Abstract
The mouse fpgs gene uses two distantly placed promoters to produce functionally distinct isozymes in a tissue-specific pattern. We queried how the P1 and P2 promoters were differentially controlled. DNA methylation of the CpG-sparse P1 promoter occurred only in tissues not initiating transcription at this site. The P2 promoter, which was embedded in a CpG island, appeared open to transcription in all tissues by several criteria, including lack of DNA methylation, yet was used only in dividing tissues. The patterns of histone modifications over the two promoters were very different: over P1, histone activation marks (acetylated histones H3 and H4 and H3 trimethylated at K4) reflected transcriptional activity and apparently reinforced the effects of hypomethylated CpGs; over P2, these marks were present in tissues whether P2 was active, inactive, or engaged in assembly of futile initiation complexes. Since P1 transcriptional activity coexisted with silencing of P2, we sought the mechanism of this transcriptional interference. We found RNA polymerase II, phosphorylated in a pattern consistent with transcriptional elongation, and only minimal levels of initiation factors over P2 in liver. We concluded that mouse fpgs uses DNA methylation to control tissue-specific expression from a CpG-sparse promoter, which is dominant over a downstream promoter masked by promoter occlusion.
In mammals, circulating folates are monoglutamate forms, which are best viewed as the transport forms of this vitamin (12, 54). After passage into peripheral cells, folates are converted to poly-γ-glutamate derivatives by the enzyme folylpoly-γ-glutamate synthetase (FPGS) (5, 52, 60). Without this metabolic trapping mechanism, mammalian cells die for lack of the end products of folate metabolism (48). FPGS is also necessary for the action of most antifolates (16, 63), and point mutations in FPGS are a common mechanism for tumor cell resistance to these drugs (51, 75). The distribution of FPGS in tissues of the mouse follows two patterns: it is found in all normal tissues with a dividing cell compartment, such as bone marrow and small intestine as well as tumors, and it is expressed in two differentiated tissues, liver and kidney (4, 21, 70). The enzyme made in dividing cells results from transcription exclusively from a promoter located immediately adjacent to the body of the gene, with subsequent maturation of a transcript containing sequences from exons 1 to 15 (69). A second isozyme is made in liver and kidney as the result of transcription from a promoter located ca. 10 kb upstream; the mature mRNA for this isozyme links sequences from two upstream exons (A1a and A1b) to exons 2 to 15, splicing out exon 1 in the process (62, 70). The two isozymes differ only in the sequence of the most N-terminal peptide, but the enzyme in dividing cells is tightly regulated by feedback inhibition by folate polyglutamates, whereas that in liver and kidney is much less sensitive to feedback control (1). Thus, the mouse has evolved a dual promoter transcriptional mechanism to ensure the tissue-specific production of two similar enzymes: one spares the mouse from loss of folates during cellular turnover, and the other allows the storage of higher levels of folates in liver and kidney (1).
The tissue specificity of mammalian gene expression is determined by the levels of trans-acting factors, as well as by epigenetic events that allow both the long-term control and the flexibility required by development. Both sets of factors are programmed in the germ line for precise gene expression in every cell in the organism. Two major epigenetic mechanisms in mammals are methylation of cytosines in CpG dinucleotides and posttranslational modifications (PTMs) of histone H3 and H4 tails (6). This “histone code” (30, 57) and DNA methylation are thought to act coordinately (6, 36). The role of CpG methylation in gene silencing has been extensively studied, and some patterns have become evident (32, 38). The CpG dinucleotide occurs in human DNA at frequencies far less than expected, but regions of genomic DNA containing relatively high densities of CpGs (CpG islands) are very often associated with the 5′ regions of mammalian genes (10, 42, 66). The CpG islands associated with the promoter regions of housekeeping or broadly expressed genes are typically unmethylated (7, 9). For some CpG island promoters that have a tissue-specific pattern of expression, the CpG islands remain unmethylated in tissues in which the gene is transcriptionally silent, as well as in tissues in which the gene is transcriptionally active (10, 11, 33, 50, 58, 73). Results based on the technique of restriction landmark genome scanning indicated that about 5% of mouse genes contained CpG islands which were methylated in a tissue-specific manner, and, for several of these cases, DNA methylation correlated with tissue-specific gene silencing (65). While there are only a few reports of DNA methylation apparently causative of tissue-specific silencing in normal tissues (24), such methylation-specific silencing has been commonly observed following derangement of DNA methylation during neoplastic transformation and tumor progression (31). The role of CpG methylation in the control of gene expression in the ≈40% of human genes that do not contain CpG islands is less well defined (2, 32). Although it is commonly thought that tissue-specific DNA modification patterns are responsible for tissue-specific gene expression, there is little direct evidence linking DNA methylation at CpG-poor promoters with transcriptional repression (32). Recently, high-resolution bisulfite sequencing of regions of human chromosomes 6, 20, and 22 revealed a proportion of CpG-sparse promoters whose methylation status reflected transcriptional repression (18). Interestingly, the P1 promoter in the mouse fpgs gene lies in a region characterized by a sparse distribution of CpG dinucleotides, while the P2 promoter lies in a CpG island, so that the tissue-specific transcription of the fpgs gene involves coordination of events at both types of promoters.
In this study, we investigated how the two promoters of the mouse fpgs gene are controlled to accomplish this pattern of tissue-specific expression. We conclude that the upstream CpG-sparse promoter is made accessible or inaccessible by coordinated DNA and histone modifications but that tissue specificity of initiation at the downstream CpG island promoter is independent of DNA methylation. From detailed chromatin immunoprecipitation (ChIP) mapping studies across the length of the 20-kb mouse fpgs gene, we found patterns of histone H3 and H4 acetylation and trimethylation of histone H3 at lysine 4 (H3K4me3) that reflected DNA hypomethylation but not necessarily transcriptional activity. We present evidence that elongating RNA polymerase II (RNAPII) complexes accumulate over the nucleosome-depleted P2 promoter sequences in mouse liver, limiting the assembly of transcriptional initiation complexes at P2. Hence, mouse fpgs represents an example of transcriptional interference in an endogenous gene.
MATERIALS AND METHODS
Isolation of the mouse fpgs locus from a 129/Sv bacterial artificial chromosome (BAC) library.
The mouse genomic 129/Sv Down-to-the-Well BAC Library (Incyte Biologicals) was screened by PCR using primers spanning fpgs intron 13 (5′-CCAACGTGACAGAGGTTTCATCC-3′ and 5′-TTGGAGCTAGCGTGTTTCTCAGC-3′). A BAC clone containing 150 to 175 kb of mouse genomic sequence was isolated that included the entire fpgs locus. HindIII and BglII fragments that hybridized with exons A1a/A1b and exon 1 cDNA probes were subcloned into pGEM (Promega). Further restriction digest analysis, Southern blotting, and sequencing determined the location of the exon A1a/A1b or exon 1 sequences within the subclones. Two HindIII fragments of 8.5 kb and 7 kb were identified that contained the exon A1a/A1b and the exon 1 regions, respectively (Fig. 1A). Subsequent direct sequencing of the BAC clone demonstrated that these HindIII fragments were contiguous.
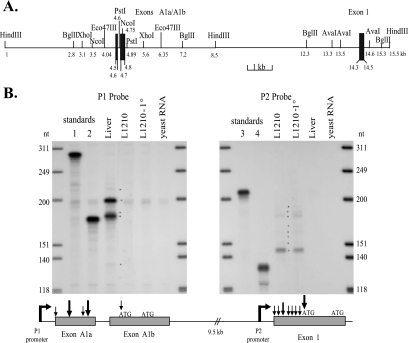
FIG. 1.
Transcriptional start site mapping over the two mouse fpgs promoters. (A) Map of the mouse fpgs locus showing the placement of exons A1a, A1b, and 1 (filled boxes) on two contiguous HindIII fragments. (B) Determination of the transcriptional start sites of the P1 and P2 promoters by RPA performed with mRNA from DBA-2 mouse liver, L1210 cultured cells, and L1210 primary (1°) leukemia cells. The first and last lanes show end-labeled DNA molecular weight markers. Four RNA standards were generated to allow the precise size determination of the protected fragments in the data lanes: in standard lanes 1 and 3 are protected fragments of 285 nt and 206 nt, respectively, from the hybridization of the P1 and P2 probes with an in vitro transcribed sense complement of each probe. To standard lane 2 was added a protected fragment of 173 nt, representing the sequence of exons A1a and A1b from nt −53 to nt +120 (relative to the first translational start site in exon A1b); in standard lane 4 is a protected fragment of 120 nt, representing the sequence of exon 1 from nt +19 to nt +138 (relative to the first translational start site in exon 1). Arrows in the lower diagram indicate the location of transcriptional start sites mapped in the figure; major (bold) and minor transcriptional start sites located within exons A1a and A1b and exon 1 sequences are noted.
