Abstract
Recent discoveries of NAD-mediated regulatory processes in mitochondria have documented important roles of this compartmentalized nucleotide pool in addition to energy transduction. Moreover, mitochondria respond to excessive nuclear NAD consumption arising from DNA damage-induced poly-ADP-ribosylation because poly(ADP-ribose) (PAR) can trigger the release of apoptosis-inducing factor from the organelles. To functionally assess mitochondrial NAD metabolism, we overexpressed the catalytic domain of nuclear PAR polymerase 1 (PARP1) and targeted it to the matrix, which resulted in the constitutive presence of PAR within the organelles. As a result, stably transfected HEK293 cells exhibited a decrease in NAD content and typical features of respiratory deficiency. Remarkably, inhibiting PARP activity revealed PAR degradation within mitochondria. Two enzymes, PAR glycohydrolase (PARG) and ADP-ribosylhydrolase 3 (ARH3), are known to cleave PAR. Both full-length ARH3 and a PARG isoform, which arises from alternative splicing, localized to the mitochondrial matrix. This conclusion was based on the direct demonstration of their PAR-degrading activity within mitochondria of living cells. The visualization of catalytic activity establishes a new approach to identify submitochondrial localization of proteins involved in the metabolism of NAD derivatives. In addition, targeted PARP expression may serve as a compartment-specific “knock-down” of the NAD content which is readily detectable by PAR formation.
NAD in mitochondria is important to maintain the vital functions of these organelles as well as the energy supply of the entire cell. For example, ATP synthesis in oxidative phosphorylation is primarily fueled by NADH, which is generated by reduction of NAD+ in the Krebs cycle. In addition to its critical role in energy transduction, NAD also participates in a variety of regulatory pathways, which include the cleavage of nicotinamide from NAD+ and thereby necessitates continuous resynthesis of the nucleotide (7, 10, 45). This requirement has been recognized, for example, for the nuclear NAD pool. However, the existence of NAD-degrading regulatory processes within mitochondria has only recently been conclusively demonstrated.
Several classes of enzymes use NAD+ as a substrate in signaling processes. NAD+ glycohydrolases generate ADP-ribose and cyclic ADP-ribose, which both participate in intracellular calcium signaling by either activating calcium channels in the plasma membrane (27) or acting as a ligand of the ryanodine receptor (28, 29), respectively. Recently, a family of protein deacetylases, the sirtuins, was identified which requires NAD+ as cosubstrate (15, 35, 37, 42). NAD+ accepts the acetyl moiety under release of nicotinamide, thereby forming O-acetyl ADP-ribose (OAADPR), which is suspected to have regulatory functions, too (18). Moreover, NAD+ serves as precursor for covalent protein modifications, mono- and poly-ADP-ribosylation. Mono-ADP-ribosylation represents the transfer of a single ADP-ribose unit onto an acceptor protein (12, 16, 19, 50), whereas in poly-ADP-ribosylation reactions, polymers consisting of up to 200 ADP-ribose units are covalently linked to the target protein (3, 23). Nuclear poly-ADP-ribosylation by poly(ADP-ribose) polymerase 1 (PARP1) has been identified as a major NAD+-consuming reaction, at least when the enzyme is catalytically activated by binding to sites of DNA damage. Indeed, extensive DNA damage can lead to cell death and cellular NAD depletion owing to PARP1 activity (2, 8, 10, 11, 56). The nature of PARP1-mediated cell death is not well understood. It might be the result of a decreased capacity to generate ATP. On the other hand, the process involves characteristic features of apoptosis such as nuclear translocation of apoptosis-inducing factor (AIF) from mitochondria (25, 58). AIF relocation depends on PARP1 activity and appears to be mediated by direct interaction of PAR with mitochondria (57). Under less drastic conditions a major function of PARP1 is to facilitate DNA repair, possibly by recruiting repair factors to the sites of damage via auto-poly-ADP-ribosylation (30, 31). At least larger polymers of ADP-ribose are rather short-lived owing to the activity of PAR glycohydrolase (PARG), which cleaves the polymer eventually into single ADP-ribose units (53). Numerous splice variants are expressed from a single PARG gene (33, 34), of which only the full-length protein is targeted to the nucleus due to a strong nuclear localization signal in exon I (34). Indeed, PARG activity is predominantly detected in cytoplasmic fractions (34, 39, 55), which is in apparent contradiction to the nuclear localization of PARP1, the clearly predominant PARP isoform (4, 43, 47). In contrast to PARP1, the PARG gene is essential, since PARG−/− mice die during early embryonic development (26). Interestingly, transgenic mice expressing PARG from a disrupted gene lacking exons II and III were viable (13). Most of the cellular PARG activity of these mice was detected in the mitochondrial fraction (13). A detailed analysis of PARG transcripts revealed novel splice variants. According to immunocytochemical studies, at least one isoform was clearly associated with mitochondria, although the suborganellar localization, and consequently its function, has remained unknown (33). A recent report suggested the dissociation of a PARG isoform from mitochondria following PARP1 activation in neuronal cells (44).
Another enzyme, ADP-ribosylarginine hydrolase-like protein 2, alternatively referred to as ADP-ribosylhydrolase 3 (ARH3), was recently also shown to possess PARG activity (40). The protein shares only very little sequence similarity with PARG. ARH3 belongs to the family of dinitrogenase reductase-activating glycohydrolase-related proteins (17). Unlike PARG, ARH3 also efficiently cleaved OAADPR into ADP-ribose and acetate (41) and could therefore have a function in the deactivation of this potential messenger molecule. ARH3 has been detected in cytosolic and nuclear fractions (40). However, we have previously predicted a mitochondrial localization for ARH3 on the basis of its potential N-terminal mitochondrial targeting sequence (36).
At least two NAD-mediated regulatory pathways have been established within mitochondria. Glutamate dehydrogenase represents a specific target for reversible mono-ADP-ribosylation (24). The modification is catalyzed by SIRT4, a mitochondrial member of the sirtuin family, and causes inhibition of glutamate dehydrogenase activity (20). It was further demonstrated that this mode of glutamate dehydrogenase regulation controls insulin secretion from the pancreas (1, 20). Another mitochondrial sirtuin, SIRT3, is involved in the regulation of acetyl-coenzyme A synthase 2 (ACS2), a mitochondrial isoform. Acetylation of ACS2 inhibits enzyme activity, and this modification is reversed by SIRT3, an NAD-dependent deacetylase (22, 48).
The presence of these NAD-mediated pathways within the organelles suggests a functional importance of mitochondrial NAD in addition to energy transduction. Indeed, mammalian mitochondria appear to possess an autonomous NAD metabolism, as indicated by the absence of an exchange of the nucleotide across the inner membrane and the presence of an NAD biosynthetic enzyme, nicotinamide mononucleotide adenylyltransferase 3, in the organelles (6). On the other hand, there is growing evidence that NAD or its derivatives might serve to transmit information between the mitochondria and the nucleus, as exemplified by PAR-mediated AIF translocation or the association of PARG with mitochondria.
