Abstract
Loading of export factors onto mRNAs is a key step in gene expression. In vertebrates, splicing plays a role in this process. Specific protein complexes, exon junction complex and transcription/export complex, are loaded onto mRNAs in a splicing-dependent manner, and adaptor proteins such as Aly/REF in the complexes in turn recruit mRNA exporter TAP-p15 onto the RNA. By contrast, how export factors are recruited onto intronless mRNAs is largely unknown. We previously showed that Aly/REF is preferentially associated with intronless mRNAs in the nucleus. Here we show that Aly/REF could preferentially bind intronless mRNAs in vitro and that this binding was stimulated by RNA helicase UAP56 in an ATP-dependent manner. Consistently, an ATP binding-deficient UAP56 mutant specifically inhibited mRNA export in Xenopus oocytes. Interestingly, ATP activated the RNA binding activity of UAP56 itself. ATP-bound UAP56 therefore bound to both RNA and Aly/REF, and as a result ATPase activity of UAP56 was cooperatively stimulated. These results are consistent with a model in which ATP-bound UAP56 chaperones Aly/REF onto RNA, ATP is then hydrolyzed, and UAP56 dissociates from RNA for the next round of Aly/REF recruitment. Our finding provides a mechanistic insight into how export factors are recruited onto mRNAs.
Eukaryotic cells are separated by the nuclear envelope into two major compartments, the nucleus and the cytoplasm. This compartmentalization requires transport through the nuclear pore complexes, the channels for material exchange across the nuclear envelope barrier (reviewed in reference 40). The vast majority of RNA species, following their synthesis and processing in the nucleus, are exported to the cytoplasm. Different RNA species, such as tRNAs, U snRNAs, mRNAs, and rRNAs, utilize distinct export pathways, i.e., distinct sets of export factors (reviewed in reference 5). Accumulating evidence shows that the pathway of RNA export can influence the fate of a given RNA in the cytoplasm (5), indicating the biological importance of the choice of RNA export pathway. However, how export factors of each pathway are loaded onto the corresponding RNA is largely unknown.
The major exporter for mRNAs is the TAP-p15 heterodimer (10, 16, 33). TAP is one of the few non-importinβ family transport receptors known to date. Although TAP itself can bind RNA, its recruitment to mRNAs requires adaptor proteins. RNA binding adaptor proteins, including Aly/REF and shuttling SR proteins, should first be recruited to mRNA, and these adaptor proteins in turn recruit mRNA exporter TAP-p15 heterodimer onto the RNA through protein-protein interactions (12, 13, 31, 37, 39, 41).
In yeast (Saccharomyces cerevisiae), the recruitment of adaptor proteins is coupled to transcription. Aly/REF (Yra1 in yeast) together with the DEXD-box RNA helicase UAP56 (Sub2 in yeast) interacts with the transcription elongation complex (THO complex), forming a larger protein complex (1, 38). Owing to this complex formation, Aly/REF and UAP56 are recruited onto the nascent mRNA transcript during transcription elongation, and hence this larger protein complex is termed TREX (transcription/export) complex (1, 38). In vertebrates, however, splicing rather than transcription plays an important role in loading these adaptor proteins onto RNA. Aly/REF and UAP56 are loaded onto mRNAs in a splicing-dependent manner as components of exon junction complex and TREX complex (3, 17, 20, 25). Interaction between Aly/REF and UAP56 plays an important role in this recruitment (23, 36).
However, splicing is not essential for recruiting export factors onto mRNAs. mRNAs produced from genes without introns (intronless mRNAs) are also exported efficiently. A certain class of intronless mRNAs contains specific RNA elements, e.g., the intronless transport element in histone H2A mRNA (14). Such RNA elements function as high-affinity binding sites for specific RNA binding adaptor proteins (12, 13). Another class of intronless mRNAs, which apparently does not contain such specific elements, still can recruit adaptor proteins such as Aly/REF. It is already well known that Aly/REF interacts with mRNAs in a splicing-independent manner, since Aly/REF plays a role in the export of intronless mRNAs without specific elements (6, 18, 26, 29, 31). We previously showed that a stretch of unstructured RNA region of a certain length (>300 nucleotides [nt]) can function as a splicing-independent feature to recruit Aly/REF onto RNA (26). Although the cap structure has recently been shown to be able to recruit Aly/REF (3, 27), this structure is stimulatory only for mRNA export. Thus, how Aly/REF is recruited to intronless mRNAs is largely unknown. More specifically, the contribution of other components, such as RNA helicase UAP56, in the export of intronless mRNAs has not been explored.
In this study, we focused on the role of UAP56 in the recruitment of Aly/REF onto intronless mRNAs and, based on the results, proposed a model of how UAP56 recruits Aly/REF onto intronless mRNAs. Although UAP56 has motifs of DExD-box RNA helicases and an RNA-unwinding activity (34), our findings suggest that UAP56 functions as an ATP-dependent molecular chaperone for RNA-Aly/REF complex formation rather than as an RNA helicase that unwinds RNA secondary structures or displaces proteins from RNA (4).
MATERIALS AND METHODS
DNA constructs and recombinant proteins.