Mapping of transcriptional start sites.
To map the transcriptional start sites of exons A1a/A1b and exon 1 by RNase protection assays (RPAs), plasmid clones containing the genomic sequences upstream of exons A1a/A1b (P1) and exon 1 (P2) were used to generate riboprobes and standard RNAs by in vitro transcription. The P1 probe construct was created by amplifying the exon A1a/A1b cDNA sequence, using primers 5′-CTGGACTGTGCACAGGATG-3′ and 5′-GGAGGCAGTCTTAGCTTCGTAAGC-3′, and cloning the PCR product into pCRII (Invitrogen). The cDNA clone was digested at an ApaLI site in exon A1a and an EcoRV site in the vector. The 4.1-kb linearized cDNA and vector sequence was isolated for subsequent ligation to the promoter sequence. A genomic fpgs clone subcloned as above from the 129/Sv BAC was digested with ApaLI and EcoRV to isolate a 127-bp fragment of P1 sequence and exon A1a sequence. This 127-bp fragment was ligated to the 4.1-kb ApaLI/EcoRV fragment to link the promoter and cDNA, without the intervening intron sequence between exons A1a and A1b. Transcripts containing exon 1 were detected using a construct created by amplifying the genomic sequence upstream of and including exon 1 (5′-CTCGGATTGGTGGGTCTTAGG-3′ and 5′-CTGATACTCCATGCCCGGC-3′) and ligating the PCR product into pCRII. The P1 and P2 constructs were linearized with EcoRV and BsaHI, respectively, and the riboprobe RNA transcripts were generated using [γ-32P]UTP (Perkin Elmer) and either SP6 or T7 RNA polymerase. The sense complements of the P1 and the P2 probes were in vitro transcribed and hybridized to the labeled riboprobes for use as standards for the mobility of a single molecular species that perfectly matches the probe. The riboprobes were hybridized to mouse liver and L1210 mRNA as previously described (70). Primary L1210 leukemia cells that had never been placed in culture were used in some experiments; they were obtained from DBA-2 mice carrying the tumor as an intraperitoneal implant.
DNase I hypersensitivity assays.
L1210 cells (1 × 108) were pelleted, washed with phosphate-buffered saline, and resuspended in lysis buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% Igepal, 0.15 mM spermine, and 0.5 mM spermidine). The nuclei were pelleted (284 × g for 10 min at 4°C), washed, and resuspended in 1 ml of DNase I digestion buffer (1 mM CaCl2, 10 mM Tris-Cl, pH 7.4, 15 mM NaCl, 60 mM KCl, 0.15 mM spermine, 0.5 mM spermidine) to which 0 to 18 μg/ml DNase I (grade DN-EP; Sigma Chemical Co.; dissolved in 20 mM Tris-Cl, pH 7.5, and 1 mM MgCl2) was added. The DNase I digestion (5 min at 25°C) was quenched with stop buffer (10 mM EDTA and 1 mM EGTA), and 1% sodium dodecyl sulfate. The samples were digested with 75 μg of proteinase K at 37°C overnight, the genomic DNA was isolated by organic extractions (phenol [pH 7.6]-chloroform [1:1] and chloroform), and the RNA was degraded with RNase A (20 μg; 2 h at 37°C). The extractions were repeated, and the samples were precipitated and resuspended in H2O. The samples were digested with HindIII overnight, and the DNA concentration of each sample was determined. Ten micrograms of each sample was run on 0.5% agarose gels and transferred to nylon membranes. PCR-generated sequences from the 5′ ends of the exon A1a and exon 1 HindIII fragments were random-primed with [γ-32P]dATP and [γ-32P]dCTP (Perkin Elmer) and used to probe the membranes.
Primary mouse tissues (liver, kidney, and brain) were excised and homogenized in buffer A (15 mM Tris-Cl, pH 7.6, 60 mM KCl, 15 mM NaCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.15 mM spermine) in a Dounce homogenizer to reduce the activity of native DNases. Single-cell suspensions were washed with buffer A and lysed in buffer A plus 0.5% Igepal. Nuclei were washed in buffer A and resuspended in buffer A plus 3 mM CaCl2 and 75 mM NaCl, to which DNase I was added. The remaining steps were followed as above.
ChIP-histone modifications.
The ChIP procedure was adapted from previously described methods (14, 40). Briefly, L1210 cells (2 × 106 to 3 × 106 cells per immunoprecipitation) or 300 mg of diced liver or brain tissue was fixed in 1% HCHO at 25°C. Liver and brain samples were washed in phosphate-buffered saline and dispersed into a single-cell suspension in a Dounce homogenizer and passed through sterile gauze to remove tissue clumps. Cells were washed, lysed in a 25 mM Tris buffer, pH 7.5 (containing 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, and 0.5% sodium deoxycholate), and sonicated using a bath sonicator (Diagenode) until 100- to 300-bp DNA fragments were obtained as visualized on an agarose gel. Sonicated lysates were precleared for 1 h at 4°C in a 50% lysis buffer-protein G-Sepharose bead (Amersham Biosciences) slurry previously blocked with 0.05 mg/ml sonicated lambda DNA and 0.05 mg/ml bovine serum albumin. The lysates were rotated at 4°C overnight with 5 μg of antibodies against the following: total histone H3 (ab1791; Abcam), monomethyl-H3K27 (H3K27me1) (item 07448; Upstate), dimethyl-H3K27 (H3K27me2) (07452; Upstate), H3K27me3 (07449; Upstate), H3K9me3 (07442; Upstate), H3K4me3 (07473; Upstate), acetyl-H3 (H3Ac) (06599; Upstate), H4Ac (06598; Upstate), H3K9Ac (07352; Upstate), and immunoglobulin G (IgG) (12-371; Upstate). Ten micrograms of antibody against H3K36me3 (ab9050; Abcam) was also used. Protein-DNA complexes were eluted from the beads and cross-links were reversed at 65°C. The immunoprecipitated DNA was precipitated with ethanol and resuspended in Tris-EDTA (TE) buffer; 1 μl of each dissolved sample was added to PCRs. PCR primer pairs amplified regions of genomic DNA every 1.5 to 2.0 kb across the length of the fpgs gene. Real-time PCR quantitations of immunoprecipitated DNA and total input DNA were performed as previously described (14), except 0.1% of the supernatant from the no-antibody adsorption reaction was used as input DNA.
Methylation-specific endonuclease mapping.
Mouse liver, brain, kidney, and L1210 genomic DNA (60 μg) were digested with HpaII (30 U/μg) at 37°C overnight. Half of the DNA was then digested with MspI (2 U/μg) overnight. Digests were analyzed by Southern blotting using 5 μg of DNA/lane and exons A1a or exon 1 cDNA sequences as probes.
Bisulfite sequencing.
Mouse tissue and L1210 genomic DNA (10 μg) were digested with HindIII at 37°C overnight and chemically deaminated as previously described (49). Two-microgram aliquots of the digests were heat denatured (95°C for 10 min) and subsequently chemically denatured (0.32 M NaOH at 50°C for 15 min). Fifty microliters of 2% low-melting-point agarose was added to each sample, and 10-μl drops were added to chilled mineral oil. The solidified agarose beads were transferred to 0.5 ml of freshly made bisulfite reaction mixture (5 M Na2S2O5, pH 5.0, 0.58 mM hydroquinone) and incubated at 50°C for 4 h protected from light. The beads were washed six times with 1 ml of TE buffer for 10 min at room temperature and incubated with 0.2 M NaOH at 37°C for 30 min, followed by three washes with TE buffer. The beads were then added directly to PCR mixtures with sense and antisense primers designed without CpG dinucleotides and chosen to amplify the bisulfite-modified sequence of the top strand. The PCR products were reamplified using nested primers. The resulting products were separated by agarose gel electrophoresis, and the gel-isolated products were cloned into pCRII (Invitrogen). Ten clones were sequenced for each PCR-amplified region.
ChIP-RNAPII and transcription factor studies.