The present study further substantiates this notion and establishes, rather unexpectedly, the localization of a mitochondrial PARG isoform and at least the majority of ARH3 in the matrix of these organelles. As a test system, we engineered a 293 cell line which expresses a truncated, catalytically active PARP1 construct in the mitochondrial matrix which constitutively generates PAR. Therefore, the localization of the PAR-degrading enzymes within the matrix could be based on the demonstration of their catalytic activity, in addition to imaging techniques. Moreover, the developed cell model is the first to permit manipulation of only the mitochondrial NAD pool in living cells. Biochemical characterization confirmed a significant decrease of total cellular NAD content, lowered mitochondrial membrane potential, and a compensatory increase of the glycolytic activity in cells expressing the mitochondrial PARP1 construct.
(Parts of this work represents the partial fulfillment of the requirements for the doctoral thesis of S. Kernstock at the University of Hamburg, Germany.)
MATERIALS AND METHODS
Chemicals, reagents, and media.
Unless otherwise noted, all chemicals and reagents were of analytical grade and purchased from Sigma-Aldrich and Merck. Reagents other than fetal bovine serum (Biochrom) and G418 (PAA Laboratories GmbH) were from Cambrex Cooperation, Nunc, or Invitrogen/Gibco. PJ34 was obtained from Calbiochem. Fluorescence-conjugated secondary antibodies were of highly cross-adsorbed quality: R-phycoerythrin donkey anti-mouse immunoglobulin (Ig) F(ab)2 fragment from Dianova; and Alexa Fluor 594 goat anti-mouse Ig, Alexa Fluor 647 goat anti-mouse Ig, and Alexa Fluor 633 goat anti-rabbit Ig from Invitrogen/Molecular Probes. The fluorescent dye JC-1 and ProLong Gold antifade reagent were from Invitrogen/Molecular Probes, enhanced chemiluminescence reagent and horseradish peroxidase-conjugated goat anti-mouse antibody were from Pierce or GE Healthcare/Amersham Biosciences. Rabbit anti-FLAG antibody was from Sigma-Aldrich; mouse anti-PAR antibody (10H) and mouse anti-myc antibody (9E10) were from hybridoma cell culture supernatants. DNA modifying and restriction enzymes were purchased from Fermentas or New England Biolabs; oligonucleotide synthesis was done by BioTeZ GmbH or MWG.
Generation of eukaryotic expression vectors.
To generate a vector targeting the C-terminal NAD+ binding domain (amino acids [aa] 572 to 1014) of PARP1 to the mitochondria, the corresponding portion of full-length PARP1 cDNA was ligated into pCMV/myc/mito (Invitrogen) (CMV is cytomegalovirus) via its PstI/SalI sites. Subsequently, cDNA encoding enhanced green fluorescent protein (EGFP; Clontech Laboratories, Inc.) was inserted into the vector via the PstI site. A vector encoding mitochondrial EGFP was prepared by replacing the GFP encoding cDNA from pCMV/myc/mito/GFP (Invitrogen) with the cDNA encoding EGFP using PstI/NotI sites of the vector and PstI/Bsp120I sites for the insert. A vector encoding an ARH3-d2eGFP fusion protein was constructed by cloning the cDNA encoding full-length ARH3 into pd2eGFP-N1 (Clontech Laboratories, Inc.) using NheI/EcoRI. Using QuikChange mutagenesis (Stratagene), the vector pCDNA5/FRT/TO was mutated to restore the NheI site in the multiple cloning site, and the insert coding ARH3-d2eGFP was recloned from pd2eGFP-N1_ARH3 into pCDNA5/FRT/TO. Using QuikChange mutagenesis (Stratagene), the FLAG epitope was fused to the C terminus of ARH3 in pd2eGFP-N1_ARH3 followed by a stop codon, thus eliminating the expression of EGFP. Vectors encoding PARG with a deletion of residues 1 to 460 [PARG(Δ1-460)] and of residues 1 to 477 [PARG(Δ1-477)] were generated by ligating the corresponding portions of full-length PARG cDNA (kindly provided by Claudia Keil, Berlin, Germany) into pFLAG-CMV-5a (Sigma-Aldrich) via its EcoRI/SalI sites. All cloned cDNAs were amplified from vectors harboring full-length cDNAs using Pfu polymerase. Recognition sites for restriction enzymes, the Kozak sequence for the PARG and ARH3 encoding vectors, and a start codon for PARG(Δ1-477) and the FLAG-epitope for ARH3 were included in the primer sequences.
PCR analysis.
Identification of the ARH3 transcript was done by PCR from cDNA prepared from 293 cells using the ARH3-specific primers ARH3 exon V forward (5′-CATGGAGGAGCGTCCATACT) and ARH3 exon VI reverse (5′-GGGCCAGGATGTCTGTCTC). Exon composition of the transcript encoding endogenous mitochondrial PARG isoforms was determined by using the PARG-specific primer PARG exon Ia forward (5′-AATTGCAGAAGCAGGCAGCGG) in combination with PARG exon IV reverse (TTACTGTGTGATTGGCAGATGG), PARG exon V reverse (AGGTTTAGGAACTTCTCCTGC), PARG exon VI reverse (GTGCAGTCTGAATGAGCTCCC), and PARG exon V/VI reverse (GCTCACCATTCTCATCTTCCAC).
Cell culture.
293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal calf serum (FCS), 2 mM glutamine, and penicillin/streptomycin. HeLaS3 cells were cultivated in Ham's F12 medium supplemented with 10% FCS and penicillin-streptomycin. Flp-In T-REx 293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) tetracycline-free FCS, 2 mM l-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 1× minimal essential medium nonessential amino acids (Invitrogen/Gibco) and gentamicin, zeocin, and blasticidin. Transient transfection of eukaryotic cells was performed for 24 to 48 h using Effectene reagent (Qiagen) according to the recommendations of the manufacturer. For generation of stably transfected 293 lines, 800,000 cells were seeded onto a six-well plate and transfected by the Ca3(PO4)2 method 1 day later. Twenty-four hours posttransfection, cells were distributed onto four 10-cm dishes following selection in the presence of G418 (550 μg/ml). Clones were isolated after 10 days of selection, and monoclonal cell lines were subsequently generated by another round of selection. Stably transfected 293 lines (termed 293mitoEGFP and 293mitoPARP) were maintained in medium containing 100 μg/ml G418. For generation of a stably transfected inducible cell line, Flp-In T-REx 293 cells were cotransfected with pCDNA5/FRT/TO_ARH3d2eGFP and pOG44, a Flp recombinase expression vector, using FuGENE6 (Roche Diagnostics GmbH). Stably transfected cells were selected and maintained with medium containing hygromycin B (200 μg/ml) and blasticidin (15 μg/ml).
Determination of medium pH and lactate concentration.
A total of 2.25 ×105 cells was seeded in 2 ml of medium in the cavity of a six-well plate. After indicated cultivation times (see Fig. 3B) the medium was removed from the cells, and the pH was measured. Lactate determination was performed by a photometric assay according to Vahsen et al. (54), which is based on monitoring the reduction of 2,6-dichlorophenol-indophenol. Prior to measurement the medium was diluted 100-fold with fresh medium.