The UAP56-GET (K95E) mutant was constructed by PCR-based mutagenesis. UAP56-LAT (S228L) cDNA was a gift from Elisa Izaurralde. DNA fragments for Flag-tagged UAP56 wild type and mutants were PCR amplified from the corresponding cDNAs and cloned into pGEX-6p. Aly/REF was also cloned in pGEX-6p. Glutathione S-transferase (GST)-Flag-tagged UAP56 and GST-Aly/REF were expressed and purified with glutathione beads by the standard procedure. When necessary, the expressed GST fusion proteins were subsequently treated with PreScission protease (2 U/μl; GE Healthcare) to remove the GST sequence. Flag-UAP56 and Aly/REF recombinant proteins were further purified by MonoQ and heparin-Sepharose chromatography, respectively.
In vitro RNA binding assay.
32P-labeled in vitro-transcribed RNAs were mixed with recombinant proteins in 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 1.6 mM MgCl2, 0.1 mM EDTA, 6% glycerol at 30°C for 15 min, and immunoprecipitation with anti-Aly/REF antibody (11G5) (reference 17) or anti-Flag antibody (M2; Sigma) was performed as follows. Antibodies were individually bound to Protein A-Sepharose beads (Amersham Biosciences) on a rotating platform at 4°C for 1 h. The above-described mixture of RNA and recombinant proteins was incubated with the antibody-bound beads at 4°C for 1 h. After the beads were washed 5 times with RSB100N buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40), they were incubated in HomoMix (50 mM Tris [pH 7.4], 5 mM EDTA, 1.5% sodium dodecyl sulfate [SDS], 300 mM NaCl, 1.5 mg/ml proteinase K [Nakalai Tesque]) for 30 min at 50°C to elute RNA in the supernatant. RNA was recovered from the supernatant by phenol-chloroform extraction and ethanol precipitation. The recovered RNA was analyzed by denaturing polyacrylamide gel electrophoresis (PAGE). When HeLa nuclear extract (HNE) was employed, HNE was added to the final concentration of 7.5% in the same buffer and incubated at 30°C for 30 min, and immunoprecipitation was performed.
ATP binding assay.
Recombinant proteins (1 μg) and 3,000 Ci/mmol [α-32P]ATP (1 μl, Amersham) were incubated in 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 1.6 mM MgCl2, 0.1 mM EDTA, 6% glycerol in a volume of 20 μl at 30°C for 10 min. The reaction mixture was then placed on a piece of Parafilm and irradiated with 254-nm UV light (Funa-UV-Linker FS-800; Funakoshi, Tokyo, Japan) at a distance of ∼5 cm for 10 min. The cross-linked protein was analyzed by SDS-PAGE and autoradiography.
GST pull-down.
GST or GST-Aly/REF (0.2 μM) was incubated with FLAG-UAP56 (2 μM) in 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 1.6 mM MgCl2, 0.1 mM EDTA, 6% glycerol, and 0.5 mg/ml RNase A at 30°C for 15 min in the presence or absence of 2 mM nucleotide. The mixture was then mixed with glutathione beads at 4°C for 60 min, and bound material was recovered and analyzed by Western blotting with anti-Flag antibody (M2).
ATPase assay.
The ATPase assay of UAP56 was performed essentially as described previously (19), except that the reaction was done in 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 1.6 mM MgCl2, 0.1 mM EDTA, 6% glycerol at 30°C for 60 min and terminated by adjusting the mixture to a final concentration of 0.75% SDS, 2.5 mM EDTA, 0.75 mg/ml proteinase K.
RNA microinjection and RNA immunoprecipitation.
RNA microinjection into Xenopus oocytes and RNA immunoprecipitation were performed as previously described (26). Analysis and quantitation of RNA bands were performed with BAS-2500 (Fujifilm) and Image Gauge version 3.45 (Fujifilm).
RESULTS AND DISCUSSION
Aly/REF can preferentially bind to mRNAs in a splicing-independent manner, and its binding is stimulated by ATP in HNE.
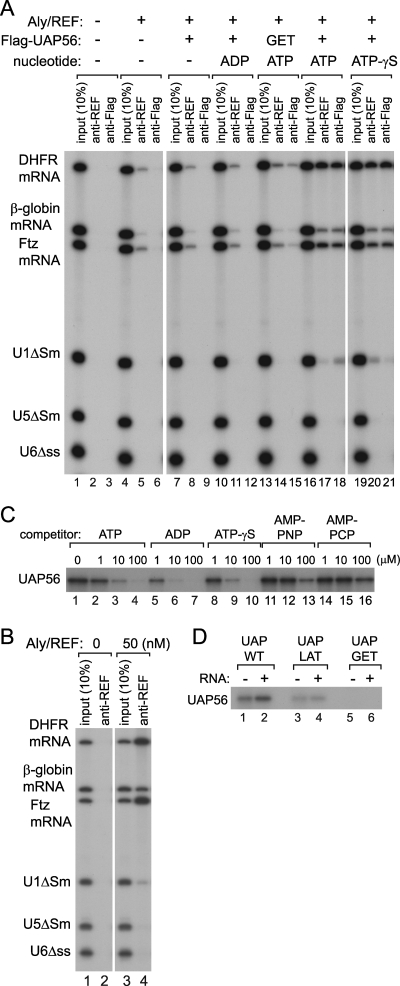
We previously showed that Aly/REF is preferentially associated with mRNAs in the nuclei of Xenopus oocytes (26, 29). Consistent with this, when a mixture of 32P-labeled in vitro-transcribed RNAs containing two intronless mRNAs (DHFR and Ftz mRNAs), CTE (constitutive transport element) (reference 32, e.g.), and U1 and U6 snRNAs was injected into the nuclei of Xenopus oocytes, the endogenous Xenopus Aly/REF protein preferentially bound to the two intronless mRNAs in the nucleus, as judged by immunoprecipitation with a monoclonal anti-Aly/REF antibody (Fig. 1A).