The ChIP protocol used to study the histone PTMs was applied to study the distribution of RNAPII over smaller, overlapping segments of the fpgs promoters and the distribution of transcription factors near P2. Cross-linked DNA fragments were sonicated down to 100 to 300 bp. Lysates were rotated at 4°C overnight with 8 μg of anti-RNAPII (05-623; Upstate), 4 μg of anti-IgG (12-371; Upstate), or 5 μg of antibodies against phosphoserine 5 peptide from RNAPII (ab5131; Abcam), phosphoserine 2 peptide from RNAPII (ab5095; Abcam), TATA-binding protein (TBP) (sc-273; Santa Cruz), TAFIIp250 (sc-17134; Santa Cruz), TFIIB (sc-225; Santa Cruz), negative elongation factor A (NELF-A) (sc-23599; Santa Cruz), or SP1 (sc-59; Santa Cruz). The antibody used to recognize total RNAPII binds both phosphorylated and unphosphorylated forms of the C-terminal domain (CTD) repeated peptide, according to the manufacturer. The specificities of the antibodies against RNAPII phosphorylated serine 5 CTD peptide and phosphorylated serine 2 CTD peptide were determined by incubating L1210 cell lysates and each antibody with increasing amounts of the serine 2 phosphopeptide (ab12793; Abcam), serine-5 phosphopeptide (18488; Abcam), and unphosphorylated CTD peptide (12795; Abcam) under our ChIP conditions (see Fig. S3 in the supplemental material). Increasing levels of each phosphopeptide progressively blocked the ChIP signal using antibody against that phosphopeptide, but the other phosphopeptide or the unphosphorylated CTD peptide was ineffective even at 10 μg per immunoprecipitation. The P1 fpgs promoter region was divided into five segments, using primers that amplified nucleotides −379 to −119, −126 to 30, 7 to 153, 121 to 288, and 287 to 449 relative to the upstream transcriptional start site. The P2 fpgs promoter was divided into six fragments amplifying nucleotides −698 to −469, −475 to −333, −340 to −167, −194 to −52, −83 to 165, and 59 to 232 relative to the transcriptional start site.
RESULTS
Mapping of the transcriptional start sites at the P1 and P2 promoters.
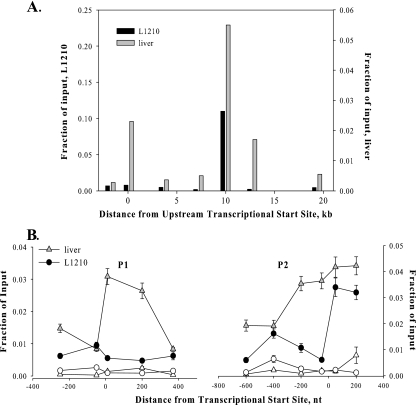
RPAs were used to define the transcriptional start sites at the P1 and P2 promoters of the mouse fpgs gene. Using mRNA from L1210 cells to protect a probe for the P2 region, multiple minor start sites were detected upstream of the first ATG in exon 1 (Fig. 1B, right panel), a profile very similar to that previously found with the equivalent human fpgs promoter (22). In contrast, fpgs transcripts that use P2 were not detected in mouse liver by these RPAs (Fig. 1B). This agrees with a previous report (70) that the P2 promoter was heavily used in mouse dividing tissues and tumors but that a very low percentage (< 5%) of fpgs transcripts in mouse liver initiated at P2. The multiple start sites at P2 in mouse L1210 cells were not unexpected, since P2 is a TATA-less promoter.
Two major transcriptional start sites were found in the fpgs P1 promoter (Fig. 1B, left panel). This promoter was heavily used in liver, but was not used in L1210 leukemic cells propagated either in culture or as an in vivo tumor (Fig. 1B). The distribution of start sites in mouse liver fell almost exclusively in what was previously defined as exon A1a by cDNA cloning (60) and 5′ rapid amplification of cDNA ends (69) experiments.
Mapping of chromatin structure near the two fpgs promoters.
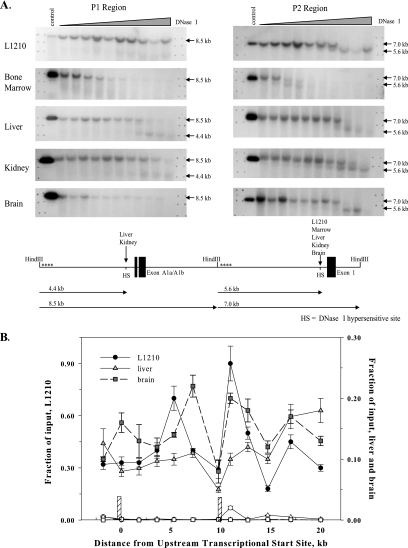
The accessibility of the fpgs promoter regions to proteins was determined from cleavage patterns in genomic DNA after exposure of intact nuclei to DNase I. At low levels of DNase I digestion, the full-length 8.5-kb HindIII fragment hybridized to the probe for the P1 region but, with increasing amounts of DNase I, a smaller fragment (4.4 kb) was detected in DNA extracted from renal and hepatic nuclei (Fig. 2A, left panels). This band was not seen in DNA extracted from mouse L1210 leukemia cells and mouse bone marrow, two dividing cell types that do not utilize the P1 promoter, nor was it observed in DNA from brain nuclei, which do not transcribe the fpgs gene from either promoter. When the P2 region was examined using a probe from the downstream HindIII fragment, a DNase hypersensitivity site was detected in all the tissues examined, i.e., L1210 cells, bone marrow, liver, kidney, and brain (Fig. 2A, right panels). The DNase hypersensitivity sites found for each promoter mapped to positions about 130 to 150 bp upstream of the transcriptional start sites defined as diagrammed in Fig. 1B. This implied that the chromatin configuration 130 to 150 bp upstream of P1 allowed access to this region in only those tissues that utilize this promoter (liver and kidney) but that the chromatin encompassing the P2 promoter was accessible in all tissues studied, whether or not this promoter was transcriptionally active.
FIG. 2.
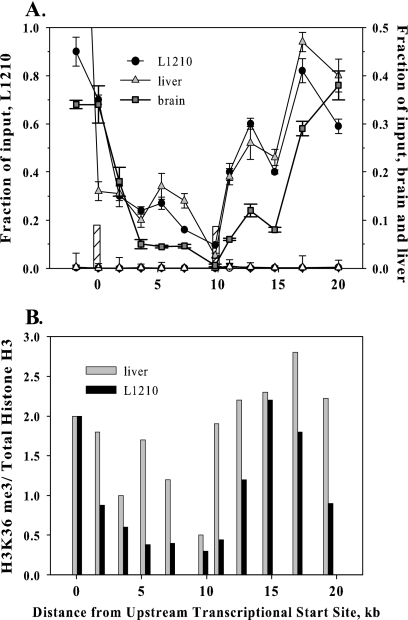
Chromatin density over the mouse fpgs locus. (A) A prominent DNase I hypersensitivity site is present in the P1 region only in tissues in which the promoter is active; the P2 region is accessible to DNase I in expressing and nonexpressing tissues. Nuclei were incubated with increasing concentrations of DNase I for 5 min at 25°C. Genomic DNA was extracted, digested with HindIII, and run on an agarose gel. Blots were probed with PCR-generated sequence at the 5′ ends of the HindIII fragments (asterisks in lower diagram). The bands in the control lanes indicate the gel mobility of the full-length HindIII fragments. The location of the DNase I hypersensitive sites (HSs) and the tissues containing such sites are indicated in the lower diagram. (B) ChIP determination of the level of histone H3 over the fpgs gene. Chromatin from mouse liver (triangles), brain (squares), or L1210 leukemia cells (circles) was cross-linked, sonicated, and immunoprecipitated with either a pan-H3 antibody (filled symbols) or a nonspecific IgG (open symbols). The content of DNA for various segments of the fpgs locus was determined by real-time PCR using primers spaced 250 to 300 nt apart; symbols are placed at the middle of each PCR fragment. Input represents amplified product from 0.1% of the starting material. The positions of the two DNase hypersensitivity sites are shown as the crosshatched bars. The experiments in this and all subsequent ChIP figures were performed at least twice, and the data are from a representative experiment.
The density of nucleosomes over the entire 20-kb fpgs locus was examined by ChIP using antibodies against histone H3. Previous studies have mapped histone H3 density over the length of a highly expressed human gene transcribed from a single promoter (72). In that study, the ChIP signal for total histone H3 reached a minimum over the promoter region, representing a low density of nucleosomes immediately before the transcriptional start site (72). The pattern of histone H3 decorating the span of the fpgs gene was more complex (Fig. 2B). The density of histone H3 reached a local minimum over the P2 promoter in L1210 cells, mouse liver, and brain and colocalized with the DNase hypersensitivity site mapped to this P2 region in these tissues (Fig. 2A and B). Somewhat surprisingly, a similar histone H3 local minimum was not seen in liver over the P1 region, in spite of the DNase hypersensitivity site over that region (Fig. 2B). Overall, the amount of histone H3 decorating the fpgs gene in mouse liver increased continuously over the span of the gene, with a notable minimum over P2. Interestingly, the histone H3 pattern in all three tissues was quite similar in spite of the differences in transcriptional activity at P1 and P2 among them (Fig. 2B).
Histone acetylation concurs with transcriptional initiation over P1 but not over P2.