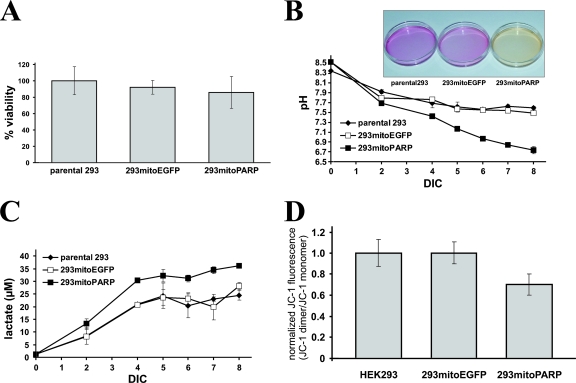
FIG. 3.
Decrease of mitochondrial NAD by mitoPARP expression resembles respiratory deficiency compensated by enhanced glycolysis. (A) Cells were seeded into 96-well plates, and the viability was determined by MTT assays. Data are shown as means ± standard deviations of three experiments, each performed in triplicate. (B) Decrease in medium pH after long-term incubation. Data are presented as means ± standard deviations of three independent experiments. The inset shows representative culture dishes of the cell lines after 7 days in culture (DIC). (C) Increase in medium lactate levels after long-term incubation of the cell lines. Cell culture supernatants from panel B were subjected to determination of the lactate concentration. (D) 293mitoPARP cells exhibit a weaker mitochondrial membrane potential. Cells were loaded with JC-1, and fluorescence spectra were recorded before and after uncoupling the mitochondrial membrane potential by adding FCCP. Relative mitochondrial membrane potentials are expressed as the ratios between fluorescence intensities at 595 nm and 534 nm before and after FCCP treatment and were normalized to the data obtained for parental 293 cells. Data represent means ± standard deviations of three experiments.
Cell viability.
Cell viability was determined using an MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay. A total of 1 × 105 to 2 × 105 cells/well were seeded on collagen-coated 96-well plates in 200 μl of medium. After 24 and 48 h the medium was replaced with 200 μl of medium supplemented with 0.3 mg/ml MTT followed by 3 h of incubation at 37°C. Accumulated formazan was dissolved in 150 μl of dimethyl sulfoxide and incubated for 2 h at room temperature (RT) and then absorbance was measured at 600 nm. The data obtained for untreated control cells were subtracted from the values obtained for MTT-treated cells. Viability was expressed as the ratio of data obtained from cells cultivated for 48 h and cells grown for 24 h and set to 100% for parental 293 cells.
Protein determination, SDS-PAGE, and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were performed according to standard procedures; enhanced chemiluminescence was used for detection. Protein determination of cell lysates prepared with 20 mM Tris (pH 7.4), 150 mM NaCl, 1% (vol/vol) Triton X-100, 1 mM EDTA, complete EDTA-free protease inhibitor cocktail (Roche Diagnostics GmbH), and 3 mM Pefabloc SC (Carl Roth GmbH &Co., Karlsruhe, Germany) as lysis buffer was done using a bicinchoninic protein assay kit (Pierce).
Determination of the mitochondrial membrane potential.
A total of 1.5 × 106 cells were sedimented, resuspended in 1 ml of Hanks balanced salt solution (HBSS) supplemented with 5 μg/ml JC-1, and incubated for 10 min at 37°C. After being washed once with ice-cold HBSS, cells were taken up in 1 ml of ice-cold HBSS and protected from light. Fluorescence spectra of cells before and after uncoupling the mitochondrial membrane potential with 2 μM carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP) for 10 min were recorded using a Perkin Elmer LS50B luminescence photometer with and excitation wavelength of 490 nm and emission wavelengths of 510 to 610 nm. The fluorescence intensities at 534 nm (JC-1 monomer) and 595 nm (JC-1 dimer) of untreated control cells were subtracted from the data obtained for JC-1-treated cells before and after uncoupling of the mitochondrial membrane potential by FCCP. Changes of the mitochondrial membrane potential were expressed as the difference in the ratios of fluorescence intensities at 595 nm and 534 nm before and after FCCP treatment. Data were normalized to those obtained for parental 293 cells.
Immunocytochemistry.
Cells grown on coverslips were fixed with ice-cold 4% (vol/vol) formaldehyde in phosphate-buffered saline (PBS) for 30 to 45 min and subsequently permeabilized for 15 min using 0.2 to 0.5% (vol/vol) Triton X-100 in PBS. After a blocking step with complete medium for 1 h at RT, cells were incubated with primary antibodies diluted in complete medium overnight at 4°C. Cells were washed once with 0.1% (vol/vol) Triton X-100 in PBS and twice with PBS, followed by a 1-h incubation at RT with secondary antibody diluted 1:250 to 1:1,000 in full medium. After nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole), the cells were washed once with 0.1% (vol/vol) Triton X-100 in PBS and four times with PBS, and the coverslips were subsequently mounted onto slides. Images were taken using the 405-, 488-, 594-, and 633-nm laser lines of a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems) equipped with a 63× oil immersion objective (numerical aperture, 1.40). Apotome images were taken using a Zeiss Axiovert 200 M microscope equipped with Apotome for optical sectioning and 63× Plan Apochromat oil immersion objective (numerical aperture, 1.25).
RESULTS
We reasoned that expression of PARP1 endowed with an established mitochondrial matrix targeting signal would render a situation of increased NAD turnover within the organelles. Moreover, since the catalytic activity of the construct would be detectable as PAR accumulation, this model system would also be suitable to reveal PAR-degrading activity within the matrix.