FIG. 1.
Recruitment of Aly/REF onto intronless mRNAs. (A) A mixture of 32P-labeled in vitro-transcribed RNAs containing two intronless mRNAs (DHFR and Ftz mRNAs), CTE, U1ΔSm, and U6Δss snRNAs was injected into the nuclei of Xenopus oocytes. The nuclear fraction was prepared after 1 h, and immunoprecipitation (IP) was performed with anti-Aly/REF monoclonal antibody (anti-REF, 11G5; ImmuQuest) (17) or control antibody (anti-Myc, 9E10; Sigma). RNA precipitated with each antibody was recovered and analyzed by denaturing PAGE and autoradiography. The input lanes contained 10% of the input mixture. (B) The same 32P-labeled RNA mixture as for panel A, except that tRNAPhe was supplemented, was incubated in 7.5% HNE, 12 mM HEPES-KOH (pH 7.9), 60 mM KCl, 1.6 mM MgCl2, 0.1 mM EDTA, 6% glycerol at 30°C for 30 min in the absence (−) or presence (+) of 2 mM ATP. After the incubation, immunoprecipitation was performed and the coprecipitated RNA was analyzed as described for panel A.
To test if this is also the case in vitro, we next performed in vitro RNA-protein binding assays. A 32P-labeled RNA mixture similar to that used for Fig. 1A was incubated with HNE at 30°C for 30 min. After the incubation, the endogenous Aly/REF protein in HNE was precipitated with a monoclonal anti-Aly/REF antibody, and the coprecipitated RNA was analyzed by denaturing PAGE (Fig. 1B). The endogenous Aly/REF protein in HNE appeared to bind preferentially to intronless mRNAs, although the binding was inefficient under this condition, due to the limited amount of Aly/REF protein in the limited amount of HNE used in the assay (Fig. 1B, lane 2). Interestingly, this Aly/REF binding to intronless mRNAs was stimulated by the presence of ATP (Fig. 1B, lanes 4 to 6). RNA binding of purified recombinant Aly/REF protein alone, without HNE, was not affected by the presence of ATP (data not shown). These results suggested that the binding preference of Aly/REF for mRNAs in vivo could be recapitulated in vitro at least to some extent and that a factor(s) in HNE stimulated the binding of Aly/REF to intronless mRNAs in an ATP-dependent manner.
Loading of Aly/REF onto intronless mRNAs by UAP56 is dependent on ATP but not ATP hydrolysis in vitro.
Since it was already shown that Aly/REF interacts with DExD-box RNA helicase UAP56 (23, 36), a good candidate for the ATP-dependent stimulatory factor in HNE was UAP56. To test this possibility, we added recombinant Flag-tagged UAP56 protein in RNA binding experiments with recombinant Aly/REF (Fig. 2A). Note that HNE was not used in these experiments and that the same 32P-labeled RNAs as in Fig. 1A were employed, except that CTE was omitted but intronless β-globin mRNA and U5ΔSm RNA were supplemented.
FIG. 2.
Recruitment of Aly/REF onto RNA by UAP56. (A) The same 32P-labeled RNA mixture as in Fig. 1A, except that CTE was omitted but intronless β-globin mRNA and U5ΔSm RNA were supplemented, was incubated with (+) or without (−) recombinant Aly/REF (10 nM) and recombinant Flag-tagged UAP56 (10 nM), either wild type (+) or GET mutant (GET, K95E), in either the absence (−) or presence (+) of 2 mM ADP, ATP, or ATP-γS at 30°C for 15 min. After the incubation, immunoprecipitation was performed with anti-Aly/REF antibody (anti-REF, 11G5; ImmuQuest) or anti-Flag antibody (M2; Sigma) and the coprecipitated RNA was analyzed as described for Fig. 1. (B) The same experiment as in panel A, lanes 1, 2, 4, and 5, except that a higher concentration (50 nM) of Aly/REF was employed. (C) Recombinant Flag-UAP56 (1 μg) and 3,000 Ci/mmol [α-32P]ATP (1 μl; Amersham) were incubated in the absence or presence of increasing concentrations of unlabeled ATP, ADP, ATP-γS, AMP-PNP, and AMP-PCP as competitors and irradiated with UV light as described in Materials and Methods. The cross-linked protein was analyzed by SDS-PAGE and autoradiography. (D) A UV cross-linking experiment similar to that shown in panel C was performed with wild-type UAP56 (UAP-WT), UAP56 LAT mutant (UAP-LAT, S228L), or UAP56 GET mutant (UAP-GET, K95E), without nucleotide competitors, in the absence (−) or presence (+) of yeast RNA (1 mg/ml; Sigma).