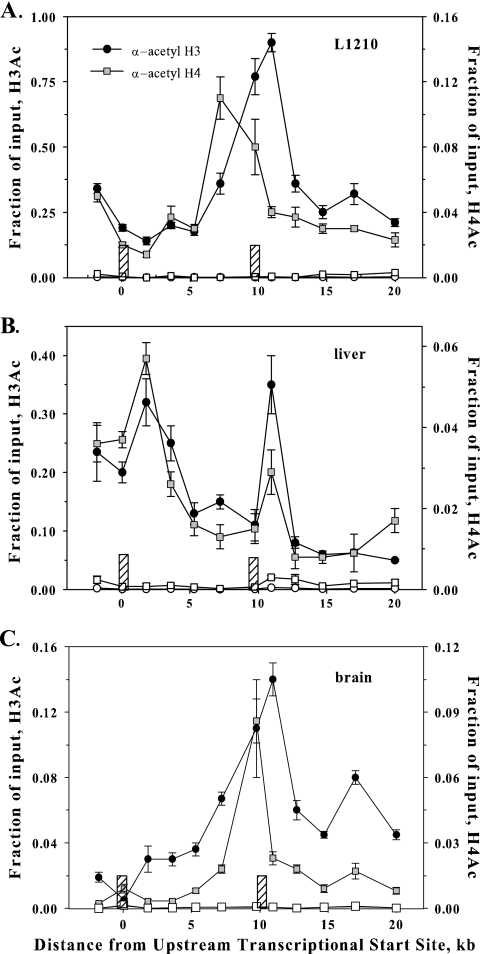
Previous studies in Saccharomyces cerevisiae and human cells indicate that active promoters have substantial acetylation of lysines in the N-terminal peptides of histones H3 and H4 (45, 55). When we probed the histones decorating the entire length of the fpgs gene, including segments containing P1 and P2 promoters, by ChIP using antibodies against H3Ac and H4Ac, we found a higher level of complexity in leukemic cells, liver, and brain (Fig. 3). As expected, abundant acetylation of both H3 and H4 was detected in mouse liver over P1 (Fig. 3B), but in L1210 cells, the nucleosomes over this region were not enriched in H3Ac and H4Ac (Fig. 3A). H3 and H4 acetylation was also a characteristic in nucleosomes over P2 in L1210, which initiates at this promoter (Fig. 3A). The regions of DNA over which H3 and H4 were acetylated extended substantially downstream of P1 in liver and on both sides of P2 in L1210 cells. Interestingly, H3 and H4 acetylation was also found over P2 in mouse liver (Fig. 3B), although the span of genomic DNA over which histone acetylation was found over P2 in liver was much narrower than seen in L1210 cells. This difference was repeatable and was seen with an antibody to H3K9Ac as well as the H3Ac and H4Ac antibodies shown in Fig. 3. Remarkably, in mouse brain, the pattern of H3 and H4 acetylation was almost identical to that in L1210 cells (Fig. 3C). Since histone acetylation is thought to mark an open promoter, finding H3 and H4 acetylation over P2 in mouse liver and brain raises the question of why P2 is not actively firing in these tissues. Clearly, a lack of transcriptional initiation at P2 in liver was not due to the absence of H3Ac and H4Ac in regional nucleosomes.
FIG. 3.
Distribution of H3Ac and H4Ac along the fpgs locus in mouse tissues expressing transcript from P1 or P2. Chromatin from L1210 leukemia cells (A), mouse liver (B), and mouse brain (C) was cross-linked, sonicated, and immunoprecipitated with either an antibody against H3Ac, H4Ac, or a nonspecific IgG (open symbols). The content of DNA for various segments of the fpgs locus was determined by real-time PCR as described in the legend of Fig. 2B. The positions of the two DNase hypersensitivity sites are shown as the cross-hatched bars. α, anti.
DNA cytosine methylation near the P1 and P2 promoters.
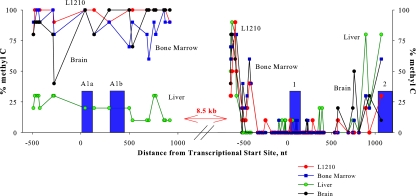
The distribution of CpG dinucleotides in the regions of mouse DNA surrounding the two fpgs promoters differed significantly. The P1 promoter was in a region in which CpG dinucleotides were sparsely represented (19 dinucleotides distributed over the region from −650 to +900 nucleotides [nt], relative to the first transcriptional start site), whereas the P2 promoter was embedded in a CpG island (%GC, 58.9; observed CpG/expected CpG, 0.71, distributed over the 1,000 nt centered on the first transcriptional start site), as defined previously (67). Initial experiments using HpaII, which is methylation sensitive, and MspI, which cleaves at the same site as HpaII (CCGG) regardless of cytosine methylation, suggested a tissue-specific difference in CpG methylation at P1 and little or no difference at P2 (see Fig. S1 in the supplemental material). Bisulfite sequencing demonstrated strikingly different patterns over the two fpgs promoters (Fig. 4). DNA from mouse liver showed an undermethylated state over P1, whereas L1210 cells, bone marrow, and brain DNA had virtually quantitative methylation at each CpG over this same region. In striking contrast, the CpG island surrounding P2 was completely unmethylated in all tissues studied, including normal and neoplastic tissues in which the P2 promoter was transcriptionally active (L1210 cells and marrow); liver, in which transcripts initiating at P2 were barely detectable; and brain, in which the entire fpgs gene was transcriptionally inactive (Table 1). Interestingly, the P2 promoter showed an unmethylated state starting from the position previously found to contain transcription factor binding sites controlling this promoter (17, 61) and continuing until just before the second exon. We note that, over the differentially methylated P1, H3 and H4 acetylation correlates with transcriptional activity but, over the unmethylated CpG island-P2 promoter region, H3 and H4 acetylation and transcriptional activity were independent. It is known that methyl-CpGs recruit specific binding proteins that, in turn, recruit histone deacetylases (34), but the acetylation at P2 suggests that CpG islands or nucleosomes around CpG islands recruit histone acetyltransferases.
FIG. 4.
CpG methylation in the regions of the mouse fpgs promoters in various tissues. CpG methylation in the P1 promoter region correlates with transcriptional activity; the P2 promoter region is unmethylated regardless of activation. Genomic DNA was treated with 5 M sodium bisulfite, amplified by PCR, and cloned, and the sequences of 10 clones were determined for each region. At the position of each CpG dinucleotide in the genomic sequence, the percentage of clones containing a methylated cytosine is represented in the graph, plotted relative to the positions of exons A1a, A1b, and 1 (filled boxes).
TABLE 1.
Quantitative reverse transcription-PCR determination of FPGS expression from P1 and P2 in mouse L1210 cells, liver, and braina
| Tissue type | Amt of FPGS in the indicated promoter region (pg of mRNA ± SD)
|
|
|---|---|---|
| P1 | P2 | |
| L1210 | <0.0001 | 0.15 ± 0.014 |
| Liver | 0.21 ± 0.0054 | 0.001 ± 0.0001 |
| Brain | <0.0001 | <0.0001 |
Total RNA (5 mg) was reverse transcribed using Superscript III in a total volume of 20 μl, and 1 μl was used in real-time PCR assays using primers specific for exons A1a/A1b to exon 3 (P1) or exon 1 to exon 3 (P2). Standard curves for quantitation were constructed using plasmids carrying cDNAs corresponding to the products of transcription from P1 and P2, respectively.
Histone H3 methylation marks over P1 and P2.
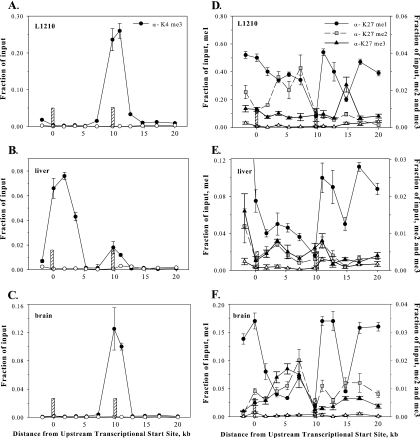
In an effort to understand the differential control of P1 and P2 in different tissues, we compared the patterns of histone H3 methylation over the length of the fpgs gene in liver and leukemic cells with the pattern expected at an expressed, single promoter (72). H3K4me3 formed a distinct peak beginning over P1 in liver and P2 in L1210 cells; these peaks continued for 3 to 4 kb after the transcriptional initiation site (Fig. 5A and B). This would concur with the observation that H3K4me3 marks the 5′ ends of many active genes in yeast (55) and human cells (45). Interestingly, a second smaller peak of H3K4me3 was found over P2 in mouse liver (Fig. 5B). To understand this second peak over P2 in mouse liver, the distribution of H3K4me3 was studied in mouse brain, where both promoters are silent (Fig. 5C). The pattern was exactly the same as that seen in L1210 cells, in spite of the fact that transcription at P2 was high in L1210 and undetectable in mouse brain (Table 1). Hence, whereas the H3K4me3 mark over the P1 promoter exactly concurs with transcriptional activity, there is no such relationship over the CpG island-embedded P2. The inactive P2 in liver was clearly in a different chromatin context than the inactive P2 in brain.