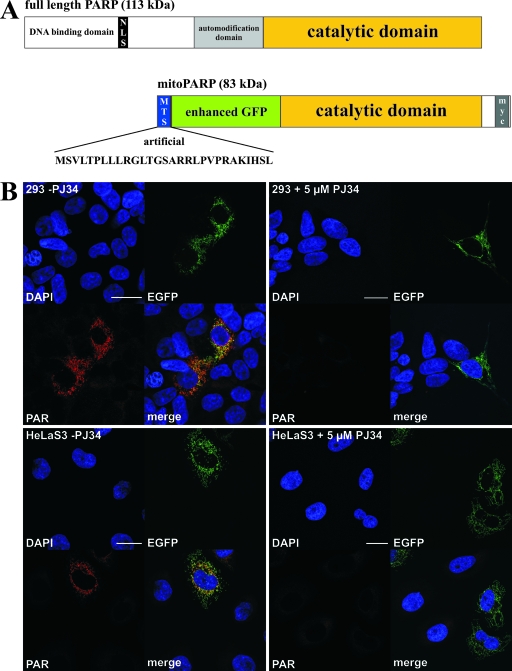
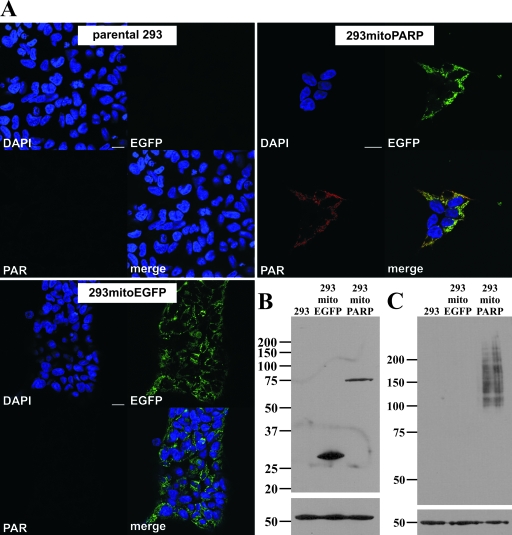
Full-length PARP1 contains a strong nuclear localization signal. However, it has been demonstrated that PARP1 lacking both the N-terminal DNA binding and auto-modification domains (including the nuclear localization signal) is active and retains the capacity to catalyze auto-modification, although at a considerably lower rate (32, 51). Therefore, a vector was constructed encoding a truncated PARP1 fused to EGFP and directed to the mitochondrial matrix by an established N-terminal targeting sequence. The resultant protein was termed mitoPARP (Fig. 1A). Transfection of the plasmid into human cells followed by immunocytochemistry using the 10H antibody revealed accumulation of mitochondrial PAR colocalizing with the intrinsic green fluorescence of the mitoPARP construct independently of the cell type (Fig. 1B, left panels). Continued incubation of the cells with a 5 μM concentration of the PARP inhibitor PJ34, starting 30 min prior to transfection, completely abolished formation of detectable PAR but did not influence the expression of the mitoPARP construct, as indicated by the intrinsic green fluorescence (Fig. 1B, right panels). Having confirmed the suitability of the mitoPARP construct, we next wished to investigate the cellular consequences of the increased consumption of mitochondrial NAD. We generated stably transfected monoclonal 293 cell lines expressing either the mitoPARP construct (293mitoPARP) or a construct lacking the PARP1 portion (293mitoEGFP) as a control. The expression level of the mitoPARP construct was lower than expression of mitochondrial EGFP in control cells (Fig. 2B). Nevertheless, mitochondrial PAR accumulation as detected by immunocytochemistry was present in all 293mitoPARP cells, whereas both parental 293 cells and 293mitoEGFP cells were PAR negative (Fig. 2A). Accordingly, Western blot analysis revealed PAR only in the 293mitoPARP cell lysate (Fig. 2C). The viability of 293mitoPARP cells was unaffected as long as they were provided with fresh medium (Fig. 3A). However, incubation of 293mitoPARP cells over an extended period of time without a medium change revealed faster acidification (Fig. 3B) and a concomitant increase of the lactate concentration in the medium (Fig. 3C). In fact, faster acidification of 293mitoPARP cells was readily detectable by the color change of phenol red in the culture medium during long-term incubation without a medium change (Fig. 3B, inset). Next, we analyzed the possible influence of the mitochondrial PARP activity on the mitochondrial membrane potential using the fluorescent cationic dye JC-1. The higher the membrane potential, the greater is the amount of the dye that accumulates within mitochondria, increasing dimer formation, which is detected by a shift of the fluorescence peak from ∼530 nm to ∼595 nm (14). Fluorescence of JC-1-treated cells was recorded at these wavelengths and related to the values obtained after uncoupling the mitochondria by the addition of FCCP. Compared to control cells, the mitochondrial membrane potential of 293mitoPARP cells was substantially diminished (Fig. 3D). These results suggest that 293mitoPARP cells have a decreased mitochondrial activity, which is likely due to a deficit of NAD within the organelles. Indeed, high-performance liquid chromatography analyses of deproteinized cell extracts showed that the total cellular NAD content decreased from 4.2 to 3.4 nmol/mg of protein in the 293mitoPARP cells. Interestingly, the redox ratio (NAD+/NADH) was also shifted (from 4.5 to 2.8), in accordance with the notion that only NAD+, but not NADH, is a substrate for PARP1. However, the cells were obviously able to compensate for mitochondrial insufficiency by increasing glycolytic activity, similar to situations when the respiratory chain is inhibited or otherwise impaired.
FIG. 1.
PAR accumulation in mitochondria mediated by a targeted PARP1 construct. (A) Molecular architecture of the generated mitoPARP construct. The C-terminal 443 aa residues of full-length PARP1 (upper panel) harboring the catalytic domain were C-terminally fused to EGFP and endowed with the established N-terminal mitochondrial targeting sequence (MTS) of cytochrome c oxidase subunit VIII (lower panel). NLS, nuclear localization signal. (B) Images from confocal laser scanning of 293 cells and HeLaS3 24 h after transient transfection with mitoPARP-encoding vector in the absence (−; left panels) and presence (+; right panels) of 5 μM PJ34. The fluorescence revealed nuclei (DAPI), the mitoPARP protein (containing functional EGFP), and PAR (by indirect immunocytochemistry using 10H antibody). Bar, 20 μm.
FIG. 2.
Constitutive presence of PAR in mitochondria of stably transfected 293 cells. (A) Images from confocal scanning of 293 cells stably transfected with mitoPARP-encoding vector (293mitoPARP panels) after indirect immunocytochemistry using 10H antibody to detect PAR. Mitochondrial PAR accumulation colocalizing with the intrinsic EGFP fluorescence of the synthetic PARP construct was detectable in all cells. 293 cells stably transfected with a construct lacking the PARP portion of the construct (293mitoEGFP panels) were EGFP positive but PAR negative, whereas in parental 293 cells (parental 293 panels) neither green fluorescence nor immunoreactivity for PAR was detected. Bar, 20 μm. (B) Forty micrograms of total protein from the indicated cell lines was separated by 10% SDS-PAGE. Western blotting was performed using myc antibody. (C) Forty micrograms of total protein of the indicated cell lines was separated by 7% SDS-PAGE. PAR was revealed by Western blotting using the 10H antibody. β-Tubulin was used as a loading control in panels B and C.
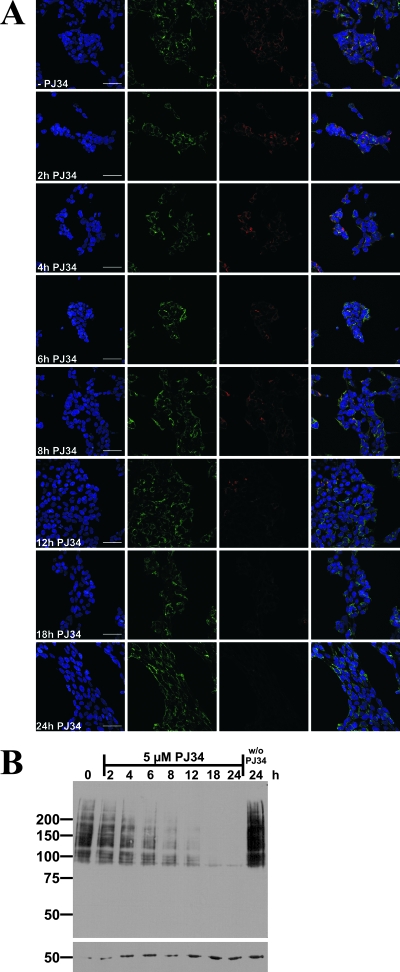
The constitutive decrease of the mitochondrial NAD content implied by the results above suggested that PAR generated by the mitoPARP construct is not persistent but, rather, subject to continuous turnover. To address this point, 293mitoPARP cells were treated with 5 μM PJ34 for up to 24 h. As shown in Fig. 4, the amount of polymer was steadily reduced, with virtually none left after 18 h of PARP inhibition. These results were similar when polymers were detected by immunocytochemistry (Fig. 4A) or Western blotting (Fig. 4B). Despite a rising cell count over a period of 24 h (as indicated by the tubulin signal), the amount of polymer decreased in the presence of PJ34 as opposed to an increase in the absence of the PARP inhibitor (Fig. 4B).