When recombinant Aly/REF (10 nM) alone was employed, precipitation of the three intronless mRNAs was observed, whereas precipitation of the three non-mRNAs was not observed (Fig. 2A, lanes 4 and 5). This is more evident when a higher amount of recombinant Aly/REF (50 nM) was employed (Fig. 2B), suggesting that the Aly/REF protein per se may have an intrinsic property of binding preferentially to mRNAs. Although the molecular mechanism underlying this preferential binding of Aly/REF will be investigated in the future studies, we did not pursue it further in this study.
When recombinant Flag-tagged UAP56 (10 nM) was added in addition to Aly/REF (10 nM), the RNA binding of Aly/REF was unaffected (Fig. 2A, compare lanes 4 and 5 with 7 and 8). However, when the same amount of UAP56 was supplied together with ATP (2 mM), the RNA binding of Aly/REF was dramatically stimulated, with the preference for binding to mRNAs still maintained (Fig. 2A, lanes 16 and 17). Note that Flag-UAP56 also bound to mRNAs directly or indirectly, as judged by the immunoprecipitation with anti-Flag antibody, behaving similarly to Aly/REF (Fig. 2A, lane 18). Stimulation was not observed when ADP was supplied instead of ATP (lanes 10 to 12). Interestingly, however, when ATP-γS, a slowly hydrolyzing ATP analog, was supplied, similar stimulation was observed (lanes 19 to 21). This was not due to contamination of ATP in the ATP-γS preparation, since the enhancing effects of ATP and ATP-γS were equally decreased at limited concentrations (data not shown). These results suggested that UAP56 possessed an activity to stimulate RNA binding of Aly/REF when it was bound to ATP and that ATP hydrolysis was not critical for the activity under these experimental conditions.
When 32P-labeled ATP was incubated with UAP56 and the mixture was irradiated with UV light (254 nm), UAP56 was cross-linked to ATP (Fig. 2C, lane 1) and the cross-linking was abolished by competition with increasing amounts of unlabeled ATP (Fig. 2C, lanes 2 to 4), confirming that UAP56 can bind ATP. ADP and ATP-γS also competed with efficiency comparable to that of ATP (Fig. 2C, lanes 5 to 10), indicating that these two nucleotides can also bind UAP56. When AMP-PNP and AMP-PCP were used as competitors in the same assay, they hardly affected the ATP binding of UAP56 (Fig. 2C, lanes 11 to 16). These nonhydrolyzable ATP analogs bound UAP56 with affinities at least 10 times lower than that of ATP. Consistent with this, the two ATP analogs could not activate UAP56 in the experiment, as in Fig. 2A (data not shown).
To further confirm that the ATP-bound form of UAP56 is essential for the stimulation of RNA binding of Aly/REF, we constructed several UAP56 mutants which may be defective in ATP binding (4). When two candidate mutants, the UAP-LAT (S228L in motif III) and UAP-GET (K95E in motif I) mutants, were chosen and subjected to the ATP binding assay as in Fig. 2C, UAP-LAT was partially defective in ATP binding (Fig. 2D, lanes 3 and 4), while UAP-GET was completely defective (lanes 5 and 6). Although the addition of RNA stimulated the ATP binding of wild-type UAP56 (lanes 1 and 2), ATP binding of UAP-GET was not observed even in the presence of RNA (lane 6). Consistent with this, the UAP-GET protein did not have stimulatory activity for RNA binding of Aly/REF even in the presence of ATP (Fig. 2A, lanes 13 to 15).
An ATP binding-deficient UAP56 mutant inhibits mRNA export in Xenopus oocytes.
Although UAP-GET was defective in stimulation of RNA binding of Aly/REF, it bound to Aly/REF with the same efficiency as wild-type UAP56 (see Fig. 4B, lanes 6 and 12). Therefore, it was anticipated that UAP-GET protein would exert a dominant negative effect on mRNA export. This possibility was then tested in RNA microinjection experiments with Xenopus oocytes.
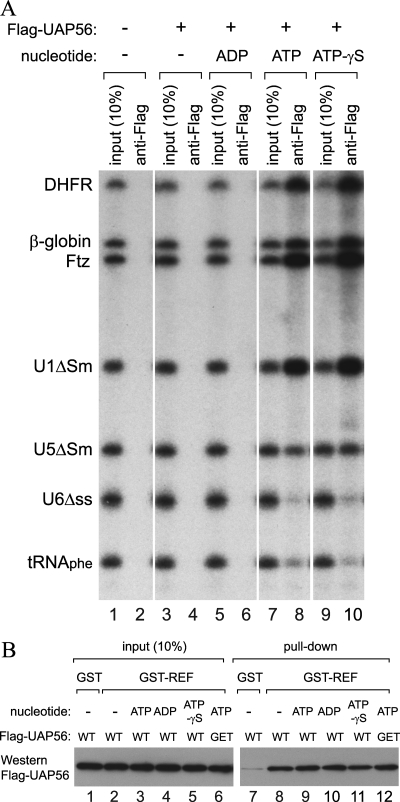
FIG. 4.