FIG. 5.
Distribution of histone H3 methylated at lysines 4 or 27 along the fpgs locus in mouse tissues expressing transcript from P1 (liver), P2 (L1210), or from neither promoter (brain). Chromatin from L1210 cells (A and D), mouse liver (B and E), and mouse brain (C and F) was cross-linked, sonicated, and immunoprecipitated with either an antibody against H3K4me3 (A, B, and C) or against H3K27me1, H3K27me2, or H3K27me3 (D, E, and F). Closed symbols are defined on panel A for panels A, B, and C and on panel D for panels D, E, and F. For each immunoprecipitation, a separate nonspecific IgG control was used (open symbols); in panels D, E, and F, IgG data were plotted on the more sensitive ordinate. The content of DNA for various segments of the fpgs locus was determined by real-time PCR as described in the legend of Fig. 2B. The positions of the two DNase hypersensitivity sites are shown as the crosshatched bars.
Methylation of H3K9 and H3K27 have been correlated with repression of transcription in yeast and mammalian cells (64) and, more recently, H3K27me1 and H3K9me3 have been found in the coding regions of mammalian genes (71). When we scanned the entire length of the fpgs gene in mouse liver and leukemic cells for H3K9me3, this modification was not detected (data not shown). Interestingly, H3K9me2 was not present over P1 or P2 in liver or L1210, but both H3K9me2 and H3K9me3 were found over the silenced globin promoter in liver and L1210 cells (see Fig. S2 in the supplemental material). H3K27me1 was present across the fpgs gene in both liver and L1210 cells (Fig. 5D and E), in agreement with recent studies that indicate H3K27me1 as a marker of actively expressing genes in euchromatin (72); however, this mark was also abundant over the fpgs locus in brain (Fig. 5F). The minimum of H3K27me1 found at P2 in all three tissues colocalized with the DNase hypersensitivity site in this region (Fig. 5D, E, and F). The H3K27me3 level was low across the span of this gene in all three tissues studied. Interestingly, H3K27me2 reached a discernible peak between the two promoters in L1210 cells. The loss of this ChIP signal in L1210 cells after P2 may indicate either the absence of H3K27me2 at nucleosomes decorating the active downstream region in leukemic cells, or it might indicate the presence of a second PTM across this region that masks the epitope recognized by the H3K27me2 antibody, e.g., phosphorylated H3 at S28.
Methylation of histone H3 at K36 in yeast has been linked to regions of transcriptional elongation and was shown to participate in the recruitment of the histone deacetylase Rpd3 downstream of the promoter (15, 35, 37). The subsequent absence of H3Ac groups in the transcribed region appears to suppress transcriptional initiation at downstream cryptic promoters in yeast (15, 35). Hence, H3K36me3 would be a probable candidate for the suppression of activation of P2 in mouse liver. We mapped the abundance of H3K36me3 across the fpgs gene to determine whether the spatial distribution of this PTM down the fpgs locus would suggest such a causal role (Fig. 6). This did not seem to be the case, since H3K36me3 was present throughout the body of the fpgs gene, except for a distinct minimum at P2 in both liver and L1210 cells (Fig. 6A), in fact coincident with the peak of H3Ac and H4Ac in both tissues. The level of H3K36me3 near P2 was lower than could be explained by the decreased level of total H3 over this region, as best evidenced by the ratio of ChIP signal for H3K36me3 to total H3 (Fig. 6B). The density of H3K36me3 then built up toward the 3′ end of the fpgs gene in both tissues, similar to the higher distribution of this mark toward the 3′ end of other yeast and mammalian genes (3, 55, 72). Hence, we could conclude that H3K36me3 was not directly involved in tissue-specific suppression of transcriptional initiation at P2 because its level was low across this region in mouse liver. The H3K36me3 level was quite low across the body of the fpgs gene in brain from +3 to +15 kb, consistent with the transcriptional inactivity of fpgs in this tissue and the role of H3K36me3 in transcriptional elongation (15, 35).
FIG. 6.
Distribution of H3K36me3 along the fpgs locus in mouse tissues. (A) Chromatin from mouse liver, mouse brain, and mouse L1210 leukemia cells was cross-linked, sonicated, and immunoprecipitated with either an antibody against H3K36me3 or IgG (open symbols). The content of DNA for various segments of the fpgs locus was determined by real-time PCR as described in the legend of Fig. 2B. The positions of the two DNase hypersensitivity sites are shown as the crosshatched bars. (B) The ratio of ChIP signal for H3K36me3 to that for total histone H3. The amount of H3K36me3 is expressed as a ratio of the ChIP signal for total histone H3 to determine whether the minimum in the H3K36me3 signal seen in panel A reflects total H3 density or a lack of methylation at H3K36 on the regional nucleosomes.
Overall, several aspects of the pattern of histone modifications over P2 in liver were intriguing: an anomalous second peak of H4Ac, H3Ac, and H3K4me3 and a distinct minimum of H3K36me3 beyond what was expected of the depleted nucleosome density over this region. In an effort to understand these patterns, we focused our attention on the components of the transcriptional complexes present across P2 in liver.
RNAPII has a bimodal distribution pattern along the fpgs gene in mouse liver.
We used ChIP experiments to map the occupancy of RNAPII at intervals along the 20-kb fpgs gene and found striking differences between the patterns seen in mouse liver and leukemic cells (Fig. 7A). In L1210 cells, there was a single segment of DNA that coprecipitated with RNAPII, namely, that centered on the transcriptional start site at P2. In liver, surprisingly, there were two peaks, one centered on P1 and a second higher peak beginning at P2. Notably, the occupancy of RNAPII continued past P2 in liver and was spread over a larger region than the peak over the initiating promoters, P2 in L1210 cells or P1 in liver.
FIG. 7.
Residency of RNAPII over fragments of the two mouse fpgs promoters in mouse liver and L1210 cells. (A) The amount of DNA from regions spaced along the fpgs gene immunoprecipitated by an antibody to RNAPII was quantitated by real-time PCR as described in the legend of Fig. 2B. The position of P1 is plotted at 0 kb and P2 is at 10 kb on the abscissa. In panel B, the DNA regions immediately surrounding P1 and P2 were fine mapped by ChIP using the same antibody. Nonspecific binding was determined by immunoprecipitation with IgG in liver (open triangles) and L1210 (open circles). The primer set plotted at 0 in panel A is also plotted at 0 for the P1 graph in panel B. The primer set plotted at 10 kb in panel A is plotted at 0 for the P2 graph in panel B. Data are plotted in panel B at the midpoint of each amplified region.
The residence of RNAPII over the two promoters was fine mapped using five PCR fragments covering 828 bp of the P1 promoter and six fragments over 938 bp of the P2 promoter; extensive sonication was used to limit DNA fragments to 100 to 300 bp. The RNAPII level over P1 was very low in L1210 cells but was still present at a level significantly higher than the IgG controls across this region (Fig. 7B, left graph). In striking contrast, RNAPII occupancy was much greater in mouse liver near P1 fragments, peaking immediately 3′ of the transcriptional start site. The same pattern was seen for L1210 cells near the P2 promoter as seen in liver near P1: polymerase occupancy peaked at or slightly after the transcriptional initiation site (Fig. 7B, right graph). However, in mouse liver, RNAPII was resident over the entire P2 region, increasing before the DNase I hypersensitivity site defined in Fig. 2 and remaining high over the P2 transcriptional start site and substantially downstream. Given that transcripts in mouse liver that initiate from P2 were not detectable (70) (Table 1), it was most likely that the RNAPII over the P2 promoter represented enzyme incorporated into elongation complexes that had originated at P1.
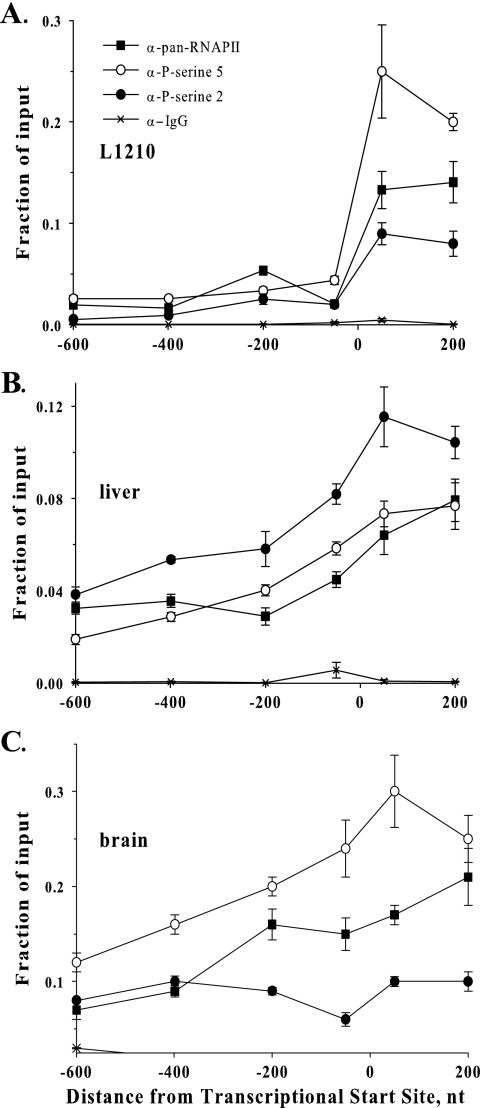
The CTD of RNAPII over P2 is phosphorylated at serine 2 in liver.