FIG. 4.
PARP inhibition in 293mitoPARP cells reveals mitochondrial PAR degrading activity. (A) Images from confocal laser scanning of 293mitoPARP cells after incubation with 5 μM PJ34 for the indicated time periods and subsequent immunocytochemistry using 10H antibody. −PJ34, no PJ34. Bar, 50 μm. (B) Western blot analysis of 293 mitoPARP cells after incubation with 5 μM PJ34 for the indicated time periods. Cells before PJ34 incubation and after 24 h incubation in the absence of PJ34 were used as a control. The increase in PAR in the control cells is due to proliferation. Immunostaining of β-tubulin (used as a loading control) confirmed the steady increase of total protein amounts owing to cell proliferation.
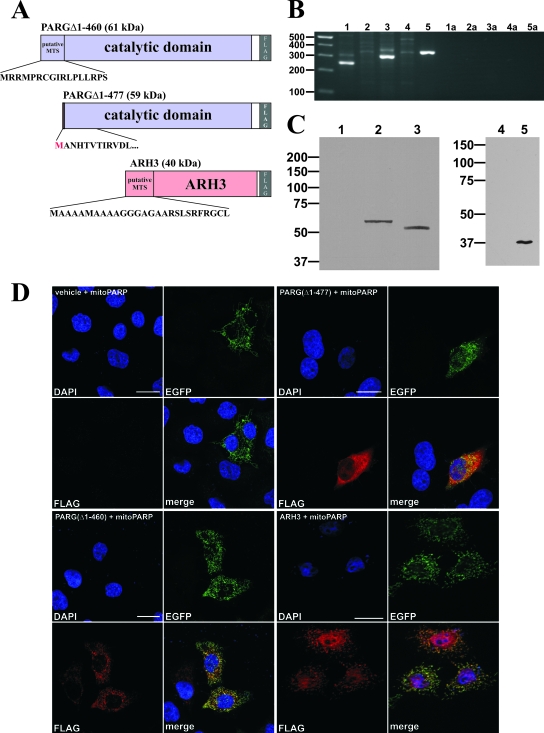
These experiments clearly indicated the presence of a PAR-degrading activity in the mitochondrial matrix. In order to address this issue, we constructed expression vectors encoding predicted mitochondrial isoforms of ARH3 and PARG (Fig. 5A), i.e., full-length ARH3 and PARG(Δ1-460) based on a proposed murine splice variant that lacks exons II and III (13). PARG(Δ1-460) contains a putative N-terminal mitochondrial targeting sequence (38) which originates from an alternative translational start in exon IV. While this study was under way, an mRNA encoding this isoform was indeed detected in wild-type mice (33). A control vector encoding a PARG similar to PARG(Δ1-460) but lacking the putative mitochondrial targeting sequence was also prepared (Fig. 5A). The protein expressed from this construct was termed PARG(Δ1-477).
FIG. 5.
A PARG isoform and ARH3 localize to mitochondria and are expressed in 293 cells. (A) Molecular architecture of the generated PARG and ARH3 constructs. The C-terminal 516 aa residues of full-length PARG (upper panel) harboring both the catalytic domain and an N-terminal predicted mitochondrial targeting sequence (MTS) were N-terminally fused to a FLAG epitope. Similarly, full-length ARH3 was endowed with a C-terminal FLAG tag (lower panel). As control, a PARG construct lacking the predicted targeting sequence was generated (middle panel). An additional methionine was added to the N terminus to enable translation. (B) PCR amplification products from HEK293 cDNA using specific primers for mitochondrial PARG isoforms (lane 1, exon Ia forward and exon IV reverse; lane 2, exon Ia forward and exon V reverse; lane 3, exon Ia forward and exon VI reverse; lane 4, exon Ia forward and exon V/VI reverse) and ARH3 (lane 5, exon V forward and exon VI reverse). The lack of specific amplicons in lanes 2 and 4 confirms the absence of exon V from the mitochondrial PARG isoform. Lanes 1a to 5a represent water controls (no DNA added) to lanes 1 to 5, respectively. (C) Western blot analyses of total cell lysates from 293 cells (lanes 1 and 4) and cells overexpressing PARG(Δ1-460) (lane 2), PARG(Δ1-477) (lane 3), and ARH3 (lane 5), using a FLAG antibody. (D) HeLaS3 cells were transiently cotransfected with mitoPARP-encoding vector and a vector encoding either PARG(Δ1-477) (panels in upper right quadrant), PARG(Δ1-460) (panels in lower left quadrant), or ARH3 (panels in lower right quadrant). As a control, empty plasmid was cotransfected (vehicle; panels in upper left quadrant). Cells were subjected to immunocytochemistry using FLAG antibody to stain PARG or ARH3. The mitoPARP protein was detected by its intrinsic EGFP fluorescence. Bar, 20 μm.
We first tested whether the mRNAs encoding ARH3 or the mitochondrially associated PARG isoform were present in the parental 293 cells because their corresponding enzyme activities could possibly account for PAR degradation in the mitochondria of the 293mitoPARP cells. Indeed, both mRNAs were expressed as revealed by reverse transcription-PCR (Fig. 5B). Several combinations of primers were used to specifically detect the mitochondrion-associated PARG isoform. The results also confirmed the absence of exon V in the human form, as previously noted (33).
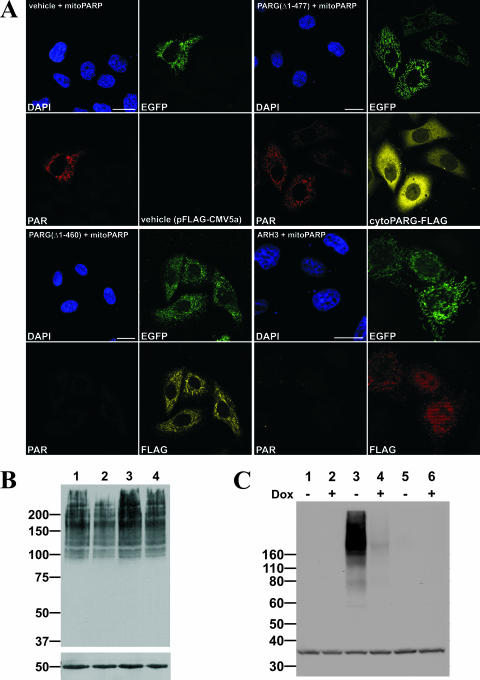
Western blot analyses using FLAG antibody confirmed the synthesis of the encoded proteins following transient transfection of the plasmids into parental 293 cells (Fig. 5C). Expression of the proteins along with the mitoPARP construct in HeLaS3 cells following immunocytochemistry using FLAG antibody established a colocalization of PARG(Δ1-460) with the intrinsic green fluorescence of the mitochondrial PARP construct, whereas PARG(Δ1-477) was distributed throughout the cytoplasm (Fig. 5D). In contrast to the PARG(Δ1-460) construct, ARH3 expression was observed in not only the mitochondria but also the nucleoplasm. Next, we analyzed whether the overexpressed proteins were capable of degrading PAR in the mitochondria, which would imply their localization within the matrix. As shown in Fig. 6A, PAR in mitochondria was detectable in HeLaS3 cells expressing both the mitochondrial PARP construct and PARG(Δ1-477), which localizes to the cytoplasm. In contrast, mitochondrial polymers were no longer detectable when the cells were cotransfected with the vector encoding mitoPARP along with vectors encoding either mitochondrial PARG(Δ1-460) or ARH3 (Fig. 6A).