Interaction of UAP56 with RNA and Aly/REF. (A) The same 32P-labeled RNA mixture as in Fig. 2A but supplemented with tRNAPhe was incubated with (+) or without (−) recombinant Flag-UAP56 (100 nM) in either the absence (−) or presence (+) of 2 mM ADP, ATP, or ATP-γS at 30°C for 15 min. After the incubation, immunoprecipitation was performed with anti-Flag antibody and the coprecipitated RNA was analyzed as described for Fig. 1. (B) Recombinant Flag-UAP56 wild type (WT) or GET mutant (GET) was incubated with GST alone (GST) or GST-Aly/REF (GST-REF) in the presence of RNase A and in the absence or presence of 2 mM ATP, ADP, or ATP-γS, and GST pull-down was performed as described in Materials and Methods. Flag-UAP56 proteins were visualized by Western blotting with anti-Flag antibody (M2).
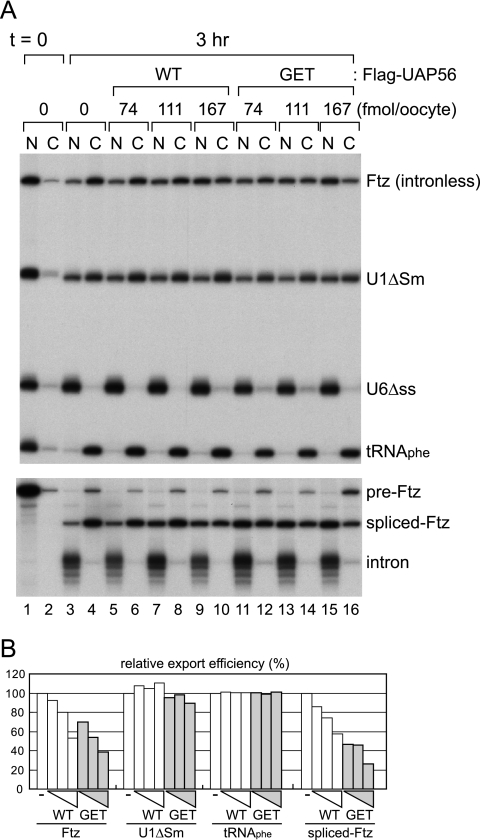
32P-labeled in vitro-transcribed RNAs were microinjected into the nuclei of Xenopus oocytes in either the absence or presence of three different amounts of recombinant Flag-UAP56 wild-type or GET mutant proteins (Fig. 3A; quantitation is shown in Fig. 3B). As was already reported, the proper amount of UAP56 is critical for mRNA export in vivo (23). Consistent with that report, excess wild-type UAP56 inhibited the export of spliced Ftz mRNA in a dose-dependent manner, whereas the same amount of protein hardly affected export of U1 or tRNA (Fig. 3A, lanes 1 to 10; quantitation is shown in Fig. 3B). Note that excess wild-type UAP56 also inhibited the export of intronless Ftz mRNA. More importantly, UAP-GET mutant protein also specifically inhibited both spliced and intronless Ftz mRNAs more strongly than the wild-type protein. The injection of higher doses of wild-type UAP56 or UAP-GET mutant led to progressive export inhibition of U1ΔSm RNA (data not shown). These results indicated that the UAP56 protein and its ATP binding are critical for export of not only spliced mRNAs but also intronless mRNAs. It should be pointed out that splicing of pre-Ftz was not affected by the UAP56 proteins, although UAP56 was originally identified as a splicing factor (7).
FIG. 3.
Effect of UAP56 proteins on RNA export. (A) A 32P-labeled RNA mixture containing intronless Ftz mRNA (Ftz), U1ΔSm, U6Δss RNAs, and tRNAPhe (top) or 32P-labeled pre-Ftz RNA (bottom) was microinjected into the nuclei of Xenopus oocytes with or without three doses of either UAP56 wild-type (WT) or UAP56-GET mutant (GET) proteins. RNA was extracted from nuclear (N) and cytoplasmic (C) fractions immediately (lanes 1 and 2) or 3 h (lanes 3 to 16) after injection and analyzed by 8% denaturing PAGE followed by autoradiography. (B) Quantitation of relative RNA export efficiency shown in panel A.
UAP56 binds to RNA in an ATP-dependent manner.
Previous work suggested that UAP56 interacts with Aly/REF and that this interaction was critical for loading Aly/REF onto spliced mRNAs (23). However, how UAP56 assists the loading of Aly/REF onto RNA, especially intronless mRNAs, has remained largely unknown. We therefore next attempted to obtain mechanistic insight into how UAP56 and its ATP binding assist the loading of Aly/REF onto RNA. One possibility was that the ATP-bound form of UAP56 simultaneously binds Aly/REF and RNA, thereby helping Aly/REF to bind to RNA. To test this possibility, we first examined the RNA binding activity of UAP56 itself in an RNA immunoprecipitation experiment similar to that shown in Fig. 2A (Fig. 4A). UAP56 bound RNA efficiently in the presence of ATP or ATP-γS but not at all in the presence of ADP or in the absence of nucleotides (Fig. 4A). Note that the RNA binding specificity of UAP56 was broader than that of Aly/REF (compare Fig. 4A with 2B). UAP56 bound efficiently to mRNAs and U1, somewhat less efficiently to U5, and weakly to U6 and tRNA (Fig. 4A).