It has been shown that phosphorylation of serine 5 residues in the repeated YSPTSPS peptide heptad in the CTD of RNAPII is present throughout transit of a gene but is maximal on RNAPII in an initiation complex (25, 39). Phosphorylation of serine 2 in this motif accumulates as the elongation complex travels through a transcribed sequence and is highest over the cleavage/polyadenylation signal (25, 39). We followed phosphorylation of the CTD of RNAPII at serine 5 and serine 2 over the P2 fpgs promoter as an index of whether the RNAPII detected there was binding in a futile initiation complex or was engaged in elongating a transcript initiated upstream. The phosphorylation of RNAPII over these fragments in L1210 cells (Fig. 8A) was taken as representative of the pattern in an initiation complex: serine 5 phosphorylation per molecule of RNAPII was highest near nt +1, the transcriptional start site, while serine 2 phosphorylation was lower and proportional to the level of RNAPII throughout this region. The distribution of RNAPII across P2 was also detectable in mouse brain (Fig. 8C), in agreement with a recent tiling study across regions of the human genome that indicated that even inactive promoters are usually attempting to load initiation complexes (27) (see Discussion). As in L1210 cells, the RNAPII present across the brain P2 was phosphorylated at serine 5 to a higher degree and serine 2 phosphorylation was minimal, indicating that these complexes were indeed futile initiation complexes. The phosphorylation of RNAPII detected over this region was quite different in mouse liver (Fig. 8B): serine 5 phosphorylation per molecule of enzyme was much lower than that seen in L1210 cells and brain throughout this region, while serine 2 phosphorylation was quite high, and, interestingly, reached a peak over the fragment that included nt +1. We interpreted this as direct evidence that the RNAPII detected over P2 in liver was a component of an actively transcribing elongation complex.
FIG. 8.
Phosphorylation of RNAPII at serine 2 and 5 detected over fragments of the P2 fpgs promoter. ChIP walking studies were performed using an antibody against RNAPII (pan-RNAPII), antibodies raised against the CTD heptad (YSPTSPS) repeat of RNAPII phosphorylated (P) at serine 2 and serine 5, and an antibody raised against IgG as a nonspecific control. Five or six fragments were amplified over the P1 and P2 fpgs promoters, respectively. Quantitation was performed using real-time PCR as described in the legend of Fig. 2B. The data points were plotted at the midpoint of each amplified region. Symbols for all panels are according to panel A. α, anti.
Evidence that elongating RNAPII over P2 blocks assembly of transcriptional initiation complexes in liver.
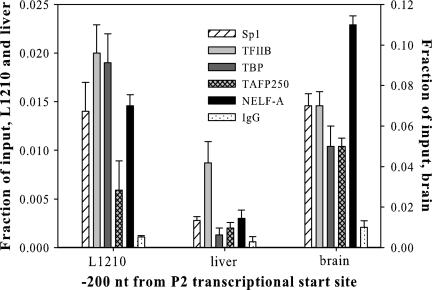
Transcription from the mouse fpgs P2 promoter has been shown to depend on several Sp1 sites (17). A typical Sp1-driven promoter involves binding of TBP to Sp1 with subsequent recruitment of other components of the preinitiation complex (PIC) (44). We assessed the binding of components of the PIC to fpgs P2. As shown in Fig. 9, in L1210 cells and mouse brain, Sp1 and TBP were loaded onto the region of DNA immediately upstream of the transcriptional initiation site characterized by the DNase hypersensitivity site (nt −200) as did TFIIB, TAFIIp250, and the NELF-A protein. These proteins persisted on the P2 locus past the point of initiation (nt +200) (data not shown). In striking contrast, the Sp1 level was very low at nt −200 in liver, and TBP was not detectable. Levels of TAF250 and NELF-A were also very low. We concluded that the binding of several proteins needed for assembly of a successful PIC was being actively prevented at P2 in liver. This is consistent with a promoter interference mechanism whereby the elongating RNAPII complexes initiating at P1 were physically occluding the DNA over the P2 region in liver.
FIG. 9.
Presence of transcription factors over the mouse fpgs P2 promoter in L1210 cells, liver, and brain. ChIP analysis was performed using antibodies directed against the sequence-specific transcription factor Sp1 and general transcription factors TFIIB, TBP, TAFp250, and the NELF-A protein. Quantitation was performed by real-time PCR using primers at −379 and −119 relative to the P2 transcriptional start site.
DISCUSSION
The mouse fpgs gene is transcribed from two alternative promoters, one of which (P2) drives transcription in normal dividing cells types and in tumors while the other (P1) controls transcription predominantly in two differentiated tissues, liver and kidney. We sought to explain how these two promoters are regulated in a virtually black and white manner, that is, tissues using P2 never initiate at P1, and tissues that initiate at P1 have a very limited, although detectable, usage of P2. Our analysis leads to the conclusion that activity at P1 correlates with the presence or absence of CpG methylation, making fpgs one of the few known cases in which methylation inversely correlates with expression in nonneoplastic tissues at a CpG-sparse promoter. In contrast, CpG methylation did not play a role in tissue-specific initiation from P2. A fairly exhaustive analysis of some of the most studied histone PTMs showed patterns across the span of this 20-kb gene only partly predictable from previous studies. However, the real surprise in our results is that RNAPII does not seem to have access to P2 for transcriptional initiation when P1 is active, despite the open chromatin context and lack of CpG methylation in the P2 region. Hence, mouse fpgs is a clear case of transcriptional interference in an endogenous mammalian gene.
We questioned whether the two promoters of the mouse fpgs gene were being differentially activated or differentially repressed. The CpG dinucleotides of the P1 promoter were methylated in tissues in which P1 was silent, but, surprisingly, regional P1 nucleosomes were not methylated at H3K9, a common repressive PTM. Lysine 9 methylation of H3 is tightly linked to DNA methylation and transcriptional repression in some systems, particularly in Neurospora (23, 68), which has led to the proposal that DNA methylation is targeted to genes that have been silenced by other mechanisms (8). Methyl-CpGs are binding sites for proteins which, in turn, serve as scaffolds for the recruitment of histone deacetylases (34, 53) and chromatin remodeling complexes (28) associated with transcriptional silencing. In the absence of the expected repressive histone H3K9 modification over P1, our results suggest that DNA methylation is the prime determinant of tissue-specific chromatin condensation and repression of the P1 promoter. Our analysis of epigenetic marks over the active P1 promoter in mouse liver showed several histone PTMs associated with active genes, i.e., H3Ac, H4Ac, and H3K4me3, coincident with hypomethylation of regional CpG dinucleotides. We concluded that, in adult dividing tissues of the mouse, the P1 fpgs promoter was locked in a transcriptionally inactive configuration by DNA methylation without the expected coincident H3K9me2 and H3K9me3 and that transcriptional activity at P1 in liver was reinforced by a series of histone PTMs commonly associated with transcriptional activity. Although it is difficult to separate out cause and effect in vivo without causing a primary change to perturb the system, the precedent literature suggests that the histone PTMs associated with transcriptional initiation are present because of the presence of RNAPII initiation complexes rather than being causative of transcriptional initiation. This interpretation is supported by previous studies in yeast, which demonstrated the recruitment of enzymes that add the H3K4me3, H4Ac, and H3Ac marks by the serine 5-phosphorylated CTD of RNAPII and by transcription factors during assembly of initiation complexes (43). Further support for this concept was furnished by recent studies by Cedar and his colleagues, who showed that genetic deletion of DNA methylation in mouse embryonic fibroblasts caused reexpression of a number of genes and also resulted in the appearance of H3 and H4 acetylation and H3K4 methylation over their promoters (41).