FIG. 6.
PARG(Δ1-460) and ARH3 degrade PAR in mitochondria. (A) Images from confocal laser scanning of HeLaS3 cells transiently cotransfected with vector encoding mitoPARP and either PARG(Δ1-460) harboring a predicted mitochondrial targeting sequence (panels in lower left quadrant) or ARH3 (panels in lower right quadrant). Control cells were cotransfected with mitoPARP-encoding vector and empty plasmid (vehicle; panels in upper left quadrant) and a vector encoding PARG(Δ1-477), which lacks the predicted targeting sequence (panels in upper right quadrant). Bar, 20 μm. (B) Stably transfected 293mitoPARP cells were either transfected with vectors encoding PARG(Δ1-460) (lane 2), PARG(Δ1-477) (lane 3), and empty plasmid (lane 4) or not transfected (lane 1) and subjected to Western blot analysis using 10H antibody. Fifty micrograms of total protein was separated by 7% SDS-PAGE; β-tubulin was used as loading control. (C) Doxycycline-induced (+) and noninduced (−) Flp-In T-REx 293 cells expressing ARH3 under control of a tetracycline-inducible promoter were transfected with the vector encoding mitoPARP (lane 3 and 4). Control cells were transfected with empty vector (lane 5 and 6) or left untransfected (lane 1 and 2). For Western blot analysis of PAR, 50 μg was separated by 4 to 12% SDS-PAGE. As a loading control eukaryotic translation initiation factor 2α was used.
To further substantiate the degradation of intramitochondrial PAR by PARG(Δ1-460), we used the stably transfected 293mitoPARP cells, which constitutively contain mitochondrial PAR. Following transient transfection with the PARG constructs, cell lysates were subjected to Western blot analysis. Although transient transfection affects only a subpopulation of the cells, the amount of polymer was notably decreased upon expression of mitochondrial PARG(Δ1-460). Comparing lane 2 in Fig. 6B to the controls (lanes 1, 3, and 4), the amount of detectable polymer was reduced. The presence of PAR was unaffected in response to transient transfection of the vector encoding cytoplasmic PARG(Δ1-477) (Fig. 6B, lane 3).
To verify the mitochondrial activity of ARH3, we made use of stably transfected T-REx cell lines expressing ARH3 under control of a doxycycline-inducible promoter. When these cells were transfected with the mitoPARP construct, they exhibited intense staining for PAR in Western blot analyses (Fig. 6C, lane 3). However, when ARH3 expression was induced with doxycycline during transfection, the amount of polymer was reduced to the limit of detection (Fig. 6C, lane 4).
DISCUSSION
This study establishes the presence of two independent PAR-degrading activities within the matrix of mitochondria. Thereby, it adds an important new angle to NAD metabolism in these organelles as well as to the metabolism of PAR. The utility of PAR generation in mitochondria has provided a tool to demonstrate the functional activity of ARH3 and a PARG isoform within the matrix. Moreover, it served to visualize specific downregulation of the mitochondrial NAD content. Targeted enzyme expression might indeed emerge as a powerful method to modulate the available amount of small molecules whose concentration cannot be easily adjusted (e.g., pharmacologically) in a living cell, let alone an organelle. For example, varying NAD kinase expression led to a 15-fold difference in the cytoplasmic NADP(H) content (46).
In normal cells, the formation of detectable amounts of PAR is catalyzed by nuclear PARP1 in response to genotoxic stress, and this occurs transiently. Thus, our initial observation that expression of PARP1 activity within mitochondria is followed by the formation of detectable polymers within this compartment was somewhat surprising because PARP1-mediated cell death has often been associated with energy depletion due to increased consumption of NAD (2, 8, 11, 56). The truncated PARP1 construct used here lacks the DNA-binding domain and consequently cannot be activated by DNA. It exhibits a catalytic activity far lower than that of the DNA-activated full-length enzyme (32, 51). Nevertheless, the accumulation of mitochondrial polymers was similar to that found in the nucleus after genotoxic treatment yet did not appear to have severely toxic effects. The increased dependence of 293mitoPARP cells on glycolysis and the decreased membrane potential indicated a lack of NAD in these mitochondria, but the increased NAD consumption by mitochondrial expression of PARP activity evidently did not lower the cellular NAD content to an extent sufficient to induce cell death. In contrast to plants (9) and the yeast Saccharomyces cerevisiae (52), a direct exchange between mitochondrial and cytosolic NAD is not known for human cells. It is therefore unlikely that the deficiency in mitochondrial NAD in 293mitoPARP cells is compensated by NAD from other cellular compartments. Instead, a decrease in mitochondrial NAD might be compensated by upregulation of glycolysis or de novo synthesis of this nucleotide within the organelles.
Of note, the products of the two identified genes encoding PAR-degrading enzymes in mammals reside within both the nucleus and mitochondria. While the requirement of this activity in the nucleus is obvious due to the transiently high activity of PARP1, the physiological relevance of PAR-degrading activity in the mitochondrial matrix is puzzling. Recent reports suggested that PAR might be the trigger to release AIF from the mitochondria whereupon it translocates to the nucleus (5, 57). According to our observations, uptake of the polymers into mitochondria is not required during PARP1-mediated apoptosis. The question then arises as to what the role of PAR-degrading activities within these organelles could be. Although the presence of poly-ADP-ribosylation in mitochondria has been considered (49), conclusive evidence supporting this possibility is lacking. However, at least for ARH3, the generation of a potential alternative substrate within mitochondria has been demonstrated. Namely, OAADPR, which is cleaved by ARH3 to ADP-ribose and acetate (41), is formed during the deacetylation of ACS2 by SIRT3 within the matrix of mitochondria (22, 48). It would indeed be interesting to study the possibility of a functional relationship between ARH3 and this regulatory mechanism. On the other hand, PARG activity has so far been associated only with PAR metabolism. Indeed, the PAR-degrading activity of PARG appears to be vital, as indicated by the rescue of a lethal phenotype in embryonic trophoblast stem cells of PARG knockout mice by 3-aminobenzamide, a PARP inhibitor (26). Therefore, assigning a functional role to a PARG isoform in mitochondria must, inevitably, be speculative at this point. Poly-ADP-ribosylation might indeed take place within mitochondria. One could argue, however, that the ARH3 activity would be sufficient to degrade the small, and in normal cells, essentially undetectable amounts of polymers. On the other hand, the intramitochondrial PARG activity might explain why possible PAR synthesis in this organelle has so far escaped detection. Second, at least the mitochondrial PARG isoform could possess a hitherto unrecognized catalytic activity which might be required within the organelles. To our knowledge, there is no indication in the literature which would support this possibility. Neither full-length PARG (41) nor in vitro expressed truncated PARG (data not shown) catalyzed any measurable OAADPR cleavage.