We also examined the effect of nucleotides on the protein-protein interaction between UAP56 and Aly/REF. Recombinant Flag-UAP56 protein was pulled down by recombinant GST-Aly/REF protein in the absence or presence of different nucleotides (Fig. 4B). The interaction of UAP56 with Aly/REF was not affected by the presence of ADP, ATP, or ATP-γS (lanes 2 to 5 and 8 to 11). UAP-GET mutant protein that was defective in ATP binding also bound to Aly/REF with the same efficiency as wild-type UAP56. These results indicated that ATP binding does not affect the protein-protein interaction between UAP56 and Aly/REF. Nevertheless, these results are consistent with a scenario in which the ATP-bound form of UAP56 simultaneously binds Aly/REF and RNA and assists Aly/REF binding to RNA.
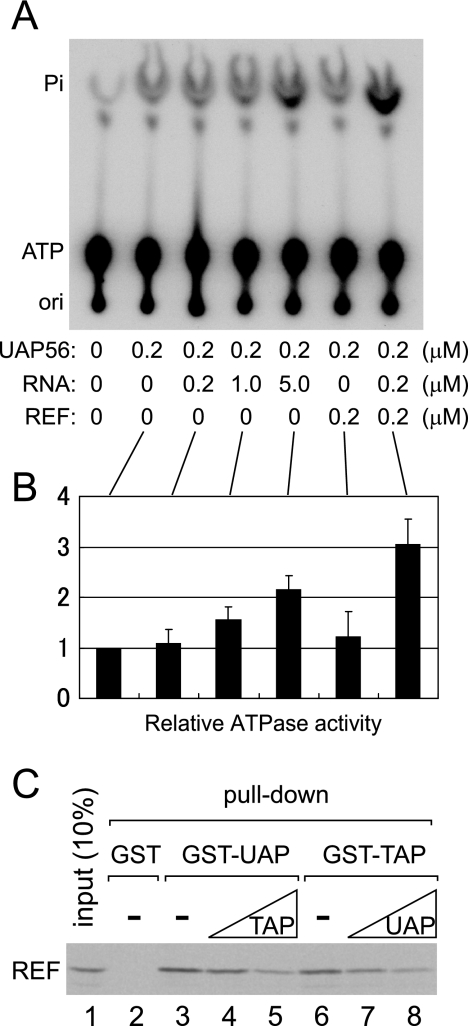
ATP hydrolysis by UAP56 is cooperatively stimulated by RNA and Aly/REF.
Although ATP hydrolysis was not critical for the activity of Aly/REF loading onto RNA under our experimental conditions, it is likely that hydrolysis of ATP bound to UAP56 takes place in vivo. To obtain a clue regarding the role of ATP hydrolysis on UAP56, we next examined the ATPase activity of UAP56 in vitro. UAP56 possessed a basal ATPase activity, and the activity was stimulated by increasing amounts of RNA (100-nt RNA derived from β-globin mRNA) (Fig. 5A and B) (references 34 and 35). However, when a small amount (0.2 μM) of RNA was employed, the stimulation of ATPase activity was hardly observed (1.1-fold) (Fig. 5A and B). When Aly/REF protein, which should bind to UAP56, was supplied (0.2 μM), the stimulation was very modest (1.2-fold) (Fig. 5A and B) (34). However, when both RNA (0.2 μM) and Aly/REF (0.2 μM) were supplied, the ATPase activity of UAP56 was cooperatively enhanced (3.0-fold) (Fig. 5A and B), suggesting that ATP hydrolysis may take place when ATP-bound UAP56 binds simultaneously to RNA and Aly/REF in vivo (see discussion below).
FIG. 5.
ATPase activity of UAP56. (A) ATPase activity of recombinant Flag-UAP56 was measured as described in Materials and Methods, with or without a 100-nt RNA derived from the β-globin mRNA sequence and/or recombinant Aly/REF. Pi, released phosphate; ori, origin of chromatography. (B) Quantitation of relative ATPase activities from four independent experiments as described for panel A. Averages and standard errors are shown. (C) 35S-labeled Aly/REF protein produced in the E. coli T7 S30 system (Promega) was incubated with GST alone (GST; 300 pmol), GST-UAP56 (GST-UAP; 300 pmol), or GST-TAP231 (GST-TAP; 10 pmol) (12, 15) in the presence of RNase A and in the absence (lanes 2, 3, and 6) or presence (lanes 4, 5, 7, and 8) of recombinant TAP231 (10 and 100 pmol; lanes 4 and 5, respectively) or UAP56 (300 and 3,000 pmol; lanes 7 and 8, respectively), and GST pull-down was performed as described for Fig. 4B. 35S-labeled Aly/REF protein was visualized by fluorography.
Interaction of UAP56 with Aly/REF competes with interaction of TAP-p15 with Aly/REF.