The P2 region was devoid of DNA methylation in all tissues studied whether or not this promoter was used to initiate transcription (Fig. 4). This makes the P2 fpgs promoter similar to the several housekeeping genes with CpG islands in and around their promoters that are not differentially methylated in expressing and nonexpressing tissues (2, 33). It should be noted that the P2 region of the fpgs gene is in a very different context in tissues which use P1 and in those which do not: in the latter, P2 is a fairly simple promoter, akin to the CpG island promoter of a housekeeping gene; in the former, P2 is in the middle of a 20-kb gene. With this perspective, it is interesting that the histone PTMs near P2 show quite distinct patterns in different tissues. In either a tissue in which P2 was exclusively used (L1210 cells) or one in which the entire fpgs locus was silent (brain), there was a broad peak of H3Ac and H4Ac centered on the hypersensitivity site at this promoter, but there was a much more compact peak of these acetylated nucleosomes over P2 in liver. Likewise, a substantial peak of H3K4me3 centers on P2 in L1210 and brain, but a distinctly smaller peak was found over this same region in liver. Previous genome-wide mapping studies have found H3Ac, H4Ac, and H3K4me2 over CpG island promoters whether they were active or silent (59, 74). Both P1 and P2 of the mouse fpgs gene would have scored positive for histone acetylation in such studies, but the data of Fig. 3 suggest that the cause or the effects of these modifications over P2 in liver and L1210 are different. In a very important recent paper (27), a genome-wide analysis of human embryonic stem cells found that the promoters of most protein-encoding genes were associated with nucleosomes containing H3K4me3 and H3Ac and were also bound to RNAPII, while only a fraction of these transcriptionally primed promoters produced mature full-length transcripts. That study also described a subset of genes that were excluded from assembly of futile transcriptional initiation complexes. In our study, we have found an example of each type of promoter: P2, at which transcriptional complexes form and result in mature transcript in L1210 cells but form futile complexes in mouse brain, and P1, which only allows the assembly of initiation complexes in tissues generating mature transcript from this promoter. The difference between these two is that P1 appears to be controlled by DNA methylation, and P2 is not; perhaps DNA methylation was also the distinguishing characteristic in the whole genome study of Guenther et al. (27).
We conclude that the silencing of P2 in mouse brain and mouse liver represents different phenomena; in brain, P2 and P1 are completely silenced whereas in liver low levels of initiation can be detected from P2. The fact that futile complexes assemble at P2 in brain makes it more likely that the absence of such complexes at P2 in liver is due to an active exclusion process.
In mouse liver, there were abundant transcripts from the fpgs gene, all initiating at the P1 promoter (Fig. 1B). Hence, a mechanism was in place to prevent the assembly of RNAPII with the requisite initiation factors at the P2 promoter, in spite of active traverse of transcriptional elongation complexes through this region of genomic DNA. How was this happening? Literature precedent suggested a promoter interference mechanism whereby activity at the P1 promoter prevents transcriptional initiation at the P2 promoter (19, 20, 26, 47, 56). In higher eukaryotes, this method of transcriptional repression has been described in the unr and N-ras genes, which are arranged in tandem; transcription of the unr gene interferes with initiation at the N-ras promoter which is spaced only 150 nt downstream of the unr termination signal (13). Similarly, deletion of an embryonic β-like globin gene, Ey, caused the activation of a second β-like globin gene, βho, which is located immediately downstream of the deleted gene (29). However, the mouse fpgs gene represents an interesting case in which transcriptional interference is used to coordinate two potentially active promoters, spaced a fairly large distance (10 kb) apart. Our analysis of the P2 promoter in liver indicates that RNAPII elongation complexes are residing over an extended length of the fpgs genomic locus, covering the nucleosome-deficient DNase-hypersensitive region before P2 and continuing downstream, physically occluding initiation at P2. Occlusion mechanisms have been proposed to involve the decreased binding of transcription factors, such as Sp1, or through steric obstruction of DNA binding regions required for transcription factor interactions (26, 29, 46). We concluded that the mouse fpgs gene represents an example of this mechanism. It is interesting that polymerase accumulation occurs exactly coincident with the start of the unmethylated P2 CpG island, suggesting that it is the presence of such a domain or some factor recruited to this sequence that slows polymerase transit.
Overall, we conclude that the mouse fpgs gene uses at least two mechanisms, epigenetic marking of the P1 promoter and occlusion of the P2 promoter, to ensure that each of two isoforms of FPGS are expressed as needed in dividing and select differentiated tissues. We are currently creating a mouse in which the P1 mouse fpgs promoter is deleted by homologous recombination to determine whether and why this mechanism is requisite for the survival of rodents.
Supplementary Material
Acknowledgments
We thank Viswanathan Ramakrishnan for his generous assistance with statistical analysis. We also thank B. Ann Woodard for her excellent technical assistance.
This work was supported in part by grants CA-39687 and CA-106630 from the National Institutes of Health, Department of Health and Human Services.
Footnotes
Published ahead of print on 12 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andreassi, J. L., II, and R. G. Moran. 2002. Mouse folylpoly-gamma-glutamate synthetase isoforms respond differently to feedback inhibition by folylpolyglutamate cofactors. Biochemistry 41226-235. [DOI] [PubMed] [Google Scholar]
- 2.Antequera, F., and A. Bird. 1993. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. USA 9011995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., R. Schneider, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 28017732-17736. [DOI] [PubMed] [Google Scholar]
- 4.Barredo, J., and R. G. Moran. 1992. Determinants of antifolate cytotoxicity: folylpolyglutamate synthetase activity during cellular proliferation and development. Mol. Pharmacol. 42687-694. [PubMed] [Google Scholar]
- 5.Baugh, C. M., and C. L. Krumdieck. 1971. Naturally occurring folates. Ann. N. Y. Acad. Sci. 1867-28. [PubMed] [Google Scholar]
- 6.Bernstein, B. E., A. Meissner, and E. S. Lander. 2007. The mammalian epigenome. Cell 128669-681. [DOI] [PubMed] [Google Scholar]
- 7.Bestor, T. H., G. Gundersen, A. B. Kolsto, and H. Prydz. 1992. CpG islands in mammalian gene promoters are inherently resistant to de novo methylation. Genet. Anal. Technol. Appl. 948-53. [DOI] [PubMed] [Google Scholar]
- 8.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 166-21. [DOI] [PubMed] [Google Scholar]
- 9.Bird, A. 1992. The essentials of DNA methylation. Cell 705-8. [DOI] [PubMed] [Google Scholar]
- 10.Bird, A., M. Taggart, M. Frommer, O. J. Miller, and D. Macleod. 1985. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 4091-99. [DOI] [PubMed] [Google Scholar]
- 11.Bird, A. P., M. H. Taggart, R. D. Nicholls, and D. R. Higgs. 1987. Non-methylated CpG-rich islands at the human alpha-globin locus: implications for evolution of the alpha-globin pseudogene. EMBO J. 6999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird, O. D., V. M. McGlohon, and J. W. Vaitkus. 1965. Naturally occurring folates in the blood and liver of the rat. Anal. Biochem. 1218-35. [DOI] [PubMed] [Google Scholar]
- 13.Boussadia, O., F. Amiot, S. Cases, G. Triqueneaux, H. Jacquemin-Sablon, and F. Dautry. 1997. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 42020-24. [DOI] [PubMed] [Google Scholar]
- 14.Bronder, J. L., and R. G. Moran. 2003. A defect in the p53 response pathway induced by de novo purine synthesis inhibition. J. Biol. Chem. 27848861-48871. [DOI] [PubMed] [Google Scholar]
- 15.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 16.Chabner, B. A., C. J. Allegra, G. A. Curt, N. J. Clendeninn, J. Baram, S. Koizumi, J. C. Drake, and J. Jolivet. 1985. Polyglutamation of methotrexate. Is methotrexate a prodrug? J. Clin. Investig. 76907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, J., P. Hayes, K. Roy, and F. M. Sirotnak. 2000. Two promoters regulate transcription of the mouse folylpolyglutamate synthetase gene three tightly clustered Sp1 sites within the first intron markedly enhance activity of promoter B. Gene 242257-264. [DOI] [PubMed] [Google Scholar]