The observed partial nuclear localization of ARH3 is in accordance with previous reports although our experiments indicate that, when overexpressed as C-terminally FLAG-tagged protein, it mainly localizes to mitochondria. Thus, akin to PARG, ARH3 may also have a differentially regulated cellular distribution. Previous studies revealed PARG activity to occur predominantly in the cytoplasm (34, 39, 55), whereas the full-length protein is exclusively found in the nucleus (34). Moreover, PARG isoforms were reported to redistribute in response to genotoxic stress and the ensuing PARP-1 activation (21). Although in vitro PARG has been observed to degrade PAR much more efficiently than ARH3 (40), our results indicate that in mitochondria of living cells, ARH3 is capable of efficient PAR degradation.
In conclusion, our investigations have identified two proteins of NAD metabolism which are present in the mitochondrial matrix of human cells. While for ARH3 a proposed function is the cleavage of OAADPR generated during protein deacetylation, the presence of a PARG isoform in this compartment raises critical questions regarding the cellular PAR metabolism. The studies presented here support an important role for mitochondrial NAD in addition to energy conversion and provide a model system to explore the functions of this pool in living cells.
Acknowledgments
We are indebted to M.-H. Ramírez-Hernández for her important contributions to the design and initial experiments of this study. The confocal imaging was performed at the Molecular Imaging Center (Fuge, Norwegian Research Council), University of Bergen.
M.N., M.Z., and F.K.-N. designed this study; M.N. established the mitoPARP system and performed the experiments with the PARG constructs. S.K. applied the mitoPARP system to ARH3 and developed the ARH3 expression systems. All authors analyzed data. M.N. and M.Z. wrote the paper.
This work was supported by funds from the Norwegian Research Council (172219/V40) and the Deutsche Forschungsgemeinschaft (ZI 541/4-2 to M.Z. and No310/6 to F.K.-N.). S.K. is the recipient of a stipend from the Studienstiftung des Deutschen Volkes.
Footnotes
Published ahead of print on 8 November 2007.
REFERENCES
- 1.Ahuja, N., B. Schwer, S. Carobbio, D. Waltregny, B. J. North, V. Castronovo, P. Maechler, and E. Verdin. 22 August 2007. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J. Biol. Chem. doi/ 10.1074/jbc.M705488200. [DOI] [PubMed]
- 2.Alano, C. C., W. Ying, and R. A. Swanson. 2004. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J. Biol. Chem. 27918895-18902. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Gonzalez, R., and M. K. Jacobson. 1987. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry 263218-3224. [DOI] [PubMed] [Google Scholar]
- 4.Ame, J. C., C. Spenlehauer, and G. de Murcia. 2004. The PARP superfamily. Bioessays 26882-893. [DOI] [PubMed] [Google Scholar]
- 5.Andrabi, S. A., N. S. Kim, S. W. Yu, H. Wang, D. W. Koh, M. Sasaki, J. A. Klaus, T. Otsuka, Z. Zhang, R. C. Koehler, P. D. Hurn, G. G. Poirier, V. L. Dawson, and T. M. Dawson. 2006. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA 10318308-18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, F., C. Lau, M. Dahlmann, and M. Ziegler. 2005. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 28036334-36341. [DOI] [PubMed] [Google Scholar]
- 7.Berger, F., M. H. Ramirez-Hernandez, and M. Ziegler. 2004. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem. Sci. 29111-118. [DOI] [PubMed] [Google Scholar]
- 8.Berger, N. A. 1985. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat. Res. 1014-15. [PubMed] [Google Scholar]
- 9.Bykova, N. V., and I. M. Møller. 2001. Involvement of matrix NADP turnover in the oxidation of NAD-linked substrates by pea leaf mitochondria. Physiol. Plant. 111448-456. [DOI] [PubMed] [Google Scholar]
- 10.Chiarugi, A. 2002. Poly(ADP-ribose) polymerase: killer or conspirator? The “suicide hypothesis” revisited. Trends Pharmacol. Sci. 23122-129. [DOI] [PubMed] [Google Scholar]
- 11.Cipriani, G., E. Rapizzi, A. Vannacci, R. Rizzuto, F. Moroni, and A. Chiarugi. 2005. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J. Biol. Chem. 28017227-17234. [DOI] [PubMed] [Google Scholar]
- 12.Corda, D., and M. Di Girolamo. 2003. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 221953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes, U., W. M. Tong, D. L. Coyle, M. L. Meyer-Ficca, R. G. Meyer, V. Petrilli, Z. Herceg, E. L. Jacobson, M. K. Jacobson, and Z. Q. Wang. 2004. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 247163-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cossarizza, A., M. Baccarani-Contri, G. Kalashnikova, and C. Franceschi. 1993. A new method for the cytofluorimetric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1). Biochem. Biophys. Res. Commun. 19740-45. [DOI] [PubMed] [Google Scholar]
- 15.Denu, J. M. 2005. The Sir 2 family of protein deacetylases. Curr. Opin. Chem. Biol. 9431-440. [DOI] [PubMed] [Google Scholar]
- 16.Di Girolamo, M., N. Dani, A. Stilla, and D. Corda. 2005. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2724565-4575. [DOI] [PubMed] [Google Scholar]
- 17.Glowacki, G., R. Braren, K. Firner, M. Nissen, M. Kuhl, P. Reche, F. Bazan, M. Cetkovic-Cvrlje, E. Leiter, F. Haag, and F. Koch-Nolte. 2002. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 111657-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubisha, O., L. A. Rafty, C. L. Takanishi, X. Xu, L. Tong, A. L. Perraud, A. M. Scharenberg, and J. M. Denu. 2006. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J. Biol. Chem. 28114057-14065. [DOI] [PubMed] [Google Scholar]
- 19.Haag, F., and F. Koch-Nolte. 1997. The vertebrate gene family of mono(ADP-ribosyl)transferases. Proposal for a unified nomenclature. Adv. Exp. Med. Biol. 419459-462. [PubMed] [Google Scholar]
- 20.Haigis, M. C., R. Mostoslavsky, K. M. Haigis, K. Fahie, D. C. Christodoulou, A. J. Murphy, D. M. Valenzuela, G. D. Yancopoulos, M. Karow, G. Blander, C. Wolberger, T. A. Prolla, R. Weindruch, F. W. Alt, and L. Guarente. 2006. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126941-954. [DOI] [PubMed] [Google Scholar]
- 21.Haince, J. F., M. E. Ouellet, D. McDonald, M. J. Hendzel, and G. G. Poirier. 2006. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim. Biophys. Acta 1763226-237. [DOI] [PubMed] [Google Scholar]
- 22.Hallows, W. C., S. Lee, and J. M. Denu. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 10310230-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassa, P. O., S. S. Haenni, M. Elser, and M. O. Hottiger. 2006. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70789-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrero-Yraola, A., S. M. Bakhit, P. Franke, C. Weise, M. Schweiger, D. Jorcke, and M. Ziegler. 2001. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 202404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, S. J., T. M. Dawson, and V. L. Dawson. 2004. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol. Sci. 25259-264. [DOI] [PubMed] [Google Scholar]
