Google Scholar]
- 24.Kuiper, G. G., E. Enmark, M. Pelto-Huikko, S. Nilsson, and J. A. Gustafsson. 1996. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. USA 935925-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98193-205. [DOI] [PubMed] [Google Scholar]
- 26.Lowrey, P. L., and J. S. Takahashi. 2004. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 5407-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews, J., and J. A. Gustafsson. 2003. Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol. Interv. 3281-292. [DOI] [PubMed] [Google Scholar]
- 28.McArthur, A. J., M. U. Gillette, and R. A. Prosser. 1991. Melatonin directly resets the rat suprachiasmatic circadian clock in vitro. Brain Res. 565158-161. [DOI] [PubMed] [Google Scholar]
- 29.Merrow, M., K. Spoelstra, and T. Roenneberg. 2005. The circadian cycle: daily rhythms from behaviour to genes. EMBO Rep. 6930-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metz, R. P., X. Qu, B. Laffin, D. Earnest, and W. W. Porter. 2006. Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev. Dyn. 235263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers, M. J., J. Sun, K. E. Carlson, G. A. Marriner, B. S. Katzenellenbogen, and J. A. Katzenellenbogen. 2001. Estrogen receptor-beta potency-selective ligands: structure-activity relationship studies of diarylpropionitriles and their acetylene and polar analogues. J. Med. Chem. 444230-4251. [DOI] [PubMed] [Google Scholar]
- 32.Morani, A., R. P. Barros, O. Imamov, K. Hultenby, A. Arner, M. Warner, and J. A. Gustafsson. 2006. Lung dysfunction causes systemic hypoxia in estrogen receptor beta knockout (ERβ−/−) mice. Proc. Natl. Acad. Sci. USA 1037165-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson, S., S. Makela, E. Treuter, M. Tujague, J. Thomsen, G. Andersson, E. Enmark, K. Pettersson, M. Warner, and J. A. Gustafsson. 2001. Mechanisms of estrogen action. Physiol. Rev. 811535-1565. [DOI] [PubMed] [Google Scholar]
- 34.Oishi, K., K. Miyazaki, K. Kadota, R. Kikuno, T. Nagase, G. Atsumi, N. Ohkura, T. Azama, M. Mesaki, S. Yukimasa, H. Kobayashi, C. Iitaka, T. Umehara, M. Horikoshi, T. Kudo, Y. Shimizu, M. Yano, M. Monden, K. Machida, J. Matsuda, S. Horie, T. Todo, and N. Ishida. 2003. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 27841519-41527. [DOI] [PubMed] [Google Scholar]
- 35.Oishi, K., H. Shirai, and N. Ishida. 2005. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARα) in mice. Biochem. J. 386575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Leary, E. S., E. R. Schoenfeld, R. G. Stevens, G. C. Kabat, K. Henderson, R. Grimson, M. D. Gammon, and M. C. Leske. 2006. Shift work, light at night, and breast cancer on Long Island, New York. Am. J. Epidemiol. 164358-366. [DOI] [PubMed] [Google Scholar]
- 37.Osterlund, M. K., M. R. Witt, and J. A. Gustafsson. 2005. Estrogen action in mood and neurodegenerative disorders: estrogenic compounds with selective properties-the next generation of therapeutics. Endocrine 28235-242. [DOI] [PubMed] [Google Scholar]
- 38.Panda, S., and J. B. Hogenesch. 2004. It's all in the timing: many clocks, many outputs. J. Biol. Rhythms 19374-387. [DOI] [PubMed] [Google Scholar]
- 39.Pettersson, K., and J. A. Gustafsson. 2001. Role of estrogen receptor beta in estrogen action. Annu. Rev. Physiol. 63165-192. [DOI] [PubMed] [Google Scholar]
- 40.Prosser, R. A. 1999. Melatonin inhibits in vitro serotonergic phase shifts of the suprachiasmatic circadian clock. Brain Res. 818408-413. [DOI] [PubMed] [Google Scholar]
- 41.Reppert, S. M., and D. R. Weaver. 2002. Coordination of circadian timing in mammals. Nature 418935-941. [DOI] [PubMed] [Google Scholar]
- 42.Reppert, S. M., and D. R. Weaver. 2001. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63647-676. [DOI] [PubMed] [Google Scholar]
- 43.Roenneberg, T., and M. Merrow. 2003. The network of time: understanding the molecular circadian system. Curr. Biol. 13R198-207. [DOI] [PubMed] [Google Scholar]
- 44.Rosenfeld, M. G., and C. K. Glass. 2001. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 27636865-36868. [DOI] [PubMed] [Google Scholar]
- 45.Schuur, E. R., L. A. McPherson, G. P. Yang, and R. J. Weigel. 2001. Genomic structure of the promoters of the human estrogen receptor-alpha gene demonstrate changes in chromatin structure induced by AP2γ. J. Biol. Chem. 27615519-15526. [DOI] [PubMed] [Google Scholar]
- 46.Srinivasan, V., M. Smits, W. Spence, A. D. Lowe, L. Kayumov, S. R. Pandi-Perumal, B. Parry, and D. P. Cardinali. 2006. Melatonin in mood disorders. World J. Biol. Psychiatry 7138-151. [DOI] [PubMed] [Google Scholar]
- 47.Stauffer, S. R., C. J. Coletta, R. Tedesco, G. Nishiguchi, K. Carlson, J. Sun, B. S. Katzenellenbogen, and J. A. Katzenellenbogen. 2000. Pyrazole ligands: structure-affinity/activity relationships and estrogen receptor-alpha-selective agonists. J. Med. Chem. 434934-4947. [DOI] [PubMed] [Google Scholar]
- 48.Tanimoto, K., H. Eguchi, T. Yoshida, K. Hajiro-Nakanishi, and S. Hayashi. 1999. Regulation of estrogen receptor alpha gene mediated by promoter B responsible for its enhanced expression in human breast cancer. Nucleic Acids Res. 27903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Triqueneaux, G., S. Thenot, T. Kakizawa, M. P. Antoch, R. Safi, J. S. Takahashi, F. Delaunay, and V. Laudet. 2004. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 33585-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiik, A., B. Glenmark, M. Ekman, M. Esbjornsson-Liljedahl, O. Johansson, K. Bodin, E. Enmark, and E. Jansson. 2003. Oestrogen receptor beta is expressed in adult human skeletal muscle both at the mRNA and protein level. Acta Physiol. Scand. 179381-387. [DOI] [PubMed] [Google Scholar]