- 18.Eckhardt, F., J. Lewin, R. Cortese, V. K. Rakyan, J. Attwood, M. Burger, J. Burton, T. V. Cox, R. Davies, T. A. Down, C. Haefliger, R. Horton, K. Howe, D. K. Jackson, J. Kunde, C. Koenig, J. Liddle, D. Niblett, T. Otto, R. Pettett, S. Seemann, C. Thompson, T. West, J. Rogers, A. Olek, K. Berlin, and S. Beck. 2006. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 381378-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eggermont, J., and N. J. Proudfoot. 1993. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 122539-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eszterhas, S. K., E. E. Bouhassira, D. I. Martin, and S. Fiering. 2002. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol. 22469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freemantle, S. J., and R. G. Moran. 1997. Transcription of the human folylpoly-gamma-glutamate synthetase gene. J. Biol. Chem. 27225373-25379. [DOI] [PubMed] [Google Scholar]
- 22.Freemantle, S. J., S. M. Taylor, G. Krystal, and R. G. Moran. 1995. Upstream organization of and multiple transcripts from the human folylpoly-gamma-glutamate synthetase gene. J. Biol. Chem. 2709579-9584. [DOI] [PubMed] [Google Scholar]
- 23.Fuks, F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15490-495. [DOI] [PubMed] [Google Scholar]
- 24.Futscher, B. W., M. M. Oshiro, R. J. Wozniak, N. Holtan, C. L. Hanigan, H. Duan, and F. E. Domann. 2002. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 31175-179. [DOI] [PubMed] [Google Scholar]
- 25.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greger, I. H., F. Demarchi, M. Giacca, and N. J. Proudfoot. 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 261294-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harikrishnan, K. N., M. Z. Chow, E. K. Baker, S. Pal, S. Bassal, D. Brasacchio, L. Wang, J. M. Craig, P. L. Jones, S. Sif, and A. El-Osta. 2005. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 37254-264. [DOI] [PubMed] [Google Scholar]
- 29.Hu, X., S. Eszterhas, N. Pallazzi, E. E. Bouhassira, J. Fields, O. Tanabe, S. A. Gerber, M. Bulger, J. D. Engel, M. Groudine, and S. Fiering. 2007. Transcriptional interference among the murine beta-like globin genes. Blood 1092210-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 31.Jones, P. A., and S. B. Baylin. 2007. The epigenomics of cancer. Cell 128683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 2931068-1070. [DOI] [PubMed] [Google Scholar]
- 33.Jones, P. A., M. J. Wolkowicz, W. M. Rideout III, F. A. Gonzales, C. M. Marziasz, G. A. Coetzee, and S. J. Tapscott. 1990. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc. Natl. Acad. Sci. USA 876117-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19187-191. [DOI] [PubMed] [Google Scholar]
- 35.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123593-605. [DOI] [PubMed] [Google Scholar]
- 36.Keshet, I., J. Lieman-Hurwitz, and H. Cedar. 1986. DNA methylation affects the formation of active chromatin. Cell 44535-543. [DOI] [PubMed] [Google Scholar]
- 37.Kizer, K. O., H. P. Phatnani, Y. Shibata, H. Hall, A. L. Greenleaf, and B. D. Strahl. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 253305-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 3189-97. [DOI] [PubMed] [Google Scholar]
- 39.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in a chromatin environment. Methods 19425-433. [DOI] [PubMed] [Google Scholar]
- 41.Lande-Diner, L., J. Zhang, I. Ben-Porath, N. Amariglio, I. Keshet, M. Hecht, V. Azuara, A. G. Fisher, G. Rechavi, and H. Cedar. 2007. Role of DNA methylation in stable gene repression. J. Biol. Chem. 28212194-12200. [DOI] [PubMed] [Google Scholar]
- 42.Larsen, F., G. Gundersen, R. Lopez, and H. Prydz. 1992. CpG islands as gene markers in the human genome. Genomics 131095-1107. [DOI] [PubMed] [Google Scholar]
- 43.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 44.Li, L., S. He, J. M. Sun, and J. R. Davie. 2004. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 82460-471. [DOI] [PubMed] [Google Scholar]
- 45.Liang, G., J. C. Lin, V. Wei, C. Yoo, J. C. Cheng, C. T. Nguyen, D. J. Weisenberger, G. Egger, D. Takai, F. A. Gonzales, and P. A. Jones. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA 1017357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429571-574. [DOI] [PubMed] [Google Scholar]
- 47.Martens, J. A., P. Y. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 192695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McBurney, M. W., and G. F. Whitmore. 1974. Characterization of a Chinese hamster cell with a temperature-sensitive mutation in folate metabolism. Cell 2183-188. [DOI] [PubMed] [Google Scholar]
- 49.McDonald, L. E., and G. F. Kay. 1997. Methylation analysis using bisulfite genomic sequencing: application to small numbers of intact cells. BioTechniques 22272-274. [DOI] [PubMed] [Google Scholar]
- 50.McKeon, C., H. Ohkubo, I. Pastan, and B. de Crombrugghe. 1982. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell 29203-210. [DOI] [PubMed] [Google Scholar]
- 51.Mini, E., G. Pizzorno, M. Coronnello, J. J. McGuire, T. Mazzei, P. Periti, and J. R. Bertino. 1988. Impaired methotrexate polyglutamylation in a human leukemia cell line resistant to short-term, high-dose methotrexate. Pharmacol. Res. Commun. 20445-446. [DOI] [PubMed] [Google Scholar]
- 52.Moran, R. G., W. C. Werkheiser, and S. F. Zakrzewski. 1976. Folate metabolism in mammalian cells in culture. I. Partial characterization of the folate derivatives present in L1210 mouse leukemia cells. J. Biol. Chem. 2513569-3575. [PubMed] [Google Scholar]
- 53.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393386-389. [DOI] [PubMed] [Google Scholar]
- 54.Noronha, J. M., and V. S. Aboobaker. 1963. Studies on the folate compounds of human blood. Arch. Biochem. Biophys. 101445-447. [DOI] [PubMed] [Google Scholar]
- 55.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 56.Proudfoot, N. J. 1986. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature 322562-565. [DOI] [PubMed] [Google Scholar]
- 57.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406593-599. [DOI] [PubMed] [Google Scholar]
- 58.Recillas-Targa, F., and S. V. Razin. 2001. Chromatin domains and regulation of gene expression: familiar and enigmatic clusters of chicken globin genes. Crit. Rev. Eukaryot. Gene Expr. 11227-242. [PubMed] [Google Scholar]
- 59.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenblatt, D. S., V. M. Whitehead, N. Vera, A. Pottier, M. Dupont, and M. J. Vuchich. 1978. Prolonged inhibition of DNA synthesis associated with the accumulation of methotrexate polyglutamates by cultured human cells. Mol. Pharmacol. 141143-1147. [PubMed] [Google Scholar]
- 61.Roy, K., M. G. Egan, S. Sirlin, and F. M. Sirotnak. 1997. Posttranscriptionally mediated decreases in folylpolyglutamate synthetase gene expression in some folate analogue-resistant variants of the L1210 cell. Evidence for an altered cognate mRNA in the variants affecting the rate of de novo synthesis of the enzyme. J. Biol. Chem. 2726903-6908. [DOI] [PubMed] [Google Scholar]
- 62.Roy, K., K. Mitsugi, and F. M. Sirotnak. 1997. Additional organizational features of the murine folylpolyglutamate synthetase gene. Two remotely situated exons encoding an alternate 5′ end and proximal open reading frame under the control of a second promoter. J. Biol. Chem. 2725587-5593. [DOI] [PubMed] [Google Scholar]
- 63.Schilsky, R. L., B. D. Bailey, and B. A. Chabner. 1980. Methotrexate polyglutamate synthesis by cultured human breast cancer cells. Proc. Natl. Acad. Sci. USA 772919-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75243-269. [DOI] [PubMed] [Google Scholar]
- 65.Song, F., J. F. Smith, M. T. Kimura, A. D. Morrow, T. Matsuyama, H. Nagase, and W. A. Held. 2005. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc. Natl. Acad. Sci. USA 1023336-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takai, D., and P. A. Jones. 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA 993740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takai, D., and P. A. Jones. 2003. The CpG island searcher: a new WWW resource. In Silico Biol. 3235-240. [PubMed] [Google Scholar]
- 68.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414277-283. [DOI] [PubMed] [Google Scholar]
- 69.Taylor, S. M., S. J. Freemantle, and R. G. Moran. 1995. Structural organization of the human folypoly-gamma-glutamate synthetase gene: evidence for a single genomic locus. Cancer Res. 556030-6034. [PubMed] [Google Scholar]
- 70.Turner, F. B., J. L. Andreassi II, J. Ferguson, S. Titus, A. Tse, S. M. Taylor, and R. G. Moran. 1999. Tissue-specific expression of functional isoforms of mouse folypoly-gamma-glutamate synthetase: a basis for targeting folate antimetabolites. Cancer Res. 596074-6079. [PubMed] [Google Scholar]
- 71.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19381-391. [DOI] [PubMed] [Google Scholar]
- 72.Vakoc, C. R., M. M. Sachdeva, H. Wang, and G. A. Blobel. 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 269185-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valadez-Graham, V., S. V. Razin, and F. Recillas-Targa. 2004. CTCF-dependent enhancer blockers at the upstream region of the chicken alpha-globin gene domain. Nucleic Acids Res. 321354-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber, M., I. Hellmann, M. B. Stadler, L. Ramos, S. Paabo, M. Rebhan, and D. Schubeler. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39457-466. [DOI] [PubMed] [Google Scholar]
- 75.Zhao, R., S. Titus, F. Gao, R. G. Moran, and I. D. Goldman. 2000. Molecular analysis of murine leukemia cell lines resistant to 5, 10-dideazatetrahydrofolate identifies several amino acids critical to the function of folylpolyglutamate synthetase. J. Biol. Chem. 27526599-26606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.