In yeast, interaction of UAP56 with Aly/REF competes with interaction of TAP with Aly/REF (36), and therefore UAP56 dissociation should be required for TAP-p15 recruitment. To examine whether this is also the case in higher eukaryotes, we next performed in vitro protein-protein interaction experiments with human proteins (Fig. 5C). 35S-labeled Aly/REF protein was efficiently pulled down by recombinant GST-UAP56 (lane 3) or GST-TAP231 (lane 6) (references 12 and 15) but not by GST alone (lane 2). Since 35S-labeled Aly/REF protein was produced in an Escherichia coli translation system, these interactions must be direct. When the GST pull-down experiments were performed in the presence of increasing amounts of recombinant TAP231 or UAP56, these interactions decreased in a dose-dependent manner (lanes 4, 5, 7, and 8). These results suggest that the interaction of UAP56 with Aly/REF indeed competes with the interaction of TAP with Aly/REF in the human system as is the case with the yeast system.
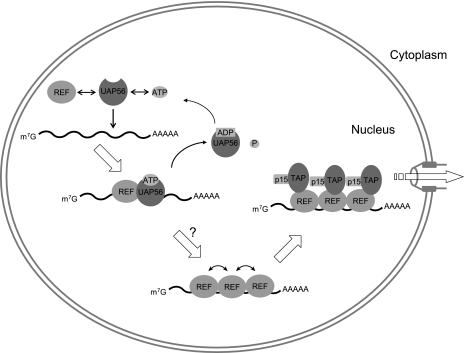
Model of how UAP56 may recruit Aly/REF onto intronless mRNAs.
Based on the results obtained in this study, we propose a model of how UAP56 may recruit Aly/REF onto intronless mRNAs (Fig. 6). Note that for the sake of simplicity, this model shows Aly/REF as the only adaptor protein to recruit TAP-p15, although this is not the case. ATP-bound UAP56 recruits Aly/REF onto the RNA by binding simultaneously to Aly/REF and RNA. The activation of the RNA binding activity of UAP56 by ATP (Fig. 4A) should be essential for this recruitment. Although Aly/REF per se can bind RNA, the heterodimeric complex of ATP-bound UAP56 and Aly/REF should be able to bind RNA more efficiently. Probably the binding specificity of the heterodimer to RNA is not restricted to mRNAs, given the results in Fig. 4A. When the dimeric complex binds RNA, the ATPase activity of UAP56 is activated (Fig. 5) and ATP hydrolysis may therefore take place. Since ADP-bound UAP56 has no affinity for RNA (Fig. 4), ATP hydrolysis should trigger the dissociation of UAP56 from RNA, leaving Aly/REF behind on the RNA (Fig. 6). Under our experimental conditions, we do not clearly observe dissociation of UAP56 from RNA and Aly/REF (Fig. 2). Although the ATPase activity of UAP56 is greatly stimulated by RNA and Aly/REF (Fig. 5), it appears not to be strong enough to quantitatively hydrolyze ATP for the quick release of UAP56 from RNA. However, this dissociation should take place in vivo, since interaction of UAP56 with Aly/REF competes with the interaction of TAP-p15 with Aly/REF (Fig. 5C), and therefore UAP56 dissociation should be required for TAP-p15 recruitment. Another factor(s) that further stimulates the ATPase activity of UAP56 should exist in vivo.
FIG. 6.
A model of export factor recruitment to intronless mRNAs. Note that for the sake of simplicity, this model shows Aly/REF as the only adaptor protein to recruit TAP-p15, although this is not the case. See the text for details.
The dissociated UAP56 should be ATP loaded again for the next round of Aly/REF loading onto RNA. This loading cycle may repeat, and multiple Aly/REF proteins may be loaded onto a single RNA molecule (Fig. 6). Since ATP-bound UAP56 binds RNA with broad specificity (Fig. 4), Aly/REF may first be loaded onto a wide range of RNA species by UAP56. However, Aly/REF would remain only on mRNAs and dissociate from other RNA species due to its preference for binding to mRNAs. The molecular mechanism of this RNA binding preference is not known. It is plausible to think that other mRNA binding proteins may stabilize the association of Aly/REF with mRNAs in vivo, but our result shown in Fig. 2 suggests that Aly/REF may have intrinsic activity to preferentially bind to mRNAs. Although the mechanism of this preference needs to be investigated, it is possible that Aly/REF proteins are stabilized on mRNAs by interacting with each other on the RNA (Fig. 6) (31). Multiple copies of TAP-p15 may then be recruited, leading to efficient mRNA export to the cytoplasm (Fig. 6). Thus, in this model, UAP56 functions as an ATP-dependent molecular chaperone for RNA-Aly/REF complex formation rather than as an RNA helicase, although UAP56 has motifs of DExD-box RNA helicases and an RNA-unwinding activity in vitro (34). Needless to say, our finding does not exclude the possibility that UAP56 also functions as an RNA helicase that unwinds RNA secondary structures or displaces proteins from RNA (4). This mode of action is reminiscent of the mechanism in which the ATP-bound clamp loader, a AAA+ ATPase, chaperones the sliding DNA clamp onto the replicating DNA and ATP hydrolysis ejects the clamp loader from the newly formed DNA-clamp complex (reviewed in reference 28). Probably more reminiscent is the function of the SMN (survival motor neuron) complex, which chaperones complex formation between U snRNAs and Sm proteins (2). The fascinating common feature between the SMN-Sm and UAP56-Aly/REF systems is that although both Sm and Aly/REF proteins per se can specifically bind to U snRNAs and mRNAs, respectively, they both require an energy-dependent molecular chaperone for efficient recruitment onto RNA.