- 26.Koh, D. W., A. M. Lawler, M. F. Poitras, M. Sasaki, S. Wattler, M. C. Nehls, T. Stoger, G. G. Poirier, V. L. Dawson, and T. M. Dawson. 2004. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 10117699-17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolisek, M., A. Beck, A. Fleig, and R. Penner. 2005. Cyclic ADP-ribose and hydrogen peroxide synergize with ADP-ribose in the activation of TRPM2 channels. Mol. Cell 1861-69. [DOI] [PubMed] [Google Scholar]
- 28.Lee, H. C. 1997. Mechanisms of calcium signaling by cyclic ADP-ribose and NAADP. Physiol. Rev. 771133-1164. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H. C., R. Aarhus, R. Graeff, M. E. Gurnack, and T. F. Walseth. 1994. Cyclic ADP ribose activation of the ryanodine receptor is mediated by calmodulin. Nature 370307-309. [DOI] [PubMed] [Google Scholar]
- 30.Malanga, M., and F. R. Althaus. 2005. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 83354-364. [DOI] [PubMed] [Google Scholar]
- 31.Masson, M., C. Niedergang, V. Schreiber, S. Muller, Menissier-de Murcia, J., and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 183563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendoza-Alvarez, H., and R. Alvarez-Gonzalez. 2004. The 40 kDa carboxy-terminal domain of poly(ADP-ribose) polymerase-1 forms catalytically competent homo- and heterodimers in the absence of DNA. J. Mol. Biol. 336105-114. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, R. G., M. L. Meyer-Ficca, C. J. Whatcott, E. L. Jacobson, and M. K. Jacobson. 2007. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp. Cell Res. 3132920-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer-Ficca, M. L., R. G. Meyer, D. L. Coyle, E. L. Jacobson, and M. K. Jacobson. 2004. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297521-532. [DOI] [PubMed] [Google Scholar]
- 35.Michan, S., and D. Sinclair. 2007. Sirtuins in mammals: insights into their biological function. Biochem. J. 4041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller-Dieckmann, C., S. Kernstock, M. Lisurek, J. P. von Kries, F. Haag, M. S. Weiss, and F. Koch-Nolte. 2006. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. USA 10315026-15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11437-444. [DOI] [PubMed] [Google Scholar]
- 38.Oei, S. L., C. Keil, and M. Ziegler. 2005. Poly(ADP-ribosylation) and genomic stability. Biochem. Cell Biol. 83263-269. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi, S., M. Kanai, S. Hanai, F. Uchiumi, H. Maruta, S. Tanuma, and M. Miwa. 2003. Subcellular localization of poly(ADP-ribose) glycohydrolase in mammalian cells. Biochem. Biophys. Res. Commun. 307915-921. [DOI] [PubMed] [Google Scholar]
- 40.Oka, S., J. Kato, and J. Moss. 2006. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281705-713. [DOI] [PubMed] [Google Scholar]
- 41.Ono, T., A. Kasamatsu, S. Oka, and J. Moss. 2006. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. USA 10316687-16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onyango, P., I. Celic, J. M. McCaffery, J. D. Boeke, and A. P. Feinberg. 2002. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 9913653-13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto, H., P. A. Reche, F. Bazan, K. Dittmar, F. Haag, and F. Koch-Nolte. 2005. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poitras, M. F., D. W. Koh, S. W. Yu, S. A. Andrabi, A. S. Mandir, G. G. Poirier, V. L. Dawson, and T. M. Dawson. 2007. Spatial and functional relationship between poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in the brain. Neuroscience 148198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollak, N., C. Dölle, and M. Ziegler. 2007. The power to reduce: pyridine nucleotides—small molecules with a multitude of functions. Biochem. J. 402205-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollak, N., M. Niere, and M. Ziegler. 13 September 2007. NAD kinase levels control the NADPH concentration in human cells. J. Biol. Chem. doi/ 10.1074/jbc.M704442200. [DOI] [PubMed]
- 47.Schreiber, V., F. Dantzer, J. C. Ame, and G. de Murcia. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7517-528. [DOI] [PubMed] [Google Scholar]
- 48.Schwer, B., J. Bunkenborg, R. O. Verdin, J. S. Andersen, and E. Verdin. 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA 10310224-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scovassi, A. I. 2004. Mitochondrial poly(ADP-ribosylation): from old data to new perspectives. FASEB J. 181487-1488. [DOI] [PubMed] [Google Scholar]
- 50.Seman, M., S. Adriouch, F. Haag, and F. Koch-Nolte. 2004. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr. Med. Chem. 11857-872. [DOI] [PubMed] [Google Scholar]
- 51.Simonin, F., L. Hofferer, P. L. Panzeter, S. Muller, M. G. de, and F. R. Althaus. 1993. The carboxyl-terminal domain of human poly(ADP-ribose) polymerase. Overproduction in Escherichia coli, large scale purification, and characterization. J. Biol. Chem. 26813454-13461. [PubMed] [Google Scholar]
- 52.Todisco, S., G. Agrimi, A. Castegna, and F. Palmieri. 2006. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2811524-1531. [DOI] [PubMed] [Google Scholar]
- 53.Ueda, K., J. Oka, S. Naruniya, N. Miyakawa, and O. Hayaishi. 1972. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys. Res. Commun. 46516-523. [DOI] [PubMed] [Google Scholar]
- 54.Vahsen, N., C. Cande, J. J. Briere, P. Benit, N. Joza, N. Larochette, P. G. Mastroberardino, M. O. Pequignot, N. Casares, V. Lazar, O. Feraud, N. Debili, S. Wissing, S. Engelhardt, F. Madeo, M. Piacentini, J. M. Penninger, H. Schagger, P. Rustin, and G. Kroemer. 2004. AIF deficiency compromises oxidative phosphorylation. EMBO J. 234679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winstall, E., E. B. Affar, R. Shah, S. Bourassa, I. A. Scovassi, and G. G. Poirier. 1999. Preferential perinuclear localization of poly(ADP-ribose) glycohydrolase. Exp. Cell Res. 251372-378. [DOI] [PubMed] [Google Scholar]
- 56.Ying, W., C. C. Alano, P. Garnier, and R. A. Swanson. 2005. NAD+ as a metabolic link between DNA damage and cell death. J. Neurosci. Res. 79216-223. [DOI] [PubMed] [Google Scholar]
- 57.Yu, S. W., S. A. Andrabi, H. Wang, N. S. Kim, G. G. Poirier, T. M. Dawson, and V. L. Dawson. 2006. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. USA 10318314-18319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, S. W., H. Wang, M. F. Poitras, C. Coombs, W. J. Bowers, H. J. Federoff, G. G. Poirier, T. M. Dawson, and V. L. Dawson. 2002. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297259-263. [DOI] [PubMed] [Google Scholar]