It should be pointed out that not only Aly/REF and UAP56 but also the entire TREX and exon junction complexes are recruited onto the mRNAs if they are produced through splicing (21, 25). It will be interesting to examine whether other components of these entire complexes are also recruited onto intronless mRNAs in vivo. It should also be pointed out that the ATPase activity of UAP56 may be fully activated in the context of the entire complexes. Finally, UAP56 is essential for the export of most mRNAs, but this is not the case with Aly/REF in Drosophila melanogaster and Caenorhabditis elegans (8, 9, 11, 22, 24, 30). Therefore, UAP56 must have additional functions beyond recruiting Aly/REF. Further experiments will be required to elucidate the entire scenario of recruitment of export factors onto mRNAs.
Acknowledgments
We thank Makoto Kitabatake for suggestions for and criticisms of this work. We also thank Elisa Izaurralde for providing us with the unpublished UAP56-LAT (S228L) mutant plasmid.
This work was supported by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Tokyo, Japan, and grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan. I.T. was supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Published ahead of print on 5 November 2007.
REFERENCES
- 1.Abruzzi, K. C., S. Lacadie, and M. Rosbash. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 232620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battle, D. J., M. Kasim, J. Yong, F. Lotti, C. K. Lau, J. Mouaikel, Z. Zhang, K. Han, L. Wan, and G. Dreyfuss. 2006. The SMN complex: an assembly machine for RNPs. Cold Spring Harbor Symp. Quant. Biol. 71313-320. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, H., K. Dufu, C. S. Lee, J. L. Hsu, A. Dias, and R. Reed. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 1271389-1400. [DOI] [PubMed] [Google Scholar]
- 4.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 36717-37. [DOI] [PubMed] [Google Scholar]
- 5.Cullen, B. R. 2003. Nuclear RNA export. J. Cell Sci. 116587-597. [DOI] [PubMed] [Google Scholar]
- 6.Erkmann, J. A., R. Sanchez, N. Treichel, W. F. Marzluff, and U. Kutay. 2005. Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA 1145-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 111864-1872. [DOI] [PubMed] [Google Scholar]
- 8.Gatfield, D., and E. Izaurralde. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 111716-1721. [DOI] [PubMed] [Google Scholar]
- 10.Gruter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1649-659. [DOI] [PubMed] [Google Scholar]
- 11.Herold, A., L. Teixeira, and E. Izaurralde. 2003. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 222472-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11837-843. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7899-905. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y., K. M. Wimler, and G. G. Carmichael. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 181642-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, Y., T. A. Yario, and J. A. Steitz. 2004. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 1019666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katahira, J., K. Strasser, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 182593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, V. N., J. Yong, N. Kataoka, L. Abel, M. D. Diem, and G. Dreyfuss. 2001. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 202062-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korneeva, N. L., E. A. First, C. A. Benoit, and R. E. Rhoads. 2005. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 2801872-1881. [DOI] [PubMed] [Google Scholar]
- 20.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 204987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 196860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longman, D., I. L. Johnstone, and J. F. Caceres. 2003. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413644-647. [DOI] [PubMed] [Google Scholar]
- 24.MacMorris, M., C. Brocker, and T. Blumenthal. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9847-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, S., R. Das, H. Cheng, E. Hurt, N. Dorman, and R. Reed. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 191512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuyama, K., I. Taniguchi, N. Kataoka, and M. Ohno. 2004. RNA length defines RNA export pathway. Genes Dev. 182074-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nojima, T., T. Hirose, H. Kimura, and M. Hagiwara. 2007. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 28215645-15651. [DOI] [PubMed] [Google Scholar]
- 28.O'Donnell, M., and J. Kuriyan. 2006. Clamp loaders and replication initiation. Curr. Opin. Struct. Biol. 1635-41. [DOI] [PubMed] [Google Scholar]
- 29.Ohno, M., A. Segref, S. Kuersten, and I. W. Mattaj. 2002. Identity elements used in export of mRNAs. Mol. Cell 9659-671. [DOI] [PubMed] [Google Scholar]
- 30.Rehwinkel, J., A. Herold, K. Gari, T. Kocher, M. Rode, F. L. Ciccarelli, M. Wilm, and E. Izaurralde. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11558-566. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 981030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saavedra, C., B. Felber, and E. Izaurralde. 1997. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol. 7619-628. [DOI] [PubMed] [Google Scholar]
- 33.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 163256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen, J., L. Zhang, and R. Zhao. 2007. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J. Biol. Chem. 28222544-22550. [DOI] [PubMed] [Google Scholar]
- 35.Shi, H., O. Cordin, C. M. Minder, P. Linder, and R. M. Xu. 2004. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl. Acad. Sci. USA 10117628-17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413648-652. [DOI] [PubMed] [Google Scholar]
- 37.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417304-308. [DOI] [PubMed] [Google Scholar]
- 39.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Seraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran, E. J., and S. R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell 1251041-1053. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407401-405. [DOI] [PubMed] [Google Scholar]